Abstract
The crenarchaeal Acidianus two-tailed virus (ATV) undergoes a remarkable morphological development, extracellularly and independently of host cells, by growing long tails at each end of a spindle-shaped virus particle. Initial work suggested that an intermediate filament-like protein, p800, is involved in this process. We propose that an additional chaperone system is required, consisting of a MoxR-type AAA ATPase (p618) and a von Willebrand domain A (VWA)-containing cochaperone, p892. Both proteins are absent from the other known bicaudavirus, STSV1, which develops a single tail intracellularly. p618 exhibits ATPase activity and forms a hexameric ring complex that closely resembles the oligomeric complex of the MoxR-like protein RavA (YieN). ATV proteins p387, p653, p800, and p892 interact with p618, and with the exception of p800, all bind to DNA. A model is proposed to rationalize the interactions observed between the different protein and DNA components and to explain their possible structural and functional roles in extracellular tail development.
INTRODUCTION
Acidianus two-tailed virus (ATV) is a member of the crenarchaeal Bicaudaviridae family that was isolated at Pozzuoli, Italy, from a hot, acidic spring with temperatures of 85°C to 93°C and a pH of 1.5, conditions under which its host, Acidianus convivator, grows optimally (13, 23). The only other characterized bicaudavirus is the Sulfolobus tengchongensis spindle-shaped virus, STSV1, which shares a similar morphology and some homologous proteins with ATV (31). Both viruses exhibit fusiform bodies with tails of variable length, at both ends for ATV and at one end for STSV1. ATV, in contrast to all other known viruses, including STSV1, undergoes an extracellular morphological transformation that is independent of host cells (13, 23). Virus particles are released from the host as tail-less fusiform structures, and within an hour to a few days, depending on temperature conditions, two tails develop in an irreversible process (13). The only other known examples of extracellular viral morphogenesis comprise initial steps of infection or final steps in particle assembly and budding, and these are triggered on the cell surface of the host (2, 22). ATV tails are assumed to facilitate virion attachment to host cell walls or membranes under harsh environmental conditions where there is a low density of potential host cells (23).
Exceptionally for a crenarchaeal virus, ATV shows both a lysogenic and a lytic reproduction cycle. Infection of host cells grown under optimal conditions results in lysogeny that can be interrupted and transformed into a lytic stage by environmental stress factors. For example, ATV propagation can be induced by lowering the culture temperature from 85°C to 75°C (23).
ATV and STSV1 have circular double-stranded DNA genomes of 62,730 and 75,294 bp and contain 72 and 74 predicted open reading frames (ORFs), respectively. ATV virions carry at least 11 proteins, whereas STSV1 has only 5. In both virions, the homologous DNA-binding proteins p131 of ATV and p40 of STSV1 dominate and are considered to be the primary DNA folding proteins (8, 31).
Of the identified structural proteins of ATV, only p618 produces a significant functional match in sequence databases: it is predicted to be an AAA ATPase (ATPases associated with diverse cellular activities). AAA proteins show remarkable functional diversity and often perform chaperone-like functions that facilitate assembly or disassembly of protein complexes (27). Although AAA proteins are highly conserved within their ATPase domains, other protein regions typically show little sequence similarity within the family. These regions usually comprise either protease domains or domains mediating interactions with other proteins or nucleic acids (18).
Viruses typically utilize host cell components to ensure efficient DNA replication, protein biosynthesis, and virion assembly, and it is generally considered that cellular chaperones facilitate the folding of virion capsid proteins and participate in virus budding (28). The host-independent tail development of ATV virions that occurs after their release inevitably requires a major rearrangement of the virion proteins, and we infer that chaperones could facilitate this reassembly process. Although STSV1 shows a similar morphology to that of ATV, it does not grow tails extracellularly and lacks potential chaperone proteins in its virions (31).
Our working hypothesis was that some ATV virion-specific proteins are directly involved in extracellular tail development and that the putative AAA ATPase p618 (23) plays an important role in this process. Here we present a detailed biochemical characterization of p618, which corresponds to the RavA (YieN) subfamily of the MoxR-type AAA ATPases (5, 26), and examine its interactions with other ATV virion proteins that are mainly absent from STSV1 in order to gain insights into the molecular mechanisms involved in extracellular tail development. Four protein interaction partners were identified, some of which also interacted with one another, including p892, carrying a von Willebrand factor A (VWA) domain, the filamentous protein p800, and two related DNA-binding proteins, p387 and p653. The possible roles of these proteins in extracellular tail development of ATV are considered, with a proposed central role for p618 and p892.
MATERIALS AND METHODS
Plasmids and bacterial strains.
Escherichia coli DH5α was used for cloning and site-directed mutagenesis, and Rosetta (DE3) was the host strain for the expression vectors based on the pET28a (Novagen, Madison, WI) or pGEX-6P (GE Healthcare, St. Chalfont, United Kingdom) plasmid. ORFs were amplified by PCR from a whole-genome shotgun library of ATV. The p618 forward primer, 5′-ACAGTCGACATGTCTCAGCAACCTTC-3′, carried an additional SalI restriction site. The reverse primer, 5′-TGTCGGCCGAGCTTGCGAGGCTGATG-3′, had an EagI restriction site that was used for cloning. The PCR product was introduced into a modified pET28a vector, pET28d, generated by cleavage with NcoI and BamHI and religation after formation of blunt ends with Klenow DNA polymerase.
Site-directed mutagenesis was performed according to the QuikChange protocol (Stratagene, La Jolla, CA) with the forward primers 5′-GGAGATAGAGGAGTCGCGGCAACCATGTTGATCG-3′ for p618K62A and 5′-GCAAAGGTAGCGTTCATAGATCAGATCTTCTATACTTCGG-3′ for p618E128Q. Reverse primers had the reverse and complementary sequences to these primers.
A glutathione S-transferase (GST) fusion construct of p618 was generated from the PCR product of p618 after it was cut with SalI and EagI and ligated into the linearized pGEX6P-3 vector. p618 DNA was inserted into pET28a, cleaved with NdeI and AhdI, and ligated to a linker oligomer coding for the additional amino acids MASASA in order to obtain p618ΔN, covering amino acids 328 to 618. AhdI and XhoI cleavage of the pET28d-p618 construct, blunting of both ends by use of Klenow DNA polymerase, and religation of the plasmid yielded the expression vector for p618ΔC (amino acids 1 to 327).
A GST-tagged fusion protein of p892 was generated using the primers 5′-GGAATTCCATATGAGCGTGGACCAAGACG-3′ and 5′-GGAATTTCGAGTTAATGAACAAGCCTCTTCAGC-3′. The PCR product was then introduced into the EcoRI and XhoI sites of the pGEX6P-1 vector.
Two sets of primers (5′-CAGGGATCCATGGTGGCTCAGGAAATGGC-3′ and 5′-GAGCTCGAGACTGAGCTTTACTGTGTTG-3′ for p387 and 5′-TTTCTCGAGATGCAACCGTGGGAGTATG-3′ and 5′-AGACGGCCGAACTGAGCTTTACTGTG-3′ for p653) were designed to amplify the coding sequences of p387 and p653, which were inserted into the pET28d vector after cleavage with BamHI and XhoI and with XhoI and EagI, respectively.
p800 was cloned as an N-terminally tagged polyhistidine fusion protein by use of the primer pair 5′-GGAATTCCATATGTCCAGCAATTATGTGC-3′ and 5′-GGAATTCTCGAGTCAGACCCTTATTAATCGAGC-3′. The PCR product was inserted into the NdeI and XhoI sites of the pET28a vector.
Sequence analysis and alignment.
Protein domain architectures were analyzed using the Pfam (http://pfam.sanger.ac.uk) and SMART (http://smart.embl-heidelberg.de/smart/set_mode.cgi?GENOMIC=1) databases. Multiple sequence alignments were generated by ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/). BLASTP searches were used for sequence comparison, and COILS (http://www.ch.embnet.org/software/COILS_form.html) was used to predict coiled-coil regions. DNA-binding sites were predicted using DNAbindR (http://turing.cs.iastate.edu:8080/PredDNA/predict.html) and BindN (http://bioinfo.ggc.org/bindn/).
Expression and purification of recombinant proteins.
Rosetta (DE3) cells transformed with pET28a and pET28d constructs were grown in LB medium supplemented with 30 μg/ml kanamycin and 25 μg/ml chloramphenicol at 37°C. Expression was induced by adding 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and was carried out overnight at room temperature. Proteins were purified by affinity chromatography using either Ni-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen, Hilden, Germany) or glutathione (GSH)-Sepharose (GE Healthcare) according to the manufacturers' protocols, including an additional heating step at 70°C for 10 min for the polyhistidine-tagged proteins. DNA contaminations were removed by using high-salt conditions during purification. If necessary, all proteins were further purified by size exclusion chromatography (SEC) using a Superdex 200 (10/300) gel filtration column (GE Healthcare). The polyhistidine-tagged constructs were detected in Western blots by use of a penta-His antibody (Qiagen) and an alkaline phosphatase-conjugated goat anti-mouse secondary antibody (Sigma-Aldrich, St. Louis, MO), using the 5-bromo-4-chloro-3-indolylphosphate (BCIP)–nitroblue tetrazolium (NBT) reagent (Sigma-Aldrich).
p618 expression in Sulfolobus solfataricus.
For expression of N-terminally polyhistidine-tagged p618 in S. solfataricus, the shuttle vector pEXA-2 was used (9). Ten milliliters of S. solfataricus InF1 cells carrying the desired plasmid was inoculated into 500 ml of SCV selection medium and grown for 4 days at 78°C to an optical density at 600 nm (OD600) of 1.0. Cells were collected by centrifugation at 4,200 × g and resuspended in 2.5 liters of medium supplemented with 0.2% d-arabinose instead of sucrose to induce expression of p618. After 3 days, the cells were harvested at an OD600 of 0.5. The protein was purified according to the protocol described above, omitting the heat shock step, and the cell lysate was incubated with 1 ml of compact Ni-NTA agarose for 1 h at room temperature. The yield was about 0.1 mg/liter of purified protein.
Size exclusion chromatography.
p618 (60 μM), p618K62A (12 μM), and p618E128Q (50 μM) were incubated for 30 min on ice in buffer B (20 mM HEPES, 100 mM NaCl, pH 7.5) in the presence or absence of 1 mM ATP and 2.5 mM MgCl2. One hundred microliters of each reaction mixture was then applied to a Superdex 200 (10/300) gel filtration column (GE Healthcare) at a flow rate of 0.5 ml/min. Absorbance was recorded at 280 nm, and eluted fractions were analyzed by SDS-PAGE.
SEC-MALLS.
Thirty microliters of p618E128Q was loaded onto a 15-ml Shodex KW804-500A (Phenomenex, Torrance, CA) high-performance liquid chromatography (HPLC) column at a concentration of 4 mg/ml and a flow rate of 0.5 ml/min. The column was equilibrated with 20 mM NaH2PO4, pH 7.2, 150 mM NaCl, 0.2 mM ATP, and 2 mM MgCl2. Multiangle laser light scattering (MALLS) analysis was performed continuously on the column eluate at 20°C. For detection, a miniDAWN Treos triple-angle light scattering detector and an Optilab rEx differential refractometer (Wyatt Technology Corp., Santa Barbara, CA) were used. The data were processed with ASTRA 5.3.15 software (Wyatt Technology Corp.).
Cross-linking.
Purified p618 was cross-linked with 0.02% glutaraldehyde at a protein concentration of 0.17 mg/ml in buffer B containing 2.5 mM MgCl2 and 1 mM either ATP or AMP-PNP or no nucleotide. After nucleotide binding for 20 min at room temperature and 30 min of cross-linking either at room temperature or at 37°C, the reaction was quenched by addition of 50 mM Tris-HCl, pH 8.0. The cross-linked complexes were analyzed on a NuPAGE Novex 3 to 8% Tris-acetate gel, pH 7.0 (Invitrogen, Paisley, United Kingdom), with Mark12 (Invitrogen) as an unstained standard.
Electron microscopy.
p618E128Q oligomers and p800 were isolated by size exclusion chromatography at 4°C. The Superdex 200 (10/300) column (GE Healthcare) was equilibrated with 50 mM NaH2PO4, 300 mM NaCl, pH 8.0, 0.2 mM ATP, and 2.5 mM MgCl2. The elution fraction of p800 was kept at 4°C overnight. The sample was then incubated for 5 min either at room temperature or at 50°C, and the proteins were adsorbed to a carbon-coated copper grid at a concentration of 0.01 mg/ml and stained with 2% uranyl acetate. Images were recorded using a Tecnai G2 (FEI, Eindhoven, Netherlands) transmission electron microscope equipped with a charge-coupled device (CCD) camera at an acceleration voltage of 120 kV.
Coprecipitation experiments.
Pulldown experiments were performed with GST fusion proteins of p618, p387, or p892 immobilized to GSH-Sepharose beads (GE Healthcare) in buffer B supplemented with 0.1% Triton X-100, 2.5 mM MgCl2, 1 mM ATP, and 0.2 to 0.3 mg/ml of p618, p618ΔN, p618ΔC, p387, p653, or p800. Increasing concentrations of p892 (0.1 mg/ml to 0.3 mg/ml) were used for a competition pulldown experiment with p387 and p892 competing for binding to GST-p618. GST conjugated to GSH beads and 0.25 mg/ml bovine serum albumin (BSA) served as controls. The reaction mixture was centrifuged for 1 min at 2,000 × g after incubation for 1 h, and 7.5-μl samples of the supernatant and pellet fractions were analyzed by SDS-PAGE, using a PAGE Ruler prestained protein ladder (Fermentas, St. Leon-Rot, Germany) as a molecular size marker. Binding was detected by Western blotting using a penta-His antibody as primary antibody.
ATPase assay.
ATPase activity was determined colorimetrically by a malachite green-based assay as described previously (14), but without quenching in 0.6 M perchloric acid, which did not affect the readout, and the color reaction was terminated by addition of an equal volume of 7.8% (wt/vol) H2SO4. Assays were performed in triplicate in microtiter plates at 60°C for 5 min with 0.01 mg/ml of p618 in buffer B and then placed on ice to terminate the reaction. ATP concentrations ranging from 0.0625 mM to 3 mM and 2.5 mM MgCl2, MnCl2, CdCl2, CoCl2, CaCl2, CuCl2, NiCl2, or FeCl2 were used. Release of inorganic phosphate was estimated by comparing the absorbance at 650 nm with a standard curve obtained using KH2PO4. The temperature dependence of the ATPase activity was analyzed at an ATP concentration of 1 mM and at temperatures ranging from 50°C to 80°C in 5°C steps. The effects of p892 and p387 on the ATPase activity of p618 were analyzed at 55°C, 65°C, and 75°C, using 1 mM ATP substrate with a 5-fold molar excess of p892 or p387.
Electrophoretic mobility shift assay.
p618 was tested for DNA binding at increasing protein concentrations (0 to 20 μM) in the presence of 2.5 mM ATP and 6.7 mM MgCl2. The DNA-binding activities of p387 and p653 were investigated at 50°C for 25 min with 1.5 pmol of radiolabeled DNA in buffer B with 0.4 mM dithiothreitol (DTT) and 2% (vol/vol) glycerol. Concentrations of 0 to 5 μM p387, 0 to 2.5 μM p653, and 0 to 12 μM p892 were used. p387 (2.5 μM) was incubated with increasing concentrations of p618, up to a p387/p618 molar ratio of 1:2, and 12 μM p892 was incubated with equimolar amounts and with a 2-fold molar excess of p618. A double-stranded DNA (dsDNA) template of 147 bp was generated by PCR and 5′-end labeling using [32P]ATP (Perkin Elmer, Waltham, MA) and T4 polynucleotide kinase (Invitrogen). Protein-bound and free DNAs were separated in an 11% nondenaturing acrylamide gel, and DNA bands were visualized autoradiographically.
RESULTS
Eleven bicaudaviral virion proteins were identified earlier for ATV, and five were identified for STSV1 (13, 31). Of these, two of the most abundant ATV proteins (p618 and p387), together with p653, a paralog of p387, are absent from STSV1. p892, originally not identified as an ATV virion protein, is also absent from STSV1. The ATV-specific proteins were expressed in E. coli carrying either a polyhistidine or GST tag and were examined for their properties and interactions.
ATPase activity of p618.
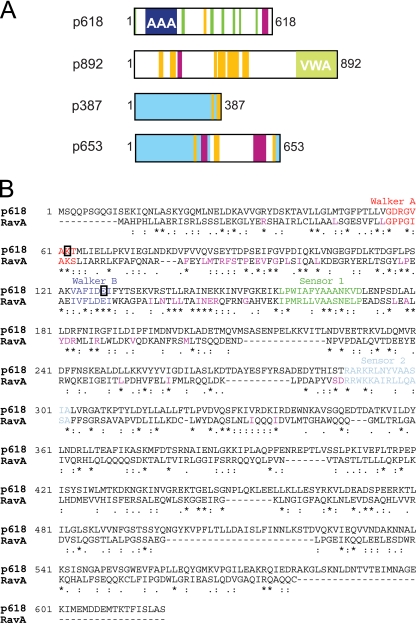
The best sequence match for p618 was a MoxR-like ATPase from the crenarchaeon Picrophilus torridus DSM 9790. Within the MoxR AAA+ ATPases, which comprise at least seven subfamilies, p618 could be assigned to the RavA (YieN) subfamily (26). Like RavA, p618 consists of two predicted domains: an N-terminal AAA module covering amino acids 51 to 192 and containing the Walker A and B motifs required for phosphate binding and nucleotide hydrolysis (11) and a poorly conserved C-terminal region exhibiting coiled-coil motifs (Fig. 1 A and B). p618 and RavA exhibit 41% sequence similarity and 28% identity. Many sequences and motifs characteristic of RavA proteins are present in p618, including a conserved tryptophan or phenylalanine residue within the Walker B motif and a preceding Leu-Pro sequence of unknown function. Moreover, the more commonly used glycine residue in the Walker A sequence is replaced by an alanine, as occurs in RavA (25) (Fig. 1B).
Fig. 1.
Properties of ATV virion proteins. (A) Domain structures of p618, p892, p387, and p653. Structural domains and sequence motifs are color coded as follows: blue, AAA domain; green, disordered regions; light green, VWA domain; yellow, low-complexity regions; purple, coiled-coil elements; and light blue, shared domain of unknown function in p387 and p653. (B) Sequence alignment of p618 and RavA showing degrees of similarity. Conserved residues within the RavA subfamily (25) are shown in purple.
To test for ATPase activity, we generated presumed loss-of-function mutations within the Walker motifs. A catalytic glutamate residue downstream from the Walker B motif was replaced by a glutamine, and a conserved lysine in the Walker A motif was changed to an alanine. Walker B motif mutations have commonly been used to create so-called substrate traps for ATPases by blocking nucleotide hydrolysis but not binding, whereas Walker A motif mutations typically eliminate ATP binding and thereby inactivate the ATPase.
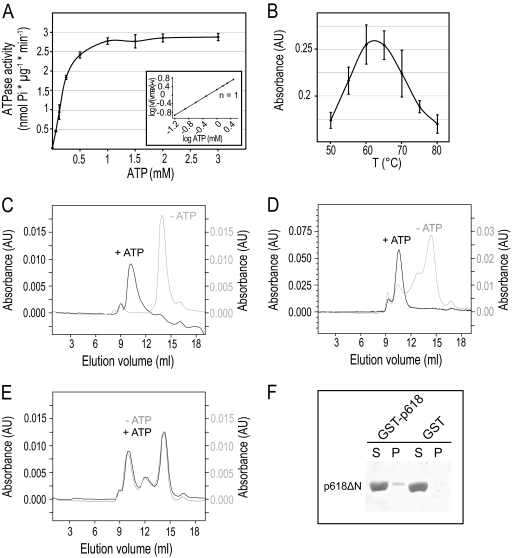
Wild-type p618 was active in ATP hydrolysis, with a Km value of 0.55 mM and a maximum velocity of 4.55 nmol/(min·μg) (Fig. 2 A). A Hill coefficient of 1.0 indicates the absence of cooperative binding at increasing ATP concentrations, whereas both Walker A and B mutants were inactive in ATP hydrolysis (see Fig. S1A in the supplemental material). As expected for a hyperthermophilic viral protein, the ATPase activity of p618 showed a bell-shaped curve with a high-temperature optimum of about 60°C (Fig. 2B), although this lies below the optimal growth temperature of the host (85°C). Since this difference could have been due to the heterologous protein expression in E. coli, with a consequent lack of stabilizing posttranslational modifications, we also expressed p618 in the archaeon S. solfataricus P2, which is phylogenetically close to the host of ATV, but no change was detected in the temperature optimum of the ATPase activity (see Fig. S1B in the supplemental material).
Fig. 2.
Properties of p618. (A) Measurement of ATPase activity. p618 (0.01 mg/ml) was incubated at 60°C with increasing amounts of ATP. Release of inorganic phosphate was determined by malachite green assay. The inset shows a Hill plot, with a Hill coefficient of 1. (B) Temperature dependence of ATPase activity was analyzed in the presence of 1 mM ATP at the indicated temperatures. (C to E) Oligomerization of p618 was analyzed by size exclusion chromatography of 60 μM p618 (C), 50 μM p618E128Q (D), and 12 μM p618K62A (E) in the presence or absence of 1 mM ATP monitored at 280 nm. (F) Coprecipitation experiments were performed with GST-p618 immobilized on GSH-Sepharose beads (GST-p618) and 0.2 mg/ml of p618ΔN in the presence of 1 mM ATP. GST conjugated to GSH beads (GST) and 0.25 mg/ml BSA served as controls. Samples (7.5 μl) of the supernatant (S) and pellet (P) fractions were analyzed by Western blotting using a penta-His antibody as primary antibody.
ATP hydrolysis requires the presence of divalent metal ions, and the conserved aspartate residue of the Walker B motif coordinates the divalent cation, usually Mg2+, as a cofactor, thereby ensuring efficient binding of ATP. We demonstrated that in the absence of Mg2+, no significant enzymatic activity was detectable (see Fig. S1C in the supplemental material). However, the requirement for divalent cations was not stringent; Mg2+ could be replaced by Mn2+, Co2+, or Cu2+ without causing a large decrease in ATPase activity. This is consistent with the properties of another AAA ATPase, from the hyperthermophilic archaeon Thermococcus kodakarensis KOD1, perhaps reflecting the fact that terrestrial hot springs are rich in transition metal ions (6).
Oligomerization of p618.
A common feature of diverse members of the AAA ATPase family is their capacity to generate oligomers, generally hexameric rings, that constitute the biologically active form. AAA ATPases act either by translocating a peptide or nucleic acid substrate through the central pore of the hexameric ring or by remodeling the substrate molecule, as described for metal chelatases (16, 27, 29). The oligomeric state of p618 was investigated by cross-linking and size exclusion chromatography, also performed in combination with multiangle laser light scattering (MALLS-SEC). Size exclusion chromatography established that p618 forms oligomeric complexes in the presence of ATP (Fig. 2C). The apparent molecular mass was estimated to be 434 kDa by MALLS-SEC analysis, consistent with hexameric ring formation (see Fig. S2A in the supplemental material). Formation of a hexameric complex was also supported by cross-linking experiments with nucleotide-bound p618 and glutaraldehyde (see Fig. S2B in the supplemental material). Cross-linking efficiency was slightly diminished when the nonhydrolyzable ATP analogue AMP-PNP was used (see Fig. S2B in the supplemental material), possibly due to a reduced affinity of the enzyme for this substrate.
In the absence of ATP, the assembly state of p618 exhibited some heterogeneity at high protein concentrations, consistent with a protein concentration-dependent self-association of p618. This effect was slightly more pronounced when the Walker B mutant was used for size exclusion chromatography (Fig. 2D). Although the Walker A mutant was not expected to oligomerize, as it was deficient in ATP binding, this mutant also formed hexamers (Fig. 2E), demonstrating that p618 assembly is stimulated by but not dependent on ATP.
Intramolecular interaction of p618.
While ATPase domains form characteristic hexameric rings of AAA proteins, adjacent domains are typically involved in substrate and/or cofactor binding. Size exclusion chromatography of the isolated N-terminal ATPase domain of p618 demonstrated that higher-order oligomers were not formed (see Fig. S2C in the supplemental material). It is possible that the C-terminal domain contributes to hexamer formation, as seen for the valosin-containing protein (VCP) p97, another AAA ATPase, where nucleotide-dependent structural changes of the ATPase domain extend to adjacent domains, suggesting that intramolecular interactions occur between the ATPase region and neighboring domains (4). Coprecipitation experiments performed with the C-terminal domain of p618 (amino acids 328 to 618) and a GST fusion protein of p618 revealed an intramolecular interaction between the two components (Fig. 2E). The recently solved structure of RavA, however, supports the view that the C-terminal domain interacts with the central region of p618 (5).
Electron microscopic analysis of p618 hexamers.
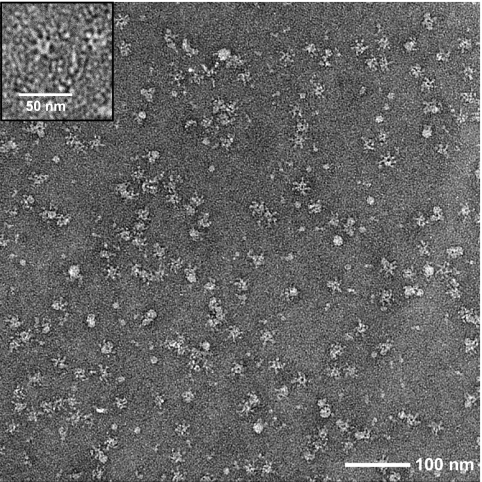
The oligomeric state of p618 was examined by negative-stain electron microscopy. The Walker B mutant was selected because mutation of the Walker B motif often results in a stronger substrate interaction which stabilizes the higher-order oligomeric state (3). Oligomerization was induced by adding ATP to the purified protein, and although some heterogeneity was observed, many particles exhibited a 6-fold symmetry axis (Fig. 3). The hexameric rings had a diameter of about 17 nm and a central cavity diameter of approximately 4.5 nm. Protrusions radiating from the central ring of the particles yielded a diameter of about 25 nm. In this respect, the oligomeric structures closely resembled those observed for RavA (5, 25). The arm-like densities may correspond to the C-terminal domain of p618.
Fig. 3.
Hexameric ring structure of p618E128Q. Oligomeric complexes of the Walker B mutant of p618 were isolated by size exclusion chromatography at 4°C in the presence of 0.2 mM ATP and analyzed by electron microscopy after negative staining with uranyl acetate. The inset shows a detailed view of a hexameric ring.
Interaction with p892.
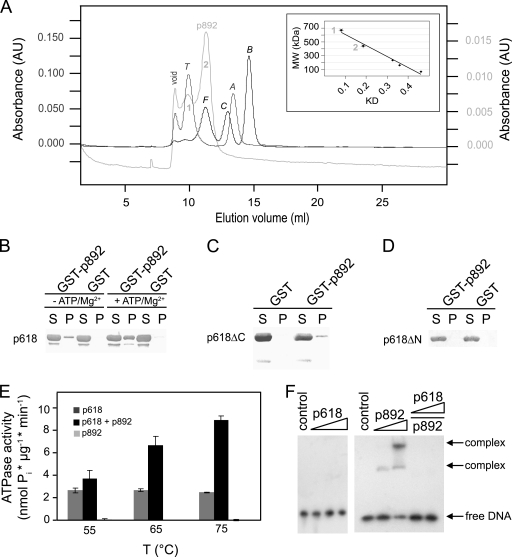
p892 carries a highly acidic central domain of about 390 amino acids containing almost 30% glutamate but, remarkably, no aspartate residues. This acidic stretch also leads to retarded mobility in SDS-PAGE gels due to reduced SDS binding, and p892 migrates at an apparent molecular mass of approximately 130 kDa, while the actual mass is 103 kDa. Moreover, p892 contains a predicted C-terminal VWA domain (Fig. 1A) which exhibits a conserved metal ion-dependent adhesion site (MIDAS) motif following the consensus sequence DxSxS at amino acid position 729. VWA domains can coordinate divalent cations via the MIDAS motif and often mediate protein-protein interactions (29). p892, like p618, oligomerizes and generates tetramers and hexamers (Fig. 4 A), with molecular masses estimated from the distribution coefficients (KD) of 460 kDa and 640 kDa, respectively.
Fig. 4.
Oligomerization and protein-protein interactions of p892. (A) Oligomerization of p892. The oligomeric state of p892 was analyzed by size exclusion chromatography and monitored at 280 nm. Thyroglobulin (T), ferritin (F), catalase (C), aldolase (A), and BSA (B) were used as standard proteins. Molecular masses of 640 kDa and 460 kDa were estimated from the calibration curve (inset) for the first and second elution peaks, consistent with hexamer and tetramer formation, respectively. (B to D) p892 interactions with p618. Deletion mutants of p618 lacking either the C-terminal (p618ΔC) or N-terminal (p618ΔN) domain were used to map the binding site on p618. Coprecipitation experiments were performed with GST-p892 immobilized on GSH-Sepharose beads (GST-p892) and 0.2 mg/ml of p618 (B), p618ΔC (C), and p618ΔN (D) in the presence of 1 mM ATP. GST conjugated to GSH beads (GST) and 0.25 mg/ml BSA served as controls. Supernatant (S) and pellet (P) fractions were analyzed by Western blotting. (E) The effect of p892 on the ATPase activity of p618 was measured at 650 nm. The ATPase assay was performed at the indicated temperatures in the presence of 1 mM ATP and a 5-fold molar excess of p892. p892 stimulated ATPase activity of p618 by 3.6-fold at 75°C. (F) Binding of p618 (0 to 20 μM), p892 (6 and 12 μM), and the p618-p892 complex to DNA was investigated at 50°C for 25 min with 1.5 nmol of labeled DNA (147 bp). p618 was added to p892 in an equimolar amount and a 2-fold molar excess in the last two lanes. The negative control contained substrate DNA only. Protein-bound and free DNAs were separated by electrophoresis on an 11% nondenaturing acrylamide gel and were visualized by autoradiography.
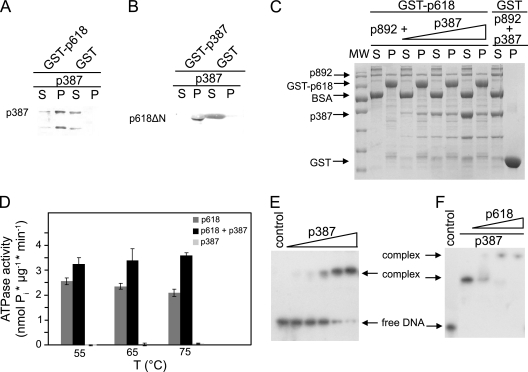
Pulldown experiments demonstrated complex formation between p618 and p892 (see Fig. 6B), with an increased affinity in the presence of ATP and Mg2+. The proteins interacted via the N-terminal ATPase domain of p618 (Fig. 4C), while no interaction was observed with the isolated C-terminal domain (Fig. 4D). The ATPase domain of p618 contains a short peptide sequence with an essential acidic residue (RYD) at position 35 that constitutes a binding motif for the VWA domain (24, 30) and probably mediates the interaction with p892. Addition of p892 enhanced the ATPase activity of p618, and at a 5:1 molar ratio of p892 to p618, there was a 2.5-fold stimulation of activity at 60°C and a 3.6-fold increase at 75°C (Fig. 4E). Many AAA proteins act on DNA as helicases, transcription factors, or repair enzymes (19, 27). p892 was found to interact with DNA, whereas p618 and the p892-p618 complex did not (Fig. 4F).
Fig. 6.
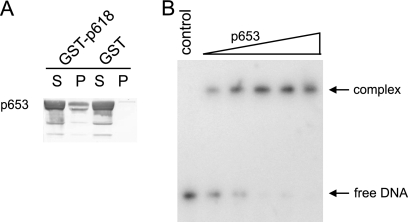
p653 protein and DNA interactions. (A) Coprecipitation experiments were performed with GST-p618 beads and 0.2 mg/ml of p653, using GST beads and BSA as controls. Supernatant (S) and pellet (P) fractions were analyzed by Western blotting. (B) DNA binding of p653 was investigated at 50°C for 25 min with 1.5 nM DNA (147 bp) and 0 to 2.5 μM p653. DNA without protein served as a negative control. Protein-bound and free DNAs were separated by electrophoresis on an 11% nondenaturing acrylamide gel and were visualized by autoradiography.
Interaction with p387 and p653.
The ATV virion protein p387 shares an amino acid sequence identity of 61.5% with p653. It also shows a high sequence similarity to p362, but the latter was not identified as a structural protein (23). p653 carries an extra C-terminal 290 amino acids (Fig. 1A), indicating a unique function of p653. No significant matches were found for these three proteins in public sequence databases. Pulldown experiments with p387 and p653 demonstrated strong binding of both to p618 (Fig. 5 A and 6 A) but no interaction with one another (data not shown). p387 spontaneously degraded into a protein fragment which comprised the binding site for p618 and lacked the N-terminal region (Fig. 5A). Furthermore, p387 interacted with the C-terminal domain of p618 (Fig. 5B) and did not compete with p892 for the binding site on p618 (Fig. 5C). Moreover, p387 stimulated the ATPase activity of p618 but was less effective than p892 (Fig. 5D).
Fig. 5.
p387 protein-protein interactions. (A) Coprecipitation experiments were carried out with p387 and GST-p618. (B) The binding site of p387 on p618 was mapped using a p618 fragment lacking the N-terminal domain (p618ΔN). Assays were performed with 0.2 mg/ml of target proteins, and GST conjugated to GSH beads and BSA served as controls. (C) A competition pulldown experiment with p618 and p892 and with increasing concentrations of p387 (0.1 to 0.3 mg/ml) was performed as described above. Supernatant (S) and pellet (P) fractions were analyzed either by SDS-PAGE with Coomassie staining or by Western blotting (A and B). A molecular size standard (MW) was used to estimate the apparent molecular protein masses. (D) The effect of a 5-fold molar excess of p387 on the ATPase activity of p618 was estimated at 650 nm at the indicated temperatures in the presence of 1 mM ATP. (E and F) Electrophoretic mobility shift assays. (E) DNA binding of p387 was investigated at 50°C for 25 min with 1.5 nM DNA (147 bp) and increasing concentrations of protein (0 to 5 μM p387). (F) Binding of p618 to the DNA-p387 complex (molar ratio of p618 to p387, 0.5 to 2) resulted in the formation of a multimolecular complex. The negative controls contained substrate DNA only. Protein-bound and free DNAs were separated by electrophoresis on an 11% nondenaturing acrylamide gel and were visualized by autoradiography.
p387 and p653 both copurified with E. coli DNA, indicating a DNA-binding activity with low sequence specificity (data not shown). Neither protein carries detectable helix-turn-helix (HTH) or leucine zipper motifs, but they exhibit a number of sequence motifs potentially involved in DNA binding. Most of these sites occur within predicted unstructured regions. Strong binding of p387 and p653 to dsDNA was observed (Fig. 5E and 6B). Moreover, p618 interacted with a p387-DNA complex, creating a slower-migrating band of higher molecular weight in the mobility shift assay (Fig. 5F).
Interactions with p800.
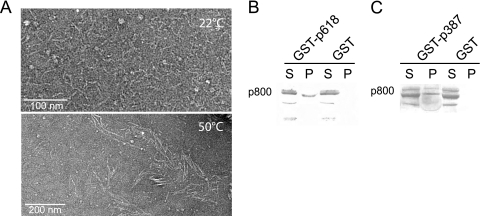
p800 contains multiple heptad repeats characteristic of coiled-coil structures and intermediate filaments, and it tends to self-aggregate to form filaments suggestive of a possible role in tail development (23). At room temperature, p800 assembled spontaneously into filaments, which were distributed homogeneously over the carbon-coated electron microscopy grid (Fig. 7 A). Moreover, the capacity to form larger filamentous structures increased at higher temperatures (Fig. 7A). In pulldown assays, p800 interacted weakly but specifically with both p618 and p387 (Fig. 7B and C).
Fig. 7.
Assembly and protein-protein interactions of p800. (A) Temperature dependence of the assembly of p800 into filaments. p800 was purified by size exclusion chromatography, kept overnight at 4°C, and then incubated for 5 min at either 22°C or 50°C. The filaments were analyzed by electron microscopy after negative staining with uranyl acetate. (B and C) Interaction of p800 with p618 and p387. Coprecipitation experiments with GST-p618 (B) and GST-p387 (C) were performed with 0.3 mg/ml p800. GST beads and BSA were used as controls. Supernatants (S) and beads as the pellet fraction (P) were analyzed by Western blotting.
DISCUSSION
MoxR ATPases belong to the AAA+ ATPase family and are implicated in chaperone-like functions in the assembly, disassembly, or activation of protein complexes (8). Several ATPases of this family have been shown to be required for the maturation of proteins or protein complexes (20, 26). The ATV protein p618 is a member of the RavA (YieN) subfamily of the MoxR AAA ATPases, which carry an N-terminal AAA+ module and a more variable C-terminal region (5, 26). We infer, therefore, that p618 also functions as a molecular chaperone within the virion and that it catalyzes and facilitates assembly of protein complexes involved in extracellular tail development. This proposal is reinforced by the presence of coiled-coil elements in the C-terminal region of p618, which are likely to constitute substrate binding determinants (17). The presence of chaperones in virions is highly unusual, even though they are occasionally encoded in viral genomes. An exception occurs for a eukaryotic closterovirus, where the tails of filamentous beet yellows virions (BYV) harbor a virus-encoded homolog of the molecular chaperone Hsp70h which mediates tail assembly and virion translocation intracellularly (1, 21).
MoxR ATPases usually cooperate with proteins carrying VWA domains to generate a chaperone complex for folding and/or activating proteins (26). Genes for the two proteins often form an operon, and sometimes the AAA and VWA protein domains are fused. RavA, the closest homolog to p618, interacts with the VWA domain-containing protein ViaA (25). The gene for p618 lies close to that for the VWA domain-containing protein p892. Thus, our observation that p618 and p892 interact specifically strongly reinforces the chaperone concept. Although p892 was not identified as a major virion component (23), a low concentration would be sufficient for a catalytic cochaperone.
p892 binds to the ATPase domain of p618 and stimulates its ATPase activity (Fig. 4C and E), possibly by stabilizing the hexameric state. Since p892 also assembles into a hexameric complex, it seems likely that p618 and p892 generate a two-ring structure similar to that of the MoxR-like and VWA domain-containing subunits of the magnesium chelatases BchI and BchD (15). Consistent with such an interaction, p892 functions as a regulator of p618 and stimulates its ATPase activity at temperatures above its intrinsic optimum of 60°C. Thus, at 85°C, where ATV tails develop most quickly (23), p618 is active only when p892 is bound as a cochaperone. The conserved MIDAS motif within the VWA domain of p892 suggests a role for divalent cations in this process.
The findings that p892 binds to DNA and that the p892-DNA complex dissociates when p618 is present (Fig. 4F) show that p618 is recruited and activated by p892 only in the immediate proximity of DNA, where most of the other identified interaction partners can be found. Electron microscopic studies of the ATV tails revealed a putative channel for DNA (23), and it is likely that DNA enters cells via the tails, as in many tailed viruses. This inference receives support from the observation that both ATV and the other characterized bicaudavirus, STSV1, attach to cells via the tips of their tails (12, 31). p653, unlike p387, was found to be present in the virion tails (12). Moreover, since it exhibits a higher affinity for DNA than either p387 or the nucleocapsid protein p131 (7), it is a candidate for facilitating viral DNA movement into the tails.
Earlier ATV studies suggested that the structural protein p800, a minor tail component, functions as a tail protrusion protein, possibly in a modified form, because it has a high content of coiled-coil structures typical of intermediate filament proteins, and moreover, the recombinant p800 protein generated filaments similar in size to structures observed in ATV tails (23). Therefore, based on our results, we propose a hypothetical model for the extracellular tail development of ATV where the filamentous p800 fibers function as a structural scaffold and the multiple heptad coiled-coil motifs within the p800 sequence serve as binding determinants for other virion proteins. Moreover, we infer that the observed interaction of p387 and p800 may reflect an inhibitory role of p387 on filament development. p892 activates p618, which in turn interacts with the highly abundant protein p387. Chaperone activity of p618 then causes dissociation of the p387-p800 complex, thereby allowing polymerization of p800. Given that both p387 and p892 bind to DNA, viral DNA is implicated, at least indirectly, in both of these steps.
At high temperatures, isolated p800 assembled spontaneously into longer filaments (Fig. 7A), and it is predicted that the final lengths of the filaments and the virion tails depend on the amount of ATP that is available within the virions. It remains unclear how the virion stores ATP. In aquatic hot springs, free ATP is very unstable, so it is likely that the virion encapsulates a pool of cellular ATP upon extrusion from the host cell. Variations in the size of this pool could explain the observed variability in virion tail length (23).
Like ATV, the other bicaudavirus, STSV1, contains a protein (p37) of similar size to p800 that is also rich in coiled-coil heptad elements. Although p37 shows little sequence similarity to p800, it is also a likely candidate for the scaffold protein of the single tail of STSV1. However, STSV1 develops a single tail intracellularly (31), and therefore a virus-encoded chaperone system like that described for ATV would be unnecessary. It could be replaced intracellularly by MoxR ATPases and VWA domain proteins encoded by adjacent genes in genomes of members of the Sulfolobales (10), such that genomic DNA of STSV1 is directed into growing virion tails, assisted by cellular chaperones of host cells.
In conclusion, the work presented here constitutes the first attempt to determine the molecular processes involved in the exceptional extracellular tail development of the ATV virion. We infer that the MoxR ATPase p618 and the VWA protein p892 play a crucial role in the dynamics of this process. Future work will focus on the relative locations of virion proteins within the tail structure and on the factors involved in the initial steps of tail development, including a predicted role for viral DNA.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gisle Vestergaard for providing the plasmid p800-pET28a and Shiraz A. Shah for help with bioinformatic analyses.
This work was supported by the EU Marie Curie SOLAR research training network and by grants from the Danish Natural Science Research Council (grant 272-08-0391) and the Danish National Research Council.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 2 March 2011.
REFERENCES
- 1. Alzhanova D. V., Napuli A. J., Creamer R., Dolja V. V. 2001. Cell-to-cell movement and assembly of a plant closterovirus: roles for the capsid proteins and Hsp70 homolog. EMBO J. 20:6997–7007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bamford D. H., Caldentey J., Bamford J. K. 1995. Bacteriophage PRD1: a broad host range DSDNA tectivirus with an internal membrane. Adv. Virus Res. 45:281–319 [DOI] [PubMed] [Google Scholar]
- 3. Bosl B., Grimminger V., Walter S. 2005. Substrate binding to the molecular chaperone Hsp104 and its regulation by nucleotides. J. Biol. Chem. 280:38170–38176 [DOI] [PubMed] [Google Scholar]
- 4. DeLaBarre B., Brunger A. T. 2005. Nucleotide dependent motion and mechanism of action of p97/VCP. J. Mol. Biol. 347:437–452 [DOI] [PubMed] [Google Scholar]
- 5. El Bakkouri M., et al. 2010. Structure of RavA MoxR AAA+ protein reveals the design principles of a molecular cage modulating the inducible lysine decarboxylase activity. Proc. Natl. Acad. Sci. U. S. A. 107:22499–22504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fukui T., Eguchi T., Atomi H., Imanaka T. 2002. A membrane-bound archaeal Lon protease displays ATP-independent proteolytic activity towards unfolded proteins and ATP-dependent activity for folded proteins. J. Bacteriol. 184:3689–3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goulet A. 2009. Caractérisation structurale et fonctionnelle de protéines de virus d'archées extrêmophiles: des repliements originaux et une nouvelle lignée virale. Ph.D. thesis. Université de la Méditerranée (Aix-Marseille II), Marseille, France [Google Scholar]
- 8. Goulet A., et al. 2010. Getting the best out of long-wavelength X-rays: de novo chlorine/sulfur SAD phasing of a structural protein from ATV. Acta Crystallogr. D Biol. Crystallogr. 66:304–308 [DOI] [PubMed] [Google Scholar]
- 9. Gudbergsdottir S., et al. 2011. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol. Microbiol. 79:35–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guo L., et al. 2011. Genome analyses of Icelandic strains of Sulfolobus islandicus: model organisms for genetic and virus-host interaction studies. J. Bacteriol. 193:1672–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hanson P. I., Whiteheart S. W. 2005. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell. Biol. 6:519–529 [DOI] [PubMed] [Google Scholar]
- 12. Häring M. 2003. Isolierung und Characterisierung neuer Viren von Hyperthermophilen. Ph.D. thesis. Universität Regensburg, Regensburg, Germany [Google Scholar]
- 13. Häring M., et al. 2005. Virology: independent virus development outside a host. Nature 436:1101–1102 [DOI] [PubMed] [Google Scholar]
- 14. Kagami O., Kamiya R. 1995. Nonradioactive method for ATPase assays. Methods Cell Biol. 47:147–150 [DOI] [PubMed] [Google Scholar]
- 15. Lundqvist J., et al. 2010. ATP-induced conformational dynamics in the AAA+ motor unit of magnesium chelatase. Structure 18:354–365 [DOI] [PubMed] [Google Scholar]
- 16. Lupas A. N., Martin J. 2002. AAA proteins. Curr. Opin. Struct. Biol. 12:746–753 [DOI] [PubMed] [Google Scholar]
- 17. Martin J., Gruber M., Lupas A. N. 2004. Coiled coils meet the chaperone world. Trends Biochem. Sci. 29:455–458 [DOI] [PubMed] [Google Scholar]
- 18. Neuwald A. F., Aravind L., Spouge J. L., Koonin E. V. 1999. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9:27–43 [PubMed] [Google Scholar]
- 19. Ogura T., Wilkinson A. J. 2001. AAA+ superfamily ATPases: common structure-diverse function. Genes Cells 6:575–597 [DOI] [PubMed] [Google Scholar]
- 20. Pelzmann A., et al. 2009. The CoxD protein of Oligotropha carboxidovorans is a predicted AAA+ ATPase chaperone involved in the biogenesis of the CO dehydrogenase [CuSMoO2] cluster. J. Biol. Chem. 284:9578–9586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peremyslov V. V., et al. 2004. Complex molecular architecture of beet yellows virus particles. Proc. Natl. Acad. Sci. U. S. A. 101:5030–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pettit S. C., Everitt L. E., Choudhury S., Dunn B. M., Kaplan A. H. 2004. Initial cleavage of the human immunodeficiency virus type 1 GagPol precursor by its activated protease occurs by an intramolecular mechanism. J. Virol. 78:8477–8485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Prangishvili D., et al. 2006. Structural and genomic properties of the hyperthermophilic archaeal virus ATV with an extracellular stage of the reproductive cycle. J. Mol. Biol. 359:1203–1216 [DOI] [PubMed] [Google Scholar]
- 24. Ruoslahti E. 1996. RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 12:697–715 [DOI] [PubMed] [Google Scholar]
- 25. Snider J., et al. 2006. Formation of a distinctive complex between the inducible bacterial lysine decarboxylase and a novel AAA+ ATPase. J. Biol. Chem. 281:1532–1546 [DOI] [PubMed] [Google Scholar]
- 26. Snider J., Houry W. A. 2006. MoxR AAA+ ATPases: a novel family of molecular chaperones? J. Struct. Biol. 156:200–209 [DOI] [PubMed] [Google Scholar]
- 27. Snider J., Thibault G., Houry W. A. 2008. The AAA+ superfamily of functionally diverse proteins. Genome Biol. 9:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sullivan C. S., Pipas J. M. 2001. The virus-chaperone connection. Virology 287:1–8 [DOI] [PubMed] [Google Scholar]
- 29. White S. R., Lauring B. 2007. AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic 8:1657–1667 [DOI] [PubMed] [Google Scholar]
- 30. Whittaker C. A., Hynes R. O. 2002. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell 13:3369–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xiang X., et al. 2005. Sulfolobus tengchongensis spindle-shaped virus STSV1: virus-host interactions and genomic features. J. Virol. 79:8677–8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.