Abstract
We report the discovery of mRNA 5′-leader trans-splicing (SL trans-splicing) in the chordates. In the ascidian protochordate Ciona intestinalis, the mRNAs of at least seven genes undergo trans-splicing of a 16-nucleotide 5′-leader apparently derived from a 46-nucleotide RNA that shares features with previously characterized splice donor SL RNAs. SL trans-splicing was known previously to occur in several protist and metazoan phyla, however, this is the first report of SL trans-splicing within the deuterostome division of the metazoa. SL trans-splicing is not known to occur in the vertebrates. However, because ascidians are primitive chordates related to vertebrate ancestors, our findings raise the possibility of ancestral SL trans-splicing in the vertebrate lineage.
Keywords: RNA splicing, SL trans-splicing, chordate/vertebrate evolution
mRNA 5′-leader trans-splicing is a mode of gene expression reported in several organisms in which the original 5′ ends of pre-mRNAs are discarded and are replaced by the 5′ segment of a spliced leader (SL) RNA (Bonen 1993; Blumenthal 1995; Davis 1996). The function of SL trans-splicing is not clear in every case and may vary. Multiple roles have been proposed, including mediation of mRNA stability or translatability (Maroney et al. 1995), resolution of polycistronic pre-mRNAs (Agabian 1990; Blumenthal 1995), and production of functional mRNAs from RNA polymerase I transcripts (Lee and Van der Ploeg 1997). In some organisms, only a subset of mRNAs undergo SL trans-splicing but in others, most or all do (Agabian 1990; Bonen 1993; Davis 1996). SL trans-splicing occurs alongside of the conventional cis-splicing process that removes introns from pre-mRNAs (Bonen 1993; Blumenthal 1995; Mair et al. 2000). There are mechanistic parallels between SL trans-splicing and conventional cis-splicing, including the use of the same set of nucleotide sequence features to mark splice donor and acceptor sites, and a strong resemblance of SL RNAs to spliceosomal U snRNAs (Agabian 1990; Bonen 1993; Nilsen 1993). These similarities imply a close evolutionary relationship between cis-splicing and SL trans-splicing, but the nature of this relationship and the overall evolutionary history of SL trans-splicing are not clear, in part because the phylogenetic distribution of SL trans-splicing has not been clearly delineated.
The known phylogenetic distribution of SL trans-splicing is uneven and includes several protist and metazoan groups (Bonen 1993; Blumenthal 1995; Davis 1996). It was first discovered in a protist group, the trypanosomes (Campbell et al. 1984; Kooter et al. 1984; Milhausen et al. 1984), then subsequently in two protosotome metazoan phyla, Nematoda (Krause and Hirsh 1987) and Platyhelminthes (flatworms) (Rajkovic et al. 1990) and in Euglena, a protist distantly related to trypanosomes (Tessier et al. 1991). SL trans-splicing has not been reported in advanced protostome phyla, that is, the arthropods, annelids, or molluscs, nor among the deuterostomes, the great division of the metazoa that includes chordates/vertebrates. However, because each discovery of SL trans-splicing was a fortuitous result of a detailed study of particular genes/mRNAs, and because extensive studies along these lines have been carried out in only a small number of organisms, the true phylogenetic range of SL trans-splicing is unknown.
Here we report the discovery of SL trans-splicing among the deuterostomes, in the ascidian Ciona intestinalis, a chordate. This finding considerably extends the known phylogenetic range of SL trans-splicing. Moreover, because ascidians are primitive chordates related to vertebrate ancestors (Berrill 1955; Katz 1983), SL trans-splicing in ascidians raises the possibility of ancestral SL trans-splicing in vertebrate evolution.
Results
A common 5′ sequence on multiple Ciona mRNAs
Evidence for trans-splicing in Ciona emerged during studies of a muscle gene encoding the contractile regulatory protein troponin I (TnI). We determined the complete sequence of body-wall muscle TnI mRNA by cDNA cloning (MacLean et al. 1997) and 5′–RACE analysis (see Materials and Methods). Surprisingly, database searching revealed that the first 16 nucleotides of the TnI mRNA sequence were similar or identical to the first 16 nucleotides of 3 other Ciona mRNAs (Fig. 1), including the CiMDFa mRNA encoding a MyoD-like muscle transcription factor (Meedel et al. 1997). The existence of a common sequence at the 5′-ends of diverse mRNA species is characteristic of SL trans-splicing (Bonen 1993; Blumenthal 1995; Davis 1996).
Figure 1.
A common 5′-sequence in several Ciona mRNAs. (A) A common 16-nucleotide sequence (bold) at the 5′-end of TnI mRNA, determined by 5′-RACE, and three additional Ciona mRNAs found by BLAST (Altschul et al. 1997) search of the GenBank database with the TnI 5′-sequence. The first 35 nucleotides of each mRNA are shown; dots indicate identity with the TnI mRNA sequence. Ci-zeta mRNA (GenBank AJ002142) obtained from ovary, encodes a proteasome subunit (Marino et al. 1999). CiMDFa mRNA (Genbank U80079) is expressed in larval tail muscle and adult body-wall muscle (Meedel et al. 1997). Cs-Endo-1 mRNA (GenBank AB024925) is a maternal mRNA from oocytes of Ciona savignyi (Imai et al. 1999). (B) RT–PCR amplification of TnI mRNAs with TnI-specific leftward priming using the SL primer for rightward priming. (Lane 1) Size markers (pBR322 HaeIII digest; top bands 587–434 bp, middle bands 267–184 bp, bottom bands ≤124 bp). (Lanes 2,4) Heart RNA template; (lanes 3,5) body-wall muscle RNA template; (lane 6) no RNA. Reverse transcriptase was omitted in lanes 4 and 5. Products of 381 bp (heart) and 240 bp (body-wall muscle) are expected; the size difference reflects tissue-specific alternative RNA splicing (MacLean et al. 1997).
Because the various mRNA 5′-end sequences in Figure 1 had been obtained by use of the same 5′–RACE technique, we confirmed the presence of the common 5′ sequence (henceforth called the SL sequence) by an independent method on the basis of RT–PCR amplification with a leftward gene-specific primer for TnI, coupled with a rightward primer containing the SL sequence (SL primer). Products of the expected distinct sizes were produced from alternatively spliced body-wall muscle and heart TnI mRNAs (Fig. 1B). Sequence analysis of the body-wall muscle product showed that the SL primer had primed rightward synthesis on TnI cDNA molecules immediately upstream of mRNA nucleotide 17, exactly as expected on the basis of 5′-RACE sequence data of Figure 1A (data not shown). Similar experiments with CiMDF(MyoD) also generated a body-wall muscle RT–PCR product of the predicted size that hybridized with a CiMDF probe (data not shown). These results confirmed independently the presence of the SL sequence established originally by 5′-RACE analysis.
To explore the possibility that additional Ciona mRNAs might contain the 5′ SL sequence, we designed an RT–PCR protocol to amplify any poly(A)-containing mRNA that also contained the 5′ SL sequence. Reverse transcription of the entire mRNA population was primed by oligo(dT) linked 3′ to an arbitrary anchor sequence, and subsequent amplification was based on leftward priming with the anchor sequence and rightward priming with the SL primer. Heterogeneous products ranging from ∼0.7 to 2.5 kb were produced from body-wall muscle and heart RNA, with prominent bands at 1.6, 1.0, and 0.8 kb among the body-wall muscle products (Fig. 2A) DNA from gel regions including the 1.0- and 0.8-kb bands was recovered and cloned. Sequence analysis of three randomly chosen insert-containing clones revealed three different and apparently complete mRNAs (GenBank AF237689–AF237691). In each case, the presence of poly(A) at one end, with an AATAAA poly(A) addition signal 20 bp upstream, served to identify the coding strand. In each case, the first ATG triplet on the coding strand was in a Kozak translation initiation consensus (Kozak 1991) context, CANNATG, and initiated an ORF of 190–250 codons encoding a protein related to previously characterized proteins (see legend to Fig. 2). As shown in Figure 2B, in each RT–PCR product, the SL primer sequence was found upstream of the ORF and separated from it by one or more in-frame stop codons. This finding establishes that the SL primer had primed rightward synthesis within the 5′-untranslated mRNA sequence — as expected for priming from the 5′-end.
Figure 2.
Additional mRNAs containing the SL sequence. (A) Amplification of multiple mRNAs by RT–PCR with SL and oligo(dT)-based primers. (Lane 1) Size markers [λ DNA, HindIII and EcoRI digest; in kilobases, from top to bottom, 21, 5.1/5.0, 4.2, 3.5, 2.0/1.9, 1.6, and (not visible) 1.4, 0.95, 0.83, and 0.5]. (Lanes 2,3) Body-wall muscle RNA template; (lanes 4,5) heart RNA template; (lane 6) no RNA. Reverse transcriptase was omitted in lanes 3 and 5. (B) 5′-untranslated sequences of three apparently complete mRNAs (GenBank AF237689–-AF237691) recovered by cloning DNA from the 1.0 and 0.8-kb bands in A, lane 2. The SL sequence is bolded; not shown is the BamHI site engineered at the 5′-end of the SL primer. The overlined ATG codons initiate ORFs of the lengths indicated, each terminated by a TAA stop codon and followed by an apparently complete 3′-untranslated sequence. In-frame stop codons within the 5′-untranslated sequences are underlined. The mRNA 1 ORF encodes a protein resembling HR-29 (Takagi et al. 1993), a myofibrillar protein from body wall muscle of the ascidian Halocynthia roretzi (46% identity over 207 aligned residues). The mRNA 2 ORF encodes a novel protein containing 17 PTDAVTL repeats resembling the mucin heptad [PTE(E/V)(P/T)TV] repeats of mammalian zonadhesins (Gao and Garbers 1998). The mRNA 3 ORF encodes a protein resembling vertebrate 27-kd heat-shock protein, hsp27 (Cooper and Uoshima 1994) (42% identity over 177 aligned residues).
Figures 1 and 2, together, show that at least seven Ciona mRNAs contain the 5′ SL sequence, and, given the heterodisperse nature of the RT–PCR products in Figure 2A, there may be more.
Gene structure consistent with SL trans-splicing
Analysis of TnI gene structure provided additional evidence consistent with SL trans-splicing. Upon isolation and sequence analysis of the Ciona TnI gene, we found exons accounting for the entire length of the TnI mRNA except for the first 16 nucleotides. Proceeding upstream from the TnI ATG start codon, the genomic DNA sequence aligned with the mRNA sequence until an AG dinucleotide immediately upstream of mRNA nucleotide 17, beyond which the sequences diverged sharply (Fig. 3). AG is the near-universal splice acceptor intron/exon boundary sequence for both cis- and trans-splicing (Bonen 1993; Blumenthal 1995; Davis 1996). Moreover, two additional features of vertebrate splice acceptor sites (Kramer 1996) were also present, that is, an A residue within the branchpoint consensus sequence YNCTRAY located 16 bp upstream of the AG dinucleotide, and an intervening pyrimidine-rich sequence. Thus, the gene structure is consistent with the possibility of trans-splicing of the SL sequence onto mRNA nucleotide 17. Alternatively, the SL sequence could represent a conventional cis-spliced exon located farther upstream. However, sequence analysis of 5429 bp of DNA upstream of the ATG start codon in one TnI allele (GenBank AF237979) and 2067 bp in another (GenBank AF237978) did not reveal the hypothetical 16-bp exon, nor any smaller microexon that might encode any 3′ part of the missing 16-nucleotide sequence.
Figure 3.
Comparison of TnI mRNA 5′-sequence and corresponding genomic DNA. The first 35 nucleotides of the mRNA are shown; the SL sequence is bolded and a vertical line marks its junction with the rest of the 5′-untranslated sequence. The sequences of two genomic DNA alleles, originating from Atlantic (A allele) or Pacific (P allele) coast animals, are shown. (The mRNA sequence derives from Atlantic coast animals.) Dots in the genomic DNA sequence show identity with the mRNA sequence (except at the left ends, where they signify additional upstream DNA). Differences between the A and P alleles are shown in lower case; a 4-base deletion in the P allele is shown by dashes. The AG dinucleotide present in the genomic DNA at the point of mRNA/genomic DNA sequence divergence is indicated by asterisks adjacent to the vertical line. A third asterisk marks the A residue in the branch point consensus sequence YRCTRAY.
The overall genomic DNA organization near the 5′-end of the CiMDF(MyoD) gene resembles that of the TnI gene (T.H. Meedel and J. Lee, unpubl.). Relevant similarities include (1) matching of genomic DNA and mRNA sequences upstream of the ATG translation start codon until an AG dinucleotide immediately upstream of mRNA nucleotide 18, and divergence of the sequences 5′ of this point [CiMDF nucleotide 17, numbered as in the cDNA sequence (Meedel et al. 1997), was absent from the genomic DNA sequence, presumably reflecting an allelic polymorphism], (2) presence of a near-consensus branchpoint sequence 16 bp upstream of the AG dinucleotide, and (3) absence of any upstream exon encoding all or part of the SL sequence within at least 1.4 kb of the ATG initiation codon. Thus, two genes known to produce mRNAs containing the 5′ SL sequence have structural features consistent with trans-splicing.
Promoter activity of DNA lacking the SL sequence
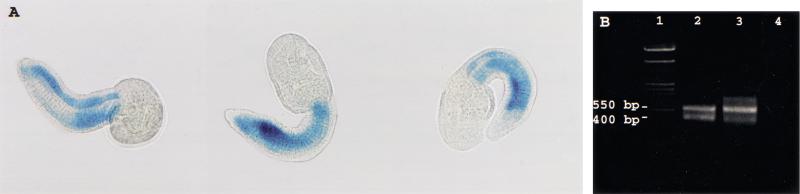
Although all of the foregoing data were consistent with trans-splicing, it remained possible that the SL sequence could be derived from cis-splicing of identical or near-identical far-upstream exons in the TnI and CiMDF genes and in the other genes that give rise to SL-ended mRNAs. However, we were able to eliminate this hypothesis by showing that far-upstream DNA is not required for muscle-specific expression of a TnI gene construct, nor for the generation of mRNA transcripts containing the SL sequence. We prepared a Ciona TnI β-galactosidase (β-gal) reporter gene construct, CiTnILacZ(−1.5), driven by 1454 bp of DNA upstream of the ATG initiation codon, in which promoterless β-gal-coding sequences were ligated to the 5′-untranslated sequence 23 nucleotides upstream of the ATG start codon (corresponds to nucleotide 58 of the TnI mRNA) (see Materials and Methods). It is important to note that this construct does not include any DNA encoding the SL sequence. Following electroporation (Corbo et al. 1997) into Ciona intestinalis zygotes, development was permitted for 12 h, at which time the embryos are at the tailbud stage and have elaborated specialized structures and cell types including the tail with its notochord and flanking rows of muscle cells. Embryos were fixed and stained to reveal β-gal expression. We found that the CiTnILacZ(−1.5) construct directed β-gal expression specifically in tail muscle cells (Fig. 4A) in a high percentage, >50%, of normally developing electroporated embryos. A control reporter construct, otherwise identical but lacking the Ciona genomic DNA segment, showed no detectable expression. Other studies have established that DNA constructs carrying diverse tissue-specific transcriptional control elements show appropriate cell-type-specific expression following electroporation into Ciona embryos (Corbo et al. 1997). These results indicate the presence of a functional muscle-specific promoter within the ∼1.5-kb cloned segment of the TnI gene, despite the absence of any sequence corresponding to the first 16 bases (the SL sequence) of the mature TnI mRNA. Effective muscle-specific expression was also obtained with a β-gal construct containing 1.4 kb of upstream DNA from the CiMDF gene, likewise lacking the SL sequence (T.H. Meedel and J. Lee, unpubl.). These results are consistent with trans-splicing of the SL sequence, but are not compatible with the hypothesis of cis-splicing of a far-upstream exon. Under a cis-splicing regime, DNA constructs that completely lack the first exon and upstream DNA of a gene would also lack the promoter (which is at the 5′-end of the first exon), yet both TnI and CiMDF genes contain functional muscle-specific promoters within DNA segments that lack the first exon (the SL sequence).
Figure 4.
In vivo expression and SL trans-splicing of a chimeric TnI/β-gal mRNA from a TnI/β-gal gene construct lacking the SL sequence. (A) Expression of β-gal, revealed by X-Gal staining (blue), in tail muscle of embryos 12 h following introduction of CiTnILacZ(−1.5) DNA into zygotes by electroporation. (B) RT–PCR amplification of β-gal mRNA with β-gal-specific leftward priming, and rightward priming with the SL primer. (Lane 1) Size markers (MBI Fermentas ladder mix, 10–0.1 kb; top visible band, 3 kb, bottom visible band 500 bp). (Lanes 2,3) RT–PCR products from two different batches of transfected embryos. The template in lane 4 was tRNA, used as a carrier in embryo RNA isolations. A 550-bp product, the size predicted for SL-ended β-gal mRNA, was produced from both embryo batches. [Production of this product required the presence of both primers and did not occur when CiTnILacZ(−1.5) plasmid DNA was used as the amplification template. An additional product of ∼400 bp seen in lanes 2 and 3 required only the β-gal-specific primer and was produced in control amplifications of CiTnILacZ(−1.5) plasmid DNA; it apparently results from rightward mis-priming by the β-gal-specific primer upstream of its normal leftward priming site.] (C) DNA sequence of 550-bp RT–PCR product. The 550-bp product (as in B) was recovered and sequenced using the SL primer (right) and β-gal-specific primer (left). The leftward sequence confirmed the presence of the SL primer (bold) immediately upstream of TnI mRNA nucleotide 17. Not shown is the BamHI site engineered at the 5′-end of the SL primer. Sequences deriving from the β-gal reporter gene are shown in lower case.
Trans-splicing of TnI/β-gal chimeric transcripts
Expression of the CiTnILacZ(−1.5) construct in vivo allowed us to show trans-splicing of an artificial mRNA substrate. Because it contains the TnI mRNA trans-splice acceptor site at nucleotide 17, the TnI/β-gal chimeric mRNA transcribed from the CiTnILacZ(−1.5) DNA construct could be subject to the same trans-splicing reaction as is the natural TnI mRNA. Therefore , we used an RT–PCR approach to ask whether β-gal mRNA molecules expressed in transfected embryos contained the SL sequence. RNA extracted from transfected embryos was reverse transcribed with a β-gal-specific primer and PCR amplification was carried out with the β-gal primer for leftward synthesis and the SL primer for rightward synthesis. A product of the expected size (550 bp) was obtained (Fig. 4B), and sequence analysis revealed exactly the structure predicted (Fig. 4C). β-gal sequences were found linked to the TnI 5′-untranslated sequence at nucleotide 58, and the SL primer was present in the sequence and located immediately upstream of TnI mRNA nucleotide 17. [In control PCR experiments, amplification of CiTnILacZ(−1.5) plasmid DNA with the same primers did not produce the 550-bp product, showing that the SL primer does not coincidentally misprime on the unrelated sequence present in the cloned TnI genomic DNA immediately upstream of nucleotide 17.] The presence of the SL sequence in mRNA transcribed from a gene construct that does not itself encode that sequence is a convincing demonstration of trans-splicing
A Ciona SL-likeRNA
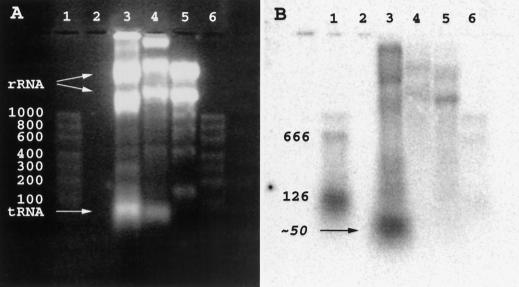
The occurrence of SL trans-splicing implies a splice donor SL RNA. To characterize Ciona SL RNA, we prepared body-wall muscle RNA by a procedure that did not involve salt precipitation, as this step in our standard RNA isolation procedure removes most small RNAs, and all known SL RNAs are small (<150 nucleotides). Northern blot analysis revealed a 50 ± 15 nucleotide RNA species, migrating ahead of the tRNA band, that hybridized with an oligonucleotide complementary to the SL sequence (Fig. 5, lane 3) No similar hybridization was seen in vertebrate (quail) muscle RNA prepared in a similar fashion (Fig. 5, lane 4), or in salt-precipitated Ciona body-wall muscle RNA (lane 5). In comparison with the hybridization signal generated by known amounts of SL-containing RNAs produced by in vitro transcription of cloned mRNA 3 (see Fig. 2), the abundance of the ∼50-nucleotide RNA was estimated to be on the order of 0.1% of the total RNA mass, representing a severalfold molar excess over the total mRNA population.
Figure 5.
Northern blot detection of Ciona SL RNA. (A) Fluorescence of ethidium bromide stained gel. (B) Autoradiography following transfer to nylon membrane and hybridization with a 5′-32P-labeled oligonucleotide complementary to the SL sequence. (Lanes 1,6) An RNA marker set (100–1000 nucleotide sizes indicated), to which has been added either 50 ng (lane 1) or 5 ng (lane 6) each of 126-nucleotide, and 666-nucleotide SL-containing in vitro transcripts of a plasmid encoding mRNA 3 (see Fig 2). (Lane 2) Blank; (lane 3) Ciona body-wall muscle RNA (not salt precipitated); (lane 4) quail muscle RNA (not salt precipitated); (lane 5) Ciona body-wall muscle RNA (salt precipitated). Large and small subunit rRNA and tRNA bands are indicated in A. Because the samples had not been salt precipitated, lanes 3 and 4 also contain genomic DNA (near sample wells).
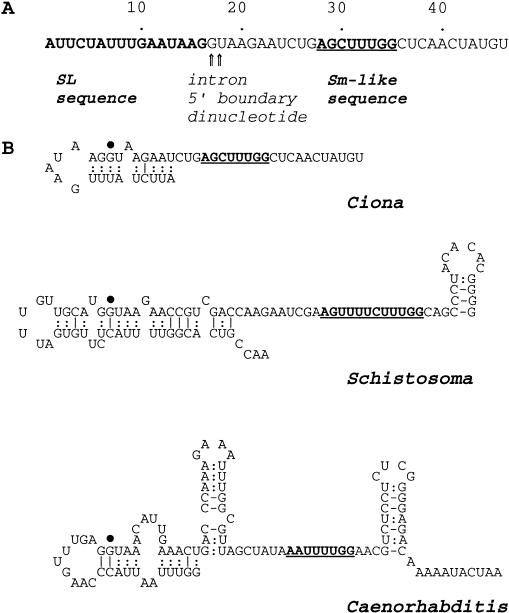
The candidate SL RNA was cloned in an approach based on in vitro polyadenylation with poly(A) polymerase (Tessier et al. 1991), followed by RT–PCR amplification using the same primers we used to amplify SL-containing mRNAs in Figure 2. A single abundant RT–PCR product of ∼100 bp was generated, and cloning and sequence analysis showed that the majority of the molecules (4/5) represented a single 46-nucleotide RNA species, consistent with the ∼50-nucleotide size of the candidate SL RNA (the polyadenylation/amplification process makes the PCR product 44 nucleotides longer than the original template RNA). The sequence (Fig. 6A) shows the principal feature expected of an SL RNA, that is, the presence of the intron 5′ boundary dinucleotide GU immediately following the SL sequence. The 46-nucleotide RNA does not show extensive similarity to known SL RNA sequences, however, previous studies have shown that SL RNAs from different phyla have little direct sequence similarity (Bruzik et al. 1988; Davis 1996). Nonetheless, SL RNAs from different phyla do share several features. In all known SL RNAs, the intron 5′ boundary GU dinucleotide is preceded by G and, except for the minor SL RNAs of Caenorhabditis (Ross et al. 1995), followed by A. Moreover, the exon/intron boundary is universally associated with a predicted secondary structure in which both G residues are involved in the downstream leg of a stem-loop structure near the 5′-end of the RNA (Bruzik et al. 1988; Rajkovic et al. 1990; Davis 1996). As shown in Figure 6, the Ciona 46-nucleotide RNA contains the conserved G/GUA boundary sequence (in a longer sequence G/GUAAGAA shared with the SL RNA of the flatworm Schistosoma), and is present in a predicted stem-loop structure similar to that predicted for other SL RNAs. An additional feature shared by the Ciona 46-nucleotide RNA and all known SL RNAs (Bruzik et al. 1988; Rajkovic et al. 1990; Davis 1996) is the presence within the intron moiety of a sequence resembling the consensus-binding site for Sm proteins (bold and underlined in Fig. 6).
Figure 6.
Sequence of Ciona SL RNA and predicted secondary structure comparison with SL RNAs of the flatworm Schistosoma and nematode Caenorhabditis. (A) Sequence of the 46-nucleotide Ciona SL RNA. 3′-poly(A), added before amplification and cloning, has been removed from the sequence; the original RNA may have contained one or more A residues at the 3′-end. The SL sequence is bolded and the Sm-like sequence is underlined and bolded [the canonical Sm consensus is RA(U)nGR]. Arrows mark the intron 5′-boundary GU dinucleotide. (B) Predicted secondary structures. The Ciona SL RNA structure was generated by mfold 3.0 (Mathews et al. 1999) and the Schistosoma and Caenorhabditis structures are from Davis (1996); in all structures the Sm-like sequence (bold, underlined) was constrained to be single stranded. In terms of ΔG, the Ciona structure shown was among the top two or three generated by calculations based on 22°C or 37°C and was within 1 kcal/mole of the optimal solution. Three-H-bond base pairs (i.e., G–C) are indicated by lines, and 2-H-bond base pairs (i.e., A–U and G–U) are indicated by paired dots. A single large dot marks the first G of the intron moiety.
Previously characterized SL RNAs are longer (90–140 nucleotides) than the Ciona 46-nucleotide RNA in both exon (22–51 vs. 16 nucleotides) and intron (54–109 vs. 30 nucleotides) moieties, and contain a predicted stem-loop structure downstream of the Sm sequence (Bruzik et al. 1988; Rajkovic et al. 1990; Davis 1996). In many, but not all cases, another stem-loop is present within the intron upstream of the Sm sequence (e.g., Caenorhabditis SL1 in Fig. 6B). The functional importance of the predicted intron stem-loops is not clear. Chemical modification studies of nematode SL RNA indicate a functional role in trans-splicing for purines in the upstream but not the downstream stem-loop (Hannon et al. 1992), however, the SL RNA of Schistosoma does not contain a similar upstream stem-loop (Rajkovic et al. 1990). Stem-loops may quantitatively facilitate Sm protein binding to U snRNAs (Jarmalowski and Mattaj 1993; Hinz et al. 1996) but they are not essential (Raker et al. 1999). As Figure 6 shows, the 46-nucleotide Ciona SL RNA is more like the relatively simple SL RNA of Schistosoma than the more complex classical SL RNA secondary structure represented by Caenorhabditis SL1.
Discussion
Our results establish the occurrence of mRNA 5′-leader (SL) trans-splicing in the ascidian chordate Ciona intestinalis and identify a probable SL RNA splice donor.
The discovery of SL trans-splicing in the deuterostomes, a major division of the metazoa, shows that SL trans-splicing has a considerably broader phylogenetic range than was known from previous studies (Bonen 1993; Davis 1996). This, and a recent report concerning the class Cestoda within the Platyhelminthes (Brehm et al. 2000), are the first reports to extend the known range of SL trans-splicing in almost a decade (Tessier et al. 1991).
Evidence for SL trans-splicing in Ciona
We found that at least seven different Ciona mRNAs, including TnI and CiMDF(MyoD) mRNAs, contain a short common sequence at the 5′-ends — a hallmark of SL trans-splicing. Moreover, the TnI and CiMDF(myoD) genes were found to have the two structural features expected of SL trans-spliced genes; (1) the SL sequence itself was not present in the gene, and (2) the mRNA sequence immediately adjacent to the SL sequence was found to correspond to the beginning of an exon, with the characteristic genomic features of a splice acceptor site. In addition, TnI and CiMDF(MyoD) β-gal reporter constructs showed effective muscle-specific expression in embryo transfection experiments, indicating the presence of functional tissue-specific promoters, despite the absence of the SL sequence. Moreover, we demonstrated the presence, in transfected embryos, of TnI/β-gal chimeric mRNAs that contain the SL sequence spliced onto the expected splice acceptor site in the TnI 5′-untranslated sequence. Finally, we identified a 46-nucleotide RNA in Ciona body-wall muscle having the expected properties of the predicted splice donor SL RNA.
Extent and role of ascidian SL trans-splicing
We do not know whether all, or only a subset, of Ciona genes/mRNAs are subject to SL trans-splicing; the seven trans-spliced mRNAs we have identified are a minimum number. These mRNAs are collectively expressed in a wide range of tissues and developmental stages. The biological role of SL trans-splicing in ascidians is currently unknown; proposals for other organisms include mediation of mRNA stability or translatability (Maroney et al. 1995), resolution of polycistronic pre-mRNAs (Agabian 1990; Blumenthal 1995), and production of functional mRNAs from RNA polymerase I transcripts (Lee and Van der Ploeg 1997).
Evolutionary implications
The occurrence of SL trans-splicing in both protists and metazoa suggests either that it is an ancestral eukaryotic character or that it has arisen independently in multiple organismal lineages (Bonen 1993). Our discovery of SL trans-splicing in a new division of the metazoa favors the ancestral-character hypothesis, because as the number of phyla known to carry out SL trans-splicing increases, the independent-origins hypothesis becomes increasingly less parsimonious. The shared features of known SL RNAs, including the Ciona 46-nucleotide RNA, are consistent with the possibility of descent from a common ancestral RNA, presumably an Sm-binding U snRNA (Bruzik et al. 1988), however, this would not preclude the possibility that the same or similar snRNAs might have independently acquired a trans-splicing role in different organismal lineages. Detailed studies of the trans-splicing mechanism in each group may be required to establish whether SL trans-splicing is an orthologous process throughout its phylogenetic range.
Because ascidians are thought to be related to vertebrate ancestors (Berrill 1955; Katz 1983), our findings raise the possibility that vertebrates could be descended from organisms that carried out SL trans-splicing. In extant vertebrates, SL trans-splicing is unknown, although other forms of trans-splicing have been reported, including the specific joining of particular pre-mRNAs (Li et al. 1999), and exon duplication by trans-splicing between two RNA molecules transcribed from the same gene (Caudevilla et al. 1998; Akopian et al. 1999). However, mammalian cells are able to trans-splice Caenorhabditis SL RNA (Bruzik and Maniatis 1992), which is consistent with the idea of vertebrate descent from SL trans-splicing ancestors, and also suggests the possibility of ongoing vertebrate SL trans-splicing that has not yet been discovered. Nonetheless, given the intense genetic research effort in vertebrates in comparison with ascidians, to have escaped detection implies that vertebrate SL trans-splicing likely occurs to a lesser extent, if at all.
Ascidian TnI and CiMDF(MyoD) genes, among others, are trans-spliced, but extensive studies of the vertebrate TnI and MyoD gene families, including detailed characterization of transcriptional start sites (Baldwin et al. 1985; Edmondson et al. 1992; Hinterberger et al. 1992; Tapscott et al. 1992; Ausoni et al. 1994; Corin et al. 1994) have revealed no SL trans-splicing, but only the conventional default mechanism of mRNA 5′-end formation. Thus, TnI and MyoD genes have either lost SL trans-splicing during vertebrate evolution, or acquired it during ascidian evolution. Functional studies indicate that, given the presence of SL RNA, it is a necessary and sufficient condition for SL trans-splicing that the target pre-mRNA contain a splice acceptor site upstream of the 5′-most splice donor site (Conrad et al. 1991, 1993). It follows that, during evolution, SL trans-splicing of a pre-mRNA could be lost either by mutational loss of the trans-splice acceptor site or by acquisition of a new splice donor site upstream of it in the transcript. The latter event would create a conventional cis-spliced intron within the gene's 5′-untranslated sequence. In view of their possible derivation from SL trans-spliced ancestral genes, it is of interest that vertebrate TnIfast and TnIslow genes contain such introns (Baldwin et al. 1985; Corin et al. 1994). Although other scenarios could also account for the evolution of 5′-untranslated introns, the active possibility of ancestral SL trans-splicing increases the range of evolutionary hypotheses that can be considered in this, and perhaps other aspects of vertebrate gene structure/organization.
Materials and methods
Animals and DNA/RNA preparation
All nucleic acid materials were derived from animals collected at the Sandwich Marina (Cape Cod, MA) except for a λ phage genomic DNA library kindly provided by R. Zeller (University of California, San Diego; see below), which was produced from animals collected in coastal southern California. Collection, maintenance of animals, and fertilization and embryonic development were as described (Meedel et al. 1997). Standard preparation of high molecular weight (salt precipitated) RNA, and of genomic (sperm) DNA were as described (Meedel and Hastings 1993). To prepare non-salt-precipitated RNA the salt-precipitation step was omitted.
5′-RACE analysis of TnI mRNA
The 5′-end of TnI mRNA was amplified from heart and body-wall muscle RNA by use of the Clontech AmpliFinder 5′RACE kit, with TCGGCAGAGATCCATGA and AGTGGATCCGC TGAGTGGCTCAAGTCGTTGGCT as the gene-specific leftward reverse transcription and amplification primers, respectively. This amplified 427-bp (from heart RNA) or 286-bp (from body-wall muscle RNA) products that were gel recovered and subjected to cycle sequencing. Both products had identical 5′-untranslated sequences, including the first 16 nucleotidess at the 5′-end.
RT–PCR analysis
Gene-specific leftward priming on TnI, CiMDF, and β-gal mRNAs was with GCTCAAGTCGTTGGCTTAG, ACTCAT TCCCGATCCGAAACTC, and AGTAACAACCCGTCGGAT TCTCC, respectively, and the SL primer GGATCCGATTC TATTTGAATAAG was used for rightward priming. For generic amplification of poly(A)+ mRNAs containing the SL sequence, reverse transcription was primed with an arbitrary T-tailed oligonucleotide GAATTCTACCTCAGAGGAGTCATATTTTTT TTTTTTT. Leftward PCR priming was done with the same sequence, but lacking 12 of the 13 T-residues at the 3′-end, and rightward priming was done with the SL primer. Gel bands were recovered, cloned in the pCR2.1 T/A vector (Invitrogen), and three randomly chosen clones were sequenced on both strands.
Cloning and analysis of TnI genomic DNA
Two approaches were used to isolate TnI genomic DNA upstream of that isolated previously (MacLean et al. 1997). Inverse PCR [IPCR, (Ochman et al. 1988)] using TnI gene-specific primers produced a 2.8-kb product containing 2067 bp of TnI DNA upstream of the ATG initiation codon (P. Pannunzio and K.E.M. Hastings, unpubl.). The IPCR product was subcloned and sequenced and its validity confirmed by genomic DNA amplification between the farthest upstream sequence and previously known TnI sequence downstream of the ATG start codon. Additional upstream DNA was obtained by hybridization screening of a λ ZAP Express (Stratagene) phage library (from Robert Zeller) of partial Sau3A-cut Ciona intestinalis DNA using the IPCR product as probe. Several TnI genomic clones were obtained, and one that included 5429 bp of DNA upstream of the ATG codon was completely sequenced on both strands. The genomic phage clone and IPCR product sequences clearly corresponded throughout their ∼2-kb overlap, although allelic differences, which are common in Ciona genes (Meedel and Hastings 1993; MacLean et al. 1997; Meedel et al. 1997) were evident [the IPCR and phage-cloned sequences derive from Atlantic (Cape Cod) and Pacific (southern California) coast animals, respectively].
Introduction of β-gal reporter constructs into embryos
CiTnILacZ(−1.5) contains TnI DNA from the subcloned IPCR product (see above) extending from a KpnI site to a Bpu1102I site, 1454 and 23 bp upstream of the ATG start codon, respectively. The blunted Bpu1102I site was ligated to the SmaI end of a 5-kb SmaI/EcoRI fragment from pRSVZ (MacGregor et al. 1987), which contains 7 bp of linker sequence, 38 bp of 5′-untranslated sequence, and the first 31 codons from Drosophila alcohol dehydrogenase mRNA, linked to Escherichia coli LacZ β-gal sequences and SV40 splicing and polyadenylation sites. Introduction of DNA constructs into Ciona intestinalis zygotes was by electroporation (Corbo et al. 1997) and following development at 18°C for 12 h embryos were fixed and stained with X-Gal. For RNA isolation, batches of electroporated embryos were examined and sorted at 12 h and pools of ∼25 normally developing embryos were frozen in 0.77 M mannitol with 20 ug of yeast tRNA. After thawing, RNA was recovered by phenol/chloroform extraction and ethanol precipitation and samples representing one-eighth of the total yield from a batch of embryos were reverse-transcribed with the β-gal-specific primer (see above) prior to PCR amplification with the β-gal primer and the SL primer.
Northern blot analysis
RNA samples separated by electrophoresis on 2% agarose gels (Thomas 1980) were vacuum-blotted to Zeta-Probe membrane (BioRad) and hybridized and washed at 37°C [as in Rajkovic et al. (1990)] with ACCTTATTCAAATAGAAT (complementary to SL sequence and adjacent intron 5′ boundary dinucleotide) labeled previously with T4 polynucleotide kinase and [γ-32P]ATP. Autoradiography was by PhosphorImaging. RNA markers were the MBI Fermentas low-molecular weight RNA marker set. SL-containing RNAs of 126 and 666 nucleotides were produced by T7 RNA polymerase transcription of plasmid containing the mRNA 3 insert (see Fig. 2) in pCR2.1, previously digested with Hin6I or HincII, respectively.
Sequence analysis of SL RNA
Non-salt-precipitated body-wall muscle RNA (3 μg) was 3′ polyadenylated with 3.5 units of poly(A) polymerase (Life Technologies) in 40-μl reactions containing 225 mM NaCl, 50 mM TrisHCl (pH 8), 10 mM MgCl2, 2.5 mM MnCl2, 0.25 mM ATP, and 0.5 mg/ml BSA for 30 min at 37°C. The polyadenylated RNA was subjected to RT–PCR as described above (see, RT–PCR analysis) and the single abundant product, ∼100 bp, was recovered following agarose gel electrophoresis and cloned into pCR2.1. Five clones were picked randomly and sequenced, and four corresponded to the 46-nucleotide SL RNA sequence shown in Figure 6; there were no base substitutions, although 3′-truncations of 2 (2 clones) and 4 nucleotides (1 clone) were observed. The fifth clone contained an unrelated sequence. Parallel analysis of five clones derived from trace amounts of similar-sized RT–PCR products made from RNA that had not been polyadenylated gave a heterogeneous collection of five different sequences.
Acknowledgments
We thank Robert Zeller for providing animals and a genomic DNA library, Pierre Pannunzio for his contribution to IPCR product isolation, and Jamie and Nancy Lee for their hospitality while T.H.M. was Visiting Scientist at the Mayo Clinic, Scottsdale, Arizona. K.E.M.H. is a Killam Scholar of the Montreal Neurological Institute. Research on muscle genes in the authors' laboratories was supported by grants from the Natural Sciences and Engineering Research Council of Canada, the Medical Research Council/Canadian Institutes of Health Research, and Rhode Island College.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL khastings@mni.mcgill.ca; FAX (514) 398-1509.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.865401.
References
- Agabian N. Trans splicing of nuclear pre-mRNAs. Cell. 1990;61:1157–1160. doi: 10.1016/0092-8674(90)90674-4. [DOI] [PubMed] [Google Scholar]
- Akopian AN, Okuse K, Souslova V, England S, Ogata N, Wood JN. Trans-splicing of a voltage-gated sodium channel is regulated by nerve growth factor. FEBS Lett. 1999;445:177–182. doi: 10.1016/s0014-5793(99)00126-x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausoni S, Campione M, Picard A, Moretti P, Vitadello M, De Nardi C, Schiaffino S. Structure and regulation of the mouse cardiac troponin I gene. J Biol Chem. 1994;269:339–346. [PubMed] [Google Scholar]
- Baldwin AS, Jr, Kittler EL, Emerson CP., Jr Structure, evolution, and regulation of a fast skeletal muscle troponin I gene. Proc Natl Acad Sci. 1985;82:8080–8084. doi: 10.1073/pnas.82.23.8080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrill NJ. The origin of vertebrates. Oxford, UK: Clarendon Press; 1955. [Google Scholar]
- Blumenthal T. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 1995;11:132–136. doi: 10.1016/s0168-9525(00)89026-5. [DOI] [PubMed] [Google Scholar]
- Bonen L. Trans-splicing of pre-mRNA in plants, animals, and protists. FASEB J. 1993;7:40–46. doi: 10.1096/fasebj.7.1.8422973. [DOI] [PubMed] [Google Scholar]
- Brehm K, Jensen K, Frosch M. mRNA trans-splicing in the human parasitic cestode Echinococcus multilocularis. J Biol Chem. 2000;275:38311–38318. doi: 10.1074/jbc.M006091200. [DOI] [PubMed] [Google Scholar]
- Bruzik JP, Maniatis T. Spliced leader RNAs from lower eukaryotes are trans-spliced in mammalian cells. Nature. 1992;360:692–695. doi: 10.1038/360692a0. [DOI] [PubMed] [Google Scholar]
- Bruzik JP, Van Doren K, Hirsh D, Steitz JA. Trans splicing involves a novel form of small nuclear ribonucleoprotein particles. Nature. 1988;335:559–562. doi: 10.1038/335559a0. [DOI] [PubMed] [Google Scholar]
- Campbell DA, Thornton DA, Boothroyd JC. Apparent discontinuous transcription of Trypanosoma brucei variant surface antigen genes. Nature. 1984;311:350–355. doi: 10.1038/311350a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudevilla C, Serra D, Miliar A, Codony C, Asins G, Bach M, Hegardt FG. Natural trans-splicing in carnitine octanoyltransferase pre-mRNAs in rat liver. Proc Natl Acad Sci. 1998;95:12185–12190. doi: 10.1073/pnas.95.21.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R, Thomas J, Spieth J, Blumenthal T. Insertion of part of an intron into the 5′ untranslated region of a Caenorhabditis elegans gene converts it into a trans-spliced gene. Mol Cell Biol. 1991;11:1921–1926. doi: 10.1128/mcb.11.4.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R, Liou RF, Blumenthal T. Conversion of a trans-spliced C. elegans gene into a conventional gene by introduction of a splice donor site. EMBO J. 1993;12:1249–1255. doi: 10.1002/j.1460-2075.1993.tb05766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper LF, Uoshima K. Differential estrogenic regulation of small M(r) heat shock protein expression in osteoblasts. J Biol Chem. 1994;269:7869–7873. [PubMed] [Google Scholar]
- Corbo JC, Levine M, Zeller RW. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development. 1997;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- Corin SJ, Juhasz O, Zhu L, Conley P, Kedes L, Wade R. Structure and expression of the human slow twitch skeletal muscle troponin I gene. J Biol Chem. 1994;269:10651–10659. [PubMed] [Google Scholar]
- Davis RE. Spliced leader RNA trans-splicing in metazoa. Parasitology Today. 1996;12:33–40. doi: 10.1016/0169-4758(96)80643-0. [DOI] [PubMed] [Google Scholar]
- Edmondson DG, Cheng TC, Cserjesi P, Chakraborty T, Olson EN. Analysis of the myogenin promoter reveals an indirect pathway for positive autoregulation mediated by the muscle-specific enhancer factor MEF-2. Mol Cell Biol. 1992;12:3665–3677. doi: 10.1128/mcb.12.9.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Garbers DL. Species diversity in the structure of zonadhesin, a sperm-specific membrane protein containing multiple cell adhesion molecule-like domains. J Biol Chem. 1998;273:3415–3421. doi: 10.1074/jbc.273.6.3415. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Maroney PA, Yu YT, Hannon GE, Nilsen TW. Interaction of U6 snRNA with a sequence required for function of the nematode SL RNA in trans-splicing. Science. 1992;258:1775–1780. doi: 10.1126/science.1465612. [DOI] [PubMed] [Google Scholar]
- Hinterberger TJ, Mays JL, Konieczny SF. Structure and myofiber-specific expression of the rat muscle regulatory gene MRF4. Gene. 1992;117:201–207. doi: 10.1016/0378-1119(92)90730-d. [DOI] [PubMed] [Google Scholar]
- Hinz M, Moore MJ, Bindereif A. Domain analysis of human U5 RNA: Cap trimethylation, protein binding, and spliceosome assembly. J Biol Chem. 1996;271:19001–19007. doi: 10.1074/jbc.271.31.19001. [DOI] [PubMed] [Google Scholar]
- Imai K, Satoh N, Satou Y. Identification and characterization of maternally expressed genes with mRNAs that are segregated with the endoplasm of early ascidian embryos. Int J Dev Biol. 1999;43:125–133. [PubMed] [Google Scholar]
- Jarmalowski A, Mattaj IW. The determinants of Sm protein binding to Xenopus U1 and U5 snRNAs are complex and non-identical. EMBO J. 1993;12:223–232. doi: 10.1002/j.1460-2075.1993.tb05648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz MJ. Comparative anatomy of the tunicate tadpole, Ciona intestinalis. Biological Bulletin. 1983;164:1–27. [Google Scholar]
- Kooter JM, De Lange T, Borst P. Discontinuous synthesis of mRNA in trypanosomes. EMBO J. 1984;3:2387–2392. doi: 10.1002/j.1460-2075.1984.tb02144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of vertebrate mRNA sequences: Intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- Krause M, Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Van der Ploeg LH. Transcription of protein-coding genes in trypanosomes by RNA polymerase I. Annu Rev Microbiol. 1997;51:463–489. doi: 10.1146/annurev.micro.51.1.463. [DOI] [PubMed] [Google Scholar]
- Li BL, Li XL, Duan ZJ, Lee O, Lin S, Ma ZM, Chang CC, Yang XY, Park JP, Mohandas TK, et al. Human acyl-CoA:cholesterol acyltransferase-1 (ACAT-1) gene organization and evidence that the 4.3-kilobase ACAT-1 mRNA is produced from two different chromosomes. J Biol Chem. 1999;274:11060–11071. doi: 10.1074/jbc.274.16.11060. [DOI] [PubMed] [Google Scholar]
- MacGregor GR, Mogg AE, Burke JF, Caskey CT. Histochemical staining of clonal mammalian cell lines expressing E. coli β galactosidase indicates heterogeneous expression of the bacterial gene. Somat Cell Mol Genet. 1987;13:253–265. doi: 10.1007/BF01535207. [DOI] [PubMed] [Google Scholar]
- MacLean DW, Meedel TH, Hastings KEM. Tissue-specific alternative splicing of ascidian troponin I isoforms. Redesign of a protein isoform-generating mechanism during chordate evolution. J Biol Chem. 1997;272:32115–32120. doi: 10.1074/jbc.272.51.32115. [DOI] [PubMed] [Google Scholar]
- Mair G, Shi H, Li H, Djikeng A, Aviles HO, Bishop JR, Falcone FH, Gavrilescu C, Montgomery JL, Santori MI, et al. A new twist in trypanosome RNA metabolism: cis-splicing of pre-mRNA. RNA. 2000;6:163–169. doi: 10.1017/s135583820099229x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino R, De Santis R, Giuliano P, Pinto MR. Follicle cell proteasome activity and acid extract from the egg vitelline coat prompt the onset of self-sterility in Ciona intestinalis oocytes. Proc Natl Acad Sci. 1999;96:9633–9636. doi: 10.1073/pnas.96.17.9633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney PA, Denker JA, Darzynkiewicz E, Laneve R, Nilsen TW. Most mRNAs in the nematode Ascaris lumbricoides are trans-spliced: A role for spliced leader addition in translational efficiency. RNA. 1995;1:714–723. [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Meedel TH, Hastings KEM. Striated muscle-type tropomyosin in a chordate smooth muscle, ascidian body-wall muscle. J Biol Chem. 1993;268:6755–6764. [PubMed] [Google Scholar]
- Meedel TH, Farmer SC, Lee JJ. The single MyoD family gene of Ciona intestinalis encodes two differentially expressed proteins: Implications for the evolution of chordate muscle gene regulation. Development. 1997;124:1711–1721. doi: 10.1242/dev.124.9.1711. [DOI] [PubMed] [Google Scholar]
- Milhausen M, Nelson RG, Sather S, Selkirk M, Agabian N. Identification of a small RNA containing the trypanosome spliced leader: A donor of shared 5′ sequences of trypanosomatid mRNAs? Cell. 1984;38:721–729. doi: 10.1016/0092-8674(84)90267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW. Trans-splicing of nematode premessenger RNA. Annu Rev Microbiol. 1993;47:413–440. doi: 10.1146/annurev.mi.47.100193.002213. [DOI] [PubMed] [Google Scholar]
- Ochman H, Gerber AS, Hartl DL. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkovic A, Davis RE, Simonsen JN, Rottman FM. A spliced leader is present on a subset of mRNAs from the human parasite Schistosoma mansoni. Proc Natl Acad Sci. 1990;87:8879–8883. doi: 10.1073/pnas.87.22.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raker VA, Hartmuth K, Kastner B, Lührmann R. Spliceosomal UsnRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol Cell Biol. 1999;19:6554–6565. doi: 10.1128/mcb.19.10.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross LH, Freedman JH, Rubin CS. Structure and expression of novel spliced leader RNA genes in Caenorhabditis elegans. J Biol Chem. 1995;270:22066–22075. doi: 10.1074/jbc.270.37.22066. [DOI] [PubMed] [Google Scholar]
- Takagi T, Yasunaga H, Nakamura A. Structure of 29-kDa protein from ascidian (Halocynthia roretzi) body wall muscle. J Biochem. 1993;113:321–326. doi: 10.1093/oxfordjournals.jbchem.a124046. [DOI] [PubMed] [Google Scholar]
- Tapscott SJ, Lassar AB, Weintraub H. A novel myoblast enhancer element mediates MyoD transcription. Mol Cell Biol. 1992;12:4994–5003. doi: 10.1128/mcb.12.11.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier LH, Keller M, Chan RL, Fournier R, Weil JH, Imbault P. Short leader sequences may be transferred from small RNAs to pre-mature mRNAs by trans-splicing in Euglena. EMBO J. 1991;10:2621–2625. doi: 10.1002/j.1460-2075.1991.tb07804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PS. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci. 1980;77:5201–5205. doi: 10.1073/pnas.77.9.5201. .. [DOI] [PMC free article] [PubMed] [Google Scholar]