Abstract
Xenotropic murine leukemia virus-related virus (XMRV) is a gammaretrovirus recently isolated from human prostate cancer and peripheral blood mononuclear cells (PBMCs) of patients with chronic fatigue syndrome (CFS). We and others have shown that host restriction factors APOBEC3G (A3G) and APOBEC3F (A3F), which are expressed in human PBMCs, inhibit XMRV in transient-transfection assays involving a single cycle of viral replication. However, the recovery of infectious XMRV from human PBMCs suggested that XMRV can replicate in these cells despite the expression of APOBEC3 proteins. To determine whether XMRV can replicate and spread in cultured PBMCs even though it can be inhibited by A3G/A3F, we infected phytohemagglutinin-activated human PBMCs and A3G/A3F-positive and -negative cell lines (CEM and CEM-SS, respectively) with different amounts of XMRV and monitored virus production by using quantitative real-time PCR. We found that XMRV efficiently replicated in CEM-SS cells and viral production increased by >4,000-fold, but there was only a modest increase in viral production from CEM cells (<14-fold) and a decrease in activated PBMCs, indicating little or no replication and spread of XMRV. However, infectious XMRV could be recovered from the infected PBMCs by cocultivation with a canine indicator cell line, and we observed hypermutation of XMRV genomes in PBMCs. Thus, PBMCs can potentially act as a source of infectious XMRV for spread to cells that express low levels of host restriction factors. Overall, these results suggest that hypermutation of XMRV in human PBMCs constitutes one of the blocks to replication and spread of XMRV. Furthermore, hypermutation of XMRV proviruses at GG dinucleotides may be a useful and reliable indicator of human PBMC infection.
INTRODUCTION
Xenotropic murine leukemia-related retrovirus (XMRV) was recently isolated from prostate tumor patients with a homozygous R462Q mutation of the antiviral interferon-inducible endoribonuclease RNase L gene (56). Several reports confirmed the presence of XMRV in human prostate cancer cells (2, 46); however, others have reported little (11) or no (1, 16) association of XMRV in prostate cancer samples. XMRV has also been linked to chronic fatigue syndrome (CFS) (28); in this study, 68 of 101 CFS patients (67%) but only 8 of 218 healthy donors (3.7%) were found to be XMRV positive. Infectious virus was isolated from blood of infected individuals, and this virus could be transmitted from peripheral blood mononuclear cells (PBMCs), T cells, and B cells to a highly permissive prostate cancer cell line (LNCaP). Cell-free transmission was also suggested, since XMRV was transmitted from patient plasma to LNCaP cells (28). However, several studies from Europe (Netherlands and United Kingdom) (10, 13, 58), China (19), and recently the United States (55) have been unable to confirm an association of XMRV with CFS. Furthermore, several studies have recently demonstrated that contamination of human samples with mouse DNA is a frequent occurrence (21, 36, 39, 44, 50), emphasizing the importance of ruling out contamination as a possible cause of the reported association between XMRV and prostate cancer or CFS. However, none of these studies presented evidence that the human samples that were found to be XMRV positive were contaminated with mouse DNA.
Innate host restriction factors inhibit replication of various viruses, including human immunodeficiency virus type 1 (HIV-1) and murine leukemia virus (MLV) (15, 27, 30). In humans, several apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3 (APOBEC3) proteins, including A3G, A3F, A3H, and A3B, have been implicated in inhibition of retroviral replication (7, 8, 26, 47). Of these, A3G (47) and A3F (26, 59, 61) appear to be critically important for inhibition of HIV-1 replication. In addition, other host restriction factors, such as Trim5α proteins (45, 54) and tetherin/BST-2/CD317 (34, 57), have been shown to inhibit HIV-1 replication at different stages of the viral life cycle. In the absence of Vif, A3G and A3F are incorporated into newly formed virus particles, leading to deamination of cytidines in minus-strand DNA and resulting in G-to-A hypermutation of the viral genome (4, 15, 24, 26, 30, 60); in addition, A3G and A3F have been shown to inhibit viral DNA synthesis and integration (3, 18, 29, 31, 32, 35, 48).
Recently, we and others reported that XMRV replication can be potently inhibited by human host restriction factors A3G, A3B, A3F, tetherin/BST-2/CD317, and murine APOBEC3 (mA3) (5, 14, 37, 52). To evade these restriction factors, HIV-1 encodes accessory proteins Vif and Vpu, which have been reported to counteract A3G/A3F and tetherin, respectively (12, 34, 47, 53, 57). However, XMRV does not express these accessory proteins. XMRV replication is potently inhibited by A3G, A3B, and A3F when the virus is produced in transiently transfected cells that express these proteins, and the virus undergoes a single cycle of replication. In addition, XMRV is hypermutated in A3G/A3F-expressing T cell lines. However, these studies did not address whether XMRV could replicate and spread in human PBMCs despite these inhibitory factors, either by utilizing an unknown mechanism of evasion or by replicating in subpopulations of cells that express low levels of host restriction factors.
Here, we report that XMRV replication and spread is potently inhibited in cultured human PBMCs and CEM cells, but not in CEM-SS cells. However, G-to-A hypermutated proviruses were detected in cultured PBMCs, indicating a low level of reinfection of PBMCs with XMRV that was subjected to cytidine deamination by one or more APOBEC3 proteins. Since PBMCs express A3G and A3F, but not A3B (23, 38), the results suggest that A3G- and/or A3F-mediated hypermutation constitutes one of the blocks to efficient replication and spread of XMRV in these cells. Moreover, infectious XMRV could be recovered from PBMCs infected with XMRV by coculturing with a canine indicator cell line. Thus, PBMCs may potentially act as a source of infectious XMRV that spreads to cells expressing low levels of the A3 proteins or other restriction factors. In addition, A3G-mediated hypermutation at GG dinucleotides may be a useful marker of human PBMC infection.
MATERIALS AND METHODS
Cells, plasmids, and virus production.
The T lymphoid cell lines CEM and CEM-SS and prostate carcinoma cell lines 22Rv1, LNCaP, and DU145 were obtained from the American Type Culture Collection (ATCC) and maintained as previously described (37). PBMCs obtained from healthy donors at the National Institutes of Health Clinical Center were isolated by fractionation through a Histopaque-1077 (Sigma-Aldrich) density gradient. Isolated cells were activated with 5 μg/ml phytohemagglutinin (PHA) and 200 U/ml interleukin-2 (IL-2) for 3 days, washed, and maintained in medium containing IL-2. Infectious XMRV was harvested from 22Rvl cells and filtered through a 0.45-μm filter, and XMRV RNA copy numbers were determined by using quantitative real-time reverse transcription-PCR (RT-PCR) for detection of XMRV env as described previously (20).
Generation of canine indicator cells for detection of infectious XMRV.
D17 canine osteosarcoma cells were transfected with the previously described MLV-based plasmid pMS2-FP-GF-no3R (6). Briefly, the plasmid contains modified long terminal repeats (LTRs) in which the sequences coding for C- and N-terminal fragments of green fluorescent protein (GFP; FP and GP) were inserted after U3 in the 5′- and 3′-LTR, respectively. The two GFP fragments share a 85-bp stretch of homology that is able to direct minus-strand transfer, resulting in the reconstitution of full-length gfp upon reverse transcription of this packaged RNA. The hygromycin phosphotransferase B gene (hygro), under the control of a simian virus 40 (SV40) promoter, is located after MLV Ψ, replacing all intervening MLV sequences. The R and U5 in the 3′-untranslated region is replaced by the SV40 terminator. Over 8,000 colonies of transfected cells were selected in medium containing hygromycin at a final concentration of 120 μg/ml, pooled, and named DIG cells (D17 indicators of gammaretroviruses).
XMRV infection and replication.
CEM and CEM-SS cells (1 × 106 cells) as well as PHA-activated PBMCs (6 × 106 or 1 × 107 cells) were infected with XMRV obtained from 22Rv1 cells at different dilutions. The XMRV RNA copies were quantified in the virus stock by using quantitative real-time RT-PCR, and different viral RNA copy numbers (104, 105, 106, 107, 108, and 1010) were used to infect cells. The cells were washed 6 h after infection and maintained in 6 ml of medium. Two milliliters of culture medium was harvested every day, frozen at −80°C, and replaced with fresh medium.
Quantitative real-time PCR.
To quantify XMRV RNA copy numbers, culture supernatants containing virus were collected and filtered through a 0.45-μm filter, and viral RNAs were isolated using a QIAamp viral RNA minikit (Qiagen). To avoid DNA contamination, the isolated RNA was treated for 30 min with DNase I, using the Turbo DNA-free kit (Ambion). To quantify the RNA copy numbers, quantitative real-time RT-PCR was performed using the LightCycler 480 RNA master hydrolysis probes reaction mixture (Roche) and the primers and probes described previously for detection of XMRV env (20). A serial 10-fold dilution of VP62 plasmid (9), kindly provided by Robert Silverman, was used to generate a standard curve.
To determine XMRV proviral copy numbers, infected CEM and CEM-SS cells were harvested at day 3 and day 13 after infection, and genomic DNA was isolated using a DNeasy blood and tissue kit (Qiagen). Genomic DNA (500 ng) was used to quantify the proviral copy numbers by using primers that detect XMRV gag as previously described (20). Quantitative real-time PCR was performed in duplicate and normalized to the CCR5 gene by using primers and probes as previously described (33).
Recovery of infectious XMRV from PHA-activated PBMCs.
To recover infectious virus from XMRV-infected PBMCs, 8 × 105 DIG cells were cocultivated with PBMCs (1 × 105 cells) that were infected with culture supernatant containing 104, 105, 106, 107, 108, or 1010 XMRV RNA copies and harvested 7 days postinfection. Cells were transferred to a 10-cm dish 3 days postinfection and maintained for 25 days. After 14 and 25 days, cells were harvested and the percentage of GFP-expressing cells was determined by fluorescence-activated cell sorting (FACS) analysis.
G-to-A hypermutation of XMRV proviruses in infected PBMCs.
Genomic DNA from XMRV-infected PBMCs was isolated with the DNeasy blood and tissue kit (Qiagen), and a 1.2-kb XMRV fragment was amplified, cloned, and sequenced as described previously (37).
Western blot analysis of A3G and A3F expression in PHA-activated PBMCs.
A3G and A3F expression was analyzed by Western blotting as previously described (37). Endogenous A3G and A3F proteins were detected by using an anti-A3G apo-C17 antibody (1:5,000 dilution) and rabbit anti-human A3F (C-18; 1:500 dilution; AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH), anti-apoC17 was from Klaus Strebel, and anti-human APOBEC3F was obtained from Michael Malim. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was detected using anti-GAPDH antibody (Abcam) and served as a loading control.
RESULTS
XMRV replication is restricted in CEM cells but not in CEM-SS cells.
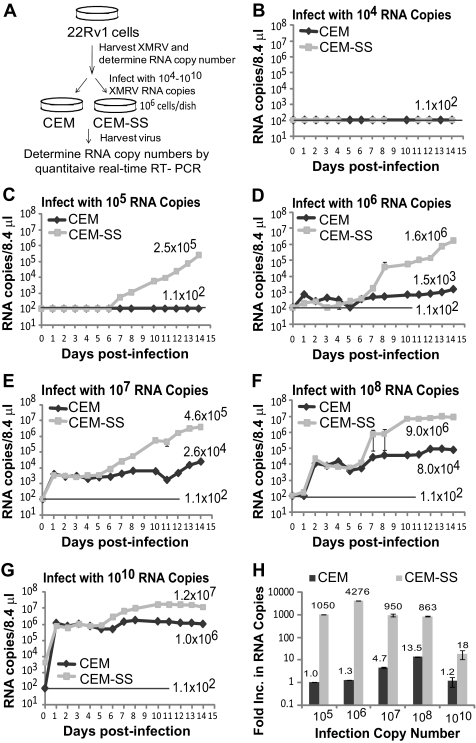
The protocol used to compare the kinetics of replication and spread of XMRV in A3G/A3F-expressing CEM and A3G/A3F-deficient CEM-SS cells is outlined in Fig. 1A. Infectious XMRV was harvested from prostate carcinoma cell line 22Rv1. The 22Rv1 cell line has been shown to produce large amounts of a virus that is virtually identical to XMRV, originally found in prostate cancer samples (22, 56). We quantified the amount of XMRV harvested from the 22Rv1 cell line by using real-time RT-PCR to detect XMRV env RNA as previously described (20). The virus stock was diluted, and different amounts of XMRV RNA copies (104, 105, 106, 107, 108, or 1010) were used to infect 1 × 106 CEM or CEM-SS cells. Culture supernatants from the infected cells were collected every day for 14 days, and the XMRV particles present were quantified using real-time RT-PCR.
Fig. 1.
Replication of XMRV in CEM and CEM-SS cells. (A) Schematic overview of the experimental protocol. Supernatant containing XMRV was harvested from 22Rv1 cells, and XMRV RNA copies in the supernatant were determined by quantitative real-time RT-PCR using primers and a probe for a region of XMRV env. Culture supernatants containing different amounts of XMRV RNA copies were then used to infect 1 × 106 CEM or CEM-SS cells. At 6 h postinfection, cells were extensively washed, and culture supernatants were harvested every day from days 0 to 14. XMRV RNA copy numbers were determined by quantitative real-time RT-PCR. (B to G) Replication kinetics of XMRV 22Rvl in CEM and CEM-SS cells infected with 22Rv1 culture supernatant containing 104 XMRV RNA copies (B), 105 XMRV RNA copies (C), 106 XMRV RNA copies (D), 107 XMRV RNA copies (E), 108 XMRV RNA copies (F), or 1010 XMRV RNA copies (G). The viral RNA in the supernatant of infected cells was isolated, and XMRV RNA was detected by quantitative real-time RT-PCR. The results are plotted as copies of viral XMRV RNA per 8.4 μl of culture supernatant of infected cells. The detection limit for quantitative RT-PCR was 1.1 × 102 copies/8.4 μl (indicated as a solid line). A representative experiment from two independent experiments is shown. Error bars indicate standard deviations. (H) Determination of increases in viral RNA copies in CEM and CEM-SS cells 14 days postinfection. The increase in viral RNA copies produced from CEM and CEM-SS cells infected with different amounts of 22Rvl XMRV are shown in panels B to G. The average number of RNA copies per 8.4 μl for days 12 to 14 was divided by the average number of RNA copies per 8.4 μl from days 1 to 3.
The amounts of XMRV produced from the infected CEM and CEM-SS cells are shown in Fig. 1B to G. Neither CEM nor CEM-SS cells infected with 22Rv1 culture supernatant containing 104 RNA molecules produced detectable amounts of XMRV (detection limit, ∼1.1 × 102 copies/8.4 μl) (Fig. 1B). XMRV particles were detected in both CEM and CEM-SS cells infected with 105, 106, 107, 108, or 1010 viral RNA copies (Fig. 1C to G, respectively); virus production at the beginning of infection (days 1 to 3) was dependent on the amount of XMRV used to infect the cells, and there was no difference in the virus production between CEM and CEM-SS cells (Fig. 1B to G).
Virus production increased to detectable levels in infected CEM-SS cells starting around days 5 to 7, whereas viral production either did not increase or increased only slightly in CEM cells infected with 105 to 1010 XMRV RNA copies (Fig. 1C to G). Virus production did not increase in either CEM-SS or CEM cells infected with 104 RNA copies, indicating that exposure to culture supernatant containing 104 XMRV RNA copies did not result in an infection event (Fig. 1B). In contrast, while no virus production could be detected in CEM cells infected with 105 RNA copies, infection was clearly established in the CEM-SS cells, as evidenced by the dramatic increase in virus production from less-than-detectable levels at day 6 to 2.5 × 105 copies per 8.4 μl at day 14 (Fig. 1C). A similar increase in virus production was observed in CEM-SS cells infected with 106, 107, and 108 RNA copies, whereas less than a 10-fold increase was observed in CEM cells (Fig. 1D, E, and F). In CEM-SS cells infected with 1010 RNA copies, a modest 18-fold increase in virus production was observed, and no increase was observed in CEM cells (Fig. 1G).
To compare the increases in virus production, we compared the average number of viral RNA copies in culture supernatants from days 1 to 3 and days 12 to 14 to determine the increase in RNA copies (Fig. 1H). The results showed a remarkable increase in virus production in CEM-SS cells infected with 105, 106, 107, and 108 RNA copies; the increase in virus production ranged from ∼863-fold to ∼4,276-fold. In contrast, there was only a modest increase in virus production in CEM cells, which reached a maximum of ∼14-fold in cells infected with 108 RNA copies. Only an ∼18-fold increase in virus production was detected in CEM-SS cells infected with 1010 RNA copies, suggesting that a large proportion of the cells were infected with the input virus, and a plateau was reached after all the cells were infected, resulting in superinfection interference. This result is consistent with the estimation that exposure of CEM or CEM-SS cells to 105 RNA copies results in 1 to 10 infection events (compare Fig. 1B and C). Thus, infection with 1010 RNA copies of input virus would result in up to 1 × 106 infection events, and a large proportion of the 1 × 106 CEM-SS cells would be infected with input virus; consequently, most of the cells would be infected after a small increase in virus production, and superinfection interference would be established, suppressing any additional increase in virus production.
An increase in virus producer cells does not account for the increase in virus production.
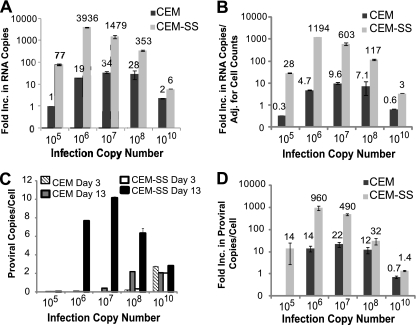
To determine whether an increase in virus producer cells could have contributed to the modest (∼14-fold) increase in virus production observed in CEM cells (Fig. 1H), we repeated the experiment described for Fig. 1 and quantified the virus producer cells every 2 days. The fold increase in virus production was determined by dividing the average RNA copies present on days 13 to 15 with the average RNA copies present on days 1 to 3 (Fig. 2A). The results were similar to those observed in Fig. 1H; the increase in viral production ranged from ∼6- to ∼3,936-fold in CEM-SS cells and up to 34-fold in CEM cells. In this experiment, the increase in viral load in CEM-SS cells after infection with 105 RNA copies was 77-fold, compared to 1,050-fold in Fig. 1H. Since <10 cells were likely to be infected after infection with 105 RNA copies, the variability in viral production may be due to stochastic events that led to greater variation in the number of initially infected cells or loss of virus production from some of the initially infected cells.
Fig. 2.
Increase in virus production and proviral copy numbers in infected CEM and CEM-SS cells. (A) CEM and CEM-SS cells were infected as described for Fig. 1A, and the increase in virus production was determined as described for Fig. 1H. (B) The increase in virus production from panel A was adjusted to the increase in cell numbers. (C) Increase in provirus copy number per cell. Infected cells were collected at day 3 and day 13, and genomic DNA was isolated. Equal amounts of DNA (500 ng) were used to detect XMRV proviral copies by quantitative real-time PCR with XMRV gag primers and probes. XMRV copy numbers were normalized to CCR5 copy numbers. The error bars indicate standard deviations of quadruplicate measurements of a representative experiment of two independent experiments. (D) Increase in XMRV proviral copies per cell from day 3 to day 13. The proviral copy numbers obtained from day 13 were divided by the proviral copy numbers from day 3 to determine the increase in proviral copy numbers per cell.
To determine the extent to which the number of virus-producing cells affected virus production, the increases in virus production were adjusted for the numbers of cells in culture (Fig. 2B). The increase in virus production in CEM and CEM-SS cells was generally reduced by 2- to 4-fold after adjusting for the number of virus-producing cells, but the CEM cells still exhibited modest increases in virus production (up to 10-fold). Therefore, despite the presence of A3G and A3F, XMRV replication was modestly increased in CEM cells infected with 106, 107, and 108 RNA copies.
We also analyzed the extent of XMRV replication in CEM and CEM-SS cells by isolating genomic DNA from day 3 and day 13 after infection and determining the number of XMRV proviral copies present by quantitative real-time PCR (Fig. 2C). The proviral copies per cell were very low at day 3 in all infections except in infections with 1010 RNA copies and were greatly increased at day 13, reaching a maximum of ∼10 in CEM-SS cells. In both CEM and CEM-SS cells infected with 1010 RNA copies, the proviral copies per cell were ∼2 at day 3 and day 13.
The fold increase in proviral copy numbers was determined by comparing the proviral copies per cell present at day 3 and day 13 (Fig. 2D). In CEM-SS cells, the proviral copies per cell increased 14-, 960-, 490-, and 32-fold after infection with 105, 106, 107, and 108 RNA copies, respectively, which were generally similar to the increases in virus production (Fig. 2B and D, compare light gray bars). Proviral copy numbers per cell did not significantly increase in CEM and CEM-SS cells infected with 1010 RNA copies, suggesting that nearly all of the cells were infected by day 3, and superinfection interference was established. In CEM cells, the proviral copies per cell increased 14-, 22-, and 12-fold in cells infected with 106, 107, and 108 RNA copies, respectively. In these cells, virus production increased 4.7-, 9.6-, and 7.1-fold, suggesting that the increase in proviral copies per cell was ∼2- to 3-fold greater than the increase in virus production. This result is consistent with the expectation that most of the proviruses in the CEM cells would be hypermutated and would be unable to produce viral particles.
XMRV replication is severely restricted in PHA-activated human PBMCs.
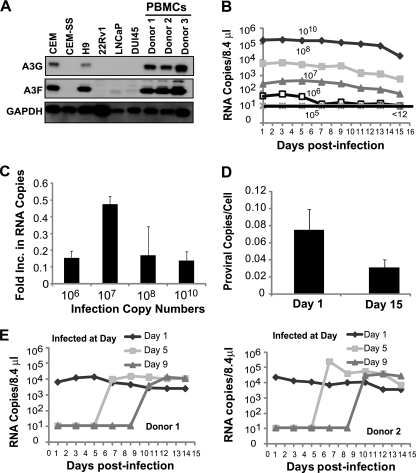
Since XMRV replication and spread was restricted in A3G/A3F-expressing CEM cells, we sought to determine the extent to which XMRV can replicate and spread in A3G/A3F-expressing human PBMCs. We first compared the steady-state levels of A3G and A3F proteins in PHA-activated PBMCs obtained from three different donors to the levels present in human T cell line and prostate cancer cell lines (Fig. 3A). As expected, A3G and A3F expression was clearly detected in CEM and H9 cells but not CEM-SS cells, and A3G was not detectable in prostate cancer cell lines 22Rv1, LNCaP, and DU145. Compared to CEM cells, A3F expression was similar in H9 cells and substantially lower in LNCaP, 22Rv1, and DU145 cells. A3G and A3F were expressed in PBMCs from all three donors at levels similar to or higher than those in CEM cells.
Fig. 3.
Replication of XMRV in PBMCs. (A) A3G and A3F expression in T cell lines (CEM, CEM-SS, and H9), prostate cancer cell lines (22Rv1, LNCaP, and DU145), and PBMCs from three different donors. Western blotting employed an equal amount of protein (10 μg); A3G or A3F protein expression was detected using anti-A3G (C-17) or anti-A3F (C-18) antibodies. GAPDH served as a loading control. (B) PBMCs (1 × 107 cells) from donor 2 were infected with different amounts of XMRV as described for Fig. 1A, and culture supernatants were harvested every 2 days. XMRV RNA copies were determined as described for Fig. 1A. Results are plotted as XMRV RNA copy number per 8.4 μl of virus supernatant. A representative experiment of three independent experiments from three different donors is shown. (C) Increase in viral production in PBMCs. The increase in viral production was determined by dividing the average RNA copy number from days 13 to 15 by the average RNA copy number from days 1 to 3. The averages of three independent experiments using three different donors are shown. The error bars indicate standard errors of the means. (D) Proviral copy numbers per cell in infected PBMCs from three donors. Infected PBMCs were harvested at day 1 and day 15, genomic DNA was isolated, and equal amounts of DNA were used to detect XMRV gag and CCR5 by quantitative real-time PCR. Error bars represent the standard errors of the means. (E) Infection of PBMCs (1 × 107 cells) from donor 1 (left) and donor 2 (right) on days 1, 5, and 9 after activation. PBMCs were infected on the specified days postactivation with XMRV (108 RNA copies) harvested from 22Rv1 cells. Culture supernatants were harvested every second day, and XMRV RNA was detected by quantitative real-time RT-PCR. The results are plotted as copies of viral XMRV RNA per 8.4 μl of culture supernatant of infected cells.
To determine the extent of XMRV replication and spread in A3G/A3F-expressing PBMCs, 22Rv1 culture supernatants containing 105, 106, 107, 108, and 1010 XMRV RNA copies were used to infect 1 × 107 PBMCs from donor 2 (Fig. 3B). Consistent with the results obtained with CEM cells, no virus was detected in PBMCs infected with culture supernatants containing 105 XMRV RNA copies, suggesting that this amount of virus is not sufficient to establish a productive infection in PBMCs. In culture supernatants of PBMCs infected with 106 XMRV RNA copies, a low level of virus was detected, which became undetectable after 7 days. In culture supernatants of PBMCs infected with 107, 108, and 1010 XMRV RNA copies, detectable levels of virus were present early in infection, which ranged from 2 × 102 to 2 × 105 RNA copies per 8.4 μl of culture supernatant. Importantly, we did not observe any increase in viral RNA copies in the infected PBMCs during the next 15 days, indicating that there was little or no virus replication and spread. To quantify viral replication and spread, we compared the average RNA copy numbers from days 1 to 3 and days 13 to 15 (Fig. 3C). Viral RNA copy numbers did not increase, since the ratios of the viral RNA copy numbers on days 13 to 15 to days 1 to 3 were less than 1 (0.14 to 0.5), indicating that XMRV replication and spread in activated human PBMCs is severely restricted.
We also compared the proviral copy numbers per cell in PBMCs infected with 1010 XMRV RNA copies from day 1 and day 15 (Fig. 3D). The proviral copy numbers per cell in the three donors on day 1 averaged ∼0.07, compared to ∼2 proviruses per CEM cell after infection with 1010 XMRV RNA copies; since 10-fold more PBMCs (1 × 107) were infected compared to CEM cells (1 × 106), infection of PBMCs was ∼3-fold less efficient (∼2 × 106 proviruses in CEM cells versus ∼7 × 105 proviruses in PBMCs). The proviral copy numbers per cell did not increase in the PBMCs from day 1 to day 15 and actually decreased from 0.07 to 0.03 proviral copies/cell, but the decrease was not statistically significant (P = 0.15; t test); these results indicated that XMRV replication and spread is severely restricted in PBMCs. Viral production from the PBMCs was approximately 10-fold lower than in CEM cells, which is in general agreement with the ∼3-fold reduction in infection efficiency.
Murine leukemia viruses require cell division to integrate into the target cell chromosomes and complete viral replication (25, 41). To determine whether XMRV replication was restricted in PBMCs because of inefficient cell division, we infected PBMCs on days 1, 5, and 9 after activation and monitored virus production (Fig. 3E). Similar virus production was observed after infection on all 3 days, indicating that equivalent numbers of cells on days 1, 5, and 9 were susceptible to XMRV infection. Thus, the severe restriction to virus replication and spread in the PBMCs was not because of the absence of cells susceptible to XMRV infection.
Hypermutation of XMRV proviruses in PHA-activated PBMCs.
To analyze the effects of A3G/A3F expression on XMRV replication in PBMCs, we harvested XMRV-infected cells at day 15, isolated total cellular DNA, PCR amplified and cloned a 1.2-kb fragment from the pol region of XMRV proviral DNA, and sequenced the cloned PCR products (Fig. 4). Proviral sequences were recovered from PBMC cultures infected with 1010 RNA copies from the three different donors described for Fig. 3; of the 38, 40, and 46 sequences obtained at day 15 from donors 1, 2, and 3, respectively, 2, 6, and 18 (∼5 to 39%) sequences were hypermutated. Furthermore, most of the G-to-A substitutions occurred in the GG dinucleotide context, which is characteristic of A3G-mediated hypermutation. The detection of these hypermutated proviruses in PBMCs indicated that some of the PBMCs were infected with virus that was produced from A3G-expressing cells. A few of the sequences also had multiple G-to-A mutations in the GA dinucleotide context, which is characteristic of A3F-mediated hypermutation. These results indicated that XMRV produced from the PBMCs packaged A3G or A3F into the virions and reinfected the PBMCs, resulting in G-to-A hypermutation in the context of GG or GA dinucleotides, respectively.
Fig. 4.
G-to-A hypermutation of XMRV proviral DNA in PBMCs. PBMCs infected with XMRV containing 1010 RNA copy numbers were harvested at day 15 from three different donors, and genomic DNA was isolated. A 1.2-kb fragment (nucleotides 2465 to 3660) of the viral DNA was amplified by PCR, cloned, and sequenced. The Hypermut color code was used to indicate the mutations (42): red, GG to AG; cyan, GA to AA; green, GC to AC; magenta, GT to AT; yellow, gaps; black, all other mutations. The total numbers of single-nucleotide substitutions are shown in the table below the sequences.
A sensitive assay for detection of replication-competent virus in XMRV-infected cells.
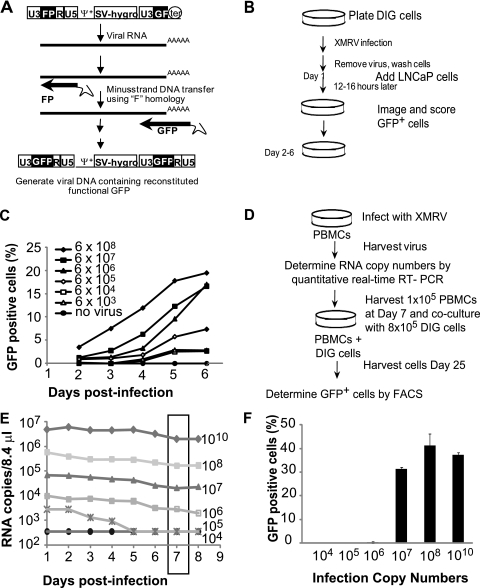
To determine whether infectious XMRV can be recovered from infected PBMCs or from other human cells, we developed a sensitive assay for recovery and detection of replication-competent XMRV. The strategy for XMRV detection and the construction of the DIG cells is illustrated in Fig. 5A. D17 canine osteosarcoma cells were transfected with the previously described MLV-based plasmid pMS2-FP-GF-no3R. This vector contains modified LTRs, in which partially redundant portions of the gfp gene are located in each LTR such that the gfp gene is reconstituted upon reverse transcription. The plasmid includes the MLV packaging signal (Ψ), the hygromycin phosphotransferase B gene (hygro), which is expressed from the SV40 promoter, and the FP portion derived from the 3′ end of gfp in the left LTR and the GF portion derived from the 5′ end of gfp in the right LTR. In addition, the vector contains all other cis-acting sequences required for retroviral replication but lacks gag, pol, and env, and thus can only be packaged by a helper virus that has infected the same cell. XMRV and MLV packaging sequences (Ψ) share ∼70% homology, and XMRV has previously been shown to package MLV-based vectors. During reverse transcription of the MS2 vector, the middle F portion of gfp, which is present in both LTRs and has replaced the R region of the MLV LTR, is used to carry out the minus-strand DNA transfer step of reverse transcription. This results in functional reconstitution of the gfp gene and expression of functional GFP. A D17 cell line that was constructed to stably express the MS2 vector was named the DIG cell line. When XMRV infects DIG cells, many of the newly assembled XMRV particles will package the DIG RNA instead of XMRV RNA, resulting in GFP-positive cells in subsequent rounds of infection.
Fig. 5.
Characterization of DIG cells and isolation of replication-competent XMRV from infected PBMCs. (A) Structure of MLV-based vector pMS2. FP, the 3′ 462-bp fragment of GFP; GF, the 5′ 350-bp fragment of GFP; Ψ+, the extended MLV packaging signal; SV-hygro, hygromycin phosphotransferase B gene under the control of the SV40 promoter; ter, SV40 termination signal. GFP was reconstituted upon F region-mediated minus-strand strong-stop transfer during reverse transcription. (B) Overview of the experimental procedure used to test the DIG XMRV reporter cell line. D17 cells stably transfected with the pMS2 vector were infected with serial 10-fold dilutions of XMRV or no XMRV. LNCaP cells were added 1 day postinfection. The cells were imaged and scored for GFP daily beginning at 2 days postinfection. (C) DIG cells were infected with serial 10-fold dilutions of XMRV or no XMRV. LNCaP cells were added 1 day postinfection. The cells were analyzed by high-content imaging and scored for GFP daily beginning at 2 days postinfection. Spread of XMRV through coculture of DIG and LNCaP cells is represented by the percentage of cells that expressed GFP. RNA copy numbers of the XMRV stock used to infect the DIG cells were determined by quantitative real-time RT-PCR. A representative experiment of three independent experiments is shown. (D) Protocol for recovering replication-competent XMRV from infected PBMCs in DIG cells. PBMCs (6 × 106) were infected with different XMRV as described for Fig. 3A, and virus production was quantified by real-time RT-PCR. At 7 days postinfection, 1 × 105 PBMCs were cocultured with 8 × 105 DIG cells. Virus recovery was detected by the percentage of GFP-positive cells. (E) Virus production in PBMCs infected with different amounts of XMRV. At 7 days postinfection, 1 × 105 infected PBMCs were collected from each infection and added to 8 × 105 DIG cells in six-well dishes. (F) At 3 days postcocultivation, the cells were transferred to a 10-cm dish, and the percentages of GFP-positive cells were determined after 25 days using FACS analysis. The averages of two independent experiments using two different donors are shown. Error bars represent the standard deviations.
The protocol used to demonstrate the sensitivity of XMRV detection using the DIG cells is outlined in Fig. 5B. Briefly, we infected the DIG cells with culture supernatants from 22Rv1 cells containing 6 × 103 to 6 × 108 RNA copies. After 5 h, the virus-containing medium was removed, and the cells were washed with fresh medium. The infected cells were further incubated for 12 to 16 h, at which point 3 × 103 LNCaP cells were added (day 1), to ensure that highly infectible cells were present in the culture. Other experiments have indicated that the D17 cells can be efficiently infected with XMRV (14) (data not shown). The infected cells were monitored every day for 5 days (days 2 to 6) by using high-content imaging (ImageXpress; Molecular Devices), and the percentage of GFP-positive cells was determined. A representative result of three independent experiments (Fig. 5C) shows that XMRV replication and spread were detectable in all cultures that were infected with XMRV.
Recovery of replication-competent XMRV from infected PBMCs.
To detect replication-competent XMRV from infected PBMCs, we cococultured DIG cells with PBMCs that were infected with different amounts of XMRV and monitored GFP expression in the DIG cells after 14 and 25 days (Fig. 5D). Activated PBMCs (6 × 106) from donor 2 were infected with different amounts of XMRV RNA copies, as described earlier (Fig. 3), and virus replication was monitored (Fig. 5E). Similar to donor 2 (Fig. 3B), no increase in virus production was observed after 8 days (Fig. 5E). In fact, the virus production slightly decreased over the course of the experiment, which corresponded with a decrease in the total number of live cells in the cultures. Virus production in the PBMCs infected with 105 RNA copies decreased to undetectable levels 5 days after infection. At 7 days postinfection, 1 × 105 infected PBMCs were cocultured with 8 × 105 DIG cells for 25 days. After 14 days (data not shown) and 25 days, DIG cells were harvested and the percentage of GFP-positive cells was determined by FACS analysis (Fig. 5F). Additional studies are required to determine the minimum amount of time required for recovery of infectious virus using the DIG cells. PBMCs infected with 1010, 108, and 107 XMRV RNA copies generated XMRV that infected the DIG cells. Subsequent spread of the MLV reporter vector in the DIG cells increased the proportion of GFP-positive cells to 31 to 41%. However, infectious XMRV could not be recovered from PBMCs infected with 104, 105, or 106 XMRV RNA copies.
DISCUSSION
Recently, we and others showed that XMRV replication is restricted by A3G, A3F, and A3B as well as human tetherin/BST2/CD317 in a single-cycle assay (5, 14, 37, 52). On the other hand, Lombardi et al. reported detection and recovery of infectious XMRV from PBMCs of chronic fatigue syndrome patients by cocultivation with LNCaP cells (28). Here, we compared viral replication in human PBMCs and T cell lines and observed that XMRV replication and spread was severely restricted in A3G/A3F-positive PBMCs and CEM cells, but not in A3G/A3F-negative CEM-SS cells. In fact, XMRV replication was more potently restricted in the PBMCs than in the CEM cells, since virus production increased modestly in CEM cells (up to 14-fold) but did not increase in the PBMCs, suggesting that other mechanisms of restriction, such as tetherin-mediated inhibition of virus release, may contribute to more potent restriction of viral replication (14, 34, 57). Although A3B was shown to inhibit XMRV replication in single-cycle assays (14), two recent studies have shown that PHA-activated PBMCs express little or no A3B (23, 38). Therefore, it is unlikely that A3B is contributing to the restriction of XMRV replication in PBMCs. However, it is also possible that XMRV infection is less efficient in PBMCs than in CEM cells, and as a consequence, the inhibitory effects of A3G are more potent in PBMCs than in CEM cells.
Our observation that XMRV replication and spread is severely restricted in PHA-activated PBMCs is a novel finding that has not so far been reported. These results are also inconsistent with the report by Lombardi et al. (28), which showed that after activation with PHA and IL-2, nearly all of the PBMCs from XMRV-positive patients become reactive to a monoclonal antibody against the MLV p30 Gag protein; these studies suggested that XMRV replicated and spread efficiently in CFS patients and was present in most of the activated PBMCs (28). In future studies, it will be important to compare the replication and spread of XMRV in PBMCs isolated from normal control donors and CFS patients.
Despite the potent restriction, the PBMCs could be infected with XMRV, as recently shown (17), and the infected PBMCs could produce virus particles; this is consistent with the mechanism of A3G/A3F-mediated inhibition of viral replication, which does not block virus entry or production but greatly reduces the infectivity of the virions (47, 61). We also observed G-to-A hypermutation of XMRV proviruses in PBMCs, indicating that virus produced from the PBMCs could reinfect the cells and generate proviruses; since there was no detectable increase in virus production, most of these reinfection events probably resulted in abortive infection or the generation of highly mutated proviral genomes that were unable to produce viral particles. The extent of hypermutation was variable between different donors and very low for donor 1, suggesting that replication and spread of XMRV in PBMCs from some donors may be very inefficient. These results indicate that hypermutation of XMRV proviral genomes at GG nucleotides, which indicate A3G substrate specificity, may serve as a useful marker for infection and replication of XMRV in human PBMCs.
We recovered replication-competent XMRV from 1 × 105 infected human PBMCs that were initially infected with 1010, 108, or 107 XMRV RNA copies by cocultivation with the DIG cells. We observed that there were approximately 0.07 XMRV proviral copies per cell in PBMCs infected with 1010 RNA copies; we estimate that of the 1 × 105 PBMCs that were cocultivated with the DIG cells, 7,000, 70, and 7 cells were initially infected upon infection with 1010, 108, and 107 XMRV RNA copies, respectively. This estimate suggests that cocultivation with as few as ∼7 PBMCs that were initially infected resulted in the rescue of replication-competent XMRV. Thus, the DIG cell line can be used for sensitive detection and isolation of XMRV or other gammaretroviruses that can serve as a helper virus for the MLV-based vector in the DIG cells.
Although XMRV replication and spread in PHA-activated PBMCs was severely restricted, PBMCs could serve as a reservoir of replication-competent XMRV and facilitate infection of A3G/A3F-deficient cells. A3G/A3F mRNA levels are similar in different T cell subsets (23, 38), but their expression levels were shown to be lower in monocytes; high levels of A3G mRNA expression have also been reported in lung, ovaries, spleen, and thymus (23, 38). However, as shown here and in previous studies (37, 52), A3G expression levels are very low in prostate cancer cells, suggesting that XMRV might replicate more efficiently in these cells. Although 22Rv1, LNCaP, and DU145 cells all expressed detectable levels of A3F, these levels were generally lower than those in CEM cells and did not appear to be sufficient to restrict XMRV replication. Although the XMRV promoter activity in lymphocytes has not been determined, we were able to readily detect virus production from the infected cells, indicating that XMRV RNA and proteins are expressed in the PBMCs. It has been reported that the XMRV promoter is responsive to androgen stimulation, and XMRV transcription is robust in LNCaP prostate tumor cells, suggesting that XMRV transcription may be higher in prostate tissues than in PBMCs (40).
We and others have shown that the anti-HIV-1 drugs zidovudine, tenofovir, and raltegravir are potent inhibitors of XMRV replication (20, 37, 43, 49, 51). Although there is no evidence to indicate that XMRV infection and replication contribute to the pathogenesis of chronic fatigue syndrome or prostate cancer, clinical trials are being contemplated to determine the effectiveness of antiviral drugs for the treatment of these diseases. We believe it is necessary to have reliable and sensitive assays for XMRV detection and replication before the efficacy of any antiviral therapy can be assessed. Our results show that monitoring virus production from PBMCs is not a reliable indicator of virus replication and spread in PBMCs; moreover, the fact that we observed little or no replication in PBMCs implies that it may be very difficult to monitor any potential inhibitory effects of antiviral drugs by evaluation of XMRV levels in peripheral blood.
In summary, our results show that XMRV replication and spread is severely restricted in PBMCs, but these cells can serve as a reservoir for generation of infectious virus that can potentially spread to cells that express low levels of these restriction factors. Additionally, hypermutation of XMRV at GG dinucleotides may be a useful marker for detection of human PBMC infection.
ACKNOWLEDGMENTS
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
Footnotes
Published ahead of print on 16 February 2011.
REFERENCES
- 1. Aloia A. L., et al. 2010. XMRV: a new virus in prostate cancer? Cancer Res. 70:10028–10033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arnold R. S., et al. 2010. XMRV infection in patients with prostate cancer: novel serologic assay and correlation with PCR and FISH. Urology 75:755–761 [DOI] [PubMed] [Google Scholar]
- 3. Bishop K. N., Holmes R. K., Malim M. H. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 80:8450–8458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bishop K. N., et al. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392–1396 [DOI] [PubMed] [Google Scholar]
- 5. Bogerd H. P., Zhang F. D. B. P., Cullen B. R. 2011. Human APOBEC3 proteins can inhibit xenotropic murine leukemia virus-related virus infectivity. Virology 410:234–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheslock S. R., Anderson J. A., Hwang C. K., Pathak V. K., Hu W. S. 2000. Utilization of nonviral sequences for minus-strand DNA transfer and gene reconstitution during retroviral replication. J. Virol. 74:9571–9579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dang Y., et al. 2008. Human cytidine deaminase APOBEC3H restricts HIV-1 replication. J. Biol. Chem. 283:11606–11614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doehle B. P., Schafer A., Cullen B. R. 2005. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology 339:281–288 [DOI] [PubMed] [Google Scholar]
- 9. Dong B., et al. 2007. An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc. Natl. Acad. Sci. U. S. A. 104:1655–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Erlwein O., et al. 2010. Failure to detect the novel retrovirus XMRV in chronic fatigue syndrome. PLoS One 5:e8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fischer N., et al. 2008. Prevalence of human gammaretrovirus XMRV in sporadic prostate cancer. J. Clin. Virol. 43:277–283 [DOI] [PubMed] [Google Scholar]
- 12. Gabuzda D. H., et al. 1992. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J. Virol. 66:6489–6495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Groom H. C., et al. 2010. Absence of xenotropic murine leukaemia virus-related virus in UK patients with chronic fatigue syndrome. Retrovirology 7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Groom H. C., Yap M. W., Galao R. P., Neil S. J., Bishop K. N. 2010. Susceptibility of xenotropic murine leukemia virus-related virus (XMRV) to retroviral restriction factors. Proc. Natl. Acad. Sci. U. S. A. 107:5166–5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harris R. S., et al. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809 [DOI] [PubMed] [Google Scholar]
- 16. Hohn O., et al. 2009. Lack of evidence for xenotropic murine leukemia virus-related virus(XMRV) in German prostate cancer patients. Retrovirology 6:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hohn O., et al. 2010. No evidence for XMRV in German CFS and MS patients with fatigue despite the ability of the virus to infect human blood cells in vitro. PLoS One 5:e15632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Holmes R. K., Koning F. A., Bishop K. N., Malim M. H. 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J. Biol. Chem. 282:2587–2595 [DOI] [PubMed] [Google Scholar]
- 19. Hong P., Li J., Li Y. 2010. Failure to detect xenotropic murine leukaemia virus-related virus in Chinese patients with chronic fatigue syndrome. Virol. J. 7:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hong S., et al. 2009. Fibrils of prostatic acid phosphatase fragments boost infections with XMRV (xenotropic murine leukemia virus-related virus), a human retrovirus associated with prostate cancer. J. Virol. 83:6995–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hue S., et al. 2010. Disease-associated XMRV sequences are consistent with laboratory contamination. Retrovirology 7:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Knouf E. C., et al. 2009. Multiple integrated copies and high-level production of the human retrovirus XMRV (xenotropic murine leukemia virus-related virus) from 22Rv1 prostate carcinoma cells. J. Virol. 83:7353–7356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koning F. A., et al. 2009. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J. Virol. 83:9474–9485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lecossier D., Bouchonnet F., Clavel F., Hance A. J. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 25. Lewis P. F., Emerman M. 1994. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J. Virol. 68:510–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liddament M. T., Brown W. L., Schumacher A. J., Harris R. S. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:1385–1391 [DOI] [PubMed] [Google Scholar]
- 27. Lochelt M., et al. 2005. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. U. S. A. 102:7982–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lombardi V. C., et al. 2009. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science 326:585–589 [DOI] [PubMed] [Google Scholar]
- 29. Luo K., et al. 2007. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J. Virol. 81:7238–7248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mangeat B., et al. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103 [DOI] [PubMed] [Google Scholar]
- 31. Mbisa J. L., et al. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 81:7099–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mbisa J. L., Bu W., Pathak V. K. 2010. APOBEC3F and APOBEC3G inhibit HIV-1 DNA integration by different mechanisms. J. Virol. 84:5250–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mbisa J. L., Delviks-Frankenberry K. A., Thomas J. A., Gorelick R. J., Pathak V. K. 2009. Real-time PCR analysis of HIV-1 replication post-entry events. Methods Mol. Biol. 485:55–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Neil S. J., Zang T., Bieniasz P. D. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430 [DOI] [PubMed] [Google Scholar]
- 35. Newman E. N., et al. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166–170 [DOI] [PubMed] [Google Scholar]
- 36. Oakes B., et al. 2010. Contamination of human DNA samples with mouse DNA can lead to false detection of XMRV-like sequences. Retrovirology 7:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Paprotka T., et al. 2010. Inhibition of xenotropic murine leukemia virus-related virus by APOBEC3 proteins and antiviral drugs. J. Virol. 84:5719–5729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Refsland E. W., et al. 2010. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 38:4274–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Robinson M. J., et al. 2010. Mouse DNA contamination in human tissue tested for XMRV. Retrovirology 7:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rodriguez J. J., Goff S. P. 2010. Xenotropic murine leukemia virus-related virus establishes an efficient spreading infection and exhibits enhanced transcriptional activity in prostate carcinoma cells. J. Virol. 84:2556–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roe T., Reynolds T. C., Yu G., Brown P. O. 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 12:2099–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rose P. P., Korber B. T. 2000. Detecting hypermutations in viral sequences with an emphasis on G→A hypermutation. Bioinformatics 16:400–401 [DOI] [PubMed] [Google Scholar]
- 43. Sakuma R., Sakuma T., Ohmine S., Silverman R. H., Ikeda Y. 2010. Xenotropic murine leukemia virus-related virus is susceptible to AZT. Virology 397:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sato E., Furuta R. A., Miyazawa T. 2010. An endogenous murine leukemia viral genome contaminant in a commercial RT-PCR kit is amplified using standard primers for XMRV. Retrovirology 7:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sayah D. M., Sokolskaja E., Berthoux L., Luban J. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569–573 [DOI] [PubMed] [Google Scholar]
- 46. Schlaberg R., Choe D. J., Brown K. R., Thaker H. M., Singh I. R. 2009. XMRV is present in malignant prostatic epithelium and is associated with prostate cancer, especially high-grade tumors. Proc. Natl. Acad. Sci. U. S. A. 106:16351–16356 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47. Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650 [DOI] [PubMed] [Google Scholar]
- 48. Shindo K., et al. 2003. The enzymatic activity of CEM15/APOBEC-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J. Biol. Chem. 278:44412–44416 [DOI] [PubMed] [Google Scholar]
- 49. Singh I. R., Gorzynski J. E., Drobysheva D., Bassit L., Schinazi R. F. 2010. Raltegravir is a potent inhibitor of XMRV, a virus implicated in prostate cancer and chronic fatigue syndrome. PLoS One 5:e9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Smith R. A. 2010. Contamination of clinical specimens with MLV-encoding nucleic acids: implications for XMRV and other candidate human retroviruses. Retrovirology 7:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Smith R. A., Gottlieb G. S., Miller A. D. 2010. Susceptibility of the human retrovirus XMRV to antiretroviral inhibitors. Retrovirology 7:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stieler K., Fischer N. 2010. Apobec 3G efficiently reduces infectivity of the human exogenous gammaretrovirus XMRV. PLoS One 5:e11738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Strebel K., Klimkait T., Martin M. A. 1988. A novel gene of HIV-1, vpu, and its 16-kilodalton product. Science 241:1221–1223 [DOI] [PubMed] [Google Scholar]
- 54. Stremlau M., et al. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427:848–853 [DOI] [PubMed] [Google Scholar]
- 55. Switzer W. M., et al. 2010. Absence of evidence of xenotropic murine leukemia virus-related virus infection in persons with chronic fatigue syndrome and healthy controls in the United States. Retrovirology 7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Urisman A., et al. 2006. Identification of a novel gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2:e25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57. Van Damme N., et al. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. van Kuppeveld F. J., et al. 2010. Prevalence of xenotropic murine leukaemia virus-related virus in patients with chronic fatigue syndrome in the Netherlands: retrospective analysis of samples from an established cohort. BMJ 340:c1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wiegand H. L., Doehle B. P., Bogerd H. P., Cullen B. R. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang H., et al. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zheng Y. H., et al. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78:6073–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]