Abstract
Natural killer (NK) cells are important innate effector cells controlled by an array of activating and inhibitory receptors. Some alleles of the inhibitory killer-cell immunoglobulin-like receptor KIR3DL1 in combination with its HLA class I ligand Bw4 have been genetically associated with slower HIV-1 disease progression. Here, we observed that the presence of HLA-B Bw4 was associated with elevated frequencies of KIR3DL1+ CD56dim NK cells in chronically HIV-1-infected individuals from the rural district of Kayunga, Uganda. In contrast, levels of KIR2DL1+ CD56dim NK cells were decreased, and levels of KIR2DL3+ CD56dim NK cells were unchanged in infected subjects carrying their respective HLA-C ligands. Furthermore, the size of the KIR3DL1+ NK cell subset correlated directly with viral load, and this effect occurred only in HLA-B Bw4+ patients, suggesting that these cells expand in response to viral replication but may have relatively poor antiviral capacity. In contrast, no association with viral load was present for KIR2DL1+ and KIR2DL3+ NK cells. Interestingly, chronic HIV-1 infection was associated with an increased polyfunctional response in the NK cell compartment, and, upon further investigation, KIR3DL1+ CD56dim NK cells exhibited a significantly increased functional response in the patients carrying HLA-B Bw4. These results indicate that chronic HIV-1 infection is associated with increased NK cell polyfunctionality and elevated levels of KIR3DL1+ NK cells in Ugandans carrying the HLA-B Bw4 motif.
INTRODUCTION
NK cells are innate lymphocytes that play a significant role in the control of viral infections, including HIV-1 (18, 27). NK cells can suppress HIV-1 replication via direct cytolysis of infected cells (1, 4, 7, 21) and through production of CC-chemokines such as MIP-1β and RANTES because these chemokines reduce virus entry through competitive inhibition of coreceptor binding (19, 31, 40). In addition, activated NK cells are an innate source of gamma interferon (IFN-γ) and tumor necrosis factor alpha (TNF-α) that contribute to the recruitment and activation of the adaptive immune response. The NK cell response is probably most important in early control of HIV viremia before the onset of the adaptive CD8 T cell response. However, it is likely that the antiviral activity of NK cells contributes to control of HIV replication throughout infection.
Recently, NK cell research in the HIV field has focused on the killer-cell immunoglobulin-like receptors (KIRs), a group of activating and inhibitory receptors that may regulate the immune response to pathogens or cellular transformations. There are 17 KIR genes coding for nine inhibitory receptors, six activating receptors, and two pseudogenes which are not expressed (35). Over 30 KIR haplotypes exist that can be divided into groups based on absence (haplotype A) or presence (haplotype B) of activating KIRs (43). In HIV-1 infection, there has been interest in the KIRs and their HLA class I ligands, because KIR and HLA genes are highly polymorphic and because certain KIR-HLA interactions could influence differences between individuals in HIV-1 disease progression (12).
The two KIR genes KIR3DL1 and KIR3DS1, which are alleles of the same locus, and the inhibitory and activating receptors they encode are associated with slower HIV-1 disease progression when found in combination with their HLA ligand (5, 34, 36, 37). However, the mechanism behind this effect is not yet well understood. KIR3DL1 and probably also KIR3DS1 recognize HLA Bw4 allotypes with the nonpolar amino acid, isoleucine (Bw4-80I), and, to a lesser extent, with the polar amino acid threonine at position 77 to 80 (Bw4-80T) (1, 11). East African populations have low frequencies of the KIR3DS1 allele and high frequencies of KIR3DL1 alleles and HLA-B with the Bw4 motif, particularly with an isoleucine at position 80, compared to other populations globally (43). Similarly, the inhibitory KIR2DL2 and KIR2DL3 gene products are alleles of the same locus and recognize HLA-C group C1 molecules and show a more balanced distribution but favor KIR2DL3 expression in East Africa. The KIR2DL1 gene is constitutively expressed across all populations, and the receptor it codes for recognizes HLA-C group C2 molecules (43). Expression of KIRs is genetically controlled (33), and the role of self-major histocompatibility complex (MHC) molecules in NK cell KIR repertoire formation is controversial (3, 50).
In this study, we have investigated the influence of HLA ligands and HIV-1 infection status and viral load on the expression of certain KIRs and function in the NK cell compartment in a cohort from the rural district of Kayunga in Uganda (17, 26). This cohort allowed analysis of NK cell KIR2DL1, KIR2DL3, and KIR3DL1 expression in humans with untreated chronic HIV-1 infection in the context of the rural East African environment. Furthermore, we investigated the link between viral burden, frequency of KIR-expressing NK cells, and NK cell function in these patients. The data are discussed in relation to the previously published protective effect of the KIR3DL1-Bw4-80I combination in progression to AIDS and the ability of the NK cell compartment to adapt to a chronic infection.
MATERIALS AND METHODS
Patients and cells.
Study participants aged 19 to 48 years were from a community-based cohort in the Kayunga district, Uganda (26). Peripheral blood mononuclear cells (PBMCs) were isolated from acid citrate dextrose (ACD)-anticoagulated whole blood within 6 h of collection by centrifugation through Ficoll-Hypaque (Pharmacia, Sweden) using Leucosep tubes (Greiner Bio-One, Germany) at 800 × g for 15 min and cryopreserved as previously described (39). The study was approved by institutional review boards both in the United States and Uganda. The human erythroleukemia cell line K562 (American Type Culture Collection, Manassas, VA) was maintained in complete medium.
Diagnostic testing.
HIV-1 testing was performed on all participants as previously described (16). Briefly, HIV-1 enzyme-linked immunosorbent assay (ELISA) screening was performed using a Genetic Systems rLAV (Bio-Rad Laboratories, Redmond, WA) ELISA. Reactive samples were further tested using a Vironostika HIV-1 Microelisa System (Organon Teknika, Durham, NC) and confirmed with a Genetic Systems HIV-1 Western Blot (Bio-Rad Laboratories, Redmond, WA). Viral load was measured using an Amplicor HIV-1 Monitor test, version 1.5 (Roche Diagnostics, Indianapolis, IN), in the standard mode. Absolute B, NK, and T cell counts were performed on whole blood using MultiTEST four-color reagent and TruCount tubes and analyzed using MultiSET software (Becton Dickinson, San Jose, CA).
KIR and HLA typing.
Subjects were genotyped for KIR3DL1/KIR3DS1 and KIR2DL2/KIR2DL3 by a sequence-specific priming (SSP) real-time PCR assay validated with International Histocompatibility Working Group panels, as previously described (30). HLA-A was genotyped by a sequence-specific priming (SSP) real-time PCR assay allowing the discrimination of Bw4 alleles. HLA-B Bw4 and Bw6 were genotyped by an SSP real-time PCR assay allowing the discrimination of Bw4 alleles having isoleucine at position 80. HLA-C was genotyped by an SSP real-time PCR assay allowing the discrimination of group 1 (C1) or group 2 (C2) alleles. Each typing reaction was a multiplex real-time PCR designed to target one ligand-specific region and one nonpolymorphic region for standardization. Samples were run in a 384-well plate format, read automatically by a 7900HT Fast Real-time PCR System (Applied Biosystems, Foster City, CA) and analyzed with Sequence Detection Software, version 2.2.2.
NK cell analysis by flow cytometry.
For phenotypic analysis cryopreserved specimens were thawed and washed, and counts were performed with Guava ViaCount reagent on a Guava PCA machine (Guava Technologies, Hayward, CA). PBMCs were distributed into 96-well U-bottom plates, washed, and subsequently stained with combinations of Aqua Live Dead Stain, anti-CD4 Qdot605 (both from Invitrogen, Carlsbad, CA), anti-CD3 energy-coupled dye (ECD), anti-KIR3DL1/KIR3DS1 (KIR3DL1/DS1) phycoerythrin (PE) (clone z27) (both from Beckman Coulter, Brea, CA), anti-CD14 allophycocyanin (APC)-H7, anti-CD16 Pacific Blue, anti-CD19 APC-H7, anti-CD56 PE-Cy7, anti-KIR3DL1 fluorescein isothiocyanate (FITC) (clone DX9), anti-KIR2DL2/DL3/DS2 PE (clone DX27) (all from BD Biosciences, San Jose, CA), anti-KIR2DL3 APC (clone 1800701), and anti-KIR2DL1 FITC (clone 143211) (both from R&D Systems, Minneapolis, MN).
For functional analysis, PBMCs were incubated with or without K562 cells for 18 h at 37°C in a 96-well U-bottom plate in the presence of anti-CD107a FITC (BD Biosciences), brefeldin A (Sigma, St. Louis, MO), and monensin (Becton Dickinson). After incubation, cells were washed and stained with Aqua Live-Dead Stain, blocked, and stained for surface markers CD3, CD4, CD14, CD16, CD19, and CD56 as above. In the KIR-specific functional assay, anti-KIR3DL1 FITC (clone DX9) was used in place of CD107a and stained with the other surface markers. Cells were next washed, fixed with Cytofix/Cytoperm, permeabilized using Perm/Wash (Becton Dickinson), stained with anti-MIP-1β PE and anti-IFN-γ APC for 30 min, washed, and fixed again in 2% formaldehyde. Samples were run on an LSRII instrument (Becton Dickinson, San Jose, CA) and analyzed using FlowJo software, version 9.1 (Tree Star, Ashland, OR) (17, 25).
Statistical analysis.
Statistical analysis was performed using GraphPad Prism software, version 5.0a for Mac OSX (GraphPad Software, La Jolla, CA). Comparisons between groups were performed using Fisher's exact test or chi-square test for categorical data and a nonparametric Mann-Whitney U test for continuous data. Associations between outcomes were determined by Spearman's rank correlation. For paired observations a paired t test was used. P values of <0.05 were considered statistically significant. Polychromatic flow cytometry analysis and presentation of distributions was performed using SPICE, version 5.1 (http://exon.niaid.nih.gov/spice) (42).
RESULTS
KIR2DL2, KIR2DL3, KIR3DL1/DS1, and HLA class I genotype in Ugandans.
To investigate the role of KIR and HLA class I regulation of human NK cells in HIV-1 infection, we studied a cohort from the Kayunga district in Uganda (17, 26). Fifty-two chronically HIV-1-infected and antiretroviral therapy (ART)-naïve subjects as well as 29 HIV-1-negative subjects were included. Cryopreserved PBMCs were used to identify carriers of KIR2DL2, KIR2DL3, KIR3DL1, and KIR3DS1 and their HLA ligands using a real-time PCR assay (Table 1) (30). The representation of KIR2DL2 homozygosity was 13% of HIV-1-uninfected and 12% HIV-1-infected participants, while 48% of the HIV-1-uninfected participants and 55% of the HIV-1-infected group were homozygous for KIR2DL3. In HIV-1-uninfected participants, homozygous HLA-C C1 genotype frequency was 4%, and homozygous HLA-C C2 frequency was 15%. In HIV-1-infected participants, homozygous HLA-C C1 genotype frequency was 24%, and homozygous HLA-C C2 frequency was 34%. Heterozygous HLA-C group C1 and C2 was observed in 81% and 42% of the uninfected and infected HIV-1 participants, respectively. No difference was observed between HIV-1-infected and uninfected participants for KIR2DL2/DL3 gene frequency; however, HLA-C genotype was significantly different between the two groups (P = 0.004), with markedly higher representation of heterozygous HLA-C group C1 and C2 in the HIV-1-uninfected group. There was no difference in the HIV-1 viral load or CD4 T cell absolute counts based on HLA-C genotype (data not shown).
Table 1.
Study population genotype statistics
| Allele(s) and genotype | No. (%) of subjects in the population |
|
|---|---|---|
| HIV-1 negative (n = 29) | HIV-1 positive (n = 52) | |
| KIR2DL2 and KIR2DL3 | ||
| KIR2DL2/KIR2DL2 | 3 (13) | 6 (12) |
| KIR2DL2/KIR2DL3 | 9 (39) | 16 (33) |
| KIR2DL3/KIR2DL3 | 11 (48) | 27 (55) |
| KIR3DL1 | ||
| KIR3DL1/KIR3DL1 | 21 (88) | 45 (90) |
| KIR3DL1/KIR3DS1 | 2 (8) | 4 (8) |
| KIR3DS1/KIR3DS1 | 1 (4) | 1 (2) |
| HLA-A | ||
| Bw4−/Bw4− | 23 (85) | 39 (76) |
| Bw4+/Bw4− | 4 (15) | 12 (24) |
| Bw4+/Bw4+ | 0 (0) | 0 (0) |
| HLA-B | ||
| Bw6/Bw6 | 5 (20) | 21 (40) |
| Bw6/Bw4-80I | 11 (44) | 16 (31) |
| Bw4-80I/Bw4-80I | 7 (28) | 10 (19) |
| Bw4-80T/Bw4-80I | 1 (4) | 1 (2) |
| Bw6/Bw4-80T | 1 (4) | 3 (6) |
| Bw4-80T/Bw4-80T | 0 (0) | 1 (2) |
| HLA-C | ||
| C1/C1 | 1 (4) | 12 (24) |
| C1/C2 | 21 (81) | 21 (42) |
| C2/C2 | 4 (15) | 17 (34) |
A homozygous KIR3DL1 genotype accounted for 90% and 88% of the HIV-1-infected and HIV-1-uninfected participants, respectively. The HLA-B Bw4-80I genotype was present in 52% of the HIV-infected and 76% of the uninfected subjects, while 8% of the HIV-1-uninfected and 10% of the HIV-1-infected individuals carried the KIR3DL1 ligand HLA-B Bw4-80T. Fifteen percent of the HIV-1-uninfected and 24% of the HIV-1-infected study population carried the HLA-A Bw4 genotype, and no participants were homozygous for HLA-A with a Bw4 motif. There were no statistically significant differences between HIV-1-infected and uninfected participants with regard to KIR3DL1/DS1 genotype or corresponding HLA-A or HLA-B class I ligands. There was no difference in the HIV-1 viral load or CD4 T cell absolute counts based on HLA-A or HLA-B genotype (data not shown).
HIV-1 infection is associated with increased levels of KIR3DL1+ CD56dim NK cells and decreased levels of KIR2DL1+ CD56dim NK cells in patients carrying their respective HLA ligands.
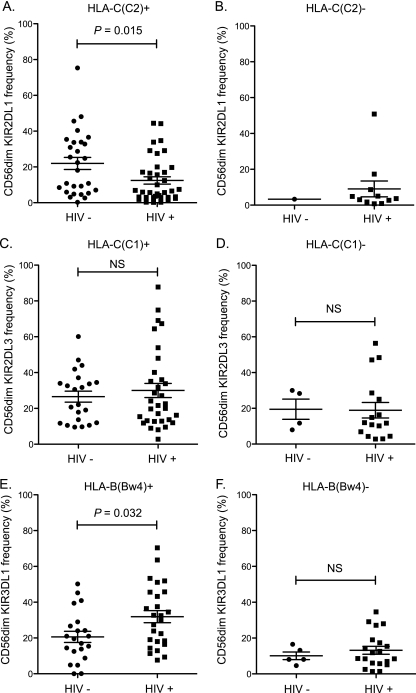
Cryopreserved specimens were thawed, washed, and phenotyped using nine-color flow cytometry panels. NK cells were identified through the expression of CD16 and CD56, and the analysis was focused on the total CD56dim NK cell population regardless of CD16 expression based on the low frequency of CD56dim CD16− NK cells and because similar results were observed between the CD16+ and CD16− NK CD56dim cell subsets. Surface expression of KIRs was identified and compared between infected and uninfected Ugandans and analyzed with regard to the presence of HLA licensing ligands. HIV-1-infected patients expressing HLA-C group C2 displayed lower frequency of KIR2DL1 expression on CD56dim NK cells than uninfected controls (P = 0.015) (Fig. 1A). Unfortunately, there was only one HIV-1-uninfected individual with KIR2DL1+ NK cells in the absence of their HLA ligand, so no comparison could be made to HIV-1-infected HLA-C group C1-homozygous individuals (Fig. 1B). No differences were observed for KIR2DL3 expression between HIV-1-infected or uninfected participants in the presence of their HLA-C ligands (Fig. 1C) or absence of their HLA-C ligands (Fig. 1D).
Fig. 1.
HLA-B Bw4 is associated with increased KIR3DL1+ NK cell frequency, while HLA-C group C2 is associated with decreased KIR2DL1+ NK cell frequency. Frozen PBMCs were thawed and stained using several multicolor flow panels to assess KIR phenotype. Plots are pregated on CD56dim CD3− CD14− CD19− live lymphocytes. (A) Vertical scatter plot comparing frequency of CD56dim NK cells expressing KIR2DL1 (clone 143211) in HLA-C group C2-positive subjects with (n = 37) or without (n = 28) HIV-1 infection. (B) Vertical scatter plot comparing frequency of CD56dim NK cells expressing KIR2DL1 in HLA-C group C2-negative subjects with (n = 11) or without (n = 1) HIV-1 infection. (C) Vertical scatter plot comparing frequency of CD56dim NK cells expressing KIR2DL3 (clone 180071 and DX27) in HLA-C C1-positive subjects with (n = 32) or without (n = 22) HIV-1 infection. (D) Vertical scatter plot comparing frequency of CD56dim NK cells expressing KIR2DL3 in HLA-C C1-negative subjects with (n = 16) or without (n = 4) HIV-1 infection. (E) Vertical scatter plot comparing frequency of CD56dim NK cells expressing KIR3DL1 in HLA-B Bw4-positive subjects with (n = 26) or without (n = 21) HIV-1 infection. (F) Vertical scatter plot comparing frequency of CD56dim NK cells expressing KIR3DL1 in HLA-B Bw4-negative subjects with (n = 20) or without (n = 5) HIV-1 infection. NS, nonsignificant.
The size of the NK cell subset expressing the KIR3DL1 receptor, as determined by double-positive staining with monoclonal antibodies (MAbs) z27 and DX9, varied between subjects. Interestingly, the percentage of CD56dim NK cells expressing KIR3DL1 was significantly higher in HIV-1-infected than in uninfected subjects with at least one HLA-B Bw4 ligand, either Bw4-80I or Bw4-80T (P = 0.033) (Fig. 1E). Notably, this effect was not observed in the HLA-B Bw6 group (Fig. 1F) although there was a trend toward a higher frequency of the KIR3DL1 subset. Furthermore, focusing solely on the HIV-1-infected group, individuals who possessed one of the KIR3DL1 ligands through HLA-B Bw4 had a significantly higher frequency of CD56dim KIR3DL1+ NK cells than those who were homozygous for HLA-B Bw6 (P < 0.001).
The frequency of KIR3DL1+ CD56dim NK cells is directly associated with HIV-1 load in HLA-B Bw4 carriers.
Within the HIV-infected group of HLA-B Bw4 carriers, KIR3DL1 expression ranged from 7.6% up to 70.4% of CD56dim NK cells. This prompted us to compare the relative size of the KIR3DL1+ NK cell subset with viral load. Interestingly, in Bw4 carriers there was a direct correlation between viral load and the percentage of KIR3DL1+ CD56dim NK cells (P < 0.001) (Fig. 2 A). This pattern was not present in Bw4 noncarriers (Fig. 2D). KIR2DL3 expression ranged from 2.8% to 87.8% of CD56dim NK cells. However, there was no correlation to viral load in either subjects carrying at least one HLA-C group C1 allele (Fig. 2B) or subjects without an HLA-C group C1 allele (Fig. 2E). KIR2DL1 expression ranged from 0.2% to 50.8% of CD56dim NK cells but also showed no correlation to viral load, irrespective of HLA-C group C1 or C2 expression (Fig. 2C and F).
Fig. 2.
Association between HIV-1 load and frequency of KIR+ NK cells in carriers with or without their corresponding HLA-ligand. (A and D) Spearman rank correlation between HIV-1 load and percentage of CD56dim NK cell KIR3DL1 expression in Bw4-80I carriers and noncarriers, respectively. (B and E) Spearman rank correlation between HIV-1 load and percentage of CD56dim NK cell KIR2DL3 expression in HLA-C (C1) carriers and noncarriers, respectively. (C and F) Spearman rank correlation between HIV-1 load and percentage of CD56dim NK cell KIR2DL1 expression in HLA-C (C2) carriers and noncarriers, respectively.
Because the HIV-1 load is both a source of antigen and a driver of disease progression, we next assessed possible correlations with CD4 T cell counts. CD4 T cell counts were negatively correlated to HIV-1 viral load in this cohort (rho = −0.379; P = 0.007). In patients carrying HLA-B Bw4 there was no correlation between KIR3DL1+ CD56dim NK cells and CD4 counts (data not shown), suggesting that KIR3DL1 expression in NK cells is not directly associated with CD4 levels despite the correlation with viral load (Fig. 2A). While the mean fluorescence intensity (MFI) of KIR3DL1 on CD56dim NK cells showed no association with HIV-1 viral load, we did observe a trend toward a positive correlation with CD4 absolute counts, but only in the Bw4 carriers (rho = 0.324; P = 0.070). In contrast, in Bw4 noncarriers there was, surprisingly, a significant inverse relationship between KIR3DL1+ CD56dim NK cells and CD4 absolute counts (rho = −0.465; P = 0.039), suggesting that the size of this subset may be elevated in the absence of the Bw4 motif in more advanced disease (data not shown). Additionally, an inverse relationship was observed between KIR2DL3+ CD56dim NK cells in the absence of HLA-C C1 and CD4 absolute counts (rho = −0.520; P = 0.039) (data not shown). There was no relationship between CD4 absolute counts and KIR2DL3+ CD56dim NK cells in the presence of their HLA-C ligand. Likewise, KIR2DL1+ CD56dim NK cells, irrespective of HLA-C genotype, showed no correlation to absolute CD4 counts. We also examined the overall number of inhibitory KIRs with ligands present, as well as the number of ligands for inhibitory KIRs, and observed no relationship with KIR expression, HIV-1 viral load, CD4 absolute count, or NK cell function. Together, these data indicate an association between HIV-1 replication and expansion of KIR3DL1+ NK cells when the KIR3DL1 ligand is present, and this response is different from CD56dim NK cells in the absence of their HLA ligand KIR3DL1+.
Chronic HIV-1 infection is associated with an increased frequency of polyfunctional CD56dim NK cells.
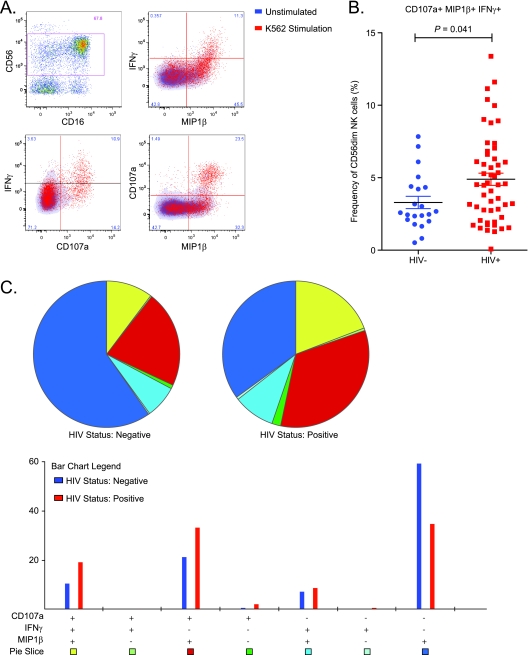
We next investigated the NK cell activity in peripheral blood samples using intracellular cytokine staining for IFN-γ and MIP-1β and staining for CD107a as a measure of degranulation (Fig. 3 A). We measured the CD56dim NK cell activity without target cells as a gauge of the endogenous level of activity and subtracted this from the CD56dim NK cell response after stimulation with K562 target cells to obtain a measure of the response to a HLA-negative target cells. Coexpression of CD107a, IFN-γ, and MIP-1β, a measure of polyfunctionality in response to K562 cells, was higher in HIV-1-infected donors than in seronegative individuals (P = 0.041) (Fig. 3B). The median frequency of triple-positive cells in the HIV-1-infected group was 4.5% (0.1 to 13.4%) of CD56dim NK cells while the frequency of response in the uninfected group was 2.6% (0.5 to 7.8%) of CD56dim NK cells.
Fig. 3.
Chronic HIV-1 infection is associated with increased polyfunctionality in NK cells. Frozen PBMCs were thawed and stimulated in the presence or absence of K562 cells at an effector/target ratio of 5:1, and functional responses were detected using multiparameter flow cytometry. (A) Representative pseudo-color histogram on CD56dim NK cell discrimination. Combinations of IFN-γ, MIP-1β, and CD107a coexpression in CD56dim NK cells after an 18-h assay with or without K562 cell stimulation are shown. Fluorescence-activated cell sorting graphs represent unstimulated (blue) density plots overlaid with K562-stimulated (red) dot plots. Boolean analysis of the three functions was performed, and unstimulated function was subtracted from the K562-stimulated condition. (B) Vertical scatter plot comparing frequency of CD56dim NK cells coexpressing IFN-γ, MIP-1β, and CD107a in people with (n = 52) or without (n = 21) HIV-1 infection. (C) SPICE software was used to graphically present the overnight responses against K562 (with unstimulated responses subtracted out) for HIV-1-infected (n = 52) and uninfected (n = 21) individuals. Pie chart shows the distribution of a single function (blue), two functions (in green, light blue, and red), and triple functions (yellow) corresponding to the Boolean subsets in the bar graph below. Pie arcs show the relative amount of each individual function: IFN-γ (yellow), MIP-1β (green), and CD107a (red). Bar chart shows the possible combinations of three functions on the x axis and the percentage of distinct functional populations within the CD56dim NK cells on the y axis.
The distribution of CD56dim NK cell responses was evaluated using a Boolean analysis of the three functions measured, CD107a, IFN-γ, and MIP-1β, in response to K562 stimulation in the HIV-1-infected and uninfected groups (Fig. 3C). Interestingly, in addition to the triple-positive functional NK cells, a CD56dim NK cell functional profile of CD107a+, IFN-γ-, and MIP-1β+ was also significantly higher in HIV-1-infected donors than in seronegative individuals (P = 0.014). There was no difference in representation of the CD107a− IFN-γ+ MIP-1β+ subset between the HIV-1-infected and uninfected groups. The CD107a− IFN-γ− MIP-1β+ subset dominated the NK cell response in the HIV-1-uninfected group. Together, these data suggest a greater level of polyfunctionality in the NK cell response in HIV-1-infected subjects than in uninfected controls.
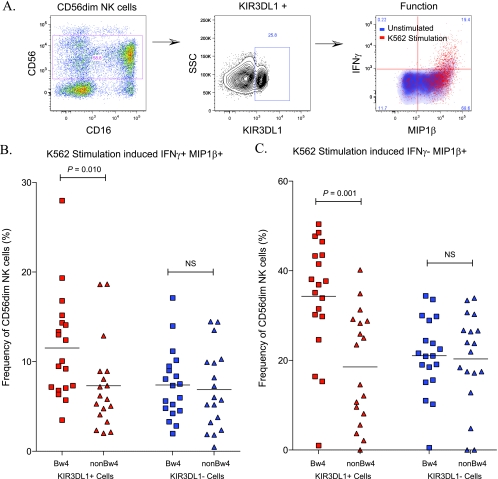
Higher responsiveness in KIR3DL1+ CD56dim NK cells in HIV-1-infected HLA-B Bw4 carriers than in noncarriers.
In order to determine the KIR3DL1+ CD56dim NK cell-specific response to K562 stimulation, we conducted our functional NK cell assay using intracellular cytokine staining for IFN-γ and MIP-1β and included KIR3DL1 (DX9) in place of the degranulation marker CD107a (Fig. 4 A). As before, K562-stimulated responses were corrected for background using unstimulated cells. The IFN-γ+ MIP-1β+ CD56dim NK cell bifunctional responses were measured in HLA-B Bw4 carriers (n = 12) and noncarriers (n = 12) (Fig. 4B). KIR3DL1+ CD56dim NK cells, in the presence of Bw4, showed a higher frequency of IFN-γ and MIP-1β coexpression than KIR3DL1− CD56dim NK cells in the same individuals (P < 0.001). Furthermore, the bifunctional IFN-γ and MIP-1β response in KIR3DL1+ CD56dim NK cells was higher in the Bw4-expressing group of patients than in the non-Bw4 group (P = 0.008). Thus, HLA-B Bw4 licenses a higher level of bifunctional response in KIR3DL1+ NK cells in HIV-1-infected Ugandans.
Fig. 4.
KIR3DL1+ CD56dim NK cells have a higher functionality in HLA-B Bw4+ individuals than participants homozygous for HLA-B Bw6. Frozen PBMCs were thawed and stimulated in the presence or absence of K562 cells at an effector/target ratio of 5:1, and functional responses were detected using multiparameter flow cytometry. (A) Representative pseudo-color histogram of CD56dim NK cell discrimination. IFN-γ and MIP-1β on KIR3DL1+ CD56dim NK cells after an 18-h assay with or without K562 cell stimulation are shown. Fluorescence-activated cell sorting graphs represent unstimulated (blue) density plots overlaid with K562-stimulated (red) dot plots. (B) Vertical scatter plot comparing coexpression of IFN-γ and MIP-1β both in KIR3DL1-expressing (red) and -nonexpressing (blue) CD56dim NK cells. Study participants with the HLA-B Bw4 motif (squares) are compared to individuals homozygous for HLA-B Bw6 (triangles). (C) Vertical scatter plot comparing MIP-1β only production in KIR3DL1-expressing (red) and -nonexpressing (blue) CD56dim NK cells. Study participants with the HLA-B Bw4 motif (squares) are compared to individuals homozygous for HLA-B Bw6 (triangles). NS, nonsignificant.
DISCUSSION
In this study we have examined several KIRs in combination with their cognate HLA ligands with particular focus on NK cell function in East Africa where both host and viral genomes are highly polymorphic. Examining KIR2DL2/DL3, KIR3DL1/DS1, HLA-A, and HLA-B, we find no difference between distribution of KIR genes, HLA-A, or HLA-B with a Bw4 motif between HIV-positive and -negative participants. Interestingly, when we looked at HLA-C, we observed an underrepresentation of heterozygous group C1/C2 in HIV-positive individuals compared to HIV-negative participants, but our data are limited in size and should be investigated further. With regard to NK cell phenotype, we found that HLA-B Bw4 is associated with an increased KIR3DL1+ NK cell frequency in this rural HIV-1 infected Ugandan population. The size of the KIR3DL1+ NK cell subset correlated directly with viral load, suggesting that a large KIR3DL1+ NK subset is not associated with viral control. HIV-1 infection was also associated with an increase in NK cell polyfunctionality, as measured by CD56dim NK cell coexpression of the functional markers CD107a, IFN-γ, and MIP-1β. Furthermore, HIV-1 infection was associated with an increased function of the KIR3DL1+ CD56dim NK cells specifically in the patients carrying HLA-B Bw4. These results suggest that the presence of HLA-B Bw4 influences the size and function of the KIR3DL1+ NK cell subset and that this subset changes in response to chronic HIV-1 infection.
It is clear that NK cell effector function is tightly regulated by self-MHC class I recognition (28). However, the role of MHC molecules in NK cell KIR repertoire formation is controversial (3, 50). Primary education, or development of KIR repertoires in the presence or absence of MHC molecules, happens early in NK development within the bone marrow, but there is a growing body of evidence to show that further development and maturation can happen in the periphery based on changes in the MHC environment (44). The present data indicate that the size of the KIR3DL1+ CD56dim NK cell subset in HIV-1-infected Ugandans is substantially greater in those who carry HLA-B Bw4 than in those who do not. The finding that this effect is most clear in HIV-1-infected subjects supports the notion that the KIR3DL1+ subset expands in response to chronic HIV burden. This apparent expansion might result from preferential survival or proliferation of these cells in the presence of HLA-B Bw4 molecules as, overall, the CD56dim NK cell compartment is reduced in HIV-1 infection (17). It might to some extent also depend on the loss of other KIR+ subsets as exemplified by the decrease in KIR2DL1+ NK cells in this cohort. The observed decrease in KIR2DL1+ NK cells may be consistent with the recent observation of decreasing KIR2DL1+ and KIR2DS1+ NK cell populations concurrent to increases in HIV-1 viral load (49). Overall, absolute NK cell counts are not decreased in chronic HIV-1 infection (17), so different NK cell subsets are responding differentially to the inflammatory environment.
HIV-1 is known to downregulate HLA-A and -B molecules to avoid CD8 T cell recognition (14, 32) and will thus probably create “missing-self” conditions on the surface of infected cells with the potential to activate NK cells in vivo (14, 28). We observe a direct positive correlation between the size of the CD56dim KIR3DL1+ NK cell subset and HIV-1 load, and this correlation exists only in subjects positive for HLA-B Bw4. This finding may suggest that the NK cell compartment adapts to chronic viremia by expanding the NK cell subset “licensed” by KIR3DL1 in the presence of its ligand. Although the activating signals and receptors involved in such a process are unknown, these findings are in line with recent findings concerning expansion of specific NK cell populations and generation of NK cell “memory” upon murine cytomegalovirus infection (45). More recently, in a North American human clinical study, Alter et al. found Bw4-dependent expansion of KIR3DL1+ NK cells during acute HIV-1 infection that persists into the chronic phase, whereas no significant effect by HLA class I was seen in uninfected individuals (2). Our finding of a positive correlation between levels of KIR3DL1+ NK cells and HIV viral load might seem to indicate that these NK cells have little antiviral effect or are even detrimental to viral control. One possible explanation could be that HIV-1 activates NK cells in two phases, similar to reports in the murine model where murine cytomegalovirus (MCMV) infection was characterized by an early acute phase of nonspecific NK proliferation followed by a later phase of Ly49H+ NK cell-specific proliferation (15, 51). One could speculate that the possible protective effect of KIR3DL1 may be exerted early in HIV-1 infection and that the expanded KIR3DL1+ NK population in Bw4+ hosts may just reflect the activated nature of the immune system observed in chronic infection and fail to exert viral control. Alternatively, NK cells could reflect a similar positive relationship that has been reported between HIV-specific Env or Nef CD8 T cell frequency and viral load (6) although this expansion should not be confused with adaptive antigen specificity. The extent to which KIR3DL1+ NK cells may help control virus and provide protection needs further investigation.
In addition to variable frequency, high and low fluorescence intensity of KIR3DL1 staining was observed in the CD56dim NK cells, suggesting that multiple alleles are present in this population based on differential antibody staining intensity as previously reported (23, 33, 47). Unfortunately, we were unable to determine KIR3DL1 alleles prevalent in this population. Norman et al. found that KIR3DL1 alleles associated with high expression and potent function were more common in sub-Saharan Africans than in other populations (38). Early reports on the association of slower progression to AIDS for KIR3DL1 in combination with HLA-Bw4 were strongest for KIR3DL1*004 (37). Although KIR3DL1*004 is primarily retained in the endoplasmic reticulum, some receptor is transported to the surface of the cell to transmit an inhibitory signal (46). Another recent study showed no protection from HIV-1 infection based on KIR3DL1*004 and HLA-Bw4 carriage (41). We do not think that KIR3DL1*004 is common in this population, based on a low frequency of this gene in a similar Ugandan cohort (data not shown). Interpreted in the context of these findings, the relative expansion of the KIR3DL1+ subset we have observed in the presence of HLA-B Bw4 may result in part from the qualities of KIR3DL1 variants in this East African population, and it will be important to define these alleles in future studies.
The cognate ligand for KIR3DL1 is HLA-B Bw4 allotypes with an amino acid substitution of isoleucine or threonine at position 80, and carriage of Bw4 is very high in Africa (43). Furthermore, certain HLA-B alleles such as B*27, B*57, and B*58 containing the 80I motif are associated with more favorable outcomes in HIV-1 disease progression (24, 36, 37) or are even associated with a reduced risk of HIV-1 infection (8). In contrast to previous findings (13, 20), we did not observe an association between HLA-B Bw4 homozygosity and lower viral loads or preserved CD4 T cell absolute counts. Due to specimen limitations, we were unable to fully genotype the HLA-B alleles present in this cohort. Other data from cohorts in Kampala would suggest that carriage of the B*27 and B*57 alleles is relatively low in Uganda, while the Bw4-80I alleles B*49, B*53, and B*58 are more frequent (29). Yet another study, however, suggests slightly different frequencies of B*27, B*57, and B*58 in Kampala (10), in line with the heterogeneity of populations in this region. Taken together, these data suggest an expanded CD56dim KIR3DL1+ NK cell subset with increased functionality in HIV-1-infected subjects in the context of HLA-B alleles more common in Uganda and with unknown influence on HIV-1 disease progression.
Despite several publications showing associations of favorable outcome in HIV-1 infection with KIR3DL1 in combination with HLA-B Bw4, the mechanism behind this protective effect is still unclear. A recent study in HIV-1-uninfected subjects, characterized the NK cells from individuals with high-affinity KIR3DL1 in combination with the Bw4 allele HLA-B*57 and found increased function, including CD107a, IFN-γ, and TNF-α in response to MHC-negative target cells (9). Similar to the work of Boulet et al. (9), our data show increased function within the KIR3DL1+ cells, but it is important to note that our data represent findings in HIV-1-infected patients with various amounts of viral burden and disease states. In addition, our findings include functional data on MIP-1β. CC chemokines such as MIP-1β produced by NK cells can reduce HIV-1 target cell entry through blocking of CCR5 (19, 31, 40). We observe enhanced MIP-1β and IFN-γ production in the CD56dim NK cell compartment in Bw4 carriers. High levels of MIP-1β production in combination with the increased antiviral cytokine IFN-γ might contribute to the lower rate of HIV-1 disease progression observed in patients carrying both Bw4-80I and alleles of KIR3DL1 (5, 34, 36, 37). Tiemessen et al. have shown that in postpartum women from South Africa, NK cell IFN-γ and/or interleukin-2 (IL-2) production in response to HIV-1 Env or Reg peptide sets was associated with lower viral load and higher CD4 T cells counts (48). Alternatively, CD8+ T cells expressing a functional profile of CD107a+ and MIP-1β+ independent of IFN-γ are associated with viral inhibition in in vitro assays (22). Our data indicate that this functional profile is increased in CD56dim NK cells in response to HIV-1 infection, but it does not associate with HIV-1 viral load or CD4 T cell absolute counts. Further studies will be required to examine these functions in greater detail in relation to HIV-1 disease progression.
In summary, we have observed that the presence of HLA-B Bw4 is associated with elevated frequencies of KIR3DL1+ CD56dim NK cells in chronically HIV-1-infected Ugandans. Furthermore, the size of the KIR3DL1-expressing CD56dim NK cell subset directly correlates with viral load, and, importantly, this occurs only in Bw4 patients. Finally, there is increased polyfunctionality in HIV-1-infected Ugandans compared to uninfected controls, and KIR3DL1+ CD56dim NK cells show increased IFN-γ and MIP-1β production in Bw4 carriers as opposed to noncarriers. These results suggest that the presence of Bw4 directs an expansion of functional KIR3DL1+ NK cells that may contribute to the previously observed epidemiological association of a licensed KIR3DL1 phenotype and slow disease progression. However, the positive correlation between KIR3DL1+ NK cells and viral replication may also be interpreted to suggest that KIR3DL1+ NK cells have poor antiviral capacity in Bw4+ hosts.
ACKNOWLEDGMENTS
We thank the Kayunga cohort volunteers for their valuable participation and cooperation during the conduct of this study and the Makerere University Walter Reed Project Staff for their continued dedication to the development of a safe and effective HIV-1 vaccine. We thank Galit Alter, Jakob Michaelsson, Niklas Björkström, and Hans-Gustaf Ljunggren for discussions and critical reading of the manuscript.
This work was supported by the U.S. Army Medical Research and Materiel Command and its Cooperative Agreement (W81XWH-04-02-0005) with the Henry M. Jackson Foundation for the Advancement of Military Medicine and with an interagency agreement (1Y-A1-26-42-07) with the Division of AIDS, National Institute of Allergy and Infectious Diseases, and the National Institutes of Health. The work was also supported by the Swedish Research Council, the Swedish Physicians against AIDS Foundation, the Swedish Cancer Foundation, and Karolinska Institutet.
The views and opinions expressed herein do not necessarily reflect those of the U.S. Army, the Department of Defense, or the National Institutes of Health.
We declare that we have no competing financial interests.
Footnotes
Published ahead of print on 16 March 2010.
REFERENCES
- 1. Alter G., et al. 2007. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J. Exp. Med. 204:3027–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alter G., et al. 2009. HLA class I subtype-dependent expansion of KIR3DS1+ and KIR3DL1+ NK cells during acute human immunodeficiency virus type 1 infection. J. Virol. 83:6798–6805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andersson S., Fauriat C., Malmberg J. A., Ljunggren H. G., Malmberg K. J. 2009. KIR acquisition probabilities are independent of self-HLA class I ligands and increase with cellular KIR expression. Blood 114:95–104 [DOI] [PubMed] [Google Scholar]
- 4. Bandyopadhyay S., et al. 1990. Natural killer cell-mediated lysis of T cell lines chronically infected with HIV-1. Clin. Exp. Immunol. 79:430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barbour J. D., et al. 2007. Synergy or independence? Deciphering the interaction of HLA Class I and NK cell KIR alleles in early HIV-1 disease progression. PLoS Pathog. 3:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Betts M. R., et al. 2001. Analysis of total human immunodeficiency virus (HIV)-specific CD4+ and CD8+ T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 75:11983–11991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bonaparte M. I., Barker E. 2004. Killing of human immunodeficiency virus-infected primary T-cell blasts by autologous natural killer cells is dependent on the ability of the virus to alter the expression of major histocompatibility complex class I molecules. Blood 104:2087–2094 [DOI] [PubMed] [Google Scholar]
- 8. Boulet S., et al. 2008. A combined genotype of KIR3DL1 high expressing alleles and HLA-B*57 is associated with a reduced risk of HIV infection. AIDS 22:1487–1491 [DOI] [PubMed] [Google Scholar]
- 9. Boulet S., et al. 2010. HIV protective KIR3DL1 and HLA-B genotypes influence NK cell function following stimulation with HLA-devoid cells. J. Immunol. 184:2057–2064 [DOI] [PubMed] [Google Scholar]
- 10. Cao K., et al. 2004. Differentiation between African populations is evidenced by the diversity of alleles and haplotypes of HLA class I loci. Tissue Antigens 63:293–325 [DOI] [PubMed] [Google Scholar]
- 11. Carr W. H., Pando M. J., Parham P. 2005. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J. Immunol. 175:5222–5229 [DOI] [PubMed] [Google Scholar]
- 12. Carrington M., Martin M. P., van Bergen J. 2008. KIR-HLA intercourse in HIV disease. Trends Microbiol. 16:620–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carrington M., O'Brien S. J. 2003. The influence of HLA genotype on AIDS. Annu. Rev. Med. 54:535–551 [DOI] [PubMed] [Google Scholar]
- 14. Cohen G. B., et al. 1999. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 10:661–671 [DOI] [PubMed] [Google Scholar]
- 15. Dokun A. O., et al. 2001. Specific and nonspecific NK cell activation during virus infection. Nat. Immunol. 2:951–956 [DOI] [PubMed] [Google Scholar]
- 16. Eller L. A., et al. 2007. Large-scale human immunodeficiency virus rapid test evaluation in a low-prevalence Ugandan blood bank population. J. Clin. Microbiol. 45:3281–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eller M. A., et al. 2009. Elevated NK cell activity despite altered functional and phenotypic profile in Ugandans with HIV-1 clade A or clade D infection. J. Acquir. Immune Defic. Syndr. 51:380–389 [DOI] [PubMed] [Google Scholar]
- 18. Fauci A. S., Mavilio D., Kottilil S. 2005. NK cells in HIV infection: paradigm for protection or targets for ambush. Nat. Rev. Immunol. 5:835–843 [DOI] [PubMed] [Google Scholar]
- 19. Fehniger T. A., et al. 1998. Natural killer cells from HIV-1+ patients produce C-C chemokines and inhibit HIV-1 infection. J. Immunol. 161:6433–6438 [PubMed] [Google Scholar]
- 20. Flores-Villanueva P. O., et al. 2001. Control of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proc. Natl. Acad. Sci. U. S. A. 98:5140–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fogli M., et al. 2008. Lysis of endogenously infected CD4+ T cell blasts by rIL-2 activated autologous natural killer cells from HIV-infected viremic individuals. PLoS Pathog. 4:e1000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Freel S. A., et al. 2010. Phenotypic and functional profile of HIV-inhibitory CD8 T cells elicited by natural infection and heterologous prime/boost vaccination. J. Virol. 84:4998–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gardiner C. M., et al. 2001. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J. Immunol. 166:2992–3001 [DOI] [PubMed] [Google Scholar]
- 24. Gaudieri S., et al. 2005. Killer immunoglobulin-like receptors and HLA act both independently and synergistically to modify HIV disease progression. Genes Immun. 6:683–690 [DOI] [PubMed] [Google Scholar]
- 25. Gonzalez V. D., et al. 2008. Application of nine-color flow cytometry for detailed studies of the phenotypic complexity and functional heterogeneity of human lymphocyte subsets. J. Immunol. Methods 330:64–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guwatudde D., et al. 2009. Relatively low HIV infection rates in rural Uganda, but with high potential for a rise: a cohort study in Kayunga District, Uganda. PLoS One 4:e4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Iannello A., Debbeche O., Samarani S., Ahmad A. 2008. Antiviral NK cell responses in HIV infection: I. NK cell receptor genes as determinants of HIV resistance and progression to AIDS. J. Leukoc. Biol. 84:1–26 [DOI] [PubMed] [Google Scholar]
- 28. Karre K. 2008. Natural killer cell recognition of missing self. Nat. Immunol. 9:477–480 [DOI] [PubMed] [Google Scholar]
- 29. Kijak G. H., et al. 2009. HLA class I allele and haplotype diversity in Ugandans supports the presence of a major east African genetic cluster. Tissue Antigens 73:262–269 [DOI] [PubMed] [Google Scholar]
- 30. Koehler R. N., et al. 2009. High-throughput genotyping of KIR2DL2/L3, KIR3DL1/S1 and their HLA class I ligands using real-time PCR. Tissue Antigens 74:73–80 [DOI] [PubMed] [Google Scholar]
- 31. Kottilil S., et al. 2003. Innate immunity in human immunodeficiency virus infection: effect of viremia on natural killer cell function. J. Infect. Dis. 187:1038–1045 [DOI] [PubMed] [Google Scholar]
- 32. Le Gall S., et al. 1998. Nef interacts with the mu subunit of clathrin adaptor complexes and reveals a cryptic sorting signal in MHC I molecules. Immunity 8:483–495 [DOI] [PubMed] [Google Scholar]
- 33. Li H., Pascal V., Martin M. P., Carrington M., Anderson S. K. 2008. Genetic control of variegated KIR gene expression: polymorphisms of the bi-directional KIR3DL1 promoter are associated with distinct frequencies of gene expression. PLoS Genet. 4:e1000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lopez-Vazquez A., et al. 2005. Interaction between KIR3DL1 and HLA-B*57 supertype alleles influences the progression of HIV-1 infection in a Zambian population. Hum. Immunol. 66:285–289 [DOI] [PubMed] [Google Scholar]
- 35. Marsh S. G., et al. 2002. Nomenclature for factors of the HLA system, 2002. Tissue Antigens 60:407–464 [DOI] [PubMed] [Google Scholar]
- 36. Martin M. P., et al. 2002. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 31:429–434 [DOI] [PubMed] [Google Scholar]
- 37. Martin M. P., et al. 2007. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 39:733–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Norman P. J., et al. 2007. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat. Genet. 39:1092–1099 [DOI] [PubMed] [Google Scholar]
- 39. Olemukan R. E., et al. 2010. Quality monitoring of HIV-1-infected and uninfected peripheral blood mononuclear cell samples in a resource-limited setting. Clin. Vaccine Immunol. 17:910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Oliva A., et al. 1998. Natural killer cells from human immunodeficiency virus (HIV)-infected individuals are an important source of CC-chemokines and suppress HIV-1 entry and replication in vitro. J. Clin. Invest. 102:223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Parsons M. S., et al. 2010. Mind the gap: lack of association between KIR3DL1*004/HLA-Bw4-induced natural killer cell function and protection from HIV infection. J. Infect. Dis. 202(Suppl. 3):S356–S360 [DOI] [PubMed] [Google Scholar]
- 42. Roederer M., Nozzi J. L., Nason M. C. 2011. SPICE: Exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A 79:167–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Single R. M., et al. 2007. Global diversity and evidence for coevolution of KIR and HLA. Nat. Genet. 39:1114–1119 [DOI] [PubMed] [Google Scholar]
- 44. Sun J. C. 2010. Re-educating natural killer cells. J. Exp. Med. 207:2049–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sun J. C., Beilke J. N., Lanier L. L. 2009. Adaptive immune features of natural killer cells. Nature 457:557–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Taner S. B., et al. 2010. Interactions of NK cell receptor KIR3DL1*004 with chaperones and conformation-specific antibody reveal a functional folded state as well as predominant intracellular retention. J. Immunol. 186:62–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thomas R., et al. 2008. Novel KIR3DL1 alleles and their expression levels on NK cells: convergent evolution of KIR3DL1 phenotype variation? J. Immunol. 180:6743–6750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tiemessen C. T., et al. 2010. Natural killer cells that respond to human immunodeficiency virus type 1 (HIV-1) peptides are associated with control of HIV-1 infection. J. Infect. Dis. 202:1444–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wong A. H., et al. 2010. Alterations in natural killer cell receptor profiles during HIV type 1 disease progression among chronically infected South African adults. AIDS Res. Hum. Retroviruses 26:459–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yawata M., et al. 2006. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J. Exp. Med. 203:633–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yokoyama W. M. 2005. Specific and nonspecific natural killer cell responses to viral infection. Adv. Exp. Med. Biol. 560:57–61 [DOI] [PubMed] [Google Scholar]