Abstract
Previous studies identified clones of the U937 promonocytic cell line that were either permissive or nonpermissive for human immunodeficiency virus type 1 (HIV-1) replication. These clones were investigated further in the search for host restriction factors that could explain their differential capacity to support HIV-1 replication. Among known HIV-1 restriction factors screened, tripartite motif-containing protein 22 (TRIM22) was the only factor constitutively expressed in nonpermissive and absent in permissive U937 cells. Stable TRIM22 knockdown (KD) rescued HIV-1 long-terminal-repeat (LTR)-driven transcription in KD-nonpermissive cells to the levels observed in permissive cells. Conversely, transduction-mediated expression of TRIM22 in permissive cells reduced LTR-driven luciferase expression by ∼7-fold, supporting a negative role of TRIM22 in HIV-1 transcription. This finding was further confirmed in the human T cell line A3.01 expressing TRIM22. Moreover, overexpression of TRIM22 in 293T cells significantly impaired basal and phorbol myristate acetate-ionomycin-induced HIV-1 LTR-driven gene expression, whereas inhibition of tumor necrosis factor alpha-induced viral transcription was a consequence of lower basal expression. In agreement, TRIM22 equally inhibited an LTR construct lacking the tandem NF-κB binding sites. In addition, TRIM22 did not affect Tat-mediated LTR transactivation. Finally, these effects were independent of TRIM22 E3 ubiquitin-ligase activity. In the context of replication-competent virus, significantly higher levels of HIV-1 production were observed in KD-nonpermissive versus control nonpermissive U937 cells after infection. In contrast, lower peak levels of HIV-1 replication characterized U937 and A3.01 cells expressing TRIM22 versus their control transduced counterpart. Thus, nuclear TRIM22 significantly impairs HIV-1 replication, likely by interfering with Tat- and NF-κB-independent LTR-driven transcription.
INTRODUCTION
The U937 promonocytic cell line, characterized by the constitutive expression of CD4 and CXCR4 (but not CCR5), has been broadly utilized to dissect viral and/or host determinants modulating CXCR4-dependent (X4) human immunodeficiency virus type 1 (HIV-1) replication. Phenotypically and functionally distinct subsets of U937 cells were earlier obtained by limiting dilution cloning and investigated for their susceptibility to HIV-1 infection and replication (2, 6, 9, 23). Two clear-cut clusters of cell clones were identified according to their capacity to efficiently or inefficiently support X4 HIV-1 replication (23) that were earlier defined as “plus” and “minus” clones (reviewed in reference 1) and are here referred to as “permissive” and “nonpermissive” U937 cells, respectively.
Concerning the potential determinants at the basis of their differential capacity to support HIV-1 replication, no differences were reported between permissive and nonpermissive cells in terms of expression of CD4 and CXCR4 (7, 44), whereas a constitutive release of the proinflammatory cytokine tumor necrosis factor alpha (TNF-α) characterized permissive cells and was shown to boost virus replication and CXCR4 expression (7). Strikingly, incubation of nonpermissive cells with 1α,25-dihydroxy-vitamin D3 (vitamin D3) reverted their phenotype to that of permissive cells that were insensitive to this agent (7). However, neither the mechanism of action of vitamin D3 nor the cause of restricted HIV-1 replication in the nonpermissive U937 cells have been clearly defined. In this regard, either constitutively expressed or interferon (IFN)-induced factors capable of restricting early steps of the HIV-1 life cycle have received considerable attention in recent years. In particular, potent cellular restriction factors such as tripartite motif 5α (TRIM5α) and apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC3G/-3F) have emerged as preventing early, preintegration steps in the HIV-1 life cycle (31, 50, 65). In addition, the deaminase activity of APOBEC3A isoforms was shown to be induced by IFN stimulation of monocytes and macrophages (69). More recently, BST-2/Theterin was identified as a factor that holds mature HIV-1 particles from being released from infected cells (71). Its expression is induced by IFN-α (34), as earlier hypothesized (56). Indeed, IFN-α activation leads to HIV-1 retention at the plasma membrane in the absence of the accessory HIV-1 protein Vpu (47). In this regard, early reports suggested that endogenously produced IFN-α contributes to restricted HIV-1 viral replication in U937 promonocytic cells (40), while it was later suggested to interfere with HIV-1 long-terminal-repeat (LTR)-driven transcription (25). Among other IFN-induced genes, TRIM22, also known as Staf50 (33, 70), was originally identified in a screening for IFN-inducible genes in the human B lymphoblastoid Daudi cell line and was reported to inhibit HIV replication at transcriptional, posttranscriptional, or posttranslational levels following transfection in different target cells (3, 70).
The aim of our study was to investigate whether any known HIV-1 restriction factors could be related to the differential permissiveness for HIV-1 replication of U937 cell clones. We report here that, among other potential candidates, only TRIM22 was selectively and constitutively expressed in the nucleus of nonpermissive, but not of permissive U937 cells. To identify the HIV-1 life cycle step(s) potentially affected by TRIM22, stable TRIM22 knockdown (KD) nonpermissive cells and transduced (TD) TRIM22 expressing permissive cells were then obtained after transduction with lentiviral vectors expressing either a TRIM22 specific small hairpin RNA (shRNA) or with TRIM22-expressing vector, respectively. Furthermore, this TRIM22-mediated restriction was confirmed also in a human T cell line stably expressing the protein. Indeed, TRIM22 interfered with both basal and phorbol myristate acetate (PMA)-ionomycin-induced LTR-driven activity but failed to interfere with either TNF-α- or Tat-dependent transcriptional transactivation. Furthermore, inhibition occurred independently of its E3 ubiquitin-ligase activity and of the tandem NF-κB binding site within the viral promoter. Thus, TRIM22 acts as a nuclear factor capable of negatively influencing HIV-1 transcription in a stimulus-specific manner.
MATERIALS AND METHODS
Cell lines.
U937 permissive and nonpermissive cells were maintained in RPMI 1640 supplemented with glutamine (2 mmol/liter), penicillin (100 U/ml), and streptomycin (100 μg/ml) from BioWhittaker, Inc. (Walkersville, MD), and with 10% fetal bovine serum (FBS; HyClone Europe, Ltd., Cramlington, United Kingdom)-complete RPMI 1640. 293T human epithelial kidney cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with glutamine (2 mmol/liter), penicillin (100 U/ml), streptomycin (100 μg/ml), and 10% FBS (HyClone Europe) (complete DMEM). A3.01 cells were originally obtained by subcloning of the acute lymphocytic leukemia CEM cell line and were shown to be highly susceptible to X4 HIV-1 infection and replication (22).
Plasmids.
Most HIV-1 vectors and plasmids were previously described (46). The pNL4-3.Luc/GFP.R−-Env− plasmid contains two frameshifts that render the pNL4-3 infectious molecular clone Env− Vpr−, whereas the nef-encoding region is replaced by either luciferase (Luc) or enhanced green fluorescent protein (EGFP) reporter genes as described by Nathaniel Landau (27) and obtained from the NIH AIDS Research and Reference Program, Division of AIDS, National Institute of Allergy and Infectious Disease, National Institutes of Health. pFSGW is an HIV-1-based transfer vector with EGFP expression control by the spleen focus-forming virus (SFFV) LTR. pFSGW was generated by replacing the ubiquitin promoter in pFUGW (provided to J. Luban by David Baltimore [California Institute of Technology]) (38), with the U3 region of the SFFV LTR, which was amplified by PCR from the pCSGW plasmid (16) and inserted into the PacI and XbaI sites of pFUGW. pALPS is a Luc-expressing HIV-1-based transfer vectors derived from pFSGW, described above. pMD2.G is a cytomegalovirus (CMV)-driven expression plasmid that encodes the vesicular stomatitis virus envelope protein VSV-G, and psPax2 is the packaging vector expressing HIV Gag-Pol; both were generously provided by Didier Trono (Ecole Polytechnique Fédérale de Lausanne) to J. Luban. pLKO.1/TRIM22shRNA lentiviral vector contains an shRNA targeting TRIM22 (catalog no. RHS3979-9574742 [Open Biosystems, Huntville, AL]; GenBank accession no. NM_006074; sequence, 5′-CGG AGC ACT CAT CTA CAA GTT CTC GAG AAC TTG TAG ATG AGT GCT CCG-3′). pLKO.1/randomshRNA nonsilencing control was kindly provided by Davide Gabellini, San Raffaele Scientific Institute, Milan, Italy. TRIM22-expressing transfer vector pAIP-TRIM22 was obtained by cloning the TRIM22 gene obtained from U937 nonpermissive cells (GenBank accession no. HQ_842635) into the pAIP transfer vector that expresses TRIM22 as part of a bicistronic RNA driven by the SFFV LTR promoter, containing the encephalomyocarditis virus internal ribosome entry site and a puromycin resistance cassette. In order to obtain a TRIM22-expressing vector, the TRIM22 coding sequence was amplified from a mix of IFN-β- and lipopolysaccharide-stimulated monocyte-derived dendritic cells (MDDC) and monocyte-derived macrophages (MDM) cDNA and was cloned between the NotI and XbaI sites of the pcDNA3.1(+) vector (Invitrogen, Carlsbad, CA). 3X-Flag-TRIM22 and the 3X-Flag-TRIM22 RING deletion mutant (ΔRING) were previously described (21). The TRIM22-C15A/C18A mutant, in which two conserved cysteine residues in the RING domain (C15/C18) were replaced by two alanine residues (C15A/C18A), was generated by PCR substitution using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. PCR amplification was performed with the primers 5′-GAG AAG GAG GTG ACC GCC CCC ATC GCC CTG GAG CTC C-3′ (forward) and 5′-GGA GCT CCA GGG CGA TGG GGG CGG TCA CCT CCT TCT C-3′ (reverse). The HIV-1 LTR-Luc reporter and the pcDNA-Tat vector were kindly provided by Ben Berkhout, University of Amsterdam. The HIV-1 ΔNF-κB LTR-Luc reporter was kindly provided by Sebastien Nisole, Institut Cochin, Université Paris Descartes, Paris, France. The pcDNA3.1-Luc reporter was kindly provided by Daniele Zacchetti, San Raffaele Scientific Institute, Milan, Italy.
Vector production.
Vectors were produced by transfection of 293T cells seeded at 6 × 105 cells/ml in six-well plastic plates (Falcon; Becton Dickinson Labware, Lincoln Park, NJ) by using Lipofectamine 2000 (Invitrogen). HIVGFP and HIVLuc vectors were produced by cotransfection of pMD2.G and pNL4-3.GFP.R−-Env−/pNL4-3.Luc.R−-Env−, respectively, at a 1:7 ratio. minHIVGFP and minHIVLuc vectors were generated by cotransfection of pMD2.G, psPax2, and either pFSGW or pALPS, respectively, at 1:3:4 ratios. VSV-pLKO.1/TRIM22shRNA and VSV-pLKO.1/randomshRNA vectors were produced by cotransfecting pMD2.G, psPax2, and either pLKO.1/TRIM22shRNA or pLKO.1/randomshRNA, respectively, at 1:3:4 ratios. Similarly, VSV-G-Transfer-TRIM22 virus was obtained by cotransfecting pMD2.G, psPax2, and pAIP-TRIM22 at 1:3:4 ratios. Vector containing supernatants were harvested 48 h posttransfection, cleared by centrifugation, filtered (0.45-μm pore size; Millex-HV polyvinylidene difluoride; Millipore, Carrigtwohill, Ireland), and stored at −80°C.
Generation of stable TRIM22 KD and of TRIM22 stably expressing cells.
In order to downregulate TRIM22 expression, nonpermissive U937 cells were seeded in six-well plates at 6 × 105 cells/ml in complete RPMI 1640 and transduced with VSV-pLKO.1/TRIM22shRNA or VSV-pLKO.1/randomshRNA vector-containing supernatants at a 1:1 ratio with complete RPMI 1640. After 24 h, a half-volume of the culture medium was replaced with fresh vector-containing supernatant. At 72 h after the second transduction, the cells were subjected to selection with puromycin (2 μg/ml; Sigma-Aldrich, St. Louis, MO) for 2 weeks. The levels of TRIM22 expression were verified by Western blotting as described below.
In order to express TRIM22, permissive U937 cells and A3.01 human T cell line were transduced twice with Transfer-TRIM22 or empty control vectors at 24-h intervals replacing culture medium with vector-containing supernatant at a 1:1 ratio. At 72 h after the second transduction, the cells were subjected to puromycin (2 μg/ml for U937 and 1 μg/ml for A3.01; Sigma-Aldrich) selection. TRIM22 expression was verified by Western blot analysis as described below.
Infections.
For single cycle infection, U937-derived permissive and nonpermissive cells, A3.01 cells, as well as their mock-transduced counterparts, were seeded at 5 × 105 cells/ml in 96-well plates in complete RPMI 1640 and infected with serial dilutions of either HIVGFP/HIVLuc or minHIVGFP/Luc vectors. The percentage of GFP+ cells and relative light units (RLU) were evaluated at 72 h posttransduction by fluorescence-activated cell sorting analysis and Luc assay (Promega, Madison, WI), respectively.
For multiple-round infections, U937 permissive and nonpermissive cells and A3.01 cells were exposed to the laboratory-adapted X4 HIV-1IIIB/LAI at the multiplicities of infection (MOIs) of 1 and 0.1, respectively. After 1 h of virus adsorption at 37°C, the cells were resuspended in complete RPMI 1640 and seeded at 5 × 105 cells/well in triplicate in 48-well tissue culture plates (Falcon). Culture supernatants were harvested every 3 to 4 days and stored at −80°C until tested for Mg2+-dependent reverse transcriptase (RT) activity assay, as previously described (74).
Quantification of mRNAs by real-time PCR.
Total RNA was extracted from permissive and nonpermissive U937 cells by using a TRIzol Plus RNA purification kit, followed by DNase I treatment (Invitrogen). cDNA was synthesized from total RNA (1 μg) using a SuperScript first-strand synthesis system (Invitrogen) with random hexamers. Semiquantitative PCR was performed on 50 ng of cDNA with the primer pair, described in detail in Table 1, and the SYBR green PCR master mix (Applied Biosystems, Foster City, CA). To normalize the mRNA expression, the human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene was amplified. Relative standard curves for all targets and the normalizer were obtained using serially diluted (from 50 to 0.016 ng) total cDNA obtained from nonpermissive U937 cells stimulated with IFN-α (1,000 U/ml) for 24 h. The nanogram amounts of target and normalizer DNA were calculated from the relative standard curves by interpolation. All reactions were performed with an ABI 7700 Prism instrument (Applied Biosystems) using the following thermal cycling conditions: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min.
Table 1.
Primer pairs used to quantify mRNAs
| Gene | Orientationa | Primer sequence (5′–3′) |
|---|---|---|
| APOBEC3F | F | TTCCACTTTAAAAACCTACGCAAA |
| R | TGGTGCTTTACAACTTCCATGGT | |
| APOBEC3G | F | GGCTCCACATAAACACGGTTTC |
| R | AAGGGAATCACGTCCAGGAA | |
| APOBEC3A | F | GCATCCGGGCCCAGACACTT |
| R | GGTGCCATTGTCCAGGCGCT | |
| BST-2 | F | AGGTCCGTCCTGCTCGGCTT |
| R | TCCAGAGGCCCTTCTCCGGC | |
| hTRIM5ab | F | AGGAGTTAAATGTAGTGCT |
| R | ATAGATGAGAAATCCATGGT | |
| hTRIM19 | F | TGCCAGGCGGAAGCCAAGTG |
| R | CCTGGCAGATGGGGCACTGC | |
| hTRIM37 | F | GCATACCAGTGTGGGCGGGT |
| R | TTGAAGCTGAGATCTTCAGGGCCTA | |
| hTRIM22 | F | GGGTGGACGTGATGCTGAA |
| R | TCACTTGTCTCTGATCCACAGAAATA | |
| GAPDH | F | CCACCCATGGCAAATTCC |
| R | TGGGATTTCCATTGATGATGACAAG |
Forward (F) and reverse (R) primers are indicated for each gene.
Sewram et al. (60).
Western blotting.
Nuclear, cytoplasmic, and whole-cell extracts (WCE) from U937 permissive and nonpermissive cells and A3.01 cells were prepared as previously described (10, 32). Samples were subjected to SDS-PAGE, transferred to nitrocellulose membrane by electroblotting, and blotted with a rabbit polyclonal antibody (Ab) raised against TRIM22, as described previously (26). Blots with anti-p84 monoclonal Ab (MAb; Abcam, Cambridge, MA) and anti-α-tubulin Ab (Sigma-Aldrich) were used as controls for purity of the nuclear and cytoplasmic fractions, respectively. An anti-actin Ab (Sigma-Aldrich) was used as a normalizer for WCE.
Detection of integrated HIV-1 DNA by Alu-PCR.
To assess levels of integrated viral DNA, the Alu-PCR protocol described by Tan et al. (68) was used with minor modifications. In brief, a nested PCR assay was carried out with two sets of Alu-LTR primers and a probe. Genomic DNA (100 ng) was first amplified with AccuPrime Taq DNA polymerase high fidelity (Invitrogen) in the presence of the primer pair AluI sense (5′-TCCCAGCTACTGGGGAGGCTGAGG-3′) and LM667 antisense (5′-ATGCCACGTAAGCGAAACTCTGGCTAACTAGGGAACCCACTG-3′) in a DNA thermal cycler (Perkin-Elmer, Waltham, MA). The thermal cycling conditions for the first-round PCR were 95°C for 5 min, 94°C for 15 s, 55°C for 30 s, and 68°C 3 min, followed by 68°C for 10 min. After the initial PCR, a real-time PCR was performed by using an aliquot equivalent to 1/10 of the 25-cycle PCR product with the second-round LTR primers and TaqMan probe. The sequences of the second set of primers and probes are λT sense (5′-ATGCCACGTAAGCGAAACTCTGGCTAACTAGGGAACCCACTG-3′), antisense LR (5′-TCCACACTGACTAAAAGGGCTTGA-3′), and ZXF-P probe (5′-FAM-TGTGACTCTGGTAACTAGCGCTCCCTCAGACCC-TAMRA-3′). As a control, every sample was also amplified with a primer pair and probe targeting mitochondrial DNA: MitoF (5′-ACCCACTCCCTCTTAGCCAATATT-3′), MitoR (5′-GTAGGGCTAGGCCCACCG-3′), and MitoP (5′-FAM-CTAGTCTTTGCCGCCTGCGAAGCA-TAMRA-3′). The real-time PCR thermal cycling conditions for the second-round PCR were as follows: 50°C for 2 min and 95°C for 10 min, followed by 50 cycles of 95°C for 15 s and 60°C for 90 s. Standard curves for both target and normalizer were obtained by nested PCR, as described above, using serially diluted genomic DNA (from 500 to 0.4 ng) extracted from permissive U937 cells infected with HIVGFP virus and maintained in culture for 4 weeks prior to total DNA extraction. Nanograms of integrated HIV-1 and mitochondrial DNA were calculated from the standard curves. The normalized amounts (ng) of integrated provirus were obtained by multiplying the amount (ng) of target provirus interpolated from the relative standard curve by a mitochondrial DNA standardization factor. This factor was obtained by dividing each value of mitochondrial DNA (ng) by the maximum value. In order to exclude contamination of unintegrated HIV-1 DNA, the same DNA samples were analyzed by HIV-1 gag real-time PCR, as previously described (73).
TRIM22 transfection in 293T cells.
TRIM22-expressing plasmids (pcDNA3.1-TRIM22, p3X-Flag-TRIM22, or p3X-Flag-TRIM22-C15A/C18A) were transfected at increasing ratios (from 1:1 to 1:10), together with a fixed amount (5 ng) of HIV-1 LTR-Luc or CMV-driven pcDNA3.1-Luc reporter plasmids. In each case, the total amount of DNA transfected was kept constant by the addition of empty vector. When required, cells were cotransfected with pcDNA-Tat (ranging from 0.5 ng to 7.8 pg) or were stimulated 24 h after transfection with TNF-α (10 ng/ml; R&D Systems, Minneapolis, MN) or 10 nM PMA (Sigma Chemical Corp.) plus ionomycin (Sigma Chemical Corp.) at 1 μM for 16 h. LTR activation was measured 48 h after transfection by a Luc assay system (Promega) according to the manufacturer's instructions.
Immunofluorescence.
HEK293 cells were transfected using FUGENE6 (Roche) with hemagglutinin (HA)-tagged TRIM22 expression construct (10 ng) in a 24-well plate and fixed after 24 h. TRIM22 was stained using mouse MAbs to the HA tag (1:2,000 dilution; Santa Cruz, Santa Cruz, CA), followed by secondary Abs to mouse immunoglobulin conjugated to Alexa Fluor 568 (1:5,000 dilution; Invitrogen). The nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole). The images were acquired by using a Nikon TE2000 microscope equipped with Piezo driven 60× oil Nikon objective lens (NA 1.4) with z-spacing of 0.2 μm and processed using the Volocity software package (Perkin-Elmer). z-Sections were combined to obtain an extended focus image.
Statistical analysis.
All statistical analysis was performed using the Prism GraphPad software v4.0 (GraphPad Software). Comparison between two groups was performed using the unpaired Student t test or the Mann-Whitney test depending on the distributions of values, analyzed with the Kolmogorov-Smirnov test for normality, while one-way analysis of variance (ANOVA) was used to compare more than two groups. Comparison of the log10 50% effective concentration (EC50) values of nonlinear regression curve fits was performed by using the F-test. P values of <0.05 were considered significant.
RESULTS
Only the nonpermissive subset of U937 cells constitutively expresses endogenous TRIM22 in the nucleus.
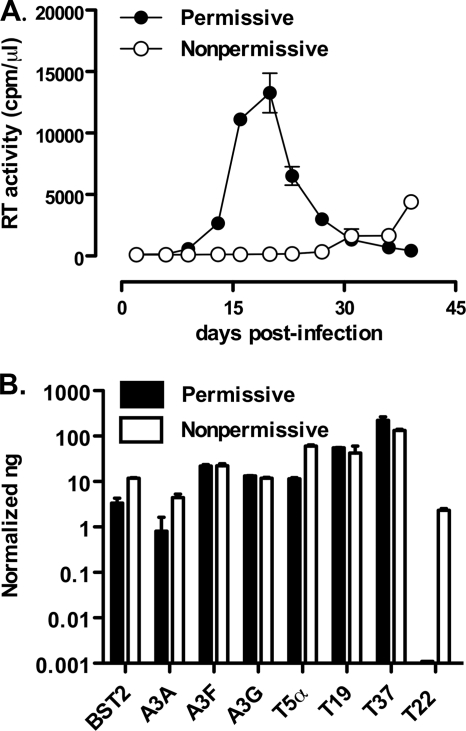
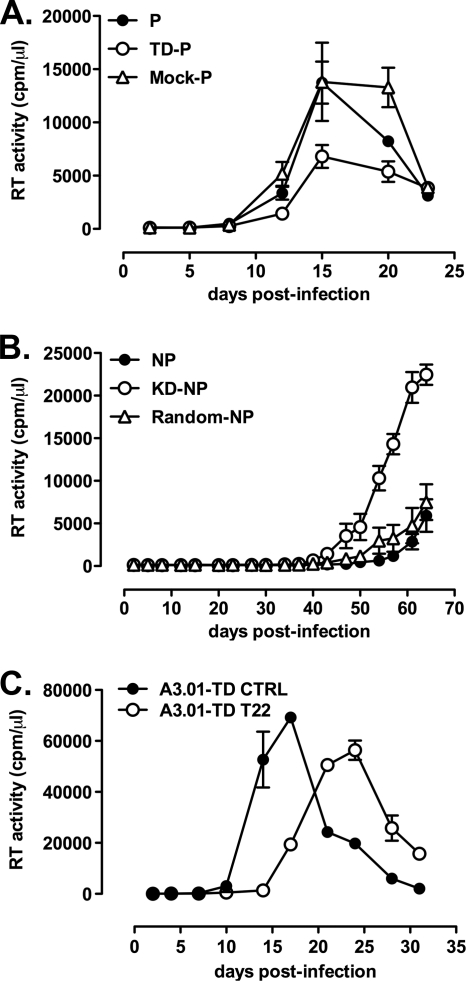
Permissive and nonpermissive U937 cells differ for their efficient and inefficient capacity to support X4 HIV-1 replication, respectively (7, 23) (Fig. 1A). Indeed, peak virus replication in permissive cells is typically observed ca. 20 days postinfection, followed by a rapid decline due to HIV-1-induced cytopathicity; in contrast, no detectable virus replication was usually observed, at least by RT activity in the culture supernatants, up to 30 or more days postinfection in nonpermissive cells (Fig. 1A).
Fig. 1.
Nonpermissive, but not permissive, U937 cell lines characterized by inefficient HIV-1 replication selectively express endogenous TRIM22. (A) Efficient and inefficient X4 HIV-1 replication in U937 cell lines. Permissive (•) and nonpermissive (○) cells were infected with HIV-1IIIB at an MOI of 1. The kinetics of viral replication were determined by measuring the RT activity in the supernatant of infected cultures harvested every 3 to 4 days. The data are expressed as means ± the standard errors of the mean (SEM; n = 3 replicas) of one representative of two independent experiments. (B) Semiquantitative real-time PCR was performed on total cDNA from unstimulated permissive and nonpermissive cell clones, normalized for GAPDH expression. The results are expressed as the normalized ng obtained through extrapolation from relative standard curves for each mRNA established from total cDNA of IFN-α treated nonpermissive cells. The results are expressed as means ± the SEM of one representative of three independent experiments performed in duplicates.
Since early reports suggested that endogenous IFN-α interferes with HIV expression in U937 cells (25), we investigated the steady-state expression levels of IFN-inducible factors, such as APOBEC3G (A3G), A3F, A3A, BST-2, hTRIM5α, TRIM19, TRIM37, and TRIM22 in both subsets of unstimulated U937 cells by semiquantitative real-time PCR. No significant differences in terms of mRNA levels of most of these factors were observed between the two subsets of U937 cells (Fig. 1B). In sharp contrast, nonpermissive cells clearly expressed TRIM22 mRNA, whereas permissive cells were devoid of its expression. Nonpermissive cells were estimated to express 2.33 ± 0.21 normalized ng of TRIM22 mRNA, whereas permissive cells were negative (Fig. 1B). Comparable basal levels of TRIM22 mRNA were confirmed in two other independent nonpermissive U937 cell clones (data not shown).
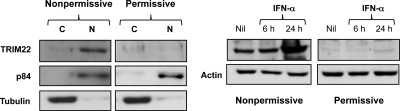
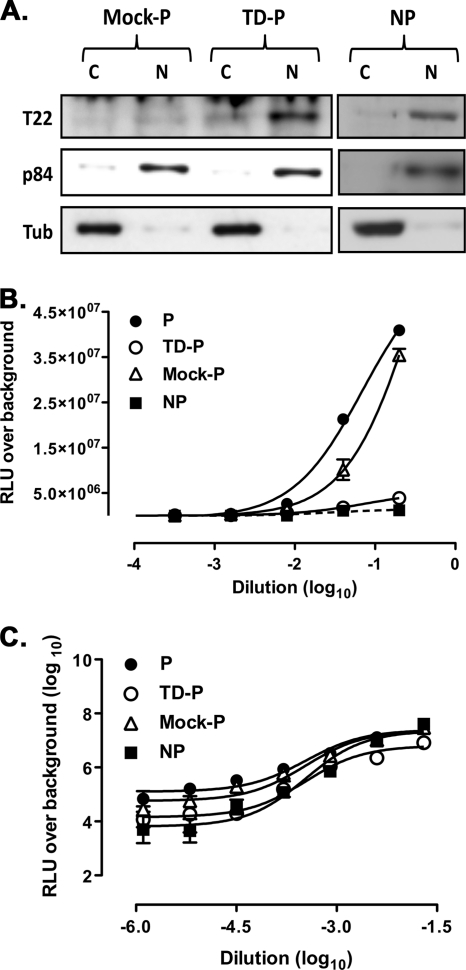
We next verified whether differences in TRIM22 expression occurred at the protein level and in which cellular compartment (nucleus versus cytoplasm). Indeed, basal expression of TRIM22 protein was observed in nonpermissive U937 cells, whereas it was not detectable in permissive cells. Furthermore, TRIM22 expression in nonpermissive U937 cells was readily detected in the nuclear fraction but only weakly in the cytoplasmic fraction (Fig. 2, left panel). Thus, TRIM22 is a suitable candidate to explain the restricted profile of nonpermissive U937 cells.
Fig. 2.
Nuclear expression and IFN-α induction of TRIM22 in nonpermissive, but not permissive, U937 cell lines. Selective TRIM22 expression was observed in the nucleus (N) but not in the cytoplasm (C) of U937 nonpermissive cells (left panel). Cytoplasmic and nuclear fractions of nonpermissive and permissive U937 protein extracts were subjected to SDS-PAGE and blotted with a polyclonal rabbit serum raised against TRIM22. Immunoblots against cytoplasmic α-tubulin and nuclear p84 antigens served as controls of the efficient cell compartment separation. The WCE obtained from nonpermissive and permissive U937 cells were obtained after 6 and 24 h of stimulation with 1,000 U of IFN-α/ml, evaluated by SDS-PAGE, and blotted with a polyclonal rabbit serum raised against TRIM22 (right panel). Actin was used as a control.
We next determined whether TRIM22 expression could be modulated in both permissive and nonpermissive U937 cells by stimulation with IFN-α, previously shown to induce activation of the Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway in both cell subsets (11). WCE were prepared 6 and 24 h after IFN-α stimulation, and TRIM22 expression was analyzed by Western blotting. IFN-α stimulation (1,000 U/ml) induced overexpression of TRIM22 protein in nonpermissive cells, whereas no evidence of protein expression was observed in permissive cells (Fig. 2, right panel).
Permissive and nonpermissive U937 cells differentially support HIV-1 LTR-driven transcription.
Nuclear expression of TRIM22 in the nonpermissive U937 cells provided support for the hypothesis that it could interfere with HIV-1 transcription, as also previously suggested (70). In order to investigate whether this subset of U937 cells would be characterized by impaired HIV-1 transcription, a feature thus far not associated with the nonpermissiveness of these cells, two VSV-G lentiviral vectors with a reporter gene under the control of the LTR of HIV-1 (clade B) or of the SFFV LTR, as a control, were produced, and U937 permissive and nonpermissive cells were then transduced with serial dilutions of these VSV-G vectors. The reporter gene expression was measured 72 h later.
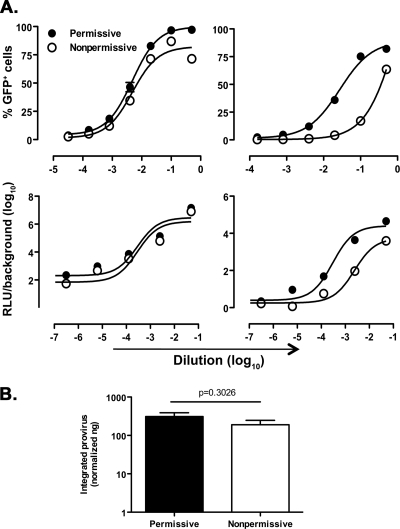
Cell transduction by vectors with SFFV LTR-dependent GFP or Luc expression (minHIVGFP/Luc) yielded almost undistinguishable curves in both permissive and nonpermissive U937 cells. In contrast, transduction with either HIV-GFP or HIV-Luc showed that basal gene transcription was 7- to 10-fold less efficient in nonpermissive than in permissive cells (Fig. 3A, upper and lower right panels, respectively; the log EC50 values and P values are reported in Table 2). This observation suggests that a selective defect in constitutive HIV-1 LTR-dependent transcription characterizes nonpermissive U937 cells.
Fig. 3.
Selective impairment of HIV-1 LTR-driven gene expression in nonpermissive U937 cells. (A) Permissive and nonpermissive U937 cells were transduced with serially diluted minHIVGFP, HIVGFP, minHIVLuc, or HIVLuc single-round reporter viruses. The percentages of GFP+ cells or RLU were measured 72 h after transduction. Titration curves were obtained by nonlinear regression curve fit using Prism GraphPad software v4.0. The results are representative of three independently performed experiments, expressed as means ± the SEM (n = 3 replicas). The 50% efficacy concentrations expressed in log10 EC50 and their respective P values are reported in Table 2. (B) Similar levels of proviral HIV DNA integration in permissive and nonpermissive U937 cells infected with HIVGFP were observed. Semiquantitative Alu-PCR was performed on genomic DNA extracted from permissive and nonpermissive U937 cells infected with VSV-G-pseudotyped HIVGFP at the dilution 1:10, which yields a major discrepancy in the percentage of GFP+ cells between permissive and nonpermissive cells. Cells were maintained in culture for 4 weeks before testing; mitochondrial DNA was used as a normalizer. The results are expressed as the normalized ng of integrated transfer vector as described in Materials and Methods. The results were obtained from six independent infections for the permissive and five independent infections for nonpermissive cells and are expressed as means ± the SEM. The P value was calculated using an unpaired Student t test.
Table 2.
Impaired HIV-1 LTR-driven gene expression in nonpermissive cellsa
| Virus | Mean log10 EC50 ± SEMb |
Pc | |
|---|---|---|---|
| Permissive | Nonpermissive | ||
| minHIVGFP | −2.32 ± 0.04 | −2.32 ± 0.09 | 0.9750 |
| HIVGFP | −2.13 ± 0.03 | −0.05 ± 0,19 | <0.0001 |
| minHIVLuc | −2.72 ± 0.09 | −2.76 ± 0.11 | 0.8466 |
| HIVLuc | −3.52 ± 0.09 | −2.69 ± 0.07 | <0.0001 |
The percentage of GFP+ cells or RLU was measured at 72 h after infection. Log10 EC50s were calculated by using a nonlinear regression curve fit of the Prism GraphPad software v.4.0.
The results are expressed as the mean log10 values of calculated the EC50s (n = 3).
P values were determined by use of the F-test comparing the best-fit EC50s.
In order to investigate whether these differences were due to a defective proviral integration into nonpermissive cells, semiquantitative Alu-PCR was performed on genomic DNA extracted from both subsets of U937 cells infected with HIV-GFP at a dilution corresponding to the point of major discrepancy in terms of percentage of GFP+ cells, i.e., (log10 dilution) = −1 (Fig. 3A, upper right panel). The cells were cultivated for 4 weeks prior to DNA extraction in order to eliminate residual nonintegrated HIV-1 DNA. The efficiency of proviral integration was indeed similar in both permissive and nonpermissive U937 cell lines (Fig. 3B), as confirmed by quantitative real-time PCR of the total HIV-1 DNA (data not shown). Overall, these findings indicate, for the first time, that the restricted pattern of HIV-1 replication typical of nonpermissive U937 cells is associated with a postintegration defect at the level of basal HIV-1 LTR-driven transcription.
TRIM22 KD rescues HIV-1 LTR-driven transcription in U937 nonpermissive cells.
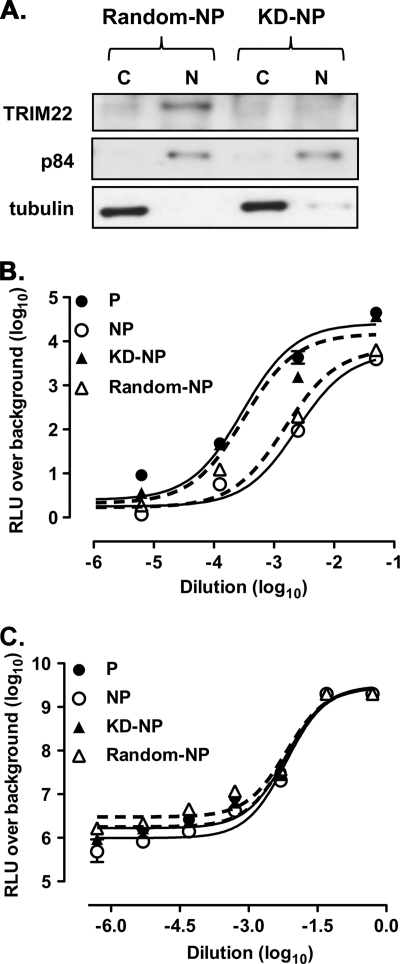
In order to investigate the potential involvement of TRIM22 in the inefficient basal HIV-1 LTR-dependent transcription observed in U937 nonpermissive cells, a stable cell line in which TRIM22 expression was significantly decreased (KD-nonpermissive) was generated by transduction of U937 nonpermissive cells with a lentiviral vector expressing specific TRIM22 shRNA (Fig. 4A). A cell line stably expressing a scrambled nonsilencing shRNA was generated as a control (random-nonpermissive [Random-NP]). No differences in cell viability or proliferation, as measured by [H3]thymidine incorporation, were observed between untransduced (control), random-nonpermissive and KD-nonpermissive (Fig. 4A, KD-NP) cells.
Fig. 4.
Knockdown of TRIM22 in nonpermissive U937 cells rescues HIV-1 LTR-driven transcription. (A) Stable expression of TRIM22-specific shRNA efficiently inhibits TRIM22 expression in nonpermissive U937 cells. Immunoblots with a polyclonal antibody against TRIM22 of nuclear (N) and cytoplasmic (C) extracts obtained from both random-nonpermissive (Random-NP) and KD-nonpermissive (KD-NP) U937 cells. (B) Rescue of HIV-1 LTR gene expression in KD-nonpermissive cells. Permissive and nonpermissive cells, as well as KD-nonpermissive and random-nonpermissive cells (KD-NP and Random-NP, dashed lines) were infected with serial dilutions of HIVLuc. (C) Comparable levels of SFFV LTR-driven gene expression was observed in permissive and nonpermissive cells, as well as in KD-nonpermissive and random-nonpermissive cells (KD-NP and Random-NP, dashed lines), infected with serial dilutions of minHIVLuc. RLU were quantified at 72 h postinfection, and the EC50s were calculated by using a nonlinear regression curve fit of the Prism GraphPad software v4.0. The results were obtained from a single experiment representative of three independently performed and are expressed as means ± the SEM (n = 3 replicas).
Control permissive and nonpermissive U937 cells, as well as the KD-nonpermissive and random-nonpermissive counterparts, were infected with serial dilutions of HIV-Luc vector, and the RLU over background levels were plotted against the dilutions of retroviral vector used. Both random and untransduced nonpermissive U937 cells showed similar levels of HIV-Luc expression (P = 0.06, F-test comparing the log10 EC50s of the fit curves) that were significantly lower than those observed in permissive U937 cells (P = 0.0004 for permissive versus random-nonpermissive; P < 0.0001 for permissive versus nonpermissive, F-test comparing the log10 EC50s of the fit curves [Fig. 4B]). In contrast, KD-nonpermissive cells showed an increased efficiency of viral transcription to levels indistinguishable from those of control permissive cells (P = 0.91, F-test comparing the log10 EC50s of the fit curves) and significantly higher than those of control and random-nonpermissive cells (P < 0.0001, F-test comparing the Log10 EC50s of the fit curves). In contrast, when reporter gene expression was dependent on the SFFV-LTR, the levels of transcription were superimposable in all cells tested (Fig. 4C; P = 0.95, F-test comparing the log10 EC50s of the fit curves).
Thus, the constitutive expression of TRIM22 in U937 nonpermissive cells likely accounts for their defective capacity to support basal HIV-1 LTR-driven gene expression.
Expression of TRIM22 impairs HIV-1 LTR-driven gene expression in permissive cells.
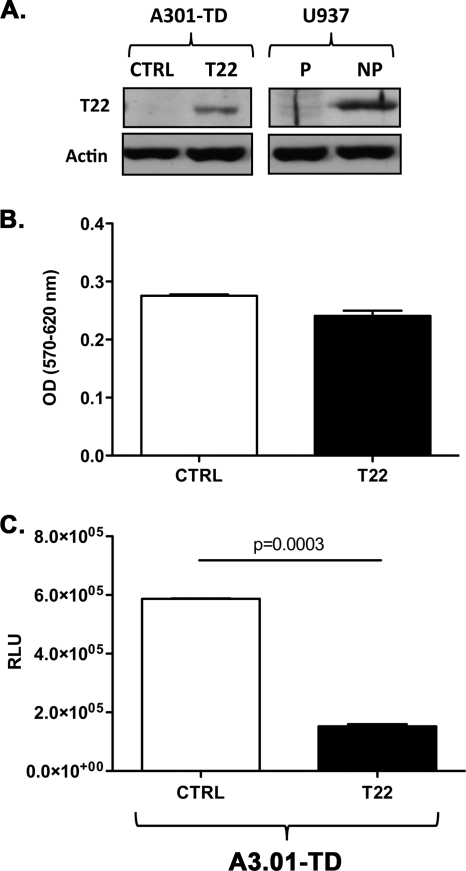
Since U937 permissive cells do not constitutively express detectable levels of TRIM22, we established a stable TRIM22-expressing permissive U937 cell line (TD-permissive) by means of cell transduction with a TRIM22 vector. As a control, permissive U937 cells were transduced with an empty vector harboring only the puromycin resistance cassette (mock-permissive). No differences in cell viability or proliferation, measured by [H3]thymidine incorporation, were observed in untransduced (control) and mock-permissive or TD-permissive cells (data not shown). Nuclear TRIM22 protein expression was indeed observed in TD-permissive cells, but not in control permissive cells, at levels comparable to those observed in nonpermissive U937 cells (Fig. 5A).
Fig. 5.
Transduction of TRIM22 in permissive U937 cells selectively represses HIV-1 LTR-driven luciferase expression. (A) TRIM22 (T22) expression in permissive (P) U937 cells was obtained by cell transduction with a TRIM22-expressing lentiviral vector (TD-P), as shown by immunoblotting of cytoplasmic (C) and nuclear (N) extracts, followed by blotting with an anti-TRIM22 polyclonal Ab. Nonpermissive (NP) cells are shown as a control. (B) TRIM22 inhibits HIV-1 transcription in TRIM22-expressing permissive cells. (C) Comparable levels of SFFV LTR-driven gene expression were observed in permissive and nonpermissive cells, as well as in TD-permissive and mock-permissive cells (TD-P and mock-P), infected with serial dilutions of minHIVLuc. The RLU were quantified at 72 h after infection, and EC50s were calculated by using the nonlinear regression curve fit of the Prism GraphPad software v4.0. The results were obtained from a single experiment representative of three independently performed and are expressed as means ± the SEM (n = 3 replicas).
Both transduced and control permissive cells were then infected with serial dilutions of the HIV-Luc reporter virus. TD-permissive cells showed a significant reduction in HIV-1 LTR-driven reporter gene expression in comparison to both control and mock-permissive cells (Fig. 5B; P < 0.0001, F-test comparing the log10 EC50s of the fit curves). In contrast, comparable levels of gene expression were observed in permissive cells transduced with the SFFV-LTR-driven reporter gene (Fig. 5C; P = 0.8, F-test comparing the log10 EC50s of the fit curves).
Thus, TRIM22 expression selectively represses HIV-1 LTR-driven transcription also in the background of U937 permissive cells, thereby mimicking the dichotomous pattern of efficient and inefficient HIV replication typical of permissive and nonpermissive U937 cell clones.
Expression of TRIM22 in A3.01 human T cells significantly impairs HIV-1 LTR-driven transcription.
In order to investigate whether the inhibitory effects of TRIM22 could be reproduced in a nonmonocytic background, the human CEM-derived CD4+/CXCR4+ T cell line A3.01 was transduced with TRIM22-expressing versus control vectors and then infected with HIV-Luc reporter virus supernatant containing 1.6 × 104 cpm equivalents of RT activity. TRIM22-transduced A3.01 cells efficiently expressed the protein compared to the control cells transduced with an empty vector (Fig. 6A; A3.01-TD T22 versus A3.01-TD CTRL), although not to the levels of U937 nonpermissive cells (Fig. 6A; A3.01-TD T22 versus U397 NP). TRIM22 expression did not alter cell viability as assessed by an MTT assay (Fig. 6B; optical density [OD] of 0.24 ± 0.008 for A3.01-TD T22 versus an OD of 0.28 ± 0.002 for A3.01-TD CTRL [P = 0.1, unpaired Student t test]). It is noteworthy that TRIM22 expression in A3.01 cells leads to an average (3.9 ± 0.19)-fold decrease in the HIV-1 LTR-driven gene expression compared to their control transduced counterparts (Fig. 6C; 152,200 ± 7,100 RLU for A3.01-TD T22 versus 587,100 ± 1,500 RLU for the A3.01-TD CTRL [P = 0.0003, unpaired Student t test]). Therefore, TRIM22-mediated restriction of HIV-1 transcription is not confined to the monocytic background but can be observed also in T cells.
Fig. 6.
Transduction of TRIM22 in A3.01 T cells impairs HIV-1 LTR-driven luciferase expression. (A) TRIM22 (T22) expression in A3.01 cells was obtained by cell transduction with a TRIM22-expressing lentiviral vector (A3.01-TD T22), as shown by immunoblotting of WCE, followed by blotting with an anti-TRIM22 polyclonal Ab. Cells transduced with an empty vector (A3.01-TD CTRL), as well as the U937 permissive (P) and nonpermissive (NP) cells, are shown as a control. (B) Cell viability was measured by an MTT assay in transduced A3.01 cells. (C) TRIM22 inhibits HIV-1 transcription in TRIM22-expressing A3.01 cells infected with 16,000 cpm of minHIVLuc/well. The RLU were quantified at 48 h after infection. The results were obtained from a single experiment representative of three independently performed and are expressed as means ± the SEM (n = 3 replicas). The P value was calculated using an unpaired Student t test.
TRIM22 selectively inhibits basal HIV-1 transcription.
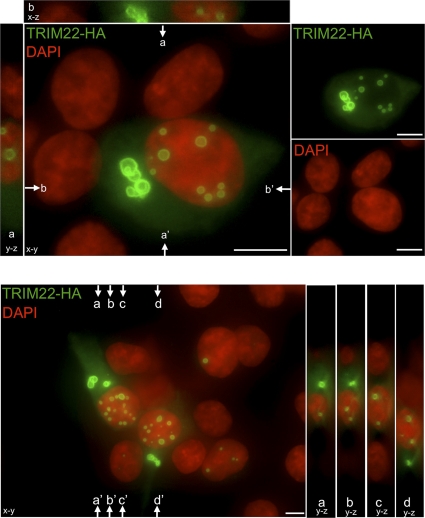
In order to investigate the mechanism by which TRIM22 downregulates HIV-1 transcription, kidney epithelial 293T cells (devoid of endogenous TRIM22 expression, data not shown) were cotransfected with increasing amounts of TRIM22-expressing vector, together with a fixed amount (5 ng) of HIV-1 LTR-Luc reporter construct. Immunofluorescence staining indicated that TRIM22 forms both nuclear and cytoplasmic bodies (Fig. 7), as previously shown for other TRIM proteins after cell transfection or transduction (57). Consistent with our observation in U937 cells, HIV-1 LTR-driven gene expression was decreased by 2.9-fold in unstimulated 293T cells with increasing TRIM22 expression levels (Fig. 8A; 94,070 ± 9,360 RLU for a 10:1 ratio of TRIM22 to LTR-Luc versus 259,730 ± 13,560 RLU for an LTR-Luc-only control [P < 0.0001, Mann-Whitney test]).
Fig. 7.
TRIM22 forms nuclear and cytoplasmic bodies. HA-tagged TRIM22 was visualized by immunofluorescence 24 h after transfection. TRIM22-HA- and DAPI-stained nuclei were pseudocolored green and red, respectively. The main images represent the extended focus of an entire z-stack. To illustrate the localization of TRIM22 within nuclei, y-z and x-z slices, indicated by arrows, are presented next to images. Two representative fields are shown. Size bars, 10 μm.
Fig. 8.
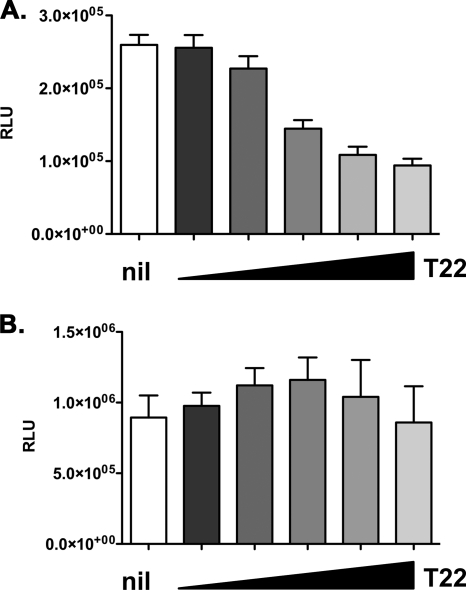
TRIM22 selectively inhibits basal HIV-1 transcription. 293T cells were cotransfected with an HIV-1 LTR-Luc (A) or a CMV-driven Luc (B) reporter, together with increasing amounts of TRIM22-expressing vector (T22, wedges), reaching a maximum of 10-fold excess with respect to the reporter plasmid. RLU were quantified 48 h posttransfection. White columns represent basal reporter activity in the absence of TRIM22 (nil). The results are from three independent experiments performed in triplicates each and expressed as means ± the SEM.
As controls of specificity, 293T cells were transfected with a reporter construct in which the Luc gene was driven by the CMV promoter, together with increasing amounts of TRIM22-expressing plasmid (Fig. 8B). Conversely to the HIV-1 LTR, TRIM22 had no inhibitory effects on this other viral promoter.
TRIM22 selectively inhibits induction of HIV-1 transcription.
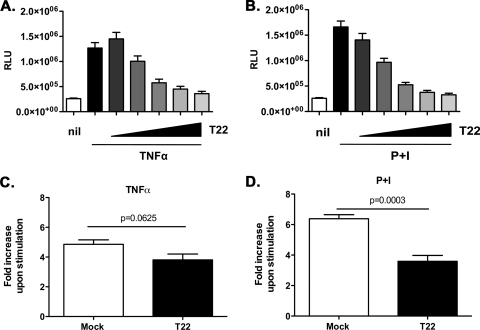
Both TNF-α and PMA, either alone or in combination with ionomycin (P+I), are known to activate HIV-1 transcription by induction of NF-κB and, in the case of PMA, of additional transcription factors (39, 41). Therefore, TRIM22-expressing and control 293T cells were stimulated 24 h after transfection with either TNF-α or P+I. TNF-α and P+I induced HIV-1 LTR transcription by 4.9- and 6.4-fold over background, respectively (Fig. 9A and B). Increasing amounts of TRIM22 led to a decrease of TNF-α-stimulated HIV-1 LTR-driven gene expression to almost a basal level (Fig. 9A; 359,780 ± 46,190 RLU for 10:1 ratio of TRIM22 to LTR-Luc versus 1.27e + 006 ± 109,990 RLU for LTR-Luc only [P < 0.0001, Mann-Whitney test]). When cells were stimulated with P+I, a 5.3-fold decrease was observed at the highest TRIM22 versus HIV-1 LTR-Luc ratio (Fig. 9B; 325,63 ± 32,19 RLU for 10:1 ratio of TRIM22 to LTR-Luc versus 1.66e + 006 ± 116,49 RLU for the LTR-Luc-only control [P < 0.0001, Mann-Whitney test]).
Fig. 9.
TRIM22 selectively inhibits the induction of HIV-1 transcription. 293T cells were cotransfected with an HIV-1 LTR-Luc reporter, together with increasing amounts of TRIM22-expressing vector (T22, wedges), reaching a maximum of 10-fold excess in respect to the HIV LTR reporter. At 24 h posttransfection, cells were stimulated with 10 ng of TNF-α/ml (A) or 10 nM PMA and 1 μM ionomycin (P+I) (B) for 16 h. The RLU were quantified at 48 h posttransfection. White columns represent basal reporter activity in the absence of TRIM22 (nil), while the first black bar represents LTR activation upon stimulation, in the absence of TRIM22. Fold inductions upon TNF-α (C) or P+I (D) stimulation were compared in the absence (Mock, white bar) or in the presence of the highest amount of TRIM22 (T22, black bar). The results are from three independent experiments performed in triplicates each and are expressed as means ± the SEM. The P values were calculated using the Mann-Whitney test.
In order to dissect whether the inhibitory effects of TRIM22 on TNF-α- and P+I-induced LTR activation could be a consequence of impaired basal transcription (Fig. 8A), we examined the fold increase in Luc activity observed upon stimulation both in mock- and TRIM22-transfected cells at the highest TRIM22 versus HIV-1 LTR-Luc ratio. Interestingly enough, TNF-α stimulation led to a fold induction of the HIV-1 LTR in TRIM22-transfected cells that was comparable to that of mock-transfected controls (Fig. 9C; a fold increase of 4.86 ± 0.3 in the control cells versus a fold increase of 3.81 ± 0.39 in TRIM22-transfected cells [P = 0.063, Mann-Whitney test]). This finding suggests that the inhibition observed in TRIM22-expressing cells upon TNF-α stimulation (Fig. 9A) could be a reflection of the impaired basal HIV-1 LTR activity rather than a specific interference with TNF-α-mediated LTR induction, namely, occurring through the NF-κB signaling cascade (55). In support of this interpretation, TNF-α was no longer able to induce transcriptional activation driven by NF-κB-deleted LTR (data not shown) that was nonetheless inhibited by TRIM22, as later discussed.
In contrast, P+I stimulation of HIV-1 LTR-driven gene expression remained significantly lower in TRIM22-expressing cells compared to controls even when normalized in terms of fold induction versus baseline (Fig. 9D; a fold increase of 6.39 ± 0.26 in the control cells versus a fold increase of 3.59 ± 0.39 in TRIM22-transfected cells [P = 0.0003, Mann-Whitney test]), suggesting that the P+I-induced signaling cascade could be specifically targeted by TRIM22.
These effects were not related to potential toxicity of TRIM22, since no differences in cell viability were observed in 293T cells transfected with increasing amounts of TRIM22-expressing vector (data not shown). Furthermore, transfection of 293T cells with a plasmid expressing TRIM34, another TRIM family member located within the same genetic cluster as TRIM22 and TRIM5α (59), described thus far to possess only weak, if any, antiretroviral activity (37, 81), or a GFP-expressing vector, did not interfere with either basal or TNF-α- or P+I-induced HIV-1 LTR-driven gene expression (data not shown).
Thus, TRIM22 impaired both basal and P+I-induced HIV-1 LTR activity but did not interfere with TNF-α-dependent HIV-1 LTR transcription, at least in 293T cells.
NF-κB and Tat-independent restriction of HIV-1 LTR-driven gene expression by TRIM22.
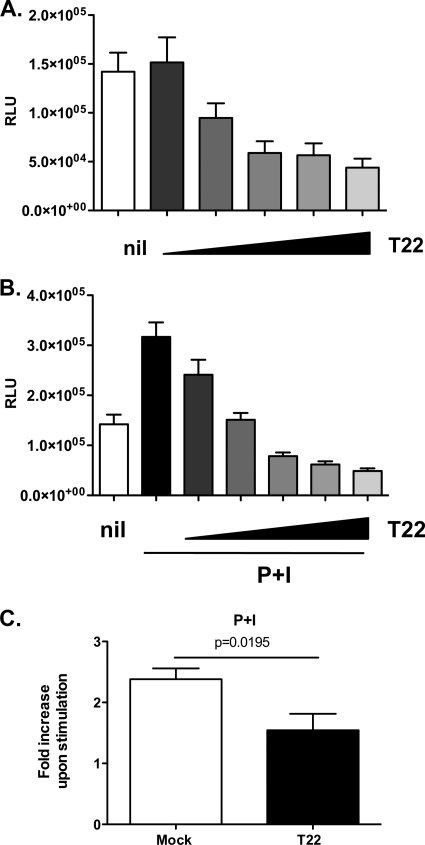
Among the transcription factors activated by P+I costimulation, AP-1, NF-κB, and NFAT are of particular relevance for the early activation of HIV-1 LTR transcription (14, 41). The tandem NF-κB binding region located within the core enhancer region of the HIV-1 LTR is a key mediator of NF-κB, as well as NFAT-dependent activation of transcription (4, 51). To investigate which element(s) is required for TRIM22-mediated inhibition of HIV-1 LTR-driven gene expression in cells stimulated with P+I, 293T cells were cotransfected with increasing amounts of TRIM22-expressing plasmid, together with a mutant LTR reporter construct lacking the tandem NF-κB binding region (Fig. 10). TRIM22 expression still led to a 3.2-fold decrease in basal LTR-driven gene expression (Fig. 10A; 43,880 ± 9,210 RLU for a 10:1 ratio of TRIM22 to ΔNF-κBLTR-Luc versus 142,020 ± 19,550 RLU for the ΔNF-κBLTR-Luc-only control [P = 0.0003, Mann-Whitney test]), whereas P+I stimulation of this mutant HIV-1 LTR reporter led to a 2.4-fold increase in the absence of TRIM22 versus a 6.4-fold increase observed with the wild-type LTR (Fig. 10B and 9B; respectively). TRIM22-expressing cells showed an up to 6.5-fold decrease in P+I-induced transcription (Fig. 10B; 48,840 ± 5,290 RLU for a 10:1 ratio of TRIM22 to ΔNF-κBLTR-Luc versus 316,930 ± 28,990 RLU for the ΔNF-κBLTR-Luc-only control [P < 0.0001, Mann-Whitney test]), suggesting that the tandem NF-κB binding region within the HIV-1 LTR is not required for TRIM22-mediated restriction of basal and P+I-induced activation, as discussed above. Consistently, P+I stimulation in TRIM22-transfected cells resulted in significantly lower levels of upregulation than what was observed in mock-transfected controls (Fig. 10C; a fold increase of 2.38 ± 0.18 in control cells versus a fold increase of 1.54 ± 0.27 in TRIM22-transfected cells [P = 0.02, unpaired Student t test]). Overall, these results indicate that TRIM22 interferes with basal and P+I-induced HIV-1 LTR transcription independently of NF-κB activation.
Fig. 10.
TRIM22 inhibits the activation of HIV-1 LTR independently of the tandem NF-κB site. 293T cells were cotransfected with an HIV-1 ΔNF-κB LTR-Luc reporter plasmid, together with increasing amounts of TRIM22-expressing vector (T22, wedges), reaching a maximum of 10-fold excess in respect to the HIV LTR reporter. At 24 h posttransfection, the cells were either left unstimulated (A) or were stimulated with 10 nM PMA and 1 μM ionomycin (P+I) (B) for 16 h. The RLU were quantified at 48 h posttransfection. White columns represent the basal reporter activity in the absence of TRIM22 (Nil), while the first black bar in panel B represents LTR activation upon stimulation, in the absence of TRIM22. (C) The fold induction upon P+I stimulation was compared in the absence (Mock, white bar) or in the presence of the highest amount of TRIM22 (T22, black bar). The results are from three independent experiments performed in triplicates each and are expressed as means ± the SEM. The P value was calculated using an unpaired Student t test.
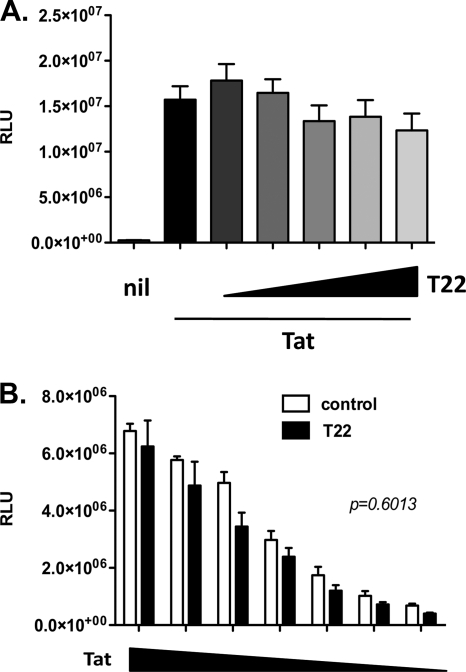
Finally, TRIM22 had no effect on Tat-dependent LTR-driven transcription (Fig. 11A; 1.23e + 007 ± 1.86e + 006 RLU for 10:1 ratio of TRIM22 to LTR-Luc versus 1.57e + 007 ± 1.48e + 006 RLU for the LTR-Luc-only control [P = 0.19, Mann-Whitney test]) even when decreasing concentrations of Tat-expressing plasmids were transfected (Fig. 11B, T22 versus control [P = 0.6, unpaired Student t test]).
Fig. 11.
Tat-mediated HIV-1 LTR transactivation is insensitive to TRIM22. (A) 293T cells were cotransfected with an HIV-1 LTR-Luc reporter plasmid and a fixed amount of Tat-expressing vector, together with increasing amounts of TRIM22-expressing vector (T22, wedges), reaching a maximum of 10-fold excess with respect to the HIV LTR. The RLU were quantified at 48 h posttransfection. White columns represent the basal reporter activity in the absence of TRIM22 (Nil), while the first black bar represents LTR activation upon transactivation, in the absence of TRIM22. (B) 293T cells were cotransfected with an HIV-1 LTR-Luc reporter plasmid and a 10-fold excess of control (control, white bars) or TRIM22-expressing (T22, black bars) vector and decreasing quantities of Tat-expressing vector (Tat, wedges). The RLU were quantified at 48 h posttransfection. The results are from three independent experiments performed in triplicates each and expressed as means ± the SEM. The P value was calculated using an unpaired Student t test.
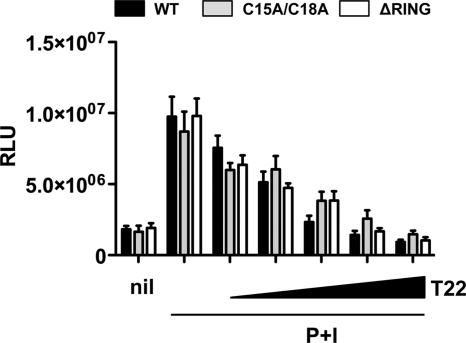
TRIM22-mediated restriction of LTR-driven gene expression is independent of its E3 ubiquitin ligase activity.
TRIM22 can exert antiviral effects also on viruses not belonging to the Retroviridae family, such as hepatitis B virus (24) and encephalomyocarditis virus (21). In both cases, the TRIM22-mediated antiviral effect was dependent on the N-terminal “really interesting new gene” (RING) finger domain harboring an E3 ubiquitin-ligase activity (19). In order to investigate whether TRIM22-mediated inhibition of HIV-1 LTR-driven gene expression was dependent on its E3 ubiquitin-ligase activity, 293T cells were cotransfected with the HIV-1 LTR-Luc reporter plasmid and increasing amounts of vectors expressing either the TRIM22 wild type (WT) or a TRIM22 mutant lacking the RING domain (ΔRING) or containing cysteine-to-alanine point mutations in positions 15 and 18 (C15A/C18A) previously shown to impair its E3 ubiquitin ligase activity (3, 19). The cells were then stimulated with P+I 24 h after transfection. However, no significant differences were observed between WT and the mutant TRIM22 constructs in terms of inhibition of LTR activation induced by P+I (Fig. 12, P = 0.99, one-way ANOVA).
Fig. 12.
TRIM22 inhibits P+I-induced HIV-1 LTR-driven gene expression independently of its E3-ubiquitin ligase activity. 293T cells were cotransfected with an HIV-1 LTR-Luc reporter plasmid and increasing amounts of either wild-type TRIM22-expressing vector (black bars, WT), TRIM22 harboring a functionally defective RING-domain (gray bars, C15A/C18A), or TRIM22 with the RING domain deleted (white bars, ΔRING). At 24 h posttransfection, the cells were stimulated with 10 nM PMA and 1 μM ionomycin (P+I) for 16 h, and the RLU were quantified at 48 h posttransfection. The first columns represent basal reporter activity in the absence of TRIM22 (nil), while the second pair of columns represents LTR activation upon stimulation, in the absence of TRIM22. The results are from three independent experiments performed in triplicates each and expressed as means ± the SEM.
Thus, unlike what observed in other viral infections, TRIM22 inhibition of HIV-1 LTR-driven gene expression occurs independently of its E3 ubiquitin-ligase activity.
TRIM22 inhibits HIV-1 replication in U937 and A3.01 cells.
In order to investigate whether the suppressive effects of TRIM22 on HIV transcription affected HIV-1 replication, both control and transduced permissive and nonpermissive U937 cell lines were infected with HIV-1IIIB/LAI at an MOI of 1. In parallel, transduced TRIM22-expressing and control A3.01 cells were infected at an MOI of 0.1. The kinetics of viral replication were monitored by determining the RT activity in the culture supernatants up to 70 days postinfection.
Neither TRIM22 nor random-KD altered the surface expression of CD4 and CXCR4 in nonpermissive U937 cells (data not shown). TD-permissive cells did not show differences in terms of kinetics of HIV-1 replication. Nevertheless, although not reaching statistical significance, their peak RT activity was reproducibly lower than that of control and mock-permissive cells (Fig. 13A; 6,800 ± 1,070 cpm/μl for TD-permissive versus 13,740 ± 1,970 cpm/μl for permissive and 13,810 ± 3,670 cpm/μl for mock-permissive at day 15 postinfection; n = 3 replicas). Conversely, KD-nonpermissive U937 cells showed both accelerated kinetics of HIV-1 replication and significantly increased peak levels of virus production versus control nonpermissive cells (22,460 ± 1,190 cpm/μl for KD-nonpermissive, 7,490 ± 2,10 cpm/μl for random-nonpermissive, and 5,920 ± 1,910 cpm/μl for nonpermissive [P < 0.0001 for KD-nonpermissive versus nonpermissive and versus random-nonpermissive, one-way ANOVA at day 64 postinfection]; Fig. 13B; n = 3 replicas). No significant differences in the kinetics and RT activity levels at the peak of virus replication were observed between either mock-TD or random-nonpermissive and control permissive and nonpermissive cells, respectively. Similarly to permissive U937 cells, TRIM22 overexpression in the A3.01 cell line showed a significant delay in the kinetics of HIV-1 replication (Fig. 13C; peak RT activity, 69,220 ± 618 cpm/μl for A3.01-TD CTRL versus 56,340 ± 2,680 cpm/μl for A3.01-TD T22 [P = 0.043, unpaired Student t test]).
Fig. 13.
TRIM22 inhibits HIV-1 replication in U937 and A3.01 cells. (A) Expression of TRIM22 decreases the efficiency of X4 HIV-1 replication in permissive cells (TD-P). (B) Suppression of TRIM22 expression promotes accelerated kinetics and increased peak levels of X4 HIV-1 replication in nonpermissive U937 cells (KD-NP). (C) TRIM22 overexpression leads to delayed viral replication in A3.01 cells (A3.01-T22) compared to mock-transduced controls (A3.01-TD CTRL). Infection supernatants were harvested at different times postinfection, and the RT activity was measured. The results were obtained from a single experiment representative of two independently performed and are expressed as means ± the SEM (n = 3 replicas).
Thus, the endogenous TRIM22 expression in U937 nonpermissive cells significantly contributes to their defective capacity to support HIV-1 replication. Moreover, expression of TRIM22 in a T cell line led to reduced transcription and viral replication further emphasizing its antiretroviral potential in critical cell targets of HIV-1 infection.
DISCUSSION
Although previous studies have suggested that cell transfection of TRIM22 could lead to a transcriptional repression of the HIV-1 LTR (8, 70), we provide here the first direct evidence of a relevant role of endogenous, nuclear TRIM22 in the control of HIV-1 LTR-dependent gene expression and virus replication. TRIM22 inhibited HIV-1 LTR-driven transcription and virus replication in a model represented by isogenic U937 promonocytic cell lines characterized by efficient (permissive) or inefficient (nonpermissive) capacity to support productive HIV-1 replication, respectively. Among other known host restriction factors, only TRIM22 was differentially expressed in the nuclei of nonpermissive cells, with permissive cells lacking detectable mRNA and protein expression. A strong restriction in both virus replication and HIV-1 LTR-dependent transcription characterized nonpermissive cells. When endogenous expression of TRIM22 was stably inhibited in nonpermissive cells by shRNA, both increased levels of HIV-1 transcription and virus replication were observed. Conversely, stable expression of TRIM22 in permissive cells resulted in impaired HIV-1 transcription and replication. These effects were selective for the HIV-1 LTR since no differences were observed when gene expression was SFFV LTR-driven. These antiviral effects on HIV-1 transcription and replication were confirmed upon infection of the human CD4+ T cell line A3.01 transfected with a TRIM22 expressing plasmid. In addition, both basal and P+I-induced LTR activation were inhibited in 293T cells transfected with increasing amounts of TRIM22-expressing plasmids. In contrast, TRIM22 did not interfere with TNF-α-induced, NF-κB-dependent, transcriptional activation or with Tat-mediated induction of viral expression.
Infection of mononuclear phagocytes (MP) at different stages of their differentiation is a crucial event in the pathogenesis of HIV-1 disease. Among several in vitro models of MP infection, the U937 promonocytic cell line has been broadly utilized. In this regard, two phenotypically and functionally distinct subsets of U937 cell clones have been described as “plus” and “minus,” redefined here as permissive and nonpermissive cells, in that they are endowed with efficient and inefficient capacities to support productive HIV-1 infection, respectively. The differential capacity of permissive and nonpermissive U937 cells to support X4 HIV-1 replication has been associated with several features, including cathepsin G-dependent cleavage of NF-κB p65 occurring exclusively in nonpermissive cells after nuclear extraction (23) and constitutive secretion of TNF-α in permissive cells (6), most likely contributing in concert with differential TRIM22 expression to the overall viral replication kinetics characterizing these U937 cellular subsets. Of interest, stimulation of nonpermissive U937 cells with vitamin D3 fully reverted HIV-1IIIB/LAI replication kinetics to the levels observed in the U937 permissive cells that were unaffected by the compound (7). The mechanism of action of vitamin D3 has remained largely unexplained, although it increased the levels of CXCR4-expression and viral entry (7). However, TRIM22 expression was not decreased in nonpermissive cells or induced in permissive cells after vitamin D3 stimulation (data not shown), as also reported independently (8). Interestingly, a recent report has demonstrated that vitamin D receptor-dependent signaling activates the HIV-1 LTR in U937 cells by both classical and nonclassical mechanisms (49), potentially involving molecular pathways able to overcome the TRIM22-mediated block.
Recently, Barr et al. demonstrated that TRIM22 transfection of human epithelial HeLa and osteosarcoma HOS-CD4-CXCR4 cells blocked the release of HIV-1 particles in the culture supernatant (3). In addition, these researchers observed that IFN-β stimulation of HOS-CD4-CXCR4 cells led to the upregulation of endogenous TRIM22 expression and inhibition of HIV-1 virion release (3). However, IFN-α/-β stimulation is known to block virion release in several cell lines, including U937-derived U1 cells (56), as a consequence of the upregulation of BST-2/tetherin (48, 53), making it difficult to single out the effect of TRIM22 per se at this late step of the HIV-1 life cycle. Independently, TRIM22 expression following transduction of human osteosarcoma U20S and 143B cells resulted in a strong reduction of cytoplasmic accumulation of Gag proteins compatible with a mechanism of transcriptional repression. We also observed a diminution of HIV-1 p24 Gag levels in the supernatant of human embryonic kidney 293T cells 48 h after cotransfection of TRIM22 and of an HIV-1 Gag expression plasmid (data not shown).
Thus, TRIM22 might affect both HIV-1 transcription or virion assembly and release as a function of the cell type and of its subcellular localization. In this regard, TRIM22 has been reported to reside either in the cytoplasm (28, 43) or in the nucleus (19, 24, 63, 64), likely accomplishing diverse biological effects. In the present study, we show that TRIM22 was largely found in the nuclear fraction in nonpermissive U937 cells, in agreement with its mechanism of interference on HIV-1 transcription. When expressed in HEK293 cells, both nuclear and cytoplasmic bodies were readily observed, a finding in agreement with previous reports (19, 24, 28, 43, 63, 64).
TRIM22-mediated transcriptional inhibition in U937 cells was specific for the HIV-1 LTR in that SFFV LTR-driven transcription was not affected by TRIM22. These observations indicate that the HIV-1 LTR harbors specific sequences required for TRIM22 inhibitory effects. In this regard, TRIM22, as well as other TRIM family members, has not yet been credited with the capacity to directly interact with DNA (29, 76, 77), whereas several host transcription factors, including NF-κB, NFAT-1, AP-1, Sp1, STAT5, and others (15, 20, 35, 45, 72), could be direct or indirect targets of TRIM22-mediated transcriptional inhibition of the HIV-1 LTR. In accordance also with the insensitivity of the CMV promoter, known to be mainly regulated by the NF-κB activation cascade (30, 42), in 293T cells and with the lack of net suppressive effects on TNF-α-driven transcription of the HIV-1 LTR, we indeed observed that TRIM22 antiviral activities were independent of the tandem NF-κB binding element within the core enhancer region of the viral promoter.
Concerning the lack of inhibition of Tat-dependent viral transcription by TRIM22, it should be underscored that TRIM22 inhibition seems specific of the HIV-1 viral promoter and likely occurs at the level of HIV-1 transcriptional initiation rather than elongation. Nevertheless, we cannot exclude a potential involvement of Tat as a component of the catalytically inactive P-TEFb/7SK snRNP complex (18), recruited to the promoter most likely as a preassembled complex and through interactions with Sp1 or other basal transcription factors.
In contrast to Tat and the NF-κB-dependent pathways, P+I-induced signaling was found to be sensitive to the inhibitory effects of TRIM22. In this regard, P+I can trigger signal pathways leading to activation of a broad array of transcription factors, including AP-1 and NFAT (13), that, together with constitutively expressed Sp1, play a crucial role in the early triggering of HIV-1 transcription (36, 58, 67, 78, 80). Interestingly, P+I-mediated activation of the viral LTR has been reported to strongly depend on an intact RBEIII region at position −120 of the viral promoter, harboring the binding site for RBF-2 (USF1/2-TFII-I) (41).
Among other features, TRIM22 possesses an E3 ubiquitin ligase domain. E3 ubiquitin ligases are involved both in nonlysosomal protein degradation and in many facets of cell biology unrelated to proteolysis, including transcription (5, 61, 75). In general, polyubiquitination of proteins results in proteasome-mediated protein degradation, whereas mono-ubiquitination may mediate other regulatory functions (5). Recently, TRIM22 E3 ubiquitin ligase activity has been shown to mediate TRIM22 antiviral activity against EMCV (21) and HBV (24). Furthermore, a mutant of TRIM22 lacking the ubiquitin ligase activity failed to interfere with HIV particle release (3). In their report, Barr et al. demonstrated that the effect of TRIM22 was abolished by mutation of functional amino acids (Cys15 and Cys18) of its RING finger domain, suggesting that functional ubiquitin ligase activity is also required for TRIM22-mediated antiviral activity against HIV-1. In contrast, we have demonstrated here that TRIM22-mediated inhibition of HIV-1 LTR-driven gene expression is independent of its E3 ubiquitin ligase activity since the functionally inactive TRIM22 mutant is still able to strongly inhibit Luc expression in 293T cells upon P+I stimulation. Thus, it is possible that this enzymatic function could play a role when TRIM22 has a prevalent cytoplasmic rather than nuclear distribution.
Besides the N-terminal RING domain, other distinct protein motifs characterize TRIM22 and other closely related members. In particular, the C-terminally located B30.2 domain, widely recognized as a major player in TRIM5α-mediated retroviral restriction since it harbors the retroviral capsid-binding motifs (52, 66, 79), has recently been involved in the cellular localization of TRIM22 (63). In addition, the B-box and Coiled-Coil domains of several TRIM proteins have also been described to mediate homo- and heteromultimerization (17), suggesting that TRIM22 might interact with other related TRIMs to mediate some aspect of its antiretroviral action. Ongoing studies aimed at identifying which TRIM22 domains are required for its anti-HIV-1 effect will bring further insight to the molecular mechanisms involved in this restriction.
In conclusion, we have provided evidence that TRIM22 is an endogenous inhibitor of HIV-1 transcription significantly influencing viral replication in an isogenic model of permissive and nonpermissive U937 promonocytic cells. Importantly, we also demonstrate that these antiviral effects are not confined to the U937 cellular context, since TRIM22 expression leads to impaired HIV-1 transcription and replication also in a human T cell line (A3.01) and in kidney epithelial cells (293T), highlighting its antiviral potential in relevant targets of HIV-1 infection. Once its physiological expression in relevant primary target cells for HIV infection is defined, as recently shown for primary human peripheral blood mononuclear cells (PBMC) (54) and as previously investigated at the RNA level in primary human monocyte-derived macrophages and PBMC (12), TRIM22 could represent a potential target for pharmacological approaches aimed at reactivating latent HIV infection in order to expose these cells to immunological surveillance and curtailing the viral reservoirs unaffected by current antiretroviral therapy. Furthermore, a recent report has shown that TRIM22 expression, but not huTRIM5α expression, in the PBMC of HIV-1 recent seroconverters positively correlated with type I IFN and CD4+ T cell counts, whereas a negative correlation was shown between TRIM22 expression and plasma viral load, emphasizing the potential role of TRIM22 as an antiviral response molecule in vivo (62). Thus, TRIM22 could be harnessed for antiviral control in the critical acute phase of HIV infection and used to counteract disease progression in infected individuals.
ACKNOWLEDGMENTS
This study was supported in part by grant 2008-2230 from Fondazione CARIPLO, Milan, Italy; the Fondation Dormeur; and SNF grant 3100A0-128655 from the Swiss National Science Foundation.
We declare that no competing interests exist.
We thank L. Galli for useful discussion on statistical analysis.
Footnotes
Published ahead of print on 23 February 2011.
REFERENCES
- 1. Alfano M., Rizzi C., Poli G., Biswas P. 2007. HIV-1 Infection of promonocytic U937 and U1 cell lines: clonal distribution of innate restriction factors, p. 89–114 In Herbein G. (ed.), Macrophage and HIV infection. Transworld Research Network, Kerala, India [Google Scholar]
- 2. Asjo B., et al. 1987. Susceptibility to infection by the human immunodeficiency virus (HIV) correlates with T4 expression in a parental monocytoid cell line and its subclones. Virology 157:359–365 [DOI] [PubMed] [Google Scholar]
- 3. Barr S. D., Smiley J. R., Bushman F. D. 2008. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS Pathog. 4:e1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bates D. L., et al. 2008. Crystal structure of NFAT bound to the HIV-1 LTR tandem κB enhancer element. Structure 16:684–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ben-Neriah Y. 2002. Regulatory functions of ubiquitination in the immune system. Nat. Immunol. 3:20–26 [DOI] [PubMed] [Google Scholar]
- 6. Biswas P., et al. 2001. Tumor necrosis factor-alpha drives HIV-1 replication in U937 cell clones and upregulates CXCR4. Cytokine 13:55–59 [DOI] [PubMed] [Google Scholar]
- 7. Biswas P., et al. 1998. 1,25-Dihydroxyvitamin D3 upregulates functional CXCR4 human immunodeficiency virus type 1 coreceptors in U937 minus clones: NF-κB-independent enhancement of viral replication. J. Virol. 72:8380–8383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bouazzaoui A., et al. 2006. Stimulated trans-acting factor of 50 kDa (Staf50) inhibits HIV-1 replication in human monocyte-derived macrophages. Virology 356:79–94 [DOI] [PubMed] [Google Scholar]
- 9. Boulerice F., et al. 1992. Differential susceptibilities of U-937 cell clones to infection by human immunodeficiency virus type 1. J. Virol. 66:1183–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bovolenta C., et al. 1999. Constitutive activation of STATs upon in vivo human immunodeficiency virus infection. Blood 94:4202–4209 [PubMed] [Google Scholar]
- 11. Bovolenta C., et al. 1999. A selective defect of IFN-gamma- but not of IFN-alpha-induced JAK/STAT pathway in a subset of U937 clones prevents the antiretroviral effect of IFN-gamma against HIV-1. J. Immunol. 162:323–330 [PubMed] [Google Scholar]
- 12. Carthagena L., et al. 2009. Human TRIM gene expression in response to interferons. PLoS One 4:e4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen J., Malcolm T., Estable M. C., Roeder R. G., Sadowski I. 2005. TFII-I regulates induction of chromosomally integrated human immunodeficiency virus type 1 long terminal repeat in cooperation with USF. J. Virol. 79:4396–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Colin L., Van Lint C. 2009. Molecular control of HIV-1 postintegration latency: implications for the development of new therapeutic strategies. Retrovirology 6:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crotti A., et al. 2007. Naturally occurring C-terminally truncated STAT5 is a negative regulator of HIV-1 expression. Blood 109:5380–5389 [DOI] [PubMed] [Google Scholar]
- 16. Demaison C., et al. 2002. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum. Gene Ther. 13:803–813 [DOI] [PubMed] [Google Scholar]
- 17. Diaz-Griffero F., et al. 2009. A B-box 2 surface patch important for TRIM5α self-association, capsid binding avidity, and retrovirus restriction. J. Virol. 83:10737–10751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. D'Orso I., Frankel A. D. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat. Struct. Mol. Biol. 17:815–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duan Z., Gao B., Xu W., Xiong S. 2008. Identification of TRIM22 as a RING finger E3 ubiquitin ligase. Biochem. Biophys. Res. Commun. 374:502–506 [DOI] [PubMed] [Google Scholar]
- 20. Duh E. J., Maury W. J., Folks T. M., Fauci A. S., Rabson A. B. 1989. Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-κB sites in the long terminal repeat. Proc. Natl. Acad. Sci. U. S. A. 86:5974–5978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eldin P., et al. 2009. TRIM22 E3 ubiquitin ligase activity is required to mediate antiviral activity against encephalomyocarditis virus. J. Gen. Virol. 90:536–545 [DOI] [PubMed] [Google Scholar]
- 22. Folks T., et al. 1985. Characterization of a continuous T-cell line susceptible to the cytopathic effects of the acquired immunodeficiency syndrome (AIDS)-associated retrovirus. Proc. Natl. Acad. Sci. U. S. A. 82:4539–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Franzoso G., et al. 1994. A family of serine proteases expressed exclusively in myelo-monocytic cells specifically processes the nuclear factor-κB subunit p65 in vitro and may impair human immunodeficiency virus replication in these cells. J. Exp. Med. 180:1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gao B., Duan Z., Xu W., Xiong S. 2009. Tripartite motif-containing 22 inhibits the activity of hepatitis B virus core promoter, which is dependent on nuclear-located RING domain. Hepatology 50:424–433 [DOI] [PubMed] [Google Scholar]
- 25. Gazzolo L., Mace K. 1990. Regulation of HIV1 replication in promonocytic U937 cells. Res. Virol. 141:259–265 [DOI] [PubMed] [Google Scholar]
- 26. Gongora C., Tissot C., Cerdan C., Mechti N. 2000. The interferon-inducible Staf50 gene is downregulated during T cell costimulation by CD2 and CD28. J. Interferon Cytokine Res. 20:955–961 [DOI] [PubMed] [Google Scholar]
- 27. He J., et al. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705–6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Herr A. M., Dressel R., Walter L. 2009. Different subcellular localizations of TRIM22 suggest species-specific function. Immunogenetics 61:271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hong S. J., Chae H., Lardaro T., Hong S., Kim K. S. 2008. Trim11 increases expression of dopamine beta-hydroxylase gene by interacting with Phox2b. Biochem. Biophys. Res. Commun. 368:650–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hummel M., et al. 2001. Allogeneic transplantation induces expression of cytomegalovirus immediate-early genes in vivo: a model for reactivation from latency. J. Virol. 75:4814–4822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huthoff H., Malim M. H. 2005. Cytidine deamination and resistance to retroviral infection: toward a structural understanding of the APOBEC proteins. Virology 334:147–153 [DOI] [PubMed] [Google Scholar]
- 32. Kajaste-Rudnitski A., et al. 2006. The 2′,5′-oligoadenylate synthetase 1b is a potent inhibitor of West Nile virus replication inside infected cells. J. Biol. Chem. 281:4624–4637 [DOI] [PubMed] [Google Scholar]
- 33. Kajaste-Rudnitski A., et al. Restriction factors of retroviral replication: the example of tripartite motif (TRIM) protein 5α and 22. Amino Acids 39:1–9 [DOI] [PubMed] [Google Scholar]
- 34. Kawai S., et al. 2008. Interferon-alpha enhances CD317 expression and the antitumor activity of anti-CD317 monoclonal antibody in renal cell carcinoma xenograft models. Cancer Sci. 99:2461–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kinoshita S., et al. 1997. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity 6:235–244 [DOI] [PubMed] [Google Scholar]
- 36. Kinter A. L., et al. 2001. Interleukin-6 and glucocorticoids synergistically induce human immunodeficiency virus type-1 expression in chronically infected U1 cells by a long terminal repeat independent posttranscriptional mechanism. Mol. Med. 7:668–678 [PMC free article] [PubMed] [Google Scholar]
- 37. Li X., et al. 2007. Unique features of TRIM5α among closely related human TRIM family members. Virology 360:419–433 [DOI] [PubMed] [Google Scholar]
- 38. Lois C., Hong E. J., Pease S., Brown E. J., Baltimore D. 2002. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295:868–872 [DOI] [PubMed] [Google Scholar]
- 39. Luznik L., Kraus G., Guatelli J., Richman D., Wong-Staal F. 1995. Tat-independent replication of human immunodeficiency viruses. J. Clin. Invest. 95:328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mace K., Duc Dodon M., Gazzolo L. 1989. Restriction of HIV-1 replication in promonocytic cells: a role for IFN-α. Virology 168:399–405 [DOI] [PubMed] [Google Scholar]
- 41. Malcolm T., Chen J., Chang C., Sadowski I. 2007. Induction of chromosomally integrated HIV-1 LTR requires RBF-2 (USF/TFII-I) and Ras/MAPK signaling. Virus Genes 35:215–223 [DOI] [PubMed] [Google Scholar]
- 42. Meier J. L., Stinski M. F. 1996. Regulation of human cytomegalovirus immediate-early gene expression. Intervirology 39:331–342 [DOI] [PubMed] [Google Scholar]
- 43. Meroni G., Diez-Roux G. 2005. TRIM/RBCC, a novel class of “single protein RING finger” E3 ubiquitin ligases. Bioessays 27:1147–1157 [DOI] [PubMed] [Google Scholar]
- 44. Moriuchi H., Moriuchi M., Arthos J., Hoxie J., Fauci A. S. 1997. Promonocytic U937 subclones expressing CD4 and CXCR4 are resistant to infection with and cell-to-cell fusion by T-cell-tropic human immunodeficiency virus type 1. J. Virol. 71:9664–9671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nabel G., Baltimore D. 1987. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326:711–713 [DOI] [PubMed] [Google Scholar]
- 46. Neagu M. R., et al. 2009. Potent inhibition of HIV-1 by TRIM5-cyclophilin fusion proteins engineered from human components. J. Clin. Invest. 119:3035–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Neil S. J., Sandrin V., Sundquist W. I., Bieniasz P. D. 2007. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe 2:193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Neil S. J., Zang T., Bieniasz P. D. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430 [DOI] [PubMed] [Google Scholar]
- 49. Nevado J., Tenbaum S. P., Castillo A. I., Sanchez-Pacheco A., Aranda A. 2007. Activation of the human immunodeficiency virus type I long terminal repeat by 1 α,25-dihydroxyvitamin D3. J. Mol. Endocrinol. 38:587–601 [DOI] [PubMed] [Google Scholar]
- 50. Nisole S., Stoye J. P., Saib A. 2005. TRIM family proteins: retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 3:799–808 [DOI] [PubMed] [Google Scholar]
- 51. Pereira L. A., Bentley K., Peeters A., Churchill M. J., Deacon N. J. 2000. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 28:663–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Perez-Caballero D., Hatziioannou T., Yang A., Cowan S., Bieniasz P. D. 2005. Human tripartite motif 5alpha domains responsible for retrovirus restriction activity and specificity. J. Virol. 79:8969–8978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Perez-Caballero D., et al. 2009. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139:499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Petersson J., et al. The human IFN-inducible p53 target gene TRIM22 colocalizes with the centrosome independently of cell cycle phase. Exp. Cell Res. 316:568–579 [DOI] [PubMed] [Google Scholar]
- 55. Poli G., Fauci A. S. 1992. The effect of cytokines and pharmacologic agents on chronic HIV infection. AIDS Res. Hum. Retrovir. 8:191–197 [DOI] [PubMed] [Google Scholar]
- 56. Poli G., Orenstein J. M., Kinter A., Folks T. M., Fauci A. S. 1989. Interferon-alpha but not AZT suppresses HIV expression in chronically infected cell lines. Science 244:575–577 [DOI] [PubMed] [Google Scholar]
- 57. Reymond A., et al. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rizzi C., et al. 2006. Pertussis toxin B-oligomer suppresses IL-6 induced HIV-1 and chemokine expression in chronically infected U1 cells via inhibition of activator protein 1. J. Immunol. 176:999–1006 [DOI] [PubMed] [Google Scholar]
- 59. Sawyer S. L., Emerman M., Malik H. S. 2007. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Pathog. 3:e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sewram S., et al. 2009. Human TRIM5α expression levels and reduced susceptibility to HIV-1 infection. J. Infect. Dis. 199:1657–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sigismund S., Polo S., Di Fiore P. P. 2004. Signaling through monoubiquitination. Curr. Top. Microbiol. Immunol. 286:149–185 [DOI] [PubMed] [Google Scholar]
- 62. Singh R., et al. 2011. Association of TRIM22 with type 1 interferon response and viral control during primary HIV-1 infection. J. Virol. 85:208–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sivaramakrishnan G., Sun Y., Rajmohan R., Lin V. C. 2009. B30.2/SPRY domain in tripartite motif-containing 22 is essential for the formation of distinct nuclear bodies. FEBS Lett. 583:2093–2099 [DOI] [PubMed] [Google Scholar]
- 64. Sivaramakrishnan G., Sun Y., Tan S. K., Lin V. C. 2009. Dynamic localization of tripartite motif-containing 22 in nuclear and nucleolar bodies. Exp. Cell Res. 315:1521–1532 [DOI] [PubMed] [Google Scholar]
- 65. Strebel K., Luban J., Jeang K. T. 2009. Human cellular restriction factors that target HIV-1 replication. BMC Med. 7:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stremlau M., Perron M., Welikala S., Sodroski J. 2005. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J. Virol. 79:3139–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Takada N., et al. 2002. RelA-associated inhibitor blocks transcription of human immunodeficiency virus type 1 by inhibiting NF-κB and Sp1 actions. J. Virol. 76:8019–8030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tan W., Dong Z., Wilkinson T. A., Barbas III C. F., Chow S. A. 2006. Human immunodeficiency virus type 1 incorporated with fusion proteins consisting of integrase and the designed polydactyl zinc finger protein E2C can bias integration of viral DNA into a predetermined chromosomal region in human cells. J. Virol. 80:1939–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Thielen B. K., et al. 2010. Innate immune signaling induces high levels of TC-specific deaminase activity in primary monocyte-derived cells through expression of APOBEC3A isoforms. J. Biol. Chem. 285:7753–7766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tissot C., Mechti N. 1995. Molecular cloning of a new interferon-induced factor that represses human immunodeficiency virus type 1 long terminal repeat expression. J. Biol. Chem. 270:14891–14898 [DOI] [PubMed] [Google Scholar]
- 71. Tokarev A., Skasko M., Fitzpatrick K., Guatelli J. 2009. Antiviral activity of the interferon-induced cellular protein BST-2/tetherin. AIDS Res. Hum. Retrovir. 25:1197–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tong-Starksen S. E., Luciw P. A., Peterlin B. M. 1987. Human immunodeficiency virus long terminal repeat responds to T-cell activation signals. Proc. Natl. Acad. Sci. U. S. A. 84:6845–6849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Vicenzi E., et al. 1999. Envelope-dependent restriction of human immunodeficiency virus type 1 spreading in CD4+ T lymphocytes: R5 but not X4 viruses replicate in the absence of T-cell receptor restimulation. J. Virol. 73:7515–7523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Vicenzi E., et al. 1994. An integration-defective U5 deletion mutant of human immunodeficiency virus type 1 reverts by eliminating additional long terminal repeat sequences. J. Virol. 68:7879–7890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Woelk T., Sigismund S., Penengo L., Polo S. 2007. The ubiquitination code: a signaling problem. Cell Div. 2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wolf D., Goff S. P. 2009. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458:1201–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yang K., et al. 2009. TRIM21 is essential to sustain IFN regulatory factor 3 activation during antiviral response. J. Immunol. 182:3782–3792 [DOI] [PubMed] [Google Scholar]
- 78. Yang X., Chen Y., Gabuzda D. 1999. ERK MAP kinase links cytokine signals to activation of latent HIV-1 infection by stimulating a cooperative interaction of AP-1 and NF-κB. J. Biol. Chem. 274:27981–27988 [DOI] [PubMed] [Google Scholar]
- 79. Yap M. W., Nisole S., Stoye J. P. 2005. A single amino acid change in the SPRY domain of human Trim5α leads to HIV-1 restriction. Curr. Biol. 15:73–78 [DOI] [PubMed] [Google Scholar]
- 80. Yedavalli V. S., Benkirane M., Jeang K. T. 2003. Tat and transactivation-responsive (TAR) RNA-independent induction of HIV-1 long terminal repeat by human and murine cyclin T1 requires Sp1. J. Biol. Chem. 278:6404–6410 [DOI] [PubMed] [Google Scholar]
- 81. Zhang F., Hatziioannou T., Perez-Caballero D., Derse D., Bieniasz P. D. 2006. Antiretroviral potential of human tripartite motif-5 and related proteins. Virology 353:396–409 [DOI] [PubMed] [Google Scholar]