Abstract
The herpes simplex virus 1 (HSV-1) UL25 gene product is a minor capsid component that is required for encapsidation, but not cleavage, of replicated viral DNA. UL25 is located on the capsid surface in a proposed heterodimer with UL17, where five copies of the heterodimer are found at each of the capsid vertices. Previously, we demonstrated that amino acids 1 to 50 of UL25 are essential for its stable interaction with capsids. To further define the UL25 capsid binding domain, we generated recombinant viruses with either small truncations or amino acid substitutions in the UL25 N terminus. Studies of these mutants demonstrated that there are two important regions within the capsid binding domain. The first 27 amino acids are essential for capsid binding of UL25, while residues 26 to 39, which are highly conserved in the UL25 homologues of other alphaherpesviruses, were found to be critical for stable capsid binding. Cryo-electron microscopy reconstructions of capsids containing either a small tag on the N terminus of UL25 or the green fluorescent protein (GFP) fused between amino acids 50 and 51 of UL25 demonstrate that residues 1 to 27 of UL25 contact the hexon adjacent to the penton. A second region, most likely centered on amino acids 26 to 39, contacts the triplex that is one removed from the penton. Importantly, both of these UL25 capsid binding regions are essential for the stable packaging of full-length viral genomes.
INTRODUCTION
The herpes simplex virus 1 (HSV-1) virion consists of a 152-kbp double-stranded DNA (dsDNA) genome that is enclosed in an icosahedral capsid, which is itself surrounded by an amorphous protein layer called the tegument, and a lipid envelope containing viral glycoproteins. Viral DNA replication generates concatemers of head-to-tail genomes in the nucleus of the infected cell (26, 27). DNA replication is concurrent with the expression of HSV late genes, including the capsid structural proteins VP5 (UL19), VP19c (UL38), VP23 (UL18), and VP26 (UL35) (reviewed in reference 24). Using the scaffold proteins encoded by the HSV-1 UL26 and UL26.5 genes, the capsid structural proteins self-assemble into a spherical capsid precursor, or procapsid (19, 31, 36). Packaging of viral genomes into procapsids requires several tightly coupled events: (i) proteolysis and expulsion of the scaffold proteins, (ii) recognition and cleavage of viral genome termini by the terminase complex, (iii) docking of the terminase-DNA complex on the capsid portal, (iv) insertion of DNA into the procapsid, and (v) sealing of the portal. The product of DNA packaging is an angular capsid that contains tightly packed viral DNA (2, 10). The mature capsid, also known as a C capsid, then exits the nucleus and is incorporated into virions.
In the absence of successful packaging, two capsid forms accumulate in the nuclei of infected cells. B capsids contain remnants of the cleaved scaffold proteins VP21, VP22a, and VP24, and A capsids are empty (10). In the current model of HSV DNA packaging, B and A capsids form when DNA packaging is terminated prior to or after insertion of DNA, respectively. DNA packaging can be blocked by null mutations of the UL6, UL15, UL17, UL25, UL28, UL32, and UL33 genes (1, 7, 15–17, 22, 25, 30). For most of the packaging genes, the null mutation completely eliminates DNA cleavage and packaging such that the DNA concatemer is not cleaved and only B capsids are formed. However, deletion of the UL25 gene results in the formation of approximately equal numbers of A and B capsids and nearly unit-length genomes that are aberrantly cleaved at one (US) end of the genome (4, 17, 29). UL25 is also required for attachment of the tegument to the capsid through its interaction with the large tegument protein UL36 and for a very early stage in infection when the viral genome uncoats (6, 20). UL25 is present on all capsid forms in increasing amounts from procapsid to B, A, and C capsids and virions (18, 28). UL25 binds to the exterior surface of the capsid and interacts with the triplex protein, VP19c, and the major capsid protein, VP5 (21). It is proposed to interact with UL17 to form the C capsid-specific component (CCSC); five copies of the CCSC surround each of the capsid vertices, where they contact triplexes and hexons that surround the penton (8, 32, 37). Much like the decoration proteins of dsDNA phages, the UL25 protein when bound may reinforce capsids to prevent loss of DNA (17, 18). Additionally, it may adopt a conformation specific to the portal vertex to maintain the packaged DNA.
The UL25 gene product is a 580-amino-acid, 62-kDa protein that contains an unstructured N-terminal tail and a flexible core of packed alpha helices, with five flexible loops extending from this structure (3). UL25 protein attaches to the external vertices of HSV-1 capsids with a capsid binding domain located within its first 50 amino acids (4, 18, 37). Based on cryo-electron microscopy (cryo-EM) reconstructions of wild-type (wt) and UL25-deficient B capsids, predictions were made that the UL25 protein would be found to occupy the penton-distal region of the UL25/UL17 heterodimer (37). Recently, we used cryo-EM and image reconstruction to visualize green fluorescent protein (GFP)-tagged UL25 bound to capsids (8). The results from these studies confirmed the modeled location of the UL25 protein in the CCSC as the region that is distal to the penton, with the N terminus (amino acids 1 to 50) of UL25 making contact with the triplex one removed from the penton. To further define the UL25 capsid binding domain, we generated recombinant viruses with either small truncations or amino acid substitutions in the UL25 N terminus. Biochemical and structural studies of these mutants demonstrate that there are two important regions within the previously defined UL25 capsid binding domain that are required for the stable packaging of full-length viral genomes.
MATERIALS AND METHODS
Cell and viruses.
African green monkey kidney cells (Vero) and the UL25 complementary cell line 8-1 were cultured as described previously (17). The HSV wt KOS strain and the UL25 deletion mutants vΔUL25 and vΔ1-50 and the UL25-GFP virus were previously described (4). UL25 mutant viruses were generated by recombination of a KOS genome contained in a bacterial artificial chromosome (BAC) (4, 11, 33, 34). The primers listed in Table 1 were used to amplify template sequences from plasmids p-EP-Kan-S (34) and p-EP-TAP-in (a gift from P. Kinchington, University of Pittsburgh). Some primers contained embedded restriction sites. To confirm mutations in UL25 mutant viruses, viral DNA was amplified by PCR using primers (5′-GCAGAAGGTCTCCGGTAATATCACC–3′ and 5′-GGTGATCAGGTACAACAGGCGCTC-3′) that flanked the UL25 open reading frame (ORF). The 2.9-kb PCR product was extracted from agarose gels and used for restriction digestion and for sequencing with primers 5′-GCGGCGGCCGCAAGACTCATTTGGATCCG-3′ and 5′-TCTCCGCGACGGCCAGAC-3′.
Table 1.
Primers used for generation of recombinant HSV BACs
| BAC | UL25 Mutation | Primer sequencea (5′-3′) |
|---|---|---|
| bFH472 | Δ1-36 | tgtgttattttcgctctccgcctctcgcagacccccgagttcccccggTAGGGATAACAGGGTAATCGATTT |
| atccaaaagtcccgggggaactcgggggtctgcgagaggcggagagcGCCAGTGTTACAACCAATTAACC | ||
| bFH471 | Δ27-37 | gaacacaggcgcttcatagtcgccgattcccgaaacttctcgcccgtctttaacctccccTAGGGATAACAGGGTAATCGATTT |
| cgccgccgtctcccgggggaggttaaagacgggcgagaagtttcgggaatcggcGCCAGTGTTACAACCAATTAACC | ||
| bFH473 | Δ40-50 | gagttcccccgggacttttggatgtcgccccaggtggtcgtcctacagTAGGGATAACAGGGTAATCGATTT |
| tgtgcgctgggcctgtaggacgaccacctggggcgacatccaaaagtcGCCAGTGTTACAACCAATTAACC | ||
| bFH480 | F26A/I27A | aggcgcttcatagtcgccgattctagaaacgccgccaccTAGGGATAACAGGGTAATCGATTT |
| gtcccgggggaactcgggggtggcggcgtttctagaatcGCCAGTGTTACAACCAATTAACC | ||
| bFH481 | F35A/W36A | gaaacttcatcacccccgagttcccccgggacgcagcgatgtcgcccTAGGGATAACAGGGTAATCGATTT |
| aggttaaagacgggcgacatcgctgcgtcccgggggaactcgggggtgGCCAGTGTTACAACCAATTAACC | ||
| bFH482 | F26A/I27A/F35A/W36A | cttcatagtcgccgattcccggaatgccgccacccccgagttcccccgggaTAGGGATAACAGGGTAATCGATTT |
| aggttaaagacgggcgacatcgctgcgtcccgggggaactcgggggtgGCCAGTGTTACAACCAATTAACC | ||
| bFH471 | NTAP | cgaccgcagttctcgtgtgttattttcgctctccgcctctcgcagATGAAGCGACGATGGAAAAAG |
| gtgttcccagacgtccagagcgtcaaatgggcagtacgggtccatGCCCAGCTTGCAGCCGCCGGA |
Uppercase letters represent the template sequence (p-EP-Kan-S or p-EP-TAP-in), lowercase letters represent HSV UL25 gene sequences, underlined letters represent restriction sites, and letters in bold represent alanine substitution mutations.
Capsid purification.
Capsids were purified as described previously (4). Briefly, Vero cells (1.5 × 108) were infected overnight (18 h at 37°C) at a multiplicity of infection (MOI) of 5 PFU/cell. Infected cells were harvested by scraping, rinsed with phosphate-buffered saline (PBS), resuspended in 20 mM Tris (pH 7.5) plus protease inhibitors (Roche), adjusted to 1% Triton X-100, and incubated for 30 min on ice. The resulting nuclei were harvested by low-speed centrifugation, resuspended in 10 ml of TNE buffer (500 mM NaCl, 10 mM Tris, 1 mM EDTA [pH 7.5]), and then sonicated to lyse the nuclei. The nuclear lysate was then cleared by low-speed centrifugation, and the resulting supernatant was layered on top of 20 to 50% sucrose (in TNE buffer) gradients (SW41 rotor at 24,000 rpm for 1 h). The positions of A, B, and C capsids were observed as light-scattering bands, with A capsids being found highest (least dense) on the gradients and C capsids being found lowest (dense fractions) on the gradients. The different capsid fractions could also be identified by the presence or absence of the scaffold protein, VP22a, since only B capsids contain the scaffold protein. The fractions were collected using a Beckman fraction recovery system apparatus (catalog number 34890; Beckman). The apparatus has a mechanism to puncture the bottoms of the tubes, and fractions are collected from bottom to top. Fractions (0.75 ml) were collected, and protein was precipitated by adding an equal volume of 16% trichloroacetic acid. Pellets were resuspended in 35 μl of 2× PAGE loading buffer (Invitrogen) supplemented with 0.4 M Tris base. Gradient fractions were subjected to 4 to 12% SDS-PAGE, and the gels were either stained with imperial blue (Pierce) to visualize capsid proteins or analyzed by immunoblotting.
Western blotting.
Protein samples were separated on a 4 to 12% SDS-polyacrylamide gel, and proteins were transferred to nitrocellulose. The nitrocellulose was washed twice in Tris-buffered saline (TBS) and incubated overnight in Rockland Near Infra Red blocking buffer (catalog number MB-070-003; Rockland Immunochemicals, Inc., Gilbertsville, PA). UL25 monoclonal antibody 25E10 (8), VP5 rabbit polyclonal antibody NC1 (5), and UL17 chicken polyclonal antibody (39) were diluted 1:5,000, 1:5,000, and 1:25,000, respectively. The diluted antibodies were reacted with the blocked nitrocellulose for 2 h at room temperature, washed five times in TBS with 0.5% Tween 20, and incubated with IRDye 800-conjugated secondary antibodies (goat anti-mouse [UL25], goat anti-rabbit [VP5], or donkey anti-chicken [UL17]; Rockland Immunochemicals) diluted 1:15,000 in Rockland Near Infra Red blocking buffer with 0.1% Tween 20. The blots were washed and scanned using an Odyssey system (Li-Cor, Lincoln NE).
Southern blotting.
Vero cells (T-175 flask) were infected with KOS or UL25 mutant viruses at an MOI of 5 PFU per cell. At 18 h postinfection, the medium was removed and the cells were washed in 1× PBS, scraped off the plate, and pelleted. The cells were lysed, and total cell DNA and DNase-resistant DNA samples were prepared as previously described (13, 29). The DNA was digested with BamHI to look for cleavage of replicated viral DNA. The digested DNA was separated by agarose gel electrophoresis, transferred to a nylon membrane, and hybridized as previously described (13). Southern blots were scanned with a Storm 840 PhosphorImager, and specific bands were quantified with ImageQuant software.
Electron microscopy of thin sections.
Vero cells (2 × 106) were infected overnight (18 h at 37°C) at an MOI of 5 PFU/cell. Cells were rinsed in 1× PBS, fixed in 2.5% glutaraldehyde for 1 h at room temperature (RT), rinsed several times with 1× PBS to remove the fixative, and harvested by scraping. The samples were stained (1% osmium tetroxide [OsO4]–1% potassium ferricyanide), sectioned, and mounted onto carbon grids for transmission electron microscopy by the Center of Biologic Imaging, University of Pittsburgh. Images were collected on an FEI T12 electron microscope (FEI, Hillsboro, OR) equipped with a Gatan UltraScan 1000 CCD camera (Gatan, Pleasanton, CA).
Cryo-EM and image reconstruction of purified capsids.
C capsids isolated from vNTAP-UL25- and vF35A/W36A-infected Vero cells and A and B capsids isolated from vF26A/I27A/F35A/W36A-infected Vero cells were prepared for imaging by cryo-EM by placing 3.5-μl aliquots of each purified capsid sample onto holey carbon EM grids and plunging them freezing into liquid ethane-propane (50:50 mix) (35) using an FEI Vitrobot Mark III instrument (85% humidity; 4.5 to 8 s of blotting). Images were collected on an FEI Tecnai TF20 microscope at magnifications of ×50,000 and ×29,000 (×30,000 calibrated) for the tagged and mutant capsids, respectively, on Kodak SO-163 film (Kodak, Rochester, NY). Micrographs were digitized on a Nikon Super CoolScan 9000 scanner (Nikon, Tokyo) at a sampling rate of 6.35 μm/pixel corresponding to 1.27 Å/pixel (×50,000) and 2.12 Å/pixel (×29,000). Particles were selected using ×3dpreprocess software, and defocus/astigmatism values were estimated using Bsoft (9, 12). An icosahedrally averaged map of each sample was calculated using AUTO3DEM (41) by refining an initial model generated from the images using the random model procedure (40, 41). The refined maps were calculated from 753, 1,760, and 331 particles to resolutions of ∼25, ∼23, and ∼28 Å for the mutant A, B and C capsids, respectively. The vNTAP-UL25 C capsid map was calculated from 4,865 particles to a resolution of ∼15 Å. Resolution values were taken at a Fourier shell correlation (FSC) of 0.5. USCF Chimera was used to visualize density maps (23). Calculation of occupancy of the CCSC-dense areas on capsids was done as previously described (8).
RESULTS
Characterization of viruses expressing UL25 with small deletions and amino acid substitutions in the N-terminal capsid binding region.
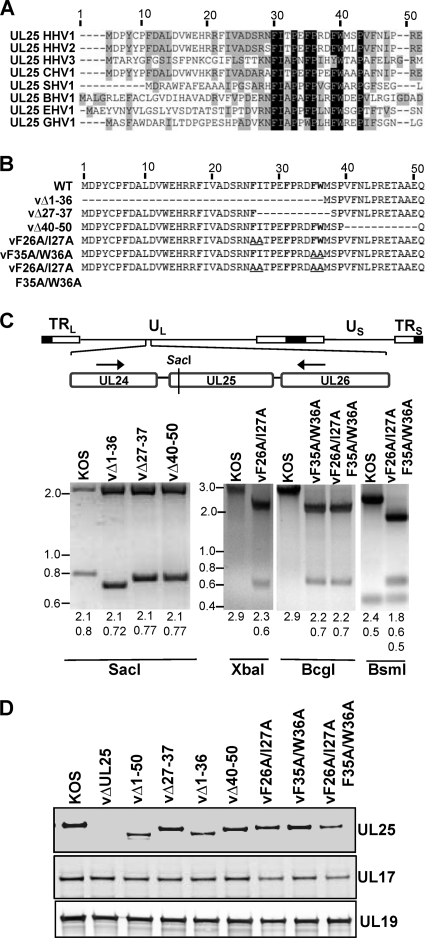
Previously, we reported that amino acids 1 to 50 of the UL25 protein are necessary and sufficient for binding of UL25 to capsids and are essential for successful DNA packaging (4). Sequence alignment of UL25 homologues from several alphaherpesviruses demonstrated that a number of residues within this N-terminal capsid binding domain are highly conserved, particularly amino acids 26 to 39 of HSV-1 (Fig. 1A) . In order to further define the capsid binding region and the role of the conserved residues in UL25 function, we generated recombinant viruses that contained defined deletions or amino acid substitutions within the first 50 amino acids of UL25. The UL25 mutations were introduced into the viral genome through the genetic manipulation of an HSV-1 (KOS) genome maintained in a BAC, as previously described (4, 11, 34). The deletions removed the coding sequences for amino acids 1 to 36, 27 to 37, and 40 to 50 of the UL25 ORF (Fig. 1B). Point mutations of the most conserved residues between amino acids 26 to 39 (F26, I27, F35, and W36) were also made in pairs (F26A and I27A or F35A and W36A) or as a quadruple mutant (F26A I27A F35A W36A).
Fig. 1.
Generation of HSV-1 mutant viruses containing small deletions or point mutations in the UL25 N terminus. (A) Alignment of UL25 amino acids 1 to 50 from eight alphaherpesviruses, including human herpesviruses 1, 2, and 3 (HHV-1,-2, and -3); chimpanzee herpesvirus 1 (CHV-1); suid herpesvirus 1 (SHV-1); bovine herpesvirus 1 (BHV-1); equine herpesvirus 1 (EHV-1); and gallid herpesvirus 1 (GHV-1). NCBI accession numbers for the above viruses are as follows: for HHV-1, ABI63487; for HHV-2, CAB06749; for HHV-3, ABE03053; for CHV-1, BAE47049; for SHV-1, CAA65011; for BHV-1, CAB01601; for EHV-1, YP_053082; and for GHV-1, BAA36558. The alignment was generated with VectorNTI software (Invitrogen). Conserved residues are highlighted in gray; identical residues are highlighted in black. The highly conserved region comprises HSV-1 amino acids F26 to P39. (B) Diagram of the HSV-1 UL25 N-terminal 50 amino acid acids with the deletion and amino acid substitution mutations that were introduced into the viral genome through the genetic manipulation of an HSV-1 (KOS) genome maintained in a recombinant BAC. Conserved residues are in bold. Deletions are represented by dashes, and alanine substitutions are underlined. (C) Confirmation of UL25 mutations in viral DNA. (Top) Diagram of the HSV-1 genome showing the 2.9-kb region amplified from viral DNA by PCR. Black arrows indicate the locations of the forward and reverse primers used for the PCR. The SacI restriction site is shown. (Bottom) PCR products were digested with the restriction enzyme SacI, XbaI, BcgI, or BsmI. SacI cleaved near the coding sequence for the UL25 N terminus, allowing visualization of small (∼30-bp) deletions within the UL25 gene. The other enzymes cleaved unique restriction sites embedded in the primers used for generating the BAC clones (Table 1). Digested PCR products were run on 2.2% agarose gel and stained with ethidium bromide. The expected sizes of the restriction fragments are listed below the gels. Molecular mass markers in kbp are shown at left. (D) Western blots of Vero cells infected with HSV-1 wt KOS or UL25 mutants were probed with monoclonal antibody 25E10 to detect UL25, a chicken polyclonal antibody to detect UL17, and a rabbit polyclonal antibody (NC1) to detect UL19.
The BACs were transfected onto UL25-complementing 8-1 cells. The recovered viruses were plaque purified, and virus stocks were prepared, on Vero or 8-1 cells. Each mutant was titrated on Vero and 8-1 cells to determine the effects of the UL25 mutations on viral replication (Table 2). KOS and the vΔ40-50 mutant showed comparable growth on Vero and 8-1 cells, while the vΔ1-36, vΔ27-37, vF26A/I27A, and vF26A/I27A/F35A/W36A mutants failed to form plaques on Vero cells. The vF35A/W36A mutant displayed an intermediate phenotype, with a 5-fold reduction in plaque formation on Vero cells compared to that of the UL25-complementing cell line with the vF35A/W36A virus, and produced very small plaques on Vero cells.
Table 2.
Virus growth assay
| Virus | Efficiency (PFU/ml) of virus plating in: |
|
|---|---|---|
| Vero cells | 8-1 cells | |
| KOS | 2 × 108 | 2 × 108 |
| vΔUL25 | <1 × 103 | 2 × 107 |
| vΔ1-36 | <1 × 103 | 5.9 × 108 |
| vΔ27-37 | <1 × 103 | 3.8 × 108 |
| vΔ40-50 | 1.9 × 108 | 2.5 × 108 |
| vF26A/I27A | <1 × 103 | 7.5 × 108 |
| vF35A/W36A | 3 × 107 | 1.5 × 108 |
| vF26A/I27A/F35A/W36A | <1 × 103 | 7.1 × 107 |
| vNTAP-UL25 | 3.4 × 107 | 3.2 × 107 |
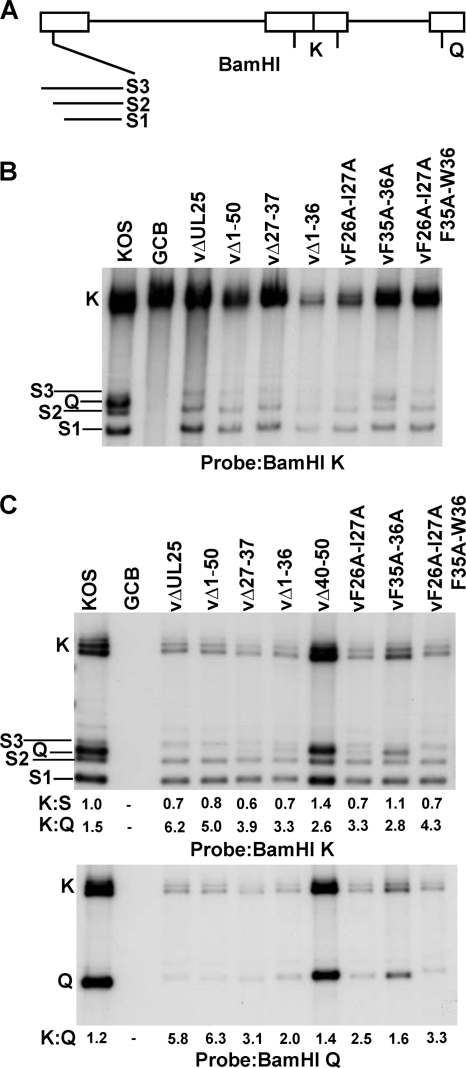
To verify the presence of the mutations in the progeny virus, viral DNA was prepared from Vero cells infected with each mutant and used in a PCR with primers flanking the UL25 ORF. The resulting 2.9-kbp PCR fragment isolated from the three deletion mutants was digested with SacI, which cleaves within a region of the UL25 ORF near its 5′ end (Fig. 1C). SacI digestion of the wt KOS PCR fragment resulted in DNA fragments of 2.1 and 0.8 kbp. The 0.8-kbp fragment was shifted to a lower molecular weight by the small deletions present in the vΔ1-36, vΔ27-37, and vΔ40-50 mutants. As a means to verify the UL25 point mutations present in the vF26A/I27A, vF35A/W36A, and vF26A/I27A/F35A/W36A viruses, we digested the 2.9-kbp PCR fragment generated from these mutants with either XbaI, BcgI, or BsmI. Sites for these restriction enzymes were included in the primers used to generate the recombinant BACs containing the F26A/I27A (XbaI), F35A/W36A (BcgI), and F26A/I27A/F35A/W36A (BsmI) mutations (Table 1 and Fig. 1C). Restriction enzyme digestion demonstrated that these restriction sites were present, resulting in appropriately sized DNA fragments for each mutant. Finally, the 2.9-kbp PCR product from each virus was sequenced to confirm that the UL25 mutants had the desired deletion or point mutations.
To determine that each mutant virus expressed a UL25 protein of the expected size, we performed immunoblotting of Vero cells infected with each mutant virus as well as KOS. Fig. 1D shows that all of the mutants produced a UL25 protein of the expected sizes. The three substitution mutants (vF26A/I27A, vF35A/W36A, and vF26A/I27A/F35A/W36A) expressed a UL25 protein that was identical in size to the wt KOS UL25 protein. The deletion mutants (vΔ1-36, vΔ27-37, and vΔ40-50) produced UL25 proteins that were slightly smaller than that of the wt due to the deletion of amino acid residues from the N-terminal region of UL25. No immunoreactive UL25 proteins were observed for the UL25 null virus, vΔUL25. Using the HSV-1 UL19 protein as a loading control, we found that all of the mutants express similar amounts of both the UL25 and UL17 proteins (Fig. 1D).
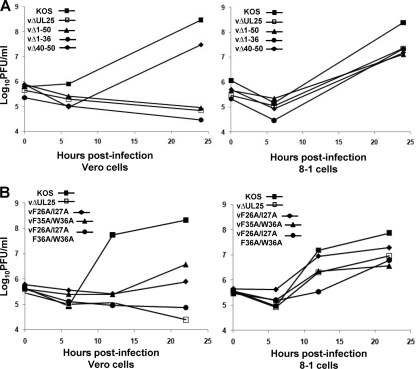
Intracellular replication of the UL25 mutants was examined by establishing single-step growth curves for each mutant in Vero and 8-1 cells. Cells were infected at an MOI of 3 with each virus, and cells were harvested at 0, 6, and 24 h postinfection (hpi) for the three deletion mutants (Fig. 2 A) or at 0, 6, 12, and 22 hpi for the three point mutants. Cells were assayed for infectious virus by a plaque assay on 8-1 cells (Fig. 2B). The Δ40-50 deletion virus produced similar amounts of virus on both Vero and 8-1 cells, while the other deletion mutants, vΔUL25, vΔ1-50, and vΔ1-36, produced virus on 8-1 cells but not on Vero cells. The three point mutants exhibited growth defects on Vero cells, with the two double mutants (vF26A/I27A and vF35A/W36A) producing some virus (titers 2- to 3-logs lower than that for KOS), while the quadruple mutant failed to replicate on Vero cells. When grown on 8-1 cells, the three point mutants all produced virus similar to what was found with the UL25 null virus, vΔUL25. Taken together, these results further defined the critical residues within the UL25 N terminus as amino acids 1 to 37 and demonstrated that deletion and substitution mutations of a conserved portion of this domain (amino acids 26 to 37) drastically impaired virus replication.
Fig. 2.
Single-step growth of KOS and UL25 mutants. Vero or 8-1 cells were infected with wt KOS or UL25 mutant viruses at an MOI of 3 at 4°C for 1 h and incubated at 37°C to the indicated time point. Cells were lysed by multiple freeze-thaws, and viral progeny were quantified by plaque assay on 8-1 cells. The results shown are averages of duplicate experiments. (A) UL25 deletion mutants. (B) UL25 double and quadruple point mutants.
Association of UL25 with purified capsids.
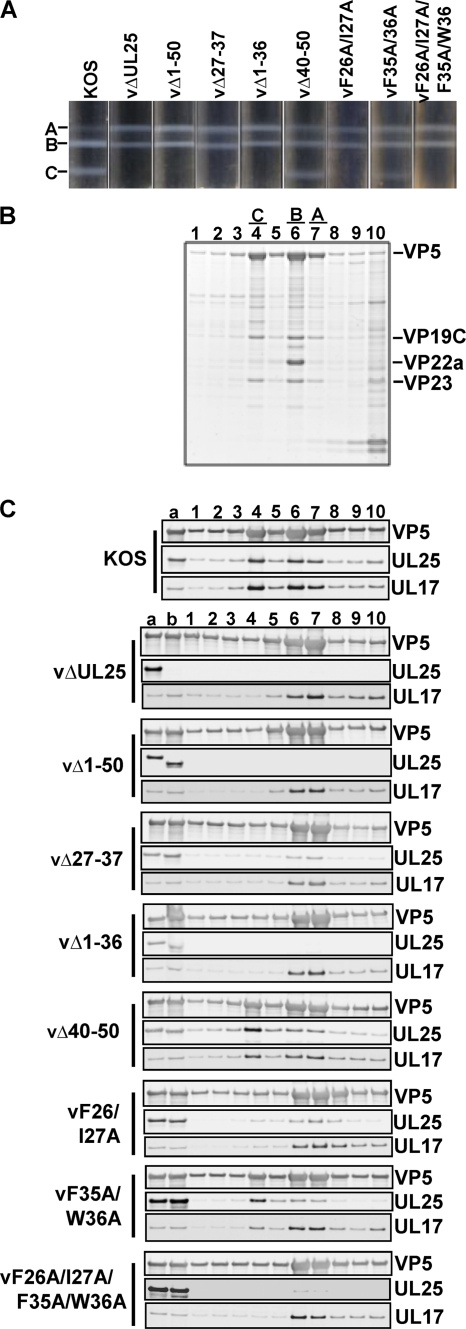
Previously, we showed that amino acids 1 to 50 of the UL25 protein are required for binding of UL25 to capsids and for DNA packaging (4). To determine what effect our new UL25 mutations had on capsid binding, we purified intranuclear capsids from Vero cells infected with wt KOS or UL25 mutant viruses. The nuclear lysates were subjected to sucrose gradient centrifugation, and capsids were detected as visible light-scattering bands (Fig. 3 A). In cells infected with wt KOS or the two UL25 mutants, vΔ40-50 and vF35A/W36A, which replicated on Vero cells, all three capsid forms were observed. The KOS gradients contained nearly equal amounts of both B and C capsids and only minor amounts of A capsids (Fig. 3A). In contrast, the gradients for the vΔ40-50 and vF35A/W36A mutants contained fewer C capsids and more A and B capsids. The lysates of the UL25 mutants that did not replicate or replicated poorly on Vero cells (vΔ1-50, vΔ1-36, vΔ27-37, vF26A/I27A, and vF26A/I27A/F35A/W36A) contained approximately equal numbers of A and B capsids but no DNA-containing C capsids. The high proportion of A capsids found along with the absence of C capsids in these UL25 mutants is identical to what was observed with the UL25 null mutant vΔUL25 (Fig. 3A).
Fig. 3.
Analysis of capsid-bound UL25. Rate-velocity sedimentation of capsids from cells infected with KOS or the indicated UL25 mutant is shown. (A) Vero cells infected with the indicated viruses at an MOI of 5 PFU per cell were harvested at 18 h postinfection, and nuclear lysates were layered onto 20 to 60% sucrose gradients and centrifuged at 24,000 rpm (SW41 rotor) for 1 h. The positions of A, B, and C capsid bands are indicated. (B) The gradient fractions from wt KOS were separated by SDS-PAGE, and the proteins were detected by Coomassie blue staining. Lane 1 is the bottom of the gradient, and lane 10 is the top. Gradient fractions that contained A, B, and C capsids are indicated. The positions of the capsid proteins (major capsid protein VP5, triplex proteins VP19C and VP23, and scaffold protein VP22a) are indicated. (C) The gradient fractions for KOS and the UL25 mutants were analyzed by SDS-PAGE, followed by immunoblotting for VP5, UL25, and UL17. Lane a, lysate from KOS-infected cells; lane b, lysate from UL25 mutant-infected cells.
To determine whether UL25 protein was present in these capsids, the sucrose gradients for KOS and the UL25 mutants were separated into 10 fractions, and the protein composition of each fraction was determined by SDS-PAGE. The stained gel of the KOS gradient is shown in Fig. 3B, and the fractions containing A (fraction 7), B (fraction 6), and C (fraction 4) capsids are indicated. The presence of capsids in these fractions was also confirmed by immunoblotting with an antibody to the major capsid protein, VP5, demonstrating that fractions 4, 6, and 7 contain elevated levels of VP5 (Fig. 3C). VP5 immunoblots of the sucrose gradient fractions for the seven UL25 mutants showed a staining pattern similar to that of KOS, with A, B, and C capsids present in fractions 7, 6, and 4, respectively (Fig. 3C). Samples were then analyzed for the presence of UL25 by immunoblotting (Fig. 3C). The KOS gradient showed that the UL25 protein was found in the fractions that contained A, B, and C capsids. A similar result was found with capsids isolated from cells infected with the vΔ40-50, vF35A/W36A, and vF26A/I27A UL25 mutants, the only three of the seven UL25 mutants that were found to produce virus on Vero cells in the single-step growth curves (Fig. 2). Immunoblotting of the gradients for the four UL25 mutants that did not replicate on Vero cells demonstrated that reduced amounts of UL25 proteins expressed by the vΔ27-37 mutant were found to associate with A and B capsids. In contrast, UL25 was not detected in any of the gradient fractions for the Δ1-50 and Δ1-36 mutants, and only a small amount of UL25 protein was found on A and B capsids of the vF26A/I27A/F35A/W36A mutant. These data demonstrate that there are two critical regions within the putative capsid binding domain (amino acids 1 to 50) of UL25. These regions include amino acids 1 to 26, which are essential for UL25 capsid binding, and a second conserved set of amino acids located between amino acids 26 and 37 that are not required for direct binding of UL25 to capsids but are necessary for optimal association with capsids.
Association of UL17 with purified capsids.
The UL25 protein forms part of an elongated molecule on the surfaces of capsids referred to as the C capsid-specific component (CCSC). The CCSC is proposed to be a heterodimer of UL25 and UL17, with five copies surrounding each of the capsid vertices. To determine if UL17 capsid association was altered with any of the UL25 mutants, the gradient fractions for each UL25 mutant were also assayed for the presence of UL17 by immunoblotting (Fig. 3C). The results show that the interaction of UL17 with capsids was not dependent on the presence of UL25, although the capsids appear to have reduced amounts of UL17 (compared to those of wt KOS) in the absence of UL25. This result was consistent with those previously described (32).
Deletion of the UL25 N terminus does not inhibit UL25 nuclear localization.
The virus-expressed UL25 protein with a deletion of amino acids 1 to 50 is present in infected-cell lysates but is not associated with capsids purified from infected-cell nuclei (4). To determine if the truncated protein was sequestered in the cytoplasm, thus preventing it from contacting intranuclear capsids, we used indirect immunofluorescence assays to track UL25 in infected cells. Vero cells were mock infected or infected with the KOS, vΔUL25, or vΔ1-50 viruses and fixed at 16 h postinfection. Cells were stained with the UL25 monoclonal antibody, 25E10, followed by staining with a Cy3-conjugated secondary antibody, and the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). The Δ1-50 UL25 protein was predominantly located in cell nuclei, and similar to the wt UL25 protein, the Δ1-50 UL25 protein was found to locate in discrete compartments within the nucleus (data not shown). These compartments are similar to assemblons or replication compartments that have been previously described and are implicated as the sites where viral DNA replication and DNA packaging take place (38). These results demonstrate that the Δ1-50 UL25 protein is stably expressed and trafficked to the nucleus and indicate that the observed defects in virus replication and DNA packaging of the UL25 mutants are due to the inability of the altered UL25 proteins to stably bind capsids.
Detection of the CCSC on the surfaces of A and B capsids isolated from the vF26A/I27A/F35A/W36A mutant and on C capsids from the vF35A/W36A mutant.
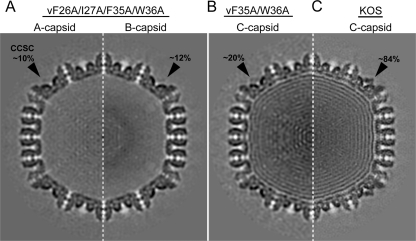
Capsids were isolated from the nuclear fraction of Vero cells infected with either the vF35A/W36A or the vF26A/I27A/F35A/W36A mutant. Cryo-EM and image processing were used to generate three-dimensional (3D) reconstructions of C capsids isolated from the former cells and A and B capsids from the latter cells (Fig. 4). The reconstructions show the presence of the CCSC (which contains UL25, putatively in complex with UL17) on all of the capsids (Fig. 4, black arrowheads), although the occupancy of the CCSC is dramatically reduced in the mutants. For the vF35A/W36A mutant, the C capsid reconstruction shows ∼20% occupancy for the UL25/UL17 complex, compared to ∼84% seen on C capsids from wt virus (Fig. 4B and C). C capsids of the quadruple mutant vF26A/I27A/F35A/W36A, although present in nuclei of infected cells as seen in thin sections (Fig. 5), could not be purified in sufficient numbers for cryo-EM, suggesting the genome is not stably incorporated. The A and B capsid reconstructions show an even lower occupancy of ∼10% for the CCSC (Fig. 4A). These results confirm that mutations in the two capsid binding regions identified here are indeed vital for capsid incorporation of UL25.
Fig. 4.
CCSC occupancy in UL25 mutant capsids. Cross-sections through density maps of the quadruple mutant A capsid (A, left half) and B capsid (A, right half), the double mutant C capsid (B), and wt virus C capsid (C). The CCSC-dense area in each map is indicated with a black arrow along with the estimated level of occupancy.
Fig. 5.
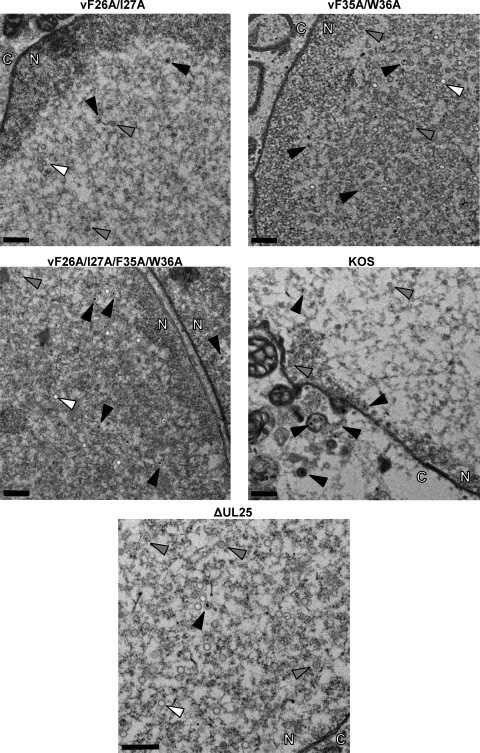
Transmission electron micrographs of thin-section preparations of virus-infected cells. Vero cells were infected with the indicated viruses at an MOI of 5, and at 18 hpi the cells were fixed and sectioned for imaging. The arrows point to A capsids (white arrows), B capsids (gray arrows), and C capsids (black arrows). The nucleus and cytoplasm are marked with N and C, respectively. Magnification of top four panels, ×4,400; magnification of bottom panel, ×6.500. Scale bars, 0.5 μm.
Transmission electron microscopy of virus-infected cells.
The previous data showed that manipulation of the UL25 N terminus did not cause mislocalization of the protein and that point mutations in the N terminus did not prevent capsid attachment. To examine whether or not the virus growth defects associated with these mutations occurred at a later stage of virion maturation, we examined thin sections of virus-infected Vero cells by transmission EM to determine the presence and locations of A, B, and C capsids. In wt KOS-infected cells, scaffold-containing B capsids and DNA-containing C capsids were visible in the nuclei (Fig. 5). C capsids were also observed as unenveloped particles in the cytoplasm, in enveloped inclusions in the cytoplasm, and in virions in the extracellular space. As previously described (4, 20), the UL25 deletion mutant, vΔUL25, produced predominately A and B capsids, but a few C capsids were also observed, and all three capsid forms were restricted to the nucleus (Fig. 5). The locations of capsids in Vero cells infected with viruses expressing the double (vF26A/I27A and vF35A/W36A) or quadruple (vF26A/I27A/vF35A/W36A) UL25 mutants closely resembled that in vΔUL25-infected cells, with the exception that there appeared to be more C capsids in the nuclei of cells infected with these mutants (Fig. 5). Although the Western blotting (Fig. 3) and cryo-EM (Fig. 4) studies show that the three substitution mutants all contain some capsid-bound UL25, the capsids made by these mutants were found exclusively in the nucleus. Previous studies with HSV and pseudorabies virus (PRV) UL25 mutants demonstrated that capsid-bound UL25 is a prerequisite for egress of mature capsids from the nucleus, and there appears to be a threshold level of UL25 per capsid that is required to trigger the nuclear-egress event (14, 20). Therefore, a block to capsid nuclear egress with the double (vF26A/I27A and vF35A/W36A) or quadruple (vF26A/I27A/vF35A/W36A) mutants may simply result from lower levels of capsid-bound UL25.
Analysis of genome cleavage in UL25 mutants.
To determine whether the viruses bearing the N-terminal UL25 mutations were impaired in DNA cleavage and packaging, we isolated total cellular DNA and DNase-resistant DNAs (packaged viral DNA) from Vero cells infected with wt and mutant viruses and subjected them to Southern blotting. Viral DNA replication generates concatemers that are cleaved into unit-length molecules and packaged into virions. The cleaved viral DNA contains free chromosomal termini, and the presence of chromosomal ends can be monitored by Southern blotting of total infected cell DNA digested with BamHI and probed with the HSV-1 BamHI K fragment. Only cleaved viral DNA with free chromosomal ends will give rise to the terminal BamHI S and Q fragments, while concatemeric DNA gives rise to only the joint-spanning BamHI K fragment (Fig. 6 A). The S fragment represents the UL terminus, and the Q fragment represents the US terminus (Fig. 6A). In nonpermissive cells infected with the UL25 null mutant, vΔUL25, the concatermeric DNA is cleaved at the L end and packaging is initiated but is not completed (4, 20). As a consequence, when total cellular DNA was digested with BamHI and the resulting Southern blots were probed with the BamHI K fragment, abundant S terminal fragments were detected but no Q terminal fragments were found (Fig. 6B).
Fig. 6.
Processing of virus DNA. (A) Schematic diagram of the HSV-1 genome showing the locations of the BamHI K, Q, and S fragments. The different sizes of the BamHI S fragment are due to the presence of one to multiple copies of the a sequence at the UL terminus. Vero cells were infected with the indicated virus at an MOI of 5 PFU per cell. At 18 h postinfection, cells were harvested and total (B) and DNase-resistant (C) DNA samples were prepared. The DNA was isolated, digested with BamHI, and subjected to Southern blotting. The blots were probed with 32P-labeled BamHI K or BamHI Q fragments of the HSV-1 genome. The ratio of the radioactivity of the BamHI joint K fragment to that of the US-end Q or UL-end S fragments was determined by quantification of the hybridizing bands with phosphorimaging software.
The Q- and S-terminal fragments were detected in Vero cells infected with the wild type or with the two UL25 mutants, vΔ40-50 (data not shown) and vF35A/W36A, that replicate on Vero cells. The different sizes of the BamHI S and K fragments are due to the presence of one to multiple copies of the a sequences, while the Q fragment contains only a single copy of the a sequence. An example of a mutant that fails to cleave replicated viral DNA is the null mutant GCB, which does not express the product of the HSV UL28 gene (30). As shown in Fig. 6B, the terminal Q and S fragments were absent from DNA isolated from Vero cells infected with GCB. All of the UL25 mutants replicated viral DNA when grown on Vero cells, as assessed by the presence of the joint K fragments, but total DNA isolated from Vero cells infected with replication-defective UL25 mutants lacked the S-terminal fragment Q (Fig. 6B). In contrast, when the DNase-resistant DNA prepared from these mutants was probed with the BamHI K fragment, low levels of the Q fragment were detected with the vΔ1-50, vΔ1-36, vΔ27-37, and vF26A/I27A mutants (Fig, 6C). When the same blot was probed with the BamHI Q fragment, the terminal S fragment was found to be present with all of the UL25 mutants, including the UL25 null mutant, vΔUL25 (Fig. 6C). The BamHI Q fragment for the vΔUL25 and vF26A/I27A/F35A/W36A mutants appears to migrate with the BamHI S3 fragment, which would account for the absence of a detectable Q fragment from these mutants when they were probed with the BamHI K fragment (Fig. 6B and C). These data indicate that full-length viral DNA was stably packaged with all of the UL25 mutants that fail to replicate on Vero cells but at greatly reduced levels compared to those found with wt HSV-1 and with the UL25 mutants (vΔ40-50 and F35A/W36A) that replicate on Vero cells. The BamHI K fragment from the GCB mutant was detected in the total DNA samples (Fig. 6B) but not in the DNase-resistant DNA samples (Fig. 6C), demonstrating that the low levels of DNase-resistant DNA detected with all of the UL25 mutants was not a result of incomplete nuclease digestion or the result of residual viral DNA from input viral genomes.
To directly examine the levels of the L- and S-terminal fragments present in the DNase-resistant samples, we measured the amounts of radioactivity in the two terminal bands and in the joint fragment K. The ratio of the amount of radioactivity in the joint to that in each terminal fragment was then calculated. In the case of the BamHI S fragments from the UL terminus, the measurement included the S1, S2, and S3 fragments. The ratio of the amount of DNase-resistant (packaged) DNA in the joint fragment to that in either the L or S terminus should be 1 if full-length viral genomes are packaged, and this was found to be the case for wt KOS (Fig. 6C). For all of the UL25 mutant viruses, the L terminus (K:S ratio) was present at levels similar to that in the wt virus (Fig. 6C). In contrast, the amount of the S terminus (K:Q ratio) was significantly reduced in all of the UL25 mutants (vΔUL25, vΔ1-36, vΔ27-37, vF26A/I27A, and vF26A/I27A/F35A/W36A) that failed to replicate on Vero cells (Fig. 6B and C). These data demonstrate that viral DNA is stably packaged in the absence of a functional UL25 protein but at much lower levels than in wt HSV-1. More importantly, it is apparent from the high K:Q ratio that the majority of the DNA that is packaged with the nonfunctional UL25 mutants is truncated at the S-terminal end of the viral genome (Fig. 6C). These results are consistent with our previous studies (4) and demonstrate that mutations that alter capsid binding of UL25 interfere with cleavage of the viral genomes at the Us terminus but do not alter the cleavage reaction at the UL terminus.
Affinity tagging the N terminus of UL25.
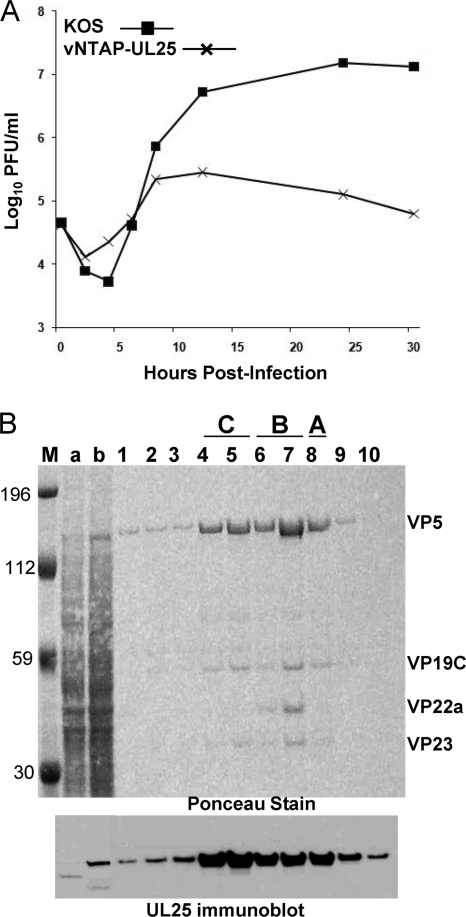
Previously, we described studies with a virus expressing a GFP-UL25 fusion protein to locate the UL25 protein on the surfaces of reconstructed capsids (8). The coding sequences for GFP were inserted between UL25 codons 50 and 51, and cryo-EM reconstructions of capsids containing the UL25-GFP protein clearly demonstrated that UL25 resided within the penton-distal region of the CCSC. To further define how the UL25 N terminus (amino acids 1 to 50) interacts with the capsid, we generated a virus, vNTAP-UL25, with a tandem affinity purification tag (TAP tag) fused to the N terminus of UL25. The TAP tag that we used for these studies consisted of a streptavidin binding peptide and a calmodulin binding peptide (InterPlay; Stratagene). To determine if addition of the TAP tag interfered with virus replication, we titrated virus stocks on Vero and 8-1 cells and calculated a single-step growth curve for vNTAP-UL25 (Table 2 and Fig. 7 A). While similar values were obtained from vNTAP-UL25 virus stock titration on Vero and 8-1 cells, the data from the growth curves were markedly different. Replication of vNTAP-UL25 was diminished compared to that of wt KOS beginning at 12 h postinfection, leading to a 2-log reduction in virus yield at 30 h postinfection. vNTAP-UL25 developed very small plaques on Vero cells that were on average 45% of the size of KOS plaques.
Fig. 7.
vNTAP-UL25. (A) Single-step growth curve. Vero cells were infected with KOS or vNTAP-UL25 at an MOI of 3 at 4°C on ice for 1 h and incubated at 37°C to the indicated time point. Cells were lysed by freeze fracture, and viral progeny in the cell lysates were quantified by titration and plaque assay on Vero cells. The results shown are averages of duplicate experiments. (B) vNTAP-UL25 protein is capsid associated. vNTAP-UL25 capsids were separated on sucrose density gradients, and the gradient fractions were analyzed by SDS-PAGE followed by immunoblotting for UL25. The blot was Ponceau stained, and the gradient fractions that contained A, B, and C capsids are indicated. The positions of the capsid proteins (major capsid protein, VP5; triplex proteins, VP19C and VP23; and scaffold protein, VP22a) are indicated. Lane 1 is the bottom of the gradient, and lane 10 is the top. Lane a, cell lysate from KOS-infected cells; lane b, cell lysate from vNTAP-UL25-infected cells. Size markers (in kilodaltons; lane M) are given.
To verify that the UL25 protein expressed by vNTAP-UL25 contained the TAP fusion, capsids were prepared from Vero cells infected with vNTAP-UL25. The capsid preparations consisted mainly of C and B capsids, with a faint A capsid band on the sucrose gradient (data not shown). The gradients were fractionated and analyzed by Western blotting for UL25. The NTAP-UL25 protein contains an additional 78 amino acids, which corresponded to an 8- to 9-kDa shift in size on SDS-PAGE (Fig. 7B, lane b). The NTAP-UL25 protein was observed on all capsid types. Thus, while insertion of the TAP tag at the UL25 N terminus had negative effects on virus replication, it did not appear to alter the ability of the protein to attach to the capsid.
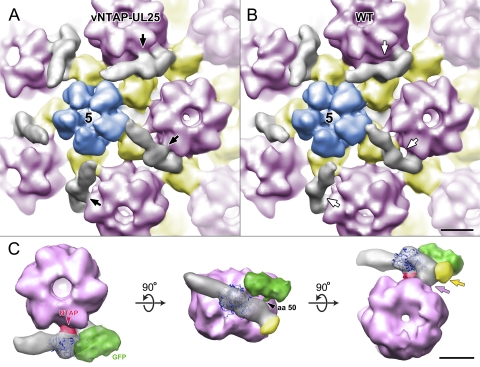
vNTAP-UL25 C capsids were analyzed by cryo-EM, and an icosahedrally averaged 3D reconstruction was generated. Figure 8 A shows a surface-rendered view of the vNTAP-UL25 C capsid map, and Fig. 8B shows a wt reconstruction. Both maps show the CCSC-dense area surrounding penton vertices (Fig. 8A and B, gray). The reconstruction of the vNTAP-UL25 C capsids shows ∼75% occupancy of the CCSC. The TAP-tagged UL25 CCSC, however, shows an additional small dense area midway along the CCSC, abutting the adjacent hexon (Fig. 8A, black arrows). No such dense area is discernible in the wt map, and we ascribe it to the N-terminal TAP tag. The region of the CCSC directly adjacent to the TAP tag (and more distal to the penton) contacts the adjacent hexon (Fig. 8C, pink arrow). This connection between the CCSC and hexon was also observed in the wt capsid. A second connection to the capsid is made at the base of the CCSC with the triplex that is once removed from the penton (Fig. 8C, yellow arrow) and is again mutual to both maps.
Fig. 8.
Location of the UL25 N terminus on the capsid surface. Comparison of the CCSC molecule on vNTAP-UL25 (A) and wt (B) C capsids. In this close-up view of the capsid vertices, the CCSC is gray. The penton is blue and labeled with a 5, as it represents the 5-fold axis of symmetry. Hexons are light pink and triplexes yellow. A black arrow indicates the position of density attributed to the TAP tag, which is present in the vNTAP-UL25 CCSC (A) but is missing (white arrows) from wt KOS CCSC (B). A region of the CCSC molecule to the right of the TAP tag makes contact with the hexon in both images. (C) Composite image of GFP- and TAP-labeled CCSC in contact with the hexon. (Left) Top view of the composite CCSC (gray), shown beside the hexon adjacent to the penton vertex. The GFP-dense area is green, and the TAP tag is dark pink. (Center) Rotation of the image shows the side view from the surface of the capsid with the triplex (yellow)-dense area that makes contact with UL25. (Right) View from underneath the hexon, looking out from the interior of the capsid. The region of UL25 between the TAP and GFP densities makes contact with the hexon (light pink) and the triplex (yellow). Atomic coordinates for residues 133 to 580 of UL25 (PDB ID, 2F5U; dark blue ribbons) have been docked within the cryo-EM-dense area.
The packed alpha-helical core of UL25 (residues 133 to 580) has been modeled to occupy the middle portion of the CCSC (9, 37) (Fig. 8C). Our earlier map of GFP-tagged UL25 revealed amino acid 50 to be located in the penton-distal portion of the CCSC, and the entire N-terminal 132 residues of UL25 are most likely also located in this region (9). As the TAP tag is fused to the N terminus of UL25, the region of the CCSC adjacent to the TAP tag (distal to the penton) must be occupied by the UL25 N terminus. Thus, we infer that the connection between the CCSC and the adjacent hexon is mediated by UL25 N-terminal amino acids 1 to 27.
In a composite image of the NTAP-UL25 CCSC, where our earlier reconstruction of GFP inserted at amino acid 50 of UL25 has been added, it can be seen that amino acid 50 of UL25 is in close proximity to the triplex once removed from the penton, and we infer that the second capsid binding domain of UL25, interacting with this triplex, is made via amino acids 26 to 39. This arrangement would place the CCSC's other occupant, putatively UL17, in the penton-proximal end of the CCSC molecule.
DISCUSSION
UL25 is a capsid-associated protein that is required for successful secondary cleavage of viral genomes from the replicated concatemer and for retention of packaged DNA inside the HSV-1 capsid. In this paper, we describe a mutational analysis of the capsid-binding domain of UL25, located within its N-terminal 50 amino acids. While deletion of amino acids 1 to 36 prevented UL25 capsid attachment, a deletion mutant missing amino acids 27 to 37 was capsid associated, indicating that the first 27 amino acids of UL25 are essential for its stable capsid binding (Fig. 3). In order to rule out the possibility that deletion of the N terminus interfered with the nuclear import of UL25, an indirect immunofluorescence assay was used to track the UL25 protein in infected cells. The UL25 protein expressed from the vΔ1-50 mutant was found to efficiently localize to the nucleus, demonstrating that this region of UL25 was not required for transport of the protein to the nucleus. Within the putative capsid binding domain, the most highly conserved residues in this region lie between amino acids 26 to 39. Deletion of these conserved amino acids (vΔ27-37) did not interfere with capsid binding (Fig. 3), but this mutant was unable to produce infectious virus (Table 2). Point mutations of four of the most highly conserved residues within this region generated mixed phenotypes. The F26A/I27A and F35A/W36A double mutants were found to replicate on Vero cells in the single-step growth studies, but the yields were >2 logs lower than what was found with wt KOS virus (Fig. 2). In contrast, the quadruple mutant, vF26A/I27A/F35A/W36A, failed to produce any virus when grown on Vero cells (Fig. 2). The phenotype of the quadruple mutant was identical to what was found with a UL25 null virus in that only A and B capsids and nearly unit-length genomes that are aberrantly cleaved at the US end of the genome were isolated from cells infected with this mutant. Western blotting of the quadruple mutant demonstrated that the UL25 protein bound to the A and B capsids.
Cryo-EM reconstructions of the purified double and quadruple mutant capsids exhibited significantly lower levels of occupancy (10 to 20%) for CCSC (UL25/UL17 complex) at capsid vertices than for wt capsids (∼84%). However, detection of CCSC on both the A and B capsids isolated from the vF26A/I27A/F35A/W36A quadruple mutant was surprising since it had been previously reported that this complex was present only on C capsids (37). It was proposed that binding sites for the CCSC were exposed on the capsid surface as a result of the increased pressure inside the capsid from the encapsidated DNA (37). We also observed the CCSC molecule on all three capsid forms isolated from both HSV-1 and PRV, with occupancy near 100% for C capsids and >50% for B capsids (F. L. Homa and J. F. Conway, unpublished data). Our measured value of 84% (instead of 100%) for wt C capsids likely results from some loss of CCSC during sample preparation, suggesting that it is easily lost without gentle preparative conditions. Therefore, it appears that binding sites for the UL25/UL17 complex are present on all three capsid forms (as a consequence of this observation, we now refer to this molecule as the capsid vertex-specific component [CVSC]). We suspect that CVSC is not tightly bound, particularly to A and B capsids, and may be inadvertently lost during sample preparation since it was only recently observed on cryo-EM reconstructions (37). Further, the outer surface of the herpesvirus capsid includes other sites away from the vertices that would appear to have the necessary subunits for CVSC to bind to—triplexes and capsomers—but no measurable density has been observed in these places. Presumably, the local configurations of capsid subunits are only precise enough around the vertices for presenting a suitable interface for the CVSC to recognize. Given the nearly maximal occupancy found on C capsids, the stable binding of CVSC to a majority of these sites appears to be critical to ensure that the full-length viral genome is retained in the capsid.
A region located in the UL25 N terminus (amino acids 1 to 27) appears to be essential for the stable binding of the CVSC to the capsid. A second region located between amino acids 26 to 39, although not essential for capsid binding, is required for efficient capsid binding and for cleavage and retention of the viral genome. The available crystallographic data on UL25 does not allow localization of these regions, since the structure omits the entire N-terminal 133 amino acids (3). Instead, we used a tagged version of UL25 bound to capsids to localize UL25 capsid binding sites. The cryo-EM 3D reconstructions indicate that the N terminus of UL25 makes contact with the hexon adjacent to the penton and that the region centered on UL25 amino acids 26 to 39 makes contact with the triplex that is once removed from the penton (Fig. 8). We have shown that the N-terminal 50 amino acids are not required for nuclear import and conclude that the evident correlation between mutation sites that affect capsid binding and their locations at or near interfaces with capsid structures suggests that the mutations grossly interfere with the interactions between UL25 and the capsid. As we indicated above, the apparent ease with which CVSC is lost from capsids and their absence from quasi-equivalent binding sites away from the vertices suggest that the binding interaction between UL25 and the capsid is not strong and may therefore easily be blocked by mutations at the interfaces.
We found that UL17 was present on the capsids from all the UL25 mutants, including those that failed to show any UL25 capsid binding (Fig. 3). Western blotting of the capsid gradients indicated that there was a slight reduction in the amount of capsid-associated UL17 with UL25 mutants that failed to bind capsids. In studies with a UL17 null virus, it was found that the B capsids produced by this virus contained about 10% of the level of UL25 found in wt B capsids (32), suggesting that binding of UL25 to capsids is dependent on the presence of UL17 and that the two proteins associate only after UL17 has bound to the capsid. Unlike UL25, the UL17 protein is required for cleavage of replicated viral DNA (25), and it remains to be determined if this function is independent of UL25 binding and formation of the CVSC dimer.
Cells infected with all of the UL25 mutants, including the UL25 null mutant, vΔUL25, contained low numbers of intranuclear C capsids (Fig. 5), an observation that has been reported previously for both the HSV and PRV UL25 null mutants (14, 20, 29). DNase protection studies demonstrated the presence of full-length viral genomes in these capsids (Fig. 6). Therefore, even in the absence of the UL25 protein some DNA packaging takes place, but as was observed in Fig. 5 and previously reported (14, 20, 29), these C capsids fail to exit the nucleus. The loop domains and conserved surface clusters of the UL25 protein have been shown to be essential for capsid nuclear egress. We observed a similar nuclear egress-deficient phenotype with the vF26A/I27A and vF35A/W36A mutants. Both mutants were found to produce virus in the single-step growth assays but at much lower levels than the wt virus. Given the very low level of occupancy of the CVSC found with the vF35A/W36A mutant, the defect with these mutants appears to be a result of the unstable binding of UL25 to the capsid, which would subsequently interfere with the nuclear egress of any capsids containing full-length viral genomes.
In summary, we identified two domains located within the first 50 amino acids of the UL25 protein that are required for its stable binding to capsids. The first domain is located within the first 27 amino acids of the UL25 protein, and the second domain resides within a cluster of conserved amino acids (residues 26 to 39). UL25 is a multifunctional protein that is required to stabilize capsids after DNA is packaged, trigger nuclear egress of the capsid once a stable DNA-containing capsid is formed, anchor the major tegument protein, UL36, to capsids, and uncoat the incoming viral genome early in infection. All herpesviruses employ a UL25 analog that most likely performs a common set of functions, suggesting that understanding how UL25 functions during HSV-1 infections will inform us how cleavage/packaging occurs in other herpesviruses and, further, that UL25 may make an excellent target for antivirals to a structural protein that is highly specific to herpesviruses.
ACKNOWLEDGMENTS
We thank Donna Stolz of the University of Pittsburgh Center for Biologic Imaging for the preparation of thin-section grids for electron microscopy. Phil Greer and Douglas Bevan are gratefully acknowledged for help with computational analysis and Jay Brown for his insightful comments on the manuscript.
This work was supported by Public Health Service grant AI060836 from the National Institutes of Health to F.L. Homa, T32 training grant AI049820 to S.K. Cockrell, and SAP 4100031302 from the Commonwealth of Pennsylvania to J. F. Conway.
Footnotes
Published ahead of print on 16 March 2011.
REFERENCES
- 1. al-Kobaisi M. F., Rixon F. J., McDougall I., Preston V. G. 1991. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology 180:380–388 [DOI] [PubMed] [Google Scholar]
- 2. Booy F. P., et al. 1991. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell 64:1007–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bowman B. R., et al. 2006. Structural characterization of the UL25 DNA-packaging protein from herpes simplex virus type 1. J. Virol. 80:2309–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cockrell S. K., Sanchez M. E., Erazo A., Homa F. L. 2009. Role of the UL25 protein in herpes simplex virus DNA encapsidation. J. Virol. 83:47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cohen G. H., et al. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34:521–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coller K. E., Lee J. I., Ueda A., Smith G. A. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J. Virol. 81:11790–11797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Conway J. F., Homa F. L. 2011. Nucleocapsid structure, assembly and DNA packaging of herpes simplex virus, p. 175–193 In Weller S. K. (ed.), Alphaheresviruses. Caister Academic Press, Norwich, United Kingdom [Google Scholar]
- 8. Conway J. F., et al. 2010. Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. J. Mol. Biol. 397:575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Conway J. F., Steven A. C. 1999. Methods for reconstructing density maps of “single” particles from cryoelectron micrographs to subnanometer resolution. J. Struct. Biol. 128:106–118 [DOI] [PubMed] [Google Scholar]
- 10. Gibson W., Roizman B. 1972. Proteins specified by herpes simplex virus. VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10:1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gierasch W. W., et al. 2006. Construction and characterization of bacterial artificial chromosomes containing HSV-1 strains 17 and KOS. J. Virol. Methods 135:197–206 [DOI] [PubMed] [Google Scholar]
- 12. Heymann J. B., Belnap D. M. 2007. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157:3–18 [DOI] [PubMed] [Google Scholar]
- 13. Homa F. L., Otal T. M., Glorioso J. C., Levine M. 1986. Transcriptional control signals of a herpes simplex virus type 1 late (γ2) gene lie within bases −34 to +124 relative to the 5′ terminus of the mRNA. Mol. Cell. Biol. 6:3652–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Klupp B. G., Granzow H., Keil G. M., Mettenleiter T. C. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J. Virol. 80:6235–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lamberti C., Weller S. K. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 72:2463–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lamberti C., Weller S. K. 1996. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology 226:403–407 [DOI] [PubMed] [Google Scholar]
- 17. McNab A. R., et al. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Newcomb W. W., Homa F. L., Brown J. C. 2006. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J. Virol. 80:6286–6294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Newcomb W. W., et al. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432–446 [DOI] [PubMed] [Google Scholar]
- 20. O'Hara M., et al. 2010. Mutational analysis of the herpes simplex virus type 1 UL25 DNA packaging protein reveals regions that are important after the viral DNA has been packaged. J. Virol. 84:4252–4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ogasawara M., Suzutani T., Yoshida I., Azuma M. 2001. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J. Virol. 75:1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Patel A. H., Rixon F. J., Cunningham C., Davison A. J. 1996. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology 217:111–123 [DOI] [PubMed] [Google Scholar]
- 23. Pettersen E. F., et al. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 24. Rajcáni J., Andrea V., Ingeborg R. 2004. Peculiarities of herpes simplex virus (HSV) transcription: an overview. Virus Genes 28:293–310 [DOI] [PubMed] [Google Scholar]
- 25. Salmon B., Cunningham C., Davison A. J., Harris W. J., Baines J. D. 1998. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 72:3779–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Severini A., Morgan A. R., Tovell D. R., Tyrrell D. L. 1994. Study of the structure of replicative intermediates of HSV-1 DNA by pulsed-field gel electrophoresis. Virology 200:428–435 [DOI] [PubMed] [Google Scholar]
- 27. Severini A., Scraba D. G., Tyrrell D. L. 1996. Branched structures in the intracellular DNA of herpes simplex virus type 1. J. Virol. 70:3169–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sheaffer A. K., et al. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stow N. D. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 75:10755–10765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tengelsen L. A., Pederson N. E., Shaver P. R., Wathen M. W., Homa F. L. 1993. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J. Virol. 67:3470–3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thomsen D. R., Roof L. L., Homa F. L. 1994. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J. Virol. 68:2442–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thurlow J. K., Murphy M., Stow N. D., Preston V. G. 2006. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J. Virol. 80:2118–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tischer B. K., Smith G. A., Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol. Biol. 634:421–430 [DOI] [PubMed] [Google Scholar]
- 34. Tischer B. K., von Einem J., Kaufer B., Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197 [DOI] [PubMed] [Google Scholar]
- 35. Tivol W. F., Briegel A., Jensen G. J. 2008. An improved cryogen for plunge freezing. Microsc. Microanal. 14:375–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trus B. L., et al. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 263:447–462 [DOI] [PubMed] [Google Scholar]
- 37. Trus B. L., et al. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol. Cell 26:479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ward P. L., Ogle W. O., Roizman B. 1996. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J. Virol. 70:4623–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wills E., Scholtes L., Baines J. D. 2006. Herpes simplex virus 1 DNA packaging proteins encoded by UL6, UL15, UL17, UL28, and UL33 are located on the external surface of the viral capsid. J. Virol. 80:10894–10899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yan X., Dryden K. A., Tang J., Baker T. S. 2007. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J. Struct. Biol. 157:211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yan X., Sinkovits R. S., Baker T. S. 2007. AUTO3DEM—an automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol. 157:73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]