Abstract
This study describes a tumor progression model for ductal pancreatic cancer in mice overexpressing TGF-α. Activation of Ras and Erk causes induction of cyclin D1-Cdk4 without increase of cyclin E or PCNA in ductal lesions. Thus, TGF-α is able to promote progression throughout G1, but not S phase. Crossbreeding with p53 null mice accelerates tumor development in TGF-α transgenic mice dramatically. In tumors developing in these mice, biallelic deletion of Ink4a/Arf or LOH of the Smad4 locus is found suggesting that loci in addition to p53 are involved in antitumor activities. We conclude that these genetic events are critical for pancreatic tumor formation in mice. This model recapitulates pathomorphological features and genetic alterations of the human disease.
Keywords: Pancreas, cancer, TGF-α, p53, Ink4a, tumor progression
Cell cycle progression is regulated by cyclin-dependent kinase (Cdk) complexes. D-type cyclins interact with Cdk4 and Cdk6 to control the progression through G1 phase (Sherr and Roberts 1999). Synthesis of D-type cyclins is induced by mitogens, and the continued synthesis throughout G1 phase depends on persistent growth factor stimulation and/or active Ras. D-type cyclins and the catalytic subunits, Cdk4/6, assemble into active complexes during early G1, followed by the activation of cyclin E–Cdk2 complexes during late G1 phase. Sequentially, Cdk4/6 and Cdk2 complexes collaborate to hyperphosphorylate the retinoblastoma gene product Rb, to initiate an E2F-dependent transcriptional program, and to determine the rate of G1 to S transition (Johnson et al. 1993). The entry into the cell cycle is further controlled by the Ink4 protein family that includes p16Ink4a and the closely related p15Ink4b, p18Ink4c, and p19Ink4d proteins (Serrano et al. 1993; Guan et al. 1994; Hannon and Beach 1994; Chan et al. 1995). The four Ink4 proteins exclusively inhibit cyclin D–Cdk4/6 holoenzymes. Furthermore, the Ink4a locus contains an overlapping gene, Arf, encoding p19Arf (Kamijo et al. 1997). Arf influences cell survival in the face of aberrant oncogenic signaling mainly as a negative regulator of the p53-destabilizing oncogene Mdm2 (Pomerantz et al. 1998; Weber et al. 1999).
K-ras mutations are frequently found in human pancreatic cancer (Almoguera et al. 1988). Oncogenic mutants of Ras remain constitutively in the active, GTP-bound form with impaired GTPase activity. Activation of Ras induces DNA synthesis in quiescent fibroblasts. The only phase in which inhibition of Ras affects cell cycle progression is G1 (Mulcahy et al. 1985). Once cells have entered S phase, Ras becomes dispensable for cell cycle progression.
In addition to activation of K-Ras, the tumor suppressor loci p53, INK4a, and SMAD4 are frequently inactivated in human pancreatic cancer (Almoguera et al. 1988; Barton et al. 1991; Caldas et al. 1994; Hahn et al. 1996). Precancerous lesions of pancreatic cancer are not definitively characterized; however, K-ras mutations and allelic inactivation of INK4a have been shown to be present in ductal hyperplasia, indicating the malignant potential of these lesions (Moskaluk et al. 1997).
Mouse models of exocrine pancreatic tumor development do not reproduce the ductal phenotype of human pancreatic adenocarcinoma but display predominantly acinar characteristics (Quaife et al. 1987). Transgenic mice overexpressing transforming growth factor (TGF)-α under the control of the rat elastase promoter (EL-TGF-α-hGH) show a transdifferentiation of acinar cells to duct-like cells, which represent premalignant lesions (Sandgren et al. 1990; Wagner et al. 1998). Here we show that these mice develop pancreatic cancer similar to the human disease regarding cellular differentiation, growth characteristics, and genetic alterations.
Results
The Ras/Erk1/2 pathway is activated in premalignant lesions in TGF-α transgenic mice
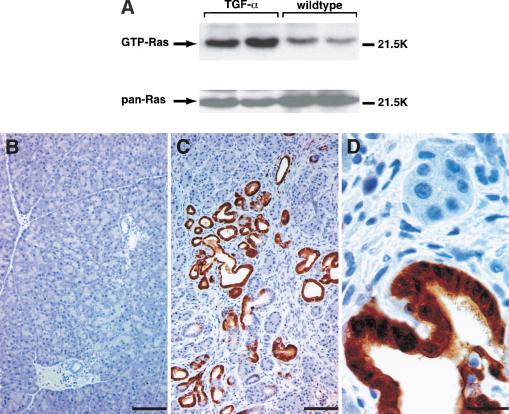
To investigate whether Ras is activated in the pancreas of TGF-α transgenic mice, we have performed a Ras-affinity precipitation assay, which selectively detects GTP-loaded Ras (Taylor and Shalloway 1996). Elevated levels of active Ras are detectable shortly after birth and persist throughout adulthood in the pancreas of TGF-α transgenic mice as compared with littermate controls (Fig. 1A). To confirm Ras downstream signaling, immunohistochemical staining for phosphorylated Erk1/2 protein was performed.
Figure 1.
(A) Ras affinity precipitation assay shows increased levels of GTP-bound Ras in pancreatic lysates of transgenic mice (TGF-α) compared with littermate controls (wild type) at the age of 180 d. Immunohistochemical staining for activated Erk1/2 protein in pancreatic tissue sections of TGF-α transgenic mice (C,D) and littermate controls (B) is shown. The brown color indicates immunoreactivity for activated (phosphorylated) Erk1/2. Bar, 100 μm (B,C) or 20 μm (D).
Figure 1C shows strong staining for phosphorylated Erk1/2 in the pancreas of TGF-α transgenic mice as compared with littermate controls (Fig. 1B). The immunoreactivity for active Erk is evident both in the nucleus and cytoplasm of duct-like cells in tubular complexes (Fig. 1D). Fibroblasts and acinar cells surrounding the tubular complexes are negative for phosphorylated Erk protein (Fig. 1D). These data show that the Ras/Erk signaling cascade is activated selectively in tubular complexes that represent the premalignant lesions in TGF-α transgenic mice.
Cyclin D1/Cdk4 activity is increased in premalignant lesions in TGF-α transgenic mice
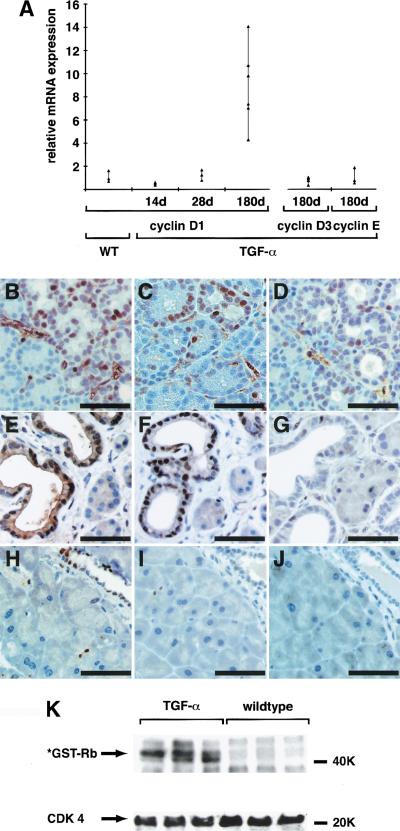
Real-time PCR analysis was performed to investigate whether Ras signaling in the pancreas of TGF-α transgenic mice leads to up-regulation of cyclin D1. Cyclin D1 mRNA is increased up to 14-fold in the pancreas of mice with developed tubular complexes (180-day-old mice). mRNA levels of cyclins D3 and E remain unchanged (Fig. 2A). In mice with developing tubular complexes (28 d), immunohistochemical analysis shows staining for cyclin D1 (Fig. 2B), Cdk4 (Fig. 2C), and cyclin E (Fig. 2D) in a few clusters of acinar cells as well as in the surrounding fibroblasts.
Figure 2.
(A) Relative mRNA levels for cyclin D1, cyclin D3, and cyclin E were evaluated in transgenic animals (TGF-α) by Real Time PCR and normalized to endogenous cyclophilin as described in Materials and Methods. Controls were performed with wild-type pancreas for each time point and target individually. One representative control is shown (WT). Immunostains were performed for cyclin D (B,E,H), Cdk4 (C,F,I), and cyclin E (D,G,J) in TGF-α transgenic mice with developing tubular complexes (at 28 d, B–D), or with developed complexes (at 180 d, E–G) and compared with wild-type controls (at 180 d, H–J). (B–D) Cyclin D, Cdk4, and cyclin E localize in small foci of developing tubular complexes and the surrounding fibroblasts. (E–G) Developed tubular complexes show strong immunoreactivity for cyclin D and Cdk4, whereas only single nuclei are positive for cyclin E. (H–J) Littermate controls show only occasional staining for either cyclin D, Cdk4, or cyclin E. (B–F) Bar, 50 μm. Autoradiograph of a representative immunocomplex kinase assay for cyclin D1/Cdk4 activity indicates increased phosphorylation of the *GST-Rb (769–921) substrate in pancreatic lysates from transgenic mice TGF-α at 180 d as compared with littermate controls (wild type).
After 180 d, expression of cyclin D and Cdk4 is significantly increased but restricted to cells within tubular complexes, whereas cyclin E is not detectable anymore (Fig. B–J). To investigate whether this up-regulation of cyclin D1 and Cdk4 results in increased activity of cyclin D1–Cdk4 complexes, immune complex kinase assays were performed. Figure 2K shows strong cyclin D1/Cdk4 activity in the pancreas of TGF-α transgenic mice, whereas GST-Rb(769–921) phosphorylation is barely detectable in the pancreas of littermate controls.
TGF-α promotes progression throughout G1, but not S phase, in premalignant lesions
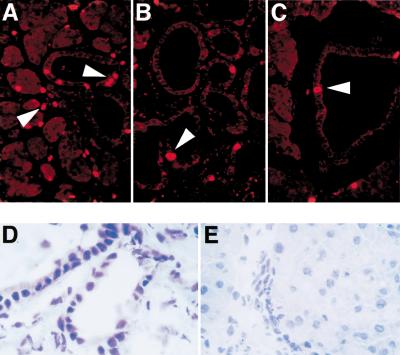
To detect cell cycle progression, immunofluorescence analysis for PCNA and BrdU labeling were performed. BrdU incorporation is predominantly localized to cells in developing tubular complexes, with labeling indices of 15.5 ±3.4% (Fig. 3A). At 180 d, fewer cells have incorporated BrdU in tubular structures, with a labeling index of 4.9 ±2.2% (Fig. 3B). Nontransgenic controls show occasional staining for BrdU in ducts (Fig. 3C). Although almost all cells within tubular complexes stain positive for cyclin D and Cdk4 (Fig. 2E,F), only few of these cells display nuclear immunoreactivity for BrdU. Similar results were obtained with PCNA staining (data not shown). Cyclin E is not induced in tubular complexes. mRNAs of cyclin E (Fig. 2A) and thymidine kinase 1 are not up-regulated in the pancreas of transgenic animals (data not shown). These data suggest that TGF-α is able to promote progression throughout G1, but not S phase, in premalignant lesions.
Figure 3.
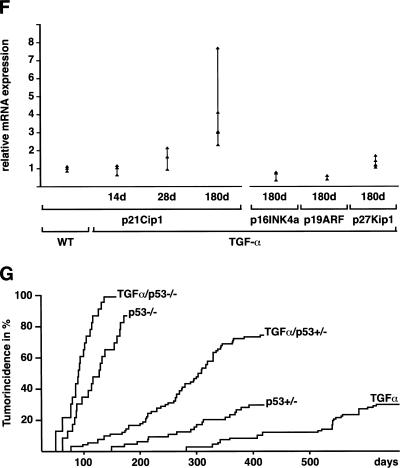
Proliferating cells are detected by immunofluorescence analysis of BrdU labeling in the pancreas of TGF-α transgenic mice with developing tubular complexes (A) or developed complexes (B) and in littermate controls (C). Arrowheads indicate BrdU-positive nuclei. (D,E) Comparison of immunohistochemical staining for p53 in the premalignant lesions in 180-day-old TGF-α transgenic mice (D) with littermate controls (E). (F) Relative mRNA levels of p21Cip1, p16Ink4a, p19Arf, and p27Kip1. Controls were performed with wild-type pancreas for each time point and target individually. One representative control is shown (WT). (G) Cumulative tumor incidence of TGF-α, TGF-α/p53+/−, TGF-α/p53−/−, p53+/−, and p53−/−. Details are described in Materials and Methods.
p53 is induced in tubular complexes and acts as a tumor suppressor in TGF-α transgenic mice
Activated Ras in primary cells is able to induce expression of cell cycle inhibitory molecules such as p16Ink4a, p21Cip1, or p53 and p19Arf. The premalignant lesions in TGF-α transgenic mice display increased nuclear immunoreactivity for p53 (Fig. 3D) as compared with controls (Fig. 3E).
p21Cip1 mRNA levels are increased more than eightfold in 180-day-old TGF-α transgenic animals compared with littermate controls (Fig. 3F). In contrast, mRNA levels of p16Ink4a, p19Arf, and p27Kip1 remain unchanged or even slightly decreased throughout adulthood (Fig. 3F). Western blot analysis confirmed the increase of p21Cip1 expression and unchanged expression of p27Kip1 in transgenic mice (data not shown).
To determine the role of p53 in tumor progression, we mated TGF-α transgenic mice to p53-deficient mice (Jacks et al. 1994). TGF-α transgenic, p53+/− and p53−/−, as well as the crossbred animals were born alive at the expected Mendelian ratio. Up-regulation of p21Cip1 mRNA is abolished in p53-null mice, although p27Kip1 mRNA is slightly induced (data not shown). TGF-α transgenic mice (31 of 107, 29%) developed malignant pancreatic tumors with a mean tumor-free survival of 410 d. Heterozygous loss of p53 resulted in an increased incidence of pancreatic tumors (66 of 90 animals, 77.3%) and decreased mean tumor-free survival to 220 d in TGF-α/p53+/− animals. All 22 TGF-α/p53−/− mice developed pancreatic tumors within 120 d after birth (Fig. 3G). p53+/− and p53−/− littermate controls developed mainly mesenchymal tumors with a frequency and latency as described previously (Fig. 3G; Jacks et al. 1994).
Tumor development in TGF-α/p53+/− and TGF-α/p53−/− mice precedes tumorigenesis in littermate control p53+/− and p53−/− animals. However, necropsy of 3 out of 90 TGF-α/p53+/− mice (3.3%) revealed both a large epithelial pancreatic tumor and extrapancreatic sarcoma (data not shown).
Pancreatic carcinomas display a ductal phenotype, grow invasively into the surrounding tissue, and metastasize
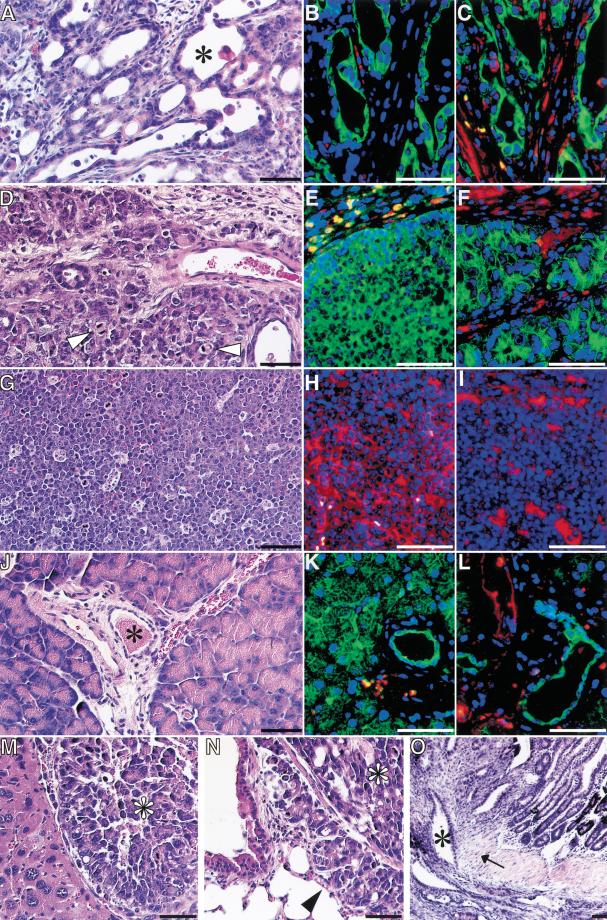
Some pancreatic tumors form ductal structures in dense connective tissue (Fig. 4A–C4); others show frequent mitotic figures surrounded by sparse fibrosis (Fig. 4D–F). p53−/− and p53+/− littermate controls develop sarcomas and lymphomas (Fig. 4G–I; Jacks et al. 1994). Furthermore, we observed malignant ascites, local invasive growth with duodenal obstruction, and metastasis to the liver and lung in TGF-α transgenic mice crossbred to p53-null mice (Fig. 4M–O). Double staining of serial sections with cytokeratin 8/18 and CD45–LCA indicates epithelial differentiation of the pancreatic tumors (Fig. 4B,E). Both pancreatic tumors are positive for the ductal marker cytokeratin 19, whereas surrounding fibroblasts express vimentin (Fig. 4C,F). In contrast, lymphoma from p53-deficient littermate controls express CD45–LCA and vimentin to a lesser degree but do not show any immunoreactivity for cytokeratin 8/18 and 19 (Fig. 4G,I). In wild-type pancreas, cytokeratin 8/18 is expressed both in acinar and ductal cells, whereas cytokeratin 19 is restricted to ducts (Fig. 4J–L).
Figure 4.
Histological analysis of two representative pancreatic tumors (A–F) is compared with thymic lymphoma (G–I) and wild-type pancreas (J–L). (A,D,G,J) Each tissue was stained with hematoxylin and eosin. (B,E,H,K) Expression of CD45-LCA (red fluorescence) and cytokeratin 8/18 (green fluorescence) was analyzed by double-immunostaining. (C,F,I,L) Expression of vimentin (red fluorescence) and cytokeratin 19 (green fluorescence) was analyzed by double-immunostaining. Note the histological appearance of pancreatic tumors with ductal structures (asterisk in A) and frequent mitotic figures (arrowhead in D). Pancreatic tumors express cytokeratin 8/18 (B,E) and 19 (C,F) in contrast with the lymphoma, which stains for CD45-LCA (H). Vimentin staining is restricted to fibroblasts and lymphoma (C,F,I). Acinar and duct cells stain for cytokeratin 8/18 in the normal pancreas (K), whereas cytokeratin 19 is restricted to ducts (asterisk in I,L). Pancreatic tumors metastasize to the liver (M) and lung (N) and infiltrate the duodenal wall (O). Metastasis (M,N) originates from the tumor shown in D–F. Bars, 100 μm in H&E stains, 50 μm in immunostains; (O) bar, 160 μm.
Pancreatic tumors in TGF-α/p53+/− mice segregate the wild-type p53 allele and acquire homozygous deletions of the Ink4a/Arf locus
Acceleration of tumor formation in TGF-α/p53+/− and TGF-α/p53−/− transgenic mice identifies the p53 protein as a rate-limiting step during tumorigenesis. To elucidate whether heterozygous p53-null mice lose the inherited wild-type p53 allele during tumorigenesis, we have applied loss of heterozygosity (LOH) analysis to laser-captured microdissected tumor specimen from F1 and F2 Cl57Bl/6–Balb/c hybrid mice informative for polymorphic genetic markers. The residual wild-type p53 allele is segregated in almost all of the tumors arising from TGF-α/p53+/− mice (Table 1). Interestingly, tumors still take more than 45 d to develop in TGF-α/p53−/− mice, suggesting that in addition to persistent activation of Ras and inactivation of p53, other genetic events have been acquired during tumorigenesis. To reveal the basis for this delayed onset, LOH analysis was performed with known tumor suppressor loci. Both alleles of the Ink4a/Arf locus were deleted in 5 out of 15 tumors, and in one case a heterozygous deletion was found, indicating a strong selection pressure for loss of functional p53 and Ink4a/Arf. In addition, we found LOH of the Smad4 locus in four tumors. The well-characterized tumor suppressor locus Rb1 was affected in only two of these tumors (Table 1).
Table 1.
Loss of heterozygosity analysis of tumors in TGF-α/p53+/− and TGF-α/p53−/− mice
|
|
Genotype
|
p53 D11Mit31, D11Mit320, D11Mit177
|
p16Ink4a D4Mit15, D4Mit145, D4Mit45
|
Smad4 D18Mit189, D18Mit210, D18Mit49
|
Rb1 D14Mit115, D14Mit69 D14Mit34
|
|---|---|---|---|---|---|
| 23 F1 | TGFα × p53+/− | LOH | no loss | LOH | no loss |
| 37 F1 | TGFα − p53+/− | LOH | biallelic loss | no loss | LOH |
| 39 F1 | TGFα − p53+/− | no loss | biallelic loss | no loss | no loss |
| 41 F1 | TGFα − p53+/− | LOH | no loss | LOH | no loss |
| 59 F1 | TGFα − p53+/− | LOH | no loss | no loss | no loss |
| 60 F1 | TGFα − p53+/− | LOH | biallelic loss | no loss | no loss |
| 63 F1 | TGFα − p53+/− | LOH | no loss | no loss | no loss |
| 64 F1 | TGFα − p53+/− | LOH | LOH | no loss | LOH |
| 70 F1 | TGFα − p53+/− | LOH | no loss | no loss | no loss |
| 73 F1 | TGFα − p53+/− | LOH | biallelic loss | LOH | no loss |
| 93 F1 | TGFα − p53+/− | LOH | no loss | no loss | no loss |
| 107 F1 | TGFα − p53+/− | LOH | no loss | no loss | no loss |
| 30 F2 | TGFα − p53+/− | LOH | no loss | not informative | no loss |
| 49 F2 | TGFα − p53+/− | not informative | biallelic loss | not informative | not informative |
| 129 F2 | TGFα − p53+/− | LOH | no loss | LOH | no loss |
Discussion
Mice overexpressing TGF-α under the control of the elastase promoter develop pancreatic tumors originating from cells within tubular complexes after 1 yr of age (Wagner et al. 1998). In the pancreas of these transgenic mice, persistent stimulation of the EGF receptor by TGF-α increases GTP loading of Ras and phosphorylation of Erk selectively in tubular structures at 180 d. At 28 d of age these tubular complexes start to develop interspersed between large areas of normal-appearing pancreas. Some cells in these areas seem to be programmed to respond positively to growth activation by Ras signaling and are positive for cyclin D1. At 180 d, numerous tubular complexes form three-dimensional accumulations of ductular structures.
Almost all of these cells show nuclear accumulation of cyclin D1. Neither acinar cells nor fibroblasts are positive for cyclin D1 or Cdk4, suggesting that increased cyclin D/Cdk4 activity is restricted to duct cells. In contrast to 28 d with few cells positive for cyclin E, nuclear accumulation of cyclin E is even less frequently observed in the pancreas at 180 d.
Moreover, no induction of other S-phase proteins such as thymidine kinase 1 or PCNA is detectable, indicating that TGF-α is able to promote progression through G1 but not S phase. These data indicate that early after birth (28 d), cells within developing tubular complexes divide, whereas later, at 180 d, most of these cells become growth-arrested. This conclusion is confirmed by BrdU labeling.
These findings recapitulate previous in vitro data that ras not only is a transforming oncogene but also can exert a negative effect on cell growth. Furthermore, it was observed that these negative effects of Ras could be overcome, in some cases, by simultaneous expression of a cooperating oncogene (Franza et al. 1986). Some more recent data provide a possible explanation for these earlier results as it has been shown that introduction of oncogenic Ras into mouse fibroblasts leads to cell cycle arrest, through induction of Cdk inhibitors p16Ink4a, p21Cip1, or p19Arf and p53 (Macleod et al. 1995; Serrano et al. 1997; Palmero et al. 1998). In contrast to p16Ink4a, which is not up-regulated in the pancreas of TGF-α transgenic mice, p21Cip1 is induced up to sevenfold at the mRNA level. Increased expression of p21Cip1 would provide a logical explanation for the G1 arrest (Sherr and Roberts 1999). The molecular basis for the induction of p21Cip1 is complex. In addition to activation through Raf/MEK/Erk, p21Cip1 is subject to transcriptional induction by p53 (el Deiry et al. 1993; Macleod et al. 1995). Nuclear accumulation of p53 is present in cells of tubular complexes, suggesting that p53 represents a candidate for mediating growth arrest in these premaligant lesions. The protein levels of p53 are determined primarily by ubiquitination and subsequent proteolytic degradation. The main ubiquitin ligase for p53 is the oncogene Mdm2. p19Arf appears to protect p53 from degradation by binding directly to Mdm2 (Pomerantz et al. 1998; Weber et al. 1999).
Although it is clear from in vitro studies that overexpression of Ras leads to the induction of p19Arf, we cannot detect an up-regulation of p19Arf in the pancreas of TGF-α transgenic mice.
Fibroblasts derived from p53−/− mice were able to overcome the G1 arrest mediated by Ras and form transformed foci (Serrano et al. 1997).
In mating experiments we show a dramatically accelerated tumor formation in TGF-α/p53−/− mice resulting in pancreatic cancer development before the age of 120 d. TGF-α transgenic mice, hemizygous for p53-deficiency, display accelerated disease, and almost 100% of these tumors segregate the wild-type p53 allele. Loss of p53 accelerates tumor formation; however, it takes 45 d to develop cancerous lesions in TGF-α transgenic mice null for p53. In the pancreas of these mice, persistent activation of Ras and cyclin D1/Cdk4 is observed shortly after birth. This suggests inactivation of additional genes coding for growth-arrest proteins to be required during tumorigenesis. LOH analysis revealed frequent homozygous losses of the Ink4a/Arf locus in TGF-α/p53+/− mice. The deletion also includes Ink4b. The Ink4a/Arf locus encodes the two related putative tumor suppressors p16Ink4a and p19Arf (Serrano et al. 1996). p16Ink4a represents a potent tumor suppressor in humans and is frequently inactivated in pancreatic neoplasms (Caldas et al. 1994). In addition, inherited mutations of exon 1α implicate p16Ink4a in familial tumor susceptibility (Gruis et al. 1995). However, the final proof of p16Ink4a as a tumor suppressor in mice awaits the specific inactivation of the p16Ink4a gene. The loss-of-function approach strongly suggested that p19Arf is the major tumor suppressor in mice (Serrano et al. 1996; Kamijo et al. 1997). Deletions of the Ink4a/Arf locus occurred in 6 of 15 tumors in TGF-α/p53+/− and TGF-α/p53−/− mice. This result was unexpected from previous findings, indicating a mutually exclusive manner of p53 and p19Arf inactivation in c-myc-induced lymphomagenesis (Eischen et al. 1999). All tumors retained other polymorphic markers, showing that the effect cannot be simply explained by chromosomal instability. These data therefore indicate that the Ink4a/Arf locus is synergistic to p53 in tumor suppression. Similarly, human pancreatic cancer involves homozygous deletion of the region 9p21-p22, resulting in inactivation of the p14Arf and p15INK4b genes (Naumann et al. 1996). Similar to human pancreatic cancer, LOH of the SMAD4/DPC4 locus occurred with a lower frequency only in our tumor model (Hahn et al. 1996). Recent data suggest functional inactivation of the TGF-β–Smad4 pathway in the presence of activated Ras, which might explain a reduced selection pressure for LOH at the Smad4 locus (Kretzschmar et al. 1999). Furthermore, the Rb1 locus was infrequently affected by LOH. This is consistent with the low number of Rb mutations in human pancreatic cancer.
The tumor spectrum of p53-deficient mice is dominated by lymphomas and sarcomas (Jacks et al. 1994). This spectrum shifted to ductal pancreatic cancer in crossbred animals. Only 3 out of 90 mice developed pancreatic tumors and sarcomas. APC-mutated (Min) mice null for p53 develop pancreatic acinar cell adenocarcinomas (Clarke et al. 1995). Our data complement this observation that p53 is critical in the protection of pancreatic epithelial cells by causing cell cycle arrest.
Carcinogenesis is a multistep process; to reach the malignant phenotype, multiple alterations affecting several levels of growth control are required. Here we report a sequence of genetic events in a murine pancreatic cancer model that recapitulates pathomorphological features and genetic alterations of the human disease.
Materials and methods
Transgenic mice
The EL-TGF-α-hGH transgenic mice (line #2261-3) and p53-deficient mice have been described (Sandgren et al. 1990; Jacks et al. 1994). Crossbreeding and inbreeding of congenic C57BL/6xEL–TGF-α–hGH transgenic and Balb/cxp53-deficient mice resulted in F1 C57BL/6-Balb/c progeny of TGF-α/p53+/− and in F2 progeny of TGF-α/p53−/− mice. Mice were sacrificed for analysis at given time points or with obvious signs of wasting disease. The tumor incidence of TGF-α (n = 107), TGF-α/p53+/− (n = 90), and TGF-α/p53−/− (n = 22) transgenic mice was monitored.
p53+/− (n = 64) and p53−/− (n = 23) littermate controls were followed up to 360 and 180 d, respectively, and evaluated for tumor development. The cumulative tumor incidence in each group was blotted as a function of the age of the animals (Origin 6.0, Northampton, MA). The mean tumor-free survival was calculated from the survival function according to Kaplan and Meier.
Activation assay for Ras, immunocomplex kinase assay, and Western blot
Activation assay for Ras was performed as described recently (Taylor and Shalloway 1996) with a modified, Mg2+-containing lysis buffer (MLB: 25 mM HEPES at pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 1% Nonidet P-40, 0.05 % sodium deoxycholate, 1 mM sodium vanadate, 10 μg/mL aprotinin, 0.5 μmol/mL leupeptin). Cdk4 activity assay was performed as described (Kerkhoff and Rapp 1997). GST-Rb(769–921) and anti-Cdk4 were purchased from Santa Cruz Biotechnology. Anti-p21Cip1 and anti-p27Kip1 were from Pharmingen.
Quantitative RNA analysis
mRNA expression was evaluated by RT–PCR analysis using Real Time PCR (TaqMan, PE Applied Biosystems) (Gibson et al. 1996). Reverse transcription of 2μg total RNA was performed in duplicate and further processed independently. PCR reaction (denaturation at 95°C for 2 min, followed by 40 cycles at 95°C for 15 sec and 60°C for 1 min; Sybr Green PCR Core Reagents, PE Applied Biosystems) was performed in triplicate and normalized to the endogenous cyclophilin mRNA level for each reaction. Target cDNA in transgenic mice was quantified using the delta-delta-CT method. Primer sequences are available on request.
Histopathological analysis
Hematoxylin/eosin and immunohistochemical analysis were performed as described previously (Wagner et al. 1998). Sections were stained with anti-p53 (1:2000, rabbit polyclonal), anti-phospho-Erk1/2 (1:200, New England Biolabs), anti-cyclin D (1:50), anti-cyclin E (1:40), anti-Cdk4 (1:50), anti-PCNA (1:200, all Santa Cruz Biotechnology), anti-cytokeratin 19 (1:200, Abcam), anti-cytokeratin 8/18 (1:400, Progen), anti-vimentin (1:40, ICN), and anti-CD45-LCA (1:50, Pharmingen). Double labeling for cytokeratin 19/vimentin and cytokeratin 8/18/CD45-LCA was performed overnight at 4°C in PBS/1.0% Triton X-100/2% BSA followed by sequential incubation with fluorochrome-labeled anti-goat (Alexa 568)/anti-mouse (Alexa 488) and anti-guinea pig (Alexa 488)/anti-rat (Alexa 568) antibodies and nuclear counterstain with Hoechst dye.
For in vivo BrdU labeling, animals were injected with BrdU (100 mg/kg body weight) intraperitoneally 3 h prior to sacrifice. Immunostaining for BrdU was performed using a monoclonal anti-BrdU antibody (1:400, Boehringer Mannheim). Labeling indices were calculated as fraction of BrdU-positive cells in relation to the total cell number and expressed as percentage ± SEM. For each time point, tubular complexes of four sections separated by 200 μm of at least three individual animals were evaluated.
Laser-captured microdissection and LOH analysis
Frozen or paraffin sections (5 μm) were applied to laser-capture microdissection (LCM; Arcturus Engineering, Mountain View, CA). Proteinase K-eluted genomic DNA was subjected to PCR amplification (1 mM dNTP, 15 pmole of each primer, 1.5 U AmpliTaqGold; PE Applied Biosystems) of the polymorphic markers located within 1–4-cM distance to the gene locus of interest. p53 (chromosome 11; 39 cM): D11Mit31 (40 cM), D11Mit320 (43cM), D11Mit177 (36cM); Ink4a/Arf (chromosome 4; 42.7 cM): D4Mit15 (42.6 cM), D4Mit145 (44.5 cM), D4Mit45 (42.5 cM); Smad4 (chromosome 18; 48 cM): D18Mit189 (48 cM), D18Mit210 (47 cM), D18Mit49 (49 cM); Rb1 (chromosome 14; 41 cM): D14Mit115 (40 cM), D14Mit69 (43 cM), D14Mit34 (40 cM). PCR products were visualized on 3% agarose or on polyacrylamide gel using fluorescence-labeled primers (ALFexpress II DNA Analysis system, Amersham Pharmacia Biotech).
Acknowledgments
We thank E. Sandgren for the generous gift of the transgenic mouse line EL-TGFα-hGH (no. 2261.3) and R.D. Palmiter for the hGH poly(A) probe. We thank R. Rittelmann for excellent technical support and S. Aigner for assistance with manuscript preparation. We thank H. Häcker and all members of the laboratory for helpful discussion. This work was in part supported by grants from the Dr. Mildred Scheel-Stiftung, Bundesministerium für Bildung und Forschung and from the Deutsche Forschungsgemeinschaft (SFB518, B6) to R.M.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL roland.schmid@medizin.uni-ulm.de; FAX 49-731-500-24302.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.184701 .
References
- Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–554. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- Barton CM, Staddon SL, Hughes CM, Hal PA, O'Sullivan C, Klöppel G, Theis B, Russell RC, Neoptolemos J, Williamson RC. Abnormalities of the p53 tumour suppressor gene in human pancreatic cancer. Br J Cancer. 1991;64:1076–1082. doi: 10.1038/bjc.1991.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldas C, Hahn SA, da Costa LT, Redston MS, Schutte M, Seymour AB, Weinstein CL, Hruban RH, Yeo CJ, Kern SE. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet. 1994;8:27–32. doi: 10.1038/ng0994-27. [DOI] [PubMed] [Google Scholar]
- Chan FK, Zhang J, Cheng L, Shapiro DN, Winoto A. Identification of human and mouse p19, a novel CDK4 and CDK6 inhibitor with homology to p16ink4. Mol Cell Biol. 1995;15:2682–2688. doi: 10.1128/mcb.15.5.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AR, Cummings MC, Harrison DJ. Interaction between murine germline mutations in p53 and APC predisposes to pancreatic neoplasia but not to increased intestinal malignancy. Oncogene. 1995;11:1913–1920. [PubMed] [Google Scholar]
- Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF–Mdm2–p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes & Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Franza BR, Jr, Maruyama K, Garrels JI, Ruley HE. In vitro establishment is not a sufficient prerequisite for transformation by activated ras oncogenes. Cell. 1986;44:409–418. doi: 10.1016/0092-8674(86)90462-9. [DOI] [PubMed] [Google Scholar]
- Gibson UE, Heid CA, Williams PM. A novel method for real time quantitative RT–PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- Gruis NA, van der Velden PA, Sandkuijl LA, Prins DE, Weaver-Feldhaus J, Kamb A, Bergman W, Frants RR. Homozygotes for CDKN2 (p16) germline mutation in Dutch familial melanoma kindreds. Nat Genet. 1995;10:351–353. doi: 10.1038/ng0795-351. [DOI] [PubMed] [Google Scholar]
- Guan KL, Jenkins CW, Li Y, Nichols MA, Wu X, O'Keefe CL, Matera AG, Xiong Y. Growth suppression by p18, a p16INK4/MTS1- and p14INK4B/MTS2-related CDK6 inhibitor, correlates with wild-type pRb function. Genes & Dev. 1994;8:2939–2952. doi: 10.1101/gad.8.24.2939. [DOI] [PubMed] [Google Scholar]
- Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Beach D. p15INK4B is a potential effector of TGF-β-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Johnson DG, Schwarz JK, Cress WD, Nevins JR. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- Kerkhoff E, Rapp UR. Induction of cell proliferation in quiescent NIH 3T3 cells by oncogenic c-Raf-1. Mol Cell Biol. 1997;17:2576–2586. doi: 10.1128/mcb.17.5.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar M, Doody J, Timokhina I, Massague J. A mechanism of repression of TGFβ/Smad signaling by oncogenic Ras. Genes & Dev. 1999;13:804–816. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod KF, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, Vogelstein B, Jacks T. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes & Dev. 1995;9:935–944. doi: 10.1101/gad.9.8.935. [DOI] [PubMed] [Google Scholar]
- Moskaluk CA, Hruban RH, Kern SE. p16 and K-ras gene mutations in the intraductal precursors of human pancreatic adenocarcinoma. Cancer Res. 1997;57:2140–2143. [PubMed] [Google Scholar]
- Mulcahy LS, Smith MR, Stacey DW. Requirements for ras proto-oncogene function during serum-stimulated growth of NIH 3T3 cells. Nature. 1985;313:241–243. doi: 10.1038/313241a0. [DOI] [PubMed] [Google Scholar]
- Naumann M, Savitskaia N, Eilert C, Schramm A, Kalthoff H, Schmiegel W. Frequent codeletion of p16/MTS1 and p15/MTS2 and genetic alterations in p16/MTS1 in pancreatic tumors. Gastroenterology. 1996;110:1215–1224. doi: 10.1053/gast.1996.v110.pm8613012. [DOI] [PubMed] [Google Scholar]
- Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature. 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee HW, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- Quaife CJ, Pinkert CA, Ornitz DM, Palmiter RD, Brinster RL. Pancreatic neoplasia induced by ras expression in acinar cells of transgenic mice. Cell. 1987;48:1023–1034. doi: 10.1016/0092-8674(87)90710-0. [DOI] [PubMed] [Google Scholar]
- Sandgren EP, Luetteke NC, Palmiter RD, Brinster RL, Lee DC. Overexpression of TGF α in transgenic mice: Induction of epithelial hyperplasia, pancreatic metaplasia, and carcinoma of the breast. Cell. 1990;61:1121–1135. doi: 10.1016/0092-8674(90)90075-p. [DOI] [PubMed] [Google Scholar]
- Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes & Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Taylor SJ, Shalloway D. Cell cycle-dependent activation of Ras. Curr Biol. 1996;6:1621–1627. doi: 10.1016/s0960-9822(02)70785-9. [DOI] [PubMed] [Google Scholar]
- Wagner M, Lührs H, Klöppel G, Adler G, Schmid RM. Malignant transformation of duct-like cells originating from acini in mice overexpressing transforming growth factor α in the exocrine pancreas. Gastroenterology. 1998;115:1254–1262. doi: 10.1016/s0016-5085(98)70098-8. [DOI] [PubMed] [Google Scholar]
- Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]