Abstract
Bombyx mori densovirus 1 (BmDNV-1), a major pathogen of silkworms, causes significant losses to the silk industry. The structure of the recombinant BmDNV-1 virus-like particle has been determined at 3.1-Å resolution using X-ray crystallography. It is the first near-atomic-resolution structure of a virus-like particle within the genus Iteravirus. The particles consist of 60 copies of the 55-kDa VP3 coat protein. The capsid protein has a β-barrel “jelly roll” fold similar to that found in many diverse icosahedral viruses, including archaeal, bacterial, plant, and animal viruses, as well as other parvoviruses. Most of the surface loops have little structural resemblance to other known parvovirus capsid proteins. In contrast to vertebrate parvoviruses, the N-terminal β-strand of BmDNV-1 VP3 is positioned relative to the neighboring 2-fold related subunit in a “domain-swapped” conformation, similar to findings for other invertebrate parvoviruses, suggesting domain swapping is an evolutionarily conserved structural feature of the Densovirinae.
INTRODUCTION
Parvoviruses are among the smallest viral pathogens. They form nonenveloped icosahedral particles with a maximum diameter of about 280 Å and have single-stranded DNA genomes. The parvovirus family has been divided into two subfamilies, the Parvovirinae, which infect vertebrates, and the Densovirinae, which infect invertebrates. Bombyx mori densovirus 1 (BmDNV-1) belongs to the Densovirinae subfamily (genus Iteravirus) (17) and causes potentially lethal flacherie disease in silkworms (28), thereby posing a major threat to silk production. So far, only CeDNV (from Casphalia extranea), DpDNV (from Dendrolimus punctatus), and BmDNV-1 belong to the genus Iteravirus, whereas BmDNV-2 and -3 have a bipartite genome with a total length of about 12.5 kb and have been excluded from the Parvoviridae and reassigned to a new family (Bidnaviridae) (31).
The 5.1-kb genome of BmDNV-1 has two overlapping genes for the nonstructural proteins in its 5′ half and a single open reading frame (ORF) that encodes the structural viral proteins (VPs) in the 3′ portion (17). The VPs are translated using probably four of five initiator codons in the ORF by a leaky scanning mechanism, resulting in four coat protein variants (VP1, 74.9 kb; VP2, 64.3 kb; major capsid protein VP3, 54.9 kb; and VP4, 51.6 kb) (17). The N-terminal part of the largest capsid protein, VP1, contains phospholipase A2 activity that is required for successful infection (7, 10, 11, 17, 36). Parvoviruses form T=1 icosahedral capsids that are assembled out of 60 nearly identical VP subunits. Each subunit consists of an eight-stranded antiparallel β-barrel known as a “jelly roll” fold (1, 13, 14, 18, 29, 30, 33–35). The same fold is utilized by the major capsid protein of many other viruses, ranging from archaeal to bacterial, plant, and animal viruses (4, 20, 27). However, parvoviruses have large insertions in loops connecting the β-strands (1, 13, 14, 18, 29, 30, 33–35). These insertions define the external surface of the virus, creating features that govern antigenicity, receptor binding, and most of the intersubunit contacts. The loops also form the structural basis of the differences between related parvoviruses. Common features of parvovirus capsids are protrusions at or around icosahedral 3-fold axes, depressions around 2-fold axes, and canyons surrounding a cylindrical pore at each 5-fold vertex. In full virions of the members of the Parvovirus genus, the 5-fold pores are occupied by a glycine-rich motif from the N-terminal region of the capsid protein (33, 35). Such glycine-rich motifs are absent in densovirus capsid proteins.
VP3 is the dominant protein among the capsid proteins of BmDNV-1 (17). Here, we determined the three-dimensional crystal structure of recombinant, empty VP3 virus-like particles (VLPs) of the silkworm parvovirus BmDNV-1 at 3.1-Å resolution. The N-terminal β-strands of 2-fold related VP3 subunits are in positions similar to those in Galleria mellonella densovirus (GmDNV) (29) and Penaeus stylirostris densovirus (PstDNV) (13). Their positions are swapped relative to the N-terminal β-strands in vertebrate parvoviruses. The BmDNV-1 structure will facilitate studies of its structure-function relationships and provide the tools for rational site-directed mutagenesis and for studying its morphogenesis and tropism. Moreover, this structure might provide a basis for the design of capsid-binding antiviral compounds (25) to protect silkworms against BmDNV-1 infections.
MATERIALS AND METHODS
Preparation of VLPs and crystallization.
The complete coding sequence of the BmDNV-1 viral coat protein VP3 was amplified by PCR using the primers BmVP3 (5′-GCTCTAGAAC AATGTCTGAA GATATAC-3′) and Bm482 (5′-CCGCTCGAGG TACGTGACTT AATGTACG-3′) and cloned into the pFastBac1 vector (Invitrogen) using the XbaI and XhoI restriction sites. The insert was sequenced and found to be identical to GenBank entry AY033435. The recombinant plasmid was then transfected into Sf9 insect cells to obtain recombinant baculovirus for the heterologous expression of BmDNV-1 VP3 (494 amino acids, 55 kDa).
Self-assembled, recombinant, empty VLPs were purified from High-Five insect cells overexpressing BmDNV-1 VP3 by gradient density centrifugation. Capsids were concentrated from the cell culture supernatant by centrifugation (4.5 h, 150,000 × g, 10°C) through a CsCl cushion (0.6 g/ml in 50 mM Tris-HCl, pH 8.7, 25 mM EDTA, 0.5% Triton X-100). The particles were further purified by sucrose gradient centrifugation (10 to 25% in 10 mM Tris-HCl, pH 7.5; 2 h, 100,000 × g, 10°C). The purity and quality of the final virus preparation were estimated by SDS-PAGE with Coomassie blue staining and cryo-electron microscopy (see Fig. S1 in the supplemental material). Two protein bands were observed. The major protein band corresponded to a molecular mass, ∼52 kDa, that was slightly smaller than the expected 55 kDa (minor band), probably due to N- or C-terminal proteolytic processing.
The purified protein capsids were crystallized using the hanging drop vapor diffusion technique. Aliquots (1.5 μl) of sample in 10 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM MgCl2, and 1 mM CaCl2 (protein concentration, 10 mg/ml) were mixed with an equal volume of reservoir solution (2% polyethylene glycol [PEG] 8000 in 10 mM Tris-HCl, pH 7.5, containing 30% glycerol), and the droplets were equilibrated against 1 ml reservoir solution at 20°C. Crystals were flash-frozen directly from the drop for X-ray data collection.
X-ray structure determination.
X-ray diffraction data were collected from a single frozen crystal (rectangular plate of about 0.3 by 0.1 by 0.02 mm) at the Advanced Photon Source (beamline BioCARS 14-ID-B; Argonne National Laboratory) (Table 1). The crystal diffracted to a 3.1-Å resolution. The diffraction data were integrated and scaled, assuming the space group to be P1 using the HKL data processing package (21) (Table 1). Only the first 900 of 1,100 frames were further processed for structure determination, reducing the multiplicity of observed reflections but perhaps avoiding radiation damage, to ensure high-quality data. Assuming one VP3 VLP per unit cell, the Matthews coefficient (VM) was 3.6 Å3/Da, corresponding to a solvent content of 65.5%. The particle orientation was determined with a self-rotation function calculated with the software program GLRF (32) using data between 5- and 3.5-Å resolution. The radius of integration was set to 150 Å. The results suggested a particle orientation given by the polar angles ψ = 49.0°, φ = 45.35°, and κ = 90.25° relative to the standard icosahedral orientation as defined by Arnold et al. (2) (XYK polar angle convention [32] and axial orthogonalization as defined by Rossmann and Blow [26].
Table 1.
X-ray data collection and refinement statistics
| Parameter | Value or model |
|---|---|
| Data collection parameters | |
| Wavelength (Å) | 1.2 |
| CCD detector | MAR CCD-165 |
| Sample-to-detector distance (mm) | 200 |
| Exposure time per frame (s) | 20 |
| Oscillation angle per frame (°) | 0.2 |
| No. of frames collected | 1,100 |
| Data reduction and refinement statisticsa | |
| Resolution range (Å) | 50–3.1 (3.21–3.10) |
| Space group | P1 |
| No. of frames used | 1–900 |
| Cell dimensions | |
| a, b, c (Å) | 245.39, 245.62, 245.74 |
| α, β, γ (°) | 59.98, 67.93, 72.27 |
| Mosaicity (°) | 0.36 |
| No. of observed reflections | 1,518,629 |
| No. of unique reflections | 789,029 (63,108) |
| Redundancy | 1.9 (1.3) |
| % completeness | 95.7 (76.6) |
| 〈I〉/〈σ(I)〉 | 16.12 (6.09) |
| Rsym ΣhΣj|Ihj−〈Ih〉|/ΣΣIhj | 0.044 (0.122) |
| Model building and refinement statisticsa | |
| Resolution range (Å) | 45–3.1 (3.24–3.10) |
| No. of residues/atoms built | 412/3,290 |
| Final R factorb | 0.209 (0.301) |
| Mean isotropic temp factor (Å2) | 18 |
| RMSD bond length (Å)/bond angle (°) | 0.0047/1.258 |
| % residues in most favored/additionally | |
| allowed/generously allowed/disallowed | |
| regions of the Ramachandran plotc | 82.5/16.9/0.3/0.3 |
Values in parentheses refer to the highest-resolution shell.
No Rfree value was calculated, because the NCS averaging implies that there is little difference between Rworking and Rfree. Because of the NCS symmetry, an independent subset of structure factors cannot be selected.
The coordinates of GmDNV (PDB accession code 1DNV) were used as an initial phasing model for molecular replacement. The model structure was oriented and positioned in the unit cell, and using 60 noncrystallographic symmetry (NCS) matrices, the initial phases were calculated with the CNS software program package (version 1.3) (5, 6). Averaging was performed with the software program AVE (RAVE package; Uppsala Software Factory [http://xray.bmc.uu.se/usf/] [15]) within an initial mask generated from the coordinates of GmDNV (converted to polyalanine) using a 7-Å radius around each atom. The phases from the model were extended from 15 to 3.1 Å in steps of one reciprocal lattice point (1/c), with 3 cycles of 60-fold NCS averaging per extension step, during which calculated structure factors were used for unobserved reflections. The software program SigmaA (23) of the CCP4 package (8) was used to include weights in the electron density calculations. Additional 10 cycles of density, averaging at 3.1-Å resolution, were performed to achieve convergence between observed and calculated structure factors. The molecular mask was improved during alternated averaging rounds and model building. The position and orientation of the protein subunits were also subjected to rigid body refinement using the CNS program. The particle position was refined by varying it in steps of 0.1° and checking for highest correlation between observed and calculated structure factors after four cycles of phase refinement using the program AVE. The final orientation of the particle was given by the polar angles ψ = 48.96°, φ = 45.35°, and κ = 90.20° relative to the standard icosahedral orientation. The unit cell dimensions and angles were subjected to a similar refinement procedure using a step size of 0.3 Å or 0.2°, respectively, resulting in no change to the unit cell parameters.
The BmDNV-1 protein structure was built into the electron density map using the software program Coot (9), starting with the polyalanine model obtained from the GmDNV coordinates. Subsequently, residues were mutated and renumbered to those of BmDNV-1 (residues 43 to 454 of a total of 494 residues) (Fig. 1). The initial R factor (calculated with the program CNS) for the manually built model was 31.5% (43.9% in the highest-resolution shell between 3.24 and 3.10 Å). Atomic positions were refined against the observed structure amplitudes for data between 45.0- and 3.1-Å resolution using the CNS program while applying strict icosahedral NCS constraints. The model was refined by manual rebuilding, alternating with coordinate and B-factor refinement in the program CNS. No water molecules were added due to the low resolution of the data. The final R factor for the model was 20.9% (30.1% in the highest-resolution shell) (Table 1). Structure analysis with the software program PROCHECK (12, 16) showed 82.5% of 360 nonglycine, nonproline residues within the most favored regions, 17.2% within allowed regions, and one residue in a disallowed region of the Ramachandran plot. The root mean square deviations (RMSD) of bond lengths and angles from idealized values were 0.0047 Å and 1.258°, respectively. Secondary structure elements were designated as defined in the program PROCHECK (Fig. 1; see also Table S1 in the supplemental material).
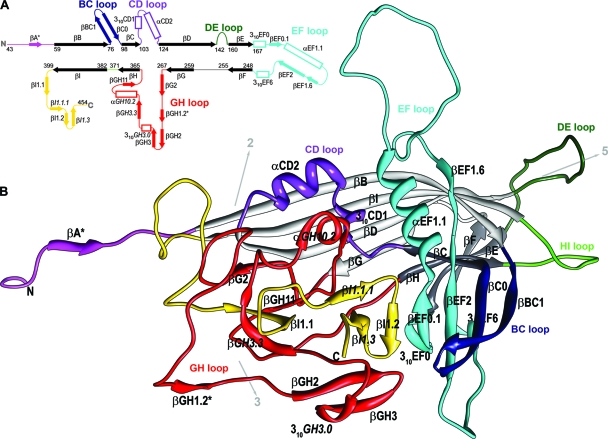
Fig. 1.
The structure of the BmDNV-1 capsid protein. (A) Schematic diagram showing the positions of secondary structural elements along the polypeptide. Numbering corresponds to the starting residues of the β-strands (solid arrows); β-strands that are part of the “jelly-roll” fold are shown in black, and those in connecting loops are shown in color. Helical elements are depicted as open rectangles. Secondary structure elements are named as defined previously (13, 35). Additional secondary structure elements designated for BmDNV-1 are labeled in italic font (see Table S1 in the supplemental material). (B) Ribbon diagram of the BmDNV-1 subunit. The BIDG and CHEF sheets of the eight-stranded β-barrel are shown in light and dark gray, respectively. The surface loops connecting the strands of the β-barrel are colored as follows: BC loop, dark blue; CD, purple; DE, dark green; EF, cyan; GH, red; and HI, green. The N-terminal region upstream of βB is shown in magenta, and the C-terminal portion downstream of βI is shown in gold. The position of icosahedral symmetry axes and their direction from the viral center are indicated by arrows. Hydrogen-bonded β structures that occur only in BmDNV-1 VLPs are marked with an asterisk. Molecular graphic images were produced using the Chimera software package (22).
Structure comparison.
The structure of BmDNV-1 was compared with those of the invertebrate parvoviruses GmDNV (PDB accession code 1DNV; 3.7-Å resolution), a pathogen of the greater wax moth, and PstDNV (PDB accession code 3N7X; 2.5-Å resolution), a shrimp parvovirus. Also, the mammalian parvoviruses canine parvovirus (CPV) (PDB accession code 4DPV; 2.9-Å resolution), adeno-associated virus 2 (AAV-2) (PDB accession code 1LP3; 3.0-Å resolution), and human parvovirus B19 (B19) (PDB accession code 1S58; 3.5-Å resolution) were used for comparison. Secondary structure elements were designated as defined in the program PROCHECK (12, 16) (see Table S1 in the supplemental material).
Protein structure accession number.
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under PDB accession number 3P0S.
RESULTS AND DISCUSSION
The crystal structure of VP3 VLPs of BmDNV-1, the first near-atomic structure of an Iteravirus, has been solved using X-ray diffraction data to 3.1-Å resolution (Fig. 1 and Table 1; see also Fig. S2 in the supplemental material). Most of the 494 residues of VP3 were traceable in the icosahedrally averaged electron density, except for 42 N-terminal and 40 C-terminal residues. The lack of ordered structure in the N-terminal region of the major coat proteins has been consistently observed for all parvovirus structures to date, most likely because of a positional variation incompatible with the icosahedral symmetry averaging imposed during structure determination. The absence of interpretable density for the C-terminal residues of VP3 may similarly be due to either positional variation or partial proteolytic processing (see Fig. S1 in the supplemental material). The ordered C terminus is located at the exterior surface of the capsid close to a 3-fold axis of the capsid. In contrast, the C termini of vertebrate parvoviruses interface with the neighboring subunits. The last four residues of the C terminus of the GmDNV coat protein are also at the surface, near the 2-fold axis, whereas the last residue of the PstDNV coat protein is located at the inner surface. The absence of the 40 C-terminal residues in the BmDNV-1 structure could therefore be due to proteolytic processing at the capsid surface (see Fig. S1 in the supplemental material). This would also explain why the estimated molecular mass of the BmDNV-1 capsid protein was somewhat lower than expected. The electron density of BmDNV-1 provided no evidence for the presence of metal ions.
The BmDNV-1 capsid protein contains an eight-stranded jelly roll motif, similar to that found in many other viral capsid proteins, consisting of two β-sheets in the BIDG and CHEF arrangement (for nomenclature, see Rossmann and Johnson [27]) (Fig. 1 and 2; see also Table S1 in the supplemental material). The ß-barrel of BmDNV-1 can be superimposed on the same motif in GmDNV with an RMSD of 1.2 Å for 69 equivalenced Cα atoms (81% of Cα atoms in the ß-barrel) and on PstDNV with an RMSD of 1.5 Å (71 Cα atoms). The β-barrel motif is in approximately the same position relative to the icosahedral symmetry axes as in other parvoviruses (Fig. 2).
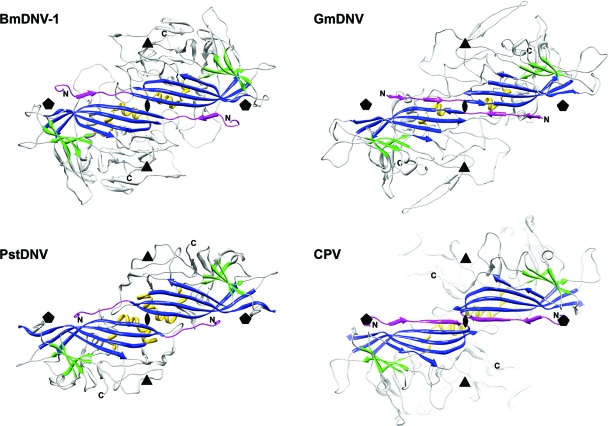
Fig. 2.
The spatial arrangement of the parvovirus core jelly roll and the N-terminal region for the capsid proteins of BmDNV-1, PstDNV, GmDNV, and CPV illustrate domain-swapping of the N terminus for the three known Densovirinae structures. Two 2-fold related symmetry mates are shown, viewed from the viral center along an icosahedral 2-fold axis. Conserved secondary structure elements of each protein subunit are colored blue (β-BIDG), green (β-CHEF), and gold (helical elements). The N-terminal region of the capsid protein, upstream of βB, including βA, is shown in magenta. The positions of icosahedral symmetry axes are indicated by polygonal symbols.
In the vertebrate parvoviruses, such as CPV, the β-BIDG sheet is extended to an antiparallel β-ABIDG sheet by the backfolded β-strand A (βA) in the N-terminal portion upstream of βB (Fig. 2). In contrast, in the Densovirus GmDNV, βB is essentially the linear extension of βA (29), producing a “domain-swapped” structure (3). Similarly, the N termini of PstDNV (13) and BmDNV-1 are pointing toward a 5-fold axis in a domain-swapped fashion relative to those of the vertebrate parvoviruses. Therefore, domain swapping is a common structural feature of the coat protein of invertebrate parvoviruses. Although the functional implications and benefits of this domain swapping are unclear, it may be necessary for structural stability by compensating for the reduced intertwining of the GH loops around the 3-fold axis (see Fig. S3 in the supplemental material). The last few N-terminal ordered residues of BmDNV-1 are pointing away from the 5-fold channel slightly toward the interior of the virus particle, similar to VLPs of B19 and PstDNV (13, 14). No density was found in the 5-fold pores that might be due to an externalization of the N termini, which may be the result of the absence of VP1 and VP2 in these BmDNV-1 VP3 VLPs.
In GmDNV, βA engages in hydrogen bonding with βB of a neighboring 2-fold-related subunit. However, in PstDNV there are no additional hydrogen bonds with the N-terminal region upstream of βB, although there are divalent metal ions that cross-link neighboring subunits. There is significant but different complementarity of shape between 3-fold-related subunits in densoviruses (see Fig. S3 in the supplemental material). There are additional hydrogen bonds between βGH1.2 and βGH2 of a 3-fold-related subunit of BmDNV-1. Furthermore, in the N-terminal region of BmDNV-1, the βΑ strand is antiparallel to βF of a neighboring 3-fold-related β-CHEF sheet rather than to the 2-fold-related β-BIDG as is the case in GmDNV. In BmDNV-1, there are only two hydrogen bonds between the side chain of glutamine 56 in the N-terminal arm and residues 66 and 67 in the βB-strand of the 2-fold-related subunit.
In most parvoviruses, the GH loop (see Fig. 1 for nomenclature) interdigitates with neighboring subunits to create trimers, thus probably providing stability (19, 24). The GH loop forms the characteristically raised structures at and around the 3-fold apices of the mammalian parvovirus capsid (1, 14, 18, 30, 33–35) and a β-annulus-type structure in GmDNV (29). The structure of the GH loop of BmDNV-1 bears little similarity to most known parvovirus structures, excepting that of GmDNV (Fig. 3). In GmDNV, a portion of the GH loop that forms the tip of the spike at the 3-fold vertices in CPV is entirely missing, whereas the part of the GH loop that forms the base of the spike is present but structurally different from that of CPV (29). In BmDNV-1, the GH loop is more intertwined with its 3-fold-related neighboring subunit than in GmDNV, but both are in contrast to PstDNV, for which this loop does not interdigitate between neighboring subunits (13) (see Fig. S3 in the supplemental material).
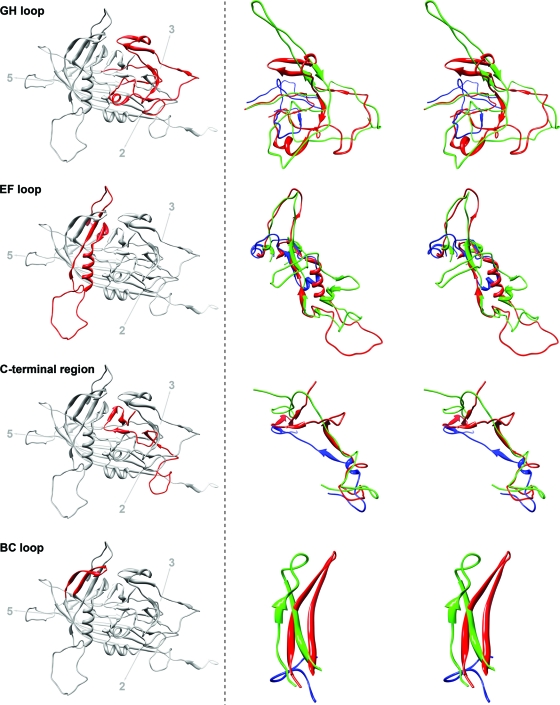
Fig. 3.
Structure comparison of loops GH, EF, BC, and the C-terminal region of BmDNV-1 with those of GmDNV and PstDNV. Left, a ribbon diagram of the BmDNV-1 capsid protein indicates the position of the particular loop (red) relative to the icosahedral symmetry axes. Right, close-up stereo views of the superpositioned loop regions of BmDNV-1 (red), GmDNV (green), and PstDNV (blue).
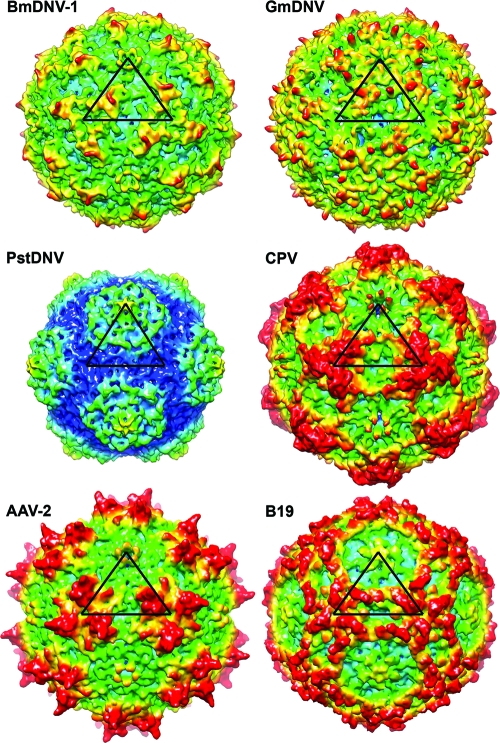
The largest portion in all parvovirus structures is made up of the loops connecting the β-barrel strands (see Fig. 1 for nomenclature). These loops form most intersubunit contacts and define the outer surfaces of the different viruses (Fig. 4). The BmDNV-1 capsid, with a maximum outer diameter of 264 Å, does not have substantial 3-fold-proximal protrusions as observed in members of the Parvovirinae subfamily, such as CPV. The protrusions on the surfaces of BmDNV-1 and GmDNV rise only slightly over the general surface, unlike the case for the Brevidensovirus PstDNV (Fig. 4).
Fig. 4.
Comparison of the BmDNV-1 protein capsid with those of other parvoviruses. Surface renderings of three-dimensional electron density maps of BmDNV-1, GmDNV, PstDNV, CPV, B19, and AAV-2 generated from atomic coordinates to 8-Å resolution. One icosahedral asymmetric unit is indicated by a black triangle. The surface is colored according to the distance from the viral center (blue, 100 Å; cyan, 107.5 Å; green, 115 Å; yellow, 122.5 Å; red, 130 Å).
The structure and length of the loop regions in BmDNV-1 show some similarity to GmDNV, but they differ greatly from the vertebrate parvoviruses, as well as the shrimp parvovirus PstDNV (Fig. 3), suggesting a smaller divergence in evolutionary development between BmDNV-1 and GmDNV than between these and the vertebrate parvoviruses, as well as PstDNV. The backbone Cα atoms of BmDNV-1 superimpose on PstDNV with an RMSD of 1.9 Å for 140 equivalenced Cα atoms (34% of all ordered BmDNV-1 residues and 47% of ordered PstDNV residues) but on GmDNV with an RMSD of 2.0 Å with 239 Cα atoms (58% for BmDNV residues and 55% of GmDNV residues). Thus, there is a far larger portion of structure that is similar between BmDNV-1 and GmDNV than there is between BmDNV-1 and PstDNV. These observations are consistent with the previous conclusion of a closer evolutionary relationship between the two insect parvoviruses BmDNV-1 and GmDNV than they have with other parvoviruses.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for the support we received from the staff of the BioCARS beamline. We are thankful to Sheryl Kelly for assistance in the preparation of this article.
Use of BioCARS Sector 14 was supported by the National Institutes of Health (NIH), National Center for Research Resources, under grant no. RR007707. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract DE-AC02-06CH11357. The work was supported by NIH grants AI 33468 to Colin R. Parrish (Cornell University) and AI 11219 to M.G.R. and support from the Natural Sciences and Engineering Research Council of Canada to P.T.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 2 March 2011.
REFERENCES
- 1. Agbandje M., McKenna R., Rossmann M. G., Strassheim M. L., Parrish C. R. 1993. Structure determination of feline panleukopenia virus empty particles. Proteins 16:155–171 [DOI] [PubMed] [Google Scholar]
- 2. Arnold E., et al. 1984. Virion orientation in cubic crystals of the human common cold virus HRV14. J. Mol. Biol. 177:417–430 [DOI] [PubMed] [Google Scholar]
- 3. Bennett M. J., Choe S., Eisenberg D. 1994. Domain swapping: entangling alliances between proteins. Proc. Natl. Acad. Sci. U. S. A. 91:3127–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benson S. D., Bamford J. K., Bamford D. H., Burnett R. M. 2004. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell 16:673–685 [DOI] [PubMed] [Google Scholar]
- 5. Brünger A. T. 2007. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2:2728–2733 [DOI] [PubMed] [Google Scholar]
- 6. Brünger A. T., et al. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905–921 [DOI] [PubMed] [Google Scholar]
- 7. Canaan S., et al. 2004. Interfacial enzymology of parvovirus phospholipases A2. J. Biol. Chem. 279:14502–14508 [DOI] [PubMed] [Google Scholar]
- 8. Collaborative Computational Project, Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 9. Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 10. Farr G. A., Zhang L. G., Tattersall P. 2005. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc. Natl. Acad. Sci. U. S. A. 102:17148–17153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Girod A., et al. 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 83:973–978 [DOI] [PubMed] [Google Scholar]
- 12. Kabsch W., Sander C. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637 [DOI] [PubMed] [Google Scholar]
- 13. Kaufmann B., et al. 2010. The structure of Penaeus stylirostris densovirus, a shrimp pathogen. J. Virol. 84:11289–11296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaufmann B., Simpson A. A., Rossmann M. G. 2004. The structure of human parvovirus B19. Proc. Natl. Acad. Sci. U. S. A. 101:11628–11633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kleywegt G. J., Read R. J. 1997. Not your average density. Structure 5:1557–1569 [DOI] [PubMed] [Google Scholar]
- 16. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. 1993. Procheck—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291 [Google Scholar]
- 17. Li Y., et al. 2001. Genome organization of the densovirus from Bombyx mori (BmDNV-1) and enzyme activity of its capsid. J. Gen. Virol. 82:2821–2825 [DOI] [PubMed] [Google Scholar]
- 18. Llamas-Saiz A. L., et al. 1997. Structure determination of minute virus of mice. Acta Crystallogr. D Biol. Crystallogr. 53:93–102 [DOI] [PubMed] [Google Scholar]
- 19. Lombardo E., Ramírez J. C., Agbandje-McKenna M., Almendral J. M. 2000. A beta-stranded motif drives capsid protein oligomers of the parvovirus minute virus of mice into the nucleus for viral assembly. J. Virol. 74:3804–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nandhagopal N., et al. 2002. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl. Acad. Sci. U. S. A. 99:14758–14763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Otwinowski Z., Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode, p. 307–326 In Carter Charles W., Jr. (ed.), Methods Enzymol., vol. 276 Academic Press, San Diego, CA: [DOI] [PubMed] [Google Scholar]
- 22. Pettersen E. F., et al. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Computat. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 23. Read R. J. 1986. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. Sect. A 42:140–149 [Google Scholar]
- 24. Riolobos L., Reguera J., Mateu M. G., Almendral J. M. 2006. Nuclear transport of trimeric assembly intermediates exerts a morphogenetic control on the icosahedral parvovirus capsid. J. Mol. Biol. 357:1026–1038 [DOI] [PubMed] [Google Scholar]
- 25. Rollinger J. M., Schmidtke M. 2011. The human rhinovirus: human-pathological impact, mechanisms of antirhinoviral agents, and strategies for their discovery. Med. Res. Rev. 31:42–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rossmann M. G., Blow D. M. 1962. The detection of sub-units within the crystallographic asymmetric unit. Acta Crystallogr. 15:24–31 [Google Scholar]
- 27. Rossmann M. G., Johnson J. E. 1989. Icosahedral RNA virus structure. Annu. Rev. Biochem. 58:533–573 [DOI] [PubMed] [Google Scholar]
- 28. Shimizu T. 1975. Pathogenicity of an infectious flacherie virus of the silkworm, Bombyx mori, obtained from sericultural farms in the suburbs of Ina city. J. Seric. Sci. Jpn. 44:45–48 [Google Scholar]
- 29. Simpson A. A., Chipman P. R., Baker T. S., Tijssen P., Rossmann M. G. 1998. The structure of an insect parvovirus (Galleria mellonella densovirus) at 3.7 Å resolution. Structure 6:1355–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Simpson A. A., et al. 2002. The structure of porcine parvovirus: comparison with related viruses. J. Mol. Biol. 315:1189–1198 [DOI] [PubMed] [Google Scholar]
- 31. Tijssen P., et al. Parvoviridae. In King A. M. Q., Adams M. J., Carstens E., Lefkowitz E. J. (ed.), Virus taxonomy: classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses, in press Elsevier, San Diego, CA [Google Scholar]
- 32. Tong L. A., Rossmann M. G. 1990. The locked rotation function. Acta Crystallogr. A 46:783–792 [DOI] [PubMed] [Google Scholar]
- 33. Tsao J., et al. 1991. The three-dimensional structure of canine parvovirus and its functional implications. Science 251:1456–1464 [DOI] [PubMed] [Google Scholar]
- 34. Xie Q., et al. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. U. S. A. 99:10405–10410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xie Q., Chapman M. S. 1996. Canine parvovirus capsid structure, analyzed at 2.9 Å resolution. J. Mol. Biol. 264:497–520 [DOI] [PubMed] [Google Scholar]
- 36. Zadori Z., et al. 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 1:291–302 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.