Abstract
Human cytomegalovirus UL103 encodes a tegument protein that is conserved across herpesvirus subgroups. Mutant viruses lacking this gene product exhibit dramatically reduced accumulation of cell-free virus progeny and poor cell-to-cell spread. Given that viral proteins and viral DNA accumulate with normal kinetics in cells infected with mutant virus, UL103 appears to function during the late phase of replication, playing a critical role in egress of capsidless dense bodies and virions. Few dense bodies were observed in the extracellular space in mutant virus-infected cells in the presence or absence of the DNA encapsidation inhibitor 2-bromo-5,6-dichloro-1-(β-d-ribofuranosyl)benzimidazole. Upon reversal of encapsidation inhibition, UL103 had a striking impact on accumulation of cell-free virus, but not on accumulation of cell-associated virus. Thus, UL103 plays a novel and important role during maturation, regulating virus particle and dense body egress from infected cells.
INTRODUCTION
The final envelopment and egress of cytomegalovirus (CMV), also known as human herpesvirus 5 (HHV-5), occur in the cytoplasm, where nucleocapsids acquire tegument and undergo final envelopment before egress to the extracellular space. A core set of common herpesvirus genes, including many tegument proteins, control these steps in evolutionarily conserved ways (34). The maturation of human CMV (HCMV), as well as another human betaherpesvirus, HHV-6A, takes place within a distinct assembly compartment (AC) that is composed of rearranged cytoplasmic organelles normally associated with the endocytic and exocytic pathways (14, 35). The HCMV AC forms during the late phase of viral replication as a cytoplasmic inclusion that has been described as a cylindrical arrangement of early endosomes and Golgi and other cytoplasmic membranes (14). Final (secondary) tegumentation and envelopment takes place within the AC as viral nucleocapsids bud into endosomal and/or Golgi apparatus-derived membranes (9, 44, 51). Envelopment is followed by egress of mature viral particles to the extracellular space and into adjacent cells, processes that are associated with cellular exocytic vesicle trafficking (9, 25, 45).
In addition to viral particles, infected cells produce noninfectious enveloped particles (NIEPs) and capsidless dense bodies (DBs) that mature within the AC and egress in parallel with virions (13, 27). Alphaherpesvirus egress has similar characteristics (48), although maturation has not been associated with a distinct AC. It is not yet clear whether envelopment of all CMV particles (virions, NIEPs, and DBs) occurs at the same or different cytoplasmic sites and whether particle egress is dependent upon viral functions that modify the exocytic pathway. Virions, DBs, and NIEPs are produced by all characterized HCMV strains, although the relative proportions vary. In contrast, only virions and DB-like light particles mature in alphaherpesviruses, herpes simplex virus 1 (HSV-1), and pseudorabies virus (PrV). The phenotypes of mutant alphaherpesviruses have suggested that (i) maturation and egress of capsid-containing and capsidless particles occur via distinct pathways (39), (ii) egress is under the control of viral proteins (20, 21), and (iii) certain mutants differentially impact release into the culture supernatant as opposed to cell-to-cell spread (19). Release and spread have both been recognized as important means of virus transmission and dissemination in viral pathogenesis.
Tegument proteins control important processes during initial infection (7, 18) to modify the environment of the newly infected cell (1, 22, 47, 55), as well as to control assembly, maturation, and egress (2, 3, 8, 12, 28, 29, 42, 46, 49, 57). The functions of most CMV tegument proteins (4, 56) remain poorly understood, although many play roles in replication based on observed replication defects for tegument gene-specific mutants (17, 58). During maturation, tegument proteins may control steps in the nucleus, such as translocation of nucleocapsids into the cytoplasm through egress of viral particles out of the cell. The current view is that tegument proteins are incorporated into virus particles in a sequential and spatially distinct order, beginning in the nucleus. In HCMV, capsid-proximal ppUL32 (pp150) nucleates an organized tegument and provides nucleocapsid stability (6, 11, 53), tegument proteins ppUL82 (pp71) and ppUL83 (pp65) are added in the nucleus (23, 24, 38), ppUL50-ppUL53 complex controls egress from the nucleus (33, 36), and ppUL99 (pp28) supports efficient tegumentation and/or envelopment (41, 46). Based on the study of viral mutants and antiviral drugs, virion maturation and egress from human fibroblasts (HF) are independent of pp65 (44) but require capsid assembly, DNA replication, encapsidation, and nucleocapsid egress to the cytoplasm, as well as translocation and final envelopment in the cytoplasmic AC. DB formation and maturation also take place in the cytoplasmic AC and require the major tegument protein, pp65, a constituent of both virions and DBs (12, 44). The final release of virions and dense bodies from cells appears to follow the cellular exocytic pathway. Mutations affecting pp150 or pp28 block the accumulation and egress of virions without impacting DB maturation. Likewise, encapsidation inhibitor 2-bromo-5,6-dichloro-1-(β-d-ribofuranosyl)benzimidazole (BDCRB) prevents the accumulation of enveloped virions without reducing the formation and egress of DB (26). Another, less well characterized tegument protein, ppUL35 (42), a ppUL82-interacting partner, influences DB formation in a way that is not yet clear. The order in which tegument proteins are added into virus particles and DBs and the manner in which these proteins orchestrate final steps of maturation and govern egress of HCMV particles remain important areas of investigation.
Genes encoding tegument proteins of alpha- and betaherpesviruses have been shown to affect nucleocapsid envelopment in the cytoplasm and virion egress from cells. The final egress of alphaherpesviruses, PrV, and bovine herpesvirus 1 (BoHV-1) is influenced by the herpesvirus-common UL7, as assessed by levels of released progeny virus (20, 43). HCMV UL103, a tegument protein (56) shares 28% amino acid sequence identity with PrV UL7 (16), is expressed late in infection (10), and may have a role augmenting viral replication based on the replication defect of insertion or substitution mutants (17, 58). To investigate the biological properties of UL103, we constructed mutant HCMV and evaluated the replication block. Consistent with observations in alphaherpesviruses, egress of HCMV virions is controlled by UL103. Moreover, UL103 function is more dramatic than UL7, providing a unique example of an HCMV function that influences cell-to-cell spread and cell-free release of all types of viral particles. This investigation reveals a common final maturation pathway for HCMV virions and DBs.
MATERIALS AND METHODS
DNA constructs and mutagenesis.
The UL103 open reading frame (ORF) was PCR amplified from HCMV Towne-BAC isolate (GenBank accession no. AY315197) and cloned in frame into EcoRI and BamHI sites of the pCMV-Tag 5A vector (Stratagene, La Jolla, CA), resulting in a c-myc epitope at the C terminus of UL103 (UL103myc). Construction of plasmid pON2999 was carried out by excising UL103-myc DNA by restriction enzymes NotI and PacI and ligating the insert into retroviral plasmid pQCXIH (Clontech Laboratories, Mountain View, CA).
Cells.
Primary foreskin-derived human fibroblasts (HF) were cultured in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) containing 4.5 g/ml glucose, 10% fetal bovine serum (catalog no. S1245OH; Atlanta Biologicals, Lawrenceville, GA), 1 mM sodium pyruvate, 2 mM l-glutamine, and 100 U/ml penicillin-streptomycin (Cellgro, Manassas, VA) at 37°C with 5% CO2. HF between passages 10 and 25 were used in all experiments. A cell line expressing UL103-myc was generated by retrovirus transduction and drug selection as previously described (5). Briefly, 293T cells were cotransfected with 8 μg of expression plasmid pON2999, 3 μg VSV-G expression plasmid, 1 μg CMV-tat expression plasmid, and 1 μg pJK3 expression plasmid (5), using Lipofectamine transfection as recommended by the manufacturer (Invitrogen, Carlsbad, CA). Cell culture supernatants were harvested and filtered through 0.45-μm cellulose acetate membrane filter (VWR International, West Chester, PA) The retrovirus-transduced cell supernatants were added together with 8 μg/ml Polybrene (Millipore, Billerica, MA) to low-passage HF. Stable UL103-expressing cells were selected using 25 μg/ml hygromycin B (Invitrogen, Carlsbad, CA).
BAC mutagenesis and recombinant viruses.
HCMV Towne-BAC isolate (GenBank accession no. AY315197) was used for generation of mutant viruses (30). Bacterial artificial chromosome (BAC) recombineering (50) was carried out using Escherichia coli strain DH10B (Invitrogen, Carlsbad, CA) containing HCMV Towne-BAC (17) and pSim6 plasmid (15). The E. coli strain was made electrocompetent by growing the bacteria at 32°C until the A600 was between 0.4 and 0.6. At this time point, cells were transferred to a 42°C shaking water bath for 15 min and immediately cooled on ice for at least 5 min. The cells were washed twice in ice-cold water, and 150 ng of a linear PCR fragment was added to suspended cells. DNA was introduced into the cells by electroporation. Kanamycin-resistant (Kanr) colonies were selected on plates containing Luria-Bertani medium (LB) and Kan). In addition to selection on LB plates containing Kan, colonies in which the Kanr-SacB cassette (Kanr cassette together with a Bacillus subtilis levansucrase [SacB] cassette) had been inserted were screened for sucrose sensitivity on 7% sucrose plates and thereafter subjected to colony PCR. The colonies were expanded in LB medium containing antibiotics, and BAC DNA was extracted according to the manufacturer's instructions (Qiagen, Valencia, CA). For construction of a partial UL103 ORF deletion mutant containing a Kanr cassette amplified from plasmid pTBE100 (a kind gift from Tim Barnett, Emory Children's Center, Atlanta, GA), the following PCR primers were used: UL103-KanMX4 F (F for forward) (5′-GAGGCCCTGATGATCCGCGGCGTGCTGGAGGTCCATACGGATTTCACTAGgttttatggacagcaagcga-3′) and UL103-KanMX4 R (R for reverse) (5′-CTACGTGTTGCGTGTTTTTTTTTCTATGATATGCGTGTCTAGTTCGCTTCgaaataccgcacagatgcgtaagg-3′). In the PCR primer sequences, nucleotides homologous to those in the Towne-BAC isolate sequence are shown in uppercase, and the nucleotides in the Kanr cassette are shown in lowercase. The deletion mutant (Δ53-750UL103, pON5020) contained the first 52 nucleotides of UL103, while nucleotides 53 to 750 were replaced with a Kanr cassette. Generation of a translation stop-frameshift UL103 mutant (UL103-Stop-F/S, pON5021) involved first replacing the entire UL103 ORF with the Kanr cassette together with a Bacillus subtilis levansucrase (SacB) cassette amplified from plasmid pTBE100 with the following primers: 5′-CACGGCAGACGAGGAGCGGCGCGGCCCAGAGCGTGTCGGCCGATTTCGAAtaggcccgtagtctgcaaatcc-3′ and 5′-CTACGTGTTGCGTGTTTTTTTTTCTATGATATGCGTGTCTAGTTCGCTTCgaaataccgcacagatgcgtaagg-3′. UL103-Stop-F/S was generated by overlap extension PCR; codon 52 of UL103 was replaced to generate a stop codon, and a nucleotide, as well as an AvrII restriction site, was inserted to introduce a translational frameshift. Namely, a primer located 50 nucleotides upstream of UL103 (5′-CACGGCAGACGAGGAGCGGCGCGGCCCAGAGCGTGTCGGCCGATTTCGAA-3′) and primer UL103 S/FS (5′GTGCACAGCAGCTCGCTGCGGTACCTAGGCCATGGAGACCAGCAGC-3′, where C and A indicate insertion of frameshift and stop codon, respectively) amplified a PCR fragment 229 bp in size. Primer located 50 nucleotides downstream of UL103 (5′-CTACGTGTTGCGTGTTTTTTTTTCTATGATATGCGTGTCTAGTTCGCTTC-3′) and primer UL103-Stop-F/S (5′-GCTGCTGGTCTCCATGGCCTAGGTACCGCAGCGAGCTGCTGTGCAC-3′ where T and G indicate insertion of the translational frameshift and stop codon, respectively) amplified a PCR fragment 667 bp in size. After gel purification of the appropriately sized PCR product according to the manufacturer's instructions (Qiagen, Valencia, CA), a second PCR using the two products as templates, together with primers located upstream and downstream of UL103, generated a PCR product 850 bp in size, which was thereafter used to replace the Kanr-SacB cassette. To make a rescue virus (UL103-R, pON5022), the mutated Stop-F/S UL103 sequence was first replaced by a Kanr-SacB cassette and then subjected to a second round of recombination by replacing Kanr-SacB with wild-type (WT) UL103.
Δ53-750UL103 was digested with restriction enzymes HindIII, SphI, and XbaI, while Δ53-750UL103 with a Kanr-SacB insert was digested with BamHI, XcmI, and SphI. UL103-Stop-F/S-BAC, UL103-R-BAC, and Towne-BAC DNAs were digested using AvrII plus NheI (see Fig. 1A), AvrII, SphI, BamHI, HindIII, and EcoRI to detect the presence of the appropriate insert and to evaluate the presence of deletions.
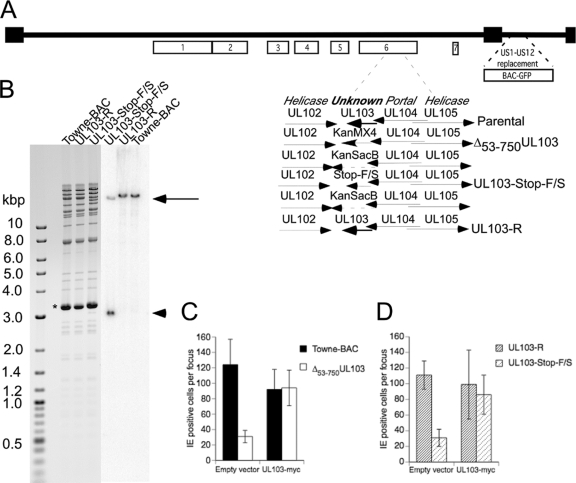
Fig. 1.
UL103 mutant virus genome structure and complementation. (A) Schematic depiction of the Towne-BAC genome, where boxes 1 to 7 identify herpesvirus-conserved gene clusters (31). The replacement of the US1-US12 region in Towne-BAC with a BAC and a GFP eukaryotic expression cassette has been previously described (30). The region encompassing UL103 in gene cluster 6 is shown expanded, with Towne-BAC, rescue, intermediate Kanr-SacB constructs, and replacement mutant viruses schematically presented. (B) Ethidium bromide-stained electrophoretically separated AvrII-NheI restriction fragment digestion products of Towne-BAC, UL103-R, and UL103-Stop-F/S Bacmid DNA (left panel) and the same samples following DNA blot hybridization to detect the UL103 ORF (right panel). Insertion of AvrII restriction sites into the UL103-Stop-F/S virus resulted in a band at 18.9 kbp (black arrow) and one at 3 kbp (black arrowhead). The thick band at 3.3 kbp (asterisk) is plasmid pSim6. (C and D) Complementation assay using UL103myc-expressing and control HF. Towne-BAC, Δ53-750UL103, UL103-R, and UL103-Stop-F/S virus infections were carried out at an MOI of 0.005 in the presence of 0.01% CMV antibody-positive pooled human IgG to prevent secondary plaque formation. Cells were fixed and stained for IE1/IE2 at 7 days postinfection (dpi), and results are graphed as the average numbers of cells per focus ± standard deviation (SD) (error bars).
DNA blot hybridization.
AvrII-NheI digestions of 2 μg of Towne-BAC, UL103-R, and UL103-Stop-F/S BAC DNA were separated on a 0.5% agarose gel and transferred to a nylon membrane (Millipore, Billerica, MA). UL103 PCR fragment from primers 5′ATGGAGGCCCTGATCCG3′ and 5′TCACTCTTCCTCTCCCCGTTCC3′ was radioactively labeled with [α-32P]dCTP (Perkin-Elmer Waltham, MA) by random priming and Klenow fragment DNA polymerase (New England BioLabs, Ipswich, MA) to fill in DNA gaps and was used as a probe.
Reconstitution of viruses.
Towne-BAC, UL103-R-BAC, Δ53-750UL103-BAC, and UL103-Stop-F/S-BAC DNA was transfected into HF as previously described (3). In addition, Towne-BAC and Δ53-750UL103-BAC DNA was also transfected into HF stably expressing UL103myc. All viruses were thereafter plaque purified.
Viral infections.
Prior to viral infection, HF seeded in 24-well plates (Corning Incorporated, Corning, NY) were grown to confluence. The medium was removed, and the cells were infected at a multiplicity of infection (MOI) of 0.3 or 3 PFU/cell. Viral titers in all inocula were verified by back titration. One hour postinfection (hpi), the cells were washed four times, and fresh medium was added to the cells. The cells and medium from infected wells were collected separately every day for 6 days and stored at −80°C until assessed for viral replication. Medium was cleared of cells by centrifugation for 2 min at 1,500 × g prior to storage, while the cells were harvested using 350 μl trypsin. Prior to sonication (Sonicator 3000; Misonix Incorporated, Farmingdale, NY) and storage at −80°C, 350 μl medium and 700 μl milk were added to cells. To determine the colocalization of UL103myc with viral and cellular antigens, UL103myc-expressing cells were infected with TownevarATCC strain virus at an MOI of 1 and fixed for indirect immunofluorescence at 3 days postinfection (dpi). Localization of UL103myc was also determined in Δ53-750UL103 virus-infected cells (MOI of 1) at day 3 postinfection.
Analysis of virus replication.
Viral titers were assessed by plaque assay for HCMV on HF. Giemsa stain was purchased from Sigma (St. Louis, MO). Results are presented as the mean ± standard deviation (SD) (n = 3), and the limit of sensitivity of the assay was 2 PFU/ml of culture medium.
IE1/IE2 fluorescent-focus assay.
HF grown on 24-well plates (Corning Incorporated, Corning, NY) were infected with a multiplicity of infection of 0.0005 PFU/cell. At 1 hpi, cells were washed and cell culture medium containing 0.01% pooled human gamma globulin (Carimune; CSL Behring, King of Prussia, PA) was added to the cells. In order to prevent secondary focus formation, fresh medium containing pooled human gamma globulin was added to cells 4 dpi. At 7 dpi, cells were fixed with 3.7% formaldehyde (EMD Chemicals, Darmstadt, Germany) and processed for indirect immunofluorescence assay (IFA) as described below. The sizes of the foci were determined by counting the number of IE1/IE2-positive (IE+) nuclei, and the results are presented as the mean ± SD (n = 9).
Indirect immunofluorescence assay.
For staining of viral and cellular antigens, uninfected and infected cells were fixed in 3.7% formaldehyde diluted in phosphate-buffered saline (PBS), for 10 min, followed by three washes with PBS. The cells were incubated in 50 mM ammonium chloride (EMD Chemicals, Darmstadt, Germany) for 10 min to quench autofluorescence and permeabilized with PBS containing 0.5% Triton X-100 (EMD Chemicals, Darmstadt, Germany) for 20 min. After three subsequent washes with PBS, the cells were incubated in blocking buffer containing 0.5% bovine serum albumin (BSA) and 5% donkey serum, diluted in PBS, for 1 h before they were incubated with the following primary antibodies: anti-IE1/IE2 (clone 8B1.2; Millipore, Billerica, MA) diluted 1:1,000; anti-pp28 (clone CH19; Virusys, Taneytown, MD) diluted 1:1,000; anti-pp65 (clone CH12; Virusys, Taneytown, MD) diluted 1:1,000; anti-golgin-97 (clone CDF4; Molecular Probes, Eugene, OR) diluted to 0.4 μg/ml; anti-GM130 (clone 35/GM130; BD Transduction Laboratories, Franklin Lakes, NJ) diluted 1:1,000; anti-EEA-1 (clone 14/EEA-1; BD Transduction Laboratories, Franklin Lakes, NJ) diluted 1:1,000; chicken anti-Myc (Gallus Immunotech Inc., Fergus, Ontario, Canada) diluted 1:5,000; and anti-CD63 (H5C6) and anti-LAMP-1 (H4A3), both obtained from the Developmental Studies Hybridoma Bank at the University of Iowa, diluted 1:1,000 and 1:100, respectively. After 1-h incubation with primary antibody, the cells were washed three times with PBS, and a secondary antibody was added to the cells. The secondary antibody was donkey anti-mouse conjugated to fluorescein isothiocyanate (FITC) (488 nm) and/or donkey anti-chicken conjugated to rhodamine (568 nm); both antibodies were diluted 1:1,000, and both were from Molecular Probes (Eugene, OR). The primary and secondary antibodies were diluted in PBS containing 0.5% BSA. The cells were incubated with Hoechst 33258 to stain for nuclei (AnaSpec Inc., San Jose, CA), diluted 1:50,000 in PBS, for 10 min. The coverslips were mounted on slides using an aqueous mounting medium (Biomeda, Foster City, CA). The entire procedure was carried out at room temperature. Confocal microscopy was carried out using an LSM510 Meta microscope.
Immunoblotting.
HF grown in 24-well plates were infected with virus at an MOI of 3. Cells were lysed in 2% SDS-containing lysis buffer and stored at −80°C. The primary antibodies used were anti-IE1/IE2 (clone 8B1.2; Millipore, Billerica, MA) diluted 1:2,000, anti-pp28 (clone CH19; Virusys, Taneytown, MD) diluted 1:2,000, anti-ppUL44 (clone 10D8; Virusys, Taneytown, MD) diluted 1:2,000, and antiactin (clone AC-74; Sigma-Aldrich, St. Louis, MO) diluted 1:5,000. The secondary antibody used was peroxidase-labeled horse anti-mouse IgG diluted 1:5,000. (Vector Laboratories, Burlingame, CA). Chemifluorescent signal was detected with ECL Western blotting detection reagents (GE Healthcare, Buckinghamshire, United Kingdom).
Quantitative PCR.
HF grown in 24-well plates were infected with virus at an MOI of 0.3. For assessment of viral DNA accumulation in cells, the cells were treated in the absence or presence of phosphonoformic acid (PFA) at 300 μg/ml (see below). DNA from cells was harvested at 24 and 48 hpi and stored at −80°C. For assessment of viral DNA in the supernatant, 200 μl of supernatant was taken and stored at −80°C. DNA from cells and supernatant was extracted using the DNeasy blood and tissue kit per the manufacturer's instructions (Qiagen, Valencia, CA). Primers amplifying IE1 exon 2 (IE1-2F [5′GGCCGAAGAATCCCTCAAAA3′] and IE1-2R [5′TCGTTGCAATCCTCGGTCA3′]) were used. Real-time PCR was performed in 7500 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA) as described previously (37).
Viral inhibitors.
BDCRB (52) was diluted in dimethyl sulfoxide (DMSO), and the viral inhibition experiment was carried out by using a concentration of 20 μM BDCRB as previously described (57). PFA was diluted in water and used at a concentration of 300 μg/ml (Sigma-Aldrich, St. Louis, MO).
Transmission electron microscopy (TEM).
HF grown in 24-well plates (Corning, NY) were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) for 1 h at room temperature and thereafter stored at 4°C prior to processing. The cells were washed in 0.1 M cacodylate buffer, postfixed with the same buffer and addition of 1% osmium tetroxide, washed and dehydrated through a graded series of ethanol to 100%, and embedded in epoxy resin. Thin sections were counterstained with uranyl acetate and lead citrate. The sections were examined with a Hitachi H-7500 TEM operated at 75 kV, and the images were captured using the digital camera system Gatan Bioscan (Pleasanton, CA). The images were acquired and analyzed with Digital Micrograph software (Pleasanton, CA).
RESULTS
Properties of UL103 mutant viruses.
Two UL103-deficient viruses were generated from Towne-BAC (17): (i) one mutant with a deletion removing most of the protein-coding sequences (Δ53-750UL103) and (ii) one mutant containing both a stop codon and a frameshift (UL103-Stop-F/S) to eliminate translation through the ORF (Fig. 1A). Mutant Δ53-750UL103 retained the amino-terminal 17 codons (out of the total 250) of UL103 to avoid disrupting the overlapping UL104 gene. The UL103 region encoding amino acids (aa) 53 to 750 was replaced with a Kanr cassette, and the insertion was verified by restriction enzyme digestion (data not shown). The rescue and UL103-Stop-F/S BACs were constructed by replacing a Kanr-SacB cassette that either reintroduced a WT ORF or terminated UL103 after aa 51 (Fig. 1A), respectively, through recombineering protocols (50). DNA blot hybridization yielded the predicted patterns for parental Towne-BAC, rescue (UL103-R-BAC), and UL103-Stop-F/S-BAC (Fig. 1B).
When recovered after transfection, UL103 mutant viruses formed small plaques on human fibroblasts (HF) compared to viruses generated from either parental (Towne-BAC) or rescue (UL103-R-BAC) constructs, indicating that mutant viruses replicated and spread poorly (data not shown). Propagation of mutant viral stocks required an average of 2 to 3 weeks longer than propagation of parental Towne-BAC or rescued UL103-R virus. In contrast, when UL103myc-expressing cells were used to support replication, Δ53-750UL103 mutant virus propagated with an efficiency similar to that of Towne-BAC virus (data not shown). To evaluate the impact of UL103 more directly, we evaluated cell-to-cell spread using a sensitive IE1/IE2 (IE) fluorescent-focus formation assay on UL103myc-expressing HF in comparison to standard HF. On UL103-expressing HF, UL103-Stop-F/S, Δ53-750UL103, and control (UL103-R and Towne-BAC) viruses formed foci with similar numbers of IE1/IE2-positive (IE+) cells, indicating similarity in cell-to-cell spread. In contrast, on HF, both mutant viruses formed small IE+ foci that averaged 30% the size of control virus foci (Fig. 1C and D). Complementation of growth on UL103-expressing cells established that the mutant virus replication defect was the direct result of failure to express functional UL103 during infection.
Replication kinetics of UL103 mutant viruses.
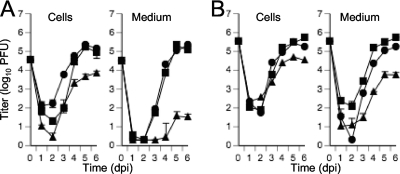
When UL103-Stop-F/S mutant virus was subjected to multistep replication conditions (MOI of 0.3) on HF, we observed a more dramatic impact on virus release than on the accumulation of cell-associated virus. As would be expected, the mutant virus replication defect was more striking under multistep conditions (MOI of 0.3) (Fig. 2A) than under single-step conditions (MOI of 3) (Fig. 2B). Cell-associated mutant virus titers were 40-fold lower and cell-free virus titers were 300-fold lower than the control virus titers. Under one-step growth conditions, mutant virus displayed 8-fold-lower cell-associated titers and 50-fold-lower cell-free titers accumulating in the culture medium compared to control viruses (Fig. 2B). The phenotype displayed by the Δ53-750UL103 mutant virus was similar to the phenotype displayed by the UL103-Stop-F/S virus in these experiments (data not shown). Mutant virus Δ53-750UL103 behaved the same whether propagated on standard or UL103myc-expressing HF, suggesting no benefit of virion-associated UL103 protein. On the basis of the results of these analyses and data shown below ascribing the UL103 defect to a late maturation stage, we did not explore the potential role of virion-associated UL103 any further. Taken together, these data indicate that UL103 influenced the accumulation of both cell-associated and cell-released virus titers, with the greatest impact on accumulation of extracellular infectious virus.
Fig. 2.
Multistep (A) (MOI of 0.3) or single-step (B) (MOI of 3) growth curves of Towne-BAC (squares), UL103-R (circles), and UL103-Stop-F/S (triangles) reconstituted viruses. The cells and medium were harvested at the indicated times postinfection and frozen at −80°C. The virus titer was determined in triplicate by plaque assay on HF and graphed as log10 mean value ± SD. The data point at 0 dpi indicates the input virus dose.
Role of UL103 during different stages of replication.
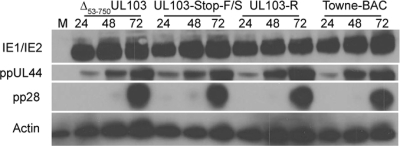
To identify the stage of replication that was affected by the disruption of UL103, viral entry, viral DNA accumulation, and viral protein levels were evaluated in UL103 mutant and control virus-infected HF. For these studies, viruses were propagated, and the properties of the virus in HF were determined. Mutant and parental viruses exhibited similar amounts of input viral DNA when evaluated in the presence of the viral DNA replication inhibitor phosphonoformic acid (PFA), consistent with a similar pattern of entry into cells (data not shown). Further, between 58 and 64% of infected cells were IE+ (MOI of 1) by IFA (data not shown), accumulating similar amounts when Δ53-750UL103, UL103-Stop-F/S, UL103-R, and Towne-BAC viruses were compared in immunoblot analyses (Fig. 3). This suggested that initial gene expression occurred independently of UL103. In addition, the levels of the β viral antigen (delayed-early antigen), ppUL44, and the γ2 (true late) viral antigen, pp28, were similar to the levels from control viruses when evaluated at 24, 48, and 72 hpi (Fig. 3), indicating that the viral gene expression cascade proceeds independently of UL103. All viruses produced similar levels of DNA at 24 and 48 hpi (data not shown), indicating that DNA replication proceeds independently of UL103. The failure of UL103 mutant viruses to impact entry, gene expression, or DNA accumulation suggests that UL103 controls a late step in replication.
Fig. 3.
Immunoblot analyses of viral proteins from HF infected with Towne-BAC, UL103-R, UL103-Stop-F/S, or Δ53-750UL103 virus and mock-infected HF (MOI of 3). Detection of IE1/IE2, ppUL44, ppUL99 (pp28), and β-actin were carried out at 24, 48, and 72 hpi. Lane M, mock-infected HF.
UL103 accumulates at a juxtanuclear site in uninfected and infected cells.
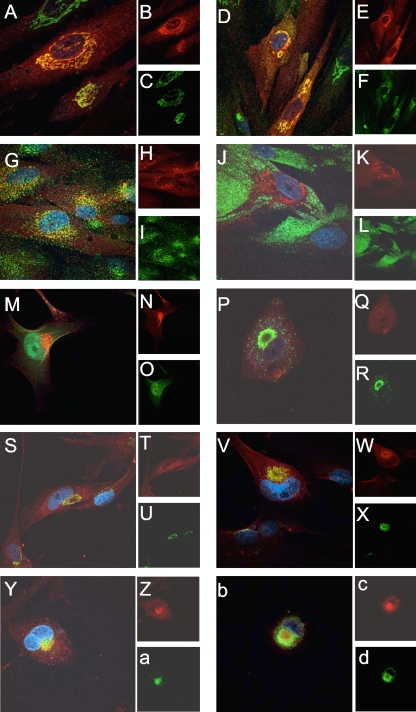
To investigate the localization of UL103 in cells, UL103myc-expressing cells were employed in colocalization studies. In uninfected cells, UL103myc antigen displayed a juxtanuclear pattern and colocalized with Golgi-resident proteins golgin-97 and GM130, but not with the early endosomal marker (EEA1) or the late endosomal marker CD63 (Fig. 4A to L). When localization was undertaken in Δ53-750UL103 mutant virus-infected cells (3 dpi), UL103myc was exclusively associated with the assembly compartment (AC) (Fig. 4M to O). Because expression of green fluorescent protein (GFP) by the Towne-BAC-derived control and mutant viruses interfered with multicolor immunofluorescence analyses, a standard laboratory strain TownevarATCC strain was employed to evaluate UL103myc localization within the AC. AC-associated antigens, including the tegument protein pp28, cellular trans-Golgi marker golgin-97, cis-Golgi marker GM130 (Fig. 4P to X), EEA-1 (Fig. 4Y to a), and CD63 (Fig. 4b to d), all accumulated in the AC late in infection and colocalized with UL103myc. Thus, pUL103 associates with the AC (41), consistent with a role in viral maturation and/or egress.
Fig. 4.
Immunofluorescence localization of cellular and viral antigens in uninfected and UL103myc-infected cells. (A to L) Immunofluorescence localization of cellular antigens in UL103myc-expressing uninfected cells using mouse anti-golgin-97 (C), mouse-anti-GM130 (F), mouse anti-EEA-1 (I), and mouse anti-CD63 (L) and donkey anti-mouse conjugated to FITC (green). UL103-myc (B, E, H, and K) was visualized with chicken anti-myc and donkey anti-chicken secondary antibody conjugated to rhodamine (red). Nuclei were stained with Hoechst 33258 (blue). Panels A, D, G, and J display merged images of Hoechst 33258, rhodamine, and FITC. (M to O) Immunofluorescence localization of UL103myc (N) at 3 dpi of Δ53-750UL103 virus-infected cells (MOI of 1) (O) using chicken anti-myc and anti-chicken secondary antibody conjugated to rhodamine (red). (M) Merged image of rhodamine, GFP fluorescence, and Hoechst 33258. (P to d) Immunofluorescence localization of cellular and viral antigens at 3 dpi of Townevar ATCC-infected cells (MOI of 1). pp28 (R), golgin-97 (U), GM130 (X), EEA-1 (a), and CD63 (d) were detected using donkey anti-mouse secondary antibody conjugated to FITC, while myc (Q, T, W, Z, and c) was detected with donkey anti-chicken secondary antibody conjugated to rhodamine. Panels P, S, V, Y, and b display merged images of Hoechst 33258, rhodamine, and FITC. UL103myc was functional in these cells, as shown in Fig. 1C and D. Original magnification obtained by confocal microscopy, ×1,000.
Roles of UL103 in release of virions and DBs.
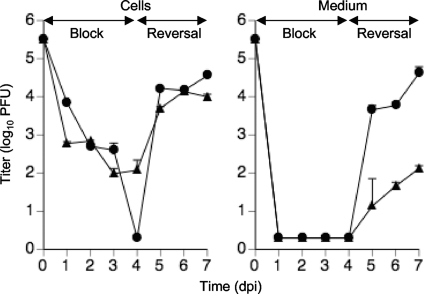
To better identify the step(s) of replication and to focus on late events, the inhibitor 2-bromo-5,6-dichloro-1-(β-d-ribofuranosyl)benzimidazole (BDCRB) (54) was employed to block viral DNA encapsidation. This reversible inhibitor prevents the translocation of nucleocapsids from the nucleus to the cytoplasm (26). We used this property to synchronize mutant and WT virus infections, thus allowing a specific focus on subsequent events of maturation and egress (Fig. 5). During BDCRB treatment through 4 dpi, accumulation of both mutant and control viruses was blocked to a similar level. After reversal of the BDCRB block, the mutant virus demonstrated a dramatic, sustained defect in release, with no reduction in the accumulation of intracellular infectious virus. When cell-free viral titers were evaluated at 1, 2, and 3 days after reversal, the mutant virus displayed on average 270-fold-lower titers compared to control (Fig. 5). This indicates that UL103 may be involved in egress after secondary envelopment.
Fig. 5.
Single-step growth curve (MOI of 3) of UL103-R (circles) and UL103-Stop-F/S (rectangles) viruses during block and reversal of BDCRB. Cells and medium were harvested individually every day for 7 days and frozen at −80°C. Virus titer was determined and graphed as log10 mean value plus SD. The data point at 0 dpi indicates the input virus dose.
As UL103 mutant virus exhibited a defect in release of infectious virus during block-reversal with BDCRB, we next investigated whether mutant and control viruses exhibited differences in DB release. DB release was evaluated by counting the numbers of GFP-positive pp65-positive (GFP+ pp65+) cells (an indicator of viral infection) and GFP-negative pp65-positive (GFP− pp65+) cells (an indicator of DB spread) during a BDCRB block. We found that rescue virus infection, in the presence of BDCRB block, promoted appearance of pp65+ cells without signs of active replication (GFP−), consistent with DB release and spread from infected (GFP+ pp65+) cells (Table 1). Importantly, mutant virus exhibited similar percentages (about 40%) of infected (GFP+ pp65+) cells, in comparison to control virus, but a much lower percentage of GFP− pp65+ cells (4%; Table 1). These results suggest that DB spread was specifically impaired in mutant virus-infected cells maintained in the presence of BDCRB. Immunoblot analyses on lysates from BDCRB-treated cells at 4 dpi (MOI of 3) indicated similar amounts of pp65 (data not shown) in mutant and control virus infections, establishing that the inability of the UL103 mutant virus to release DB was not the result of altered pp65 expression in mutant virus-infected cells.
Table 1.
Scoring pp65-positive and pp65-negative cells surrounding GFP+ virus-infected cells during BDCRB block
| Virus | GFP+ pp65+ | GFP− pp65+ |
|---|---|---|
| UL103-Stop-F/S | 40 ± 0.6a | 4 ± 0.6 |
| UL103-R | 43 ± 0.6 | 55 ± 1.7 |
Percentage of cells at 4 dpi with an MOI of 0.3 (±SD).
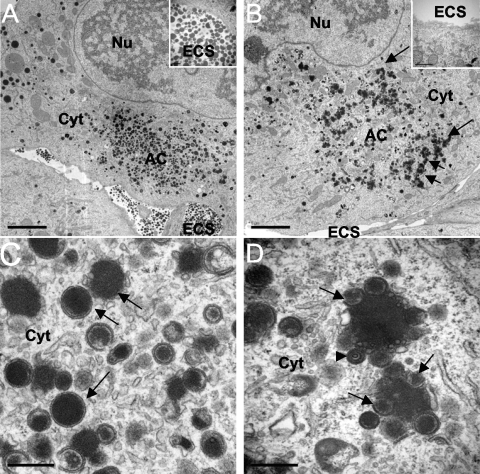
The failure to observe pp65 spread in mutant virus-infected cultures suggested a block in DB release. To confirm this possibility, ultrastructural analyses were conducted at 4 dpi in BDCRB-treated cells. More than 90 cells infected with UL103-R or UL103-Stop-F/S were evaluated. Cells infected with either mutant or rescue control virus contained numerous DB-like particles and few NIEPs in the cytoplasm. Consistent with lack of spread in the pp65 fluorescence spread assay, only a rare DB was detected in the extracellular space of UL103 mutant virus-infected cells (Fig. 6B), while this space was DB rich in control infected cells (Fig. 6A). These results confirmed evaluations of DB spread by the pp65 fluorescence spread assay (Table 1). Furthermore, the area corresponding to the AC in mutant virus-infected cells (Fig. 6D) appeared to contain less electron-dense pp65-related material compared with control infected cells, and in addition, the electron-dense material present in mutant virus-infected cells was arranged in an unexpected pattern. In control infected cells, DBs were dense, circular structures and distributed throughout the AC (Fig. 6A and C), whereas in UL103 mutant virus-infected cells, DB-like aggregates of 1 to 2 μm in diameter encircled the periphery of the AC (Fig. 6B). The inner part of these AC-associated aggregates consisted of dense material, while the outer edge consisted of structures that appeared to be DB-like, although the associated membranes appeared disrupted (Fig. 6D). Despite the morphological differences, similar levels of DB-like material were observed and similar amounts of pp65 antigen were detected by immunoblotting cells infected with rescue and mutant viruses (data not shown). In combination, the results of these analyses indicated that UL103 is necessary for efficient release of DB as well as virions.
Fig. 6.
Characterization of viral components in cells treated with BDCRB and infected with UL103-R virus (A and C) or UL103-Stop-F/S virus (B and D) at 4 dpi by TEM. The nucleus (Nu), cytoplasm (Cyt), extracellular space (ECS), and assembly compartment (AC) are indicated. The arrows in panel B point to cytoplasmic aggregates. (C and D) TEM depicting size bar of 0.5 μm. In panels C and D, the arrows point to dense bodies (DBs) and the arrowhead points to a noninfectious enveloped particle (NIEP). Bars, 5 μm (A and B) and 0.5 μm (C and D).
Distribution of progeny viral particles during release of BDCRB.
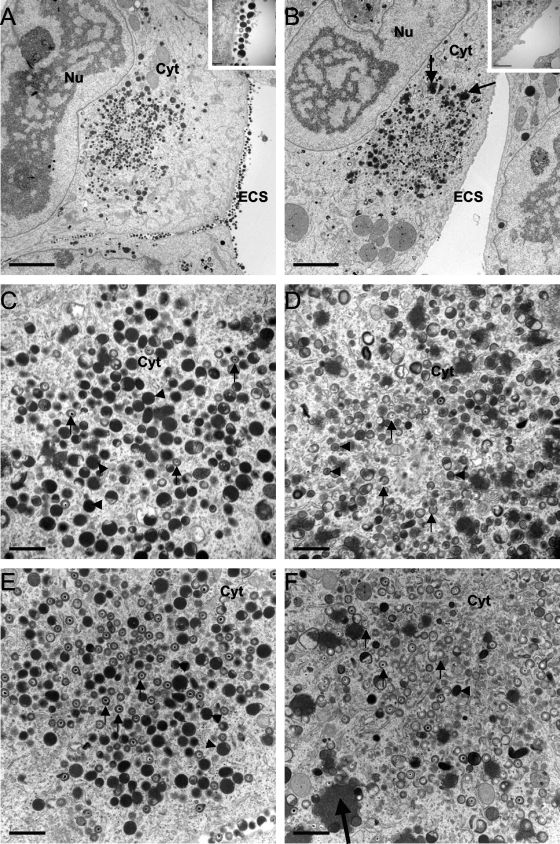
To better understand the fate of viral particles in UL103 mutant virus-infected cells, we undertook ultrastructural analyses 1 day after release (5 dpi) of BDCRB. More than 90 cells infected with each virus were evaluated. Importantly, similar levels of viral particles were observed in the cytoplasm (17 ± 7 viral particles/field; n = 9) of cells infected with either mutant or rescue virus, and similar levels of cell-associated infectivity were scored (Fig. 5). Despite these observations, mutant virus particles appeared to be in the process of envelopment or de-envelopment (Fig. 7), a pattern that was distinct from the fully enveloped particles in control virus-infected cells. Additionally, the AC in cells infected with mutant virus also appeared less electron-dense and highly vacuolized compared to control infected cells. In sharp contrast to infection with rescue virus, where large amounts of DB and viral particles were located extracellularly (Fig. 7A), only rare particles were observed extracellularly in mutant virus-infected cells (Fig. 7B). These results were completely consistent with the low levels of extracellular infectious viral particles detected in mutant virus-infected cells after release of BDCRB (Fig. 5). Similar findings were observed in the absence of BDCRB (data not shown). These findings validate our approach using this drug to synchronize infection at the late stage of genome encapsidation to more carefully examine the UL103 mutant virus phenotype. To address the possible role of UL103 as a virion stability factor, cell culture medium was collected between day 3 and day 5 postinfection, and the ratio of viral DNA to PFU was determined. The ratio of extracellular viral DNA to PFU was only about 6-fold higher for mutant virus preparations compared to WT and rescue virus (Table 2), suggesting that, once released, viral particles exhibited nearly the same infectivity independent of UL103 function. Taken together, these data demonstrate that UL103 is not required for accumulation of infectious virus but functions primarily to support viral egress from cells to the culture medium.
Fig. 7.
TEM of cells infected with UL103-R virus (A, C, and E) or with UL103-Stop-F/S virus (B, D, and F) at 5 dpi. (A to D) Virus-infected cells 1 day after the reversal (5 dpi) of BDCRB. (E and F) Virus-infected cells that were not treated with BDCRB. The small black arrows point to viral particles, and the black arrowheads point to DBs. The large black arrows in panels B and F point to aggregates. The nucleus (Nu), cytoplasm (Cyt), and extracellular space (ECS) are indicated. Bars, 5 μm (A and B) and 1 μm (C to F).
Table 2.
DNA to PFU ratio in extracellular virions
| Virus | Mean value |
|---|---|
| UL103-Stop-F/S | 5.7 ± 2.1a |
| UL103-R | 0.9 ± 0.1 |
| Towne-BAC | 1.0 ± 0.0 |
Ratio, viral DNA copy number per PFU (10−4) ± SD.
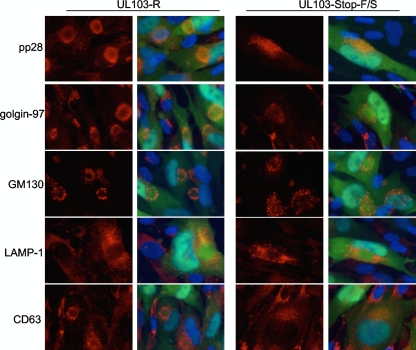
Based on TEM studies, the cytoplasmic architecture of cells infected with mutant virus appeared to differ from cells infected with rescue control virus. To further investigate the impact of mutant virus on cells, we localized viral and cellular antigens in the presence and absence of BDCRB (Fig. 8). In the presence of BDCRB, viral particles were not released from cells (Fig. 5), thus allowing us to focus on imaging primary rather than secondary infection. The pattern of LAMP-1 staining was indistinguishable and indicated that lysosomes were excluded from the AC in both UL103 mutant virus- and rescue virus-infected cells (Fig. 8). In contrast, pp28, Golgi markers, and late endosome marker were altered in mutant virus-infected cells. Thus, the staining pattern of pp28, a tegument protein that accumulates within the AC (40) and controls the final steps of maturation, suggested alterations in the AC of mutant virus-infected cells. As UL103myc colocalized with Golgi membranes in uninfected and infected cells (Fig. 4) and Golgi membranes accumulated in a characteristic ring-like pattern around the AC (14), we compared localization of the Golgi-resident proteins GM130 and golgin-97 during rescue virus and mutant virus infections and found the patterns were disorganized within mutant virus-infected cells (Fig. 8). As expected, both Golgi antigens displayed an arrangement consistent with previous observations in control virus infections (14, 49). In addition to differences in localization of pp28 and Golgi markers evident in these analyses, the staining patterns for CD63 exhibited a strikingly different pattern in mutant virus-infected cells. Thus, during rescue control virus infections, CD63 appeared in a ring-like pattern as expected (9). In contrast, CD63 appeared distributed throughout the AC in cells infected by mutant virus. Similar patterns were observed in cells that were not treated with BDCRB (data not shown). Thus, the AC became altered in the absence of UL103. The impact of UL103 on release may be associated with the manner in which other viral or host proteins are delivered to the AC.
Fig. 8.
Immunofluorescence localization of viral and cellular antigens in cells infected with UL103-R or UL103-Stop-F/S virus and treated with BDCRB. The cells were infected at an MOI of 0.3 and fixed 4 dpi. pp28, golgin-97, GM130, LAMP-1, and CD63 were visualized with donkey anti-mouse conjugated to rhodamine. Original magnification, ×1,000.
DISCUSSION
In these studies, we have revealed a role for HCMV UL103 in the egress of progeny virions, DBs, and NIEPs from infected cells, bringing to light virus modification of a pathway controlling virus release and cell-to-cell spread. The results of TEM studies along with the results of immunofluorescence and viral yield assays on infected cells suggest that tegumentation and envelopment of nucleocapsids in the AC precedes egress. In contrast to other tegument proteins that are involved in maturation steps prior to envelopment (2, 8, 46, 49), the results of our studies indicate that UL103 has the greatest impact on egress following envelopment. This phenotype will facilitate study of mechanisms controlling virion release from infected cells.
Our data are consistent with UL103 playing a role in controlling the release of progeny and in cell-to-cell spread and are consistent with prior systematic mutagenesis analyses demonstrating a decrease in HCMV replication for UL103 mutant viruses (17, 58). Here, we demonstrate that these defects were independent of the MOI, indicating that the function of UL103 cannot be supplanted by any other tegument protein. Moreover, the current studies using BDCRB block-reversal experiments to synchronize infection under high-MOI conditions rescued the defect in intracellular virion accumulation and more clearly defined UL103 as a tegument protein involved in virus release. Our experimental approach used BDCRB to delay maturation prior to encapsidation to promote continued maturation and egress. These conditions allowed the UL103 mutant virus to overcome an apparent defect in cell-associated virion accumulation. This approach more clearly highlighted the role UL103 plays during egress of viral particles.
Ultrastructural analyses undertaken to gain further insights into the role of UL103 in virus-infected cells indicated no discernible difference between mutant and rescue virus-infected cells in the number of viral particles present in the cytoplasm; however, the number of extracellular viral particles was significantly lower in mutant virus-infected cells than in rescue virus-infected cells. Thus, UL103 had no impact on the formation of intracellular viral particles, as indicated by TEM or by cell-associated virus titers, while efficient egress was dependent on this viral gene product. The ability of UL103 to enhance release of viral particles underscores the critical role of UL103 in HCMV replication.
This study has extended the understanding of HCMV particle morphogenesis and egress. It appears that formation of viral particles and DBs occurs via parallel but independent pathways that converge in the cytoplasm, where egress occurs via a common pathway. In HCMV-infected BDCRB-treated cells, DBs were formed and released extracellularly, while nucleocapsids were present in the nucleus at the step of encapsidation. These data are consistent with previous studies demonstrating that deletion of pp65 impacts DB maturation but not that of viral particles (12, 44). Similar to HCMV, assembly of DB-like HSV-1 light particles also occurs independently of virion maturation (39), which may indicate that formation and release of DBs and light particles occur by a common mechanism(s). During BDCRB block, UL103 mutant virus-infected cells exhibited a failure to release DBs. Instead, DBs accumulated in the cytoplasm with aggregates of DB-like material forming at the periphery of AC. A prior study demonstrated formation of aggregates when pp65 was tagged with an IE1 peptide labeled with a myc epitope tag (32), suggesting that the normal appearance of DB can be altered by changing egress efficiency as well as by modifying the major DB protein constituent.
Our study may suggest that the function of UL103 is conserved among other members of the herpesvirus family, as disruption of UL7, the UL103 homolog, resulted in reduced plaque sizes and reduced virus titers in cell culture and most notably, reduced titers of extracellular virus (20, 43). Given that three homologs belonging to distant members of the herpesvirus family, UL7 of pseudorabies virus (PrV) and bovine herpesvirus 1 (BoHV-1) and UL103 of HCMV, share salient features by enhancing release of viral particles, we suggest that viruses within the herpesvirus family share common viral assembly pathways during the final steps of maturation. Overall, the data indicate that UL103 is of importance in HCMV egress.
ACKNOWLEDGMENTS
We thank Hong Yi at the Robert P. Apkarian Integrated Electron Microscopy Core, Laura Bender at the Cell Imaging and Microscope Core at Winship Cancer Institute for technical assistance, William J. Kaiser for assistance with cell cultures, and Christopher Collins for assistance with DNA blotting. We also thank members of the Mocarski lab for critical reading of the manuscript. BDCRB was kindly provided by John Drach, University of Michigan. The H5C6 (CD63) and H4A3 (LAMP-1) monoclonal antibodies developed by J. Thomas August and James E. K. Hildreth were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences.
This research was supported by PHS grant RO1 AI020211.
Footnotes
Published ahead of print on 23 February 2011.
REFERENCES
- 1. Abate D. A., Watanabe S., Mocarski E. S. 2004. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J. Virol. 78:10995–11006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adamo J. E., Schroer J., Shenk T. 2004. Human cytomegalovirus TRS1 protein is required for efficient assembly of DNA-containing capsids. J. Virol. 78:10221–10229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. AuCoin D. P., Smith G. B., Meiering C. D., Mocarski E. S. 2006. Betaherpesvirus-conserved cytomegalovirus tegument protein ppUL32 (pp150) controls cytoplasmic events during virion maturation. J. Virol. 80:8199–8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baldick C. J., Jr., Shenk T. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartz S. R., Vodicka M. A. 1997. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods 12:337–342 [DOI] [PubMed] [Google Scholar]
- 6. Baxter M. K., Gibson W. 2001. Cytomegalovirus basic phosphoprotein (pUL32) binds to capsids in vitro through its amino one-third. J. Virol. 75:6865–6873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bechtel J. T., Shenk T. 2002. Human cytomegalovirus UL47 tegument protein functions after entry and before immediate-early gene expression. J. Virol. 76:1043–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blankenship C. A., Shenk T. 2002. Mutant human cytomegalovirus lacking the immediate-early TRS1 coding region exhibits a late defect. J. Virol. 76:12290–12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cepeda V., Esteban M., Fraile-Ramos A. 2010. Human cytomegalovirus final envelopment on membranes containing both trans-Golgi network and endosomal markers. Cell. Microbiol. 12:386–404 [DOI] [PubMed] [Google Scholar]
- 10. Chambers J., et al. 1999. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J. Virol. 73:5757–5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen D. H., Jiang H., Lee M., Liu F., Zhou Z. H. 1999. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology 260:10–16 [DOI] [PubMed] [Google Scholar]
- 12. Chevillotte M., et al. 2009. Major tegument protein pp65 of human cytomegalovirus is required for the incorporation of pUL69 and pUL97 into the virus particle and for viral growth in macrophages. J. Virol. 83:2480–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Craighead J. E., Kanich R. E., Almeida J. D. 1972. Nonviral microbodies with viral antigenicity produced in cytomegalovirus-infected cells. J. Virol. 10:766–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Das S., Vasanji A., Pellett P. E. 2007. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J. Virol. 81:11861–11869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Datta S., Costantino N., Court D. L. 2006. A set of recombineering plasmids for gram-negative bacteria. Gene 379:109–115 [DOI] [PubMed] [Google Scholar]
- 16. Dijkstra J. M., Fuchs W., Mettenleiter T. C., Klupp B. G. 1997. Identification and transcriptional analysis of pseudorabies virus UL6 to UL12 genes. Arch. Virol. 142:17–35 [DOI] [PubMed] [Google Scholar]
- 17. Dunn W., et al. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223–14228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feng X., Schroer J., Yu D., Shenk T. 2006. Human cytomegalovirus pUS24 is a virion protein that functions very early in the replication cycle. J. Virol. 80:8371–8378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fuchs W., Granzow H., Klopfleisch R., Klupp B. G., Mettenleiter T. C. 2006. The UL4 gene of pseudorabies virus encodes a minor infected-cell protein that is dispensable for virus replication. J. Gen. Virol. 87:2517–2525 [DOI] [PubMed] [Google Scholar]
- 20. Fuchs W., et al. 2005. The UL7 gene of pseudorabies virus encodes a nonessential structural protein which is involved in virion formation and egress. J. Virol. 79:11291–11299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fuchs W., Klupp B. G., Granzow H., Mettenleiter T. C. 1997. The UL20 gene product of pseudorabies virus functions in virus egress. J. Virol. 71:5639–5646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hayashi M. L., Blankenship C., Shenk T. 2000. Human cytomegalovirus UL69 protein is required for efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc. Natl. Acad. Sci. U. S. A. 97:2692–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hensel G., Meyer H., Gartner S., Brand G., Kern H. F. 1995. Nuclear localization of the human cytomegalovirus tegument protein pp150 (ppUL32). J. Gen. Virol. 76:1591–1601 [DOI] [PubMed] [Google Scholar]
- 24. Hensel G. M., et al. 1996. Intracellular localization and expression of the human cytomegalovirus matrix phosphoprotein pp71 (ppUL82): evidence for its translocation into the nucleus. J. Gen. Virol. 77:3087–3097 [DOI] [PubMed] [Google Scholar]
- 25. Homman-Loudiyi M., Hultenby K., Britt W., Soderberg-Naucler C. 2003. Envelopment of human cytomegalovirus occurs by budding into Golgi-derived vacuole compartments positive for gB, Rab 3, trans-Golgi network 46, and mannosidase II. J. Virol. 77:3191–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hwang J. S., et al. 2007. Identification of acetylated, tetrahalogenated benzimidazole d-ribonucleosides with enhanced activity against human cytomegalovirus. J. Virol. 81:11604–11611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Irmiere A., Gibson W. 1983. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 130:118–133 [DOI] [PubMed] [Google Scholar]
- 28. Krosky P. M., Baek M. C., Coen D. M. 2003. The human cytomegalovirus UL97 protein kinase, an antiviral drug target, is required at the stage of nuclear egress. J. Virol. 77:905–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lorz K., et al. 2006. Deletion of open reading frame UL26 from the human cytomegalovirus genome results in reduced viral growth, which involves impaired stability of viral particles. J. Virol. 80:5423–5434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marchini A., Liu H., Zhu H. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McGeoch D. J., Rixon F. J., Davison A. J. 2006. Topics in herpesvirus genomics and evolution. Virus Res. 117:90–104 [DOI] [PubMed] [Google Scholar]
- 32. Mersseman V., et al. 2008. Refinement of strategies for the development of a human cytomegalovirus dense body vaccine. Med. Microbiol. Immunol. 197:97–107 [DOI] [PubMed] [Google Scholar]
- 33. Milbradt J., Auerochs S., Marschall M. 2007. Cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. J. Gen. Virol. 88:2642–2650 [DOI] [PubMed] [Google Scholar]
- 34. Mocarski E. 2007. Comparative analysis of herpesvirus-common proteins, p. 44–58 In Arvin A., Campadelli-Fiume G., Mocarski E., Moore P. S., Roizman B., Whitley R., Yamanishi K. (ed.), Human herpesviruses. Biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 35. Mori Y., et al. 2008. Human herpesvirus-6 induces MVB formation, and virus egress occurs by an exosomal release pathway. Traffic 9:1728–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Muranyi W., Haas J., Wagner M., Krohne G., Koszinowski U. H. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854–857 [DOI] [PubMed] [Google Scholar]
- 37. Potena L., et al. 2007. Frequent occult infection with cytomegalovirus in cardiac transplant recipients despite antiviral prophylaxis. J. Clin. Microbiol. 45:1804–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Revello M. G., Percivalle E., Di Matteo A., Morini F., Gerna G. 1992. Nuclear expression of the lower matrix protein of human cytomegalovirus in peripheral blood leukocytes of immunocompromised viraemic patients. J. Gen. Virol. 73:437–442 [DOI] [PubMed] [Google Scholar]
- 39. Rixon F. J., Addison C., McLauchlan J. 1992. Assembly of enveloped tegument structures (L particles) can occur independently of virion maturation in herpes simplex virus type 1-infected cells. J. Gen. Virol. 73:277–284 [DOI] [PubMed] [Google Scholar]
- 40. Sanchez V., Greis K. D., Sztul E., Britt W. J. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sanchez V., Sztul E., Britt W. J. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J. Virol. 74:3842–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schierling K., Buser C., Mertens T., Winkler M. 2005. Human cytomegalovirus tegument protein ppUL35 is important for viral replication and particle formation. J. Virol. 79:3084–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schmitt J., Keil G. M. 1996. Identification and characterization of the bovine herpesvirus 1 UL7 gene and gene product which are not essential for virus replication in cell culture. J. Virol. 70:1091–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schmolke S., Kern H. F., Drescher P., Jahn G., Plachter B. 1995. The dominant phosphoprotein pp65 (UL83) of human cytomegalovirus is dispensable for growth in cell culture. J. Virol. 69:5959–5968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Severi B., Landini M. P., Govoni E. 1988. Human cytomegalovirus morphogenesis: an ultrastructural study of the late cytoplasmic phases. Arch. Virol. 98:51–64 [DOI] [PubMed] [Google Scholar]
- 46. Silva M. C., Yu Q. C., Enquist L., Shenk T. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77:10594–10605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stinski M., Meier J. 2007. Immediate-early CMV gene regulation and function, p. 241–263In Arvin A., Campadelli-Fiume G., Mocarski E., Moore P. S., Roizman B., Whitley R., Yamanishi K. (ed.), Human herpesviruses. Biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 48. Szilagyi J. F., Cunningham C. 1991. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J. Gen. Virol. 72:661–668 [DOI] [PubMed] [Google Scholar]
- 49. Tandon R., Mocarski E. S. 2008. Control of cytoplasmic maturation events by cytomegalovirus tegument protein pp150. J. Virol. 82:9433–9444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thomason L., et al. 2007. Recombineering: genetic engineering in bacteria using homologous recombination. Curr. Protoc. Mol. Biol. 78:1.16.1–1.16.24 [DOI] [PubMed] [Google Scholar]
- 51. Tooze J., Hollinshead M., Reis B., Radsak K., Kern H. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163–178 [PubMed] [Google Scholar]
- 52. Townsend L. B., Devivar R. V., Turk S. R., Nassiri M. R., Drach J. C. 1995. Design, synthesis, and antiviral activity of certain 2,5,6-trihalo-1-(beta-d-ribofuranosyl)benzimidazoles. J. Med. Chem. 38:4098–4105 [DOI] [PubMed] [Google Scholar]
- 53. Trus B. L., Gibson W., Cheng N., Steven A. C. 1999. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J. Virol. 73:2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Underwood M. R., et al. 1998. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J. Virol. 72:717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Upton J. W., Kaiser W. J., Mocarski E. S. 2010. Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe 7:302–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Varnum S. M., et al. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wolf D. G., Courcelle C. T., Prichard M. N., Mocarski E. S. 2001. Distinct and separate roles for herpesvirus-conserved UL97 kinase in cytomegalovirus DNA synthesis and encapsidation. Proc. Natl. Acad. Sci. U. S. A. 98:1895–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yu D., Silva M. C., Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. U. S. A. 100:12396–12401 [DOI] [PMC free article] [PubMed] [Google Scholar]