Abstract
Interruption of suppressive highly active antiretroviral therapy (HAART) in HIV-infected patients leads to increased HIV replication and viral rebound in peripheral blood. Effects of therapy interruption on gut-associated lymphoid tissue (GALT) have not been well investigated. We evaluated longitudinal changes in viral replication and emergence of viral variants in the context of T cell homeostasis and gene expression in GALT of three HIV-positive patients who initiated HAART during primary HIV infection but opted to interrupt therapy thereafter. Longitudinal viral sequence analysis revealed that a stable proviral reservoir was established in GALT during primary HIV infection that persisted through early HAART and post-therapy interruption. Proviral variants in GALT and peripheral blood mononuclear cells (PBMCs) displayed low levels of genomic diversity at all times. A rapid increase in viral loads with a modest decline of CD4+ T cells in peripheral blood was observed, while gut mucosal CD4+ T cell loss was severe following HAART interruption. This was accompanied by increased mucosal gene expression regulating interferon (IFN)-mediated antiviral responses and immune activation, a profile similar to those found in HAART-naive HIV-infected patients. Sequence analysis of rebound virus suggested that GALT was not the major contributor to the postinterruption plasma viremia nor were GALT HIV reservoirs rapidly replaced by HIV rebound variants. Our data suggest an early establishment and persistence of viral reservoirs in GALT with minimal diversity. Early detection of and therapy for HIV infection may be beneficial in controlling viral evolution and limiting establishment of diverse viral reservoirs in the mucosal compartment.
INTRODUCTION
Highly active antiretroviral therapy (HAART) is effective at suppressing HIV replication and restoring CD4+ T cell numbers in peripheral blood and has contributed to a significant decrease in morbidity and mortality of HIV-infected patients (18, 30). However, HAART has failed to eradicate viral reservoirs that have been established early in infection, leading to viral persistence and incomplete immune restoration (5, 12, 14). Interruption of HAART results in rapid viral resurgence and CD4+ T cell loss in peripheral blood (25).
Studies involving structured treatment interruption (STI) have suggested that interruption might be beneficial by allowing the development of HIV-specific responses to control viral replication and to alleviate drug toxicities (38, 44). However, interruption consistently led to viral rebound and to CD4+ T cell depletion without enhancing antiviral responses (34). Additionally, concerns have been raised about the development of drug-resistant viruses (42).
The gut-associated lymphoid tissue (GALT) harbors the majority of the body's lymphoid tissue and is an important site for host-pathogen interactions during HIV infection (6).
Our previous studies demonstrated that primary HIV infection leads to severe CD4+ T cell depletion and disruption of the gut microenvironment (16, 18). Also, CD4+ T cell restoration in GALT during therapy was shown to be modest and delayed compared to that in peripheral blood (19). Currently, no information is available on the effects of interruption of HAART on GALT.
In the present study, we evaluated longitudinal changes in HIV replication, viral evolution, and compartmentalization between GALT and peripheral blood mononuclear cell (PBMC) compartments in relation to T cell homeostasis and mucosal gene expression for three HIV-infected patients for whom HAART was initiated during primary infection. These patients opted to interrupt HAART after several months, presenting an opportunity to assess the effects of HAART interruption in GALT and to investigate the origin of rebounding virus.
MATERIALS AND METHODS
Patient characteristics and sample collection.
Patients 116, 134, and 140 initiated HAART at 6, 4, and 5 weeks postinfection, respectively (Table 1). These patients were considered at high risk of HIV infection and were being routinely monitored for HIV seropositivity. Their stage of primary HIV infection was identified due to the detection of the plasma HIV RNA load (branched-DNA assay) while they were seronegative for HIV antibody (enzyme-linked immunosorbent assay [ELISA] and Western blotting). Jejunal biopsy specimens (by upper endoscopy) and peripheral blood samples were collected prior to the initiation of HAART and during HAART (Fig. 1). The CD4+ and CD8+ T cell numbers in peripheral blood were determined during their routine clinical visits. Peripheral blood samples were also collected after the interruption of HAART from patients 116 (1 year and 6 years postinterruption), 134 (1 year and 5 years postinterruption), and 140 (9 months postinterruption). Peripheral blood CD4+ T cell numbers and HIV copy numbers in plasma (branched-DNA assay) and jejunal biopsy specimens (real-time PCR) were determined as previously described (32, 40). The sensitivity of these assays is 50 copies/ml in plasma and 50 copies/μg gut tissue RNA. Samples were collected as per the Institutional Review Board-approved protocol and processed as previously described (19, 32).
Table 1.
Clinical characteristics of HIV-infected patients
| Patient | Sexa | Age (yr) | HAART regimen |
|---|---|---|---|
| 116 | M | 35 | Abacavir, lamivudine, nelfinavir |
| 134 | M | 35 | Abacavir, lamivudine, lopinavir/ritonavir, zidovudine, |
| 140 | M | 35 | Abacavir, lamivudine, lopinavir/ritonavir, zidovudine |
M, male.
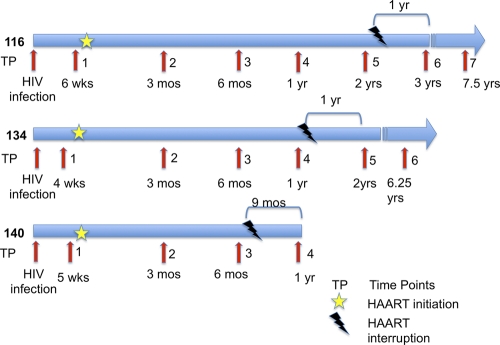
Fig. 1.
Clinical course and therapy schedule of HIV-infected patients. Patients 116, 134, and 140 initiated therapy during primary HIV infection and discontinued thereafter. Time points for the sample collection of peripheral blood and gut biopsy specimens during HIV infection are shown. Time since putative date of HIV infection is indicated under each time point arrow in weeks (wks), months (mos) (17), and years (yrs). Stars indicate when patients initiated HAART, and bolts indicate when patients interrupted HAART. The time at the top of each time line indicates how much time elapsed from therapy interruption to the next time point when samples were collected.
CD4+ T cell numbers in peripheral blood and GALT and PBMC.
Immunophenotypic analysis of T-lymphocyte subsets from PBMC and GALT was performed as previously described (18, 33, 43). Briefly, isolated cells were incubated with T cell-specific mouse anti-human monoclonal antibodies (30 min at 4°C). After incubation, cells were washed and fixed with 1% paraformaldehyde and analyzed by four-color flow cytometry (FACScalibur; Becton Dickinson). A minimum of 100,000 events was collected for each sample, and data were analyzed using the FlowJo software program (Tree Star Inc., San Carlos, CA).
RNA isolation and DNA microarray analysis.
Total RNA was extracted utilizing the Qiagen RNeasy RNA isolation kit (Qiagen, Valencia, CA). mRNA amplification, labeling, hybridization to human GeneChips U133 (Affymetrix, Santa Clara, CA), staining, and scanning were performed according to the Affymetrix gene expression analysis technical manual at the Microarray Core Facility, University of California, Davis (39).
Microarray data were analyzed using RMA-based (GeneSpring, GX; Agilent Technologies, Santa Clara, CA) algorithms. A minimum fold change of 50% (P value ≤ 0.05) was used as a cutoff criterion. Gene expression profiles in GALT of the patients following interruption of HAART (n = 3) were compared to the mucosal profiles from HIV-negative healthy controls (n = 3). Although the sample numbers were relatively small in each group, the P-value criteria chosen ensured that all reported changes were consistent within the group and that the mean fold change was not unduly influenced by large changes in any one individual. To prevent potential sampling bias within each patient, two or more biopsy samples were collected from different sites along the jejunum and pooled. Pathways and processes that were statistically enriched in genes meeting fold-change and statistical criteria were identified by Fisher's exact test (P value ≤ 0.05) in utilizing the Database for Annotation, Visualization and Integrated Discovery (DAVID) (7, 23). The changes in gut mucosal gene expression resulting from treatment interruption were further evaluated by comparing them to changes observed in previously published studies of acute- and chronic-stage HIV patients and long-term nonprogressors (LTNP) (39, 40).
PCR amplification and sequencing.
DNA was isolated from jejunum and PBMC samples using the Qiagen DNeasy blood and tissue Kit (Qiagen, Valencia, CA). Envelope's V1-C4 region was amplified using nested PCR (outer and inner primers, respectively: HIV01F [5′TATGGGGTACCTGTGTGGAAG] and HIV01R [5′TGTGAGTTGCAACAGATGCTGT] and HIV03F [5′AAAGCCATGTGTAAAATTAACC3′] and HIV03R [′5′ATGGGAGGGGGCATACATTGC3′]). PCRs were carried out in quadruplicate as previously described (26). Each 50-μl reaction mixture contained 5 μl 10× buffer (Fisher Scientific), 4 μl 2.5 mM deoxynucleoside triphosphates (dNTPs), 100 pmol respective primers, 0.5 μl of proofreading enzyme Taq polymerase, and 5 μl template. PCR products were resolved in ethidium bromide 1% agarose gels, pooled, and cloned using the pCR4-Topo TA cloning vector (Invitrogen, Carlsbad, CA). Ten to thirty clones from each sample were sequenced using Prism Dye terminator kits (ABI, Foster City, CA) at the Viral and Rickettsial Disease Laboratory of the California Department of Health Services Core, Richmond, CA (VRDL) and Davis Sequencing (Davis, CA).
RNA was extracted from plasma samples using the QIAamp viral RNA minikit and reverse transcribed using the SuperScript III First-Strand synthesis system with random hexamer primers (Invitrogen, Carlsbad, CA), followed by nested PCR using primers encompassing the V3 region (20). PCRs were carried out and processed as described for DNA. Only pretherapy and postinterruption time points were analyzed for the study. Each sample was processed on different days to avoid contamination. Even though reaction mixtures were pooled to encompass the diversity of HIV variants, the area being amplified was not large and was unlikely to give rise to a significant amount of PCR recombination. Limiting dilution to approximately one copy was performed on postinterruption plasma samples from patient 116 (based on real-time PCR viral loads) and yielded very similar sequences, indicating that PCR recombination might not have been a problem. Using a very similar molecular cloning approach for a comparable region of envelope, others found PCR misincorporation artifacts to be minimal (27). Furthermore, Jordan et al. found no difference in population structure or genetic diversity between terminal dilution cloning and conventional cloning for 14 of 17 subjects studied, demonstrating both methods are suitable for the study of viral genomic diversity (28).
Genetic compartmentalization, diversity, and divergence.
We used a nonparametric statistical test within the DnaSPv5 software package (31) for detecting genetic differentiation of subpopulations originally proposed by Hudson, Boos, and Kaplan to study population subdivision (24). The method computes all pairwise differences between groups and determines if within-group genetic distances are smaller than between-group distances using 1,000 permutations to test for statistical significance. This method has been used to analyze HIV-1 population dynamics (1, 37). The average distances within population 1 and within population 2 are computed. The distance metric is log (I + Dij), where D is the simple distance. The weighted average of these two is Ks*. Next, the sequences in the distance matrix are randomly sampled, without replacement, to create simulated subpopulations 1 and 2. A Ks* simulated is computed and compared to the observed Ks*. The fraction of times that Ks* simulated is less than or equal to Ks* observed is recorded. The null hypothesis that two viral populations (in this case through pairwise comparisons between GALT and peripheral blood) were genetically similar (i.e., constituted the same population) would be rejected if the P value of the Ks* statistic was <0.05, suggesting the presence of population subdivision and restricted gene flow (compartmentalization). If the P value for the Ks* statistic was >0.05, the null hypothesis that two viral populations were not genetically different was not rejected, thus suggesting the absence of population subdivision or compartmentalization. DnaSPv5 was also utilized to compute diversity and divergence values for the various viral populations, and paired t tests using the Sigma Plot software program (Systat Software, Inc., San Jose, CA) were performed to determine statistical significance.
Phylogenetic analysis.
Multiple sequence alignments were generated from viral envelope sequences using the BioEdit sequence alignment editor (22) with default gap parameters. Three maximum-likelihood trees describing each patient's data sets were built, and P values were generated by implementing the program Dnaml from the Phylip 3.68 software package, which estimates phylogenies from nucleotide sequences by maximum likelihood using the F84 model of nucleotide substitution, which allows the expected frequencies of the four bases to be unequal, allows the expected frequencies of transitions and transversions to be unequal, and has several ways of allowing different rates of evolution at different sites (10). Clonal duplicates from each individual sample were removed from the phylogenetic analysis but were maintained for calculating diversity and divergence values.
Microarray data accession number.
The data discussed in this paper have been deposited in NCBI's Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE28177 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE28177).
RESULTS
Increased viral replication and CD4+ T cell depletion in peripheral blood following HAART interruption.
To characterize alterations in T cell homeostasis that were a consequence of HAART interruption, we evaluated PBMC and jejunal tissue samples from three HIV-infected patients who initiated HAART during primary HIV infection and opted to interrupt treatment later due to personal reasons. All three patients displayed a striking increase in plasma viremia following HAART interruption (Fig. 2A). However, all three patients maintained CD4+ T cell counts within the range of 500 to 1,500 cells/mm3 for at least 2 years after the cessation of therapy.
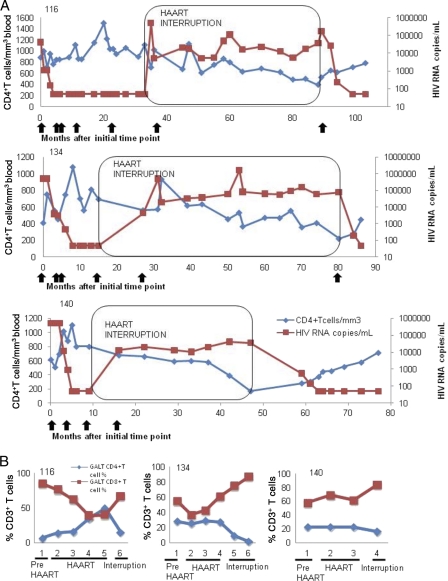
Fig. 2.
Measurement of HIV loads and CD4+ T cell numbers in longitudinal samples of peripheral blood and GALT of HIV-infected patients prior to and post-HAART interruption. (A) CD4+ T cell numbers (blue) and HIV RNA copies/ml(red) are plotted across time (in months). Arrows underneath each panel represents the time of GALT sample collection from each patient and represent time points as they are indicated in Fig. 1. The boxed area indicates the time period of therapy interruption. (B) CD4 (blue) and CD8 (red) T-cell percentages from GALT.
HAART interruption leads to severe loss of gut mucosal CD4+ T cells.
We evaluated T cell subsets in GALT prior to and following the interruption of HAART. HIV viral RNA was undetectable during the course of HAART in 2 of 3 patients, with patient 140 displaying transient viral replication in GALT. Following therapy interruption, 1 of 3 patients had an increase in viral load at different time points. In contrast to findings for peripheral blood, a severe depletion of CD4+ T cells was observed in GALT following HAART interruption. The loss in mucosal CD4+ T cells coincided with an increase in CD8+ T cells (Fig. 2B). The CD4+ T cell loss was particularly striking for patient 116, who had restored gut mucosal CD4+ T cells during HAART from 7% pre-HAART levels to normal levels (∼50%) during HAART. Patients 134 and 140 maintained GALT CD4+ T cell levels at 28% and 23%, respectively, during HAART, but preservation of CD4+ T cells was rapidly lost following therapy interruption. The magnitude of CD4+ T cell loss in GALT suggested that the gut mucosal immune system was highly susceptible to HIV infection when therapeutic control of viral replication was removed.
Gut mucosal responses to HAART interruption.
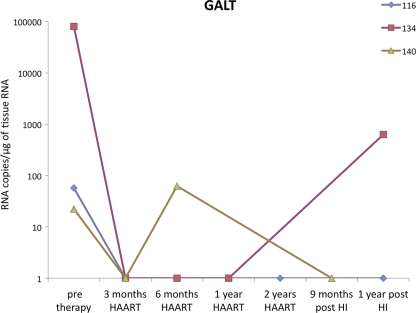
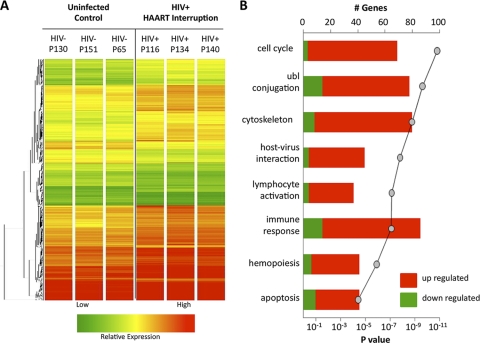
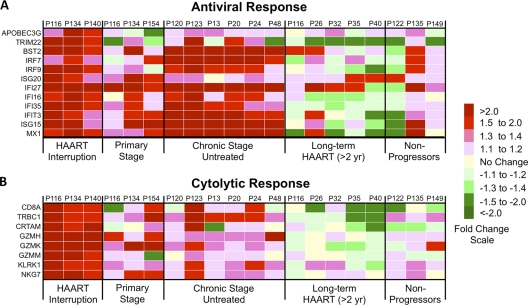
Interestingly, although the viral burden increased substantially in the peripheral blood of all patients after therapy interruption, replicating virus was not detected in GALT in 2 of the 3 patients at the time biopsy specimens were collected and was present at low levels (629 HIV copies/μg total RNA) in the remaining patient (Fig. 3). To evaluate the mechanisms of host response to viral rebound in GALT following HAART interruption, we determined gene expression profiles in the jejunal mucosa by DNA microarray analysis. Mean levels of gene transcription between the 3 HIV-infected patients with HAART interruption and 3 healthy HIV-negative controls were compared. Hierarchical clustering identified similar patterns of expression among the 912 upregulated and 346 downregulated gene transcripts (Fig. 4A). Several functional gene categories were found to be statistically (Fisher's exact score) enriched within the data set, including cell cycle regulation, ubiquitin ligase conjugation, cytoskeletal maintenance, host-virus interaction, lymphocyte activation, immune response, hemopoiesis, and apoptosis (Fig. 4B).
Fig. 3.
Measurement of HIV RNA loads in longitudinal gut biopsy specimens from HIV-infected patients. Viral RNA loads in GALT were determined by reverse transcription-PCR (RT-PCR). Time points of sample collection are shown. HI, HAART interruption.
Fig. 4.
HAART interruption leads to alterations in intestinal mucosal gene expression. (A) Hierarchical clustering of genes whose expression was altered in the intestinal mucosa of 3 patients following HAART interruption as compared to baseline gene expression in 3 healthy HIV-negative controls. (B) Pathway analysis of genes up- or downregulated in GALT following HAART interruption indicated a statistical enrichment for genes involved in the cell cycle, ubiquitin ligase conjugation, cytoskeletal activity, host virus interaction, lymphocyte activation, immune response, hemopoiesis, and apoptosis.
To characterize the relationship between the host response to HAART interruption and rebound viremia, we focused the downstream analysis of the DNA microarray data on evaluation of genes associated with immune processes. Mucosal antiviral responses appeared to be largely type 1 IFN driven (Fig. 5A) and included factors that inhibit virion budding (tetherin/BST2) and infectivity (APOBEC3G). Regulation of genes in the molecular signatures associated with HAART interruption was then evaluated in gut mucosal gene expression profiles during the acute and chronic stages of HIV infection in HAART-naive patients, HIV-infected patients with long-term HAART administration (>2 years), and long-term HIV-infected nonprogressors (LTNP). This comparative analysis indicated that the HAART interruption expression profile was most consistent with profiles of HAART-naive chronically HIV-infected patients.
Fig. 5.
Mucosal immune response-associated gene expression following HAART interruption. Fold changes in the expression of genes involved in antiviral responses (A) or cytolytic activity (B) in the intestinal mucosa following HAART interruption are shown in heat maps. The transcription profile of genes during HAART interruption was compared to the profiles for HAART-naive patients with acute or chronic HIV infection, patients with long-term HAART administration (>2 years), and long-term HIV-infected nonprogressors.
Biomarkers of CD8+ T cell cytolytic responses were also upregulated, including CD8 molecules, T cell receptors, and granzymes H, K, and M (Fig. 5B). These findings were also supported by the flow cytometric data showing increases in gut mucosal CD8+ T cell percentages (Fig. 2B). In contrast to antiviral responses, the signature of upregulated cytolytic factors was observed only following HAART interruption and not in other patient groups. Changes in the expression of genes in GALT that mediate antigen presentation, complement activity, and trafficking were detected (presented in Fig. S1 in the supplemental material).
Collectively, transcriptional signatures of the HAART interruption appear to reflect a robust ongoing gut mucosal response that might have contributed to the dampening of rebounding viremia and inhibited seeding from this compartment. However, because gut biopsy specimens were collected several months following HAART interruption, it was not possible to determine if the transcriptional signatures were also established immediately after cessation of therapy.
Low proviral diversity and divergence in gut mucosa.
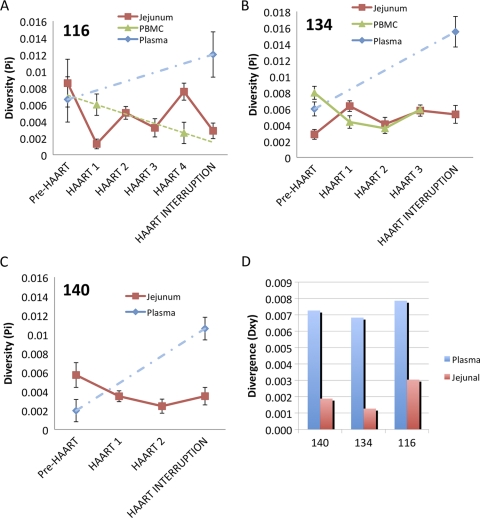
Proviral sequences from GALT and PBMC demonstrated minimal diversity (Fig. 6A to C) and viral divergence (Fig. 6D) in all patients during primary HIV infection. The viral diversity remained unchanged during HAART and at 8 to 12 months after therapy interruption. The HIV population from plasma during primary HIV infection also showed a low diversity that was comparable to low diversity values seen among proviral sequences from GALT and PBMC samples (paired t test P value = 0.719). However, plasma viral populations demonstrated a significant increase in genomic diversity (paired t test P value = 0.025) following therapy interruption compared to that of plasma viral populations from primary HIV infection prior to HAART initiation. These observations suggest that increased viral replication following therapy interruption might have supported viral divergence. However, proviral sequences in longitudinal gut mucosal samples showed a low level of diversity. Proviral sequences in GALT also demonstrated a lack of divergence from the early founder HIV sequences throughout the course of study. These findings support our observations of dampened viremia in GALT compared to the peripheral blood compartment after cessation of therapy and suggest that the gut mucosal viral reservoir may not be a significant source for rebounding systemic viremia following therapy interruption.
Fig. 6.
HIV diversity (Pi) and divergence (Dxy) in GALT and peripheral blood prior to and during HAART and at post-HAART interruption. (A to C) HIV diversity plots for patients 116, 134, and 140, respectively. Each color (see key) represents diversity values of HIV population from GALT, PBMC, and plasma compartments during the course of HIV infection. Dashed lines indicate trends from available data. (D) Divergence histogram comparing HIV sequence divergence in plasma and GALT at a pre-HAART time point compared to a post-interruption of HAART time point. Measurements represent the average number of nucleotide substitutions per site between different populations. Bars indicate standard errors.
Compartmentalization of rebound plasma HIV quasispecies following HAART interruption.
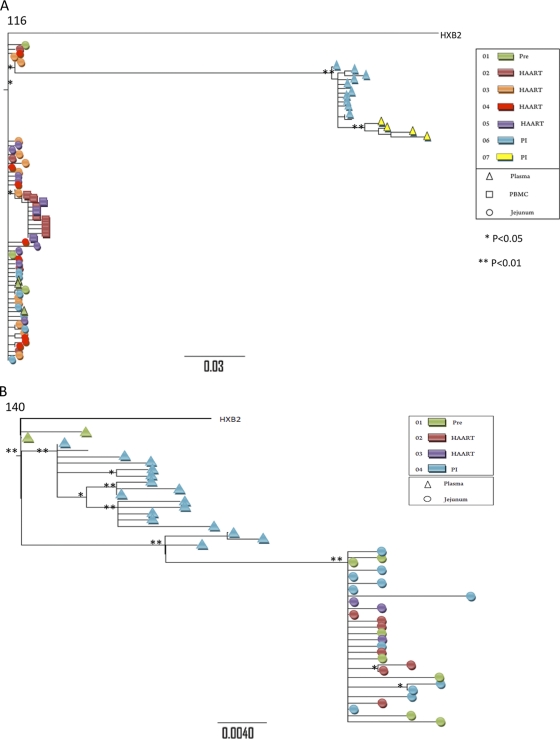
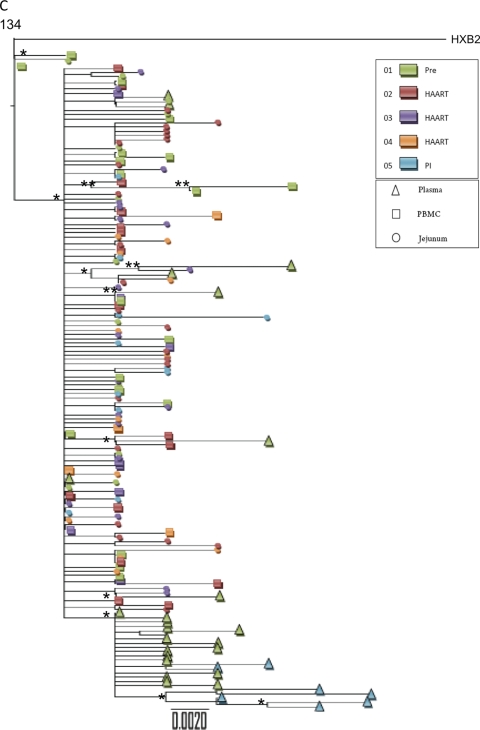
To compare the extent of gene flow between the gut mucosal and peripheral blood compartments prior to and following HAART interruption, we analyzed proviral envelope sequences from GALT and PBMC and viral sequences from plasma samples. Viral phylogenies illustrated that HIV quasispecies from plasma samples post-HAART interruption were compartmentalized from proviral sequences in GALT or PBMC (PBMC samples of patient 140 were unavailable) (Table 2 and Fig. 7). The order of sequences in phylogenetic trees does not indicate the chronology of events. Trees illustrate genetic distances of sequences from each other; therefore, in Fig. 7B, for example, plasma sequences on the left do not signify their ancestry to GALT provirus sequences on the right. These data provide further evidence that GALT might not have contributed to the rebounding plasma viremia in these patients or that the rebound virus arises from a minor viral population not sampled by the approach used here. In contrast, plasma viral sequences in patient 116 during primary HIV infection intermingled with sequences from the GALT and PBMC compartments, and gene flow analysis was consistent with panmictic viral populations, suggesting that GALT and peripheral blood compartments contributed to plasma viremia during primary HIV infection for this patient (Table 2 and Fig. 7). Thus, during primary HIV infection, GALT and PBMCs may contribute to plasma viral divergence, as suggested by the intermingling of viral sequences. However, following HAART interruption, resurging plasma viral loads may not result from seeding by those compartments.
Table 2.
P values from HIV gene flow analysis between GALT, PBMC, and plasma compartments based on envelope sequences from HIV-infected patients
| Sample type | Patient no. | Sample (nb) | P value for sequence comparison with indicated samplea (nb) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| JD 01 (3) | JD 02 (4) | JD 03 (14) | JD 04 (6) | JD 05 (8) | JD 06 (7) | BD 02 (8) | BD 05 (3) | ||||
| Plasma | 116 | PR 01 (3) | 1 NS | 0.2740 NS | 0.2040 NS | 0.5650 NS | 0.5050 NS | 0.4040 NS | 0.4500 NS | 0.9030 NS | |
| PR 06 (PIc) (13) | 0.001** | 0.001** | 0*** | 0*** | 0*** | 0*** | 0*** | 0.002** | |||
| PR 07 (PI) (4) | 0.029* | 0.026* | 0*** | 0.0020** | 0.0040** | 0.0020** | 0.0020** | 0.0220* | |||
| JD 01 (12) | JD 02 (21) | JD 03 (14) | JD 04 (18) | JD 05 (11) | BD 01 (19) | BD 02 (17) | BD 03 (8) | BD 04 (4) | |||
| 134 | PR 01 (23) | 0*** | 0*** | 0*** | 0*** | 0*** | 0*** | 0*** | 0*** | 0.0170* | |
| PR 05 (PI) (8) | 0*** | 0*** | 0*** | 0*** | 0*** | 0*** | 0*** | 0*** | 0.0130* | ||
| JD 01 (3) | JD 02 (4) | JD 03 (14) | JD 04 (6) | ||||||||
| 140 | PR 01 (2) | 0.0210* | 0.0070** | 0.1120 | 0.0110* | ||||||
| PR 04 (PI) (17) | 0*** | 0*** | 0*** | 0*** | |||||||
| JD 01 (3) | JD 02 (4) | JD 03 (14) | JD 04 (6) | JD 05 (8) | JD 06 (7) | ||||||
| PBMCs | 116 | BD 02 (8) | 0.0210* | 0.0020* | 0.00*** | 0.00*** | 0.00*** | 0.00*** | |||
| BD 05 (3) | 0.0610 NS | 0.0290* | 0.0030** | 0.0120* | 0.0300* | 0.0100* | |||||
| JD 01 (12) | JD 02 (21) | JD 03 (14) | JD 04 (18) | JD 05 (11) | |||||||
| 134 | BD 01 (19) | 0.6070 NS | 0.0090** | 0.3050 NS | 0.6710 NS | 0.6720 NS | |||||
| BD 02 (17) | 0.6830 NS | 0.0140* | 0.5340 NS | 0.9020 NS | 0.4700 NS | ||||||
| BD 03 (8) | 0.8370 NS | 0.1670 NS | 0.5640 NS | 0.7220 NS | 0.4640 NS | ||||||
| BD 04 (4) | 0.5790 NS | 0.8240 NS | 0.7130 NS | 0.6800 NS | 0.5300 NS | ||||||
Numbers represent P values from the gene flow analysis from the DnaSp software package using 1,000 permutations. Probability was obtained by the permutation test with 1,000 replicates. NS, not significant; *, 0.01 < P < 0.05; **, 0.001 < P < 0.01; ***, P < 0.001. JD, jejunal proviral sequences; BD, PBMC proviral sequences; PR, plasma viral sequences. Numbers following tissue type indicate time points as shown in Fig. 1.
n, total number of clonal sequences in each category.
PI, post-therapy interruption time point.
Fig. 7.
Phylogenetic analysis of V3 region of HIV envelope gene from peripheral blood and GALT from HIV-infected patients. HIV sequences derived from plasma (triangle), PBMC (square), or GALT (circle) from HIV-infected patients (see key for colors representing different time points) were analyzed to construct phylogenetic trees. “PI” indicates post-therapy interruption time points. Trees were constructed using the maximum-likelihood approach by running dnaml from the Phylip 3.68 package. This approach also generates P values for each node in the tree to assess significance, marked below by the use of “*” for P values between 0.01 and 0.05 and “**” for P values below 0.05. Samples from patients 116 (A), 140 (B), and 134 (C) are analyzed.
DISCUSSION
Although HAART leads to effective suppression of plasma viral loads and restoration of CD4+ T cells in peripheral blood in HIV-infected patients, it fails to eradicate established viral reservoirs (21). Viral persistence is attributed to viral latency in cellular reservoirs, ongoing replication, or poor drug penetration leading to viral sanctuaries (4, 11, 48). These viral reservoirs represent a major obstacle in the eradication of HIV-1. Furthermore, the benefits gained through HAART are rapidly lost following the interruption of therapy. A rapid rebound of the viral burden is seen in peripheral blood along with the loss of CD4+ T cells (5, 9, 13). Other studies have demonstrated a failure to lower the virological “set point” or a failure to boost or preserve durable HIV-specific immunity following treatment interruption (34, 36, 38). Our results are in support of previously reported findings that despite increased levels of viral replication, patients initiating HAART during primary HIV infection (PHI) maintained normal levels of CD4+ T cells in the peripheral blood for 1 year following HAART interruption.
Most of the previously reported studies, however, focus on viral sequences derived from plasma or peripheral blood mononuclear cells, which do not accurately describe the full scope of HIV infection in the tissue compartments. Besides the rapid and profound depletion of CD4+ T cells in GALT, van Marle et al. demonstrated that distinct HIV-1 quasispecies populate several parts of the gut (45). Furthermore, Yukl et al. found that the levels of T cell restoration and activation in HIV-infected patients differ between different parts of the gastrointestinal tract (47). It is still not resolved whether the GALT is a major contributor of viral diversity due to residual viral replication during HAART and whether it is a major source of resurging virus following treatment interruption. In this study, we evaluated longitudinal immunologic and virologic changes in the gut mucosa caused by HAART interruption and analyzed HIV sequences to determine the contribution of GALT reservoirs to systemically rebounding virus. The PBMC and GALT samples from three HIV-infected patients were collected over a span of several years, starting with primary HIV infection and continuing during HAART and following the interruption of HAART.
Longitudinal analysis of proviral sequences from both GALT and PBMC at multiple time points for each patient showed no temporal structure and uniformly low genomic diversity. It is possible that this stable pool of viral sequences may originate with infection of resting CD4+ T cells established during primary HIV infection. Previous reports of the analysis of viral quasispecies during primary acute HIV infection revealed a homogeneous population of early founder viruses, with the majority of infections being initiated by a few transmitted viral variants (29). These findings are in agreement with our results of a low-diversity viral population in GALT during primary HIV infection. Furthermore, our findings suggest that a substantial proportion of the HIV provirus harbored in CD4+ T cells during HAART and following therapy interruption may be derived from the original pool of viruses established during primary HIV infection.
In that regard, our study also highlights the potential importance of early initiation of therapy in controlling viral diversification. Previous studies have shown that HIV-induced immunopathogenesis is initiated early in HIV infection. Therefore, initiation of HAART during early HIV infection has the potential to revert or limit mucosal pathogenesis to some extent while preserving HIV-specific CD8+ T cell functions (15, 35). The clinical picture of patients who started HAART early and achieved effective viral suppression and CD4+ T cell restoration resembles the clinical outcomes in LTNPs (40). Both groups of patients maintained or restored CD4+ T cells in GALT and had increased expression of genes regulating gut mucosal regeneration and repair, as well as suppression of inflammation-related gene expression (40). Previous studies by Guadalupe et al. demonstrated that expression of genes involved in gut mucosal growth and repair was increased in patients with efficient mucosal CD4+ T-cell restoration during long-term HAART (19). In the simian immunodeficiency virus (SIV)-infected rhesus macaque model of AIDS, initiation of antiretroviral therapy (ART) during primary SIV infection led to a near-complete restoration of gut mucosal CD4+ T cells and the appearance of virus-specific polyfunctional CD8+ T cell responses (46). Moreover, early treatment of acute HIV infection has been shown to elicit narrower cytotoxic T lymphocyte (CTL) responses, stronger T-helper-cell responses, and a less-diverse virus population in peripheral blood (2). Our findings provide evidence that in HIV-infected patients initiating early HAART, low proviral diversity is maintained in both GALT and PBMC for several years, even after the interruption of HAART. Considering that viral escape mutations also lead to increased diversification, initiation of therapy in earlier stages of HIV infection may limit the ability of the virus to escape host immune response.
In agreement with previous studies (13, 36), we show that therapy interruption in HIV-infected patients resulted in a rebound of plasma viremia within 2 weeks. It is also known that HIV persistence, despite the presence of therapy, might be attributed to latent provirus in CD4+ T cells (11, 12, 21). Resurging virus after therapy interruption was dissimilar from both cell-associated HIV RNA and virus found in resting CD4+ T cells, suggesting that other compartments may have contributed to plasma viremia (5, 9). Studies of HIV evolution and viral reservoirs have shown HIV sequence compartmentalization in the central nervous system, genital tracts, parts of the gastrointestinal tract, and other tissues (3, 8, 37, 41, 45). It has also been proposed that rebounding virus comes from reactivation of long-lived latently HIV-infected cells rather than from an expansion of populations of low-level replicating virus (26). Zhang et al. reported that plasma viral rebound after therapy interruption could have originated from the latent viral reservoir in PBMC, though viral rebound in patients with incomplete viral suppression was genetically different (48). We observed that HIV sequences in plasma post-HAART interruption are genetically different from proviral sequences from GALT, suggesting that GALT is also unlikely to be the main source of rebounding virus. Previous studies have shown that various parts of the gastrointestinal tract may have various levels of viral replication and different viral variants (45, 47). Therefore, we cannot exclude the possibility that other parts of the gastrointestinal tract might be responsible for plasma viremia at post-therapy interruption. Due to the unavailability of PBMC samples at post-HAART interruption, the contribution of the PBMC to plasma viremia could not be simultaneously assessed in this study. We recognize that there is a possibility of sampling error in which an unsampled GALT cell population might be contributing to plasma viremia; however, this limitation is common to all studies of this type. The inability to show similarities between viral populations in GALT and rebounding plasma virus despite the profound depletion of GALT CD4+ T cells during interruption would also suggest that CD4+ T cell depletion in this situation is largely the result of immune-based pathogenetic events or is the result of productive infection leading to rapid T cell killing with little reseeding of cellular reservoirs. While it remains possible that a very small population of infected cells in GALT not sampled in these studies was responsible for rebounding plasma virus, based on the number of postinterruption viral sequences that were analyzed, it seems unlikely that GALT was the main contributor to plasma viremia. To exclude artifacts due to bulk PCR amplification and cloning, single-genome amplification (SGA) was performed on one of our samples for comparison to the conventional molecular cloning method, and sequences were found to be very similar or equal by both methods. Furthermore, Jordan et al. (28) performed a comparative study between standard PCR/cloning and SGA for their ability to reproduce intrapatient polymorphism of HIV-1 populations and found no significant differences.
In GALT samples collected several months after cessation of therapy, a robust increase in expression of antiviral machinery and CTL-associated molecules coincided with little or no detection of HIV RNA. While the discordance in rebound viremia between the PBMC and GALT compartments may be explained in part by renewed reductions in CD4+ T cells serving as HIV targets, gene expression profiling suggests that a vigorous mucosal response may also play a role. Previous studies examining gene expression responses in GALT to acute-stage (16) and chronic-stage (40) HIV infection in HAART-naive patients have not found the broad increases in both innate (IFN-driven) and adaptive (CTL-driven) immune responses that were observed in patients following HAART interruption. The nature of the host response combined with the lack of GALT-specific representation in the circulating HIV pool suggests that a large initial burst in virus replication with the capacity to seed systemic viremia may not have occurred in that compartment. However, this interpretation should be taken with caution due to the small number of patients analyzed, the inherent cellular heterogeneity and variation of endoscopic biopsy samples, and the variation in time intervals (6 to 12 months) between cessation of HAART and sample collection.
Additional studies, perhaps best designed with the SIV model, may be necessary to definitively address questions regarding the kinetics of rebound viremia in various compartments. Further studies are needed to delineate whether latently infected cells in GALT harbor replication-competent virus and whether such cells are inherently stable or are the product of amplification by either cell division or low-level HIV replication not reflected in the plasma virus. Given the inability of most HIV-infected patients to spontaneously control viral rebound and halt disease progression, patients are likely to require lifelong antiretroviral therapy until the perplexities of viral reservoirs are clarified. Our findings emphasize the need to evaluate both mucosal and peripheral blood compartments for changes in immune parameters and viral loads during HAART and/or during HAART interruption and suggest that starting therapy early might be important in impeding virus evolution.
Supplementary Material
ACKNOWLEDGMENTS
We thank the patients for their participation in the study and the clinic staff for their help. We also thank Xylona Bibal and Jimmy Pan for their technical assistance.
This study was supported by grants from the National Institutes of Health (DK61297 and AI43274).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 23 February 2011.
REFERENCES
- 1. Achaz G., et al. 2004. A robust measure of HIV-1 population turnover within chronically infected individuals. Mol. Biol. Evol. 21:1902–1912 [DOI] [PubMed] [Google Scholar]
- 2. Altfeld M., et al. 2001. Cellular immune responses and viral diversity in individuals treated during acute and early HIV-1 infection. J. Exp. Med. 193:169–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bull M., et al. 2009. Compartmentalization of HIV-1 within the female genital tract is due to monotypic and low-diversity variants not distinct viral populations. PLoS One 4:e7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chun T. W., et al. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183–188 [DOI] [PubMed] [Google Scholar]
- 5. Chun T. W., et al. 2000. Relationship between pre-existing viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat. Med. 6:757–761 [DOI] [PubMed] [Google Scholar]
- 6. Dandekar S. 2007. Pathogenesis of HIV in the gastrointestinal tract. Curr. HIV/AIDS Rep. 4:10–15 [DOI] [PubMed] [Google Scholar]
- 7. Dennis G., Jr., et al. 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4:P3. [PubMed] [Google Scholar]
- 8. Diem K., et al. 2008. Male genital tract compartmentalization of human immunodeficiency virus type 1 (HIV). AIDS Res. Hum. Retroviruses 24:561–571 [DOI] [PubMed] [Google Scholar]
- 9. Dybul M., et al. 2003. Genetic characterization of rebounding human immunodeficiency virus type 1 in plasma during multiple interruptions of highly active antiretroviral therapy. J. Virol. 77:3229–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Felsenstein J. 2004. PHYLIP (Phylogeny Inference Package) version 3.6. [Google Scholar]
- 11. Finzi D., et al. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512–517 [DOI] [PubMed] [Google Scholar]
- 12. Finzi D., et al. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300 [DOI] [PubMed] [Google Scholar]
- 13. Fischer M., et al. 2003. HIV RNA in plasma rebounds within days during structured treatment interruptions. AIDS 17:195–199 [DOI] [PubMed] [Google Scholar]
- 14. Furtado M. R., et al. 1999. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N. Engl. J. Med. 340:1614–1622 [DOI] [PubMed] [Google Scholar]
- 15. Ganesan A., et al. 2010. Immunologic and virologic events in early HIV infection predict subsequent rate of progression. J. Infect. Dis. 201:272–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. George M. D., Sankaran R. E. S., Dandekar S. 2005. Early antiretroviral therapy for simian immunodeficiency virus infection leads to mucosal CD4+ T-cell restoration and enhanced gene expression regulating mucosal repair and regeneration. J. Virol. 79:2709–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Greenier J. L., et al. 2005. Simian immunodeficiency virus (SIV) envelope quasispecies transmission and evolution in infant rhesus macaques after oral challenge with uncloned SIVmac251: increased diversity is associated with neutralizing antibodies and improved survival in previously immunized animals. Virol. J. 2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guadalupe M., et al. 2003. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J. Virol. 77:11708–11717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guadalupe M., et al. 2006. Viral suppression and immune restoration in the gastrointestinal mucosa of human immunodeficiency virus type 1-infected patients initiating therapy during primary or chronic infection. J. Virol. 80:8236–8247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gunthard H. F., et al. 1999. Evolution of envelope sequences of human immunodeficiency virus type 1 in cellular reservoirs in the setting of potent antiviral therapy. J. Virol. 73:9404–9412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haggerty C. M., Pitt E., Siliciano R. F. 2006. The latent reservoir for HIV-1 in resting CD4+ T cells and other viral reservoirs during chronic infection: insights from treatment and treatment-interruption trials. Curr. Opin. HIV AIDS 1:62–68 [DOI] [PubMed] [Google Scholar]
- 22. Hall A. T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis. Nucleic Acids Symp. Ser. (Oxf.) 1999(41):95–98 [Google Scholar]
- 23. Huang D. W., Sherman B. T., Lempicki R. A. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 [DOI] [PubMed] [Google Scholar]
- 24. Hudson R. R., Boos D. D., Kaplan N. L. 1992. A statistical test for detecting geographic subdivision. Mol. Biol. Evol. 9:138–151 [DOI] [PubMed] [Google Scholar]
- 25. Imamichi H., et al. 2001. Human immunodeficiency virus type 1 quasi species that rebound after discontinuation of highly active antiretroviral therapy are similar to the viral quasi species present before initiation of therapy. J. Infect. Dis. 183:36–50 [DOI] [PubMed] [Google Scholar]
- 26. Joos B., et al. 2008. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc. Natl. Acad. Sci. U. S. A. 105:16725–16730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Joos B., et al. 2005. Low human immunodeficiency virus envelope diversity correlates with low in vitro replication capacity and predicts spontaneous control of plasma viremia after treatment interruptions. J. Virol. 79:9026–9037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jordan M. R., et al. 2010. Comparison of standard PCR/cloning to single genome sequencing for analysis of HIV-1 populations. J. Virol. Methods 168:114–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Keele B. F., et al. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Levy J. A. 2009. HIV pathogenesis: 25 years of progress and persistent challenges. AIDS 23:147–160 [DOI] [PubMed] [Google Scholar]
- 31. Librado P., Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452 [DOI] [PubMed] [Google Scholar]
- 32. Macal M., et al. 2008. Effective CD4+ T-cell restoration in gut-associated lymphoid tissue of HIV-infected patients is associated with enhanced Th17 cells and polyfunctional HIV-specific T-cell responses. Mucosal Immunol. 1:475–488 [DOI] [PubMed] [Google Scholar]
- 33. Mattapallil J. J., Smit-McBride Z., McChesney M., Dandekar S. 1998. Intestinal intraepithelial lymphocytes are primed for gamma interferon and MIP-1beta expression and display antiviral cytotoxic activity despite severe CD4(+) T-cell depletion in primary simian immunodeficiency virus infection. J. Virol. 72:6421–6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ortiz G. M., et al. 2001. Structured antiretroviral treatment interruptions in chronically HIV-1-infected subjects. Proc. Natl. Acad. Sci. U. S. A. 98:13288–13293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oxenius A., et al. 2000. Early highly active antiretroviral therapy for acute HIV-1 infection preserves immune function of CD8+ and CD4+ T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 97:3382–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oxenius A., et al. 2002. Stimulation of HIV-specific cellular immunity by structured treatment interruption fails to enhance viral control in chronic HIV infection. Proc. Natl. Acad. Sci. U. S. A. 99:13747–13752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Potter S. J., et al. 2004. HIV-1 compartmentalization in diverse leukocyte populations during antiretroviral therapy. J. Leukoc. Biol. 76:562–570 [DOI] [PubMed] [Google Scholar]
- 38. Ruiz L., et al. 2001. HIV dynamics and T-cell immunity after three structured treatment interruptions in chronic HIV-1 infection. AIDS 15:F19–F27 [DOI] [PubMed] [Google Scholar]
- 39. Sankaran S., et al. 2008. Rapid onset of intestinal epithelial barrier dysfunction in primary human immunodeficiency virus infection is driven by an imbalance between immune response and mucosal repair and regeneration. J. Virol. 82:538–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sankaran S., et al. 2005. Gut mucosal T cell responses and gene expression correlate with protection against disease in long-term HIV-1-infected nonprogressors. Proc. Natl. Acad. Sci. U. S. A. 102:9860–9865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schnell G., Price R. W., Swanstrom R., Spudich S. 2010. Compartmentalization and clonal amplification of HIV-1 variants in the cerebrospinal fluid during primary infection. J. Virol. 84:2395–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schweighardt B., et al. 2002. Emergence of drug-resistant HIV-1 variants in patients undergoing structured treatment interruptions. AIDS 16:2342–2344 [DOI] [PubMed] [Google Scholar]
- 43. Smit-McBride Z., Mattapallil J. J., McChesney M., Ferrick D., Dandekar S. 1998. Gastrointestinal T lymphocytes retain high potential for cytokine responses but have severe CD4(+) T-cell depletion at all stages of simian immunodeficiency virus infection compared to peripheral lymphocytes. J. Virol. 72:6646–6656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Toulson A. R., et al. 2005. Treatment interruption of highly active antiretroviral therapy in patients with nadir CD4 cell counts >200 cells/mm3. J. Infect. Dis. 192:1787–1793 [DOI] [PubMed] [Google Scholar]
- 45. van Marle G., et al. 2007. Compartmentalization of the gut viral reservoir in HIV-1 infected patients. Retrovirology 4:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Verhoeven D., Sankaran S., Silvey M., Dandekar S. 2008. Antiviral therapy during primary simian immunodeficiency virus infection fails to prevent acute loss of CD4+ T cells in gut mucosa but enhances their rapid restoration through central memory T cells. J. Virol. 82:4016–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yukl S. A., et al. 2010. Differences in HIV burden and immune activation within the gut of HIV-positive patients receiving suppressive antiretroviral therapy. J. Infect. Dis. 202:1553–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang L., et al. 2000. Genetic characterization of rebounding HIV-1 after cessation of highly active antiretroviral therapy. J. Clin. Invest. 106:839–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.