Abstract
Ranaviruses (family Iridoviridae, genus Ranavirus) are large, double-stranded DNA (dsDNA) viruses whose replication is restricted to ectothermic vertebrates. Many highly pathogenic members of the genus Ranavirus encode a homologue of the eukaryotic translation initiation factor 2α (eIF2α). Data in a heterologous vaccinia virus system suggest that the Ambystoma tigrinum virus (ATV) eIF2α homologue (vIF2αH; open reading frame [ORF] 57R) is involved in evading the host innate immune response by degrading the interferon-inducible, dsRNA-activated protein kinase, PKR. To test this hypothesis directly, the ATV vIF2αH gene (ORF 57R) was deleted by homologous recombination, and a selectable marker was inserted in its place. The ATVΔ57R virus has a small plaque phenotype and is 8-fold more sensitive to interferon than wild-type ATV (wtATV). Infection of fish cells with the ATVΔ57R virus leads to eIF2α phosphorylation, in contrast to infection with wtATV, which actively inhibits eIF2α phosphorylation. The inability of ATVΔ57R to prevent phosphorylation of eIF2α correlates with degradation of fish PKZ, an interferon-inducible enzyme that is closely related to mammalian PKR. In addition, salamanders infected with ATVΔ57R displayed an increased time to death compared to that of wtATV-infected salamanders. Therefore, in a biologically relevant system, the ATV vIF2αH gene acts as an innate immune evasion factor, thereby enhancing virus pathogenesis.
INTRODUCTION
Ranaviruses (family Iridoviridae, genus Ranavirus) are large, double-stranded DNA (dsDNA) viruses that can infect a wide variety of ectothermic vertebrates, including amphibians, reptiles, and fish (13, 70). However, the ecological and economical impacts of ranavirus infections are currently unknown, even though ranavirus infections of lower vertebrates have increased over the past few decades (13, 70). In addition, the mechanisms that enable ranaviruses to infect such a diverse group of hosts and cause, in some cases, high rates of morbidity and mortality have not been fully uncovered. While there are major epidemics associated with ranavirus infections in threatened amphibian species, commercially valuable fish, and reptiles (1–3, 8, 15, 17–19, 22, 23, 26, 28, 31, 33, 34, 40, 42–45, 52, 63, 64, 73, 75), ranaviruses are also thought to spread with fish, amphibians, and reptiles that are moved globally for bait and food and as pets (11, 15, 29, 49, 60). Because of the economical and ecological impact that these viruses have on ectothermic vertebrates, it is essential to begin to uncover the determinants of ranavirus host range and pathogenesis.
Genomic sequencing of Ambystoma tigrinum virus (ATV), originally isolated from tiger salamanders (Ambystoma tigrinum stebbinsi) in southern Arizona (28), revealed a number of genes that may enhance viral pathogenesis based on homology to other known proteins in the database (30). One gene in particular, the ATV homologue of the eukaryotic translation initiation factor 2α (vIF2αH; ATV open reading frame [ORF] 57R), has been suggested to be important for ranavirus pathogenesis (40). In addition, we have recently shown using a heterologous vaccinia virus (VACV) system that the ATV vIF2αH may play an important role in evading the host innate immune response (L. Tripuraneni, J. K. Jankovich, M. C. Heck, J. O. Langland, and B. L. Jacobs, unpublished data). By replacing the VACV innate immune evasion gene, E3L (9, 10, 16, 36, 37, 55, 61, 62), with the ATV vIF2αH gene, there is a rapid degradation of cellular PKR, an important cellular antiviral molecule that upon activation phosphorylates the eukaryotic translation initiation factor eIF2α (38, 59), thereby inhibiting protein synthesis. Therefore, we hypothesized that in a more relevant system (i.e., ATV-salamander model system), the ATV vIF2αH gene may influence viral pathogenesis by evading the host innate immune response (i.e., degrading cellular PKR). Using methods similar to those described for generating a recombinant Bohle iridovirus (47), we have generated a knockout recombinant ATV by deleting the ATV ORF 57R and then characterized this recombinant virus in cells in culture and in a salamander model. This research offers insight into a ranavirus immune evasion pathway and suggests that there is a yet-uncharacterized innate immune response in salamanders, the natural host of ATV.
MATERIALS AND METHODS
Cells and virus.
Fathead minnow (FHM; ATCC CCL-42) cells were maintained in minimal essential medium (MEM) with Hank's salts (Gibco) supplemented with 5% fetal bovine serum (FBS) (HyClone) and 0.1 mM nonessential amino acids and vitamins (Invitrogen). Epithelioma papulosum cyprini (20) and bluegill fry (BF2; ATCC CCL-91) cells were maintained in MEM supplemented with 10% FBS and 0.1 mM nonessential amino acids and vitamins (Invitrogen). All cells were incubated at 20 to 22°C in the presence of 5% CO2. The viruses used in this study are the Ambystoma tigrinum virus (wild-type ATV [wtATV]), isolated from tiger salamanders in 1996 (28), and the recombinant virus made in this study. The ATV deletion mutant lacking the vIF2αH gene (ORF 57R) was designated ATVΔ57R.
Recombinant ATV DNA construct and PCRs.
The PCR construct used in the in vivo recombination (IVR) reaction mixture contains a 200-bp region upstream of the ATV immediate early 18K gene (pICP18) that promotes the expression of the G418 (neomycin) resistance gene (G418r). This cassette is flanked by 1.2 kbp of sequence homologous to the 5′ (left) and 3′ (right) regions surrounding the ATV vIF2αH gene. The homologous flanking arms recombine with the viral DNA, generating the recombinant virus (Fig. 1A). The following primers were used to amplify the above-mentioned regions of the ATV genome and the G418 resistance gene: vIF2-Left-forward (5′ ATTTACCCAAAAATTGCGTTTC 3′), vIF2-Left-reverse (5′ ATTTCCATATAACAGACAGTAG 3′), vIF2-Right-forward (5′ TGAAAAAAGCTCTATCGAGCAG 3′), vIF2-Right-reverse (5′ TCTCTCACGTTGAGGATAAAG 3′), G418r-forward (5′ ATGAGGATCGTTTCGCATGATTG 3′), G418r-reverse (5′ TCAGAAGAACTCGTCAAG 3′), pICP18-forward (5′ AACTAGGTCCGCCGATGAGC 3′), pICP-18-reverse (5′ GCTCATCGGCGGACCTAGTT 3′). Once individual PCR products were obtained using the forward-reverse primer sets, overlapping PCR products were then generated. The overlapping IVR PCR product was generated by first combining the pICP-18 PCR product with the G418r PCR product and adding primers pICP-18-forward and G418r-reverse to generate a 1.1-kb PCR product. This PCR product was then mixed with the 1.2-kb vIF2-left and vIF2-right PCR products and the PCR primers vIF2-Left-reverse, pICP18-forward, G418r-reverse, and vIF2-Right-reverse were added. This generated a 3.5-kb PCR product cassette (pICP18-G418r), and this product was blunt-end cloned into a pUC19-based plasmid (Invitrogen) that was renamed pJJ57. This plasmid was then used as a template to PCR amplify large quantities of PCR product used in the IVR reactions (see below) as well as for diagnostic PCRs. The generation of a recombinant ATV was confirmed by PCR amplification of the 57R locus using flanking primers 57R-forward (5′ GAGGTATATTTTTGCAAGG 3′) and 57R-reverse (5′ TCTCAAACCTTTCCAATCG 3′). For amplification of 0.5 kb of the major capsid protein (MCP), MCP4 and MCP5 primers (5′ GACTTGGCCACTTATGAC 3′ and 5′ GTCTCTGGAGAAGAAGAA 3′, respectively) developed by Mao et al. (41) were used. All PCRs described above were performed using the following PCR conditions: 50-μl reaction mixtures containing 100 ng template DNA, 0.3 μM each primer, 0.2 mM deoxynucleoside triphosphate (dNTP) (total concentration of all four nucleotides), 1 unit of Taq polymerase in 1× buffer B (Promega), and 1.5 mM MgCl2. Amplification cycles were initiated with a single cycle of 94°C for 5 min, followed by 30 cycles of 94°C (30 s), 54°C (60 s), and 72°C (60 s) and a final cycle of 72°C for 5 min. PCR products were visualized by electrophoresis on 0.8% agarose gels stained with 0.5 μg/ml ethidium bromide. All PCR products were isolated using the Wizard SV gel and PCR cleanup system according to the manufacturer's instructions (Promega).
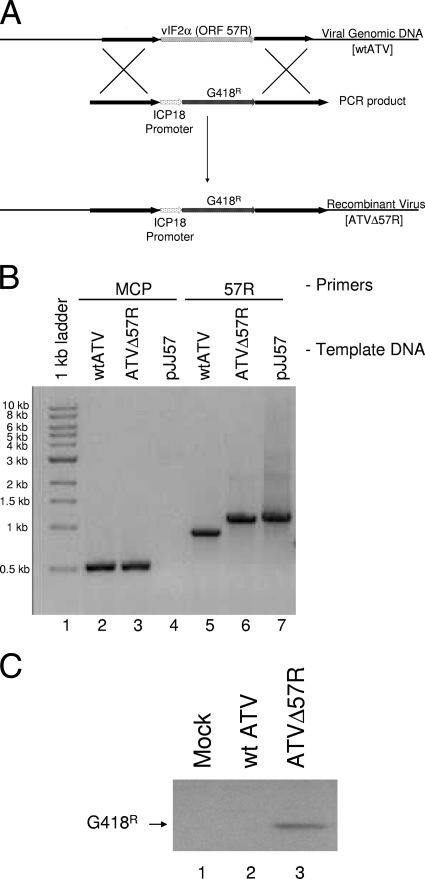
Fig. 1.
Generating a recombinant ATV. (A) ATV recombination schematic. A PCR product containing the ICP18 promoter sequence driving G418 (neomycin) resistance (G418r) was generated. This cassette was then flanked with 1,200 nucleotides of sequence up- and downstream of the ATV ORF 57R. This PCR construct was blunt-end cloned into a pUC19-based plasmid and renamed pJJ57. The plasmid was used as a template to generate linear PCR products that were transfected into wtATV-infected cells. Homologous recombination between the ATV genome and the PCR product deleted ORF 57R and replaced this ATV gene with the ICP18-G418r cassette. (B) Confirmation of recombinant ATV: PCR. To confirm that the ATV ORF 57R was deleted and replaced with the pICP18-G418r cassette, primers flanking ORF 57R were used to amplify this region of viral DNA (57R; lanes 5 to 7). pJJ57 is a plasmid containing the recombination PCR product (see Fig. 1A legend) and is used as a control. Primers specific for 0.5 kb of the ranavirus major capsid protein PCR were added as another control (MCP; lanes 1 to 4). (C) Confirmation of recombinant ATV: Western blot analysis. FHM cells were infected with wtATV or ATVΔ57R at an MOI of 5 or mock infected with medium alone. After 3 hpi, cells were harvested and proteins were isolated as described in Materials and Methods. Equal cell volumes of protein were separated by 12% SDS-PAGE, after which proteins were transferred and probed using antibodies specific to the G418-phosphotase (G418r) protein. Antibodies binding to the G418r protein were visualized by chemiluminescence.
ATV in vivo recombination.
The following protocol was adapted from Pallister et al. (47) and used successfully to knock out the ATV vIF2αH gene. Homologous recombination between PCR DNA and ATV DNA will result in a recombinant ATV in which the vIF2αH gene (ORF 57R) has been replaced by the G418r gene (Fig. 1A). Lightly confluent monolayers of BF2 cells grown in 35-mm dishes were infected with wtATV at a multiplicity of infection (MOI) of 10, and the virus was allowed to adsorb for 1 h at room temperature. While viral attachment was proceeding, 500 ng of ATV recombination PCR DNA was added to FuGene6 transfection reagent (Roche Diagnostics) according to the manufacturer's instructions. The solutions (i.e., DNA and the FuGENE6 transfection reagent mix) were incubated for 25 min at room temperature. At that time, DNA-FuGENE6 complexes were added to the infected cells and incubated at 20 to 22°C for 48 to 72 h. The cultures were then harvested and frozen at −80°C, and the virus was subsequently released by three cycles of freeze-thaw. The IVR was passaged up to four times in FHM cells in the presence of 1 mg/ml G418. Moreover, virus was plaque purified three times in the presence of G418 and then twice in the absence of G418. To confirm generation of a knockout virus, PCR and sequencing of the ATV ORF 57R region were performed (see above). In addition, Western blot analysis was performed to ensure that the G418 resistance gene was synthesized (see below).
Plaque size determination.
Plaque size phenotypes of both wtATV and ATVΔ57R were determined using the ImageQuant 5.2 software (GE Healthcare). FHM cells were infected with wtATV and ATVΔ57R. Once plaques were observed, the plates were stained with a 20% ethanol and 1% crystal violet solution and photographed, and the images were loaded into the ImageQuant program. Initially, plaques were indicated as clear loci in a monolayer of blue-stained cells. To determine plaque sizes, the images were inverted so that each plaque was dark relative to the monolayer. Ten plaques for each virus were used to determine plaque size by the relative intensity of the pixels in each plaque. Using this approach, greater pixel intensity is indicative of a larger plaque size. The pixel intensities of the 10 plaques were averaged, and the standard error was determined.
Interferon-like activity.
Interferon (IFN)-like activity was produced in FHM cells by treating them with dsRNA, as described by others (46, 58). Briefly, 250 μg of poly(I:C) was filter sterilized by passing through a 0.2-μm filter and mixed with 350 μl of FHM medium and 10.5 μl (3% final concentration) Fugene6 transfection reagent (Roche Applied Sciences). Control transfections contained no poly(I:C). This mixture was incubated at room temperature (20 to 22°C) for 20 min before being added to <50% confluent monolayers of FHM cells in a 75-cm2 tissue culture flask. The transfection mixture was rocked for 1 h, at which time 9.5 ml of medium was added. Poly(I:C)-treated and untreated FHM cells were incubated at room temperature for 24 h. After the incubation period, the medium was removed, centrifuged at 1,000 × g for 10 min to remove any cellular debris, and then frozen at −80°C until assayed for activity. IFN-like activity (here referred to as IFN) was measured by plaque reduction assay. The concentration of IFN that inhibits 50% of wtATV plaque formation is defined as 1 FHM unit of IFN. To assay for IFN activity, FHM IFN was 2-fold serially diluted and 1 ml of each IFN dilution was added to 35-mm dishes containing approximately 50% confluent monolayers of FHM cells. Mock-transfected cell medium was used as a control in these experiments. After 16 to 24 h of treatment, the IFN was removed and FHM cells were infected with ∼200 PFU of wtATV or ATVΔ57R or mock infected with medium alone. Cells were rocked for 1 h and overlaid with a 1:1 mixture of 2× MEM with 20% FBS and 1.5% methylcellulose solution. Cells were incubated at room temperature in 5% CO2 for 6 to 8 days, at which time the medium-methylcellulose mixture was removed by aspiration and cells were stained with a 20% ethanol and 1% crystal violet solution. Plaques were counted, and the percent plaque reduction was determined by dividing the number of plaques for a particular dose of IFN by the number of plaques in the mock-treated cells. Data are representative of results from multiple independent experiments.
Single-cycle growth characteristics.
FHM cells were seeded in 35-mm dishes and were pretreated the following day with 1 FHM U/ml of IFN or left untreated. After a 16- to 24-h incubation period, the cells were infected with wtATV or ATVΔ57R at an MOI of 5. Infected cells were harvested at 6 and 48 h postinfection (hpi). The titer was determined at each time point by using a standard plaque assay in E. papulosum cyprini cells and crystal violet staining. Virus yield was calculated by subtracting the final virus titer (i.e., 48 hpi) from the starting titer (i.e., 6 hpi). Data are the averages of results from multiple independent experiments. Statistical analysis was performed using the single-factor analysis of variance (ANOVA) option in SigmaStat (Systat Software, Inc.).
Cell extractions and Western blot analysis.
Mock-infected FHM cells or cells infected with either wtATV or ATVΔ57R at an MOI of 5 were collected at various times postinfection and pelleted by centrifugation at 1,000 × g at 4°C for 10 min. The cells were then lysed using 50 μl of NP-40 lysis buffer (20 mM HEPES [pH 7.5], 120 mM KCl, 5 mM Mg acetate, 1 mM dithiothreitol [DTT], 10% [vol/vol] glycerol, 0.5% NP-40). Equal cell volumes of cellular extracts were subjected to SDS-PAGE on 12% polyacrylamide gels for 1 h at 150 V. Proteins were transferred to nitrocellulose at 100 V for 45 min in 10 mM CAPS, pH 11.0, with 20% methanol and 14 mM 2-mercaptoethanal (2-ME). The blot was blocked for 1 h in 1× TBS with milk (20 mM Tris-HCl [pH 7.8], 180 mM NaCl, 3% Carnation nonfat dry milk). The blots were incubated overnight at 4°C with one of the following primary antibodies at the appropriate dilution: α-G418 phosphatase (Ab-Cam), 1:250 dilution; α-eIF2α-Pi Ser-51 (Cell Signaling), 1:1,000 dilution; α-6×His tag and α-Myc tag (Rockland), 1:1,000 dilution each. Primary antibodies were removed, and the blot was washed three times with 1× TBS containing milk for 0.5 h at room temperature. The blot was then probed with a 1:15,000 dilution of goat anti-rabbit IgG-peroxidase conjugate antibody (Sigma) for 1 h at room temperature. These secondary antibodies were then removed, and the blot was washed three times for 10 min each in 1× TBS with milk and then washed three times for 5 min each in 1× TBS without milk. The blot was developed by chemiluminescence. To ensure equal loading of proteins, extracts were separated by SDS-PAGE and Coomassie stained.
eIF2α phosphorylation assay.
FHM cells were seeded so that they were <50% confluent at the time of treatment. Cell monolayers were pretreated with 1 FHM U/ml of IFN for 16 to 24 h. Cells were infected with either wtATV or ATVΔ57R at an MOI of 5. Six hours postinfection, cells were harvested as described above, and phosphorylated eIF2α (Ser 51) was detected by Western blot analysis as described above.
35S labeling of proteins.
FHM cells were seeded so that they were <50% confluent at the time of treatment. Cell monolayers were pretreated with 1 FHM U/ml of IFN for 16 to 24 h. Cells were infected with either wtATV or ATVΔ57R at an MOI of 5. Cells were labeled with 50 μCi/ml of [35S]methionine-cysteine protein label mix (Perkin-Elmer) at 12 hpi. Thirty minutes prior to labeling, cells were washed twice with phosphate-buffered saline (PBS) and incubated with Dulbecco's modified Eagle's medium (DMEM) without l-glutamine, l-methionine, and l-cysteine (Cellgro). Starvation medium was removed, and labeling medium containing 50 μCi/ml of [35S]methionine-cystein was added for 30 min. Cytosolic extracts (36) were prepared, and de novo protein synthesis of equal cell volumes was analyzed via autoradiography.
PKZ assay.
FHM cells were seeded at ∼30% confluence in 35-mm dishes. The following day, cells were transfected with 500 ng of pcDNA3.1::PKZ or pcDNA3.1::PKZ-K199R plasmids (57) that had been incubated with Fugene6 according to the manufacturer's instructions. These plasmids contain the sequence for PKZ and a catalytically inactive mutant of PKZ and PKZ-K199R, respectively, that is expressed in transfected cells. Both PKZ and PKZ-K199R have C-terminal 6×His and c-Myc tags so that expressed proteins can be visualized by Western blot analysis. One day posttransfection, cell monolayers were infected with either wtATV or ATVΔ57R at an MOI of 5. Cell extracts were prepared at 8 hpi, and PKZ was visualized by Western blot analysis as described above. PKZ and PKZ-K199R proteins observed in the Western blots were quantified using the ImageQuant 5.2 software (GE Healthcare). The relative intensity of each PKZ protein from ATV- and ATVΔ57R-infected cells was normalized to PKZ levels of mock-infected cells. Data presented are the averages and standard errors of results from multiple independent experiments.
Pathogenesis in the salamander model.
Laboratory-reared tiger salamanders (Ambystoma tigrinum) were mock infected or infected with 104 PFU of wtATV or ATVΔ57R by bath immersion (n = 20 salamanders per treatment). The titer of virus used in these experiments is a log higher than the 50% lethal dose of wtATV (J. K. Jancovich, unpublished observations). Animals were housed in individual 56-oz Ziplock containers at 17°C with a 12-h light/dark cycle and were fed meal worms two times per week. Salamanders were monitored for signs of disease (28) and mortality. Sick salamanders were euthanized by an overdose of MS-222 (Argent Laboratories) according to IACUC protocols established at Arizona State University (1049R). Data are presented as the percent survival (number of dead salamanders/total number of salamanders per treatment) over a 50-day period. The average time to death was determined by adding the total number of days each animal survived for each treatment and dividing that number by the number of animals in each treatment.
RESULTS
Generation of a recombinant ATV.
To test the hypothesis that the ATV vIF2αH gene plays a role in host immune evasion, a mutant of ATV, deleted of the vIF2αH gene (ORF 57R), was generated by homologous recombination between viral DNA and PCR product (Fig. 1A). This virus, ATVΔ57R, was isolated by blind passage and plaque purification in the presence of 1 mg/ml G418. As wtATV does not replicate in the presence of this concentration of G418 (data not shown), recombinant ATV expressing the G418 resistance gene was able to be isolated from a mixture of wild-type and recombinant viruses. Since a PCR product was used for the in vitro recombination, the most likely G418r virus is the result of a double-recombination event deleting the ATV vIF2αH gene and inserting the G418 resistance gene in its place (Fig. 1A).
The deletion of the ATV vIF2αH gene and the insertion of the pICP18-G418r cassette were confirmed by PCR and sequencing as well as by Western blot analysis for G418r gene expression. Using primers that flanked the ATV ORF 57R, a 0.8-kb PCR product, indicative of the presence of the ATV 57R ORF (i.e., the ATV vIF2αH gene), is observed using wtATV DNA as a template (Fig. 1B, lane 5). However, with ATVΔ57R, a 1.2-kb PCR product is present, indicating the insertion of the pICP18-G418r cassette and the deletion of ATV ORF 57R (Fig. 1B, lane 6). As a control for this PCR, the pJJ57 template generated a 1.2-kb PCR product identical to the PCR product observed for the recombinant ATV (Fig. 1B, lane 7). Sequencing of these PCR products confirmed the replacement of the ATV vIF2αH gene with the pICP18-g418r cassette (data not shown). We used PCR to the viral major capsid protein as a control PCR (Fig. 1B, lanes 2 to 4). Both wtATV and ATVΔ57R generated a 0.5-kb band indicative of the presence of the MCP in these viruses (Fig. 1B, lanes 2 and 3). However, as expected, when a plasmid was used as a template in this reaction (i.e., pJJ57), no PCR product was observed (Fig. 1B, lane 4). Further confirmation of the generation of a recombinant ATV was obtained by Western blot analysis. Mock-infected cells and cells infected with wtATV did not produce a G418r protein product, as detected by Western blot analysis by using an antibody specific to the G418 phosphatase protein (Fig. 1C, lanes 1 and 2, respectively). In contrast, a band indicative of a G418r gene product was observed in cells infected with the recombinant ATVΔ57R virus (Fig. 1C, lane 3). These data confirm that a recombinant ATV lacking ORF 57R was obtained in pure culture.
Virus plaque phenotype.
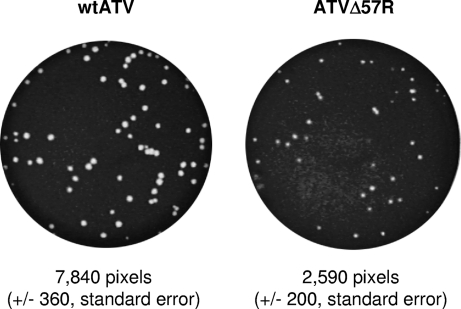
Plaque size phenotypes of both wtATV and ATVΔ57R were then determined (Fig. 2). Using pixel density (i.e., intensity) as a measure of plaque size, we determined the average number of pixels for 10 independent plaques for each virus. Wild-type ATV has an average pixel intensity of 7,840 pixels (±360, standard error), while the average size of an ATVΔ57R plaque is 2,590 pixels (±200, standard error). Therefore, wtATV has a plaque phenotype that is about 3-fold larger than that of ATVΔ57R.
Fig. 2.
Plaque size phenotype. FHM cells were infected with wtATV and ATVΔ57R until plaques were observed. Plaques were visualized by staining with a 20% ethanol and 1% crystal violet solution, and plaque sizes were determined using the ImageQuant 5.2 software (GE Healthcare). Ten plaques for each virus were used to determine plaque size by the relative intensity of the pixels in each plaque. The pixel intensities were averaged, and the standard errors were determined.
Interferon sensitivity of ATV.
Interferon (IFN) sensitivity of both wtATV and ATVΔ57R was then investigated. IFN-like activity was generated using spent culture medium (SCM) from poly(I:C)-treated and untreated FHM cells. ATVΔ57R was sensitive to a 1:32 dilution of FHM SCM, with no plaques observed at this dilution of IFN (Fig. 3A). In contrast, wtATV was 8-fold less sensitive to IFN treatment, with no plaques observed at a 1:4 dilution of the SCM (data not shown). The data show that a 50% plaque reduction of wtATV was observed at approximately a 1:64 dilution of IFN, while a 50% reduction of ATVΔ57R plaques required between a 1:256 to 1:128 dilution of IFN-containing medium (Fig. 3A and B). Untreated FHM cell SCM had no effect on the reduction of plaques for either wtATV or ATVΔ57R (data not shown). Therefore, the 1:64 dilution of FHM SCM is defined as 1 U of FHM IFN. These data suggest that wtATV is sensitive to effects of IFN, but upon deletion of the ATV eIF2α homologue IFN sensitivity increases 8-fold.
Fig. 3.
Interferon sensitivity. IFN expression was induced in FHM cells using dsRNA according to the protocol outlined in the Materials and Methods. FHM cells in 35-mm dishes were treated with 2-fold dilutions of spent culture medium (SCM) containing FHM IFN for 16 to 24 h. At that time, ∼200 PFU of either wtATV or ATVΔ57R was used to infect the treated cells. (A) Cells were stained with a 20% ethanol, 1% crystal violet solution to visualize plaques 7 days postinfection. Numbers above each image indicate the dilution of IFN in the form of spent culture medium (SCM). (B) Graphical representation of IFN sensitivity. Data for each individual experiment were normalized to the number of plaques observed in mock-treated cells. The percentage of plaque reduction was determined by dividing the number of plaques observed at each SCM dilution (i.e., volume of SCM in each culture) by the number of plaques observed in mock-infected cultures. Data presented are the averages and standard errors of results from multiple independent experiments.
Virus replication kinetics.
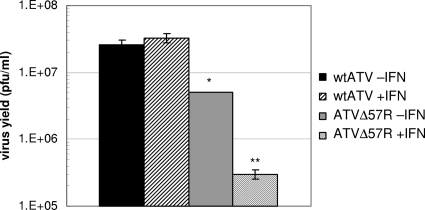
Single-cycle replication characteristics were then determined. FHM cells were treated with or without SCM containing 1 FHM unit of IFN for 16 to 24 h and then infected with either wtATV or ATVΔ57R at an MOI of 5. Cells were harvested, and titers were determined at 6 and 48 hpi. The virus yield was determined as the amount of virus amplified above the background level (i.e., titer at 48 hpi − titer at 6 hpi). Wild-type ATV replicated to similar viral titers in the presence and absence of IFN (average titers of 2.6 × 107 and 3.3 × 107 PFU/ml, respectively) (Fig. 4). The ATVΔ57R virus titers in the absence of IFN treatment (average titer of 5.1 × 106 PFU/ml) are about one-half of a log lower than that of wtATV. However, there was a statistically significant difference between ATVΔ57R replication and wtATV replication (P value of <0.05) (Fig. 4). In addition, when FHM cells were pretreated with IFN and then infected with ATVΔ57R, a statistically significant reduction in virus yield is observed compared to non-IFN-treated ATVΔ57R-infected cells (average titer of 3 × 105 PFU/ml) (P value of <0.001) (Fig. 4). These data suggest that under a single round of replication, the ATV vIF2αH gene is able to overcome the antiviral effects of IFN. Upon the deletion of this gene, the virus does not replicate above background levels nor overcome the effects of IFN treatment.
Fig. 4.
Single-cycle growth characteristics. FHM cells were either pretreated with 1 FHM unit of IFN or left untreated for 16 to 24 h. At that time, the cells were infected with either wtATV or ATVΔ57R at an MOI of 0.01. Cells and virus were harvested at 6 and 48 hpi, and virus titer was determined by plaque assay. Data are presented as total virus yield (PFU ml−1 virus titer at 48 hpi minus PFU ml−1 virus titer at 6 hpi). Data are averages of results from two independent experiments. Error bars represent standard errors. Statistical analysis was performed using the Student t test option in SigmaStat (Systat Software, Inc.). Statistically significant differences between treatments are indicated: *, comparing wtATV −IFN to ATVΔ57R −IFN (P value of <0.050); **, comparing ATVΔ57R −IFN and ATVΔ57R +IFN (P value of <0.001).
Protein synthesis and eIF2α phosphorylation.
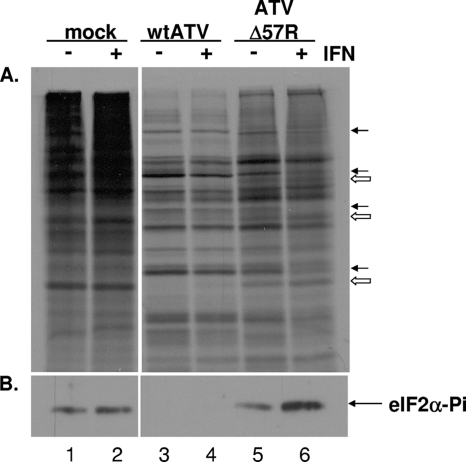
To explore a potential mechanism of action of this IFN sensitivity, a molecular study of total protein synthesis and cellular eIF2α phosphorylation was initiated. FHM cells were pretreated with or without 1 FHM unit of IFN for 16 to 24 h and then infected with either wtATV or ATVΔ57R at an MOI of 5. At 12 hpi, cells were lysed with NP-40 and proteins were isolated for Western blot analysis or labeled with 50 μCi/ml of [35S]methionine-cysteine, and total protein synthesis was observed by autoradiography after SDS-PAGE. Cellular protein synthesis was inhibited and virus-specific protein synthesis was observed in untreated and IFN-treated FHM cells infected with wtATV by 12 hpi compared to those of mock-infected cells (Fig. 5A, lanes 1, 2, 3, and 4). In contrast, some cellular protein synthesis was present (open arrows) in addition to virus-specific protein synthesis (closed arrows) in cells infected with ATVΔ57R at 12 hpi without IFN (Fig. 5A, lane 5). IFN treatment slowed down virus-specific protein synthesis in FHM cells infected with ATVΔ57R (Fig. 5A, lane 6). However, virus-specific proteins are present. Therefore, while the deletion of the ATV vIF2αH gene delayed the onset of viral protein synthesis in IFN-treated FHM cells, there is still a partial inhibition of host protein synthesis and induction of virus-specific protein synthesis.
Fig. 5.
Protein synthesis and eIF2α activation. FHM cells were pretreated with or without 1 FHM U/ml of interferon for 16 to 24 h. Cells were then mock infected or infected with wtATV or ATVΔ57R at an MOI of 5. (A) Cells were labeled with [35S]methionine-cysteine at 12 hpi. Extracts were prepared, and de novo protein synthesis of equal cell volumes was analyzed via autoradiography. Closed arrows indicate virus-specific proteins, while open arrows indicate cellular-specific proteins. (B) Cytoplasmic extracts were prepared at 12 hpi, and equal cell volumes of protein were separated by SDS-PAGE. Western blot analysis was conducted using anti-eIF2-α (pS51) (Cell Signaling). Proteins were visualized by chemiluminescence.
We then asked if the inhibition of cellular protein synthesis correlated with cellular eIF2α phosphorylation. Wild-type ATV was able to inhibit cellular eIF2α phosphorylation in the presence and absence of IFN treatment (Fig. 5B, lanes 3 and 4). In contrast, in FHM cells infected with the ATVΔ57R virus, eIF2α phosphorylation was observed without IFN treatment, and levels of eIF2α were increased in IFN-treated cells (Fig. 5B, lanes 5 and 6). eIF2α phosphorylation was observed in mock-infected cells regardless of IFN treatment in this experiment (Fig. 5B, lanes 1 and 2). However, this phenomenon was not observed in all experiments. In other experiments, mock-infected cells did not lead to eIF2α phosphorylation (data not shown). In addition, eIF2α phosphorylation was never observed at high levels after a wtATV infection (Fig. 5B, lane 3), and eIF2α phosphorylation was always observed upon an ATVΔ57R infection (Fig. 5B, lane 5). These data suggest that the ATV vIF2αH gene product inhibits cellular eIF2α phosphorylation, allowing viral protein translation to continue and inhibiting cellular protein synthesis in a virus-infected cell.
Degradation of PKZ in infected cells.
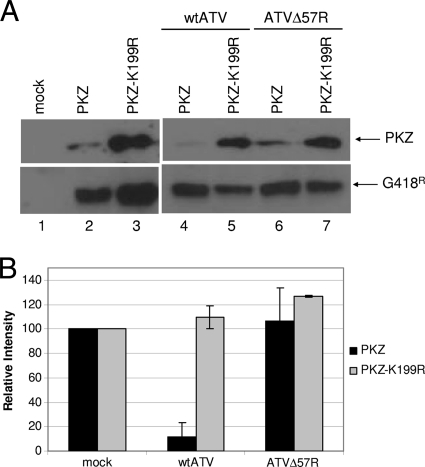
Previous studies using VACV suggest that the ATV vIF2αH functions to inhibit cellular eIF2α phosphorylation by degrading PKR (Tripuraneni et al., unpublished). However, antibodies to a fish PKR homologue were not available at the time of this study. In fish, a similar PKR-like protein, PKZ, has been indentified, and the gene encoding this protein contains Z-DNA binding domains in place of the characteristic dsRNA binding domains found in the N-terminal region of identified PKRs (5, 56, 57, 65, 74). Since there are currently no molecular reagents to study PKZ-like molecules in vivo, a plasmid containing PKZ with C-terminal 6×His and c-Myc tags was obtained (from A. Rich, MIT). FHM cells transfected with this PKZ plasmid produced detectable PKZ protein of expected molecular weight, detected by Western blot analysis, compared to mock-transfected cells (Fig. 6A, top, lanes 1 and 2). PKZ levels were decreased in FHM cells after infection with wtATV by 8 hpi (Fig. 6A, top, lane 4) compared to those in cells after infection with ATVΔ57R at the same time point (Fig. 6A, top, lane 6). These data suggest that vIF2αH plays an important role in downregulating PKZ levels. In addition, downregulation of PKZ requires an active form of the molecule. Uninfected FHM cells transfected with PKZ or with a catalytically inactive form of PKZ, PKZ-K199R, a mutation that abolishes PKZ activation, show detectable PKZ by Western blot analysis (Fig. 6A, top, lanes 2 and 3). In addition, as observed previously (57), the levels of the inactive form of PKZ (K199R mutation) are greater than that of wtPKZ in mock-infected cells (Fig. 6A, top, lane 3). Introducing the plasmid containing PKZ-K199R followed by infection with either wtATV or ATVΔ57R did not lead to PKZ downregulation (Fig. 6A, top, lanes 5 and 7, respectively). To ensure equal loading of proteins, we assayed for G418 phosphatase expression from the plasmid also expressing PKZ (Fig. 6A, bottom). In addition, equal loading of proteins was confirmed by Coomassie staining (data not shown). Quantifying the levels of PKZ in these Western blots shows a 90% decrease in PKZ levels in wtATV-infected cells compared to the levels of PKZ in ATVΔ57R-infected cells (Fig. 6B). In contrast, levels of PKZ-K199A increased above that of mock-infected cells for both wtATV and ATVΔ57R infections (Fig. 6B). Therefore, these data suggest that an active form of PKZ is required for its downregulation in this ranavirus system.
Fig. 6.
PKZ regulation by ATV. (A) PKZ assay. FHM cells were transfected with pcDNA3.1-PKZ, the active form of PKZ, or pcDNA3.1-PKZ-K199R, the inactive form of PKZ, and the cells were incubated for 24 h. At that time, cells were either mock infected or infected with wtATV or ATVΔ57R at an MOI of 5. Cells were harvested at 8 hpi, and protein extracts were obtained. Western blot analysis of equal cell volumes was analyzed using primary antibodies specific to PKZ C-terminal 6×His and Myc tags (Rockland) or anti-G418 phosphatase (Abcam) followed by chemiluminescence. (B) PKZ quantification. PKZ and PKZ-K199R proteins observed in the Western blots were quantified using the ImageQuant 5.2 software (GE Healthcare). The relative intensity of each PKZ protein from ATV- and ATVΔ57R-infected cells was normalized to PKZ levels of mock-infected cells. Data presented are the averages and standard errors of results from two independent experiments. *, nonspecific proteins or potential degradation products as described by Rothenberg et al. (57).
Pathogenesis in a salamander model.
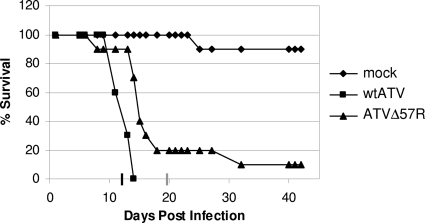
The data obtained so far suggest that the ATV vIF2αH is nonessential for replication in cells in culture and that this viral gene inhibits cellular eIF2α phosphorylation by degrading PKZ. Therefore, to determine if the ATV ORF 57R gene is essential for pathogenesis in a tiger salamander, animals were infected by bath immersion with either wtATV or ATVΔ57R or mock infected with medium alone. Wild-type ATV killed 100% of the exposed salamanders at a dose of 104 PFU in 14 days (Fig. 7). In contrast, 50% of the tiger salamanders exposed to the same dose of ATVΔ57R survived until day 14 postinfection, and two salamanders survived throughout the experiment (Fig. 7). In addition, the mean time to death of wtATV-infected salamanders was 12.5 days compared to 19.7 days for salamanders infected with ATVΔ57R. Therefore, the ATV ORF 57R gene product enhances virus pathogenesis in the virus' natural host, the tiger salamander (Ambystoma tigrinum).
Fig. 7.
Pathogenesis in the salamander model. Laboratory-reared tiger salamanders (Ambystoma tigrinum) were either mock infected or infected with 104 PFU of wtATV or ATVΔ57R by bath immersion (n = 20 salamanders per treatment). Animals were fed meal worms two times per week and monitored for signs of disease and mortality. The graph shows the percent survival (number of dead salamanders/total number of salamanders per treatment) over a 50-day period. Average times to death for wtATV and ATVΔ57R are indicated on the x axis as a black and a gray line, respectively.
DISCUSSION
Ranaviruses are important pathogens of lower vertebrates (11, 13, 70), yet our understanding of how these viruses cause disease is limited. Ranaviruses cause catastrophic disease in tiger salamanders throughout North America, frogs in Asia, and fish and reptiles throughout the world, and they are considered an emerging infectious disease (11, 13, 15, 70). In addition, ranaviruses are listed by the World Organization for Animal Health as a notifiable disease (www.oie.int/eng/en_index.htm). Therefore, understanding how these important pathogens cause disease is essential in our understanding of how to prevent or predict ranavirus infections.
It has been hypothesized that the ranavirus-specific vIF2αH gene plays a role in viral pathogenesis (40). While this study suggests a correlation between vIF2αH and pathogenesis, it is limited by comparing nonisogenic virus strains, one containing a full-length vIF2αH gene and one containing a truncated version of this gene. Therefore, comparing the pathogenesis of homologous viruses that differ only in the presence or absence of the vIF2αH gene will provide a better understanding of the role of the vIF2αH gene in pathogenesis and innate immune evasion.
This is the first published report of a recombinant ranavirus to study gene function. While our protocol to generate a recombinant ranavirus is similar to that of Pallister et al. (47), they deleted the Bohle iridovirus vIF2αH and inserted the globin gene from the cane toad (Bufo marinus). However, the goal of that study was to generate a potential biological control agent against the cane toad. In contrast, our focus was on characterizing the ranavirus vIF2αH after knocking this gene out of the virus. Therefore, together, these studies open the possibility to elucidate the function of other ranavirus nonessential genes and begin to gain insight into the mechanisms of ranavirus immune evasion and pathogenesis.
The ATVΔ57R deletion mutant displayed a small plaque phenotype but replicated to near wild-type levels in FHM cells. Therefore, the ATV ORF 57R is nonessential for virus growth in cells in culture. Cells infected with ATVΔ57R consistently phosphorylated cellular eIF2α and were 8-fold more sensitive to IFN than those infected with wtATV. The phosphorylation of cellular eIF2α correlated with the failure to downregulate PKZ. Testing pathogenesis of wtATV and ATVΔ57R viruses in the virus' natural host, tiger salamanders (Ambystoma tigrinum), showed that the deletion mutant, ATVΔ57R, is slightly less pathogenic than wtATV when tiger salamanders were infected by bath immersion. Therefore, this study shows a direct correlation between pathogenesis and the presence of the vIF2αH gene and suggests a role for the ATV vIF2αH gene in evasion of the host's innate immune response.
The battle between host and pathogen is a constant struggle. Virus replication depends on the transcription and translation of viral genes ensuring the production of progeny virus. Cell, and therefore host, survival requires mechanisms that defend against foreign attacks, and the innate immune response is the first line of defense against an invading pathogen (4, 69). Mammalian and ectothermic vertebrate innate immune responses are similar, yet there are still areas, such as the interferon (IFN)-like responses, that have yet to be fully characterized from other lower vertebrates (i.e., fish and amphibians) (39). The data obtained in this study suggest that the salamander immune system has IFN- and PKR-like molecules and that the ATV vIF2αH gene acts to overcome these antiviral responses. IFNs have been isolated and sequenced from a number of fish species (53), and IFN was identified in Xenopus (50, 51). However, at this time an IFN-like molecule has not been identified in salamanders. In addition, a PKR-like enzyme has been identified and cloned from fish cells (25), zebrafish (57), and frogs (56), although it appears frogs may have two types of PKR-like molecules. In frogs, one molecule appears similar to mammalian PKR, while the other type resembles the fish PKR-like proteins, also known as PKZ, that contain Z-DNA binding motifs in place of the dsRNA binding motifs found in mammalian PKR (56, 57). The role of the Z-DNA binding motif of PKZ has yet to be determined. The dsRNA binding motif of mammalian PKR recognizes dsRNA initiating PKR activation (14, 48, 54, 67, 72). If this model holds true for fish and amphibians, then another pathogen-associated molecular pattern (PAMP), perhaps Z-DNA or RNA, may be the key to the PKR-PAMP pathway evolution.
Viruses have evolved elaborate mechanisms to inhibit the PKR response (21, 35). These viral countermeasures have been shown to block the PKR response at virtually every step in the pathway, from activation through substrate phosphorylation. For many viruses, including vaccinia virus (VACV), herpes simplex virus type 1, Epstein-Barr virus, influenza virus, and hepatitis C virus, multiple mechanisms are encoded to evade PKR activity (35). For most of these viruses, it is unclear if these multiple PKR inhibitory mechanisms play redundant roles or are necessary to regulate PKR activity at different stages in the virus life cycle.
While many viral inhibitors of the cellular PKR response are known, at this time there are only two viruses that have been shown to degrade PKR, Rift Valley fever virus (RVFV) (24, 27) and polioviruses (6, 7). The ranavirus vIF2αH gene is the first viral gene from a dsDNA virus to regulate PKR/PKZ protein levels in infected cells. Our data presented here as well as the data characterizing the ATV vIF2αH gene using a heterologous vaccinia virus system (Tripuraneni et al., unpublished) suggest that the ATV vIF2αH gene is required for the regulation of PKR/PKZ. In addition, the data suggest that an active form of PKR/PKZ is necessary for the ATV vIF2αH gene-dependent degradation of this cellular gene. Therefore, we hypothesize that the ATV vIF2αH gene is necessary for regulation of this innate immune pathway in virus-infected cells. However, the precise mechanism of action in regulating PKR/PKZ levels by this ranavirus protein is still unclear. Future work will focus on understanding this mechanism of PKZ regulation in ATV-infected cells.
It is interesting to note that eIF2α phosphorylation occurs in mock-infected FHM cells yet cellular protein synthesis continues. We discovered that FHM cell confluence influenced cellular eIF2α phosphorylation in mock-infected cells (data not shown). Cells needed to be treated and/or infected at <50% confluence for consistency in eIF2α phosphorylation. In addition, any manipulation of these cells (e.g., transfection reagents) would increase the probability of eIF2α phosphorylation in mock-treated cells for some experiments. Therefore, it is possible that cellular stress influences cellular eIF2α phosphorylation in FHM cells (32). However, in every experiment that FHM cells were infected with wtATV, cellular eIF2α phosphorylation was not observed, and FHM cells infected with ATVΔ57R consistently had cellular eIF2α phosphorylation. It has been shown previously that in FV3-infected FHM cells, eIF2α phosphorylation is observed (12). This phenomenon may be the result of a truncated FV3 vIF2αH gene, and therefore the lack of this gene does not enable the virus to inhibit cellular eIF2α phosphorylation (66). However, even though FV3 does not inhibit eIF2α phosphorylation in infected FHM cells, viral protein synthesis was not adversely affected. In another study, cellular eIF2α phosphorylation was not observed in baby hamster kidney cells infected with FV3 (71). These observations suggest that there may be other viral proteins and/or mechanisms that shut off host protein synthesis and allow viral protein synthesis to continue when cellular eIF2α has been phosphorylated. In addition, these data suggest that alternative, yet-undescribed translational control mechanisms exist in lower vertebrates. As we begin to understand these mechanisms, it will advance our knowledge of the evolution of cellular and viral translational control mechanisms.
We hypothesized that the ATV vIF2αH gene is a pathogenesis factor that enhances ATV pathogenesis by regulating the IFN-inducible enzyme PKR. In fact, data suggesting that the ATV vIF2αH gene degrades PKR using a heterologous system (Tripuraneni et al., unpublished) was confirmed in this study by using a transfection-infection assay for PKZ in a homologous viral system. The mechanism of degradation in this homologous system has yet to be determined. Further studies using this knockout homologous viral system are required before a clearer picture of ATV innate immune evasion can be determined.
ACKNOWLEDGMENTS
This work was supported in part by the Integrated Research Challenges in Environmental Biology (IBN-9977063) and the Division of Environmental Biology (0213851) grants from the National Science Foundation.
Footnotes
Published ahead of print on 9 March 2011.
REFERENCES
- 1. Ahne W. 1988. Infectious hematopoietic necrosis, a new disease of fish in Europe. Tierarztliche Umschau. 43:239–242 [Google Scholar]
- 2. Ahne W., Bremont M., Hedrick R. P., Hyatt A. D., Whittington R. J. 1997. Iridoviruses associated with epizootic haematopoietic necrosis (EHN) in aquaculture. World J. Microbiol. Biotechnol. 13:367–373 [Google Scholar]
- 3. Allender M. C., et al. 2006. Intracytoplasmic inclusions in circulating leukocytes from an eastern box turtle (Terrapene carolina carolina) with iridoviral infection. J. Wildl. Dis. 42:677–684 [DOI] [PubMed] [Google Scholar]
- 4. Basset C., Holton J., O'Mahony R., Roitt I. 2003. Innate immunity and pathogen-host interaction. Vaccine 21:S12–S23 [DOI] [PubMed] [Google Scholar]
- 5. Bergan V., Jagus R., Lauksund S., Kileng O., Robertsen B. 2008. The Atlantic salmon Z-DNA binding protein kinase phosphorylates translation initiation factor 2 alpha and constitutes a unique orthologue to the mammalian dsRNA-activated protein kinase R. FEBS J. 275:184–197 [DOI] [PubMed] [Google Scholar]
- 6. Black T. L., Barber G. N., Katze M. G. 1993. Degradation of the interferon-induced 68,000-M(r) protein kinase by poliovirus requires RNA. J. Virol. 67:791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Black T. L., Safer B., Hovanessian A., Katze M. G. 1989. The cellular 68,000-Mr protein kinase is highly autophosphorylated and activated yet significantly degraded during poliovirus infection: implications for translational regulation. J. Virol. 63:2244–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bollinger T. K., Mao J., Schock D., Brigham R. M., Chinchar V. G. 1999. Pathology, isolation, and preliminary molecular characterization of a novel iridovirus from tiger salamanders in Saskatchewan. J. Wildl. Dis. 35:413–429 [DOI] [PubMed] [Google Scholar]
- 9. Chang H. W., Uribe L. H., Jacobs B. L. 1995. Rescue of vaccinia virus lacking the E3L gene by mutants of E3L. J. Virol. 69:6605–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang H. W., Watson J. C., Jacobs B. L. 1992. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. U. S. A. 89:4825–4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chinchar V. G. 2002. Ranaviruses (family Iridoviridae): emerging cold-blooded killers. Arch. Virol. 147:447–470 [DOI] [PubMed] [Google Scholar]
- 12. Chinchar V. G., Dholakia J. N. 1989. Frog virus 3-induced translational shut-off: activation of an eIF-2 kinase in virus-infected cells. Virus Res. 14:207–223 [DOI] [PubMed] [Google Scholar]
- 13. Chinchar V. G., Hyatt A., Miyazaki T., Williams T. 2009. Family Iridoviridae: poor viral relations no longer. Curr. Top. Microbiol. Immunol. 328:123–170 [DOI] [PubMed] [Google Scholar]
- 14. Clemens M. J., Elia A. 1997. The double-stranded RNA-dependent protein kinase PKR: structure and function. J. Interferon Cytokine Res. 17:503–524 [DOI] [PubMed] [Google Scholar]
- 15. Daszak P., et al. 1999. Emerging infectious diseases and amphibian population declines. Emerg. Infect. Dis. 5:735–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dave R. S., et al. 2006. siRNA targeting vaccinia virus double-stranded RNA binding protein [E3L] exerts potent antiviral effects. Virology 348:489–497 [DOI] [PubMed] [Google Scholar]
- 17. De Voe R., et al. 2004. Ranavirus-associated morbidity and mortality in a group of captive eastern box turtles (Terrapene carolina carolina). J. Zoo Wildl. Med. 35:534–543 [DOI] [PubMed] [Google Scholar]
- 18. Donnelly T. M., et al. 2003. What's your diagnosis? Ranavirus infection. Lab Anim. (NY) 32:23–25 [DOI] [PubMed] [Google Scholar]
- 19. Drury S. E. N., Gough R. E., Cunningham A. A. 1995. Isolation of an iridovirus-like agent from common frogs (Rana temporaria). Vet. Rec. 137:72–73 [DOI] [PubMed] [Google Scholar]
- 20. Fijan N., et al. 1983. Some properties of the Epithelioma papulosum cyprini (EPC) cell line from carp Cyprinus carpio. Ann. Virol. 134:207–220 [Google Scholar]
- 21. George C. X., et al. 2009. Tipping the balance: antagonism of PKR kinase and ADAR1 deaminase functions by virus gene products. J. Interferon Cytokine Res. 29:477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Green D. E., Converse K. A., Schrader A. K. 2002. Epizootiology of sixty-four amphibian morbidity and mortality events in the U. S. A., 1996-2001. Ann. N. Y. Acad. Sci. 969:323–339 [DOI] [PubMed] [Google Scholar]
- 23. Greer A. L., Berrill M., Wilson P. J. 2005. Five amphibian mortality events associated with ranavirus infection in south central Ontario, Canada. Dis. Aquat. Organ. 67:9–14 [DOI] [PubMed] [Google Scholar]
- 24. Habjan M., et al. 2009. NSs protein of Rift Valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J. Virol. 83:4365–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu C. Y., Zhang Y. B., Huang G. P., Zhang Q. Y., Gui J. F. 2004. Molecular cloning and characterisation of a fish PKR-like gene from cultured CAB cells induced by UV-inactivated virus. Fish Shellfish Immunol. 17:353–366 [DOI] [PubMed] [Google Scholar]
- 26. Hyatt A. D., et al. 2002. First identification of a ranavirus from green pythons (Chondropython viridis). J. Wildl. Dis. 38:239–252 [DOI] [PubMed] [Google Scholar]
- 27. Ikegami T., et al. 2009. Rift Valley fever virus NSs protein promotes post-transcriptional downregulation of protein kinase PKR and inhibits eIF2 alpha phosphorylation. Plos Pathog. 5:e1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jancovich J. K., Davidson E. W., Morado J. F., Jacobs B. L., Collins J. P. 1997. Isolation of a lethal virus from the endangered tiger salamander Ambystoma tigrinum stebbinsi. Dis. Aquat. Organ. 31:161–167 [Google Scholar]
- 29. Jancovich J. K., et al. 2005. Evidence for emergence of an amphibian iridoviral disease because of human-enhanced spread. Mol. Ecol. 14:213–224 [DOI] [PubMed] [Google Scholar]
- 30. Jancovich J. K., et al. 2003. Genomic sequence of a ranavirus (family Iridoviridae) associated with salamander mortalities in North America. Virology 316:90–103 [DOI] [PubMed] [Google Scholar]
- 31. Johnson A. J., et al. 2008. Ranavirus infection of free-ranging and captive box turtles and tortoises in the United States. J. Wildl. Dis. 44:851–863 [DOI] [PubMed] [Google Scholar]
- 32. Kimball S. R. 1999. Eukaryotic initiation factor eIF2. Int. J. Biochem. Cell Biol. 31:25–29 [DOI] [PubMed] [Google Scholar]
- 33. Langdon J. S., Humphrey J. D., Williams L. M. 1988. Outbreaks of an EHNV-like iridovirus in cultured rainbow trout, Salmo gairdneri Richardson, in Australia. J. Fish Dis. 11:93–96 [Google Scholar]
- 34. Langdon J. S., Humphrey J. D., Williams L. M., Hyatt A. D., Westbury H. A. 1986. First virus isolation from Australian fish—an iridovirus-like pathogen from redfin perch, Perca fluviatilis L. J. Fish Dis. 9:263–268 [Google Scholar]
- 35. Langland J. O., Cameron J. M., Heck M. C., Jancovich J. K., Jacobs B. L. 2006. Inhibition of PKR by RNA and DNA viruses. Virus Res. 119:100–110 [DOI] [PubMed] [Google Scholar]
- 36. Langland J. O., Jacobs B. L. 2004. Inhibition of PKR by vaccinia virus: role of the N- and C-terminal domains of E3L. Virology 324:419–429 [DOI] [PubMed] [Google Scholar]
- 37. Langland J. O., Jacobs B. L. 2002. The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology 299:133–141 [DOI] [PubMed] [Google Scholar]
- 38. Levin D., London I. M. 1978. Regulation of protein synthesis: activation by double-stranded RNA of a protein kinase that phosphorylates eukaryotic initiation factor 2. Proc. Natl. Acad. Sci. U. S. A. 75:1121–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Magnadottir B. 2006. Innate immunity of fish (overview). Fish Shellfish Immunol. 20:137–151 [DOI] [PubMed] [Google Scholar]
- 40. Majji S., et al. 2006. Rana catesbeiana virus Z (RCV-Z): a novel pathogenic ranavirus. Dis. Aquat. Organ. 73:1–11 [DOI] [PubMed] [Google Scholar]
- 41. Mao J., Tham T. N., Gentry G. A., Aubertin A., Chinchar V. G. 1996. Cloning, sequence analysis, and expression of the major capsid protein of the iridovirus frog virus 3. Virology 216:431–436 [DOI] [PubMed] [Google Scholar]
- 42. Mao J. H., Hedrick R. P., Chinchar V. G. 1997. Molecular characterization, sequence analysis, and taxonomic position of newly isolated fish iridoviruses. Virology 229:212–220 [DOI] [PubMed] [Google Scholar]
- 43. Mao J. H., Wang J., Chinchar G. D., Chinchar V. G. 1999. Molecular characterization of a ranavirus isolated from largemouth bass Micropterus salmoides. Dis. Aquat. Organ. 37:107–114 [DOI] [PubMed] [Google Scholar]
- 44. Marschang R. E., et al. 1999. Isolation and characterization of an iridovirus from Hermann's tortoises (Testudo hermanni). Arch. Virol. 144:1909–1922 [DOI] [PubMed] [Google Scholar]
- 45. Marschang R. E., Braun S., Becher P. 2005. Isolation of a ranavirus from a gecko (Uroplatus fimbriatus). J. Zoo Wildl. Med. 36:295–300 [DOI] [PubMed] [Google Scholar]
- 46. Nygaard R., Husgard S., Sommer A. I., Leong J. A., Robertsen B. 2000. Induction of Mx protein by interferon and double-stranded RNA in salmonid cells. Fish Shellfish Immunol. 10:435–450 [DOI] [PubMed] [Google Scholar]
- 47. Pallister J., et al. 2007. Bohle iridovirus as a vector for heterologous gene expression. J. Virol. Methods 146:419–423 [DOI] [PubMed] [Google Scholar]
- 48. Patel R. C., Sen G. C. 1998. Requirement of PKR dimerization mediated by specific hydrophobic residues for its activation by double-stranded RNA and its antigrowth effects in yeast. Mol. Cell. Biol. 18:7009–7019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Picco A. M., Collins J. P. 2008. Amphibian commerce as a likely source of pathogen pollution. Conserv. Biol. 22:1582–1589 [DOI] [PubMed] [Google Scholar]
- 50. Qi Z. T., Nie P. 2008. Comparative study and expression analysis of the interferon gamma gene locus cytokines in Xenopus tropicalis. Immunogenetics 60:699–710 [DOI] [PubMed] [Google Scholar]
- 51. Qi Z. T., Nie P., Secombes C. J., Zou J. 2010. Intron-containing type I and type III IFN co-exist in amphibians: refuting the concept that a retroposition event gave rise to type I IFNs. J. Immunol. 184:5038–5046 [DOI] [PubMed] [Google Scholar]
- 52. Qin Q. W., et al. 2003. Characterization of a novel ranavirus isolated from grouper Epinephelus tauvina. Dis. Aquat. Organ. 53:1–9 [DOI] [PubMed] [Google Scholar]
- 53. Robertsen B. 2006. The interferon system of teleost fish. Fish Shellfish Immunol. 20:172–191 [DOI] [PubMed] [Google Scholar]
- 54. Robertson H. D., Mathews M. B. 1996. The regulation of the protein kinase PKR by RNA. Biochimie 78:909–914 [DOI] [PubMed] [Google Scholar]
- 55. Romano P. R., et al. 1998. Inhibition of double-stranded RNA-dependent protein kinase PKR by vaccinia virus E3: role of complex formation and the E3 N-terminal domain. Mol. Cell. Biol. 18:7304–7316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rothenburg S., Deigendesch N., Dey M., Dever T. E., Tazi L. 2008. Double-stranded RNA-activated protein kinase PKR of fishes and amphibians: varying the number of double-stranded RNA binding domains and lineage-specific duplications. BMC Biol. 6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rothenburg S., et al. 2005. A PKR-like eukaryotic initiation factor 2alpha kinase from zebrafish contains Z-DNA binding domains instead of dsRNA binding domains. Proc. Natl. Acad. Sci. U. S. A. 102:1602–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Saint-Jean S. R., Perez-Prieto S. I. 2007. Effects of salmonid fish viruses on Mx gene expression and resistance to single or dual viral infections. Fish Shellfish Immunol. 23:390–400 [DOI] [PubMed] [Google Scholar]
- 59. Samuel C. E. 1979. Mechanism of interferon action: phosphorylation of protein synthesis initiation factor eIF-2 in interferon-treated human cells by a ribosome-associated kinase processing site specificity similar to hemin-regulated rabbit reticulocyte kinase. Proc. Natl. Acad. Sci. U. S. A. 76:600–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schloegal L. M., Picco A. M., Kilpatrick A. M., Davies A. J., Hyatt A. D. 2009. Magnitude of the US trade in amphibians and presence of Batrachochytrium dendrobatidis and ranavirus infection in imported North American bullfrogs (Rana catesbeiana). Biol. Conserv. 142:1420–1426 [Google Scholar]
- 61. Shors S. T., Beattie E., Paoletti E., Tartaglia J., Jacobs B. L. 1998. Role of the vaccinia virus E3L and K3L gene products in rescue of VSV and EMCV from the effects of IFN-alpha. J. Interferon Cytokine Res. 18:721–729 [DOI] [PubMed] [Google Scholar]
- 62. Shors T., et al. 1997. Complementation of vaccinia virus deleted of the E3L gene by mutants of E3L. Virology 239:269–276 [DOI] [PubMed] [Google Scholar]
- 63. Speare R., Freeland W. J., Bolton S. J. 1991. A possible iridovirus in erythrocytes of Bufo marinus in Costa Rica. J. Wildl. Dis. 27:457–462 [DOI] [PubMed] [Google Scholar]
- 64. Speare R., Smith J. R. 1992. An iridovirus-like agent isolated from the ornate burrowing frog Limnodynastes ornatus in northern Australia. Dis. Aquat. Organ. 14:51–57 [Google Scholar]
- 65. Su J. G., Zhu Z. Y., Wang Y. P. 2008. Molecular cloning, characterization and expression analysis of the PKZ gene in rare minnow Gobiocypris rarus. Fish Shellfish Immunol. 25:106–113 [DOI] [PubMed] [Google Scholar]
- 66. Tan W. G. H., Barkman T. J., Chinchar V. G., Essani K. 2004. Comparative genomic analyses of frog virus 3, type species of the genus Ranavirus (family Iridoviridae). Virology 323:70–84 [DOI] [PubMed] [Google Scholar]
- 67. Thomis D. C., Samuel C. E. 1995. Mechanism of interferon action: characterization of the intermolecular autophosphorylation of PKR, the interferon-inducible, RNA-dependent protein kinase. J. Virol. 69:5195–5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Reference deleted.
- 69. Turvey S. E., Broide D. H. 2010. Innate Immunity. J. Allergy Clin. Immunol. 125:S24–S32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Williams T., Barbosa-Solomieu V., Chinchar V. G. 2005. A decade of advances in iridovirus research. Adv. Virus Res. 65:173–248 [DOI] [PubMed] [Google Scholar]
- 71. Willis D. B., Goorha R., Chinchar V. G. 1985. Macromolecular synthesis in cells infected by frog virus-3. Curr. Top. Microbiol. Immunol. 116:77–106 [DOI] [PubMed] [Google Scholar]
- 72. Zhang F., et al. 2001. Binding of double-stranded RNA to protein kinase PKR is required for dimerization and promotes critical autophosphorylation events in the activation loop. J. Biol. Chem. 276:24946–24958 [DOI] [PubMed] [Google Scholar]
- 73. Zhang Q. Y., et al. 2001. Characterization of an iridovirus from the cultured pig frog Rana grylio with lethal syndrome. Dis. Aquat. Organ. 48:27–36 [DOI] [PubMed] [Google Scholar]
- 74. Zhu R., Zhang Y. B., Zhang Q. Y., Gui J. F. 2008. Functional domains and the antiviral effect of the double-stranded RNA-dependent protein kinase PKR from Paralichthys olivaceus. J. Virol. 82:6889–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zupanovic Z., et al. 1998. Isolation and characterization of iridoviruses from the giant toad Bufo marinus in Venezuela. Dis. Aquat. Organ. 33:1–9 [DOI] [PubMed] [Google Scholar]