Abstract
Plus-strand RNA viruses without 5′ caps require noncanonical mechanisms for ribosome recruitment. A translational enhancer in the 3′ untranslated region (UTR) of Turnip crinkle virus (TCV) contains an internal T-shaped structure (TSS) that binds to 60S ribosomal subunits. We now report that the 63-nucleotide (nt) 5′ UTR of TCV contains a 19-nt pyrimidine-rich element near the initiation codon that supports translation of an internal open reading frame (ORF) independent of upstream 5′ UTR sequences. Addition of 80S ribosomes to the 5′ UTR reduced the flexibility of the polypyrimidine residues and generated a toeprint consistent with binding to this region. Binding of salt-washed 40S ribosomal subunits was reduced 6-fold when the pyrimidine-rich sequence was mutated. 40S subunit binding generated the same toeprint as 80S ribosomes but also additional ones near the 5′ end. Generation of out-of-frame AUGs upstream of the polypyrimidine region reduced translation, which suggests that 5′-terminal entry of 40S subunits is followed by scanning and that the polypyrimidine region is needed for an alternative function that requires ribosome binding. No evidence for RNA-RNA interactions between 5′ and 3′ sequences was found, suggesting that TCV utilizes an alternative means for circularizing its genome. Combining 5′ and 3′ UTR fragments in vitro had no discernible effect on the structures of the RNAs. In contrast, when 80S ribosomes were added to both fragments, structural changes were found in the 5′ UTR polypyrimidine tract that were not evident when ribosomes interacted with the individual fragments. This suggests that ribosomes can promote an interaction between the 5′ and 3′ UTRs of TCV.
INTRODUCTION
Upon cell entry, the genome of a positive-strand RNA virus first serves as a template for the translation of proteins that are required to initiate genome amplification before serving as a transcription template for synthesis of complementary strands. Efficient translation initiation requires that viral 5′ and 3′ ends be brought into proximity, similar to requirements for translation of cellular mRNAs as specified by the closed-loop model (28, 59). Juxtaposition of the ends of most eukaryotic mRNAs is accomplished by a protein bridge that forms when the scaffolding protein eukaryotic initiation factor 4G (eIF4G) simultaneously binds to eIF4E, which interacts with the 5′-terminal cap (m7GpppN), and to the poly(A) binding protein (PABP), which interacts with the 3′ poly(A) tail (10, 19, 44, 59). The 40S ribosomal subunit, in conjunction with a tRNA-containing ternary complex, then binds to eIF4G via eIF3 and scans in the 5′-to-3′ direction until an initiation codon in a good context is encountered, which is followed by joining of the 60S subunit (43).
RNA viruses that lack the translation signals used by most cellular mRNAs for ribosome recruitment must employ alternative strategies to initiate translation. The extensive, highly folded 5′ untranslated regions (UTRs) of uncapped animal viruses contain complex structures (200 to 500 nucleotides [nt]) known as internal ribosome entry sites (IRESs), which are required for recruitment of 40S ribosomal subunits (34). Ribosome entry can be independent of translation initiation factors (eIFs), as found for hepatitis C virus (HCV) and dicistrovirus IRESs, or can require eIFs exclusive of the eIF4E cap binding protein (15, 36).
5′ UTRs of plant RNA viruses are of limited size (∼10 to 200 nt), precluding ribosome recruitment by animal virus-like IRES structures. Many plant virus 5′ UTRs contain translational enhancers (5′ TEs) comprising pseudoknots, or pyrimidine-rich or purine-rich unstructured sequences, some of which can direct translation of internal open reading frames (ORFs) in bicistronic mRNAs, a hallmark of IRES activity (24, 25, 31, 41, 54, 60, 69). Cellular mRNAs that are translated when cap-dependent translation is inhibited also contain IRESs of limited size in their 5′ UTRs (6, 15, 27, 33, 50). As with plant RNA viruses, unstructured regions may be important for passive ribosome recruitment, since high IRES activity correlates with weak secondary structure for cap-independent translation of yeast and Drosophila mRNAs (16, 18, 61, 66).
5′ IRES activity can be enhanced by interaction with specific elements in 3′ UTRs, leading to template circularization similar to that of host mRNAs (20, 21, 51, 55, 63). Protein-dependent cyclization involving the 3′ poly(A) tail and 5′ IRES has been characterized for picornaviruses (11) and Tobacco etch potyvirus, a member of the poliovirus superfamily (41, 48). 3′ sequences other than poly(A) tails can also serve as translational enhancers in IRES-mediated ribosome recruitment, which may be connected with interacting viral and cellular proteins that stimulate IRES activity (4, 32, 45, 55). Long-distance RNA-RNA interactions between 3′ elements and the 5′IRES have been described for hepatitis C virus (49) and foot-and-mouth disease virus (51) and have been proposed to be involved in the switch between translation and replication. The long-range RNA-RNA interactions that circularize the genome of capped, nonpolyadenylated dengue virus appear to play a role in replication (2, 49), while translation requires an additional bridge mediated by poly(A) binding protein interaction with an interior 3′ poly(A) sequence and likely subsequent interaction with cap-bound eIF4G (45). The growing number of examples of protein-dependent and protein-independent circularization of host and viral mRNAs strongly suggest that formation of a closed-loop structure between 5′ and 3′ ends is an evolutionary necessity to provide protection from cytoplasmic nucleases, as well as a means of ribosome recycling and to ensure that only intact RNAs are translated (17).
Tombusviruses and luteoviruses, which lack 5′ caps and 3′ poly(A) tails, contain critical translation enhancers in their 3′ UTRs known as 3′ cap-independent translation elements (CITEs) (7, 26). 3′ CITEs comprise a variety of structures, some of which have been shown to bind to specific eIFs (38, 62). The bound eIFs are then delivered to the 5′ end by a circularized template generated by kissing-loop interactions between CITE-associated hairpins and 5′ proximal hairpins (8, 9, 14, 52, 62). The genome of the tombusvirus Carnation Italian ringspot virus (CIRV) harboring a 3′ CITE from Maize necrotic spot virus is bridged by a long-distance RNA-RNA interaction that brings eIF4F bound to the 3′ CITE to the 5′ end, promoting 40S subunit entry (40). Blackcurrant reversion virus (BRV) (family Comoviridae), which contains a 5′-linked Vpg and 3′ poly(A) tail, also has a translationally important 3′ CITE and sequences that form long-distance RNA-RNA interactions, indicating that 3′ CITEs are not restricted to nonpolyadenylated viruses (23, 25).
Turnip crinkle virus (TCV) (4,054 nt) is a member of the genus Carmovirus in the Tombusviridae, a family of monopartite single-stranded plus-sense RNA viruses without 5′ caps or 3′ poly(A) tails. TCV carries five genes that are translated from the genomic (gRNA) and two subgenomic RNAs (sgRNAs) (Fig. 1 A). p28 and the translational read-through protein p88 are translated from the gRNA and constitute the virus-encoded subunits of the replicase, the p8 and p9 movement proteins are translated from the bicistronic 1.7 sgRNA, and the 1.45 sgRNA is the mRNA for the p38 coat protein (46). TCV contains an unusual 3′ CITE that synergistically enhances translation of a reporter gene in the presence of the viral 5′ UTR in vivo (46, 57). The core region of the 3′ CITE maps to positions 3810 to 3951, which contain a critical hairpin (H4) and partially overlaps an ∼100-nt internal T-shaped structure (TSS) composed of three hairpins (H4a, H4b, and H5) and two pseudoknots (Ψ3 and Ψ2) (35, 57, 71). The internally located TSS binds to the P sites of 80S ribosomes through the 60S ribosomal subunit, and this binding is important for efficient translation (57). The TSS also functions as a stable scaffold for interaction with external sequences through the large internal symmetrical loop of H5 (67). RNA-dependent RNA polymerase (RdRp) interaction with the 3′ end of TCV causes a widespread conformational shift that structurally rearranges the TSS and surrounding region, including elements required for efficient ribosome binding (67), suggesting that the RdRp may inhibit its own translation by disrupting the structure of the 3′ CITE.
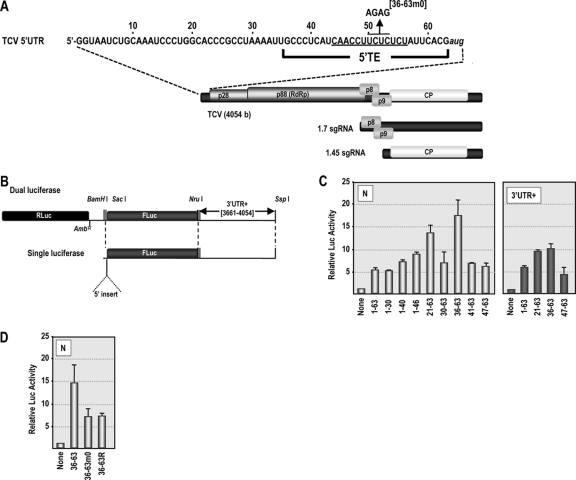
Fig. 1.
The 5′ UTR of TCV gRNA contains a pyrimidine-rich element that can promote translation of an internal ORF. (A) Genomic organization of TCV gRNA and two sgRNAs. p28 and p88, virus-encoded replicase; p8 and p9, movement proteins; CP, coat protein. The sequence of the 5′ UTR is shown. The initial location of the 5′ TE (positions 36 to 63) is indicated along with location of a mutation (36-63m0) used to determine the importance of the sequence. (B) Diagram of the single- and dual-luciferase reporter constructs. NruI and SspI restriction sites were used to linearize constructs for in vitro transcription to produce RNA fragments containing or lacking 3′ UTR sequences. Rluc, Renilla luciferase; Fluc, firefly luciferase; Amb, amber stop codon. (C) Assessment of the translation activity of the 5′ UTR and 5′ UTR fragments in RRL using the dual-luciferase constructs containing viral 5′ sequences in the absence (N) or presence (3′ UTR+) of viral 3′ sequences. Data from at least three replicate experiments are shown. Standard deviation bars are given. (D) Translational activity of the 36-63 region containing mutations within the polypyrimidine-rich region shown in panel A. 36-63R, random 28-nt sequence.
An important unanswered question is how 60S subunits tethered to the 3′ end of the viral genome might be available for translation initiating from the 5′ end. Unlike the case for 3′ CITEs of other viruses in the Tombusviridae family, no obvious sequences that might support kissing-loop or other RNA-RNA interactions are evident in TCV 5′ and 3′ UTRs. To address the question of genome circularization in TCV, we have examined the 5′ UTR for sequences that are important for translation in vitro and in vivo. We have determined that a polypyrimidine stretch near the initiation codon supported translation of an internal ORF independent of upstream 5′ UTR sequences. Addition of 80S ribosomes to the 5′ UTR reduced the flexibility of the polypyrimidine residues and generated a toeprint consistent with binding to this region. 40S subunit binding also generated the same toeprint and additional ones near the 5′ end. Generation of out-of-frame AUGs upstream of the polypyrimidine region reduced translation, which supports 5′-terminal entry of 40S subunits followed by scanning and suggests that the polypyrimidine region is needed for an alternative function that requires ribosome binding. Combining 5′ and 3′ UTR fragments in vitro had no discernible effect on the structures of the RNAs, supporting an absence of RNA-RNA interaction between these regions. In contrast, in the presence of the 3′ UTR and 80S ribosomes, structural changes were found in the 5′ UTR polypyrimidine tract that were not evident when ribosomes interacted with the individual fragments. This suggests that ribosomes may be promoting an interaction between the 5′ and 3′ UTRs of TCV.
MATERIALS AND METHODS
Construction of dual-luciferase reporter constructs for in vitro translation assays.
A dual-luciferase reporter construct, pLuci (12), containing in-frame Renilla (Rluc) and firefly (Fluc) luciferase ORFs downstream of the T7 promoter was used to assay the ability of 5′ UTR fragments to promote internal ribosome entry in rabbit reticulocyte lysates (RRL). An amber mutation was introduced directly downstream of Rluc to prevent read-through of Fluc. Various gRNA 5′ UTR segments, some containing point mutations, were inserted upstream of Fluc into the BamHI/SacI site. The 3′ end of the construct contained a downstream segment (10 nt) to which a 3′ region of TCV (positions 3661 to 4054), containing the entire 3′ UTR and 140 nt of upstream coding sequence, was added. For RRL translation assays, 1.5 μg of in vitro-transcribed uncapped RNAs containing the dual-luciferase cassette was incubated in 20 μl of RRL translation mixture (Ambion) at 30°C for 90 min. After translation, a dual-reporter assay system (Promega) was used to measure the activity of the reporter luciferases with a TD 20/20 luminometer (Turner Designs).
In vivo translation assays.
A single-luciferase reporter construct, T7-Fluc-3′UTR, was used to assay for translation in vivo. T7-Fluc-3′UTR contained an upstream segment (30 nt), adjacent to the T7 promoter, to which full-length wild-type (wt) and mutant genomic and subgenomic TCV 5′ UTRs or Cardamine chlorotic fleck virus (CCFV), Saguaro cactus virus (SCV), and Japanese iris necrotic ring virus (JINRV) 5′ UTRs were added. The 3′ end of the construct contained the 3′ region of TCV (positions 3661 to 4054). These constructs were linearized with SspI or other appropriate restriction enzyme and used as templates for T7 polymerase-driven RNA synthesis in vitro. Thirty micrograms of uncapped in vitro-transcribed single-luciferase constructs was inoculated into protoplasts along with 10 μg of uncapped transcripts containing internal control Rluc. Protoplasts were harvested at 18 h postinfection (hpi), cells lysed in 1× Passive lysis buffer (Promega), and luciferase activity assayed as described above.
Construction of TCV mutants.
Oligonucleotide-mediated site-directed mutagenesis was used to construct the 5′ UTR TCV mutants. pTCV66, which contains full-length wt TCV sequence downstream from a T7 promoter, was used as a template during PCR. PCR fragments containing 5′ UTR mutations were inserted into SspI/BsmBI-digested pTCV66. All mutants were verified by sequencing. The wt and 5′ UTR TCV mutants were linearized with SmaI and used as templates for T7 polymerase-driven RNA synthesis in vitro.
Protoplast preparation, inoculation, and RNA gel blots.
Protoplasts were prepared using callus cultures from Arabidopsis thaliana ecotype Col-0 as previously described (70). To assay for genomic accumulation of TCV, 20 μg of uncapped in vitro-transcribed TCV gRNA, wt or mutant, was inoculated into protoplasts using polyethylene glycol (PEG). Total RNA was extracted at 40 hpi and subjected to electrophoresis. RNA was transferred to a nitrocellulose membrane, and the genomic RNA was detected using a complementary 32P-labeled oligonucleotide. Levels of TCV gRNA were normalized to 26S rRNA.
Isolation of 80S ribosomes and 40S ribosomal subunits.
Yeast ribosomes (strain JD1090) were isolated as previously described (37). To isolate 40S ribosomal subunits, 80S ribosomes were resuspended in buffer D [50 mM HEPES-KOH (pH 7.6), 10 mM Mg(CH3COO)2, 0.5 M KCl, 1 mg/ml heparin, and 2 mM dithiothreitol (DTT)]. A 150-μl portion of the suspension was applied to 12.5 ml of a 10 to 30% sucrose gradient with the same buffer conditions and centrifuged at 20,000 rpm for 16 h at 4°C in a swinging-bucket SW41 rotor. The gradient was fractionated, and fractions containing 60S and 40S subunits were identified by agarose gel electrophoresis. Fractions containing 40S ribosomal subunits were collected together, dialyzed for 4 h against 50 mM HEPES-KOH (pH 7.6), 5 mM Mg(CH3COO)2, 50 mM NH4Cl, and 2 mM DTT, and concentrated on Amicon Ultra (100k) columns (Millipore). Following addition of glycerol to a final concentration of 25%, 40S ribosomal subunits were stored at −80°C.
Ribosome binding assays.
All 5′ UTR fragments consisted of a 19-nt linker sequence followed by the complete 5′ UTR. To synthesize the TCV wt 5′ UTR fragment (using a single-luciferase construct containing 5′ and 3′UTR+ insertions [5′+3′] as a template), one round of PCR was carried out with the following oligonucleotides: the 5′ oligonucleotide (5′T7-poly) contains the T7 promoter followed by a linker sequence (GTAATACGACTCACTATAGGAGATCTAGCGCTGG), and the 3′ oligonucleotide was complementary to positions 40 to 63 of TCV and included an additional guanylate from the linker positioned downstream of the TCV sequence. All PCR products were transcribed using T7 RNA polymerase followed by treatment with RQ1 RNase-free DNase (Promega) to digest the template DNA. Filter binding assays were performed as previously described (37) in 50 μl of binding buffer [80 mM Tris-HCl (pH 7.4), 160 mM NH4Cl, 11 mM Mg(CH3COO)2, 6 mM β-mercaptoethanol, 0.4 mM GTP, 2 mM spermidine, and 0.4 μg/ml of poly(U)] containing 25 pmol of ribosomes or 40S ribosomal subunits and 2 to 100 pmol of 32P-5′-end-labeled 5′ UTR fragments.
Toeprinting with purified 40S and 80S ribosomal subunits.
Transcripts containing the 5′ 120 nt of TCV gRNA were used for toeprinting assays along with salt-washed 80S ribosomes and 40S ribosomal subunits. Ribosome binding reaction mixtures contained 3 pmol of RNA and 15 pmol of 80S ribosomes or 40S ribosomal subunits in 10 μl of binding buffer [80 mM Tris-HCl (pH 7.4), 160 mM NH4Cl, 11 mM Mg(CH3COO)2, 6 mM β-mercaptoethanol, 0.4 mM GTP, 2 mM spermidine, and 0.4 μg/ml of poly(U)] and 2.8 mM cycloheximide. Reaction mixtures were incubated at 30°C for 30 min. To detect toeprints, primer extension with SuperScriptIII reverse transcriptase (Invitrogen) was performed as follows. Eight microliters of the binding reaction mixture was added to 20 μl of primer extension buffer (SSIII FS buffer; Invitrogen) [20 mM DTT, 11 mM Mg(CH3COO)2, 0.5 mM each nucleoside triphosphate (dNTP)] supplemented with 6 pmol of 32P-end-labeled primer complementary to positions 103 to 120. Reaction mixtures were heated for 2 min at 55°C, followed by incubation at 37°C for 2 min to allow annealing of the oligonucleotide. After the addition of 120 U of SuperScript III, reaction mixtures were incubated for 30 min at 55°C. After the reaction was terminated by addition of 1 μl of 4 M NaOH, the mixture was incubated at 95°C for 2 min, followed by addition of 29 μl of a 4:25 (vol/vol) mixture of unbuffered 1 M Tris-HCl and stop dye (85% formamide, 0.5× Tris-borate-EDTA [TBE], 50 mM EDTA [pH 8.0], bromophenol blue, and xylene cyanol). Transcripts were subjected to electrophoresis through 8% polyacrylamide–7 M urea sequencing gels.
RNAs for in-line probing.
Plasmids pTCV-F4, pTCV-3UTR, and pTCV-5U3U (5′+3′) were generated by inserting PCR fragments of the F4 region (positions 3859 to 4054), the complete 3′ UTR (positions 3801 to 4054), and positions 1 to 63 fused to 3801 to 4054, respectively, into pUC19 digested with EcoRI and SmaI. T7 promoter sequences were introduced at the 5′ end of each fragment during PCR amplification. Mutations in pTCV-5U3U (M1, M2, or both M1 and M2) were generated using oligonucleotide-mediated mutagenesis. Constructs were confirmed by sequencing using universal primer M13-reverse. Plasmids were linearized with SmaI and transcribed into corresponding RNA fragments with T7 RNA polymerase. TCV 5′ UTR RNA fragments were transcribed from corresponding PCR fragments containing T7 promoter sequences.
In-line probing of RNA structure.
In-line probing was performed as previously described with modifications (35, 67). Briefly, RNA transcripts of F4, TCV 5′ UTR, TCV 3′ UTR, 5′+3′, and 5′+3′-derived mutants were purified from agarose gels, dephosphorylated with Antarctic phosphatase (NEB), 5′ end labeled using T4 polynucleotide kinase (NEB) and [γ-32P]ATP, and then purified by polyacrylamide gel electrophoresis. 5′-end-labeled fragments were denatured at 75°C and slowly cooled to 25°C. Five picomoles of end-labeled RNA was incubated at 25°C in 50 mM Tris-HCl (pH 8.5) and 20 mM MgCl2 for 14 h in the absence or presence of 10 pmol of yeast 80S ribosomes or purified proteinase K. For trans interaction assays, 5 or 50 pmol of unlabeled fragments was combined with 5 pmol of labeled fragment and subjected to in-line cleavage as described above. In-line probing with RdRp utilized a TCV RdRp-maltose binding protein (MBP) fusion protein purified from Escherichia coli (67), and in-line probing was carried out at 25°C for 1 h. RNA cleavage ladders were made by incubating 5 pmol of end-labeled RNA in 1 μg yeast tRNA, 50 mM NaHCO3-Na2CO3 (pH 9.2), and 1 mM EDTA for 5 min at 95°C. RNase T1 digests were produced by incubating 10 pmol of denatured end-labeled RNA in 1 μg yeast tRNA, 20 mM sodium citrate (pH 5.0), 1 mM EDTA, 7 M urea, and 1 U RNase T1 (Ambion) for 3 min at room temperature. All reaction mixtures were ethanol precipitated, resuspended with gel loading buffer II (Ambion), heated at 95°C for 2 min, and subjected to electrophoresis through 8 M urea–8% or 12% denaturing polyacrylamide gels followed by autoradiography. At least three independent in-line probing assays were produced for each fragment. Profile differences were noted only if found in all replicate gels.
RESULTS
A 28-nt pyrimidine-rich sequence from the 5′ UTR of TCV stimulates translation of an internal ORF.
To identify sequences within the 5′ UTR of TCV that are involved in cap-independent translation, a bicistronic reporter construct that contains a 5′ ORF coding for Renilla luciferase (Rluc) and a downstream ORF coding for firefly luciferase (Fluc) (12) was used. An amber stop codon located downstream of Rluc prevents read-through into the second ORF (Fig. 1B). Various fragments from the TCV gRNA 5′ UTR were inserted upstream of Fluc, and the TCV 3′-terminal 394 nt (3′ UTR+; positions 3661 to 4054) was inserted downstream of Fluc to assay for additive or synergistic effects of 5′ segments with 3′ sequences.
Insertion of the gRNA full-length 5′ UTR (positions 1 to 63) in the absence of 3′ UTR+ (N series) resulted in a 5.4-fold increase in translation of Fluc using rabbit reticulocyte lysates (RRL) (Fig. 1C). Insertion of positions 1 to 30, 1 to 40, or 1 to 46 gave translation that was similar to that for positions 1 to 63 or slightly enhanced. Constructs containing positions 21 to 63 and 36 to 63 induced 13.6-fold and 17.5-fold increases in Fluc activity, respectively, suggesting that one or more elements in the 5′ proximal region of the 5′ UTR are repressive. Shortening the inserted fragment to positions 41 to 63 or positions 47 to 63 reduced translational enhancement to only 8.8-fold and 6.1-fold, respectively, suggesting that 5′ translational enhancement requires at least a portion of the region between positions 36 and 41 (Fig. 1C, left). When the TCV 3′ region was included with selected constructs, no further enhancement in translation levels was evident (Fig. 1C, right), indicating that the TCV 3′ CITE is not active in commercially available RRL, similar to findings for some other 3′ CITEs (39). Translational enhancement by 3′ UTR+ was also negligible when selected constructs were assayed using commercially obtained wheat germ lysates (data not shown).
Positions 36 to 63 exhibited the highest level of translational activity, and this region is tentatively designated a 5′ TE (Fig. 1A). This region contains 21/28 pyrimidines, suggesting that a pyrimidine-rich sequence may be important for enhanced translational activity. To further explore the importance of the polypyrimidine sequence for translation in vitro, four of the pyrimidines (UCUC; positions 50 to 53) were altered to AGAG (construct 36-63m0) (Fig. 1A) in the dual-luciferase N-series construct. In addition, a construct was generated in which the 36- to 63-nt insert was replaced with a randomized sequence of the same length, containing the four nucleotides in equal proportion (36-63R). Translation of RNA containing 36-63m0 or 36-63R in RRL was reduced by 2-fold compared with that of the wild-type (wt) 36-63 sequence (Fig. 1D), suggesting that the polypyrimidine region contributes to efficient 5′ TE activity in these constructs.
Mutations in the 5′ TE reduce translation and viral accumulation in vivo.
To further explore properties of the 5′ UTR and 5′ TE in a more natural setting and environment, mutations were generated in a construct containing the complete gRNA 5′ UTR upstream, and 3′ UTR+ downstream, of a single Fluc reporter gene (Fig. 1B). This construct was previously used to identify the TCV 3′ CITE in vivo, since the presence of both 5′ and 3′ regions is synergistic for Fluc translation in protoplasts (57). RNA transcripts containing two to four base alterations throughout the 5′ UTR (Fig. 2 A) were transfected into Arabidopsis protoplasts along with control Rluc transcripts, and luciferase activity was determined 18 h later. With one exception, all mutations located in the 5′ portion of the 5′ UTR maintained or enhanced wt luciferase activity levels (Fig. 2B). Mutation m3, which converted two adjacent adenylates to cytidylates, was the exception, reducing translation by 36% (Fig. 2B). m5, which contained alterations in a second A-rich element, also reduced translation similarly to m3. The ∼50% increase in translation of transcripts containing m4, m7, or m8 mutations suggests that these alterations may be disrupting the presumptive repressive element in positions 1 to 35 (Fig. 1C).
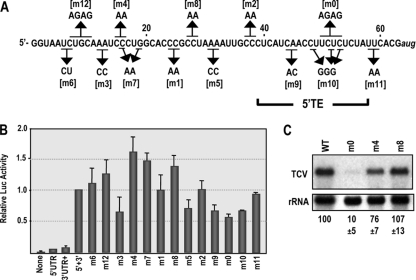
Fig. 2.
Effect of mutations in the TCV 5′ UTR on translation of a single-luciferase construct in protoplasts. (A) Positions of point mutations in the 5′ UTR. Names of mutations are bracketed. The location of the 5′ TE after completion of these experiments is shown. (B) Translational activity in protoplasts of single-luciferase constructs containing the 5′ UTR (wt and mutant) and 3′ UTR+. Luciferase was assayed at 18 h after transfection. None, no 5′ or 3′ sequences; 5′ UTR, 5′ UTR only; 3′ UTR+, 3′ region (positions 3661 to 4054) only; 5′+3′, 5′ UTR and 3′ UTR+. Data from at least three replicate experiments are shown. Standard deviation bars are given. (C) TCV gRNA containing mutations denoted above the lanes were assayed for accumulation in protoplasts at 40 h postinoculation (hpi). Levels of accumulation and standard deviations in three independent experiments are given below the lanes.
Several mutations within the central region of the 5′ TE (m0, m9, and m10) reduced translation by 32 to 44% (Fig. 2B). However, mutations located at the edges of the 5′ TE (m2 and m11), which converted two cytidylates at positions 37 and 38 to adenylates or two uridylates at positions 58 and 59 to adenylates, respectively, had no effect on translation. Based on these results, the 5′ TE resides between positions 39 and 57.
Several mutations were selected for further testing in the context of full-length TCV gRNA. m0, located within the 5′ TE, and upstream mutations m4 and m8 were incorporated into the gRNA and protoplasts inoculated with mutant or wt TCV RNA transcripts. m4, which enhanced translation of the single-luciferase reporter by 62%, reduced virus accumulation at 40 hpi by 24%. This reduction could imply that enhanced translation lowers virus accumulation fitness, e.g., by affecting the switch between translation and replication. Alternatively, m4 could cause a primary effect on replication due to 5′-end proximity and possible disruption of a cis element involved in TCV plus-strand synthesis. However, m8, which also had a beneficial effect on translation, did not affect virus accumulation in protoplasts. In contrast, m0, which reduced translation by 44%, had a strong negative effect on virus accumulation, with virus levels reduced by 90% (Fig. 2C).
Different viral 5′ UTRs can enhance translation in the presence of the TCV 3′ UTR.
To examine whether synergy between the TCV 5′ UTR and the 3′ 394 nt of TCV (3′ UTR+) is specific for TCV 5′ UTR sequences, we assayed whether 5′ UTRs from other carmoviruses, which share limited sequence similarity, could functionally replace the TCV 5′ UTR in translation assays. The 5′ UTR of Cardamine chlorotic fleck virus (CCFV) contains a similar polypyrimidine region, while the 5′ UTRs of Japanese iris necrotic ring virus (JINRV) and Saguaro cactus virus (SCV) share no sequence similarity with the TCV 5′ UTR outside a replication-required carmovirus consensus sequence (CCS) at their 5′ termini (Fig. 3 A) (13). Both CCFV and JINRV 5′ UTRs supported high levels of luciferase activity, while activity of the SCV 5′ UTR was similar to that of a random sequence (Fig. 3B). To determine if JINRV and CCFV 5′ UTRs are compatible with downstream TCV sequences for replication and translation, the 5′ UTR of TCV was replaced with the 5′ UTRs of JINRV and CCFV in full-length TCV gRNA, and the chimeric viruses were assayed for accumulation in protoplasts. The JINRV 5′ UTR was fully functional as a replacement sequence, with the chimeric virus accumulating to wt TCV levels in protoplasts (Fig. 3C), while virus containing the CCFV 5′ UTR accumulated to 19% of wt levels. These results suggest that the JINRV 5′ UTR can enhance translation mediated by the TCV 3′ end in the absence of any apparent sequence similarity (outside the CCS) with the 5′ UTR of TCV.
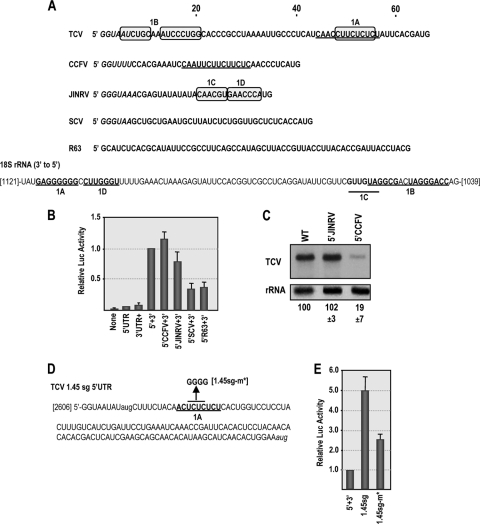
Fig. 3.
Related and unrelated viral 5′ UTRs can enhance translation in the presence of the TCV 3′ UTR. (A) Top, 5′ UTR sequences of TCV, CCFV, JINRV, and SCV. The similar polypyrimidine-rich sequences in TCV and CCFV 5′ UTRs are underlined. Carmovirus consensus sequences (G1–3A/U4–13) are in italics. The sequence of a randomized control fragment (R63) is also shown. Sequences in 5′ UTR that are complementary to one region of 18S rRNA are boxed. Bottom, partial sequence of Arabidopsis 18S rRNA in the 3′-to-5′ orientation. Locations of sequences complementary to 1A and 1B segments in the TCV 5′ UTR and to 1C and 1D sequences in the JINRV 5′ UTR are shown. Bases that can form canonical base pairs within the segments are in bold. (B) The 5′ UTR of TCV was replaced with the 5′ UTR of CCFV, JINRV, or SCV in the single-luciferase construct and luciferase activity measured in protoplasts at 18 h posttransfection. Data from at least three replicate experiments are shown. Standard deviation bars are given. (C) Accumulation of TCV gRNA in which the 5′ UTR was replaced with the 5′ UTR of JINRV or CCFV. RNA was extracted from inoculated protoplasts at 40 hpi. Levels of accumulation and standard deviations in three independent experiments are given below the lanes. (D) Sequence of the TCV 1.45 kb sgRNA. The 1A complementary sequence is underlined. The mutation generated in this region (1.45sg-m*) is shown. (E) Polypyrimidine region in the 1.45sg UTR is also important for synergistic translational enhancement. The reporter construct containing 3′ UTR+ was engineered to contain the 5′ UTR of the 1.45 sgRNA, and wt and mutant (1.45sg-m*) RNA transcripts were assayed for translation in protoplasts. Standard deviations from three independent experiments are shown.
One possible mode of ribosome recruitment is through interaction between 5′ UTR sequences and 18S rRNA sequences. The 5′ UTR of TCV contains two regions that might support interaction with 18S rRNA sequences (positions 48 to 55 in the 5′ TE and positions 5 to 20 [Fig. 3A, sequences labeled 1A and 1B]). The 18S rRNA 1A sequence (Fig. 3A, positions 1111 to 1118), which is conserved throughout eukaryotes, is known to be available for stable intermolecular base pairing (1). The JINRV 5′ UTR also contains two sequences that are complementary to the same region of the 18S rRNA (labeled 1C and 1D in Fig. 3A).
The 1.45 sgRNA 5′ UTR (Fig. 3D) contains a polypyrimidine sequence that is similar to the 5′ TE and complementary to region 1A in 18S rRNA. To test the importance of this sequence for translation, the complete 1.45 sgRNA 5′ UTR was inserted into the single-luciferase construct and translation assayed for in protoplasts. Translation was 5-fold higher with the 1.45 sgRNA 5′ UTR than with the gRNA 5′ UTR (Fig. 3E). Mutations that disrupted the sgRNA 1A sequence (1.45sg-m*) (Fig. 3D) reduced translation by 2-fold compared to transcripts synthesized from the wt construct (1.45sg-wt) (Fig. 3E). This level of reduction is similar to that found for analogous mutations in the gRNA 5′ UTR polypyrimidine sequence (Fig. 2). These results support a role for the polypyrimidine sequence in TCV translation.
40S ribosomal subunits bind to the 5′ UTR of TCV and scan to initiate translation.
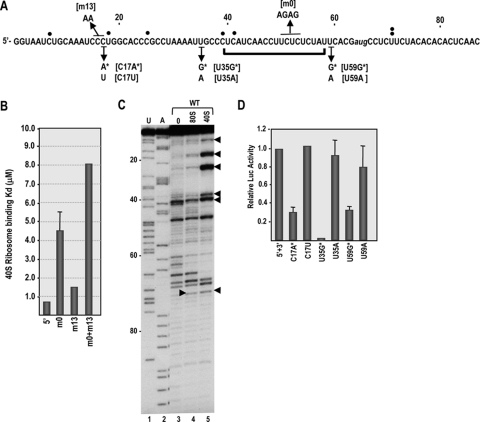
To investigate whether the polypyrimidine sequence in the 5′ TE can attract 40S ribosomal subunits, we first conducted filter binding assays using salt-washed 40S ribosomal subunits and wild-type and mutant TCV 5′ UTR fragments. The mutations assayed were the 4-nt alteration in the 5′ TE polypyrimidine sequence (m0) and a 2-nt alteration in the upstream region complementary to the rRNA 1B sequence (m13) (Fig. 4 A). Wild-type 5′ UTR transcripts bound to 40S ribosomal subunits with a dissociation constant (Kd) of 0.72 μM, while 5′ UTR mutations m0 and m13 reduced binding to the transcripts by 6.3- and 2.2-fold, respectively (Fig. 4B). 5′ UTR transcripts containing both m0 and m13 bound 40S subunits 11-fold more weakly than the wt, suggesting that altering both regions had an enhanced negative effect. The Kd for binding of salt-washed 80S ribosomes to the 5′ UTR was 2.88 μM, indicating that the 5′ end has a preference for 40S subunits.
Fig. 4.
40S ribosomal subunits bind to the 5′ UTR of TCV and scan to initiate translation. (A) TCV 5′ sequence showing the location of the 5′ TE (bracket) and mutations used for this analysis. Single filled circles denote reverse transcriptase-mediated stop sites in the presence of bound 40S subunits, with the double circles at U71 denoting the stop site found in the presence of both 40S subunits and 80S ribosomes. Open circles are new stop sites found using template containing the m0 mutations. (B) 40S ribosomal subunits bind to the 5′ UTR of TCV. Salt-washed 40S ribosomal subunits were added to radiolabeled wt and mutant 5′ UTR RNA fragments and binding affinity determined using filter binding assays. Standard deviations from three experiments are shown. (C) Toeprints with purified ribosomes and 40S ribosomal subunits. Salt-washed 80S ribosomes and 40S ribosomal subunits were mixed with RNA containing the 5′ 120 nt of TCV gRNA, and toeprints were determined by reverse transcription with SuperScriptIII. U and A, sequencing ladders; 0, no added ribosomes. Arrowheads denote locations of new polymerase stop sites in the presence of 40S subunits or 80S ribosomes. (D) Out-of-frame initiation codons upstream or downstream of the 5′ TE downregulate translation in vivo. Single-luciferase constructs containing the TCV 5′ UTR and 3′ UTR+ and mutations shown below the sequence in panel A were assayed for translation in protoplasts. Standard deviations in three independent experiments are shown.
To examine the location of ribosome entry in the 5′ UTR of TCV, we carried out toeprinting assays with 80S ribosomes and 40S subunits. In toeprinting assays, a 32P-labeled oligonucleotide is annealed downstream of a protein's presumed binding site, followed by extension of the primer by reverse transcription until polymerization is arrested when the enzyme is impeded by the bound protein. For eukaryotic ribosomes positioned at the translation initiation site, reverse transcription termination is usually 15 to 17 nt downstream of the initiation codon. Ribosome toeprinting is routinely carried out in translation-competent extracts in the presence of cycloheximide, which allows 40S subunits together with the ternary complex and required translation initiation factors to access the template and then scan to the initiation codon (42). 40S subunit scanning on unstructured templates requires initiation factor eIF1, and additional factors are needed if weak secondary structure exists. Since we were interested in initial binding sites in the absence of scanning, toeprinting was carried out using purified salt-washed 80S ribosomes and 40S ribosomal subunits.
Reverse transcription using a template containing the 5′ 120 nt of TCV gRNA in the absence of ribosomes produced a number of strong stop sites in the 5′ UTR (Fig. 4C, lane 3). The reason for the unusual number of stops is not known. Using identical conditions, reverse transcription from the 3′ end of the TCV 3′ UTR, which contains substantial secondary and tertiary structure (35), produced no discernible strong stops (V. A. Stupina and A. E. Simon, unpublished data). Toeprinting of 80S ribosomes bound to a fragment containing the 5′ 120 nt of TCV genomic RNA produced a new reverse transcriptase termination site at position U71 (Fig. 4C, rightward arrowhead), which would correspond with a ribosome positioned at the polypyrimidine tract of the 5′ TE. Toeprinting using 40S subunits also produced a termination site at position U71 as well as five strong upstream termination sites within the 5′ 42 nt (Fig. 4C, lane 5). New transcription termination sites could be caused by the bound ribosome/ribosomal subunit (i.e., a true toeprint) or by a structural change in the RNA that generates a new stop site but is unconnected to the position of the bound ribosome. If the multiple 5′-proximal stop sites represent transcription impediments by bound 40S subunits, then this would suggest that 40S subunits enter near the 5′ end and then must scan to the site of translation initiation. To test for ribosome scanning, point mutations were generated at three sites in the single-luciferase construct containing the TCV 5′ UTR and 3′ UTR+, and translation was assayed in protoplasts. The mutations created either out-of-frame initiation codons that should affect translation mediated by scanning or alternative single-base changes that controlled for other translational effects associated with altering specific sites. Two of the sites subjected to mutation were upstream of the 5′ TE (C17A and U35G), and one site was downstream of the 5′ TE (U59G) (Fig. 4A). Mutations that generated new initiation codons reduced translation by 70 to 94%, while the same site alterations had no significant effects on translation (Fig. 4D). U35G, which caused the greatest reduction in translation, contained a purine at the −3 position, which is known to be a key factor in enhancing translation initiation (30, 65). These results suggest that 40S subunit scanning is important for translation mediated by the 5′ UTR of TCV and that the primary function of the 5′ TE is apparently not for ribosome entry.
Binding of ribosomes to the polypyrimidine region of the 5′ TE.
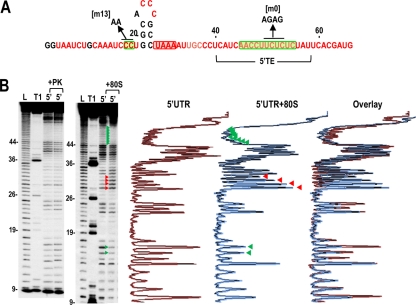
To investigate if the new reverse transcriptase stop site at position U71 represents a “true” toeprint caused by binding of 40S subunits or 80S ribosomes to the polypyrimidine track, we performed in-line probing structural analysis of the 5′ UTR in the presence and absence of added 80S ribosomes. In-line probing reports on the flexibility of each nucleotide, as only flexible (i.e., unpaired or unconstrained) residues are able to rotate such that the 2′OH, RNA backbone phosphate, and oxyanion leaving group are linear, which is required for phosphodiester bond cleavage (56). Proteins interacting with the RNA should reduce the flexibility of residues in the bound region, creating a “footprint” as well as possibly causing additional structural changes. 80S ribosomes were used for footprinting, as U71 was the only new polymerase stop site generated in the toeprinting assay, and thus the bound templates should be more uniform.
In-line cleavage of the 5′ UTR alone revealed that most residues are flexible, with the exceptions of positions 19 to 28, which are predicted to form a short hairpin (Fig. 5 A and B). Addition of 80S ribosomes to the 5′ UTR fragment enhanced the flexibility of residues at positions 30 to 33 and reproducibly reduced the flexibility of 11 consecutive residues in the 5′ TE (positions 45 to 55) (Fig. 5B; see also Fig. 9B and C). Five additional residues in the 5′ TE (positions 40 to 44) exhibited reduced flexibility in two of the three in-line probings (see, e.g., Fig. 9B and C). The reduced flexibility of a stretch of polypyrimidine residues in the 5′ TE suggests possible protection against self cleavage by ribosome binding, which is consistent with a bound 80S ribosome generating a toeprint at U71. In contrast, addition of a control protein, proteinase K, did not detectably alter the flexibility of any residue in the 5′ UTR. These results support the 5′ TE as the major interacting site for 80S ribosomes and a secondary site for interaction of 40S subunits.
Fig. 5.
In-line cleavage profiles of the 5′ UTR in the presence and absence of 80S ribosomes. (A) Structure of the 5′ UTR of TCV. The location of the 5′ TE is shown. Residues in red are flexible and susceptible to cleavage, with color intensity reflecting the degree of cleavage. Green and red boxes denote residues that have reduced or enhanced susceptibility to cleavage, respectively, in the presence of 80S salt-washed ribosomes. Locations of mutations used for ribosome binding assays are shown. (B) Autoradiograph of in-line cleavage analysis of 5′-labeled 5′ UTR in the presence and absence of proteinase K (+PK) (left) or 80S ribosomes (+80S) (right). Green and red arrowheads denote residues whose cleavage pattern was altered by the presence of ribosomes in all three replicates. L, partial hydroxide cleavage ladder; T1, partial RNase T1 digest of denatured RNA showing the location of guanylates; 5′, 5′ UTR in-line cleavage profile. Numbering is from the 5′ end of the gRNA. Densitometer tracings of the 5′ UTR and 5′ UTR + 80S ribosome lanes and an overlay of the two tracings are shown to the right. Residues with consistent differences in all three replicates (enhanced or reduced cleavages) are denoted by red and green arrowheads, respectively.
Fig. 9.
The ribosome-mediated cleavage profile of the 5′ UTR is altered in the presence of the 3′ UTR. (A) Structure of the TCV 5′ UTR. Residues in red are flexible and susceptible to cleavage, with color intensity reflecting the degree of cleavage. Green and red boxes denote residues that have reduced or enhanced susceptibility to cleavage, respectively, in the presence of 80S ribosomes alone. Red circles denote residues with enhanced cleavage in all three replicas in the presence of ribosomes and 10× unlabeled 3′ UTR. (B) Cleavage profile for the 5′ UTR in the presence and absence of 1× or 10× unlabeled 3′ UTR and 80S ribosomes. Red and green arrowheads denote residues that show enhanced or reduced cleavage, respectively, in the presence of 80S ribosomes (as also shown in Fig. 4B). Red circles denote residues with enhanced cleavage in the presence of ribosomes and 10× unlabeled 3′ UTR. Note that the cleavage profiles for the 5′ UTR and 5′ UTR plus 1× or 10× unlabeled 3′ UTR are identical, indicating no discernible RNA-RNA interactions in the absence of ribosomes. (C) An enlargement of the upper portion of the gel shown in panel B. (D) Cleavage profile for the 3′ UTR in the presence and absence of 1× or 10× unlabeled 5′ UTR and 80S ribosomes. Note no differences in the cleavage profile between lanes 3 to 5 or lanes 6 to 8.
Evidence against RNA-RNA cyclization sequences within the 5′ and 3′ UTRs of TCV.
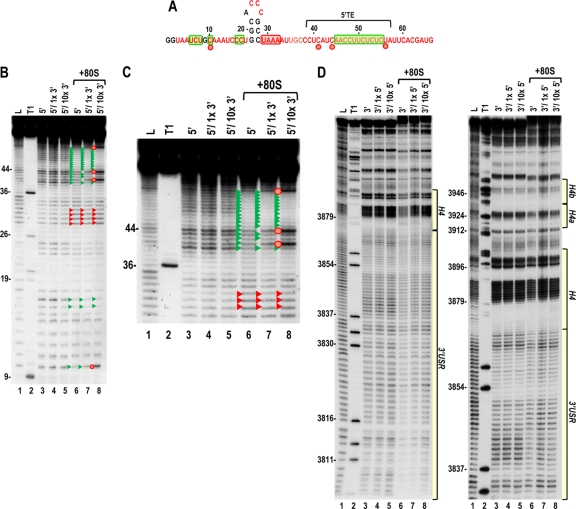
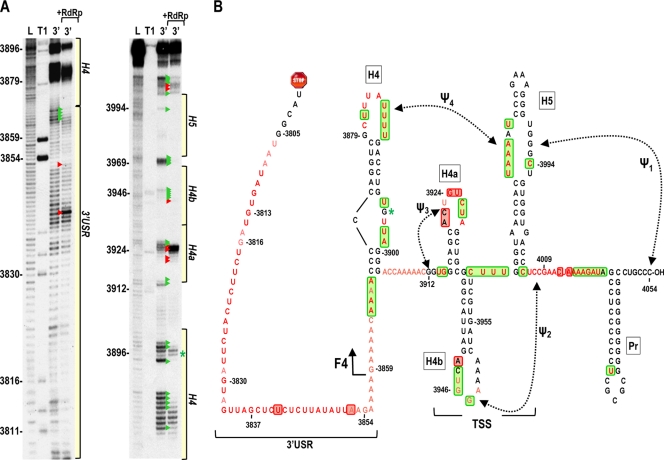
The ability of the unrelated JINRV 5′ UTR to enhance translation when associated with the 3′ UTR of TCV argues against the presence of specific canonical RNA-RNA cyclization sequences such as those found for other members of the Tombusviridae family. To further examine this possibility, we used in-line cleavage to determine if any residues lose flexibility when the 5′ and 3′ UTRs are combined (such as would occur when bases interact). Before such a study was possible, we needed to determine the cleavage profile for the complete TCV 3′ UTR (positions 3801 to 4054). In-line probing of the 3′ UTR fragment (Fig. 6 A) revealed that cleavages in the 3′-terminal 185 bases were identical to those previously found using a shorter fragment (fragment F4) containing positions 3859 to 4054 (67). The region upstream of the F4 fragment, which is critical for efficient translation (57), was substantially unstructured, with every residue producing a discernible cleavage (Fig. 6A). This region (positions 3806 to 3868) is designated the 3′ unstructured region (3′ USR).
Fig. 6.
In-line cleavage profile of the TCV 3′ UTR. (A) 5′-labeled 3′ UTR transcripts were subjected to in-line cleavage in the presence and absence of purified RdRp. A short run and a longer run of the same samples are shown. L, partial hydroxide cleavage ladder; T1, partial RNase T1 digest; 3′, 3′ UTR in-line cleavage profile; 3′ +RdRp, an equal molar amount of RdRp was added before in-line cleavage. Base numbering is from the 5′ end of TCV. Green and red arrowheads denote residues whose cleavage pattern was altered by the presence of RdRp in all replicates. The locations of the 3′ USR and hairpins shown in panel B are shown. (B) Secondary and tertiary structures of the 3′ region of TCV. Locations of the TSS, which includes hairpins H4a, H4b and H5, and two pseudoknots (Ψ2 and Ψ3), and H4 and the 3′ USR are denoted. The endpoint of the previously analyzed F4 fragment is shown. Residues in red are flexible and susceptible to cleavage, with color intensity reflecting the degree of cleavage. Green and red boxes denote residues that have reduced or enhanced susceptibility to cleavage, respectively, in the presence of RdRp. An asterisk denotes a single difference in the RdRp-mediated conformational shift in the F4 fragment compared to the full 3′ UTR. This cleavage is not present in the F4 fragment in the presence of RdRp.
RdRp, when binding to the shorter F4 fragment, induces a widespread conformational shift, including regions within the TSS that are important for efficient ribosome binding (67). To determine if the conformational shift is similar in the longer 3′ UTR fragment, RdRp was added to labeled 3′ UTR, which was then subjected to in-line cleavage. In the region shared by fragment F4 and the complete 3′ UTR, the cleavage profiles when bound to the RdRp were nearly identical (67, 68). The only exception was at position G3896 in the asymmetric loop of H4, where the cleavage in the F4 fragment is much reduced in the presence of the RdRp. While these results support our previous suggestion that the sequence within the F4 fragment constitutes an RNA domain (35, 67), altered residue flexibility upon RdRp binding was also evident at several locations in the 3′ USR, suggesting some cross talk between the 3′ USR and downstream sequences.
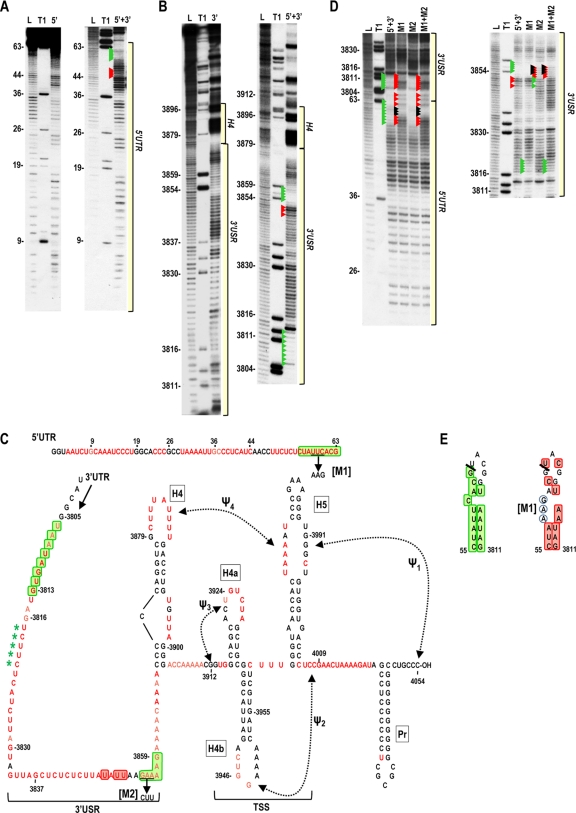
To search for possible interactions between 5′ and 3′ UTRs, a construct containing the full-length 5′ UTR linked to the complete 3′ UTR (5′+3′) was generated and the cleavage profile compared to those of the individual fragments. In-line cleavage of 5′+3′ revealed reduced residue flexibility in positions 55 to 63 in the 5′ TE and positions 3806 to 3813 and 3854 to 3859 in the 3′ USR region (Fig. 7). Since the sequences UUC within the 5′ TE and GAA in positions 3854 to 3856 could potentially pair, mutations that should, when combined, maintain compensatory pairing were generated in the two sequences (M1 and M2) in 5′+3′, and single and combined mutations were analyzed by in-line probing (Fig. 7C). As shown in Fig. 7D, altering the individual regions did not generate corresponding differences in the putative partner region, and both alterations together had an additive effect on the in-line cleavage pattern. Although an interaction between these two sequences was not supported, alteration M1 in the 5′ TE produced enhanced flexibility both locally and at positions 3806 to 3813. Examination of these two regions revealed the possible formation of a hairpin that spans the 5′ UTR/3′ UTR junction, with nearly every residue in the putative hairpin losing flexibility when the UTRs are joined in 5′+3′ (Fig. 7E, left). M1 alterations completely disrupted this putative hairpin, based on nearly all residues regaining their flexibility (Fig. 7E, right). The exceptions were the altered bases and one adjacent base (AAGA), which may be interacting with a second sequence (UCUU, positions 3817 to 3820), which exhibited reduced flexibility in the mutant fragment and is perfectly complementary with the M1 mutant region. While these results support base pairing between the 5′ TE and positions 3806 to 3813 in 5′+3′, this interaction is either not occurring or is of minor importance in the full-length virus. This conclusion is based on previous analysis of deletions generated in full-length TCV near the beginning of the 3′ UTR (positions 3802 to 3817), which had little effect on TCV accumulation in plants (5). This suggests that the interaction may be an artifact of artificially linking the 5′ and 3′ UTRs in the 5′+3′ fragment.
Fig. 7.
In-line cleavage profile of covalently linked 5′ UTR and 3′ UTR. Transcripts containing the 5′ UTR covalently linked to the 3′ UTR (5′+3′) were 5′ end labeled and subjected to in-line cleavage. (A) Comparison of 5′+3′ and 5′ UTR (5′) cleavage profiles. Residues with enhanced or reduced cleavage in 5′+3′ compared with the 5′ UTR alone are denoted by red and green arrowheads, respectively. The location of the 5′ UTR region within the 5′+3′ cleavage profile is shown. See the legend to Fig. 5 for other abbreviations. (B) Comparison of the 5′+3′ and 3′ UTR (3′) cleavage profiles. Residues with enhanced or reduced cleavage in 5′+3′ compared with the 3′ UTR alone are denoted by red and green arrowheads, respectively. The locations of 3′ USR and 3′ hairpins are indicated. Longer electrophoresis of samples did not reveal any additional changes downstream of this region (not shown). (C) Locations of residues with enhanced (red boxes) or reduced (green boxes) flexibility in 5′+3′ compared with the individual UTR fragments. See the legend to Fig. 5 for more details. Mutations (M1 and M2) generated to access a possible interaction between two sequences that had reduced flexibility in 5′+3′ are shown. Asterisks denote loss of cleavage at these positions in the M1 mutant 5′+3′ fragment. (D) In-line cleavage profile for 5′+3′ transcripts containing either M1, M2, or both mutations (M1+M2). Arrowheads in the 5′+3′ lane denote differences from the profiles of the individual fragments (from panels A and B). Red and green arrowheads in the mutant cleavage lanes denote differences from the wt 5′+3′ transcripts. Black arrowheads denote bases that were mutated. (E) Possible interaction between the 5′ and 3′ UTR regions in 5′+3′. Left, green boxes denote residues with reduced flexibility in the 5′+3′ fragment compared with the individual UTRs. The thick line denotes the junction between the 5′ and 3′ UTRs in the 5′+3′ fragment. Right, red boxes denote enhanced flexibility of residues in 5′+3′ transcripts containing M1 mutations (circled). Note that the altered sequence remains inflexible (5′AAGA) and can now pair with sequence in the 3′ UTR region of the 5′+3′ fragment (5′UCUU) that also showed reduced flexibility in the m1 fragment.
5′ and 3′ UTR fragments were next combined in trans to search for interactions. For this assay, the 5′ UTR or 3′ UTR was labeled, 1× or 10× levels of unlabeled 3′ UTR or 5′ UTR fragments were added, and the mixtures were subjected to in-line cleavage. No differences were found in the cleavage pattern for any residue in either the 5′ UTR or 3′ UTR when fragments were combined in trans (see Fig. 9B and D, lanes 3 to 5). Electrophoretic mobility shift assays also revealed no evidence for RNA-RNA interactions under conditions that clearly show such interactions in other viruses (data not shown). These results, together with the ability of the unrelated 5′ UTR of JINRV to functionally replace the 5′ UTR of TCV, suggest that circularization of the TCV genome is not achieved through RNA-RNA interactions.
Ribosome-mediated bridge between 5′ and 3′ UTRs of TCV.
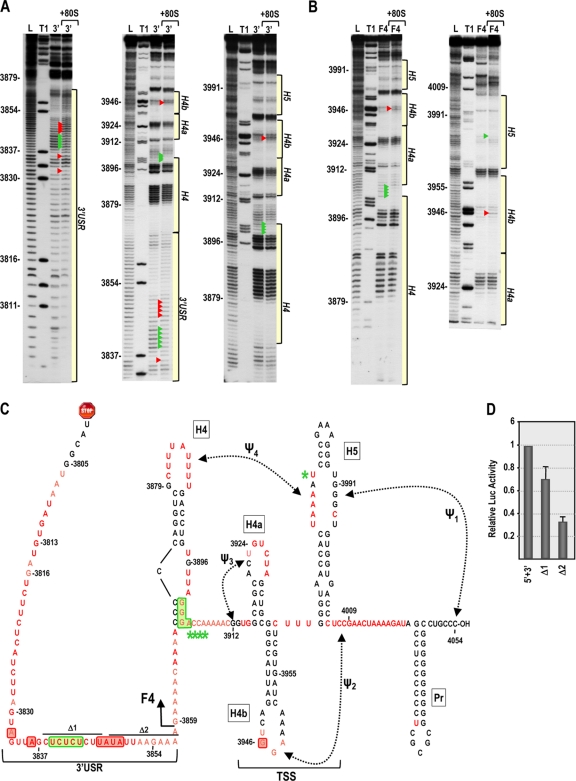
Since our current results suggest that 40S subunits bind to the 5′ UTR and since 60S subunits were previously shown to bind to the 3′ UTR (57), one possible mechanism for bridging the 3′ and 5′ ends could involve joining of ribosomal subunits to form 80S ribosomes or 80S ribosomes binding simultaneously to both 5′ and 3′ UTRs. Before examining whether ribosomes contribute to bridging the 5′ and 3′ ends of TCV, a cleavage profile was generated for 80S ribosome interaction with the TCV 3′ UTR. Unlike the widespread conformational shift induced by RdRp binding to the 3′ UTR, ribosome binding to the 3′ UTR had only a slight effect on the cleavage profile (Fig. 8 A). Two locations within the highly structured F4 domain displayed cleavage differences in the presence of 80S ribosomes: a single residue with enhanced flexibility within the TSS (position 3946 in the loop of H4b) and four residues with reduced flexibility in the lower stem and adjacent to H4. Ribosome binding to the shorter F4 fragment also resulted in enhanced cleavage at position 3946, but the four residues with reduced cleavage in the H4 region were now all adjacent to the H4 lower stem (Fig. 8B, arrowheads). Ribosome binding to fragment F4 also altered the flexibility of a single residue in the large symmetrical loop of H5 (position 3979).
Fig. 8.
In-line cleavage profiles of the 3′ UTR and fragment F4 in the presence and absence of 80S ribosomes. (A and B) In-line cleavage autoradiograph of labeled 3′ UTR (A) or fragment F4 (B) in the presence (+80S) and absence of 80S ribosomes. Green and red arrowheads denote residues whose cleavage pattern was altered by the presence of ribosomes in all three replicates. See the Fig. 5 legend for more details. (C) Structure of the 3′ UTR of TCV. Residues in red are flexible and susceptible to cleavage, with color intensity reflecting the degree of cleavage. Green and red boxes denote residues in the 3′ UTR fragment that have reduced or enhanced susceptibility to cleavage, respectively, in the presence of 80S salt-washed ribosomes. F4 shared the 80S-induced enhanced cleavage at position 3946 only, and asterisks denote residues in F4 with reduced flexibility in the presence of 80S ribosomes. Locations of deletions Δ1 and Δ2, used to assay for effect on translation, are shown. (D) Residues with altered cleavages in the 3′ USR upon ribosome binding are important for translation. The reporter construct containing the TCV 5′ UTR and 3′ UTR+ was engineered to delete the segments shown in panel C. RNA transcripts were assayed for translation in protoplasts. Standard deviations from three independent experiments are shown.
In the 3′ USR of the 3′ UTR fragment, ribosome binding reproducibly altered the cleavage profile between positions 3832 and 3849 (Fig. 8). To determine whether this region of the 3′ USR is important for translation, two deletions were generated in the single-luciferase construct containing the TCV 5′ UTR and 3′ UTR+. Δ1, which removed a pyrimidine-rich sequence at positions 3838 to 3846, reduced translation by 30%, while Δ2, which removed positions 3848 to 3856, reduced translation by 68% (Fig. 8D). This suggests that sequences within the 3′ USR with altered cleavage profiles upon ribosome binding are needed for efficient translation.
To determine if the presence of both 5′ UTR and 3′ UTR fragments affected the ribosome-associated cleavage profiles of the individual fragments, ribosomes were added to the individual fragments and to the combined fragments and the RNAs subjected to in-line cleavage (Fig. 9 B and C). As also shown in Fig. 5, the cleavage profile for 80S binding to the 5′ UTR displayed protection of most residues in the 5′ TE (Fig. 9B and C, lane 6). When labeled 3′ UTR or 5′ UTR was subjected to in-line cleavage in the presence of 1× or 10× unlabeled 5′ UTR or 3′ UTR, respectively, no differences in the cleavage pattern were observed (Fig. 9B, C, and D, compare lanes 3 to 5). When 3′ and 5′ UTR fragments were combined in the presence of ribosomes, no changes in residue flexibility in the 3′ UTR were found beyond those observed for ribosome-3′ UTR interaction as described above (Fig. 9D, compare lanes 6 to 8). In contrast, addition of 1× unlabeled 3′ UTR to labeled 5′ UTR in the presence of 80S ribosomes caused a slight enhancement in flexibility at two residues (positions 41 and 44) within the 5′ TE. When 10× 3′ UTR was added, these two residues and two additional residues (positions 10 and 56) displayed significantly enhanced cleavage (Fig. 9B and C). Position 56 is within the 5′ TE, while position 10 is in the 1B region that is complementary to 18S rRNA. This result strongly suggests that 80S binding to the 5′ TE is altered when the 3′ UTR is present, and it supports possible ribosome-mediated bridging of the 5′ and 3′ UTRs.
DISCUSSION
Cyclization of the RNA template constitutes an important step during cap-dependent translation (24, 29), allowing for protection of terminal regions of the RNA, recruitment of eIFs, and recycling of ribosomal subunits. Translational enhancement by many plant virus 3′ CITEs requires association with sequences that promote cyclization by forming long-distance RNA-RNA interactions (8, 9, 17, 23–25, 40, 49, 52, 64). In luteoviruses and tombusviruses, RNA-RNA interactions between the 5′ UTR and 3′ CITE-associated sequences enhance translation by bringing CITE-bound initiation factors to the 5′ end for correct positioning at the translation initiation site (40, 62). For the tombusvirus I-shaped 3′ CITE, the ability to bind simultaneously to both eIF4E and sequences in the 5′ UTR is necessary for recruitment of ribosomes to the 5′ end of the genome (40). Other plant RNA virus 5′ UTRs have elements with IRES-like activity as assayed using dual-reporter constructs, but the limited length of the region upstream of initiation codons suggests that small-subunit entry differs from that of animal virus IRESs (3, 22, 24, 25, 31, 41, 54, 60, 69). 5′ elements with IRES-like activity in TEV and BRV contain polypyrimidine sequences that are complementary to a region of 18S rRNA that is known to be accessible to base pairing with an mRNA template (24, 25, 54, 69). This has led to suggestions that small IRES-like elements in plant viruses may act by attracting 40S subunits through cRNA-RNA interactions, similar to the case for prokaryotic Shine-Dalgarno sequences (53).
Using a dual-luciferase construct, we determined that a fragment (positions 36 to 63) containing a polypyrimidine region enhances translation of the downstream ORF by ∼17-fold in RRL (Fig. 1C). This pyrimidine-rich sequence is also present in the 5′ UTR of the 1.45 sgRNA and is located 21 nt downstream of the p8 initiation codon in the 1.7 sgRNA. The consensus sequence for the element, CAAC/U4–13, is also located in similar positions relative to the initiation codons in the related virus Cardamine chlorotic fleck virus (CCFV), which also contains a TCV-like TSS. Mutations in the pyrimidine-rich portion of the element in the TCV 36-63 5′ UTR fragment (mutant 36-63m0) reduced translation of the downstream reporter in RRL (Fig. 1D). The same mutations incorporated into the complete 5′ UTR inserted upstream of a single reporter ORF (m0) reduced translation in protoplasts and severely affected accumulation in full-length virus (Fig. 2C). Two additional mutations in the element within the reporter construct (m9 and m10) also reduced translation in protoplasts (Fig. 2B), as did mutations in the sequence within the 5′ UTR of the 1.45 sgRNA (Fig. 4F).
Based on the in vitro and in vivo analyses of TCV 5′ UTRs, a 5′ TE lies within positions 39 to 57, the majority of which is the CAAC/U4–13 motif (positions 44 to 56). Addition of 80S ribosomes to the 5′ UTR reduced the flexibility of nearly all residues within this motif in all replicate experiments (Fig. 5B), which is likely a consequence of direct ribosome binding at this location. Ribosome binding to the motif is also supported by the finding that mutations in this sequence reduced the efficiency of ribosome binding to a 5′ UTR fragment (Fig. 4B) and is consistent with the appearance of a new reverse transcriptase stop site at U71 (Fig. 4C).
While these results suggest that the polypyrimidine sequences in the 5′ TE have IRES-like properties, other results suggest that ribosome entry is more complex. In vitro toeprinting of bound 40S subunits suggests the existence of multiple 5′ proximal binding sites besides the polypyrimidine region, and the negative effect of upstream out-of-frame initiation codons also suggests that entry is near the 5′ end followed by scanning, as was previously found for Tomato bushy stunt virus and Barley yellow dwarf virus (9, 47). Thus, the intriguing question is what the principal function of the polypyrimidine sequence within the 5′ TE is. One possibility is that the sequence serves as a pause site for scanning 40S subunits or as a direct entry site for subunits used for an activity that enhances translation, such as participation in template cyclization. The ubiquitous presence of canonical or noncanonical means of cyclizing translated mRNAs suggests that TCV must also possess a mechanism for circularizing its genome. Currently, there is no evidence that TCV supports the type of RNA-RNA interactions common for other viruses within the Tombusviridae. In-line cleavage profiles for the 5′ and 3′ UTRs did not reveal any consistent differences when the two UTRs were combined. Additionally, no long-distance interactions were detected when the two fragments were covalently joined, except for the appearance of an artificial hairpin formed from sequence spanning the junction (Fig. 7E). Replacement of the TCV 5′ UTR in a translational reporter construct and in the full-length genomic RNA with the 5′ UTR of JINRV did not significantly affect translation or virus accumulation, respectively, despite the lack of sequence similarity between the two 5′ UTRs (Fig. 3B and C). SCV, whose 5′ UTR is not compatible with the TCV 3′ UTR+, differs from JINRV and TCV by containing a stable hairpin at its 5′ terminus, and thus 40S entry may occur by a different mechanism. Altogether, these results argue against the presence of long-distance RNA-RNA interactions between TCV 5′ and 3′ UTRs as a means of template circularization.
The possibility that ribosomes may participate in cyclization of the TCV viral RNA is based on our finding that addition of the 3′ UTR alters how 80S ribosomes interact with the 5′ TE region (Fig. 9B and C). Since the cleavage profile of the 3′ UTR was not altered, this interaction does not appear to involve release of the 3′ UTR. We have recently found that a virus in a different genus, Pea enation mosaic virus, which also contains a 3′ TSS, undergoes similar structural changes at the 5′ end in the presence of 80S ribosomes and the 3′ UTR (F. Gao and A. E. Simon, unpublished data). This suggests that ribosome bridging of 5′ and 3′ sequences may not be a property exclusive to carmoviruses.
Three regions have been defined as comprising the TCV 3′ CITE: the TSS, H4, and sequence upstream of H4 (57). Our previous results indicated that the TSS was capable of binding to both 80S ribosomes and 60S ribosomal subunits, and binding was reduced when Ψ3, which mimics the amino acceptor stem, was disrupted. The current results now show that ribosome interaction with the 3′ UTR or the shorter F4 fragment has only a slight effect on residue flexibility within the TSS (Fig. 8), suggesting that binding may involve sequences that are already constrained by base pairing. Ribosome binding to the 3′ UTR also reduced flexibility at the base of H4 in a manner that was slightly different from that when binding to the shorter F4 fragment, suggesting that ribosome interaction in the region is influenced by sequence upstream of H4. While the function of H4 in translation is not currently understood, alterations in the terminal and internal loops affect both translation and replication (57, 67). The sequence upstream of H4 is now determined to be mainly unstructured, with every residue susceptible to various degrees of in-line cleavage (Fig. 8A). Previous chemical structure probing of full-length TCV gRNA that extended a short distance into this region also indicated a lack of structure (58). Interestingly, residue flexibility was reproducibly altered in a localized region within the 3′ USR upon ribosome binding (positions 3832 to 3849). Deletion of positions 3848 to 3856 reduced translation by 68% (Fig. 8D), suggesting that the event reflected in the cleavage profile change in this region is needed for efficient translation. While the 3′ USR did not enhance ribosome binding to the TSS in vitro (57), it is possible that trans-acting factors interact with the 3′ USR in vivo and influence 60S binding to the 3′ end. Alternatively, the 3′ USR may be needed for 60S transfer to, or interaction with, the 5′ end. This possibility is currently under active investigation.
ACKNOWLEDGMENTS
We acknowledge John C. McCormack for providing data for Fig. 3C and Kerong Shi for providing data for Fig. 3B and Fig. 4D.
This work was supported by a grant from the U.S. Public Health Service (GM 061515-05A2/G120CD) to A.E.S., NIH grant R01 GM058859 to J.D.D., and a grant from the American Heart Association (AHA 0630163N) to A.M. V.A.S. was supported by NIH Institutional Training Grant T32-AI51967-01.
Footnotes
Published ahead of print on 9 March 2011.
REFERENCES
- 1. Akbergenov R. Z., et al. 2004. ARC-1, a sequence element complementary to an internal 18S rRNA segment, enhances translation efficiency in plants when present in the leader or intercistronic region of mRNAs. Nucleic Acids Res. 32:239–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alvarez D. E., Lodeiro M. F., Luduena S. J., Pietrasanta L. I., Gamarnik A. V. 2005. Long-range RNA-RNA interactions circularize the dengue virus genorne. J. Virol. 79:6631–6643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Basso J., Dallaire P., Charest P. J., Devantier Y., Laliberte J. F. 1994. Evidence for an internal ribosome entry site witin the 5′ non-translated region of turnip mosaic potyvirus RNA. J. Gen. Virol. 75:3157–3165 [DOI] [PubMed] [Google Scholar]
- 4. Bradrick S. S., Walters R. W., Gromeier M. 2006. The hepatitis C virus 3′-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res. 34:1293–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carpenter C. D., Oh J. W., Zhang C. X., Simon A. E. 1995. Involvement of a stem-loop structure in the location of junction sites in viral-RNA recombination. J. Mol. Biol. 245:608–622 [DOI] [PubMed] [Google Scholar]
- 6. Chappell S. A., Edelman G. M., Mauro V. P. 2000. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc. Natl. Acad. Sci. U. S. A. 97:1536–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dreher T. W., Miller W. A. 2006. Translational control in positive strand RNA plant viruses. Virology 344:185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fabian M. R., White K. A. 2004. 5′–3′ RNA-RNA interaction facilitates cap- and poly(A) tail-independent translation of tomato bushy stunt virus mRNA—a potential common mechanism for Tombusviridae. J. Biol. Chem. 279:28862–28872 [DOI] [PubMed] [Google Scholar]
- 9. Fabian M. R., White K. A. 2006. Analysis of a 3′-translation enhancer in a tombusvirus: a dynamic model for RNA-RNA interactions of mRNA termini. RNA 12:1304–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gallie D. R. 2007. Translational control in plants and chloroplasts, p. 747–774 In Mathews M. B., Sonenberg N., Hershey J. W. B. (ed.), Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 11. Gamarnik A. V., Andino R. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74:2219–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grentzmann G., Ingram J. A., Kelly P. J., Gesteland R. F., Atkins J. F. 1998. A dual-luciferase reporter system for studying recoding signals. RNA 4:479–486 [PMC free article] [PubMed] [Google Scholar]
- 13. Guan H. C., Carpenter C. D., Simon A. E. 2000. Analysis of cis-acting sequences involved in plus-strand synthesis of a turnip crinkle virus-associated satellite RNA identifies a new carmovirus replication element. Virology 268:345–354 [DOI] [PubMed] [Google Scholar]
- 14. Guo L., Allen E. M., Miller W. A. 2001. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol. Cell 7:1103–1109 [DOI] [PubMed] [Google Scholar]
- 15. Hellen C. U. T., Sarnow P. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593–1612 [DOI] [PubMed] [Google Scholar]
- 16. Hernandez G. 2008. Was the initiation of translation in early eukaryotes IRES-driven? Trends Biochem. Sci. 33:58–64 [DOI] [PubMed] [Google Scholar]
- 17. Hernandez G., Altmann M., Lasko P. 2010. Origins and evolution of the mechanisms regulating translation initiation in eukaryotes. Trends Biochem. Sci. 35:63–73 [DOI] [PubMed] [Google Scholar]
- 18. Hernandez G., Vazquez-Pianzola P., Sierra J. M., Rivera-Pomar R. 2004. Internal ribosome entry site drives cap-independent translation of reaper and heat shock protein 70 mRNAs in Drosophila embryos. RNA 10:1783–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hinnebusch A. G., Dever T. E., Asano K. 2007. Mechanism of translation initiation in the yeast Saccharomyces cerevisiae, p. 225–268 In Mathews M. B., Sonenberg N., Hershey J. W. B. (ed.), Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 20. Isken O., et al. 2003. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J. 22:5655–5665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Isken O., Grassmann C. W., Yu H. Y., Behrens S. E. 2004. Complex signals in the genomic 3′ nontranslated region of bovine viral diarrhea virus coordinate translation and replication of the viral RNA. RNA 10:1637–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jaag H. M., et al. 2003. An unusual internal ribosomal entry site of inverted symmetry directs expression of a potato leafroll polerovirus replication-associated protein. Proc. Natl. Acad. Sci. U. S. A. 100:8939–8944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Karetnikov A., Keranen M., Lehto K. 2006. Role of the RNA2 3′ non-translated region of Blackcurrant reversion nepovirus in translational regulation. Virology 354:178–191 [DOI] [PubMed] [Google Scholar]
- 24. Karetnikov A., Lehto K. 2007. The RNA2 5′ leader of Blackcurrant reversion virus mediates efficient in vivo translation through an internal ribosomal entry site mechanism. J. Gen. Virol. 88:286–297 [DOI] [PubMed] [Google Scholar]
- 25. Karetnikov A., Lehto K. 2008. Translation mechanisms involving long-distance base pairing interactions between the 5′ and 3′ non-translated regions and internal ribosomal entry are conserved for both genomic RNAs of Blackcurrant reversion nepovirus. Virology 371:292–308 [DOI] [PubMed] [Google Scholar]
- 26. Kneller E. L. P., Rakotondrafara A. M., Miller W. A. 2006. Cap-independent translation of plant viral RNAs. Virus Res. 119:63–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Komar A. A., Hatzoglou M. 2005. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J. Biol. Chem. 280:23425–23428 [DOI] [PubMed] [Google Scholar]
- 28. Komarova A. V., Brocard M., Kean K. M. 2006. The case for mRNA 5′ and 3′ end cross talk during translation in a eukaryotic cell. Prog. Nucleic Acid Res. Mol. Biol. 81:331–367 [DOI] [PubMed] [Google Scholar]
- 29. Kozak M. 1979. Inability of circular messenger-RNA to attach to eukaryotic ribosomes. Nature 280:82–85 [DOI] [PubMed] [Google Scholar]
- 30. Kozak M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234:187–208 [DOI] [PubMed] [Google Scholar]
- 31. Levis C., Astiermanifacier S. 1993. The 5′ untranslated region of PVY RNA, even located in an internal position, enables initiation of translation. Virus Genes 7:367–379 [DOI] [PubMed] [Google Scholar]
- 32. Lourenco S., Costa F., Debarges B., Andrieu T., Cahour A. 2008. Hepatitis C virus internal ribosome entry site-mediated translation is stimulated by cis-acting RNA elements and trans-acting viral factors. FEBS J. 275:4179–4197 [DOI] [PubMed] [Google Scholar]
- 33. Macejak D. G., Sarnow P. 1991. Internal initiation of translation mediated by the 5′ leader of a cellular messenger-RNA. Nature 353:90–94 [DOI] [PubMed] [Google Scholar]
- 34. Martinez-Salas E., Pacheco A., Serrano P., Fernandez N. 2008. New insights into internal ribosome entry site elements relevant for viral gene expression. J. Gen. Virol. 89:611–626 [DOI] [PubMed] [Google Scholar]
- 35. McCormack J. C., et al. 2008. Structural domains within the 3′ untranslated region of Turnip crinkle virus. J. Virol. 82:8706–8720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Merrick W. C. 2004. Cap-dependent and cap-independent translation in eukaryotic systems. Gene 332:1–11 [DOI] [PubMed] [Google Scholar]
- 37. Meskauskas A., Petrov A. N., Dinman J. D. 2005. Identification of functionally important amino acids of ribosomal protein L3 by saturation mutagenesis. Mol. Cell. Biol. 25:10863–10874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miller W. A., Wang Z., Treder K. 2007. The amazing diversity of cap-independent translation elements in the 3′-untranslated regions of plant viral RNAs. Biochem. Soc. Trans. 35:1629–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nicholson B. L., White K. A. 2008. Context-influenced cap-independent translation of Tombusvirus mRNAs in vitro. Virology 380:203–212 [DOI] [PubMed] [Google Scholar]
- 40. Nicholson B. L., Wu B. D., Chevtchenko I., White K. A. 2010. Tombusvirus recruitment of host translational machinery via the 3′ UTR. RNA 16:1402–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Niepel M., Gallie D. R. 1999. Identification and characterization of the functional elements within the tobacco etch virus 5′ leader required for cap-independent translation. J. Virol. 73:9080–9088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pestova T. V., Kolupaeva V. G. 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 16:2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pestova T. V., et al. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 98:7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pestova T. V., Lorsch J. R., Hellen C. U. T. 2007. The mechanism of translation initiation in eukaryotes, p. 87–128 In Mathews M. B., Sonenberg N., Hershey J. W. B. (ed.), Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 45. Polacek C., Friebe P., Harris E. 2009. Poly(A)-binding protein binds to the non-polyadenylated 3′ untranslated region of dengue virus and modulates translation efficiency. J. Gen. Virol. 90:687–692 [DOI] [PubMed] [Google Scholar]
- 46. Qu F., Morris T. J. 2000. Cap-independent translational enhancement of turnip crinkle virus genomic and subgenomic RNAs. J. Virol. 74:1085–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rakotondrafara A. M., Polacek C., Harris E., Miller W. A. 2006. Oscillating kissing stem-loop interactions mediate 5′ scanning-dependent translation by a viral 3′-cap-independent translation element. RNA 12:1893–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ray S., et al. 2006. Tobacco etch virus mRNA preferentially binds wheat germ eukaryotic initiation factor (eIF) 4G rather than eIFiso4G. J. Biol. Chem. 281:35826–35834 [DOI] [PubMed] [Google Scholar]
- 49. Romero-Lopez C., Berzal-Herranz A. 2009. A long-range RNA-RNA interaction between the 5′ and 3′ ends of the HCV genome. RNA 15:1740–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sarnow P. 1989. Translation of glucose-regulated protein-78-immunoglobulin heavy-chain binding protein messenger-RNA is increased in poliovirus-infected cells at a time when cap-dependent translation of cellular messenger-RNAs is inhibited. Proc. Natl. Acad. Sci. U. S. A. 86:5795–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Serrano P., Pulido M. R., Saiz M., Martinez-Salas E. 2006. The 3′ end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA-RNA interactions with the 5′ end region. J. Gen. Virol. 87:3013–3022 [DOI] [PubMed] [Google Scholar]
- 52. Shen R. Z., Miller W. A. 2004. The 3′ untranslated region of tobacco necrosis virus RNA contains a barley yellow dwarf virus-like cap-independent translation element. J. Virol. 78:4655–4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shine J., Dalgarno L. 1975. Determinant of cistron specificity in bacterial ribosomes. Nature 254:34–38 [DOI] [PubMed] [Google Scholar]
- 54. Skulachev M. V., et al. 1999. Internal initiation of translation directed by the 5′-untranslated region of the tobamovirus subgenomic RNA I-2. Virology 263:139–154 [DOI] [PubMed] [Google Scholar]
- 55. Song Y. T., et al. 2006. The hepatitis C virus RNA 3′-untranslated region strongly enhances translation directed by the internal ribosome entry site. J. Virol. 80:11579–11588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Soukup G. A., Breaker R. R. 1999. Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5:1308–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stupina V. A., et al. 2008. The 3′ proximal translational enhancer of Turnip crinkle virus binds to 60S ribosomal subunits. RNA 14:2379–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sun X. P., Simon A. E. 2006. A cis-replication element functions in both orientations to enhance replication of Turnip crinkle virus. Virology 352:39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Svitkin Y. V., Sonenberg N. 2006. Translational control by the poly(A) binding protein: a check for mRNA integrity. Mol. Biol. 40:684–693 [PubMed] [Google Scholar]
- 60. Tanguay R. L., Gallie D. R. 1996. Isolation and characterization of the 102-kilodalton RNA-binding protein that binds to the 5′ and 3′ translational enhancers of tobacco mosaic virus RNA. J. Biol. Chem. 271:14316–14322 [DOI] [PubMed] [Google Scholar]
- 61. Terenin I. M., et al. 2005. A cross-kingdom internal ribosome entry site reveals a simplified mode of internal ribosome entry. Mol. Cell. Biol. 25:7879–7888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Treder K., et al. 2008. The 3′ cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA 14:134–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vera-Otarola J., et al. 2010. The 3′ untranslated region of the Andes hantavirus small mRNA functionally replaces the poly(A) tail and stimulates cap-dependent translation initiation from the viral mRNA. J. Virol. 84:10420–10424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wu B. D., White K. A. 1999. A primary determinant of cap-independent translation is located in the 3′-proximal region of the tomato bushy stunt virus genome. J. Virol. 73:8982–8988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xia X. H. 2007. The+4G site in Kozak consensus is not related to the efficiency of translation initiation. PLoS One 2:e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Xia X. H., Holcik M. 2009. Strong eukaryotic IRESs have weak secondary structure. PLoS One 4:e4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yuan X. F., Shi K. R., Meskauskas A., Simon A. E. 2009. The 3′ end of Turnip crinkle virus contains a highly interactive structure including a translational enhancer that is disrupted by binding to the RNA-dependent RNA polymerase. RNA 15:1849–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yuan X. F., Shi K. R., Young M. Y. L., Simon A. E. 2010. The terminal loop of a 3′ proximal hairpin plays a critical role in replication and the structure of the 3′ region of Turnip crinkle virus. Virology 402:271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zeenko V., Gallie D. R. 2005. Cap-independent translation of tobacco etch virus is conferred by an RNA pseudoknot in the 5′-leader. J. Biol. Chem. 280:26813–26824 [DOI] [PubMed] [Google Scholar]
- 70. Zhang J. C., Zhang G. H., Guo R., Shapiro B. A., Simon A. E. 2006. A pseudoknot in a preactive form of a viral RNA is part of a structural switch activating minus-strand synthesis. J. Virol. 80:9181–9191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zuo X. B., et al. 2010. Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3′ UTR of turnip crinkle virus. Proc. Natl. Acad. Sci. U. S. A. 107:1385–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]