Abstract
ICP0 is a transcriptional activating protein required for the efficient replication and reactivation of latent herpes simplex virus 1 (HSV-1). Multiple regions of ICP0 contribute its activity, the most prominent of which appears to be the RING finger, which confers E3 ubiquitin ligase activity. A region in the C terminus of ICP0 has also been implicated in several activities, including the disruption of a cellular repressor complex, REST/CoREST/HDAC1/2/LSD1. We used quiescent infection of MRC-5 cells with a virus that does not express immediate-early proteins, followed by superinfection with various viral mutants to quantify the ability of ICP0 variants to reactivate gene expression and alter chromatin structure. Superinfection with wild-type virus resulted in a 400-fold increase in expression from the previously quiescent d109 genome, the removal of heterochromatin and histones from the viral genome, and an increase in histone marks associated with activated transcription. RING finger mutants were unable to reactivate transcription or remove heterochromatin from d109, while mutants that are unable to bind CoREST activate gene expression from quiescent d109, albeit to a lesser degree than the wild-type virus. One such mutant, R8507, resulted in the partial removal of heterochromatin. Infection with R8507 did not result in the hyperacetylation of H3 and H4. The results demonstrate that (i) consistent with previous findings, the RING finger domain of ICP0 is required for the activation of quiescent genomes, (ii) the RF domain is also crucial for the ultimate removal of repressive chromatin, (iii) activities or interactions specified by the carboxy-terminal region of ICP0 significantly contribute to activation, and (iv) while the effects of the R8507 on chromatin are consistent with a role for REST/CoREST/HDAC1/2/LSD1 in the repression of quiescent genomes, the mutation may also affect other activities involved in derepression.
INTRODUCTION
Lytic replication of herpes simplex 1 (HSV-1) occurs in epithelial cells, followed by viral infection of the sensory neurons enervating the site of initial infection. The virus can establish latency in neurons, a state characterized by an almost complete lack of virus gene expression, with the exception of the latency-associated transcript (80, 81). Since latent HSV-1 DNA is not extensively methylated (53) and is found to be packaged in nucleosomes (12), it is likely that chromatin structure participates in repression of the genome and helps to control gene expression. Chromatin can modulate gene expression on a number of levels. Histones can physically impede the access of the transcriptional machinery to the DNA. Certain modifications of the histone tails can recruit repressive or activating proteins to promoters. For example, the repressive chromatin mark trimethylation of histone H3 lysine 9 (H3K9me3) has been shown to form a binding site for, and recruit, heterochromatin protein 1γ (HP1γ) (1, 54, 64).
As in latency in animal models, viral DNA was shown to be in a chromatin structure in a cell culture model of quiescence where incoming viral DNA is chromatinized in the absence of viral immediate-early (IE) proteins (33). Specifically, it was seen that in HEL cells, histones are deposited on the incoming genome by 4 h postinfection (hpi), and these histones begin to acquire trimethylation of histone H3K9. By 24 hpi, HP1γ becomes associated with viral promoters (23). This accumulation of heterochromatin is coincident with a decrease in gene expression and helps to explain the global lack of gene expression during viral latency and quiescence. It has been previously shown that latent viral genomes in vivo also contain heterochromatin (88), suggesting that the quiescent infection system mimics aspects of model latency in mice.
Periodically, latent viral DNA can reactivate and cause recurrent disease at the initial site of infection. Reactivation stimuli include various forms of cellular stress (12, 13, 56, 69, 89) and diminished immune function (34). The IE protein ICP0 is required for efficient reactivation from latency in vivo (5–7, 44, 45, 50). ICP0 is a promiscuous transactivator of gene expression (6, 7, 22, 35, 65, 70). ICP0 does not bind DNA but mediates its functions through interactions with a large number of cellular proteins, including those involved in cell cycle regulation (15, 87), transcription (15, 51), the double-stranded break repair mechanism (48, 68), and the interferon response (14, 63).
Many effects of ICP0 are possibly the result of degradation of cellular proteins through its RING finger domain (4, 19). The RING finger is a C3HC4 zinc binding region that coordinates two zinc ions and functions as an E3 ubiquitin ligase (2, 19, 23, 29). One of the major degradation targets of the RING finger domain are constituents of the ND10, or promyelocytic leukemia (PML), bodies (8, 26). Incoming viral genomes localize next to ND10 bodies, which are upregulated by interferon (24, 83), and are thought to play a role in the intrinsic cellular antiviral response (21). ICP0 colocalizes with, and causes the degradation of, the major ND10 components PML and Sp100 proteins (38, 66). The RING finger is also responsible for the degradation of the centromeric proteins CENP-A (58), CENP-B (57), and CENP-C (25). RING finger mutations that disrupt zinc binding eliminate the ability of ICP0 to degrade components of ND10 bodies, as well as other proteins, and reduce the transcriptional activation function of ICP0 (17, 55).
ICP0 has also been shown to interact with histone deacetylases in a RING finger-independent manner. ICP0 interacts with and causes the relocalization of class II histone deacetylase (HDAC) (59). C-terminal amino acids 537 to 613 of ICP0 have homology to CoREST, and amino acids 669 to 718 are involved in binding to CoREST. This allows ICP0 to interact with and disrupt the REST/CoREST/HDAC1/2/LSD1 repressor complex, leading to the relocalization of components of the complex (37, 39–41). When this region of ICP0 is mutated to abrogate CoREST binding, viral replication and degradation of PML are delayed but not completely eliminated (41). Some mutant forms of ICP0 truncated amino terminal to the CoREST homology region similarly show partial loss of activation function and partial loss of the ability to reactivate in explanted mouse ganglia (5).
Previously, we have shown that ICP0 expression is able to prevent the accumulation of histones, repressive histone modifications, and higher-order chromatin structure on the incoming viral genome, as well as cause the hyperacetylation of the few histones that do bind (33). We have also shown that provision of ICP0 can result in the removal of heterochromatin and histones H3 and H4 and result in the acetylation of remaining histones on genomes that have been repressed in relatively long-term quiescence (33a). In the present study, we have examined the relative contributions of the RING finger and CoREST regions of ICP0 to the reactivation of gene expression from quiescent HSV-1. This was correlated with the effects of provision of the individual ICP0 variants on the state of chromatin on the expressed genes.
MATERIALS AND METHODS
Cells and viruses.
Experiments were performed using MRC-5 (human embryonic lung) cells, Vero (African green monkey kidney) cells, and U2OS (human osteosarcoma) cells obtained from and propagated as recommended by the American Type Culture Collection. E11 cells were maintained as previously described (74). The viruses used in this study were the IE mutant d109 (74), wild-type HSV-1 (KOS), the ICP0-negative mutant n212 (7), the ICP0 RING finger point mutant (referred to as RF) (55), the CoREST binding mutant R8507 (41), and control mutant R8508 (41). The RF virus and the R8507 and R8508 viruses were gifts of Bernard Roizman (University of Chicago). d109 was propagated on F06 cells as described previously (74), and all other viruses were propagated on U2OS cells to minimize differences between the viral stocks.
The viruses RFBac and n518Bac were made in the following way. A KOS bacterial artificial chromosome (36) in Escherichia coli was obtained from David Leib (Dartmouth). It was transferred to E. coli GS1783 (from Greg Smith (Northwestern). The GS1783 strain contains chromosomal inducible Red recombination functions and the I-SceI restriction enzyme (85). These were used to create the intended mutants in E. coli exactly as described by Tischer et al. (85, 86). The oligonucleotides used to create the RF mutant were ACGGATGAGATCGCGCCCCACCTGCGCTGCGACACCTTCCCGCTGCAGATGAAAACCTGGATGCAATAGGGATAACAGGGTAATCGATTT and GCACAGCGGGCAGGTGTTGCGCAATTGCATCCAGGTTTTCATCTGCAGCGGGAAGGTGTCGCAGCGGCCAGTGTTACAACCAATTAACC. The RFBac mutant lacks amino acids 134 to 142 of ICP0, which are CMHRFCPC, the core of the RING finger. It also inserts 6 bp encoding a PstI site at the deleted locus for identification. The oligonucleotides used to create n518Bac were AAGAGGCGCGGGTCGGGCCAGGAAAACCCCTCCCCCCAGTCCGGCTAGTTAACTAGCCCCCCCCCTCGCGCCGGCATAGGGATAACAGGGTAATCGATTT and GTGCGTCGCCGCCCTCTTGGCCCCTGCCGGCGCGAGGGGGGGGGCTAGTTAACTAGCCGGACTGGGGGGAGGGGTTGCCAGTGTTACAACCAATTAACC. The n518Bac mutant truncates the ICP0 molecule at amino acid 518 by virtue of the insertion of translational stop codons in all three reading frames. The sequence encoding the stop codons also encodes an HpaI site, which was used for identification. The procedure of Tischer et al. (85, 86) results in the incorporation of only the intended mutations into one of the two copies of ICP0 in the KOSBac genome. Intact KOSBac DNA isolated from clones using the procedure of Birnboim and Doly (3) that was determined to contain the desired mutation by restriction enzyme analysis was used to transfect monolayers of U2OS cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Progeny from the transfection were plaque isolated and analyzed by restriction enzyme analysis for the presence of the mutations in both copies of the ICP0 gene. These isolates were plaque purified an additional 2 times on U2OS cells prior to the generation of stocks of the viruses by propagation in U2OS cells.
ChIP.
Chromatin immunoprecipitation (ChIP) was carried out as previously described (33, 76), with a few modifications. A total of 5 × 106 MRC-5 cells were seeded into 100-mm dishes (∼105 cells/cm2) and infected with d109 at a multiplicity of infection (MOI) of 10 PFU per cell at room temperature for 1 h. After adsorption, the inoculum was removed and 37°C 5% fetal bovine serum (FBS) in Dulbecco's modified Eagle's medium was added. Infected cells were maintained at 37°C for 24 h. At 24 hpi, the medium was replaced with fresh medium and infected cells were maintained at 34°C. On day 4 postinfection, the medium was again replaced with fresh medium. At day 7 postinfection, d109-infected cells were either mock superinfected or superinfected with various mutants at an MOI of 10 PFU per cell for 1 h at room temperature. After adsorption, the inoculum was aspirated and the conditioned medium (which was saved and maintained at 37°C) was replaced. This was considered time zero after superinfection. At 4 hpi, cells were treated with 1% formaldehyde for 10 min at 37°C, washed 3 times with cold phosphate-buffered saline (PBS) containing protease inhibitors (67 ng/ml aprotinin, 1 ng pepstatin, 0.16 mM TLCK [Nα-p-tosyl-l-lysine chloromethyl ketone], 1 mM phenylmethylsulfonyl fluoride [PMSF]), and scraped into PBS containing protease inhibitors. The cells were pelleted at 3,000 rpm for 10 min at 4°C, resuspended in cold sodium dodecyl sulfate (SDS) lysis buffer (100 μl per million cells) containing protease inhibitors (1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1], 4 μg/ml aprotinin, 2 μg/ml pepstatin, 0.15 mM TLCK, 0.6 mM PMSF), and incubated on ice for 30 min. The antibodies used were anti-histone H3 (Abcam ab1791), anti-histone H4 (Millipore 05-858), anti-acetyl histone H3 (Millipore 06-599), anti-acetyl histone H4 (Millipore 06-866), anti-trimethyl histone H3 lysine 9 (Millipore 07-422), anti-dimethyl histone H3 lysine 4 (Millipore 07-030), and anti-heterochromatin protein 1γ (Millipore 05-690). A no-antibody control was included in each ChIP experiment, and the value for the no-antibody control was subtracted from the immunoprecipitation results before the percent input of immunoprecipitation was calculated.
RNA isolation and reverse transcription (RT).
RNA was isolated with the Ambion RNaqueous-4PCR kit by following the included protocol. Briefly, 5 × 106 MRC-5 cells in 100-mm plates were infected at an MOI of 10 PFU of d109 per cell for 1 week and superinfected as in the ChIP experiments. RNA was harvested at the indicated time points by adding 500 μl lysis/binding buffer. Cells were scraped and vortexed. An equal volume of 67% ethanol was added, and the solution was added to a filter, which was centrifuged at 12,500 rpm at 4°C for 1 min. The bound RNA was washed with wash buffers 1 and 2/3. RNA was eluted with 60 μl 65°C elution solution. The RNA was treated with DNase I at 37°C for 30 min to degrade any residual DNA.
RT was performed by using the Ambion RT kit and following the included instructions. Two micrograms of total RNA was reverse transcribed in a reaction volume of 20 μl containing RNase inhibitor, oligo(dT) primers, 1 μl Moloney murine leukemia virus reverse transcriptase, and 2 μl 10× reaction buffer. The reaction tube was incubated at 85°C for 3 min to remove RNA secondary structure, and the RT reaction was carried out for 1 h at 44°C. After the RT reaction was complete, the reaction tube was incubated at 95°C for 10 min in order to inactivate the reverse transcriptase. Eight microliters of cDNA was diluted 1:8 by adding 40 μl DNase/RNase-free H2O for use in quantitative PCRs (qPCRs). Additionally, 1 μg RNA was diluted in a total of 60 μl DNase/RNase-free H2O for use as a negative control in qPCRs.
qPCR.
Reactions for ChIP or cDNA quantification were performed in triplicate using 2.5 μl of DNA for each reaction as described previously (76), with a few modifications. Before the 96-well reaction plate was set up, a master mix containing 0.3125 μl of each primer (stock concentration, 1 mM), 6.25 μl Applied Biosystems SYBR green super mix with 1.0 μM 6-carboxy-X-rhodamine (Bio-Rad), and 3.125 μl of water for a total of 10 μl for each reaction was made. The final reaction volume was 12.5 μl, including the DNA. The primers used for ChIP and cDNA quantification and their locations relative to the transcription start site of the gene to be analyzed are as previously published (33). d106 DNA was also included in each plate, in a standard curve of 1:10 dilutions from 250,000 to 25 copies per well, which covers the threshold cycle values for the ChIP DNA samples tested. qPCR was run on a StepOne Plus real-time PCR machine. The conditions for the run were 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 1 min. At the end of the run, a dissociation curve was completed to determine the purity of the amplified products. Results were analyzed using the StepOne v2.1 software from Applied Biosystems.
SDS-PAGE analysis.
Cultures of MRC-5 cells for SDS-polyacrylamide gel electrophoresis (SDS-PAGE) analysis were prepared similarly to those described for RNA and ChIP analyses. Cells (106) were seeded into 35-mm petri dishes and incubated at 37°C for 24 h, after which time the medium was changed to 5% FBS and the cultures were placed at 34°C. The cells were incubated for an additional 7 days with a change of medium after 3 days. The medium was then removed from the cultures and saved. Monolayers were then infected with the indicated viruses at an MOI of 10 PFU/cell as described above. The conditioned medium was then added back to the cultures, which were further incubated for an additional 4 h at 37°C. The medium was then removed, and 50 μCi of l-[35S]methionine (1,175 Ci/mmol; MP Biologicals) was added in 1 ml Tris-buffered saline (TBS) and incubated for an additional 30 min at 37°C, after which time SDS-PAGE samples were prepared from each culture as previously described (11).
SDS peptides were electrophoretically separated on a 10% polyacrylamide Tris-HCl Criterion gel (Bio-Rad Laboratories) as prescribed by the manufacturer. The electrophoretically separated SDS peptides were transferred to a nitrocellulose membrane (Protran BA85; Whatman) by using a Criterion blotter (Bio-Rad Laboratories) as prescribed by the manufacturer. The nitrocellulose membrane was then incubated in 2.5% dry milk in TBS plus 0.1% Tween 20 (TBST) at 4°C overnight. The membrane was washed briefly with TBST and then incubated in a 1:1,000 dilution of ICP0 monoclonal antibody (1112) in 0.5% dry milk in TBST for 2 h at room temperature with shaking. The antibody was then removed, and the membrane was washed with three changes of TBST for 10 min each. The membrane was then incubated in a 1:10,000 dilution of goat anti-mouse IRDye 800 (LI-COR Inc.) for 1 h. The antibody was again removed, and the membrane was washed with three changes of TBST for 10 min each, followed by a 10-min wash in TBS. The membrane was then allowed to dry, after which time it was imaged with an Odyssey infrared imager (LI-COR Inc.) to visualize ICP0. The membrane was then exposed to X-ray film to visualize all of the infected cell polypeptides.
RESULTS
d109 genomes persisting at 1 week postinfection in human fibroblasts are highly repressed, and the presence of heterochromatin is observed. In a previous study (33a), we have shown that the expression of wild-type ICP0 leads to the reduction of heterochromatin on the quiescent d109 genome and, to various extents, chromatinization. This was accompanied by increased expression from the human cytomegalovirus major IE (HCMV) promoter driving the gene for green fluorescent protein (GFP) in d109, as well as the resident tk and gC genes. Much of the work concerning mutations in ICP0 has been focused on productive infection and therefore pertains to how the activity of ICP0 prevents genome repression. Less work has been done on the relative effects of different regions of ICP0 on gene expression and chromatin structure upon reactivation from a highly repressed state. Therefore, HSV mutants expressing specific ICP0 proteins were used to superinfect quiescently infected cells and this was followed by measurements of gene expression by RT-PCR and histone abundance by ChIP.
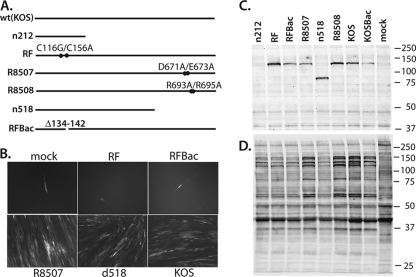
The following ICP0 mutants were used in this study (Fig. 1A). d99 has both copies of the entire ICP0 gene and promoter (74) deleted. n212 is a nonsense mutant that does not specify ICP0 function as measured by gene activation and virus growth (5). RF has multiple base mutations that change two of the metal-chelating Cys residues (C116G/C156A) in the zinc finger (55). RFBac is an independent RF mutant that lacks amino acids CMHRFCPC of the RING finger. R8507 has multiple base mutations resulting in the changes D671A/E673A, abrogating CoREST binding (41). R8508 is a mutant containing changes (R693A/R695A) that were previously shown not to affect ICP0 function (41). n518Bac is truncated at amino acid 518 by virtue of the insertion of nonsense codons in all reading frames. HSV-1 that does not express ICP0 displays a deficiency in plaque formation in noncomplementing cells. The requirement of ICP0 for growth is greatly reduced in U2OS cells (91). These cells are likely to be missing some pathway or protein required for the repression of incoming HSV-1 genomes (46). Additionally, L7 cells, which are Vero cells stably transfected with ICP0 (75), are able to complement ICP0-negative viruses. The stocks of viruses used in these experiments were grown on U2OS cells rather than L7 cells to eliminate the possibility that the wild-type ICP0 provided by L7 would be present in the virions of the mutant progeny and affect the outcome of the experiments. The titers of the stocks of d99, n212, RF, RFBac, R8507, n518Bac, R8508, KOS, and KOSBac grown on U2OS cells were determined by plaque assay on L7 cells, U2OS cells, and Vero cells (Table 1).
Fig. 1.
Viruses used in theses studies. Panel A shows a representation of the ICP0 alleles in the indicated mutants (wt, wild type). Some of these mutants were used to superinfect MRC-5 cells that were infected for 1 week with d109 (MOI = 10 PFU/cell). GFP expression was assessed 24 h later by fluorescence microcopy (B). One-week confluent cultures of MRC-5 cells were also infected with wild-type and ICP0 mutant viruses for 4 h and labeled with [35S]methionine, and SDS-PAGE samples were prepared as described in Materials and Methods. SDS peptides were electrophoretically separated, transferred to a nitrocellulose membrane, and analyzed by Western blotting for ICP0 (C). The membrane was then exposed to X-ray film for visualization of all of the infected cell polypeptides (D). The values to the right are molecular sizes in kilodaltons.
Table 1.
Titers of virus stocks used in this studya
| Virus | Titer (PFU/ml)b on: |
L7/Veroc | ||
|---|---|---|---|---|
| L7 cells | U2OS cells | Vero cells | ||
| d99 | 1.8 × 109 | 1.1 × 109 | 2.0 × 107 | 90 |
| n212 | 2.2 × 109 | 1.7 × 109 | 2.0 × 107 | 110 |
| RFBac | 6.7 × 108 | 5.7 × 108 | 1.2 × 107 | 56 |
| RF | 5.2 × 108 | 6.4 × 108 | 1.7 × 107 | 31 |
| n518Bac | 1.0 × 109 | 4.5 × 108 | 5.1 × 107 | 20 |
| R8507 | 8.3 × 108 | 9.0 × 108 | 2.9 × 108 | 2.9 |
| R8508 | 1.7 × 109 | 8.0 × 108 | 1.4 × 109 | 1.2 |
| KOSBac | 2.5 × 109 | 1.4 × 109 | 2.0 × 109 | 1.2 |
| KOS | 2.6 × 109 | 1.3 × 109 | 2.5 × 109 | 1.0 |
Viruses stocks were prepared from infected U2OS cells as described in Materials and Methods.
The titers of the indicated virus stocks were determined by plaque assay on the indicated cell lines.
The ratio of the titer on L7 cells to that on Vero cells.
Comparing the abilities of the various mutants to form plaques on L7 and Vero cells provides a relative measure of the defects caused by the various mutations in ICP0. n212, which behaves as a null mutant (d99), showed an approximately 100-fold defect in plaque formation on Vero cells relative to L7 cells. The RING finger mutants (RF and RFBac) displayed a 30- to 60-fold reduction in plating efficiency on Vero cells, while R8507 had a plaque-forming defect on Vero cells of slightly less than 3-fold. n518Bac was considerably more defective than R8507 but less so than the RING finger mutants. KOS, KOSBac, and R8508 formed plaques with essentially the same efficiency on both Vero and L7 cells, indicative of little to no defect in R8508. The following conclusions can be made from these observations. (i) The relative magnitude of the defects resulting from the RF and R8507 mutations is qualitatively consistent with previous observations (41). However, the absolute magnitude of the defects resulting from these mutations is not as great as previously seen (41). The reason for the quantitative difference is not clear; however, it is not due to making comparisons on U2OS cells, since the U2OS-to-Vero cell ratios are similar to the L7-to-Vero cell ratios. (ii) The RF mutants appear to be less defective than n212. This implies that the RF mutants may have residual activity independent of the ubiquitin ligase activity. Alternatively, the RF mutant protein may have a negative effect on virus growth in the presence of complementing activity. (iii) The region between amino acid 518 and the C terminus specifies interactions and activities that are important for activity in addition to that defective in R8507. Despite the above quantitative differences, these data are consistent with previously published studies showing that the while the carboxy-terminal region of ICP0 has consequences for ICP0 function and virus growth, the ubiquitin ligase activity specified by the RING finger is of greater consequence.
To further characterize the mutants used in this study, 1-week-old monolayers of MRC-5 cells were infected with the viruses at an MOI of 10 PFU/cell and labeled with [35S]methionine from 4.0 to 4.5 hpi. SDS-PAGE samples were prepared, and the extracts were subjected to SDS-PAGE and analyzed by Western blotting with a monoclonal antibody to ICP0 (Fig. 1C). After imaging of the filter for visualization of ICP0, it was exposed to X-ray film to visualize the infected cell polypeptides (Fig. 1D). All of the mutants tested specified the expected size ICP0 polypeptide (Fig. 1C). Antibody-reacting peptides in addition to those seen in mock-infected cells were not seen in n212-infected cells. Virus-specific polypeptides could be seen in all of the samples from the mutant virus-infected cells (Fig. 1D). The intensity of the virus-specific signal was somewhat less in more defective mutants, i.e., n212, RF, RFBac, and n518Bac. The polypeptide profiles of R8507 and R8508 were similar to those of KOS and KOSBac.
We next looked at the ability of the mutants to activate quiescent genomes. MRC-5 cells were infected with d109 and incubated in reduced serum for 1 week. The quiescent cultures were then infected with the mutants at an MOI of 10 PFU/cell. Twenty-four hours later, the cultures were visualized by GFP fluorescence. Figure 1B shows the results of mock, RF, BacRF, R8507, n518Bac, and KOS infections. A single fluorescent MRC-5 cell is shown in each of the images of mock-, RF-, and RFBac-infected cells. Consistent with previous observations (84), this was seen in fewer than 1 in 10 fields and corresponds to the d109 genomes that were not sufficiently repressed. R8507 and n518Bac activated the quiescent 109 genomes similarly to KOS, as inferred by the visual assessment of GFP expression from the HCMV IE promoter at 24 h after superinfection. These results are similar to those of a recent study that visually assessed the activation of lacZ from the HCMV IE promoter at 24 h postinduction of the gene for ICP0 (31). That study found that mutations in the C-terminal 1/3 of the ICP0 protein reduced activation by only 50%, at most, and concluded that this region is almost completely dispensable for derepression.
RNA expression from quiescent genomes upon reactivation by ICP0 mutants.
We have previously shown that gene expression from d109 in human fibroblasts decreases from 4 to 24 hpi and that this is accompanied by increased chromatinization of the genome and the formation of heterochromatin. HP1γ was not present on the d109 genomes at 4 hpi but was present at 24 h (33). Therefore, repression is established over time and the d109 genome exists in cells in different states with respect to gene expression and chromatinization. This affords the opportunity to determine the relative contributions of the different activities of ICP0 to the depression of d109 genomes at different stages of repression. Therefore, MRC-5 cells (human lung fibroblasts) were infected with d109 and subsequently mock superinfected or superinfected with the viruses in Table 1 at 7 days after d109 infection. The superinfected cultures were incubated for 4 h, and then RNA was harvested and analyzed by RT-PCR. Determining the level of induction at 4 h after superinfection by RT-PCR is more sensitive and quantitative than visual assessments made at 24 hpi. The previous study also demonstrated that the induction of the quiescent genomes begins as early as 2 hpi (33a).
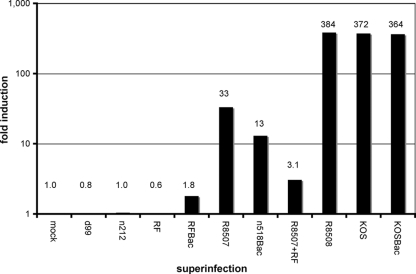
One week after d109 infection, the level of GFP mRNA was approximately 7 × 105 copies per μg of total RNA or about 7 molecules per cell (mock superinfection). The mRNA molecules are probably not uniform from cell to cell but rather concentrated in a small subpopulation of cells (Fig. 1B). The values for GFP mRNA for the remainder of the superinfections were normalized to the mock superinfection value (Fig. 2). Superinfection with R8508, KOS, or KOSBac resulted in a 364- to 384-fold increase in the level of GFP mRNA. Increases in GFP mRNA over the level in mock-superinfected cells were not observed in d99-, n212-, RF- and RFBac-infected cells. Superinfection with the R8507 and n518Bac mutants resulted in 33- and 13-fold increases in GFP mRNA, respectively. Therefore, while these mutations do not eliminate the ability of ICP0 to induce the quiescent GFP-encoding gene, they substantially reduce this activity, as measured by this assay. In addition, as the results of the plaque assay indicate (Table 1), the greater impairment seen with n518 is probably a result of eliminating multiple functions or interactions crucial to the function of ICP0 relative to R8507. For this reason, R8507 was used in the remainder of the experiments to observe effects on chromatin structure. Lastly, we entertained the possibility that the two regions of ICP0 may function independently by conducting a superinfection with both R8507 and RF. As shown in Fig. 2, the two mutants do not complement, indicating that they do not specify independent activities that could function in trans. In fact, RF appeared to be dominant to R8507.
Fig. 2.
Relative abundance of GFP mRNA in d109-infected MRC-5 cells after superinfection or coinfection. Infections, RNA isolation, cDNA preparation, and RT-PCR were performed as described in Materials and Methods. Quiescent infection by d109 was established for 1 week and followed by superinfection with the indicated mutant(s). Each value indicates the fold induction (over mock superinfection) of GFP mRNA due to the indicated superinfections.
Changes in chromatin structure of highly repressed quiescent d109 induced by ICP0 mutants.
Previously, it has been shown that ICP0 expression prevents the accumulation of higher-order chromatin structure, repressive histone modifications, and histones on viruses entering quiescence (33). We have previously shown that the expression of wild-type ICP0 can reverse these repressive events on quiescent genomes (33a). In order to determine the effects of ICP0 mutants on the chromatin structure of quiescent virus, MRC-5 cells were infected with d109 and 1 week later either mock superinfected or superinfected with n212, RF, R8507, R8508, or KOS. At 4 hpi, ChIP was performed.
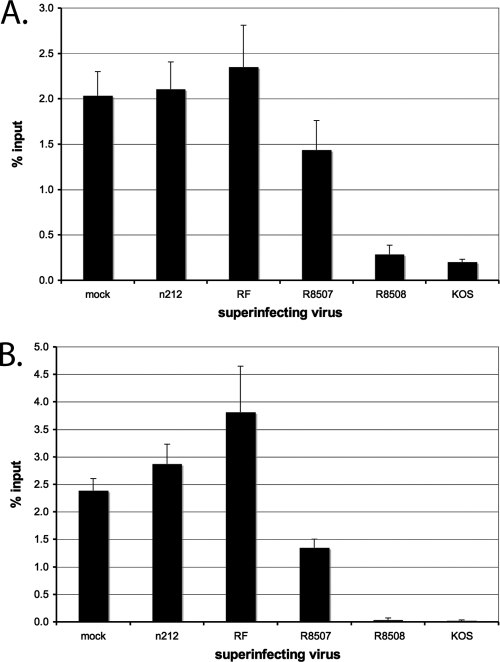
We first looked at the effects of the various mutants on HP1γ on the HCMV IE promoter on d109 (Fig. 3 A). In a previous study, HP1γ was found on approximately 0.7% of the viral genomes after 24 h in HEL cells (33). In contrast, by day 7 after d109 infection, HP1γ was bound to almost 2.5% of HCMV promoters. KOS and R8508 superinfection led to almost complete removal of HP1γ from d109, while R8507 superinfection resulted in a partial reduction of HP1γ on the d109 genome. Superinfection with the ICP0-negative mutant n212, as well as the RF mutant, did not cause the removal of HP1γ from quiescent d109 genomes. Interestingly, the RF mutant resulted in a small increase in HP1γ.
Fig. 3.
Repressive chromatin modifications associated with the HCMV promoter of d109 in MRC-5 cells after superinfection. Quiescent infection by d109 was established for 1 week. ChIP with antibodies to HP1γ (A) and H3K9me3 (B), followed by RT-PCR, was performed as described in Materials and Methods. Shown are the percentages of total genomes bound after superinfection with the indicated virus at 4 h after superinfection with the indicated virus. Error bars represent standard error of the mean from multiple experiments.
Modifications of the histone tails are associated with changes in transcriptional permissiveness, and some are able to recruit proteins involved in transcriptional activation or repression (49). Trimethylation of histone H3 lysine 9 (H3K9me3) is associated with repression and serves as a docking site for HP1γ (71, 77). Using ChIP, the relative effects of the ICP0 mutant proteins on this heterochromatin mark were assessed. R8508 and KOS superinfection resulted in complete loss of H3K9me3 from the d109 HCMV promoter (Fig. 3B). R8507 superinfection resulted in an approximately 50% decrease in H3K9me3. Superinfection with n212 or RF did not reduce the abundance of H3K9me3 from the d109 HCMV promoter relative to mock superinfection. As with HP1γ (Fig. 3A), the level of H3K9me3 was significantly increased relative to that obtained by mock superinfection.
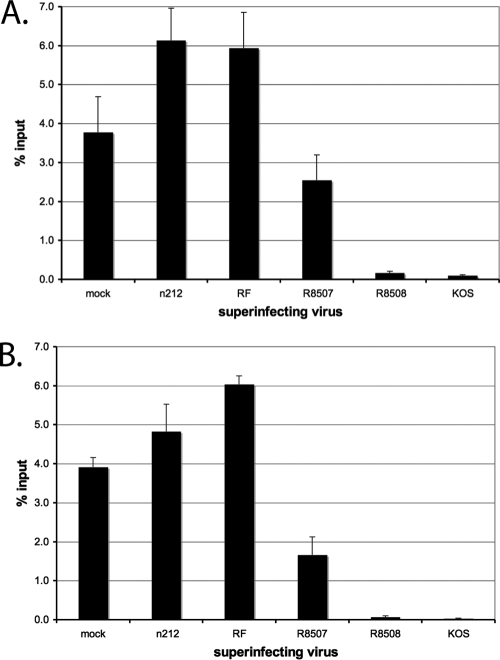
ICP0 expression prevents histone H3 association with the viral genome early in infection (9, 33). The expression of ICP0 can also reduce the abundance of H3 and H4 on highly repressed, quiescent genomes (33a). The various ICP0 mutants were tested for the ability to reduce the abundance of histones H3 and H4 from the quiescent d109 genomes (Fig. 4A). Again, 1-week-old cultures of d109-infected MRC-5 cells were superinfected with the various mutants and subsequently analyzed by ChIP. The pattern of histone H3 (Fig. 4A) and H4 (Fig. 4B) abundance approximately reflects the pattern seen with the heterochromatic marks (Fig. 3). KOS and R8508 superinfection resulted in an almost complete reduction of histone H3 and H4 levels on the d109 HCMV promoter. n212 and RF did not reduce the abundance of either histone H3 or H4. Again, R8507 superinfection resulted in a partial reduction of histone H3 (∼30%) and histone H4 (∼55%).
Fig. 4.
Binding of histones H3 and H4 to the HCMV promoter of d109 in MRC-5 cells after superinfection. ChIP with antibodies to histones H3 and H4 and RT-PCR were performed as described in the legend to Fig. 2 to determine the percentages of genomes bound by histones H3 (A) and H4 (B). Graphs show the percentage of total genomes bound at 4 h after superinfection with the indicated virus. Error bars represents the standard error of the mean from multiple experiments.
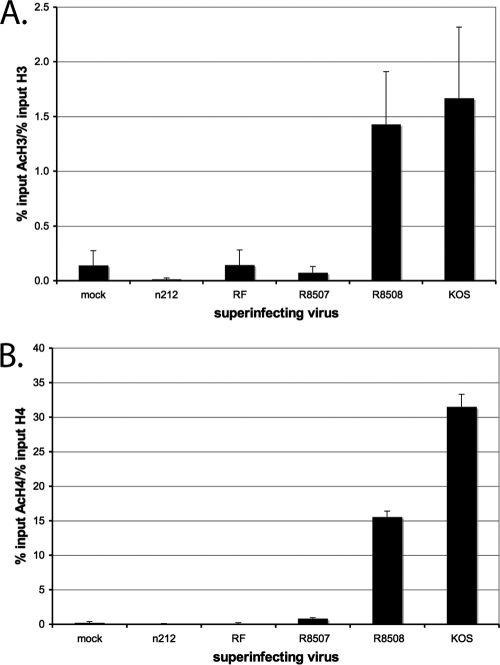
Since the R8507 mutant does not disrupt the REST/CoREST/HDAC1/2/LSD1 repressive complex (41), it was of interest to investigate the effects of the mutations in ICP0 on histone acetylation levels and histone H3 lysine 4 dimethylation. Again, ChIP was performed using antibodies to H3, H4, AcH3, AcH4, and H3K4me2 on samples prepared from d109-ininfected cells and superinfected with the indicated mutants as described for the previous ChIP experiments (Fig. 5 and 6). In order to account for the different levels of histones on d109 after superinfection with each virus, levels of AcH3 and H3K4me2 were normalized to total histone H3 and levels of AcH4 were normalized to total histone H4. Levels of AcH3 on quiescent d109 were extremely low, and superinfection with R8508 or KOS resulted in a 10-fold increase in the hyperacetylation of histone H3 (Fig. 5A). R8508 and KOS superinfections resulted in approximately 75- and 150-fold increases in H4 hyperacetylation of the d109 promoter, respectively (Fig. 5B). n212 and RF superinfections did not result in an increase in hyperacetylation of histone H3 or H4. Interestingly, R8507, which is able to activate gene expression (Fig. 2) and reduce the abundance of repressive marks (Fig. 3) and histones H3 and H4 (Fig. 4), did not result in an increase in the relative abundance of acetylated histones (Fig. 5).
Fig. 5.
Binding of hyperacetylated histone H3 (AcH3) and hyperacetylated H4 (AcH4) to the HCMV promoter of d109 in MRC-5 cells after superinfection. ChIP with an antibody to AcH3 (A) or AcH4 (B) and RT-PCR were performed as described in the legend to Fig. 2 to determine the percentage of d109 genomes bound by AcH3 or AcH4 at 4 h after superinfection with the indicated virus. AcH3 was normalized to the amount of histone H3 (A) and AcH4 was normalized to the amount of histone H4 (B) on the genome by dividing the percentage of acetylated histone by the percentage of total histone (Fig. 3). Error bars represent standard deviations from multiple experiments.
Fig. 6.
Binding of histone H3 dimethyl lysine 4 (H3K4me2) to the HCMV promoter of d109 in MRC-5 cells after superinfection. ChIP with an antibody to H3K4me2 and RT-PCR were performed as described in the legend to Fig. 2 to determine the percentage of d109 genomes bound by H3K4me2 at 4 h after superinfection with the indicated virus. H3K4me2 was normalized to the amount of histone H3 on the genome by dividing the percentage of acetylated histone by the percentage of total histone (Fig. 3). Error bars represent standard deviations from multiple experiments.
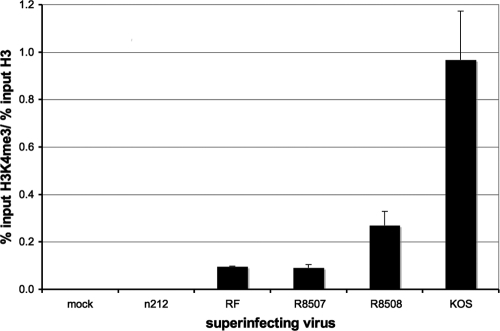
LSD1 is a component of the CoREST repressor complex, which is responsible for a removal of H3K4me2, a mark of active transcription (79). Disrupting REST/CoREST/HDAC1/2/LSD1 would presumably interfere with the demethylation of H3K4, increasing the abundance of H3Kme2. Quiescent d109 genomes had undetectable levels of H3K4me2 on the HCMV promoter, while KOS superinfection greatly increased the relative abundance of histone H3 bearing H3K4me2 (Fig. 6). n212 and RF did not greatly affect the level of H3K4me2. However, R8507 was also not able to affect the level of H3K4me2. Therefore, while R8507 is able to activate transcription and reduce heterochromatin and histone abundance in general, it is unable to affect the relative abundance of AcH3, AcH4, or H3K4me2. Interestingly, R8508 is partially defective in the removal of this mark, indicating that LSD1 is still partially active.
DISCUSSION
ICP0 is a multifunctional protein with seemingly distinct functional domains contributing to its overall effects. The E3 ubiquitin ligase function (4) of the ICP0 RING finger in exon 2 has been shown to cause the degradation of numerous proteins, including those in ND10 bodies, such as PML and SP100 (8), and affects the location of hDaxx and ATRX at theses sites (60). Both of the latter proteins are involved in the repression of incoming viral genomes (60). Additionally, the region comprising the C-terminal 250 amino acids of ICP0 has been shown to contribute to a number of different functions, including; localization to ND10, multimerization, and interaction with a number of infected cell proteins (27, 28, 42, 62, 67). Part of this region is also involved in the disruption of the transcription repressor complex REST/CoREST/HDAC1/2/LSD1 (37, 39–41). Through these and possibly other functions, ICP0 is able to prevent the repression of incoming genomes (20, 21, 43, 52, 72, 73). ICP0 also precludes the formation of heterochromatin on incoming genomes (33). In addition, once the genome is highly repressed in heterochromatin, the provision of ICP0 results in the reduction of repressive chromatin and the activation of expression (33a). Since ICP0 appears to have multiple functions, it was of interest to determine the abilities of various mutant forms of ICP0 to reactivate gene expression and cause changes in specific aspects of chromatin structure of highly repressed HSV-1.
Activation of quiescent genomes by mutant forms of ICP0.
It is clear that the regions mutated in R8507 and n518Bac are not required for activation of quiescent genomes (Fig. 1B and 2). This is in agreement with results previously obtained by Everett et al. using different mutant forms of ICP0 with changes in the C-terminal region (31). However, the approaches used in the experiment shown n Fig. 1B and in the previous study (31) rely on visual assessments, GFP fluorescence in the former and lacZ staining in the latter. In addition, the observations were both made 24 h after the initiation of ICP0 expression. In a previous study, we showed that superinfection induction of mRNA can begin as early as 2 h after superinfection. Visual assessments made at 24 h postinduction may be subject to the effects of downstream events, accumulation, and saturation of the detection mechanism. When induction was measured by the abundance of mRNA at 4 hpi, it is clear that the mutants display significant differences that are not obvious in Fig. 1B. While both R8507 and n518Bac induced quiescent genomes, appearing similar to KOS in the GFP fluorescent assay (Fig. 1B), their abilities to induce quiescent genomes relative to that of KOS, as measured by the abundance of GFP mRNA, were reduced by 11-, and 29-fold, respectively. Therefore, mutation of this region reduces the rate of reactivation of quiescent genomes.
This is not surprising, given that deletion of the C-terminal region of ICP0 has been shown to reduce the activity of ICP0 in several other assays (5–7). As previously discussed, this region encodes many possible activities that could contribute to the overall function of ICP0. The 29-fold reduction in ICP0 activity observed for n518Bac relative to the wild type may be a consequence of defects in multiple activities and interactions. Only the null and RF mutant forms were more impaired. However, the 11-fold impairment of R8507, which possesses two point mutations, may be a result of a smaller set of biochemical defects than that in n518Bac. One of these may be the ability to disrupt the REST/CoREST repressor complex (41).
As previously demonstrated (31), mutation of the RING finger motif completely eliminated ICP0 activity. However, while the levels of GFP mRNA in the RING finger motif mutant-superinfected cultures were similar to those of d99 and n212, the lowest levels were consistently observed in the RF mutant. In addition, the abundances of histones and repressive histone marks were consistently the highest levels observed (Fig. 3 and 4). When RF and R8507 were used to cosuperinfect the d109-infected fibroblasts, the resulting level of GFP mRNA decreased relative to that seen in R8507-infected cells (Fig. 2). We have also found that the level of GFP mRNA in d109-infected MRC-5 cells cosuperinfected with KOS and RF was reduced relative to that observed in KOS-superinfected cells (data not shown). These observations raise two possibilities that are not exclusive of one another, i.e., (i) the RF mutant is transdominant to wild-type ICP0 and (ii) the RF mutant can function as a corepressor. Both of these possibilities could contribute to a seemingly confounding observation in Table 1. The plaque-forming efficiency (L7/Vero) of RF is 31, while that of an ICP0 null mutant is 110, at first glance giving the impression that RF has an activity that promotes virus growth. However, there is no evidence from the molecular observations in this study or from the literature that RING finger mutants have activity. We have found that the number of genomes per L7 plaque-forming unit in an RF mutant stock is 2 to 3 times what it is for the null mutant. Further studies are needed to shed light on the implications of these observations.
Effects of mutant ICP0 proteins on chromatin.
ICP0 expression has been shown to prevent histone H3 association with the HSV-1 genome both during lytic infection and during entry into quiescence (9, 33). Quiescent virus can be found in a heterochromatic state and is associated with higher-order chromatin proteins such as HP1α and HP1γ (10, 33). Superinfection with n212 or RF had no effect on the association of HP1γ with the d109 genome, while KOS and R8508 superinfection caused almost complete removal of HP1γ. R8507 superinfection resulted in partial removal of HP1γ from d109. The same pattern was seen for H3K9me3, a mark of repressive chromatin that serves as a binding site for HP1γ. The abilities of the various ICP0 mutants to cause the removal of histone H3 and histone H4 mimicked their abilities to remove heterochromatin, with n212 and RF unable to cause histone removal, R8508 and KOS superinfection resulting in almost complete removal, and R8507 causing an intermediate removal of histones. Therefore, an ICP0 molecule that is unable to dissociate the repressive REST/CoREST complex (R8507) is able to activate quiescent genomes and can lead to the removal of repressive chromatin, albeit to a lesser extent than the wild type.
Since R8507 is deficient in disruption of the CoREST repressor complex, it follows that this would lead to a decrease in acetylated histones by the action of the HDAC1/2 portion of this complex, as well as a decrease in H3K4me2, a mark of active transcription that is removed by LSD1. Unlike with KOS and R8508, the levels of acetylated histones H3 and H4 remaining bound to the d109 genome following superinfection with R8507 did not increase relative to those found after mock, RF, and n212 superinfections. Collectively, these results are consistent with involvement of the REST/CoREST complex in the repression of HSV genomes and the idea that an activity of ICP0 relieves this repression. The significance of the REST/CoREST complex and the effects of ICP0 have not always been observed. Recently, it was shown that a dominant negative REST expressed from the virus resulted in increased virus replication in vivo (16). However, in one cell type, depletion of CoREST did not enhance the replication of ICP0 null viruses (18). It is also known that different cell types differ with respect to the extent of repression of quiescent genomes (84). Therefore, the effects of the REST/CoREST complex on HSV gene expression may be more consequential in some cell types than in others. The inability of R8507 to cause a significant increase in hyperacetylation of histones is consistent with a mechanism involving the disruption of the REST/CoREST repressor complex, or some other mechanism resulting in acetylated histones, in the derepression of quiescent or latent genomes. It may be that once heterochromatin is removed from the genome, hyperacetylation of histones and H3K4 dimethylation are important for the maintenance of a euchromatic state.
The intermediate phenotype of R8507 with respect to effects on heterochromatin may suggest that the mutation in R8507 affects not only CoREST binding but the ubiquitin ligase activity as well. R8507, which eventually causes the degradation of PML, shows a delay in this activity (41). Additionally, ICP0 mutant forms D13 and D14, which contain deletions of amino acids 633 to 680 and 680 to 720, respectively, are impaired in ND10 localization (61). These deletions overlap the portion of ICP0 required to bind CoREST, which falls between amino acids 668 and 718. Thus, it is possible that the mutation in R8507 affects not only the REST/CoREST complex but other functions of ICP0 that lead to gene activation.
In contrast, the inability of the RF mutant to remove chromatin structure from d109 is consistent with its lack of transcriptional activation. The ubiquitin ligase activity mediated by the RING finger is responsible for the degradation of the ND10 organizing proteins PML and SP100. The ND10 bodies have been implicated in viral silencing (21), and there is a significant link between components of these bodies and epigenetic regulation of transcription. Members of the HP1 family are associated with SP100, forming a link between ND10 bodies and chromatin. Both HP1 and the splice variant SP100-HMG have the ability to act as transcriptional repressors when bound to a promoter (78). Additionally, ND10 components ATRX and hDaxx can also be found at heterochromatin, interact with one another, and are members of a repressive complex that has SWI/SNF chromatin remodeling activity (47, 82, 90). Depletion of PML, SP100, ATRX, or hDaxx (30, 32, 60) can increase the transcription from ICP0-negative HSV-1 mutants, directly implicating disruption of ND10 bodies in changes in the chromatin state of quiescent viral genomes.
These studies show that the ubiquitin ligase activity of ICP0 mediated through the RING finger is necessary for the reactivation of quiescent genomes and reduction of heterochromatin. A mutation in ICP0 that inactivates its ability to disrupt REST/CoREST/HDAC1/2/LSD1 did not result in the increased appearance of acetylated histones or H3K4me2 as did wild-type ICP0. This is consistent with the involvement of the REST/CoREST complex in repression. The ability of ICP0 to disrupt this repressive complex, while not absolutely required for reactivation, may be important in maintaining a euchromatic state once heterochromatin is removed. However, the same mutant ICP0 did not lead to full removal of heterochromatin or activation of gene expression. Therefore, our results are also consistent with the possibility that the mutation that interferes with the disruption of the REST/CoREST complex also affects the other activities that result in heterochromatin removal and gene activation. Further studies are needed to address these possibilities.
ACKNOWLEDGMENTS
This work was supported by NIH grant R01 AI044821 to N.A.D. M.W.F. was funded by training grant T32 AI049820.
Footnotes
Published ahead of print on 16 March 2011.
REFERENCES
- 1. Bannister A. J., et al. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120–124 [DOI] [PubMed] [Google Scholar]
- 2. Barlow P. N., Luisi B., Milner A., Elliott M., Everett R. 1994. Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J. Mol. Biol. 237:201–211 [DOI] [PubMed] [Google Scholar]
- 3. Birnboim H. C., Doly J. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boutell C., Sadis S., Everett R. D. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cai W., et al. 1993. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J. Virol. 67:7501–7512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cai W., Schaffer P. A. 1992. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J. Virol. 66:2904–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cai W. Z., Schaffer P. A. 1989. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J. Virol. 63:4579–4589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chelbi-Alix M. K., de The H. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935–941 [DOI] [PubMed] [Google Scholar]
- 9. Cliffe A. R., Knipe D. M. 2008. Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J. Virol. 82:12030–12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coleman H. M., et al. 2008. Histone modifications associated with herpes simplex virus type 1 genomes during quiescence and following ICP0-mediated de-repression. J. Gen. Virol. 89:68–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Compel P., DeLuca N. A. 2003. Temperature-dependent conformational changes in herpes simplex virus ICP4 that affect transcription activation. J. Virol. 77:3257–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Danaher R. J., Jacob R. J., Chorak M. D., Freeman C. S., Miller C. S. 1999. Heat stress activates production of herpes simplex virus type 1 from quiescently infected neurally differentiated PC12 cells. J. Neurovirol. 5:374–383 [DOI] [PubMed] [Google Scholar]
- 13. Danaher R. J., Jacob R. J., Miller C. S. 2006. Reactivation from quiescence does not coincide with a global induction of herpes simplex virus type 1 transactivators. Virus Genes 33:163–167 [DOI] [PubMed] [Google Scholar]
- 14. Decman V., Kinchington P. R., Harvey S. A., Hendricks R. L. 2005. Gamma interferon can block herpes simplex virus type 1 reactivation from latency, even in the presence of late gene expression. J. Virol. 79:10339–10347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diao L., et al. 2005. Herpes virus proteins ICP0 and BICP0 can activate NF-kappaB by catalyzing IkappaBalpha ubiquitination. Cell Signal. 17:217–229 [DOI] [PubMed] [Google Scholar]
- 16. Du T., Zhou G., Khan S., Gu H., Roizman B. 2010. Disruption of HDAC/CoREST/REST repressor by dnREST reduces genome silencing and increases virulence of herpes simplex virus. Proc. Natl. Acad. Sci. U. S. A. 107:15904–15909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Everett R., O'Hare P., O'Rourke D., Barlow P., Orr A. 1995. Point mutations in the herpes simplex virus type 1 Vmw110 RING finger helix affect activation of gene expression, viral growth, and interaction with PML-containing nuclear structures. J. Virol. 69:7339–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Everett R. D. 2010. Depletion of CoREST does not improve the replication of ICP0 null mutant herpes simplex virus type 1. J. Virol. 84:3695–3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Everett R. D. 2000. ICP0 induces the accumulation of colocalizing conjugated ubiquitin. J. Virol. 74:9994–10005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Everett R. D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22:761–770 [DOI] [PubMed] [Google Scholar]
- 21. Everett R. D. 2006. Interactions between DNA viruses, ND10 and the DNA damage response. Cell. Microbiol. 8:365–374 [DOI] [PubMed] [Google Scholar]
- 22. Everett R. D. 1984. Trans activation of transcription by herpes virus products: requirement for two HSV-1 immediate-early polypeptides for maximum activity. EMBO J. 3:3135–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Everett R. D., et al. 1993. A novel arrangement of zinc-binding residues and secondary structure in the C3HC4 motif of an alpha herpes virus protein family. J. Mol. Biol. 234:1038–1047 [DOI] [PubMed] [Google Scholar]
- 24. Everett R. D., Chelbi-Alix M. K. 2007. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89:819–830 [DOI] [PubMed] [Google Scholar]
- 25. Everett R. D., Earnshaw W. C., Findlay J., Lomonte P. 1999. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 18:1526–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Everett R. D., et al. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Everett R. D., Maul G. G. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062–5069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Everett R. D., Meredith M., Orr A. 1999. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J. Virol. 73:417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Everett R. D., Orr A., Preston C. M. 1998. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 17:7161–7169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Everett R. D., Parada C., Gripon P., Sirma H., Orr A. 2008. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J. Virol. 82:2661–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Everett R. D., Parsy M. L., Orr A. 2009. Analysis of the functions of herpes simplex virus type 1 regulatory protein ICP0 that are critical for lytic infection and derepression of quiescent viral genomes. J. Virol. 83:4963–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Everett R. D., et al. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 80:7995–8005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ferenczy M. W., DeLuca N. A. 2009. Epigenetic modulation of gene expression from quiescent herpes simplex virus genomes. J. Virol. 83:8514–8524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a. Ferenczy M. W., DeLuca N. A. 2011. Reversal of heterochromatic silencing of quiescent herpes simplex virus type 1 by ICP0. J. Virol. 85:3424–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Freeman M. L., Sheridan B. S., Bonneau R. H., Hendricks R. L. 2007. Psychological stress compromises CD8+ T cell control of latent herpes simplex virus type 1 infections. J. Immunol. 179:322–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gelman I. H., Silverstein S. 1985. Identification of immediate early genes from herpes simplex virus that transactivate the virus thymidine kinase gene. Proc. Natl. Acad. Sci. U. S. A. 82:5265–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gierasch W. W., et al. 2006. Construction and characterization of bacterial artificial chromosomes containing HSV-1 strains 17 and KOS. J. Virol. Methods 135:197–206 [DOI] [PubMed] [Google Scholar]
- 37. Gu H., Liang Y., Mandel G., Roizman B. 2005. Components of the REST/CoREST/histone deacetylase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells. Proc. Natl. Acad. Sci. U. S. A. 102:7571–7576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gu H., Roizman B. 2003. The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc. Natl. Acad. Sci. U. S. A. 100:8963–8968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gu H., Roizman B. 2009. Engagement of the lysine-specific demethylase/HDAC1/CoREST/REST complex by herpes simplex virus 1. J. Virol. 83:4376–4385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gu H., Roizman B. 2007. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc. Natl. Acad. Sci. U. S. A. 104:17134–17139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gu H., Roizman B. 2009. The two functions of herpes simplex virus 1 ICP0, inhibition of silencing by the CoREST/REST/HDAC complex and degradation of PML, are executed in tandem. J. Virol. 83:181–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hagglund R., Roizman B. 2002. Characterization of the novel E3 ubiquitin ligase encoded in exon 3 of herpes simplex virus-1-infected cell protein 0. Proc. Natl. Acad. Sci. U. S. A. 99:7889–7894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hagglund R., Roizman B. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 78:2169–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Halford W. P., Kemp C. D., Isler J. A., Davido D. J., Schaffer P. A. 2001. ICP0, ICP4, or VP16 expressed from adenovirus vectors induces reactivation of latent herpes simplex virus type 1 in primary cultures of latently infected trigeminal ganglion cells. J. Virol. 75:6143–6153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Halford W. P., Schaffer P. A. 2001. ICP0 is required for efficient reactivation of herpes simplex virus type 1 from neuronal latency. J. Virol. 75:3240–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hancock M. H., Corcoran J. A., Smiley J. R. 2006. Herpes simplex virus regulatory proteins VP16 and ICP0 counteract an innate intranuclear barrier to viral gene expression. Virology 352:237–252 [DOI] [PubMed] [Google Scholar]
- 47. Ishov A. M., Vladimirova O. V., Maul G. G. 2004. Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX. J. Cell Sci. 117:3807–3820 [DOI] [PubMed] [Google Scholar]
- 48. Jackson S. A., DeLuca N. A. 2003. Relationship of herpes simplex virus genome configuration to productive and persistent infections. Proc. Natl. Acad. Sci. U. S. A. 100:7871–7876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jenuwein T., Allis C. D. 2001. Translating the histone code. Science 293:1074–1080 [DOI] [PubMed] [Google Scholar]
- 50. Jordan R., Schaffer P. A. 1997. Activation of gene expression by herpes simplex virus type 1 ICP0 occurs at the level of mRNA synthesis. J. Virol. 71:6850–6862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kawaguchi Y., et al. 2001. Herpes simplex virus 1 alpha regulatory protein ICP0 functionally interacts with cellular transcription factor BMAL1. Proc. Natl. Acad. Sci. U. S. A. 98:1877–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Knipe D. M., Cliffe A. 2008. Chromatin control of herpes simplex virus lytic and latent infection. Nat. Rev. Microbiol. 6(3):211–221 [DOI] [PubMed] [Google Scholar]
- 53. Kubat N. J., Tran R. K., McAnany P., Bloom D. C. 2004. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J. Virol. 78:1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lachner M., O'Carroll D., Rea S., Mechtler K., Jenuwein T. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116–120 [DOI] [PubMed] [Google Scholar]
- 55. Lium E. K., Silverstein S. 1997. Mutational analysis of the herpes simplex virus type 1 ICP0 C3HC4 zinc ring finger reveals a requirement for ICP0 in the expression of the essential alpha27 gene. J. Virol. 71:8602–8614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Loiacono C. M., Taus N. S., Mitchell W. J. 2003. The herpes simplex virus type 1 ICP0 promoter is activated by viral reactivation stimuli in trigeminal ganglia neurons of transgenic mice. J. Neurovirol. 9:336–345 [DOI] [PubMed] [Google Scholar]
- 57. Lomonte P., Morency E. 2007. Centromeric protein CENP-B proteasomal degradation induced by the viral protein ICP0. FEBS Lett. 581:658–662 [DOI] [PubMed] [Google Scholar]
- 58. Lomonte P., Sullivan K. F., Everett R. D. 2001. Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J. Biol. Chem. 276:5829–5835 [DOI] [PubMed] [Google Scholar]
- 59. Lomonte P., et al. 2004. Functional interaction between class II histone deacetylases and ICP0 of herpes simplex virus type 1. J. Virol. 78:6744–6757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lukashchuk V., Everett R. D. 2010. Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J. Virol. 84:4026–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Maul G. G., Everett R. D. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75(Pt. 6):1223–1233 [DOI] [PubMed] [Google Scholar]
- 62. Meredith M., Orr A., Elliott M., Everett R. 1995. Separation of sequence requirements for HSV-1 Vmw110 multimerisation and interaction with a 135-kDa cellular protein. Virology 209:174–187 [DOI] [PubMed] [Google Scholar]
- 63. Mossman K. L., Saffran H. A., Smiley J. R. 2000. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 74:2052–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nakayama J., Rice J. C., Strahl B. D., Allis C. D., Grewal S. I. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110–113 [DOI] [PubMed] [Google Scholar]
- 65. O'Hare P., Hayward G. S. 1985. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J. Virol. 53:751–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. O'Rourke D., Elliott G., Papworth M., Everett R., O'Hare P. 1998. Examination of determinants for intranuclear localization and transactivation within the RING finger of herpes simplex virus type 1 IE110k protein. J. Gen. Virol. 79(Pt. 3):537–548 [DOI] [PubMed] [Google Scholar]
- 67. Parkinson J., Everett R. D. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006–10017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Parkinson J., Lees-Miller S. P., Everett R. D. 1999. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73:650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Preston C. M., Nicholl M. J. 2008. Induction of cellular stress overcomes the requirement of herpes simplex virus type 1 for immediate-early protein ICP0 and reactivates expression from quiescent viral genomes. J. Virol. 82:11775–11783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Quinlan M. P., Knipe D. M. 1985. Stimulation of expression of a herpes simplex virus DNA-binding protein by two viral functions. Mol. Cell. Biol. 5:957–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rea S., et al. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593–599 [DOI] [PubMed] [Google Scholar]
- 72. Roizman B., Gu H., Mandel G. 2005. The first 30 minutes in the life of a virus: unREST in the nucleus. Cell Cycle 4:1019–1021 [DOI] [PubMed] [Google Scholar]
- 73. Roizman B., Knipe D. M., Whitley R. J. 2007. Herpes simplex viruses, p. 2501–2601In Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Strauss S. E. (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 74. Samaniego L. A., Neiderhiser L., DeLuca N. A. 1998. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J. Virol. 72:3307–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Samaniego L. A., Wu N., DeLuca N. A. 1997. The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected-cell survival in the absence of ICP4 and ICP27. J. Virol. 71:4614–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sampath P., Deluca N. A. 2008. Binding of ICP4, TATA-binding protein, and RNA polymerase II to herpes simplex virus type 1 immediate-early, early, and late promoters in virus-infected cells. J. Virol. 82:2339–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Schotta G., et al. 2002. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 21:1121–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Seeler J. S., Marchio A., Sitterlin D., Transy C., Dejean A. 1998. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc. Natl. Acad. Sci. U. S. A. 95:7316–7321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Shi Y., et al. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941–953 [DOI] [PubMed] [Google Scholar]
- 80. Spivack J. G., Fraser N. W. 1987. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J. Virol. 61:3841–3847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Stevens J. G., Wagner E. K., Devi-Rao G. B., Cook M. L., Feldman L. T. 1987. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science 235:1056–1059 [DOI] [PubMed] [Google Scholar]
- 82. Tang J., et al. 2004. A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J. Biol. Chem. 279:20369–20377 [DOI] [PubMed] [Google Scholar]
- 83. Taylor J. L., Unverrich D., O'Brien W. J., Wilcox K. W. 2000. Interferon coordinately inhibits the disruption of PML-positive ND10 and immediate-early gene expression by herpes simplex virus. J. Interferon Cytokine Res. 20:805–815 [DOI] [PubMed] [Google Scholar]
- 84. Terry-Allison T., Smith C. A., DeLuca N. A. 2007. Relaxed repression of herpes simplex virus type 1 genomes in murine trigeminal neurons. J. Virol. 81:12394–12405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tischer B. K., Smith G. A., Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol. Biol. 634:421–430 [DOI] [PubMed] [Google Scholar]
- 86. Tischer B. K., von Einem J., Kaufer B., Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197 [DOI] [PubMed] [Google Scholar]
- 87. Van Sant C., Lopez P., Advani S. J., Roizman B. 2001. Role of cyclin D3 in the biology of herpes simplex virus 1 ICP0. J. Virol. 75:1888–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wang Q. Y., et al. 2005. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. U. S. A. 102:16055–16059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wilcox C. L., Smith R. L., Freed C. R., Johnson E. M., Jr 1990. Nerve growth factor-dependence of herpes simplex virus latency in peripheral sympathetic and sensory neurons in vitro. J. Neurosci. 10:1268–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Xue Y., et al. 2003. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc. Natl. Acad. Sci. U. S. A. 100:10635–10640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yao F., Schaffer P. A. 1995. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J. Virol. 69:6249–6258 [DOI] [PMC free article] [PubMed] [Google Scholar]