Abstract
For positive-strand RNA viruses, the viral genomic RNA also acts as an mRNA directing the translation of the replicase proteins of the virus. Replication takes place in association with cytoplasmic membranes, which are heavily modified to create specific replication compartments. Here we have expressed by plasmid DNA transfection the large replicase polyprotein of Semliki Forest virus (SFV) in mammalian cells from a nonreplicating mRNA and provided a separate RNA containing the replication signals. The replicase proteins were able to efficiently and specifically replicate the template in trans, leading to accumulation of RNA and marker gene products expressed from the template RNA. The replicase proteins and double-stranded RNA replication intermediates localized to structures similar to those seen in SFV-infected cells. Using correlative light electron microscopy (CLEM) with fluorescent marker proteins to relocate those transfected cells, in which active replication was ongoing, abundant membrane modifications, representing the replication complex spherules, were observed both at the plasma membrane and in intracellular endolysosomes. Thus, replication complexes are faithfully assembled and localized in the trans-replication system. We demonstrated, using CLEM, that the replication proteins alone or a polymerase-negative polyprotein mutant together with the template did not give rise to spherule formation. Thus, the trans-replication system is suitable for cell biological dissection and examination in a mammalian cell environment, and similar systems may be possible for other positive-strand RNA viruses.
INTRODUCTION
When the RNA component of a positive-strand RNA virus enters the cytoplasm of a permissive cell, it is first translated. This primary translation commonly yields a polyprotein, which contains at least the replicase proteins of the virus. Some of the replicase proteins together with possible host factors then recruit the viral RNA to replication complexes, where RNA replication commences with the synthesis of at least one complementary negative strand, which can then be used as a template for positive-strand RNA synthesis. Newly synthesized positive strands can go through the same cycle, leading to an exponential increase in RNA synthesis during the early phase of virus replication. The replication complexes of positive-strand RNA viruses are specialized membrane structures, whose appearance and localization vary in different virus groups, but they can include different sizes of single-membrane or double-membrane vesicles, membrane invaginations, or complex membranous web structures (27, 30, 39). The formation of the replication complexes requires a number of often poorly characterized substages, including at least the direct localization or more complex transport of the replication proteins and RNA to the correct membranes, the membrane binding of the replication proteins, formation of the characteristic membrane structures, and recruitment and action of the host factors which are likely to be required for many of these stages.
The genus Alphavirus includes some 30 known members, most of which are transmitted by mosquitoes between mammalian and avian hosts (9). Alphavirus infection can be associated with neurological symptoms (e.g., Venezuelan, western, and eastern equine encephalitis viruses) or with fever, rash, and arthritis (e.g., Chikungunya virus, Ross River virus, and Sindbis virus [SIN]). The alphavirus genome is a capped and polyadenylated RNA of approximately 11.5 kb containing two open reading frames (ORFs). The four nonstructural or replication proteins (nsPs) nsP1 to nsP4 are translated from the first ORF as a polyprotein, P1234. In some members of the genus, there is an opal termination codon between nsP3 and nsP4, which leads to the production of P123 as the major product and P1234 as the minor read-through product of translation (13, 47). In the first stage of replication, the core polymerase nsP4 is separated by proteolytic cleavage from P123, and the combination P123-nsP4 mediates minus-strand RNA synthesis in so-called early replication complexes, which have a short half-life. Further processing to individual nsPs transforms these complexes into stable late replication complexes, which synthesize positive-sense genomic and subgenomic RNAs (15, 22, 42, 50). The subgenomic RNA represents the 3′ one-third of the genome and is the mRNA for the structural proteins of the virus.
Functional replication complexes are not formed, if the nsPs are expressed separately (20). In addition to being functionally important in replication, the polyprotein stage is required for the proper formation of protein-protein interactions, as well as the membrane association and targeting of the replication complexes to the correct membranes (40). nsP1, which is a peripheral membrane protein, acts as the sole membrane anchor of the replication complex (1, 46). The replication complexes are initially found on the plasma membrane and transported from there to the surfaces of large endolysosomal vacuoles (45). Morphologically, the alphavirus replication complexes are invaginations of the membrane. Each invagination, also called a spherule, is approximately 50 nm in inner diameter, and the interior of the invagination is always connected to the cytoplasm by a narrow neck-like structure (8, 17). In the models proposed (13, 41, 45), the newly synthesized positive-sense RNAs are released to the cytoplasm through the neck, and the double-stranded RNA (dsRNA) replication intermediates are protected inside the spherules. Many positive-sense RNA virus replication complexes closely resemble the alphavirus spherules, but they can be located on different membranes. Brome mosaic virus (BMV) spherules are found on the endoplasmic reticulum (ER), nodavirus spherules on mitochondria, tombusvirus-caused invaginations on peroxisomes or mitochondria, and dengue virus replication complexes at the ER (5, 29, 41, 51).
In this work, the aim was to establish a system which would allow the dissection of the different stages in the assembly of the functional alphavirus replication complex. We have expressed various replication proteins derived from Semliki Forest virus (SFV) and provided them with separate replication templates inside BHK cells. This system affords more experimental freedom than complete virus genomes or replicons, in which the production of the replication proteins is always coupled to the replication of the mRNA from which they are translated. We demonstrate that the expressed polyproteins could efficiently utilize templates provided in trans. Only in the presence of an active replicase protein together with the template, abundant native spherule structures were observed both at the plasma membrane and in endosomal structures.
MATERIALS AND METHODS
Cell culture and viruses.
BSR T7/5 (BSR) cells (4), a derivative of BHK cells stably expressing T7 RNA polymerase, were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Gibco-BRL, Grand Island, NY), 2% Bacto tryptose phosphate broth (Difco Laboratories, Detroit, MI), 1% nonessential amino acids (Gibco), 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin (Gibco), and 1 mg/ml G418 for selection of T7 polymerase expression. Cells for correlative light electron microscopy (CLEM) experiments were grown on no. 2 glass-bottom P35G-2-14-C-Grid dishes (MatTek). SFV was propagated as described previously (46), and SFV-Rluc (where Rluc represents Renilla luciferase) has been described elsewhere (36).
Plasmid constructs.
Regions encoding the wild-type replicase polyproteins originated from plasmids P123 and P1234 (28); regions encoding the polyproteins with mutated processing sites originated from plasmids P1∧2∧3 and P1∧2∧34, where the symbol ^ indicates point mutations preventing cleavage) (49). To achieve high-level expression of polyprotein constructs, the polyprotein regions were inserted after the internal ribosome entry site (IRES) element of encephalomyocarditis virus (EMCV) in pTM1 (kindly provided by Bernard Moss, National Institutes of Health). For this purpose, the region encoding amino acids 1 to 66 of P1234 was PCR amplified and cloned into the pTM1 vector so that the initiation codon for nsP1 was exactly positioned in the start site (NcoI site) of the pTM1 vector. Subsequently, the fragments encoding the remainder of the polyproteins were subcloned into this construct by using the naturally occurring EcoRV restriction site (encompassing the codons for amino acids [aa] 64 to 66 of nsP1) for the 5′ end and selected unique restriction sites from the pTM1 polylinker as 3′ cloning sites. To obtain the constructs expressing fluorescent nsP3 fusion protein, the ZsGreen marker was cloned in frame in the naturally occurring XhoI site in the C-terminal region of nsP3 (insertion between aa residues 1740 and 1741 of the polyprotein P1234). The presence of ZsGreen was denoted with a Z (as in P123Z4).
The ubiquitin fusion technique (22, 48) was used to obtain nsP4 with a native N-terminal tyrosine residue (ubi4). The construct was inserted into the NcoI site of the pTM1 vector, as described for the polyproteins. Inactive polymerase mutant nsP4GAA was achieved by replacing the NheI-AflII fragment of nsP4 with a mutation-containing fragment from pETnsP4GAA, in which it had been constructed by PCR-based mutagenesis followed by sequencing. In addition, the GAA mutation was transferred to P1234 and P1∧2∧34 polyproteins as a NotI-BlpI fragment.
The basic truncated replication template was constructed as follows. The region corresponding to 307 bp of the 5′ end of the SFV genome (based on the sequence of the infectious clone pSFV4) was first amplified by PCR using sense primer 5′-ATAAAGCTTAATACGACTCACTATAGATGGCGGATGTGTGACATACAC-3′ (the sequence of the introduced T7 RNA polymerase promoter is underlined, and the sequence corresponding to SFV is shown in boldface) and antisense primer 5′-ATAGGATCCCTCGAGCATCATTCTCCTGGAAGGCGCAC-3′. This introduced a correctly positioned T7 promoter upstream of the SFV-specific sequence and XhoI and BamHI restriction sites downstream of the sequence. The amplified insert encodes an N-terminal fragment of the nsP1 reading frame of SFV (codons 1 to 74). The insert was then cloned into pUC18 using HindIII and BamHI restriction enzymes. The 3′ end of the replication template was ordered as a synthetic DNA from Geneart AG (Germany) and consisted of (i) a short polylinker containing XhoI, ApaI, and BamHI recognition sites (inserted in frame with the reading frame encoding the N-terminal region of nsP1); (ii) a truncated subgenomic promoter of SFV4, encompassing bases from positions −37 to +51 with respect to the transcription start site; (iii) a second polylinker region containing NotI, BglII, and SalI recognition sites; (iv) a 3′-terminal fragment of the 3′ untranslated region (UTR) of SFV4 with a length of 61 bp, followed by a poly(A) tract of 69 residues; and (v) the hepatitis delta virus antisense ribozyme sequence, followed by a T7 transcription terminator. This construct was fused with the 5′ region of the template using XhoI and SacI restriction sites and designated pUC18 templ+. The overall length of the cassette was 779 bp, and the calculated length of the transcript was 601 bp, including the poly(A) tail.
The pUC18 templ+ construct was used to create additional replication templates. To obtain a template expressing an easily detectable marker, the sequence of the Renilla luciferase gene was PCR amplified with primers creating either an ApaI or NotI restriction site at the 5′ end and a BamHI or BglII restriction site at the 3′ end. The Renilla luciferase gene was inserted either in frame with the N-terminal region of nsP1 by using the first polylinker (ApaI and BamHI) in pUC18 templ+ or as a separate reading frame under the control of the subgenomic promoter by using the second polylinker (NotI and BglII) in pUC18 templ+. These templates were designated TshortNsluc (or simply Tshort) and TshortStluc, respectively. A 5′ UTR modification was introduced into the short templates in order to increase specific replication levels. ATGGCG in the very beginning of the 5′UTR was changed to ATGACG (where the changed nucleotide is underlined) by PCR-based mutagenesis.
In order to increase the length of the Tshort replication template, the gene encoding the red fluorescent protein Tomato was amplified from ptdTomato-N1 (Clontech Laboratories, Mountain View, CA) and cloned with BglII and SalI into the second polylinker site, to yield Tmed. The longest replication template, Tlong, was created by addition of a gene encoding beta-galactosidase (from pSV-beta-galactosidase control vector; Promega, Madison, WI) immediately after the termination codon of the luciferase gene to the BamHI site in reverse orientation in order to avoid any residual translation of the gene.
The cloning of the Tcis template construct was performed by amplifying P123Rluc from plasmid PSFVRlucH2 (36) with a 5′ primer containing the T7 RNA polymerase promoter and the 5′ UTR modification together with a HindIII restriction site and a 3′ primer containing a BamHI restriction site. In this construct, the luciferase gene is inserted after nsP3, as described by Pohjala et al. in 2008 (36). The amplified fragment was cloned to pUC18 templ+.
Antibodies.
Rabbit polyclonal anti-nsP1, anti-nsP2, anti-nsP3, and anti-nsP4 have been described previously (17). Mouse monoclonal antibody against dsRNA (J2) was purchased from Scicons (Hungary). Mouse monoclonal antibody against lysobisphosphatidic acid (LBPA) was kindly provided by Vesa Olkkonen (National Institute for Health and Welfare, Helsinki, Finland). Secondary antibodies conjugated with Alexa Fluor 488/568/647 were obtained from Molecular Probes (Invitrogen, Carlsbad, CA).
DNA transfection.
BSR cells were transfected with plasmids expressing polyproteins and templates under the T7 promoter. Lipofectamine LTX (Invitrogen) was used according to the manufacturer's instructions. Forty nanograms of plasmids encoding replicase proteins and 50 ng of template plasmids were cotransfected into each well of a 96-well plate. In larger plates, the amounts were scaled up according to the area. For expression periods longer than 6 h, the medium containing the transfection reagent was replaced after 4 to 6 h with regular medium.
Luciferase and cytotoxicity assays.
Transfected BSR cells on 96-well plates were collected in Renilla luciferase assay system lysis buffer as instructed by the manufacturer (Promega). The assay reagent was added prior to luminometric measurements. Cytotoxicity was measured with a CellTiter-Glo luminescent cell viability assay (Promega). The assays were always conducted with three parallel biological replicates.
Quantitative real-time PCR analysis.
Total cellular RNAs were isolated with TriZol reagent (Invitrogen), according to the manufacturer's instructions, from BSR cells transfected with the indicated combinations of plasmids. To synthesize cDNAs, 100 ng of total RNA was reverse transcribed with a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). The cDNAs were further diluted 1/10 and analyzed by SYBR green I (Roche Applied Biosciences, Basel, Switzerland) with a LightCycler 480 instrument (Roche) using primers specific for the SFV nsP1 N-terminal fragment and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) of BHK cells, designed by the universal probe library assay design center (Roche). The sequences of the primers were as follows: nsP1 forward, 5′-CGCCAAAAGATTTTGTTCCA-3′; nsP1 reverse, 5′-CCATCGTGGGTGGTTAATCT-3′; BHK GAPDH forward, 5′-ATCCCACCAACATCAAATGG-3′; and BHK GAPDH reverse, 5′-AAGACGCCAGTAGACTCCACA-3′. The expression of the template RNA was determined by the relative quantification method using GAPDH as an endogenous control. Data were collected from two separate biological experiments, which were both run twice in triplicate.
Immunofluorescence analysis and confocal microscopy.
Transfected BSR cells were analyzed by immunofluorescence microscopy as described previously (40). Briefly, samples were fixed with 4% paraformaldehyde and permeabilized with Triton X-100, followed by incubation with primary antibodies. After washes with Dulbecco's phosphate-buffered saline containing 0.2% bovine serum albumin, secondary antibodies labeled with Alexa Fluor 488/568/647 (Molecular Probes) were used. Samples were analyzed with a Leica SP5 confocal microscope using an HCX APO ×63/1.30 Corr (glycerol) CS 21 objective.
Correlative light electron microscopy.
CLEM was used to detect the spherules after plasmid transfections. Samples were fixed with glutaraldehyde for 30 min and washed. A Leica SP2 confocal microscope (HC PL APO ×20/0.7 CS air objective) was used to image the transfected cells with fluorescence as well as in the reflection mode to visualize the cells and the grid. Samples were then prepared for transmission electron microscopy (EM); chemically fixed and osmium-stained cells were processed for flat embedding and ultrathin sectioning as described previously (38). A Jeol 1200 EX II transmission electron microscope operated at 80 kV was used for relocating the same cells previously analyzed with the confocal microscope and for detecting the spherules.
Image analysis.
Confocal images were analyzed using ImageJ software (National Institutes of Health, Bethesda, MD). For three-dimensional (3D) reconstruction of the sample (see Fig. 2C), a confocal series was deconvoluted with AutoDeblur software by using the 3D Blind Deconvolution algorithm (Media Cybernetics, Inc., Bethesda, MD), and Imaris software (Bitplane AG, Zurich, Switzerland) was used to visualize the maximum projection of the cell.
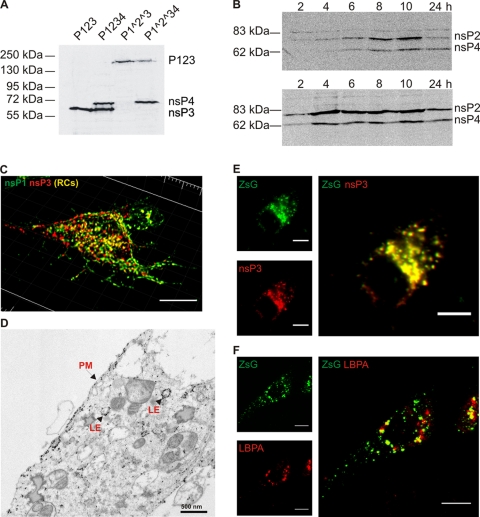
Fig. 2.
Polyprotein expression and localization. (A) BSR cells were transfected with the polyprotein constructs indicated at the top. Extracts were prepared at 24 h posttransfection and analyzed by Western blotting with a combination of antisera against SFV nsP3 and nsP4. Molecular mass markers are shown on the left, and expressed protein products are indicated on the right. (B) Accumulation of the processing products of P1234 polyprotein in BSR cells over time. The upper panel shows extracts from BSR cells transfected with the P1234 construct, while the lower panel shows BSR cells infected with SFV (50 PFU/cell). Western blotting was performed with a combination of antisera against nsP2 and nsP4. Molecular mass markers are shown on the left, and expressed protein products are indicated on the right. (C) Localization of the processing products of P1234 in BSR cells at 24 h posttransfection. Immunofluorescence staining was performed with antisera against nsP1 (green) and nsP3 (red). A 3D representation of the cell is shown. Bar, 10 μm. (D) Electron microscopy-level localization of P1234 at 16 h. Immuno-EM was performed with anti-nsP1 and anti-nsP3 antisera. In the localization studies of panels C and D, the template NSluc was cotransfected with the polyprotein constructs. PM, plasma membrane; LE, late endosome. (E) Localization at the 24-h time point of nsP3 (red) and ZsGreen (green) signals in BSR cells transfected with P123Z. (F) Colocalization on ZsGreen-positive structures (green) in P123Z expressing BSR cells with LBPA (red). In panels E and F, separate channels are illustrated on the left and the merged images are shown on the right. A single confocal optical slice from the middle of the cell is shown. Bars, 10 μm.
Immuno-EM.
Immuno-EM for locating expressed polyproteins was performed as described previously (46). Samples were immunolabeled simultaneously with anti-nsP1 and anti-nsP3 antibodies and were analyzed with a Tecnai 12 microscope.
RESULTS
Expression of replicase polyproteins.
The full-length SFV replicase polyprotein P1234 comprises 2,432 amino acid residues. We wanted to express this and other large polyproteins in mammalian cells at relatively high levels through a system which affords easy manipulation. We therefore adopted BSR T7/5 cells, a stable cell line expressing bacteriophage T7 RNA polymerase (4). One advantage of this system is that the BSR T7/5 cells are derived from BHK cells, which are highly permissive for SFV replication and which have been used in many previous studies of SFV cell biology (2, 10, 45). The polyproteins were expressed from transfected plasmids containing the EMCV IRES element downstream of the T7 promoter, to enhance translation of the presumably uncapped transcripts synthesized by the T7 polymerase. In addition to wild-type P1234, constructs expressing the shorter polyprotein P123, as well as variants in which the cleavages between nsP1/nsP2 and nsP2/nsP3 are prevented by mutations at the cleavage sites, were utilized (Fig. 1A). In some constructs, the fluorescent protein ZsGreen (abbreviated Z in polyproteins) was included in the polyproteins as a fusion with nsP3 (45), for easy detection of nsP3- and transfection-positive cells.
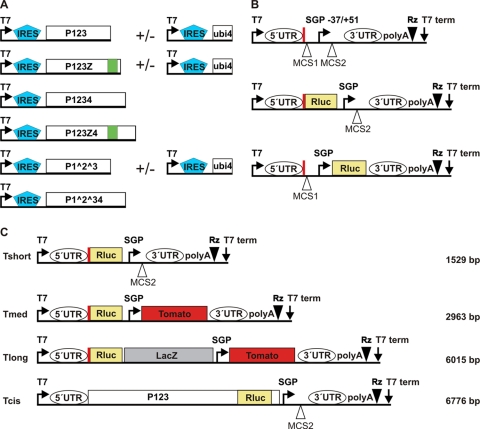
Fig. 1.
Schematic representation of the main constructs used. The constructs or their different parts are not to scale. (A) SFV replicase protein expression constructs. Polyprotein production is enhanced by the EMCV IRES element. P1234 and P123 can also be produced in forms including the fluorescent protein ZsGreen (denoted by Z and marked in green), which is fused with nsP3. In some constructs, cleavages have been prevented by point mutations, as indicated with the symbol ∧. nsP4, when it is provided separately, is initially produced as a fusion with ubiquitin (ubi4), leading to the formation of the native N terminus. In some constructs (data not shown), nsP4 polymerase activity is inhibited by an active-site mutation, denoted nsP4GAA. (B) Basic template constructs. The viral 5′ UTR, placed precisely under the T7 promoter, directs the expression of the N-terminal portion of nsP1 (74 aa, marked with a thick red vertical line), as this region also contains an important cis-acting replication element. Two multiple-cloning sites (MCS) are included. In the Nsluc constructs (in the middle), this peptide is fused with the N terminus of Renilla luciferase (Rluc). In the Stluc constructs (at the bottom), the Rluc gene is placed after the subgenomic promoter (SGP), where the structural genes of the virus would normally lie. The 3′ end of the viral 3′ UTR, including poly(A), is followed by a ribozyme (Rz) sequence. (C) Different lengths of template constructs. Rluc is produced directly from the 5′ end of the genome (Nsluc). The short template contains no additional genes (Tshort). Tmed encodes the fluorescent protein Tomato under the control of the subgenomic promoter. Tlong, in addition to the previous, contains the β-galactosidase (LacZ) gene, which is not expressed. Tcis produces the P123 polyprotein fused with Rluc and is both an expression construct and a template at the same time. The lengths of the replicating templates are given on the right.
The polyproteins were easily detected in transfected cell extracts by Western blotting. Wild-type polyproteins were efficiently cleaved into individual nsPs, whereas mutations at the cleavage sites completely prevented the cleavage of P1^2^3 (Fig. 2 A). The amount of the individual replicase proteins increased at least up to 10 h posttransfection. At 24 h, the amount of proteins was apparently reduced, which may have resulted from the loss of plasmid-based expression and continued cell division (Fig. 2B). These results were compared with those obtained during SFV infection, during which a more rapid accumulation of replicase proteins took place. The final levels were already achieved at 4 h postinfection (Fig. 2B), after which the synthesis of further nsPs is shut off in infected cells (13). The level of nsP expression during SFV infection was somewhat higher than that achieved by the T7-based system, and the difference was estimated to be approximately 2-fold at the 10-h time point.
The localization of the proteins expressed in BSR cells was examined by both light and electron microscopy. As the goal was to establish replication, even in the initial localization experiments a template construct was cotransfected with the protein expression construct to achieve conditions similar to the final experimental setup. After transfection of P1234, both nsP1 and nsP3 were found on the plasma membrane as well as in intracellular punctate structures; in many of these structures, the two proteins colocalized (Fig. 2C). As polyprotein cleavage was rapid, the majority of this was evidently true colocalization in the form of protein complexes and not the result of polyprotein staining. A significant fraction of nsP1 and nsP3 did not colocalize with each other. However, this is also the case in SFV-infected cells: a significant fraction of the polyprotein components dissociates, whereas only a fraction remains together in replication complexes (17). In SFV-infected cells, the replication complexes are found on the inner surface of the plasma membrane and on the outer surface of endosomal and lysosomal vacuoles (17). These two localizations were also demonstrated for nsP1 and nsP3 by immunoelectron microscopy in the BSR system (Fig. 2D). In constructs which included the fluorescent ZsGreen, nsP3 showed perfect colocalization with ZsGreen, as expected (Fig. 2E). Many of the intracellular vacuoles positive for ZsGreen were also positive for the lysosomal marker LBPA (Fig. 2F), indicating that they resemble replication complex-bearing vacuoles in infected cells (45). Altogether, the Western blotting and microscopic studies showed that the T7-based expression system appears to correspond well to the polyprotein expression and localization conditions achieved during SFV infection.
Replication of RNA templates.
To study whether the expressed polyproteins could form functional replication complexes, we provided in trans RNA templates containing the known sequence elements required for a full alphavirus replication cycle. These are located at the 5′ UTR, at the beginning of the nsP1-coding region, and at the end of the 3′ UTR (47). Therefore, the entire 5′ UTR and the first 222 nucleotides of the replicase ORF were included, together with 61 nucleotides at the end of the genome, followed by a poly(A) tail. Additionally, these constructs included a short subgenomic promoter (Fig. 1B). The initial relatively short (approximately 1.5-kb) model templates encoded Renilla luciferase either under the control of the subgenomic promoter (termed Stluc) or directly at the 5′ end of the genome (Nsluc). In the latter case, the luciferase protein product was fused with the N-terminal 74 amino acids of nsP1. The templates were transfected to the cells as DNA plasmids, which contained, in addition to the T7 promoter, the hepatitis delta virus ribozyme immediately after the poly(A) tail (Fig. 1B).
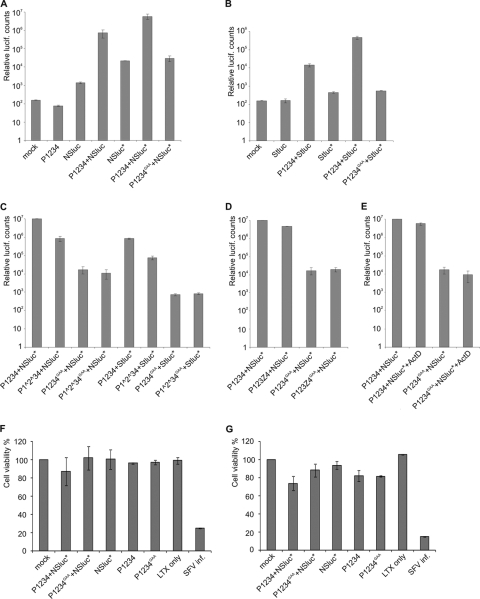
In the initial experiments, luciferase activity was examined at 6 h posttransfection. On the basis of polyprotein expression, this was expected to be an early time point at which replication complexes continue to assemble. When BSR cells were transfected with the Nsluc plasmid alone, there was an ∼10-fold increase of luciferase activity over the background of nontransfected cells, which was expected, since luciferase can be directly translated from the RNA transcribed by the T7 polymerase. Transfecting polyprotein expression constructs alone produced no increase (Fig. 3A). When the polyprotein P1234-encoding plasmid was cotransfected with Nsluc, there was a large increase in luciferase activity, approximately 500-fold compared to that with the template alone (Fig. 3A). Next, a single point mutation near the 5′ end of the template was introduced (see Materials and Methods). This was designed to open a presumed hairpin structure, as it has been shown in alphaviruses that similar mutations can enhance RNA replication (6, 18). For the mutated template, increased luciferase activity was seen with the template alone and also when P1234 was present than with the wild-type template (Fig. 3A). The effect of the mutation might therefore be to increase either translation or replication, or both. Finally, as a negative control a double mutation at the conserved GDD motif of the polymerase nsP4 active site was used, changing the two metal-chelating aspartates to alanines (designated nsP 4GAA). The mutant polymerase provided no increase in luciferase activity compared to that achieved with template alone (Fig. 3A), indicating that the increase in luciferase activity is dependent on RNA replication and is not caused by other effects of polyprotein expression, such as stabilization of the RNA templates.
Fig. 3.
Replication of DNA-derived RNA templates and its effect on cell viability. (A to E) BSR cells were transfected with the indicated polyprotein constructs together with the short template containing the Rluc gene fused with the N-terminal fragment of nsP1 (Nsluc) or under the subgenomic promoter (Stluc). An asterisk denotes the modified templates (a point mutation in the 5′ UTR). The cells were lysed at 6 h posttransfection, and the luciferase activity was measured. Standard errors of the means of two biological experiments (three replicates in each) are shown. Note the logarithmic scale. lucif, luciferase. (A and B) Specific replication of template Nsluc (A) or Stluc (B) by P1234 polyprotein. Polyprotein with a polymerase active-site mutation (designated GAA) results in background-level luciferase production. (C) Effect of uncleavable polyproteins on the replication of short templates. (D) Presence of ZsGreen in the polyprotein does not interfere with the replication of Nsluc. (E) Addition of ActD does not affect the replication of Nsluc. (F and G) Viability of the transfected cells at the 6-h (F) and 24-h (G) time points. The viability of mock-transfected cells was taken to be 100%. Note the linear scale. For SFV infection, 10 PFU/cell was used.
Similar results were obtained with the template Stluc (Fig. 3B). However, in this case the activity produced by the template alone was negligible, as the luciferase gene is preceded by the truncated nsP1 open reading frame. Coexpression of the polyprotein P1234 increased the activity ∼100-fold. The template hairpin mutation reproducibly yielded an even larger increase in the production of luciferase from the Stluc position than from the Nsluc position, although the reasons for this are not clear. This result, however, suggests that the increased luciferase activity is due to increased replication, since the engineered template mutation is not part of the subgenomic RNA. Due to the apparent increase in template activity, only the mutated template was used in further experiments. Again, the polymerase mutant P1234GAA produced no increase in luciferase activity (Fig. 3B).
When the nsP1/nsP2 and nsP2/nsP3 cleavages were blocked, there was a reduction of activity compared to that of wild-type P1234 with both templates, Nsluc and Stluc (Fig. 3C). This correlates with earlier results indicating that cleavage-deficient P123 is less active in plus-strand RNA synthesis (21). Figure 3B and C shows the functionality of the subgenomic promoter, indicating that subgenomic RNA synthesis is amenable for characterization using this system. In the current work, further experiments concentrated on the synthesis of the full-length model RNA.
We have previously shown that the inclusion of ZsGreen fused with nsP3 is fully compatible with SFV replication (45). Also in the current system, the polyprotein P123Z4 gave results very similar to those obtained with P1234 (Fig. 3D). In addition, the replication of RNA was largely unperturbed when actinomycin D (2 μg/ml) was added to the cells at 3 h posttransfection (Fig. 3E), which is a characteristic typical for viral RNA-dependent RNA replication. A sensitive cytotoxicity assay measuring cellular ATP levels was conducted both at the early 6 h time point and at 24 h posttransfection. trans-Replication or individual plasmid transfection had a minor effect on cell viability only at the late time point compared to the viability of mock-transfected cells (Fig. 3F and G), indicating that cytotoxic effects do not hamper the use of the system in cell biological experiments. The highly cytotoxic SFV infection was used as a control.
Time course of luciferase production and RNA replication with different template lengths.
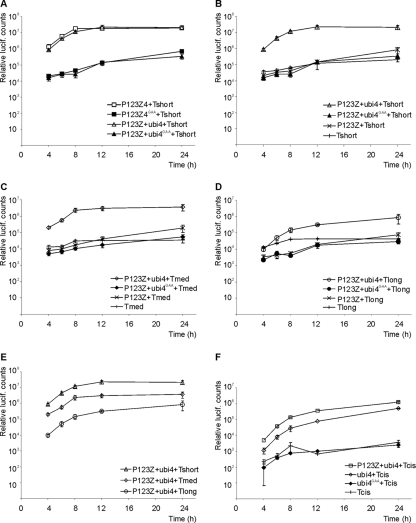
In the preceding experiments, P1234 was provided as a single polyprotein construct. While it has been established that nsP1, nsP2, and nsP3 have to be provided together as P123 to achieve replicase complex activity, assembly, and localization (20, 40), we tested the possibility of providing nsP4 expressed from a separate plasmid to introduce further flexibility into the system. A time course experiment was performed with P123Z4 and the two-plasmid combination P123Z-ubi4. The corresponding GAA mutants were used as a negative control. There was no significant difference in the accumulation of luciferase activity between the single polyprotein and the combination in which nsP4 was produced separately (Fig. 4 A). In the time course with active polymerase, luciferase accumulation was already evident at 4 h posttransfection. The activity continued to increase exponentially until 8 h posttransfection, increased more slowly at between 8 h and 12 h, and remained stable thereafter.
Fig. 4.
Time course of luciferase (lucif.) activity production. The indicated polyprotein and template plasmids were transfected to BSR cells. Cells were lysed at the time points shown, and luciferase activity was measured. Open symbols represent active replicase complexes, whereas filled symbols mark the polymerase mutant (GAA). In addition to the GAA mutants, background-level luciferase production was detected by the expression of templates alone and a combination lacking the polymerase nsP4 (P123Z-template). Standard errors of the means of two biological experiments (three replicates in each) are shown. (A) Comparison of the polyprotein P123Z4 with the two-plasmid combination P123Z-ubi4; (B to D) replication of short, medium-sized, and long templates; (E) comparison of replication of templates with different length; (F) comparison of template replication in cis with regard to the polyprotein P123 versus pure trans-replication.
In the previous experiments, a relatively short model template, Tshort (∼1.5 kb), was used. To increase the length of the template, additional genes were inserted into the template (Fig. 1C). When the longer templates were transfected with the polyprotein, luciferase activity increased more slowly and reached a smaller final value (Fig. 4B to E). With the Tlong (∼6 kb) template, the activity continued to increase after the 12-h time point but at the 24-h time point still did not reach the levels produced from the shorter templates (Fig. 4E). However, the increase above the control levels achieved by the template alone, with a polyprotein containing mutated polymerase, or in the absence of the nsP4 subunit was very clear for all the templates (Fig. 4B to D).
Although the main purpose of these experiments was to establish replication of templates provided in trans, we were also curious whether this requirement made the replication less efficient. We therefore constructed a replicating template, which also produced the polyprotein P123, including Renilla luciferase (Fig. 1C). However, the amount of luciferase produced with this template, when nsP4 was provided separately, was somewhat smaller than that produced with the comparable long template, which replicated in trans (Fig. 4F). In fact, luciferase levels could be slightly boosted by expressing additional P123Z in trans from a polyprotein expression plasmid (Fig. 4F). Therefore, it appears that the need to recruit the template in trans with respect to P123 is not a significant handicap in this system, but instead, other factors may be limiting the extent of replication.
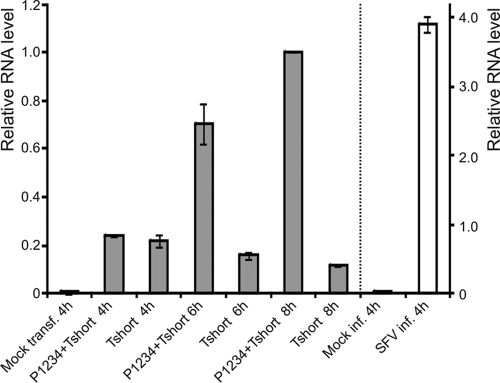
The activity generated by the expressed polyprotein and the template provided in trans was compared to the luciferase activity derived from a viral genome containing the luciferase insertion in the nonstructural reading frame (in the same position as ZsGreen in the replicase polyprotein). This virus, termed SFV-Rluc, replicates as well as wild-type SFV (36). The activity produced by P1234 with the short template reproducibly reached almost 107 relative units by 6 h posttransfection (Fig. 3) and continued to increase after that, reaching a maximal value of between 3 × 107 and 5 × 107 units by 12 h (Fig. 4A). SFV-Rluc typically yielded ∼107 relative units at 6 h postinfection (multiplicity of infection, 10), but by 12 h postinfection the activity decreased slightly, as translation of replicase proteins is shut off. To more directly assess the RNA levels produced by the trans-replication system, we carried out quantitative reverse transcription-PCR (RT-PCR) experiments comparing the replication of the short template in trans with that during regular SFV infection. In cells transfected with the short template alone, the amount of template was observed to decrease after 4 h posttransfection, which was the earliest time point examined (Fig. 5). In contrast, when the polyprotein construct for P1234 was cotransfected, the amount of RNA increased ∼5-fold between the 4-h and 8-h time points. The presence of the polyprotein could also influence the stability of the RNA, but control experiments with the GAA mutant suggest that this effect is not significant (data not shown). In SFV-infected cells at 4 h postinfection, the RNA levels measured were approximately 4 times higher than those achieved at 8 h in the trans-replication system (Fig. 5).
Fig. 5.
RNA replication quantified by RT-PCR. BSR cells were transfected (transf.) with the indicated polyprotein and template plasmids. Total cellular RNAs were isolated at different time points and analyzed with quantitative real-time PCR. SFV infection (inf.) was performed to compare the replication efficiency during infection to that of the T7-based system. The results were normalized for GAPDH and calculated with the relative expression method using the P1234-Tshort 8-h sample as a reference (relative level, 1.0). Note the different scales: the scale for the T7-based system is on the left of the dotted line (results shown with gray bars), and the scale for the infected cell samples is on the right (results shown with white bars).
Evidence for RNA replication by immunofluorescence.
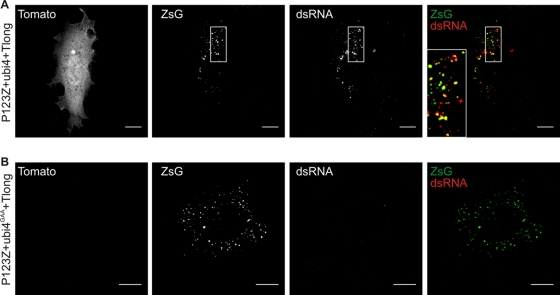
In the longer templates, the fluorescent protein Tomato was encoded by the subgenomic RNA (Fig. 1C), affording another readout of replication, which can be observed at the single-cell level. The luciferase measurements had already shown that products from the subgenomic RNA are expressed only when there is active replication (Fig. 3B). We focused on Tlong, as this template is closer in length to that of SFV genomic RNA, although it is still significantly shorter than the ∼11.5-kb genome. With Tlong, Tomato-positive cells were observed only when the polyprotein P123Z was provided together with nsP4 and the template (Fig. 6 A). While the majority of cells were positive for ZsGreen, the amount of Tomato-positive cells increased slowly over time and reached ∼20% by 24 h posttransfection, which was selected as a suitable time point for studies of the long template (see also Fig. 7A). The cells positive for Tomato were also positive for ZsGreen and for dsRNA, and the latter two markers showed colocalization in many instances (Fig. 6A). dsRNA staining is a good marker for SFV replication, indicating the location of the replication complexes (45), whereas a significant fraction of nsP3 (and thus nsP3-ZsGreen) is found outside the replication complexes (40). When the inactive polymerase was cotransfected with the template, no cells positive for Tomato or dsRNA were observed, although the transfected cells remained positive for ZsGreen (Fig. 6B). With the two-plasmid combination P123Z-nsP4, it was estimated that >90% of the cells that were positive for ZsGreen were also positive for nsP4, which is in accordance with the high level of replication achieved with this combination (Fig. 4A).
Fig. 6.
Specific replication of Tlong template assessed by Tomato expression and dsRNA staining. BSR cells were transfected with P123Z-ubi4 wild type (A) or the polymerase mutant version (B) together with Tlong template. Samples were fixed at 24 h posttransfection and processed for immunofluorescence analysis. Anti-dsRNA antibody was used to stain the active replication complexes. Separate channels are shown in gray scale, whereas the merged images on the right illustrate ZsGreen (green) and dsRNA (red) colocalization (yellow). Representative confocal optical slices from the middle of the cells are shown. The boxed area in the upper panel is zoomed in in the merged image, and it highlights the colocalization between ZsGreen and dsRNA signals. Bars, 10 μm.
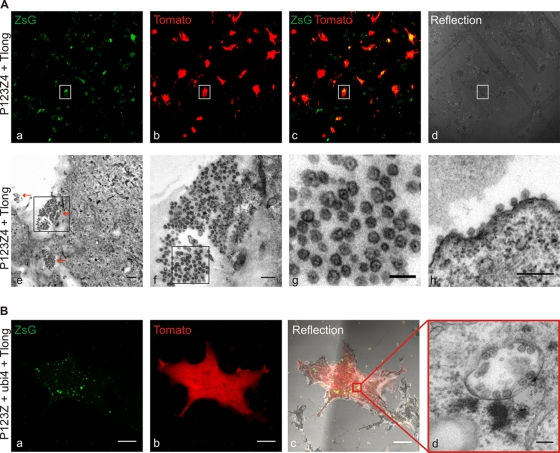
Fig. 7.
Spherule formation studied by CLEM. (A) BSR cells on MatTek grid dishes were cotransfected with P1234Z and template Tlong. The sample was fixed at 24 h posttransfection and imaged with a confocal microscope for correlation (upper panel). (a to c) ZsGreen and Tomato coexpression was taken as an indication for active replication. (d) Reflection image was acquired to visualize the grid and relocate the cells at the EM level. The boxed cell was chosen for further characterization with EM (lower panel). (e) Bottom sections of the relocated cell showed large amounts of spherules, as analyzed with TEM. Three large clusters of spherules are indicated with red arrows. Bar, 500 nm. The boxed area illustrating one large cluster of spherules at the bottom of the cell is shown in the image in panel f, in which the scale bar is 200 nm. (g) Zoomed in area of the boxed area in panel f of the clustered spherules showing a characteristic dark dot inside the structures. Bar, 100 nm. (h) Clusters of spherules were also found at the plasma membrane on the rim of the cell. Bar, 200 nm. (B) Cotransfection of P123Z-ubi4-Tlong. The sample was prepared for CLEM as described for panel A. A confocal optical slice highlights the ZsGreen-positive structures (a) and Tomato expression (b) in the cell analyzed in an EM. (c) A reflection image was used to relocate the cell at the EM level. (a to c) Bars, 10 μm. (d) A relocated cell was analyzed with EM. A representative modified endolysosome (cytopathic vacuole) with numerous spherules is illustrated. Bar, 100 nm.
Requirements for formation of replication complex spherules.
The replication complexes in SFV-infected cells, as well as in cells containing SFV replicons, are manifested as numerous membrane invaginations, termed spherules, at the inner surface of the plasma membrane and the outer surface of endolysosomal structures (8, 45). In the trans-replication system, CLEM was utilized to relocate the replication-positive, Tomato-fluorescent cells, as shown in the upper row of Fig. 7 A. Abundant spherules were detected at the bottom plasma membrane of the replication-positive cells. Interestingly, they were often found in large clusters (Fig. 7A). As these sections are parallel to the cell surface, the spherule necks are not easily visualized in these images. Spherules could also be visualized on the rim of the plasma membrane (Fig.7Ah). Similarly, the combination in which nsP4 was provided separately yielded many replication-positive cells which could be relocated (Fig. 7B). In both plasmid combinations, in addition to the plasma membrane spherules, typical intracellular vacuoles bearing spherules could be seen, as illustrated for the three-plasmid combination in Fig.7Bd.
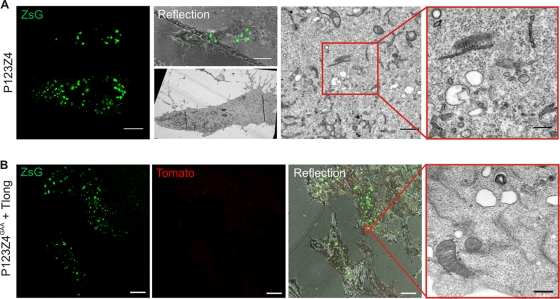
To study the viral requirements for spherule formation, we first examined cells expressing the fluorescent replicase protein P123Z4 alone in the absence of template (Fig. 8 A). Using CLEM, we found no convincing evidence for spherule formation in transfection-positive cells under these conditions. To substantiate this finding, a total of 2,156 spherules were calculated in sections derived from ∼12 positive cells from two to three independent experiments, in positive-control cells transfected with P123Z4+Tlong. In contrast, when a similar volume of positive cells expressing P123Z4 alone was examined, 11 structures that closely resembled spherules were found by visual inspection. Although spherules are rather distinct in appearance, it is noteworthy that rare individual membranous structures visually indistinguishable from spherules can also be observed in untransfected cells. In looking at large volumes of cells expressing polyprotein alone, we never observed any sign of clusters of spherules, which were often seen in replication-positive cells (Fig. 7A). Thus, P123Z4 does not have a significant ability to form spherules. Similar results were obtained when polymerase-defective P123Z4GAA was expressed together with the template Tlong (Fig. 8B). In this case, even fewer suspect, potentially spherule-like structures were observed. Therefore, our study shows that SFV nonstructural proteins by themselves cannot generate spherules. Formation of spherules required the RNA-synthesizing activity of the polymerase complex and a replication-competent RNA template.
Fig. 8.
Requirements for spherule formation. BSR cells were transfected with the polyprotein P123Z4 alone (A) or with polymerase-defective P123Z4GAA together with the template Tlong (B). The sample was fixed at 24 h posttransfection, imaged with a confocal microscope, and then prepared for CLEM. Representative images are shown. In the EM images of transfection-positive cells, intracellular vacuoles (A) or plasma membrane (B) devoid of spherules can be seen. Bars, rightmost EM images, 200 nm; confocal microscopic images, 10 μm.
DISCUSSION
We present here an efficient trans-replication system for SFV in which separately expressed replication polyproteins can utilize template molecules provided in trans. It has been well established that alphavirus replicases are capable of replicating templates in trans in helper-based systems for generating virus replicon particles (3, 25, 43). In these systems, the replicase polyproteins are produced by replicons (i.e., replicating RNAs) which lack the genes encoding structural proteins but contain all the cis-acting elements. The structural proteins are produced from separate replication-competent helper RNA molecules provided in trans. Thus, both cis-replication of the replicon RNA and trans-replication of the helper RNA are ongoing in the replicon-helper systems. A crucial feature of the system described in this work is that there is no cis-replication of the mRNA for the replicase proteins. The expression of the replicase proteins is therefore independent of any of their enzymatic activities. Thus, inactive components, such as the polymerase-negative mutant used in the current work, can be produced at wild-type protein levels and analyzed for any remaining activities or properties.
To our knowledge, the system described here is the first well-characterized, efficient, and pure (i.e., without simultaneous cis-replication of replicon RNAs) plasmid-based trans-replication system devised for a polyprotein-producing positive-sense RNA virus in mammalian cells. Polyprotein expression and RNA levels remained somewhat lower than those achieved during SFV infection (Fig. 2 and 5), but luciferase activities were equal to those observed with reporter-bearing virus, in spite of the fact that in the transfection-based system not all cells are positive for RNA replication. Interestingly, the efficiency of replication, measured by luciferase activity, fell considerably with longer templates. This might be due to multiple factors, including template stability, which have not yet been examined experimentally. However, viral proteins, dsRNA intermediates, and marker gene products could be easily visualized using microscopic techniques with all sizes of templates (Fig. 2 and 6). Previously, a trans-replication system for SIN has been developed by using recombinant vaccinia virus-based expression of replicase proteins and model templates (20, 23). This system has yielded much invaluable information on alphavirus RNA replication in intact cells and cell extracts (19, 22, 24), but the construction and use of recombinant vaccinia viruses complicate its use. No published information is available on the relative efficiency of the RNA synthesis in the vaccinia virus-based system. Vaccinia virus infection also causes massive reorganization of cellular membrane systems (30), and therefore, vaccinia virus is not easily applicable to the cell biological studies that we wished to carry out.
Alphaviruses are relatively complex viruses, possessing an intricate scheme for replicase polyprotein processing, which may hamper the establishment of trans-replication, as specific polyprotein intermediates are required for the initial stages of replication (see Introduction [22, 49]). Several nonmammalian positive-strand RNA viruses have multicomponent genomes, do not produce polyproteins, and routinely perform trans-replication. For BMV and for flock house nodavirus, it has been possible to construct efficient trans-replication systems in Saccharomyces cerevisiae yeast cells, which are rather distantly related to the normal host cells utilized by these viruses (12, 37). Similarly, efficient trans-replication is possible for the single-component genome of tomato bushy stunt virus, which has a simple set of replicase proteins with no proteolysis (35). For all these viruses, many important insights into the replication process and the host factors required have been achieved with such systems (33). We believe that our system provides the tools required for the dissection of alphavirus replication in a mammalian cell environment. We have demonstrated that the system is amenable to many variations, e.g., inclusion of various marker genes in the genome or under the subgenomic promoter, inclusion of a fluorescent marker in the replicase, and the split of replicase into two components, P123 and nsP4 (Fig. 1), while maintaining the efficiency of the system.
It may be possible to construct similar trans-replication systems for other positive-sense RNA viruses, although formidable hurdles remain to be overcome at multiple levels. For many viruses, genome replication has a strong preference for replication in cis. To some extent, this is likely to be true for alphaviruses, as suggested, e.g., by the relatively poor ability of temperature-sensitive mutants to complement each other (13). For poliovirus, trans-replication cannot really be observed in cells (34), but in cell-free replication systems, it is observed with a low efficiency (32, 44). Flaviviruses, such as Kunjin virus, also display strongly preferential replication in cis (14, 26). These preferences may be due to coassembly or colocalization requirements or needs for polyprotein precursors, all of which may be amenable to experimental manipulation, when their molecular mechanisms are better understood. Coronaviruses and arteriviruses can replicate defective interfering RNAs in trans (11, 31). They produce a very large and complicated set of replicase proteins, which may be difficult to express in cells in the absence of viral replication. However, it remains unknown whether this set of proteins could be split into two or more separate components.
We have shown here that alphavirus replication complexes can be formed in trans with a relatively high efficiency. With the long template most closely resembling the viral genome, we observed efficient formation of spherules in replication-positive cells (Fig. 7). These spherules resembled in all respects the spherules found in SFV-infected cells: they were similar in size, contained electron-dense material inside, remained connected to the cytoplasm at all times, and were found both at the plasma membrane and in intracellular vacuoles. CLEM was an important technique in order to study the transfected cells, as it allowed unambiguous identification (correlation) of individual replication-positive or transfection-positive cells. Furthermore, with CLEM we were able to focus on specific intracellular regions that were positive for ZsGreen fluorescence and therefore contained the replication proteins (Fig. 7B and 8B). By the aid of CLEM, we were able to extract data from replication-negative cells (Fig. 8) and show that the replicase protein alone or a polymerase-defective replicase provided with a template was not able to form spherules to any significant extent. Given the relatively large internal volume of mammalian cells and the significant fraction of cells that remain untransfected, substantiation of the negative results is especially important. Recently, Frolova and coworkers published evidence that SIN replicase polyproteins expressed alone in the absence of a trans-replicating template did not give rise to spherules (7). These negative data were not shown or quantified.
Interestingly, in the plant virus BMV, which belongs to the alphavirus-like superfamily, replicase protein 1a alone is capable of efficiently making spherules (41). In contrast, it was recently shown that nodavirus spherule formation requires both RNA template and active polymerase (16). Therefore, at least two pathways for spherule formation appear to exist in different virus groups: one in which viral replicase proteins (together with cellular components) are capable of membrane deformation and a second one in which, additionally, viral RNA and polymerase action both in some fashion contribute to membrane alteration. In the latter pathway, it is intriguing to speculate how the action of a polymerase could drive conformational changes or even provide mechanical force. Even though the alphavirus replicase polyprotein contains domains distantly related to BMV replicase protein 1a, currently they appear to follow different modes in regard to spherule formation. Further mechanistic insights into the different pathways of membrane deformation induced by viral components are clearly required. The experimental systems established in this work will be useful to study the requirements for spherule formation on both the viral and host cell side and to establish and dissect the substeps required for the assembly and activity of alphavirus replication complexes.
ACKNOWLEDGMENTS
This work was supported by Academy of Finland (grant 127214) and the Sigrid Jusélius Foundation. P.S. was supported in part by a fellowship from the Viikki Graduate School in Molecular Biosciences, K.H. by an Academy of Finland postdoctoral fellowship (grant 132085), and E.J. by Academy of Finland (grant 115025).
We thank Karl-Klaus Conzelmann for kindly providing the BSR T7/5 cells and Mervi Lindman for excellent technical assistance in EM. Andres Merits, Aleksei Lulla, and Valeria Lulla are acknowledged for valuable discussions, and Katri Kallio is acknowledged for help in CLEM experiments.
Footnotes
Published ahead of print on 9 March 2011.
REFERENCES
- 1. Ahola T., Lampio A., Auvinen P., Kääriäinen L. 1999. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 18:3164–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barth B. U., Wahlberg J. M., Garoff H. 1995. The oligomerization reaction of the Semliki Forest virus membrane protein subunits. J. Cell Biol. 128:283–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bredenbeek P. J., Frolov I., Rice C. M., Schlesinger S. 1993. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J. Virol. 67:6439–6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buchholz U. J., Finke S., Conzelmann K. K. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Di Franco A., Russo M., Martelli G. P. 1984. Ultrastructure and origin of cytoplasmic multivesicular bodies induced by carnation Italian ringspot virus. J. Gen. Virol. 65:1233–1237 [Google Scholar]
- 6. Frolov I., Hardy R., Rice C. M. 2001. Cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 7:1638–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frolova E. I., Gorchakov R., Pereboeva L., Atasheva S., Frolov I. 2010. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J. Virol. 84:11679–11695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Froshauer S., Kartenbeck J., Helenius A. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Griffin D. E. 2001. Alphaviruses, p. 917–963In Knipe D. M., Howley P. M. (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 10. Helenius A., Kartenbeck J., Simons K., Fries E. 1980. On the entry of Semliki Forest virus into BHK-21 cells. J. Cell Biol. 84:404–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Izeta A., et al. 1999. Replication and packaging of transmissible gastroenteritis coronavirus-derived synthetic minigenomes. J. Virol. 73:1535–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Janda M., Ahlquist P. 1993. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell 72:961–970 [DOI] [PubMed] [Google Scholar]
- 13. Kääriäinen L., Ahola T. 2002. Functions of alphavirus nonstructural proteins in RNA replication. Prog. Nucleic Acid Res. Mol. Biol. 71:187–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khromykh A. A., Sedlak P. L., Westaway E. G. 2000. cis- and trans-acting elements in flavivirus RNA replication. J. Virol. 74:3253–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim K. H., Rumenapf T., Strauss E. G., Strauss J. H. 2004. Regulation of Semliki Forest virus RNA replication: a model for the control of alphavirus pathogenesis in invertebrate hosts. Virology 323:153–163 [DOI] [PubMed] [Google Scholar]
- 16. Kopek B. G., Settles E. W., Friesen P. D., Ahlquist P. 2010. Nodavirus-induced membrane rearrangement in replication complex assembly requires replicase protein a, RNA templates, and polymerase activity. J. Virol. 84:12492–12503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kujala P., et al. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 75:3873–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kulasegaran-Shylini R., Thiviyanathan V., Gorenstein D. G., Frolov I. 2009. The 5′UTR-specific mutation in VEEV TC-83 genome has a strong effect on RNA replication and subgenomic RNA synthesis, but not on translation of the encoded proteins. Virology 387:211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lemm J. A., Bergqvist A., Read C. M., Rice C. M. 1998. Template-dependent initiation of Sindbis virus RNA replication in vitro. J. Virol. 72:6546–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lemm J. A., Rice C. M. 1993. Assembly of functional Sindbis virus RNA replication complexes: requirement for coexpression of P123 and P34. J. Virol. 67:1905–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lemm J. A., Rice C. M. 1993. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J. Virol. 67:1916–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lemm J. A., Rumenapf T., Strauss E. G., Strauss J. H., Rice C. M. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 13:2925–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li G. P., Pragai B. M., Rice C. M. 1991. Rescue of Sindbis virus-specific RNA replication and transcription by using a vaccinia virus recombinant. J. Virol. 65:6714–6723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li M. L., Stollar V. 2004. Identification of the amino acid sequence in Sindbis virus nsP4 that binds to the promoter for the synthesis of the subgenomic RNA. Proc. Natl. Acad. Sci. U. S. A. 101:9429–9434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liljeström P., Garoff H. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology (NY) 9:1356–1361 [DOI] [PubMed] [Google Scholar]
- 26. Liu W. J., Sedlak P. L., Kondratieva N., Khromykh A. A. 2002. Complementation analysis of the flavivirus Kunjin NS3 and NS5 proteins defines the minimal regions essential for formation of a replication complex and shows a requirement of NS3 in cis for virus assembly. J. Virol. 76:10766–10775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mackenzie J. 2005. Wrapping things up about virus RNA replication. Traffic 6:967–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Merits A., Vasiljeva L., Ahola T., Kääriäinen L., Auvinen P. 2001. Proteolytic processing of Semliki Forest virus-specific non-structural polyprotein by nsP2 protease. J. Gen. Virol. 82:765–773 [DOI] [PubMed] [Google Scholar]
- 29. Miller D. J., Schwartz M. D., Ahlquist P. 2001. Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J. Virol. 75:11664–11676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miller S., Krijnse-Locker J. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Molenkamp R., Rozier B. C., Greve S., Spaan W. J., Snijder E. J. 2000. Isolation and characterization of an arterivirus defective interfering RNA genome. J. Virol. 74:3156–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morasco B. J., Sharma N., Parilla J., Flanegan J. B. 2003. Poliovirus cre(2C)-dependent synthesis of VPgpUpU is required for positive- but not negative-strand RNA synthesis. J. Virol. 77:5136–5144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nagy P. D. 2008. Yeast as a model host to explore plant virus-host interactions. Annu. Rev. Phytopathol. 46:217–242 [DOI] [PubMed] [Google Scholar]
- 34. Novak J. E., Kirkegaard K. 1994. Coupling between genome translation and replication in an RNA virus. Genes Dev. 8:1726–1737 [DOI] [PubMed] [Google Scholar]
- 35. Panavas T., Nagy P. D. 2003. Yeast as a model host to study replication and recombination of defective interfering RNA of tomato bushy stunt virus. Virology 314:315–325 [DOI] [PubMed] [Google Scholar]
- 36. Pohjala L., Barai V., Azhayev A., Lapinjoki S., Ahola T. 2008. A luciferase-based screening method for inhibitors of alphavirus replication applied to nucleoside analogues. Antiviral Res. 78:215–222 [DOI] [PubMed] [Google Scholar]
- 37. Price B. D., Roeder M., Ahlquist P. 2000. DNA-directed expression of functional flock house virus RNA1 derivatives in Saccharomyces cerevisiae, heterologous gene expression, and selective effects on subgenomic mRNA synthesis. J. Virol. 74:11724–11733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Puhka M., Vihinen H., Joensuu M., Jokitalo E. 2007. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J. Cell Biol. 179:895–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Salonen A., Ahola T., Kääriäinen L. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285:139–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Salonen A., et al. 2003. Properly folded nonstructural polyprotein directs the Semliki Forest virus replication complex to the endosomal compartment. J. Virol. 77:1691–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schwartz M., et al. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505–514 [DOI] [PubMed] [Google Scholar]
- 42. Shirako Y., Strauss J. H. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 68:1874–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Smerdou C., Liljeström P. 1999. Two-helper RNA system for production of recombinant Semliki Forest virus particles. J. Virol. 73:1092–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Spear A., Sharma N., Flanegan J. B. 2008. Protein-RNA tethering: the role of poly(C) binding protein 2 in poliovirus RNA replication. Virology 374:280–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Spuul P., Balistreri G., Kääriäinen L., Ahola T. 2010. Phosphatidylinositol 3-kinase-, actin-, and microtubule-dependent transport of Semliki Forest virus replication complexes from the plasma membrane to modified lysosomes. J. Virol. 84:7543–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Spuul P., et al. 2007. Role of the amphipathic peptide of Semliki Forest virus replicase protein nsP1 in membrane association and virus replication. J. Virol. 81:872–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Strauss J. H., Strauss E. G. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Varshavsky A. 1996. The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. U. S. A. 93:12142–12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vasiljeva L., et al. 2003. Regulation of the sequential processing of Semliki Forest virus replicase polyprotein. J. Biol. Chem. 278:41636–41645 [DOI] [PubMed] [Google Scholar]
- 50. Wang Y. F., Sawicki S. G., Sawicki D. L. 1994. Alphavirus nsP3 functions to form replication complexes transcribing negative-strand RNA. J. Virol. 68:6466–6475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Welsch S., et al. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]