Abstract
Ourmia melon virus (OuMV) is the type member of the genus Ourmiavirus. These viruses have a trisegmented genome, each part of which encodes a single protein. Ourmiaviruses share a distant similarity with other plant viruses only in their movement proteins (MP), whereas their RNA-dependent RNA polymerase (RdRP) shares features only with fungal viruses of the family Narnaviridae. Thus, ourmiaviruses are in a unique phylogenetic position among existing plant viruses. Here, we developed an agroinoculation system to launch infection in Nicotiana benthamiana plants. Using different combinations of the three segments, we demonstrated that RNA1 is necessary and sufficient for cis-acting replication in the agroinfiltrated area. RNA2 and RNA3, encoding the putative movement protein and the coat protein (CP), respectively, are both necessary for successful systemic infection of N. benthamiana. The CP is dispensable for long-distance transport of the virus through vascular tissues, but its absence prevents efficient systemic infection at the exit sites. Virion formation occurred only when the CP was translated from replication-derived RNA3. Transient expression of a green fluorescent protein-MP (GFP-MP) fusion via agroinfiltration showed that the MP is present in cytoplasmic connections across plant cell walls; in protoplasts the GFP-MP fusion stimulates the formation of tubular protrusions. Expression through agroinfiltration of a GFP-CP fusion displays most of the fluorescence inside the nucleus and within the nucleolus in particular. Nuclear localization of the CP was also confirmed through Western blot analysis of purified nuclei. The significance of several unusual properties of OuMV for replication, virion assembly, and movement is discussed in relation to other positive-strand RNA viruses.
INTRODUCTION
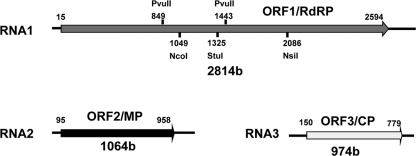
Ourmia melon virus (OuMV) is the type member of the genus Ourmiavirus. OuMV was first isolated in melon crops in Ourmia in Iran (35, 53), the only country where its presence has been documented. Since its early discovery, OuMV has raised curiosity because of its unique virion morphology compared to other plant viruses: bacilliform particles of three different lengths with pointed ends and a tubular body containing discontinuities marked by fissures (35). Evidence was shown that each genomic RNA is encapsidated independently (35). Within the same genus are two other viruses: Epirus cherry virus (EpCV) (7) and Cassava virus C (CaVC) (3). No biological vector has been implicated in plant-to-plant transmission in the field (1, 42). Recent nucleotide sequence and phylogenetic analyses have shown that the three Ourmiavirus species have a conserved genome organization (53). All have a trisegmented plus-strand RNA genome. Each segment potentially encodes only one protein (Fig. 1). The largest RNA segment contains open reading frame 1 (ORF1) which encodes a putative RNA-dependent RNA polymerase (RdRP). The RdRP is phylogenetically related to the RdRPs of viruses in the Narnaviridae, a heterogeneous group comprised chiefly of viruses which infect fungi, including the yeast Saccharomyces cerevisiae. The RNA2 ORF2 encodes a putative movement protein (MP) (circa 30 kDa). In contrast to the phylogenetic relationships for RNA1, the OuMV MP shares similarities with MPs of plant-infecting viruses of the genus Tombusvirus. Finally, RNA3 encodes the 22-kDa coat protein (CP) which has no obvious phylogenetic relationships to any protein in the database (53). The different phylogenetic origins of RNA1 and RNA2 (from a fungal virus and a plant virus, respectively) point to interkingdom virus reassortment as a possible new mechanism of viral evolution apt to induce the emergence of a novel viral genus (32, 53). To better understand the molecular mechanisms behind the Ourmiavirus life cycle, a reverse genetic system is required.
Fig. 1.
OuMV genome organization. Each encoded ORF is indicated by an arrow. The most important restriction sites used in the mutagenesis process are shown.
Here, we describe the development of a reverse genetic system for OuMV launched by a mix of three Agrobacterium tumefaciens clones harboring binary plasmids expressing each of the genomic RNAs under the control of the 35S promoter. Using this system, we provide new genetic evidence for the role of each virus-encoded protein in the infection cycle in N. benthamiana plants. Furthermore, we identified unexpected genetic requirements for virion assembly.
MATERIALS AND METHODS
Construction of full-length agroinfectious clones.
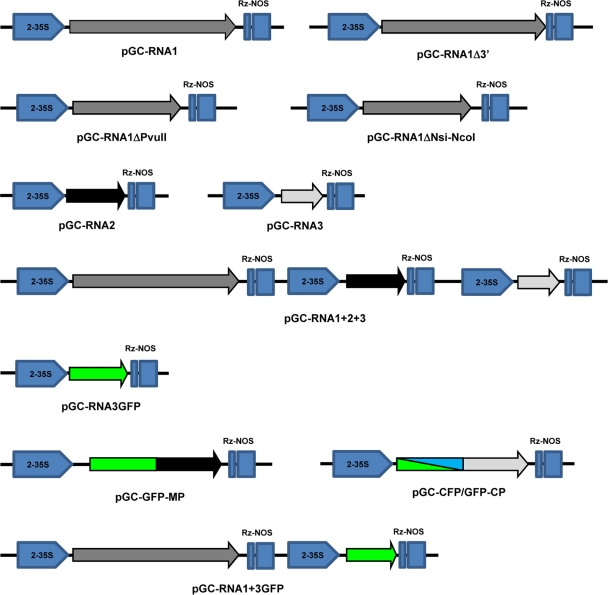
The full-length cDNAs of each of the three genomic segments was obtained through reverse transcription-PCR (RT-PCR) using as a template RNA obtained from plants infected with the OuMV isolate VE9. RNA was extracted with a Spectrum Plant Total RNA kit and buffer A (Sigma, St. Louis, MO), following the manufacturer's protocol. Reverse transcription was carried out as previously described (65). PCR was done using Phusion (Finnzymes, Espoo, Finland) as previously described and with the appropriate primer combinations: OuMV-RNA1F and OuMV-RNA1R for full-length RNA1, OuMV-RNA2F and OuMV-RNA2R for RNA2, and OuMV-RNA3F and OuMV-RNA3R for RNA3 (see Table S1 in the supplemental material). Each blunt-end segment was kinased (except for RNA1, which was amplified with phosphorylated oligonucleotides) and ligated in the StuI-SmaI-cut pJL89 (34). The three plasmids were called pGC-RNA1, pGC-RNA2, and pGC-RNA3, respectively. Each plasmid contained the full-length segment with the exact 5′ end expressed downstream from a double 35S promoter and the exact 3′ end processed by the hepatitis delta virus (HDV) ribozyme present in the plasmids. Each plasmid was transformed into the Agrobacterium tumefaciens strain C58C1 and agroinfiltrated as previously described (8, 9, 37). We then assembled plasmids that contained RNA1 plus RNA3, RNA2 plus RNA3, RNA1 and RNA2, and finally a clone containing RNA1, RNA2, and RNA3 together. The clones were assembled through PstI partial digestion and Klenow fill-in to eliminate one of the two sites upstream or downstream from the RNA-expressing construct and by inserting the 35S-RNA(X)-HDV fragment (cut with PstI), where “X” is any of the three genome sequences, in a step-by-step process. Deletion of RNA1 was derived through PvuII digestion and religation of pGC-RNA1, or, alternatively, through NsiI-NcoI digestion, Klenow treatment, and religation of the same plasmid giving pGC-RNA1ΔPvuII and pGC-RNA1ΔNsi-NcoI clones, respectively. Another deletion in RNA1 was obtained by using in the original RT-PCR the primers RNA1F and OuMV-RNA1-2613R, a reverse primer which anneals in a region of RNA1 immediately downstream from the ORF1 stop codon; this clone (pGC-RNA1Δ3′) expresses full-length ORF1 without the 3′ untranslated region (UTR; circa 200 nucleotides [nt]) present in full-length RNA1. A list of the main constructs and their schematic representation are given in Fig. 2.
Fig. 2.
Schematic representation of constructs built and used in this study. Each clone has the same pJL89-derived backbone, where double StuI/SmaI restriction digestion allows insertion of blunt-end PCR fragments derived from RT-PCR of full-length genomic segments between the double 35S promoter (2-35S) and the hepatitis delta virus ribozyme (Rz) followed by a Nos terminator. Arrow directions indicate the orientation of the viral transcripts obtained for each of the described clones. The arrows indicate each ORF encoded by the genome.
In vitro transcription and transcript inoculation.
For in vitro transcription and transcript inoculation, we scrupulously followed the protocols previously detailed (66), except that the template for transcription was obtained through gel purification of a PCR product carried out with the high-fidelity polymerase Phusion (Finnzymes) and using as a template the plasmids pGC-RNA1, pGC-RNA2, and pGC-RNA3 and the oligonucleotides RNA1-T71-F and OuMV-RNA1R for RNA1, RNA2-T71-F and OuMV-RNA2R for RNA2, and RNA3-T71-F and OuMV-RNA3R for RNA3 (see Table S1 in the supplemental material).
Site-directed mutagenesis and recombinant Ourmiavirus clones.
Site-directed mutagenesis was carried out through PCR-mediated mutagenesis with the use of Phusion under the conditions suggested by the manufacturer: an SmaI site was inserted in RNA3 at positions 162 and 768 (oligonucleotides RNA3-SmaF and RNA3-SmaR), thus deleting the coding region of the CP (originating clone, pGC-RNA3Sma); an NcoI site was inserted at the exact amino terminus of ORF2 (oligonucleotides RNA2-NcoImut-F and RNA2-NcoImut-R), thus yielding pGC-RNA2NcoI. A green fluorescent protein (GFP)-encoding sequence was amplified from clone pTG-P19/wt (where wt is wild type) (37) by using the oligonucleotides GFP-Sma-F and GFP-Sma-R and the oligonucleotides GFP-NcoI-F and GFP-NcoI-R (see Table S1 in the supplemental material). The two digested PCR fragments were inserted in the SmaI site of the pGC-RNA3Sma clone and in the NcoI site of pGC-RNA2NcoI to obtain pGC-RNA3GFP and pGC-GFP-MP, respectively. Plasmid pBin-CFP (where CFP is cyan fluorescent protein) was derived through PCR amplification of a CFP fragment (using the same oligonucleotides described above and used for GFP) and then cloned in the SmaI site of pBin61 (8) using as a template the pEF3-CFP-2 plasmid (Addgene, Inc., Cambridge, MA). An NcoI site was also inserted at the amino terminus of the CP through PCR mutagenesis by using the oligonucleotides RNA3-NcoI-F and RNA3-NcoI-R (see Table S1). CFP and GFP were both inserted in frame into the NcoI site after amplification with the oligonucleotides GFP-NcoI-F and GFP-NcoI-R (see Table S1) to obtain a CFP-CP or a GFP-CP fusion protein, yielding the plasmids pGC-GFP-CP and pGC-CFP-CP. A Myc peptide was inserted in frame to the amino terminus of the MP through direct ligation of a kinased PCR product obtained with Phusion polymerase by using the primer pairs RNA2-Myc-F and RNA2-Myc-R, yielding the plasmid pGC-RNA2myc.
Cell fractionation, nuclei purification, and Western and Northern blot analyses.
Cellular fractionation between soluble (SOL) and membrane-bound proteins (P30 fraction; from centrifugation at 30,000 × g) was performed as described previously (45). Nuclear extracts were obtained with a CelLytic PN plant nuclei isolation/extraction kit (Sigma, St. Louis, MO) according to the manufacturer's instructions following the protocol for a highly pure preparation. Western and Northern blot analyses were carried out as previously described (53), except that minus- and plus-strand probes were derived by cloning fragments of each RNA in pSC-B-amp/kan (Agilent Technologies, Santa Clara, CA). In detail, a probe for RNA1 was derived with a 5′ end 300-bp fragment obtained through PCR by using RNA1-1F and RNA1-RealR. The fragment was cloned in pSC-B/amp/kan, and both orientations were kept and cut for transcription with EcoRV. Similarly, a probe for RNA2 was obtained by using the oligonucleotides RNA2-1F and RNA2-RealR; the probe for RNA3 was assembled with a 400-bp fragment obtained with the oligonucleotides RNA3-1F and RNA3-RealR. Conditions for Myc detection by Western blot analysis were previously described (45).
Virion partial purification, whole-virus agarose gel assay, and electron microscopic analyses.
For partial virion purification, 1 g of the area of the agroinfiltrated leaves was processed using a FastPrep bead beater (MP Biomedicals, Solon, OH), placing the leaf tissue in a 15-ml Falcon tube with 5 ml of 0.5 mm zirconia beads and 8 ml of protein extraction buffer as previously described (45). The leaf extract was left at 37°C for 30 min to allow endogenous RNase activity to take place (25). Five milliliters of the extract was then mixed with an equal volume of chloroform by vortexing, and the phases were separated by centrifugation at 3,000 × g for 10 min. The supernatant was treated with 10% polyethylene glycol (PEG) (8,000 molecular weight [MW]) and 1% NaCl and precipitated for 30 min at 4°C. After centrifugation at 4,000 × g for 20 min, the pellet was resuspended in 300 ml of 0.05 M phosphate buffer (pH 7). Agarose gels for whole-virus separation were prepared and run as previously described (50). Samples were transferred to nylon or nitrocellulose membranes through capillary transfer. Western and Northern blot analyses were carried out following standard protocols as previously described (53). The negatively stained, partially purified virion preparation trapped on the grid was examined by immunosorbent electron microscopy (ISEM) techniques as previously described (43). Specific anti-CP antiserum (A253) previously described (53) was diluted 1:2,000.
Confocal microscopy observation.
Confocal microscopy was performed using a Leica TCS-SP2 microscope (Leica Microsystems, Mannheim, Germany) equipped with a long-distance 40× water immersion objective (HCX Apo 0.80). A 488-nm Ar laser line was used to excite the GFP and to collect transmitted light images of the samples. An emission window of 500 to 525 nm was used to image GFP fluorescence. CFP fluorescence was excited with the 458-nm line of the Ar laser and recorded at 450 to 510 nm. Red fluorescent protein (RFP) fluorescence was excited using the 544-nm line of an He/Ne laser and imaged at 550 to 620 nm. Subcellular localization of the fluorescent fusion proteins was analyzed in either single optical sections or z-axis projections from several overlaid sections.
Transgenic plants expressing RFP-endoplasmic reticulum (RFP-ER) and RFP-histone 2B (RFP-H2B) (18, 39) were used as controls for localization of ER and nuclei. pBin-GFP and pBINm-gfp5-ER were previously described (37, 68, 70). N. benthamiana protoplasts from agroinfected leaves were derived at 3 days postinoculation (dpi) as previously described (24).
RESULTS
Infectivity of binary recombinant plasmids and of in vitro transcribed viral RNA.
Through direct cloning of the full-length RT-PCR product of each genomic segment in pJL89 (34), we obtained clones expressing each of the three RNAs downstream from a 35S promoter, and each RNA is putatively processed by an HDV ribozyme (58) to the exact 3′ end previously determined (Fig. 2). Using oligonucleotides incorporating the T7 promoter immediately upstream from the viral genomic sequence, we derived the templates for in vitro transcription of the three genomic RNAs through PCR. Agroinfiltration and mechanical inoculation of in vitro generated positive-sense transcripts resulted in systemic infection of N. benthamiana plants when the three constructs were mixed; we also found that agroinfiltration of the single plasmid containing all three OuMV segments gave infection. All infections were confirmed by Western and Northern blot analyses (see Fig. S1 in the supplemental material), and systemic symptoms were undistinguishable from those obtained through mechanical inoculation of the original VE9 isolate (see Fig. S1).
RNA1 encodes a fully competent viral replicase.
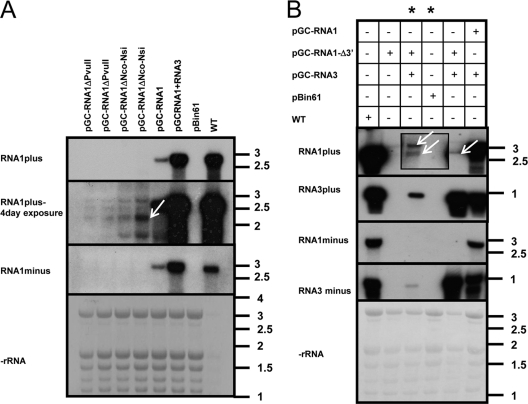
When we infiltrated N. benthamiana leaves with pGC-RNA1 designed to generate only OuMV RNA1, Northern blot analysis of infiltrated leaves showed accumulation of both minus- and plus-strand OuMV RNA1, demonstrating that agroinfiltration of RNA1 alone is sufficient for its cis replication (Fig. 3A). Deletions in RNA1 in two different regions (PvuII deletion and NsiI-NcoI deletion) abolished the ability to accumulate minus-strand RNA, thereby showing a lack of replication. This is the first genetic evidence that, indeed, RNA1 encodes a fully competent viral replicase and that this is sufficient for its own replication.
Fig. 3.
RNA1 encodes a fully competent cis- and trans-acting replicase. (A) Northern blot analysis of total RNA extracted from agroinfiltrated areas 3 days postinfiltration (dpi). The top panel shows viral plus-strand RNA evaluated with a 2-h exposure to the X-ray film; the middle panel shows the same blot exposed for 4 days to evaluate accumulation of the deleted RNA1 transcript (arrow); the bottom panels show the minus-strand RNA accumulation with a 4-day exposure and the rRNA, respectively. Each agroinfiltrated clone is detailed at the top of each lane and graphically described in Fig. 2. (B) Northern blot analysis of RNA extracted at 3 dpi with a combination of agroclones specified above the panels (except for the samples corresponding to the lanes marked by asterisks, which were instead processed 2 dpi). From top to bottom, each of the four panels shows plus-strand RNA1, plus-strand RNA3, minus-strand RNA1, and minus-strand RNA3. The bottom panel shows rRNA stained with methylene blue on the membrane. The inset replaces the original 2-day exposure with a 6-day exposure. The arrows indicate the two primary transcripts from pGC-RNA1Δ3′ (ribozyme processed and full-length). The RNA ladder bands are those from BrightStar Biotinylated RNA Millennium Markers (Ambion), and the sizes are expressed in kb.
We also constructed another plasmid designed to express the full-length coding sequence of ORF1 but lacking the 200 nt of the RNA1 3′ UTR. When this was agroinfiltrated alone and in combination with pGC-RNA3, there was no replication of RNA1. However, we did find evidence for generation of a functional replicase as RNA3 replication was observed in the doubly infiltrated tissue (Fig. 3B). This demonstrates that the OuMV RNA1-encoded RdRP is sufficient for replication of RNA3 in trans.
Virion assembly requires actively replicating RNA3.
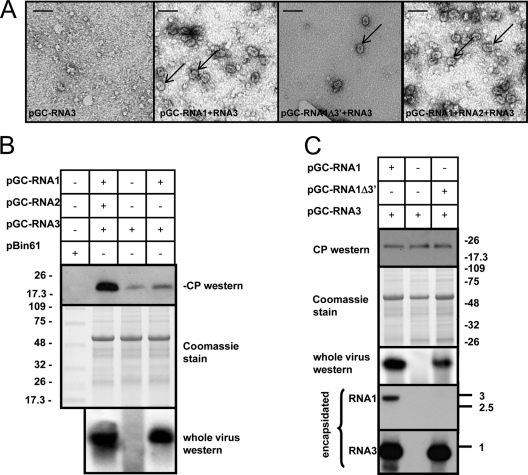
We next evaluated requirements for virus particle accumulation in the agroinfiltrated leaves. Electron microscopic observation of sap or partially purified virions failed to reveal the presence of virus particles when only pGC-RNA3 was agroinfiltrated in N. benthamiana leaves, as did ISEM (Fig. 4A). These data suggest that the mere expression of RNA3-encoded CP by itself cannot originate capsids although both the RNA3 and the CP were present in the agroinfiltrated leaf, as shown by Western and Northern blot analyses (Fig. 4B). However, particles were detected when pGC-RNA1 and pGC-RNA3 were agroinfiltrated together. These data show that RNA2 is not required for particle formation (Fig. 4A) but also suggest that actively replicating RNA3 is required. RNA1 replication in itself is not necessary for virion formation since pGC-RNA1Δ3′ lacking the RNA1 3′ UTR was able to support replication and encapsidation of RNA3 (Fig. 3B and 4C). Furthermore, Northern blot analysis of RNA extracted from the purified particles confirmed that RNA1 lacking the whole 3′ UTR was not encapsidated. This also might suggest that requirements for encapsidation are present in the 3′ UTR of RNA1 (Fig. 4C). Apparent inconsistencies between different experiments related to RNA3 and CP expression when RNA3 was agroinfiltrated alone or with an active viral RdRP could be due to the different time points of extraction after agroinfiltration. For example, in the earlier phases, when transcription still predominates, the amounts of RNA3 and CP are comparable among the various combinations (as shown in Fig. 4C); in the later phases, when viral replication predominates over the simple accumulation of processed RNA from plasmid transcription from the 35S promoter, the differences between the different treatments (with or without active viral RdRP) are much higher (see Fig. S4 in the supplemental material).
Fig. 4.
Virion formation analysis in agroinfiltrated areas at 3 days postagroinfiltration (dpi) with the combination of agroclones described in Fig. 2. (A) Electron micrograph of negatively stained ISEM preparations of partially purified virus preparation from agroinfiltrated leaves. The arrows indicate single viral particles. Bar, 50 nm. (B) The top panel shows Western blotting of SDS-PAGE-separated protein extracts from the agroinfiltrated area to detect OuMV coat protein (CP); the middle panel shows Coomassie staining for total protein extracts separated on SDS-PAGE, and the bottom panel shows a Western blot for the CP after partial virus purification and whole-virus agarose gel separation to detect virion presence. Combinations of the binary plasmids used in each agro-suspension treatment are specified at the top of the blots. (C) The three top panels are the same as in panel B; the two bottom panels show the results of Northern blot analysis for OuMV RNA1 and RNA3, respectively, extracted from partially purified virus preparations. The RNA ladder bands are those from BrightStar Biotinylated RNA Millennium Markers (Ambion), and the sizes are expressed in kb. Thermo Scientific Pierce Blue Prestained MW Marker (Pierce) was used for Western blot analysis, and the sizes of the bands are expressed in kDa.
Viral determinants for local and systemic infection in N. benthamiana plants.
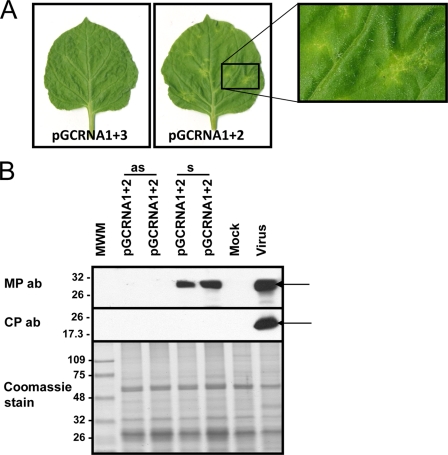
Necrotic local lesions were always visible on N. benthamiana leaves inoculated with a combination of all three RNA transcripts and/or in the agroinfiltrated area, and systemic symptoms later developed on upper leaves (Table 1; see also Fig. S2 and Fig. S3 in the supplemental material). When any of the genome segments alone were used as the inoculum, no local lesions or systemic symptoms were apparent. We next evaluated various combinations of the genome segments for their abilities to induce local lesions and systemic infection. When only pGC-RNA1 and pGC-RNA2 were used, necrotic local lesions developed on inoculated leaves (Table 1; see also Fig. S2 and Fig. S3). Interestingly, with the same plasmid mix, well-defined necrotic areas on the upper uninoculated leaves were also observed (12 dpi) (Fig. 5A). However, in contrast to inoculations with all three plasmids, when only genome segments 1 and 2 were expressed, the sizes of the necrotic areas remained limited throughout the life of the plant. In order to confirm that the predicted RNA2-encoded MP was produced in these infections, we used Western blot analysis of the symptomatic and nonsymptomatic areas of the upper leaves. In the necrotic areas, the MP was abundantly expressed, whereas the CP was absent, as expected (Fig. 5B). However, no accumulation of MP was observed in the neighboring asymptomatic areas of the leaves (Fig. 5B), showing that, although vascular transport was allowed, subsequent spread within the upper leaves did not occur. Accumulation of RNA1 in the symptomatic area was also demonstrated through Northern blot analysis (data not shown). Therefore, long-distance movement of both the RNA1 and RNA2 genomic segments through the vascular system does not strictly require the CP. But as these partial systemic infections failed to spread in the upper leaves, this suggests that the CP is instead required for efficient and complete infection of the upper leaves.
Table 1.
Overview of test results for infectivity, necrotic local response, CP accumulation, and virion formation after inoculation using different combinations of agroclones and RNA transcripts
| Inoculum type and RNA segment(s)a | Systemic infection | Necrotic local response | Virion formation in agroinflitrated leaves | CP accumulation in agroinfiltrated leaves |
|---|---|---|---|---|
| Agroclone(s) | ||||
| RNA1(+) | − | − | − | − |
| RNA2(+) | − | − | − | − |
| RNA3(+) | − | − | − | + |
| RNA1(+), RNA2(+), RNA3(+) | + | + | + | + |
| RNA1(+), RNA2(+) | +b | + | − | − |
| RNA1(+), RNA3(+) | − | − | + | + |
| RNA3(+), RNA2(+) | − | − | − | + |
| RNA1Δ3′, RNA2(+), RNA3(+) | − | − | + | + |
| RNA transcript(s) | ||||
| RNA1(+) | − | − | NAd | NA |
| RNA2(+) | − | − | NA | NA |
| RNA3(+) | − | − | NA | NA |
| RNA1(+), RNA2(+), RNA3(+) | + | + | NA | NA |
| RNA1(+), RNA2(+) | +c | + | NA | NA |
| RNA1(+), RNA3(+) | − | − | NA | NA |
| RNA3(+), RNA2(+) | − | − | NA | NA |
| RNA1Δ3′, RNA2(+), RNA3(+) | − | − | NA | NA |
+, plus strand.
Infection was limited to an area of circa 0.5-cm diameter at some vascular tissue exit sites.
Rare infection foci at some vascular tissue exit sites.
NA, not applicable.
Fig. 5.
Limited necrotic areas in upper uninoculated leaves when the plant is agroinfiltrated with a mix of RNA1- and RNA2-expressing clones. (A) Symptoms on upper uninoculated leaves at 12 days postinfiltration (dpi) with a combination of agrobacterium clones. The inset to the right shows an enlargement of the localized symptomatic area. (B) Western blotting for the presence of the OuMV MP (upper panel) and CP (middle panel). Samples were collected at 15 dpi in symptomatic (s) and asymptomatic (as) areas of an upper uninoculated leaf. Mock, inoculation with empty pBin61; virus, infection by mechanical inoculation of sap from N. benthamiana infected with OuMV VE9 isolate. The bottom panel shows Coomassie staining of the proteins loaded in the SDS-PAGE gel. Thermo Scientific Pierce Blue Prestained MW Marker (Pierce) is the molecular weight marker (MWM), and the sizes of the bands are expressed in thousands. The arrows indicate the position of the MP and CP bands in the Western blot. ab, antibody.
The expression of each genomic RNA through agroinfiltration, either alone or in all possible combinations with the remaining genomic RNAs, was evaluated locally in the agroinfiltrated areas through Northern and Western blotting (see Fig. S4 in the supplemental material); RNA1 accumulation of both positive and negative strands was higher when RNA1 was coagroinfiltrated with either RNA2 or RNA3 or both, and expression levels of both RNA2 and RNA3 and of their derived proteins were much lower when either was agroinfiltrated singly than when coinoculated with RNA1. No synergistic effect was observed for the coexpression of both RNA2 and RNA3 in the same leaf area. The upregulation of RNA1 accumulation when RNA1 was coexpressed with replicating RNA3 (as shown in Fig. 3A; see also S4) could be due to direct protection against RNA degradation due to coat protein encapsidation although other explanations, such as interference with antiviral defense pathways, namely, posttranscriptional gene silencing (PTGS), cannot be ruled out. A further noteworthy detail is the higher accumulation of CP and of RNA3 when the three segments are agroinfiltrated together as opposed to the mix of RNA1 and RNA3 only (as shown in Fig. 4A; see also Fig S4). Such upregulation could depend on either a direct synergistic effect of RNA2 or an indirect effect caused by the higher number of infected cells, due to the promotion of movement exerted by the presence of actively replicating RNA2.
The role of the RNA2-encoded ORF in cell-to-cell movement.
Taken together, the above experiments demonstrate that all three genomic RNAs and their encoded proteins are required for development of full-fledged systemic infections, but the specific role of the RNA2-encoded putative MP was still not proven. Therefore, to investigate the role of this protein, we performed microscopic analyses to assess possible MP trafficking and subcellular localization. A dilution series of standardized agrobacterial clone suspensions of pGC-GFP-MP, pGC-3GFP, pGC-3CFP, or pGC-GFP-CP (Fig. 2) were used to obtain transient expression of GFP-MP, GFP and CFP, or GFP-CP. Observation of fluorescence at 3 days postagroinfiltration showed that CFP, GFP, or GFP-CP was retained only in single cells, whereas in the same leaves GFP-MP expression was also visible in groups of cells, indicating that the chimeric protein was not localized to individual cells (Fig. 6). Our attempts to derive actively replicating virus vectors (Fig. 2) which replaced the CP with GFP failed (data not shown), but the pGC-RNA3GFP clone was still able to express GFP, presumably because of 35S-driven activity. This clone was therefore used to show that RNA2 can contribute to rendering the cell-to-cell connection (plasmodesmata or de novo tubule formation) more permeable to GFP. Using a dilution series of the agroinoculation mix to obtain single transfected cells (10−3 dilution), we observed that GFP was confined to one cell when pGC-RNA1 plus pGC-RNA3GFP was agroinfiltrated, whereas when pGC-RNA1 plus pGC-RNA3GFP was diluted into a pGC-RNA2 suspension and used for agroinoculation, strong GFP fluorescence in single cells was associated with a weaker fluorescent signal in one or more of the surrounding cells (data not shown), similar to what we observed with GFP-MP fusion. This result does not necessarily imply that virus infection can move from one cell to another in the absence of the CP, but it does suggest that RNA2-encoded MP facilitates GFP transport to adjacent cells. Taken together, these two experiments show that both the MP expressed from RNA2 and the GFP-MP fusion facilitate protein movement to neighboring cells, which is a functional feature of viral movement proteins.
Fig. 6.
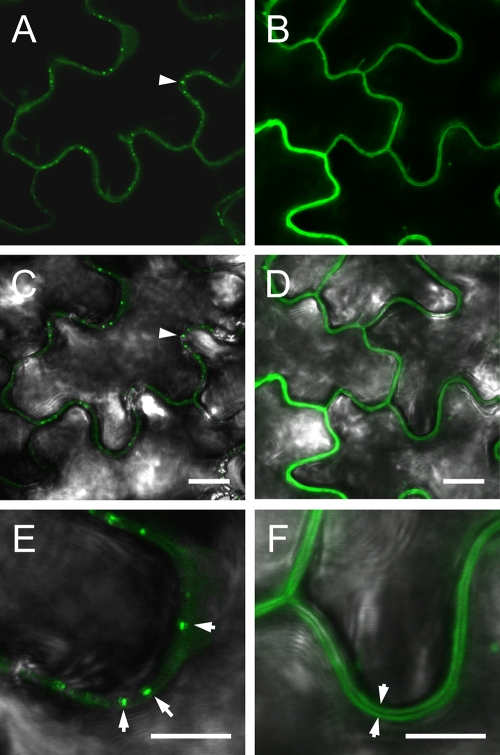
Cell-to-cell movement of GFP-MP fusion expressed in epidermal cells. Agro-suspensions precisely diluted at an optical density at 600 nm of 0.5 were agroinfiltrated in 1/10 serial dilutions until single-cell transfection was obtained. Only fully developed N. benthamiana leaves were agroinfiltrated. All pictures correspond to the 10−3 dilution. Fluorescence images (A, C, E, and G) are overlaid with the respective bright-field images (B, D, F, and H). (A and B) Localization of pBinGFP-CP fluorescence limited to the transfected epidermal cell (asterisk). The weak red signal (A and B, E and F, and G and H) derives from excitation of chloroplast autofluorescence in the mesophyll. Analogous single-cell localization (asterisk) of pBin-CFP (C and D) fluorescence is visible as well as for pBin-GFP (E and F). When pBin-GFP-MP was agroinfiltrated (G and H), fluorescence leaked from the transfected cell (asterisk) to the neighboring cells (arrowhead). Bar, 20 μm.
We next characterized the subcellular localization of the MP in vivo by visualizing fluorescence from the GFP-MP fusion in epidermal agroinfiltrated cells and protoplasts derived from agroinfiltrated leaves. Two constructs were used. The first (pBin-GFP-MP) expressed the GFP-MP fusion from the pBin61 vector, and so a canonical mRNA with a poly(A) tail was transiently transcribed in this case. The second construct (pGC-GFP-MP) expressed GFP-MP from the OuMV RNA2 clone, so the primary RNA was processed through a ribozyme. In both cases, fluorescence was detected although quantitatively the pBin-GFP-MP showed much higher fluorescence (data not shown). The epidermal cells from leaves agroinfiltrated with pGC-GFP-MP showed a weak cytoplasmic fluorescence and a striking punctate pattern within the cell walls (Fig. 7A, C, and E). Similar structures were never observed in control plants expressing the GFP alone (Fig. 7B, D, and F), where the cell wall was clearly visible as a dark line separating the fluorescent cytoplasm of adjacent cells. No obvious colocalization with RFP-ER (in transgenic plants) was observed (data not shown), indicating that pGC-GFP-MP fluorescence was not associated with the endoplasmic reticulum. Protoplasts from agroinfiltrated leaves showed brightly labeled bodies in an otherwise weakly fluorescent cytoplasm. More interestingly, this fluorescence pattern was associated with intense labeling of tubular protrusions extending from the plasma membrane into the extracellular space (Fig. 8A to D); the tubules were not present when GFP or GFP-ER constructs were used (Fig. 8E to H).
Fig. 7.
Subcellular localization of GFP-MP versus GFP in N. benthamiana leaf epidermal cells, observed 3 days postagroinfiltration. Images show localization of pGC-GFP-MP (A, C, and E) and pGC-RNA3-GFP (B, D, and F) in agroinfiltrated leaves. The fluorescence images in panels A and B are overlaid with the corresponding bright-field images in panels C and D; only the overlaid pictures are shown for the higher magnification images in panels E and F. In panels A and C the punctate pattern of MP-GFP fluorescence and its association with the cell wall are visible (arrowhead); panel E shows a particularly evident pattern (arrowheads). Localization of free GFP (B and D) to the cytoplasm clearly identifies the unlabeled cell wall (F) as a dark line separating the fluorescent cytoplasms of adjacent cells (arrowheads). Bars, 10 μm (A to D) and 5 μm (E and F).
Fig. 8.
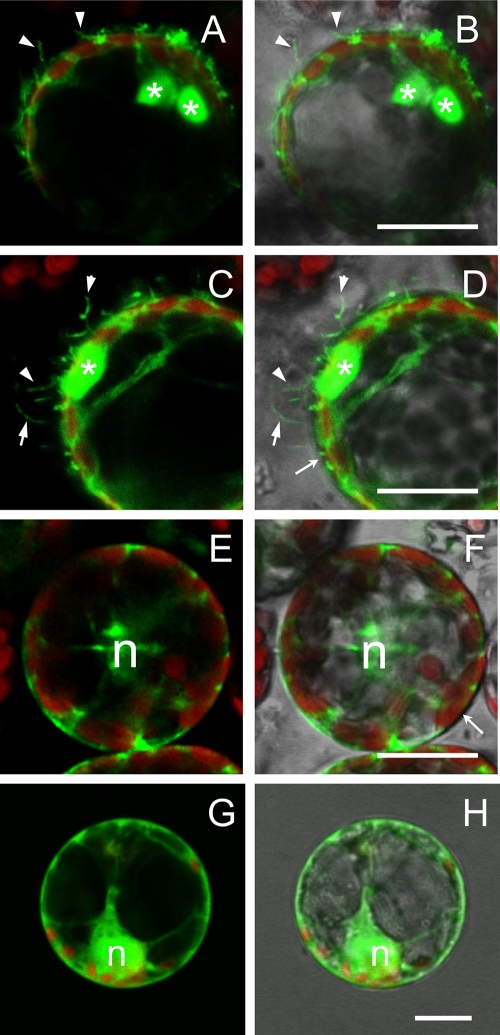
Subcellular localization of GFP-MP fusion in N. benthamiana protoplasts. (A, C, E, and G) Fluorescent signals of GFP (green) and chloroplasts (red). (B, D, F, and H) The same pictures overlaid with the corresponding bright-field images. Protoplasts transfected with the pGC-GFP-MP construct are shown in panels A to D. Besides the large aggregates visible in the cytoplasm (asterisks), GFP-MP fluorescence highlights numerous tubular protrusions of the protoplast plasma membrane. Such protrusions were never observed in protoplasts expressing the cytosolic GFP construct from the pGC-RNA3-GFP construct (E and F) or the endoplasmic reticulum-associated GFP-ER (G and H). n, nucleus. Bar, 20 μm.
CP subcellular localization.
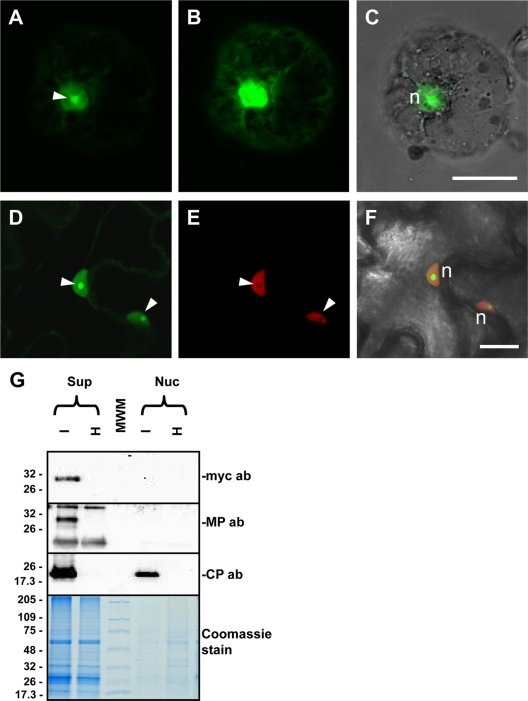
The above experiments as well as computer-assisted predictions of the OuMV-encoded RNA2 protein suggest that it is an MP. However, given the limited size of the local or systemic necrotic lesions when RNA3 was not expressed, a direct or indirect role of the CP in facilitating virus spread needed to be addressed. For this purpose we analyzed by confocal microscopy the subcellular localization of GFP when it was fused to the CP in viral RNA3 (pGC-GFP-CP) in agroinfiltrated leaves. This construct showed marked localization of fluorescence in the nucleus and, in particular, in the nucleolus (Fig. 9A to F) and putative Cajal bodies (see Fig. S5 in the supplemental material). Also, protoplasts prepared from these leaves showed greater fluorescence inside the nucleus, with a higher concentration in the nucleolus (Fig. 9A, B, and C). Our contention that the nucleolus and the Cajal bodies are indeed sites of CP accumulation is further supported by the fact that GFP-CP fluorescence highlights areas and spots inside the nucleus where H2B fused to RFP is absent (Fig. 9D, E, and F). Although H2B protein has been detected in proteome studies specific for the nucleolus (48), our results are consistent with previous studies reporting the exclusion of H2B-RFP from the nucleolus in transgenic plants expressing such a construct (39). Finally, although GFP alone is able to permeate the nucleus, this is generally not the case when the GFP is fused to nonnuclear proteins. Furthermore, this permeability does not extend to the nucleolus (see Fig. S5), suggesting that accumulation of GFP-CP inside the nucleolus is indeed due to the CP. Nuclear localization of the CP was also suggested by its presence in the purified preparations of the nuclei, where the MPs were absent (Fig. 9G). In a setting of viral infection, the MP proved to be a particularly good control for contamination of the nuclear fraction with cytoplasmic proteins since it was present in both the soluble and the P30 fraction (see next paragraph) (Fig. 10).
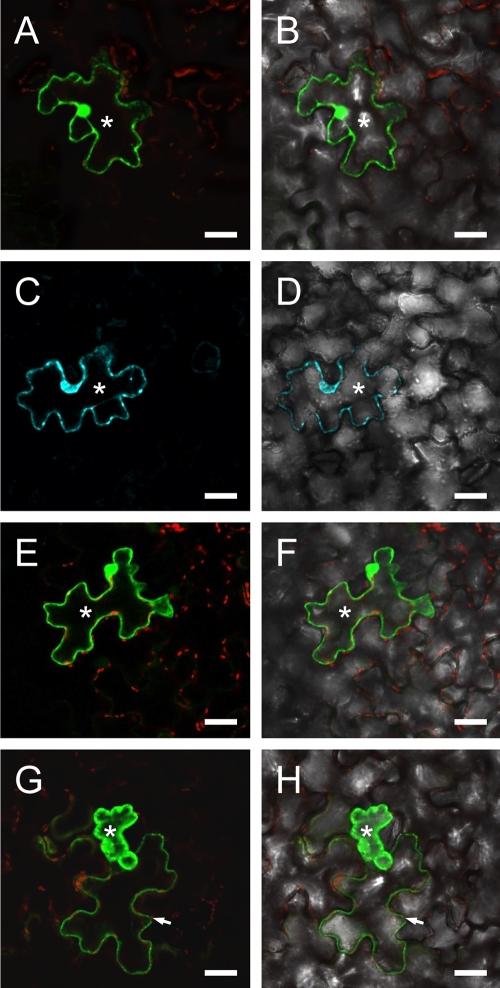
Fig. 9.
Nucleolar localization of GFP-CP fusion protein. Images show subcellular localization of GFP-CP fusion in N. benthamiana protoplasts (A to C) and epidermal cells (D to F) and Western blotting for CP and MP localization on nuclear extracts (G). (A) In protoplasts, GFP-CP shows the strongest fluorescent intensity in the nucleolus (arrowhead) although clear labeling is present in the whole nucleus. (B) A weaker signal is visible in the cytoplasm after the gain of the photo multiplier tube was increased. (C) Overlay of the image in panel A with the corresponding bright-field image to visualize cell boundaries. An analogous strong labeling of the nucleolus is visible in leaf epidermal cells. In this case, the GFP-CP construct was agroinfiltrated in transgenic plants expressing H2B-RFP. (D) GFP fluorescence, where strong nucleolar labeling is evident (arrowheads). (E) Nucleoli are also visible as dark unlabeled spots in the corresponding H2B-RFP image, due to the exclusion of H2B from the nucleolus. (F) Both fluorescent signals overlaid with the bright-field image of the same cells. n, nucleus. Bar 20 μm. (G) Western blot showing the presence of OuMV CP in a purified nuclear extract from OuMV-infected N. benthamiana plants. No MP is present in the extract, as demonstrated by the lack of labeling with both anti-MP and anti-Myc antibodies (ab) since a Myc-MP fusion was agroinfiltrated and used for the OuMV infection. Thermo Scientific Pierce Blue Prestained MW Marker (Pierce) is the molecular weight marker (MWM), and the sizes of the bands are expressed in thousands.
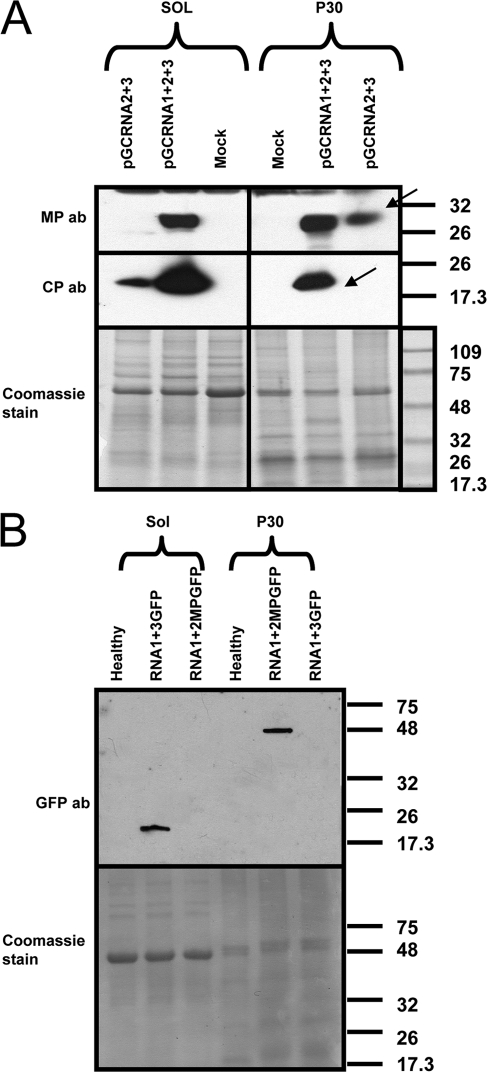
Fig. 10.
Distribution of OuMV CP and MP in subcellular fractions during active viral infection or coexpressed through agroinfiltration in the absence of RNA1. (A) Western blot of proteins from virus infection (pG-CRNA1+2+3) and coagroinfiltration of RNA2 and RNA3 (pGC-RNA2+3) in the absence of RNA1. Samples were processed at 4 dpi. Arrows indicate the specific CP and MP bands. ab, antibody. (B) Control for proper separation between cytosolic and noncytosolic fraction. GFP was expressed from pBin-GFP through agroinfiltration, and samples for Western blotting were processed at 3 dpi. GFP-MP is a fusion of GFP with the movement protein obtained from agroinfiltration of pBin-GFP-MP and processed at 3 dpi. Sol, cytosolic fraction; P30, noncytosolic, membranous fraction; mock, pBin61-infiltrated leaf; ab, antibody. Bottom panels show Coomassie staining of a replicate of the gel used for Western blotting. Thermo Scientific Pierce Blue Prestained MW Marker (Pierce) is the molecular weight marker (MWM), and the sizes of the bands are expressed in thousands.
Accumulation of MP and CP in cytosolic and noncytosolic fractions.
In order to validate the above localization observations, a cell fractionation protocol to separate soluble from membrane-bound proteins was applied to the virus-infected plants, and the CP and MP distribution in these fractions was analyzed. MP was predominantly found in the membrane fraction, whereas the CP accumulated preferentially in the soluble fraction (Fig. 10A); but both proteins were present also in the other fraction. In contrast, when the same separation protocol was applied to leaves expressing CP and MP not in a setting of a viral infection, but rather mixed through agroinfiltration in the absence of RNA1, the CP was detected only in the soluble fraction, and the MP was detected only in the membrane-enriched fraction (Fig. 10A). As a control for proper separation, GFP was found only in the soluble fraction (as expected), whereas a GFP-MP fusion was found only in the P30 fraction (Fig. 10B). In the separation protocol we used, nuclei (and all cellular organelles) are disposed through an initial low-speed centrifugation step (45).
DISCUSSION
Although discovered more than 30 years ago (35), ourmiaviruses have been carefully studied only recently (53). Surprisingly, sequence analysis of their genomes revealed no obvious relationship with any of the viruses whose sequences are deposited in publicly available databases, except for a very distant similarity in their RdRPs with those of members of the Narnaviridae, a heterogeneous family of viruses that does not include plant viruses. Furthermore, similarity was noted for the Ourmiavirus MP with those of the plant aureoviruses (53). Also, their virion morphology is unique among viruses characterized so far (35). The very distant relationships to plant viruses and their unique morphology prompted us to further characterize the details of their life cycle through a reverse genetic approach. We successfully assembled a set of full-length cDNA clones putatively transcribing in vivo each exact RNA genome segment, thanks to a 35S promoter and an HDV ribozyme to process the exact 3′ end. Agroinfiltration of a mix of the three clones efficiently reproduced an infection on N. benthamiana indistinguishable from that obtained by mechanical inoculation with the wild-type virus.
We found that OuMV local infections and necrotic local lesions can be induced only when both RNA1 and RNA2 are inoculated together, whereas either of the two genomic segments alone failed to infect plants and induce symptoms. We could rule out that this might have been due to the lack of accumulation of a functional protein in each of the two single transcript inoculations; in fact, when each of the two genome-expressing plasmids was inoculated alone, we observed direct evidence of MP accumulation for RNA2 (see Fig. S4 in the supplemental material), while for the RNA1-encoded ORF1 the accumulation of minus-strand RNA1 is indirect evidence for the accumulation of active replicase protein (Fig. 3). This suggests that direct or indirect interaction between the two proteins is indeed necessary to elicit necrotic local lesions on N. benthamiana. Furthermore, the lack of necrotic response when the coinfection was carried out with an RNA1 with a deleted 3′ UTR but which still allowed for replication of RNA2 suggests the hypothesis that necrotic response requires active replication of RNA1 (or a region in the 3′ UTR) and not simply accumulation of the RdRP and the MP together in the same cell. Necrotic response has been mapped to a single specific virus protein or an RNA domain, as in the classic case of p19 of the Tomato bushy stunt virus (TBSV) in Nicotiana tabacum (60) or in the 5′ UTR of the nepovirus Grapevine chrome mosaic virus (15). More complex models for necrotic response (particularly for a generalized lethal necrosis response), implying the requirements of more than one virus-encoded protein, have also been hypothesized (10, 38).
Arguably, the most interesting result from our study is the link between virus replication and virion assembly. Virion formation and RNA encapsidation have been studied in vitro in the Tomato mosaic virus (TMV) model since the early 1950s (16). Other virus models where the virus is able to self-assemble in vitro include the Brome mosaic virus (BMV), Turnip crinkle virus (TCV), Alfalfa mosaic virus (AMV), and Cowpea mosaic virus (CPMV) (22). Studies of virion formation in vivo are more recent, and new insights into virion in vivo assembly and packaging have been gained with the use of agroinfiltration (5). Within this context, very few model systems are under investigation, and detailed descriptions of plus-stranded isometric viruses have only recently been reviewed (52). To our knowledge, our study is the first to indicate that virion assembly of a nonenveloped plus-stranded RNA virus strictly requires the presence of RNA derived from replication (excluding RNA derived from transcription only). Coupling between replication and packaging in plus-strand RNA viruses was first shown to occur in the poliovirus (46). Two other animal viruses, flock house virus (FHV) and Kunjin virus (KUNV), were demonstrated to strictly require replication for proper packaging (28, 67). Among plant viruses, the best-studied model for packaging is the Brome mosaic virus (BMV), a tripartite plant virus where coupling between replication and packaging was also shown, with clear evidence for its role in properly selecting the encapsidated progeny RNA, excluding cellular RNA (4–6). Furthermore, a specific mutant of the N-terminal part of the CP can assemble in vivo, but not in vitro, and an interaction between the CP and the replicase was hypothesized (11). Nevertheless, for BMV, in vivo production of BMV CP and RNA3 indeed results in virion formation (5), whereas in the ourmiavirus model, virion formation itself occurs only with active replication of RNA3. In ourmiaviruses, active replication is therefore also required for the structural aspects of capsid assembly, which to our knowledge has not yet been described in nonenveloped plus-strand RNA viruses. A recent report that also hints at a role for replicase in packaging was published for the Turnip yellow mosaic virus (TYMV) (61), a virus species which cannot be assembled in vitro but requires the presence of replication vesicles induced in the outer chloroplast membrane (40).
Phylogenetic analysis has suggested that the RNA2-encoded protein could act as a movement protein (53). Furthermore, antibodies against the OuMV MP specifically reacted with virion-containing tubular structures found in the cytoplasm and across the cell walls (53). Here, we provide the first genetic evidence confirming the hypothesis that the RNA2-encoded protein is a movement protein. Lack of RNA2 in the transcript mix resulted in no visible infection foci on the inoculated leaf. Although RNA1 alone is replication competent, no symptoms developed on inoculated leaves, and it remained localized in inoculated cells. When coinoculated with RNA2, however, symptoms developed on the inoculated and distal leaves; thus, the RNA2-encoded MP facilitated cell and vascular transport of both RNA1 and RNA2 as well.
Confocal microscopy of GFP-MP fusion expression in the agroinfiltrated areas revealed that the MP is present in cytoplasmic connections across the plant cell walls; GFP-MP fusion expression also stimulates the formation of plasma membrane tubular protrusions in protoplasts derived from the agroinfiltrated area. This localization of GFP-MP in vivo is consistent with previous descriptions of tubule formation stimulated by the expression of the MP in other virus model systems (19, 26, 27, 54, 63).
Taking together the results from the genetic analysis and MP localization, we can outline a model where cell-to-cell virus movement is realized efficiently as virus particles, as previously hypothesized from electron microscopy observations (53). The CP is not strictly necessary for cell-to-cell or long-distance movement since in the presence of RNA1 and RNA2 mixed inoculation, we could detect localized infection in the inoculated and upper uninoculated leaves; however, these infections failed to develop fully in the upper leaves. In this model, the RNA3-encoded CP (likely assembled as virions) aids in efficient invasion of the mesophyll, possibly interfering with the plant antiviral defense systems (sensu latu) or in specific steps involving barriers between leaf tissues. Furthermore, the formation of membranous tubules in protoplasts expressing GFP-MP suggests that cell-to-cell movement occurs through tubules passing within highly modified plasmodesmata, as occurs with several other RNA and DNA viruses (26, 27, 54, 63). This is in apparent contrast with the phylogenetic similarity of the OuMV MP with the MPs of the aureoviruses, a group of plant viruses devoid of tubule formation and in which the CP is dispensable for cell-to-cell movement. This apparent paradox should be viewed in the context of studies which have found that different types of MPs can complement each other across functionally classified groups (2, 33, 44, 62). To our knowledge, the Ourmiavirus model would be the first tubule-forming virus that does not strictly require the CP for cell-to-cell movement and therefore shares properties with non-tubule-forming plant virus models. The fact that the CP is not strictly required for long-distance movement in vascular tissue is another feature shared by a few plant virus groups. But this property is often host specific: viruses having this property occur among the tombusviruses (14, 55, 59), the dianthoviruses (69), the hordeiviruses (23), the umbraviruses (57), the tobraviruses (64), and the pomoviruses (41). Although our data show that the CP of ourmiaviruses is not strictly required for vascular movement, its role in rendering vascular movement more efficient cannot be excluded. Further work is needed to elucidate the histological details of movement and replication in different plant tissues and organs. In addition, we cannot exclude a possible specific role of the CP in overcoming specific barriers to movement, such as phloem uploading or unloading (17), since agroinoculation itself could be a factor in overcoming such barriers.
Another interesting result here is the nucleolar localization of the OuMV CP. There is a growing body of evidence (21) for the nuclear localization of proteins encoded by plus-stranded RNA viruses (in both animals and plants) that replicate in the cytoplasm; in some cases, such viruses were demonstrated not to require a nuclear phase for their replication (21). Studies on RNA plant viruses have reported the presence of virus-encoded proteins in the nucleus: in the Cucumber mosaic virus (CMV), the silencing suppressor protein 2b displayed nuclear localization (36); the Tombusvirus p19 is also translocated by plant ALY proteins to the nucleus (13). In other RNA plant virus systems, such as sobemovirus, umbravirus, potyvirus, luteoviruses, and the Satellite panicum mosaic virus (SPMV), the CP itself is localized to the nucleus (20, 30, 47, 49, 56). To date, the function of nucleolar localization has been shown in only two systems: in umbraviruses, ORF3 localization to the nucleolus was described as being necessary for long-distance virus spread (31). In this system, the role of fibrillarin, a nuclear protein, was found to be important for the assembly of RNP particles which are transported systemically (12, 29). Our genetic analyses seem to exclude the role of the CP in long-distance movement in ourmiaviruses, at least in N. benthamiana. A second system where the role of nucleolar localization was demonstrated is the potyvirus Potato virus A (PVA), where nuclear inclusion protein a (NIa) localization to the nucleus and the nucleolus was reported to be relevant for several essential steps in the viral life cycle (51). An area of further work is the mapping of subdomains in both the MP and CP necessary for their subcellular localization and functionality. A bioinformatic approach (http://psort.hgc.jp/) showed the presence of putative nuclear localization signals in the CP sequence, which will be validated by a mutagenesis approach in future work.
In this study we also show the different separation profiles of the MP and the CP into soluble and membrane-bound fractions depending on their expression within the context of a viral infection or simply thorough coexpression in the same cell without RNA1. In OuMV-infected plants, MP and CP were found in both the membranous and soluble fractions. If active replication does not occur, such as upon agroinfiltration and transient expression, both proteins were present in only one of the two fractions. The different affinities for the differential centrifugation fractions do not imply different subcellular localizations. Further in vivo subcellular localization experiments investigating the interaction between the two proteins and the replicase are needed. A well-studied example of different subcellular localizations (and affinity for subcellular fractions) of a specific viral protein is that of the SPMV CP (49). In this system, only the whole virion is cytosolic, whereas mutations inhibiting virion formation induce CP accumulation in various other compartments, including the cell wall, nuclei, and membranes.
Overall, the compact genome of OuMV reveals a number of unique features, and a reverse genetic system in plants is a necessary preliminary step to attempt its replication in yeasts, given its similarity to yeast viruses.
Supplementary Material
ACKNOWLEDGMENTS
Giulia Crivelli was supported by a Ph.D. scholarship from the Italian Ministry of the University and Scientific Research.
We are grateful to Riccardo Lenzi and Caterina Perrone for their technical assistance and to Bryce W. Falk and J. Burgyan for reading the manuscript. Seeds of transgenic N. benthamiana plants expressing RFP-ER and H2B-RFP were a generous gift of M. M. Goodin.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 16 March 2011.
In memoriam G. Crivelli.
REFERENCES
- 1. Accotto G. P., Milne R. G. 2008. Ourmiavirus, p. 500–501In Mahy B. W., Van Regenmortel M. H. V.(ed.), Encyclopedia of virology. Elsevier, London, United Kingdom [Google Scholar]
- 2. Agranovsky A. A., et al. 1998. Beet yellows closterovirus HSP70-like protein mediates the cell-to-cell movement of a potexvirus transport-deficient mutant and a hordeivirus-based chimeric virus. J. Gen. Virol. 79:889–895 [DOI] [PubMed] [Google Scholar]
- 3. Aiton M. M., Lennon A. M., Roberts I. M., Harrison B. D. 1988. Two new cassava viruses from Africa, p. 43.In Proceedings of the 5th International Congress of Plant Pathology, Kyoto, Japan Phytopathological Society of Japan, Tokyo, Japan [Google Scholar]
- 4. Annamalai P., Rao A. L. N. 2006. Packaging of brome mosaic virus subgenomic RNA is functionally coupled to replication-dependent transcription and translation of coat protein. J. Virol. 80:10096–10108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Annamalai P., Rao A. L. N. 2005. Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: a functional role for viral replicase in RNA packaging. Virology 338:96–111 [DOI] [PubMed] [Google Scholar]
- 6. Annamalai P., Rofail F., DeMason D. A., Rao A. L. N. 2008. Replication-coupled packaging mechanism in positive-strand RNA viruses: synchronized coexpression of functional multigenome RNA components of an animal and a plant virus in Nicotiana benthamiana cells by agroinfiltration. J. Virol. 82:1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Avgelis A., Barba M., Rumbos I. 1989. Epirus cherry virus, an unusual virus isolated from cherry with rasp-leaf symptoms in Greece. J. Phytopathol. 126:51–58 [Google Scholar]
- 8. Bendahmane A., Farnham G., Moffett P., Baulcombe D. C. 2002. Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 32:195–204 [DOI] [PubMed] [Google Scholar]
- 9. Bendahmane A., Kanyuka K., Baulcombe D. C. 1999. The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11:781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burgyan J., Hornyik C., Szittya G., Silhavy D., Bisztray G. 2000. The ORF1 products of tombusviruses play a crucial role in lethal necrosis of virus-infected plants. J. Virol. 74:10873–10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Calhoun S. L., Speir J. A., Rao A. L. N. 2007. In vivo particle polymorphism results from deletion of a N-terminal peptide molecular switch in Brome mosaic virus capsid protein. Virology 364:407–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Canetta E., et al. 2008. A plant virus movement protein forms ringlike complexes with the major nucleolar protein, fibrillarin, in vitro. J. Mol. Biol. 376:932–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Canto T., Uhrig J. F., Swanson M., Wright K. M., MacFarlane S. A. 2006. Translocation of Tomato bushy stunt virus P19 protein into the nucleus by ALY proteins compromises its silencing suppressor activity. J. Virol. 80:9064–9072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dalmay T., Rubino L., Burgyan J., Russo M. 1992. Replication and movement of a coat protein mutant of cymbidium ringspot tombusvirus. Mol. Plant Microbe Interact. 5:379–383 [DOI] [PubMed] [Google Scholar]
- 15. Fernandez I., Candresse T., Le Gall O., Dunez J. 1999. The 5′ noncoding region of grapevine chrome mosaic nepovirus RNA-2 triggers a necrotic response on three Nicotiana spp. Mol. Plant Microbe Interact. 12:337–344 [DOI] [PubMed] [Google Scholar]
- 16. Fraenkel-Conrat H., Williams R. 1955. Reconstitution of active tobacco mosaic virus from its inactive protein and nucleic acid components. Proc. Natl. Acad. Sci. U. S. A. 41:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gilbertson R. L., Lucas W. J. 1996. How do viruses traffic on the “vascular highway”? Trends Plant Sci. 1:260–268 [Google Scholar]
- 18. Goodin M. M., Chakrabarty R., Banerjee R., Yelton S., DeBolt S. 2007. New gateways to discovery. Plant Physiol. 145:1100–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gopinath K., et al. 2003. Intracellular distribution of cowpea mosaic virus movement protein as visualised by green fluorescent protein fusions. Arch. Virol. 148:2099–2114 [DOI] [PubMed] [Google Scholar]
- 20. Haupt S., et al. 2005. Nucleolar localization of potato leafroll virus capsid proteins. J. Gen. Virol. 86:2891–2896 [DOI] [PubMed] [Google Scholar]
- 21. Hiscox J. A. 2007. RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 5:119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hull R. 2002. Assembly of icosahedral viruses, p. 163–168In Hull R. (ed.), Matthews' plant virology. Academic Press, San Diego, CA [Google Scholar]
- 23. Jackson A. O., Petty I. T. D., Jones R. W., Edwards M. C., French R. 1991. Molecular genetic-analysis of barley stripe mosaic-virus pathogenicity determinants. Can. J. Plant Pathol. 13:163–177 [Google Scholar]
- 24. Jones R. W., Jackson A. O., Morris T. J. 1990. Defective-interfering RNAs and elevated-temperatures inhibit replication of Tomato bushy stunt virus in inoculated protoplasts. Virology 176:539–545 [DOI] [PubMed] [Google Scholar]
- 25. Jupin I., Richards K., Jonard G., Guilley H., Pleij C. W. A. 1990. Mapping sequences required for productive replication of Beet necrotic yellow vein virus rna-3. Virology 178:273–280 [DOI] [PubMed] [Google Scholar]
- 26. Kasteel D. T. J., et al. 1996. The movement proteins of Cowpea mosaic virus and Cauliflower mosaic virus induce tubular structures in plant and insect cells. J. Gen. Virol. 77:2857–2864 [DOI] [PubMed] [Google Scholar]
- 27. Kasteel D. T. J., N. vanderWel N., Jansen K. A. J., Goldbach R. W., M. vanLent J. W. 1997. Tubule-forming capacity of the movement proteins of Alfalfa mosaic virus and Brome mosaic virus. J. Gen. Virol. 78:2089–2093 [DOI] [PubMed] [Google Scholar]
- 28. Khromykh A. A., Varnavski A. N., Sedlak P. L., Westaway E. G. 2001. Coupling between replication and packaging of flavivirus RNA: evidence derived from the use of DNA-based full-length cDNA clones of Kunjin virus. J. Virol. 75:4633–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim S. H., et al. 2007. Interaction of a plant virus-encoded protein with the major nucleolar protein fibrillarin is required for systemic virus infection. Proc. Natl. Acad. Sci. U. S. A. 104:11115–11120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim S. H., Ryabov E. V., Brown J. W. S., Taliansky M. 2004. Involvement of the nucleolus in plant virus systemic infection. Biochem. Soc. Trans. 32:557–560 [DOI] [PubMed] [Google Scholar]
- 31. Kim S. H., et al. 2007. Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J. 26:2169–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kristensen D. M., Mushegian A. R., Dolja V. V., Koonin E. V. 2010. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 18:11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lewandowski D. J., Adkins S. 2005. The tubule-forming NSm protein from Tomato spotted wilt virus complements cell-to-cell and long-distance movement of Tobacco mosaic virus hybrids. Virology 342:26–37 [DOI] [PubMed] [Google Scholar]
- 34. Lindbo J. A. 2007. High-efficiency protein expression in plants from agroinfection-compatible Tobacco mosaic virus expression vectors. BMC Biotechnol. 7:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lisa V., et al. 1988. Ourmia melon virus, a virus from Iran with novel properties. Ann. App. Biol. 112:291–302 [Google Scholar]
- 36. Lucy A. P., Guo H. S., Li W. X., Ding S. W. 2000. Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J. 19:1672–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Margaria P., Ciuffo M., Pacifico D., Turina M. 2007. Evidence that the nonstructural protein of Tomato spotted wilt virus is the avirulence determinant in the interaction with resistant pepper carrying the Tsw gene. Mol. Plant Microbe Interact. 20:547–558 [DOI] [PubMed] [Google Scholar]
- 38. Margaria P., Pacifico D., Ciuffo M., Turina M. 2008. The small genomic segment of Tomato spotted wilt virus isolate Br20 is necessary but not sufficient to induce lethal necrosis in Nicotiana benthamiana and local necrotic lesions in Nicotiana tabacum cv. White Burley. J. Plant Pathol. 90:337–343 [Google Scholar]
- 39. Martin K., et al. 2009. Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 59:150–162 [DOI] [PubMed] [Google Scholar]
- 40. Matthews R. E. F. 1991. Plant virology. Academic Press, San Diego, CA [Google Scholar]
- 41. McGeachy K. D., Barker H. 2000. Potato mop-top virus RNA can move long distance in the absence of coat protein: evidence from resistant, transgenic plants. Mol. Plant Microbe Interact. 13:125–128 [DOI] [PubMed] [Google Scholar]
- 42. Milne R. G. 2005. Ourmiavirus. In Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA [Google Scholar]
- 43. Milne R. G. 1993. Solid-phase immuno electron microscopy of virus preparations, p. 25–70In Eaton A. D. H. B. T. (ed.), Immuno-gold electron microscopy in virus diagnosis and research. CRC Press, Boca Raton, FL [Google Scholar]
- 44. Morozov S. Y., et al. 1997. Complementation of a potato virus X mutant mediated by bombardment of plant tissues with cloned viral movement protein genes. J. Gen. Virol. 78:2077–2083 [DOI] [PubMed] [Google Scholar]
- 45. Navarro B., Rubino L., Russo M. 2004. Expression of the Cymbidium ringspot virus 33-kilodalton protein in Saccharomyces cerevisiae and molecular dissection of the peroxisomal targeting signal. J. Virol. 78:4744–4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nugent C. I., Johnson K. L., Sarnow P., Kirkegaard K. 1999. Functional coupling between replication and packaging of poliovirus replicon RNA. J. Virol. 73:427–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Olspert A., Paves H., Toomela R., Tamm T., Truve E. 2010. Cocksfoot mottle sobemovirus coat protein contains two nuclear localization signals. Virus Genes. 40:423–431 [DOI] [PubMed] [Google Scholar]
- 48. Pendle A. F., et al. 2005. Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol. Biol. Cell 16:260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Qi D., Omarov R. T., Scholthof K. B. G. 2008. The complex subcellular distribution of Satellite panicum mosaic virus capsid protein reflects its multifunctional role during infection. Virology 376:154–164 [DOI] [PubMed] [Google Scholar]
- 50. Qiu W. P., Scholthof K. B. G. 2001. Genetic identification of multiple biological roles associated with the capsid protein of satellite panicum mosaic virus. Mol. Plant Microbe Interact. 14:21–30 [DOI] [PubMed] [Google Scholar]
- 51. Rajamaki M. L., Valkonen J. P. T. 2009. Control of nuclear and nucleolar localization of nuclear inclusion protein a of picorna-like Potato virus A in Nicotiana species. Plant Cell 21:2485–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rao A. L. N. 2006. Genome packaging by spherical plant RNA viruses. Annu. Rev. Phytopathol. 44:61–87 [DOI] [PubMed] [Google Scholar]
- 53. Rastgou M., et al. 2009. Molecular characterization of the plant virus genus Ourmiavirus and evidence of inter-kingdom reassortment of viral genome segments as its possible route of origin. J. Gen. Virol. 90:2525–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ritzenthaler C., Schmit A. C., Michler P., Stussigaraud C., Pinck L. 1995. Grapevine fanleaf nepovirus p38 putative movement protein is located on tubules in-vivo. Mol. Plant Microbe Interact. 8:379–387 [Google Scholar]
- 55. Rochon D. M., Johnston J. C., Riviere C. J. 1991. Molecular analysis of the cucumber necrosis virus genome. Can. J. Plant Pathol. 13:142–154 [Google Scholar]
- 56. Ryabov E. V., Kim S. H., Taliansky M. 2004. Identification of a nuclear localization signal and nuclear export signal of the umbraviral long-distance RNA movement protein. J. Gen. Virol. 85:1329–1333 [DOI] [PubMed] [Google Scholar]
- 57. Ryabov E. V., Robinson D. J., Taliansky M. E. 1999. A plant virus-encoded protein facilitates long-distance movement of heterologous viral RNA. Proc. Natl. Acad. Sci. U. S. A. 96:1212–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Scholthof H. B. 1999. Rapid delivery of foreign genes into plants by direct rub-inoculation with intact plasmid DNA of a Tomato busby stunt virus gene vector. J. Virol. 73:7823–7829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Scholthof H. B., Morris T. J., Jackson A. O. 1993. The capsid protein gene of tomato bushy stunt virus is dispensable for systemic movement and can be replaced for localized expression of foreign genes. Mol. Plant Microbe Interact. 6:309–322 [Google Scholar]
- 60. Scholthof H. B., Scholthof K. B. G., Jackson A. O. 1995. Identification of Tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a potato-virus-x vector. Plant Cell 7:1157–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shin H. I., Kim H. Y., Cho T. J. 2010. The Pro/Hel region is indispensable for packaging non-replicating Turnip yellow mosaic virus RNA, but not replicating viral RNA. Mol. Cells 29:463–469 [DOI] [PubMed] [Google Scholar]
- 62. Solovyev A. G., et al. 1997. Host-controlled cell-to-cell movement of a hybrid Barley stripe mosaic virus expressing a dianthovirus movement protein. Intervirology 40:1–6 [DOI] [PubMed] [Google Scholar]
- 63. Storms M. M. H., Kormelink R., Peters D., vanLent J. W. M., Goldbach R. W. 1995. The nonstructural NSm protein of Tomato spotted wilt virus induces tubular structures in plant and insect cells. Virology 214:485–493 [DOI] [PubMed] [Google Scholar]
- 64. Swanson M., Barker H., MacFarlane S. A. 2002. Rapid vascular movement of tobraviruses does not require coat protein: evidence from mutated and wild-type viruses. Ann. Appl. Biol. 141:259–266 [Google Scholar]
- 65. Turina M., et al. 2006. Characterization of four viral species belonging to the family Potyviridae isolated from Ranunculus asiaticus. Phytopathology 96:560–566 [DOI] [PubMed] [Google Scholar]
- 66. Turina M., Maruoka M., Monis J., Jackson A. O., Scholthof K. B. G. 1998. Nucleotide sequence and infectivity of a full-length cDNA clone of Panicum mosaic virus. Virology 241:141–155 [DOI] [PubMed] [Google Scholar]
- 67. Venter P. A., Krishna N. K., Schneemann A. 2005. Capsid protein synthesis from replicating RNA directs specific packaging of the genome of a multipartite, positive-strand RNA virus. J. Virol. 79:6239–6248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang J. B., Turina M., Stewart L. R., Lindbo J. A., Falk B. W. 2009. Agroinoculation of the Crinivirus, Lettuce infectious yellows virus, for systemic plant infection. Virology 392:131–136 [DOI] [PubMed] [Google Scholar]
- 69. Xiong Z., Kim K. H., Giesmancookmeyer D., Lommel S. A. 1993. The roles of the Red-clover necrotic mosaic virus capsid and cell-to-cell movement proteins in systemic infection. Virology 192:27–32 [DOI] [PubMed] [Google Scholar]
- 70. Yeh H. H., Tian T. Y., Medina V., Falk B. W. 2001. Green fluorescent protein expression from recombinant lettuce infectious yellows virus-defective RNAs originating from RNA 2. Virology 289:54–62 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.