Abstract
Bluetongue virus (BTV), a member of the Reoviridae family, is an insect-borne animal pathogen. Virus release from infected cells is predominantly by cell lysis, but some BTV particles are also released from the plasma membrane. The nonstructural protein NS3 has been implicated in this process. Using alternate initiator methionine residues, NS3 is expressed as a full-length protein and as a truncated variant that lacks the initial 13 residues, which, by yeast-two hybrid analyses, have been shown to interact with a cellular trafficking protein S100A10/p11. To understand the physiological significance of this interaction in virus-infected cells, we have used reverse genetics to investigate the roles of NS3 and NS3A in virus replication and localization in both mammalian and insect vector-derived cells. A virus expressing NS3 but not NS3A was able to propagate in and release from mammalian cells efficiently. However, growth of a mutant virus expressing only NS3A was severely attenuated, although protein expression, replication, double-stranded RNA (dsRNA) synthesis, and particle assembly in the cytoplasm were observed. Two of three single-amino-acid substitutions in the N-terminal 13 residues of NS3 showed phenotypically similar effects. Pulldown assay and confocal microscopy demonstrated a lack of interaction between NS3 and S100A10/p11 in mutants with poor replication. The role of NS3/NS3A was also assessed in insect cells where virus grew, albeit with a reduced titer. Notably, however, while wild-type particles were found within cytoplasmic vesicles in insect cells, mutant viruses were scattered throughout the cytoplasm and not confined to vesicles. These results provide support for a role for the extreme amino terminus of NS3 in the late stages of virus growth in mammalian cells, plausibly in egress. However, both NS3 and NS3A were required for efficient BTV growth in insect cells.
INTRODUCTION
In recent years, accumulating evidence about the nonlytic release of nonenveloped viruses has contradicted the view that the release of these viruses is a passive process due to cell lysis. Studies on small nonenveloped viruses such as simian virus 40 (8) and poliovirus (25) have indicated that these viruses may exit from the infected cells nonlytically. Our lab has also previously provided strong evidence that bluetongue virus (BTV), a member of the Reoviridae family, exits the infected cells not only by cell lysis but also by newly synthesized particles budding from the plasma membrane early in infection (16). Thus, the egress of nonenveloped viruses appears to be a complex process and requires further investigations.
BTV is vectored by insect Culicoides spp. to ruminants, causing disease in vertebrate hosts (sheep, cattle, and goats) that has considerable economic impact. Thus, BTV has the ability to replicate in both insect and mammalian hosts. In cell culture, mammalian cells infected with BTV exhibit strong cytopathic effects (CPE) after a few hours of infection. In contrast, no obvious CPE can be visualized in insect vector cell culture upon BTV infection (15, 28). The nonenveloped BTV particle is a complex icosahedral structure consisting of seven structural proteins (VP1 to VP7) that are organized in an outer capsid and an inner capsid (core) (27). The outer capsid is composed of two major proteins, VP2 and VP5, and is responsible for attachment and membrane penetration. Both are lost during endocytosis, and the inner core is subsequently released into the cytoplasm. The BTV core, which consists of the remaining five proteins and the viral genome of 10 double-stranded RNA (dsRNA) segments, then synthesizes and extrudes the transcripts of the 10 genomic RNA segments into the cytoplasm. In addition to 10 structural proteins, two major nonstructural (NS) proteins, NS1 and NS2, are also synthesized during early infection, and each plays an essential role in virus replication (23).
Unlike the nine larger RNA genomic segments, each of which encodes a single protein product, the smallest segment, S10 of BTV, encodes two nonstructural proteins, NS3 (229 residues) and NS3A (216 residues), a truncated form of NS3. NS3A lacks the first 13 residues from the N-terminal end of NS3 and is the product of a second initiation codon in the gene (11). The structures of NS3 and NS3A comprise a long N-terminal domain and a short C-terminal cytoplasmic domain connected by two transmembrane domains and a small extracellular region. These two proteins are the only glycosylated membrane proteins encoded by BTV, with a single glycosylation site in the asparagine residue located in the extracellular domain (32).
NS3 has been shown to play a critical role during virus egress. We reported previously that BTV virus-like particles expressed by recombinant baculoviruses in the presence of NS3 are efficiently released from infected Spodoptera cells (17). Further, we showed that NS3 interacts with the outer capsid proteins and that mutations in the VP2 interaction domain of NS3 completely disrupted the assembly, trafficking, and release of the virus from infected cells (3, 4, 7).
A fascinating characteristic of NS3 is that it possesses late domain motifs similar to those present in certain proteins of enveloped viruses (9, 10). Through these L domains, BTV is able to interact with the cellular protein Tsg101, which is involved in the vacuolar protein-sorting pathway (31). Tsg101 is a component of the ESCRT-I cellular exocytic pathway that has been implicated in the release of several enveloped viruses, including retroviruses such as HIV or filoviruses such as Ebola virus (12, 19). Further, the interaction with Tsg101 has been shown to be essential for the correct budding of BTV from infected cells (7, 31).
In addition to the involvement of the cellular protein Tsg101 during BTV egress, our lab has also shown that NS3 is able to interact with the host protein S100A10/p11 (also known as calpactin light chain). Using the yeast two-hybrid genetic system, it was shown that NS3 interacts with S100A10/p11 and that this interaction required the NS3 N-terminal sequences (3). S100A10/p11 is a Ca2+-insensitive member of the S100 family that in cells forms a heterotetrameric complex with two heavy chains of annexin II (also known as annexin A2) to form the calpactin complex. The calpactin complex is involved in trafficking of proteins and targeting to specific membranes (reviewed in reference 21). It is notable that the N-terminal residues of NS3 involved in S100A10/p11 interaction are not present in NS3A. The function of NS3A in virus replication or morphogenesis is still unclear. However, the second initiation site in the NS3 gene is absolutely conserved among all the BTV serotypes, suggesting that NS3A may have a particular role that has not yet been elucidated. In our previous study, several experimental strategies were used to demonstrate that the interaction of NS3 and S100A10/p11 is significant for virus replication and release (3); however, it was not possible to obtain direct evidence of the relevance of this interaction due to the lack of a reverse genetics (RG) system for BTV. Recently, we established a helper virus-independent RG system for BTV that uses T7 transcripts of virus RNA segments to facilitate the introduction of targeted mutations into the BTV genome and investigation of their effect in the context of a replicating virus.
In this study we have exploited the RG system to generate mutant viruses targeting NS3 and NS3A proteins and utilized these viruses to investigate the role of each protein in the absence of the other in virus replication and morphogenesis. In addition, we also introduced mutations targeting the sequences of NS3 that were previously found to be involved in the interaction with S100A10/p11. The detailed analysis of these mutant viruses supports the hypothesis that NS3, but not NS3A, plays a key role in virus assembly and trafficking in mammalian cells. However, both NS3 and NS3A are required for BTV trafficking from insect vector cells.
MATERIALS AND METHODS
Cell lines and virus.
BSR (derived from baby hamster kidney [BHK]) cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 5% fetal calf serum (FCS). The stable BSR/NS3 cell line was grown in DMEM–5% FCS supplemented with 7.5 μg/ml of puromycin (Sigma-Aldrich). KC cells (derived from Culicoides spp. and kindly provided by P. Mertens, Institute for Animal Health, Pirbright, United Kingdom) were maintained at 28°C in Schneider's medium supplemented with 5% FCS.
BTV serotype 1 (BTV-1) wild-type (WT) and mutant virus stocks were obtained by infecting mammalian cells at a low multiplicity of infection (MOI) and harvested when CPE was evident. Titers of viral stocks were obtained by plaque assay and expressed as PFU per ml. Viral stocks were stored at 4°C until use.
Site-directed mutagenesis.
To introduce specific mutations in the BTV-1 S10 sequence, the site-directed mutagenesis method described by Weiner et al. (30) with modifications was used; two complementary primers bearing the specific mutations were used for amplification with pUCBTV1T7S10 as a template (7). The primers used for site-directed mutagenesis were as follows: 1NS3Q7P_F, 5′-GCCATGCTATCCGGGCTGATCCCTAGGTTCGAGGAAGAAAAAAT G-3′; 1NS3Q7P_R, 5′-CATTTTTTCTTCCTCGAACCTAGGGATCAGCCCGGATAGCATGGC-3′; 1NS3R8E_F, 5′-GCCATGCTATCCGGGCTGATCCAGGAATTCGAGGAAGAAAAAATG-3′; 1NS3R8E_R, 5′-CATTTTTTCTTCCTCGAATTCCTGGATCAGCCCGGATAGCATGGC-3′; 1NS3E10A_F, 5′-CCGGGCTGATCCAAAGGTTCGCTGAGGAAAAAATGAAACATAAT C-3′; 1NS3E10A_R, 5′-GATTATGTTTCATTTTTTCCTCAGCGAACCTTTGGATCAGCCCGG-3′; 5′BTV1NS3M1, 5′-GGTTCGAGGAAGAAAAAGCTAAGCATAATCAGGAACGGGTTGAAGAG-3′; 3′BTV1NS3M1, 5′-CAACCCGTTCCTGATTATGCTTAGCTTTTTCTTCCTCGAACCTTTG-3′; 5′BTV1 NS3AM14, 5′-GTTAAAAAGTGTCGCTGCCGCTCTATCCGGCCTGATCCAAAGGTTCGAGG-3′; 3′BTV1NS3AM14, 5′-CCTTTGGATCAGGCCGGATAGAGCGGCAGCGACACTTTTTAAC-3′; 5′BTV1S10N150A, 5′-TGGCGTTTAAGCTAGCGGGTACATCAGCCGAGATACCTCAATGGTTTAA G-3′; and 3′BTV1S10N150A, 5′-TGAGGTATCTCGGCTGATGTACCCGCTAGCTTAAACGCCACGCTCATATC-3′. All generated constructs were sequenced to ensure the presence of the introduced mutations and absence of extra changes.
Transfection of mammalian cells with plasmid-derived T7 transcripts: rescue of mutant BTVs.
The T7 BTV transcripts were prepared as described previously (5) using the mMESSAGE mMACHINE T7 Ultra kit (Ambion). A total of 400 ng of each of the 10 BTV transcripts was incubated with 0.1U/μl RNasin Plus (Promega) and mixed with Lipofectamine 2000 (Invitrogen) as described previously (5). BSR cells were transfected twice with the mixture, and at 3 h after the second transfection the cells were overlaid with 1.5% agarose type VII (Sigma-Aldrich) containing 4% FCS and further incubated at 35°C for 3 to 5 days or until onset of CPE.
Genomic RNA purification and analysis.
Preparation of genomic dsRNA from cells infected with control or mutant BTVs and analysis of those samples were performed as described previously (5). cDNA was generated using specific primers, and the presence of the designed mutation in each S10 was verified by sequencing.
Coimmunoprecipitation assay.
BSR cells were infected with WT or mutant viruses as is indicated at an MOI of 0.5 to 1.0, harvested at 36 h, washed with cold phosphate-buffered saline (PBS), and resuspended in immunoprecipitation assay buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate) supplemented with a cocktail of protease inhibitors (protease inhibitor cocktail set V, EDTA free; Calbiochem). After 20 min of incubation on ice, the cell lysates were clarified and aliquots were incubated overnight with 1.5 μg of anti-S100A10 monoclonal antibody (AbCam) at 4°C. A suspension of 10% protein A-Sepharose CL4B (Sigma-Aldrich) was added to the samples and incubated for a further 3 h at 4°C. The beads were then centrifuged and washed three times with buffer containing 100 mM Tris-HCl (pH 8.0), 500 mM LiCl, 1.5% NP-40, and 0.1% bovine serum albumin. To monitor the S100A10/p11-NS3 interaction, sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting were performed using a 1:5,000 dilution of a polyclonal anti-NS3 antibody.
Confocal microscopy.
The effect of the designed mutations in NS3 on cellular localization of the protein relative to that of the S100A10/p11 protein was analyzed by confocal microscopy. Briefly, BSR cells were infected with mutant or WT viruses and fixed with 4% paraformaldehyde at 24 h postinfection. After permeabilization with 1% Triton X-100, samples were incubated with a polyclonal rabbit antibody against NS3 and a polyclonal guinea pig antibody that recognized S100A10/p11. As secondary antibodies, anti-rabbit-conjugated Alexa Fluor 488 (Sigma-Aldrich) and anti-guinea pig-conjugated Alexa Fluor 546 (Sigma-Aldrich) diluted 1:300 were used. The nuclei of the cells were stained with Hoechst 33258 (Sigma-Aldrich). Samples were analyzed under a Zeiss LSM510 confocal microscope (magnification, ×100), and images were obtained using LSM510 image browser software.
Virus growth kinetics and virus release.
For determination of virus growth curves of the mutant or WT BTVs, monolayers of mammalian BSR cells, BSR/NS3 cells, or KC cells were infected at a low MOI (0.05 to 0.1). At different time points postinfection (as indicated for each experiment), cells and supernatant were harvested and frozen and thawed twice for mechanical disruption of the cells, and the total titer was determined by plaque assay.
For virus release studies, monolayers of KC cells were infected at an MOI of 0.6 to 1 with BTV mutants or the WT as a control. After 1 h of absorption, cells were washed with medium and incubated in Schneider's medium supplemented with 2% FCS for 4 days. The supernatants were collected, and the cells were washed twice with fresh medium before being harvested. The titer of each fraction was determined by plaque assay.
Cell sectioning and electron microscopy analysis.
BSR or KC cells were infected with the indicated virus and at 24 h and 6 days postinfection, respectively, were processed for cell sectioning. Briefly, after three washes with serum-free medium, monolayers were incubated in 2.5% glutaraldehyde–2% formaldehyde for 15 min, followed by a second fixation with 2.5% paraformaldehyde–2.5% glutaraldehyde–0.1% sodium cacodylate (pH 7.4), and postfixed in 1% osmium tetroxide–0.1% sodium cacodylate. Cells were dehydrated in increasing concentrations of ethanol and embedded in epoxy resin (TAAB Laboratories Equipment Ltd.). Ultrathin sections were stained with Reynolds lead citrate (22).
RESULTS
Effect of mutations in the first or second initiation codon of S10: expression of NS3 or NS3A.
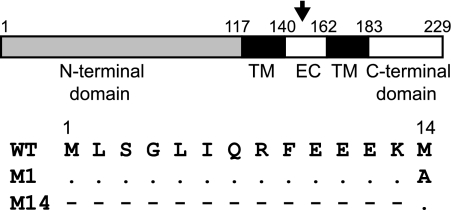
Although the presence of the second initiation codon in the S10 gene is highly conserved among different strains of BTV, very little is known about the function of the second protein, NS3A, during virus replication. In other orbiviruses, such as African horse sickness virus or equine encephalosis virus, the NS3 gene also presents a second initiation codon, although the position varies from residue 11 or 12 to 19 (26). This observation suggests that NS3A may play a particular role during virus replication. In order to determine if NS3A has a particular role in BTV replication, independent of NS3, we generated two mutant viruses segregating each initiation codon. A change was introduced in the first initiation codon in the NS3 gene to disable the ATG codon so that a mutant virus (BTVM14) would synthesize a protein from only the second initiation codon, NS3A. In a second mutant virus (BTVM1), the second ATG codon of S10 changed to alanine so that only the full-length NS3 protein, from the first initiation codon, was expressed (Fig. 1).
Fig. 1.
Schematic representation of NS3. The two cytoplasmic domains (N terminal and C terminal), the two transmembrane domains (TM), and the extracellular domain (EC) are indicated. Numbers above indicate amino acid position according to the NS3 amino acid sequence. The glycosylation site at asparagine position 150 is also shown (arrow). The changes in either the first or the second methionine residue introduced by site-directed mutagenesis are shown in the lower panel; dots indicate no change, and dashes indicate absence of expressed residues.
To generate these mutant viruses, BSR cells were transfected with the T7-derived BTV-1 single-stranded RNA (ssRNA) corresponding to segments 1 to 9 together with one of the mutant S10 transcripts. At 3 days posttransfection, clear plaques could be seen for the BTVM1 mutant, indicating that this mutation did not impede the rescue of viable virus. In contrast, no plaques were visible at 4 days posttransfection for the BTVM14 rescue experiment, suggesting that the deletion of NS3 and expression of only NS3A prevented virus recovery. We therefore used a complementing cell line that expresses NS3/NS3A constitutively (BSR/NS3 cell line) and has been used successfully for rescuing different NS3 mutant viruses previously (7). Transfection of BSR/NS3 with the complete set of transcripts, including the S10 mutant BTVM14, rendered clear plaques, indicating that NS3 provided in trans allowed virus rescue in BSR cells. The presence of the introduced mutation in the rescued viruses was confirmed by sequencing the reverse transcription-PCR (RT-PCR) products corresponding to S10 obtained from each virus. Virus stocks were amplified and used as low-MOI (∼0.05) inocula to compare growth curves to that of the wild type in both BSR and BSR/NS3 cell lines. The total virus titers at 24 and 48 h postinfection, determined by plaque assay in BSR/NS3 cells, showed that virus BTVM1 had growth kinetics similar to those of wild-type BTV in both cell lines (Fig. 2 A, left panel). However, BTVM14 failed to grow in BSR cells (Fig. 2A, right panel) but grew to titers similar to those of the wild type in BSR/NS3 cells, albeit with delayed kinetics, confirming the original rescue data. Thus, full-length NS3 is essential for productive replication in mammalian cells. However, despite there being no productive growth, some CPE was observed for all viruses in both the BSR and BSR/NS3 cell lines. To assess at what stage replication was blocked in the BTVM14 virus, BSR cells were infected with each virus at a high MOI (0.5) and cell lysates prepared at 24 h postinfection for an investigation of their protein expression profile and ability to synthesis dsRNA. Western blotting using an antibody that recognizes NS3/NS3A demonstrated that cells infected with BTVM1 expressed a protein at the same molecular mass of the upper band in the control WT infection (Fig. 2B, compare lanes 1 and 2). In contrast, cells infected with BTVM14 expressed only a protein of lower molecular mass which comigrated with the lower of the two NS3-related proteins expressed in BTV-infected cells (Fig. 2B, compare lanes 1 and 3), confirming that the individual mutant viruses express only NS3 or NS3A, as designed. In addition, dsRNA extracted from infected cells showed a profile identical to that of the wild-type virus (Fig. 2C, compare lanes 2 and 3 with lane 1). These results indicate that both mutant viruses are able to infect BSR cells, initiate protein synthesis, and progress in the replication cycle up to and including dsRNA synthesis. The block to replication demonstrated by BTVM14 is therefore downstream of these events.
Fig. 2.
Characterization of BTVM1 and BTVM14 mutant viruses. (A) The total titer at different time points postinfection for either mutant or WT viruses in complementary BSR/NS3 cells (left panel) or normal BSR cells (right panel) was determined, expressed as PFU/ml, and plotted on a logarithmic scale. (B) The expression of NS3 was assessed in cell lysates from mammalian BSR cells at 48 h postinfection with mutant virus BTVM1 (lane 2) or BTVM14 (lane 3); cell lysates from mock-infected cells (lane 4) or WT-infected cells (lane 1) were included. Lane M, molecular mass standard in kDa. Western blotting was performed using an antibody against BTV NS3. (C) Genomic dsRNA from BSR cells infected with WT virus (lane 1) or mutant virus BTVM1 (lane 2) or BTVM14 (lane 3) was purified and analyzed on a nondenaturing polyacrylamide gel.
Association with S100A10/p11.
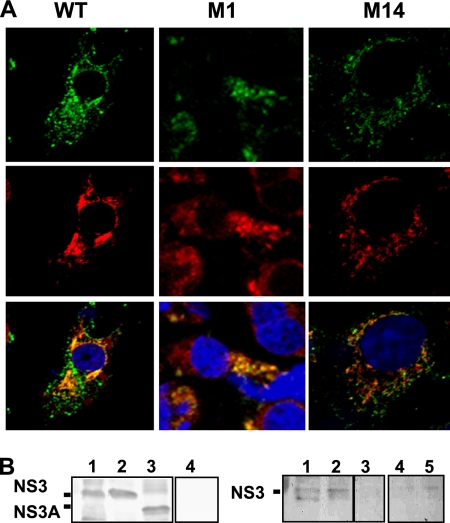
As an association between NS3 and the cellular exocytic protein S100A10/p11 component of the calpactin complex had been previously reported (3), we assessed if the changes introduced in viruses BTVM1 and BTVM14 had any effect on the interaction between NS3 and S100A10/p11. BSR cells were infected with mutant or WT viruses and processed for intracellular staining with serum specific for NS3 and an anti-S100A10/p11 antibody. Wild-type BTV showed an NS3 and S100A10/p11 distribution pattern consistent with the proteins being endoplasmic reticulum (ER) and Golgi resident, with evidence of colocalization (Fig. 3 A, left panels). Mutant virus BTVM1 showed a similar pattern, although the colocalization was somewhat less distinct (Fig. 3A, middle panels). In contrast, mutant virus BTVM14 showed a punctuate staining pattern for NS3 and no colocalization with S100A10/p11 (Fig. 3A, right panels). These data confirm the interaction between NS3 and S100A10/p11 in virus-infected cells (3) and indicate that the first 13 residues of NS3 are critical to it.
Fig. 3.
Interaction of BTV NS3 with S100A10/p11 in infected mammalian cells. (A) Colocalization of NS3 (green) with S100A10/p11 (red). BSR cells were infected with WT or mutant viruses as indicated and at 24 h postinfection were fixed, permeabilized, and processed for confocal microscopy. (B) Coimmunoprecipitation of NS3 and S100A10/p11. BSR cells were infected with WT virus (lane 1) or mutant viruses BTVM1 (lane 2) and BTV M14 (lane 3) and at 36 h postinfection were harvested and processed as described in Materials and Methods. Expression of NS3 in cell lysates was assayed by Western blotting using an antibody against NS3 (left panel). As control, a lysate from noninfected cells (lane 4) was included. The coimmunoprecipitation of NS3 WT virus (right panel, lane 1) or mutants M1 (lane 2) and M14 (lane 3) with S100A10/p11 was performed using a monoclonal antibody that recognized S100A10/p11 and analyzed by Western blotting with an antibody against NS3. As controls, a lysate from mock-infected cells (lane 4) and a lysate from cells infected with WT virus but without antibody against S100A10/p11 (lane 5) were included.
To further confirm an interaction between NS3 and S100A10/p11, cell lysates from a high-MOI infection of BSR cells were subject to pulldown with S100A10/p11 followed by SDS-PAGE and Western blotting using an anti-BTV serum. Full-length NS3 was pulled down by anti-S100A10/p11 from lysates infected with WT and BTVM1 virus (Fig. 3B, right panel, lanes 1 and 2). No pulldown was apparent for lysates prepared from BTVM14-infected cells (Fig. 3B, right panel, lane 3) although levels of NS3A in the original lysate were similar to those of NS3 in the WT- and BTVM1-infected cells (Fig. 3B, left panel, lanes 1 to 3). Thus, a failure of interaction between NS3A and S100A10/p11, as demonstrated by biochemical assay and confocal microscopy, is a characteristic of BTVM14 but not of BTVM1 or the WT virus.
Rescue of mutant viruses with single changes on the S100A10/p11 interaction domain of NS3.
Previously, it was shown that the NS3 and cellular S100A10/p11 interaction demonstrated by yeast-two hybrid analysis, protein immunoprecipitation, and competitive peptide assay was dependent on a putative helix in the 13 N-terminal residues of NS3 (3). To probe the function of the helix in the in vivo system, three mutations previously shown to disrupt the interaction, that is, glutamine 7 to proline (Q7P), arginine 8 to glutamic acid (R8E), and glutamic acid 10 to alanine (E10A), were introduced into NS3 by RG as before (Fig. 4). Following transfection of BSR cells with the T7-derived BTV-1 ssRNA, plaques were demonstrated for mutants BTVR8E and BTVE10A but only pinpoint plaques were detected in BTVQ7P. As before, all three viruses could be grown in BSR/NS3 cells. The presence of the introduced mutations in the rescued viruses was confirmed by sequencing of the RT-PCR products corresponding to S10.
Fig. 4.
Schematic representation of the substitution mutations introduced in the N-terminal end of NS3. Specific changes in the first 13 residues in each mutant (Q7P, R8E, and E10A) are indicated. Dots indicate no change.
Infection of both BSR and BSR/NS3 cells followed by plaque assay on BSR/NS3 cells revealed delayed growth kinetics for two of the three viruses in BSR cells compared with the WT (Fig. 5 A, right panel). Mutant BTVR8E grew to titers similar to those of the WT by 48 h postinfection. In contrast, the titer of mutant BTVQ7P at 48 h postinfection was about 2 logs lower than that of the WT virus at the equivalent time point, and mutant BTVE10A failed to grow in BSR cells. All mutants grew in BSR/NS3 helper cells, although mutant BTVE10A, with the most severe phenotype on BSR cells, grew with delayed kinetics (Fig. 5A, left panel). Thus, single-residue changes in the 13-residue N terminus of BTV NS3 previously shown to bind S100A10/p11 are attenuating in the context of a replicating virus.
Fig. 5.
Characterization of mutant BTVQ7P, BTVR8E, and BTVE10A viruses. (A) Complementary BSR/NS3 cells (left panel) or normal BSR cells (right panel) were infected with mutant virus BTV Q7P, R8E, or E10A or with the WT and harvested at different time points postinfection as indicated. Total titer was determined by plaque assay, expressed as PFU/ml, and plotted on a logarithmic scale. (B) The expression of NS3 was assessed by Western blotting in BSR cell lysates processed at 48 h postinfection with mutant virus Q7P (lane 1), R8E (lane 2), or E10A (lane 3). Lysate from cells infected with WT BTV (lane 4) was included as control. The number on the left indicate the molecular mass standard in kDa. (C) Genomic dsRNA purified from BSR cells infected with BTVQ7P (lane 2), BTVR8E (lane 3), or BTVE10A (lane 4) was purified and analyzed in a nondenaturing polyacrylamide gel. As a control, dsRNA from WT virus was included (lane 1).
To assess at what stage replication was arrested as a result of the single-residue changes in NS3, BSR cells were infected with each virus at high a MOI (0.5) and cell lysates prepared at 24 h postinfection for determination of protein expression and dsRNA profile as described above. Western analyses demonstrated that all three mutant viruses expressed levels of both NS3 and NS3A that were similar to those in the WT infection (Fig. 5B, compare lanes 1 to 4). Similarly, dsRNA extracted from infected cells exhibited profiles equivalent to that in wild-type virus infections (Fig. 5C). These results indicate that all three mutant viruses infect BSR cells and complete dsRNA synthesis. Their attenuated phenotype is therefore due to a defect in the later phase of the replication cycle, as described for the BTVM14 virus.
Mutant viruses containing single-residue changes in NS3 and their interaction with S100A10/p11.
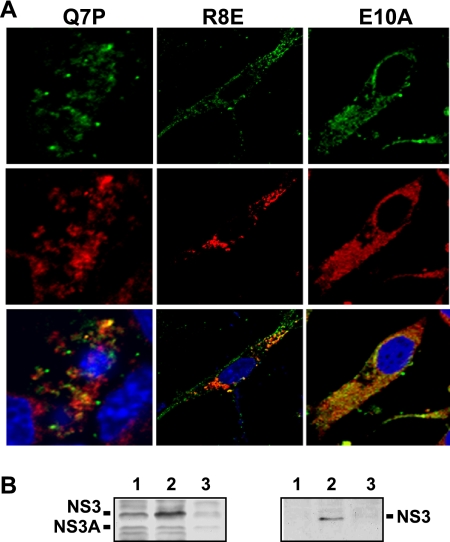
To examine further the effect of single-residue mutations in NS3 on the NS3-S100A10/p11 interaction, all three mutants were used to infect BSR cells at a high MOI and the cells were fixed at 24 h postinfection and processed for confocal microscopy. The distribution of NS3 and S100A10/p11 was analyzed by use of specific antibodies as before. BTVR8E-infected cells (with a minor defective phenotype) showed evidence of colocalization, although it was less distinct than that in the WT infection (cf. Figure 6 A with 3A). However, no distinct colocalization between NS3 and S100A10/p11 was visualized in either BTVQ7P- or BTVE10A (with a major defective phenotype)-infected cells. Cells infected with each mutant virus were also subjected to coimmunoprecipitation assay for the biochemical interaction between NS3 and S100A10/p11 as before. BTVR8E showed a clear pulldown of NS3 by S100A10/p11 (Fig. 6B, right panel, lane 2), whereas no such interaction could be shown for either mutant BTVQ7P or BTVE10A (Fig. 6B, right panel, lanes 1 and 3) despite similar levels of NS3 and NS3A expression in all lysates (Fig. 6B, left panel). These results indicate that two of the single-residue changes in the N terminus of NS3 result in lack of association with S100A10/p11 in infected cells, which correlates with an attenuated phenotype for virus replication, similar to the data shown above for BTVM14.
Fig. 6.
Effects of single substitutions in NS3 on NS3-S100A10/p11 interaction. (A) Colocalization of NS3/NS3A (green) with S100A10/p11 (red) proteins. BSR cells were infected with mutant viruses as indicated and at 24 h postinfection were processed for confocal microscopy as described in Materials and Methods. (B) Interaction of NS3 with S100A10/p11. BSR cells were infected with WT or mutant viruses and at 36 h postinfection cells were processed as described in Materials and Methods. The expression of NS3 in lysate from cells infected with BTVQ7P (lane 1), BTVR8E (lane 2), or BTVE10A (lane 3) was verified by Western blotting (left panel). The coimmunoprecipitation of NS3 using a monoclonal antibody that recognizes S100A10/p11 was analyzed by Western blotting using the antibody against NS3 (right panel).
Ultrastructural analysis of BSR cells infected with mutant BTVM1, BTVQ7P, and BTVM14 viruses.
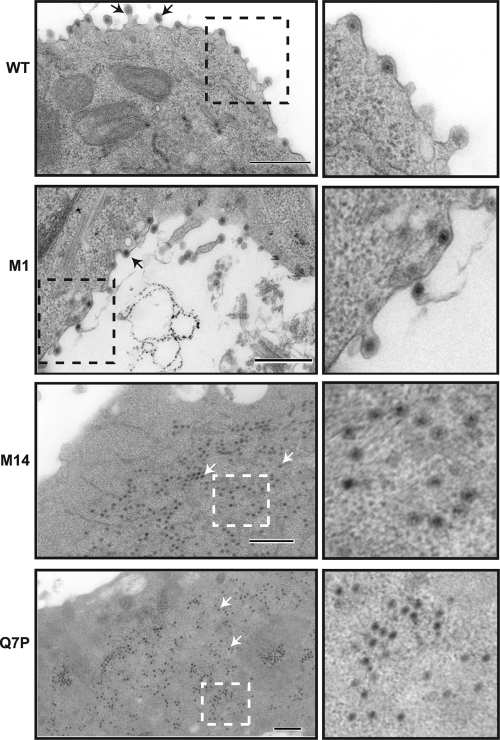
To examine the extent of virus assembly in cells infected with mutant viruses attenuated through deletion or single-residue changes in the N-terminal 13 residues of NS3, BSR cells infected at a high MOI were processed at 24 h postinfection and sectioned for electron microscopy. Cells infected with WT or BTVM1 (WT phenotype) showed virus particles with a typical membrane association and some membrane budding, as previously described (16) (Fig. 7, upper panels). In cells infected with BTVQ7P and BTVM14, however (Fig. 7, lower panels), no membrane-associated particles were observed in any of the fields examined (n = 20; two independent observers). Abundant virus particles were observed but were found exclusively within the cytoplasm. The size and appearance of the particles suggested that they were predominantly incomplete virions, indicating that virus assembly had progressed to a very late stage but had remained incomplete. These images support a hypothesis in which the interaction of the BTV protein NS3 with S100A10/p11 via its N terminus is essential for a very late stage of BTV virus assembly or release.
Fig. 7.
Ultrastructural analysis of mammalian cells infected with mutant or WT viruses. BSR cells were infected with WT, BTVM1, BTVM14, or BTVQ7P virus, and at 24 h postinfection cells were processed for sectioning. Arrows in the left panels indicate particles. An area of each section is amplified in the right panels to show the detail. Bars, 500 nm.
Virus release from vector Culicoides cells.
BTV replicates in insect vector-derived as well as mammalian cells. Unlike in mammalian cells, however, the release of the virus from insect cells appears to be primarily nonlytic, as no cytopathic effect is observed following infection. Thus, to investigate if the phenotypes demonstrated for the NS3 mutants virus described in BSR cells were also observed in insect cells, NS3 mutant viruses were used to infect Culicoides-derived (KC) cells. Growth curves of KC cells infected with each mutant virus or WT virus were determined using plaque assay on BSR/NS3 cells as before. In addition, as BTV is normally secreted from insect cells, both total and extracellular virus titers were determined. Mutant viruses replicated in KC cells, albeit with some attenuation compared to the WT. BTVM1 and BTVR8E, which had wild-type phenotypes in BSR cells, were attenuated in KC cells (Fig. 8 A). BTVM14, BTVQ7P, and BTVE10A, which were severely attenuated in BSR cells, were also most attenuated in KC cells (Fig. 8A). The relative degree of attenuation was greater when the extracellular virus was compared; while all mutants released fewer viruses into the supernatant than did the WT, BTVM1 and BTVR8E reduced virus release by approximately 5-fold whereas BTVM14, BTVQ7P, and BTVE10A reduced virus release by more than 50-fold (Fig. 8B). The fact that all mutant viruses grew in KC cells without additional NS3 provided in trans suggests that the late stages of BTV assembly and release from mammalian and insect cells are fundamentally different.
Fig. 8.
Virus growth and virus release from Culicoides-derived cells. (A) KC cells were infected with mutant BTVM1, BTVM14, BTVQ7P, BTVR8E, or BTVE10A or with WT virus, and at 1, 2, and 3 days postinfection cells and supernatant were harvested and the total titer was determined by plaque assay. (B) KC cells were infected with mutant or WT viruses, and at 4 days postinfection cell-free and cell-associated fractions were collected. The titer of each fraction was determined and the relative release calculated as the ratio of cell-free to total virus titer, normalized for the control virus, and plotted on a logarithmic scale. Bars represent the averages and standard deviations from at least two experiments.
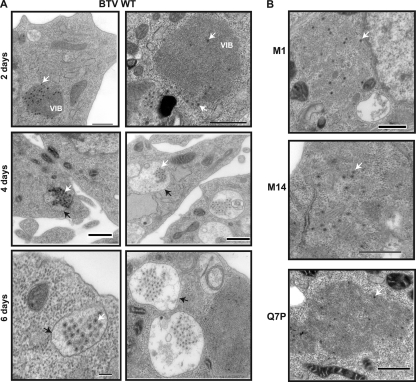
As virus release was attenuated in KC cells, the effects of the NS3 mutations on virus distribution and trafficking, in comparison to that of the WT virus, were examined by use of ultrathin sections of infected KC cells. Sections were prepared at 2, 4, and 6 days postinfection to allow for the fact that BTV replicates more slowly in insect than in mammalian cells (18). In KC cells infected with the WT virus, particles could be detected from day 2 (Fig. 9 A, upper panels) and increased in number at later time points (Fig. 9A, middle and lower panels). At 4 and 6 days postinfection, and in contrast to the results in BSR cells, particles were found predominantly associated with vesicle-like structures (Fig. 9A, middle and lower panels). No budding from the plasma membrane was seen at any time point. In contrast, KC cells infected with BTV M1, BTVM14, and BTVQ7P viruses at 6 days postinfection were found exclusively in the cytoplasm of the cells and were not associated with vesicles (compare Fig. 9A, lower panels, and B). Thus, while KC cells supported virus replication in the case of the NS3 mutations described, there was an altered route of virus trafficking compared to that for the WT, which may be consistent with reduced virus release.
Fig. 9.
Cell sectioning analysis of infected insect Culicoides cells. (A) KC cells were infected with WT virus, harvested at 2, 4, and 6 days postinfection, and processed for cell sectioning. White arrows indicate virus particles and black arrows membrane structures. (B) KC cells were infected with mutant BTVM1, BTVM14, or BTVQ7P viruses as indicated and processed at 6 days postinfection. Bar, 500 nm.
DISCUSSION
The canonical view of the release of nonenveloped viruses from infected cells is that the predominant mechanism involved is cell lysis. However, there is an increasing appreciation that, as for enveloped viruses, complex trafficking and budding strategies are also involved. In the case of BTV, both mechanisms appear to act with a preponderance of lysis during mammalian cell infection but essentially none during infection of insect cells (15, 28, 29). Nonstructural protein 3 (NS3), whose expression level varies between hosts (14), is critical to these processes. The mechanism of release may be linked to pathogenesis and spread, plausibly via CPE, as NS3 is also one of the most rapidly evolving of the BTV segments (2). Understanding the precise functions of NS3 during the virus replication cycle is therefore an important goal. Uniquely among the BTV genomic segments, the NS3-encoding segment, S10, expresses two proteins in infected cells, NS3 and NS3A. NS3A is the result of a second initiation event at methionine 14 and thus lacks the first 13 residues present in the full-length protein. The specific function of NS3A, if any, during virus infection is not known, although the methionine is conserved, suggesting that it plays an important role in the life cycle.
Previous work has shown that BTV NS3 protein interacts with Tsg101 and that this interaction is essential for virus budding from mammalian cells (7, 31). In addition, yeast two-hybrid screening identified a cellular protein, S100A10/p11, which is known to be involved in the exocytic pathway, as a binding partner and showed that the complete deletion of, or mutations in, the N-terminal 13 residues of NS3 perturbed this interaction. The impact of those mutations in virus replication, however, could not be assessed directly. Here, using the established RG system for BTV (5) and an NS3 helper cell line which enabled the isolation of conditionally lethal mutations (7), we investigated the role of the N-terminal domain of NS3 in virus release, addressing directly the specific role of NS3A.
A mutant virus expressing only NS3 (BTVM1) was able to replicate in BSR cells with near-WT kinetics, while a mutant virus expressing only NS3A (BTVM14) failed to replicate. The block in the replication cycle for BTVM14 was shown to be at a late stage, as virus infections showed virus protein expression, genome replication, and assembly into virus cores or incomplete particles. Virus budding, which was observed for WT BTV, was not observed for BTVM14, which appeared rather to accumulate in the cytosol, supporting a hypothesis that virus release, which may be coupled to the late stages of assembly, was the point of arrest. The failure of the replication cycle to progress correlated with a failure of BTVM14 to engage with S100A10/p11, as evidenced by a lack of intracellular localization and interaction demonstrated by pulldown assay. Both colocalization and pulldowns were apparent for the WT virus and mutant BTVM1. Moreover, two out of three point mutations within the N-terminal 13 residues of NS3 previously identified as preventing interaction with S100A10/p11 in vitro gave a phenotype similar to that of BTVM14 following their rescue into replicating virus through reverse genetics. These data strongly support a direct link between the ability of NS3 to interact with S100A10/p11 in vivo and virus release from infected cells. One single-residue mutant, BTVR8E, retained a near-WT phenotype despite the reported lack of interaction with S100A10/p11 in vitro (3). It is probable that the strength of association in vivo was sufficient to enable biological functionality although unable to demonstrate biochemical interaction under the in vitro conditions used, which only inadequately represent the infected cell.
Thus, NS3 interaction with S100A10/p11 is a key event in the late stages of BTV replication in BSR cells, and the first 13 residues of NS3, directly or indirectly, mediate the interaction concerned. An engagement by BTV with S100A10/p11 in infected cells appears to be physiologically reasonable, as S100A10/p11 is present in both cytosol and membranes and recent observations have shown it to be involved in the trafficking of plasma membrane proteins (6). Several cellular membrane proteins, including sodium channel Nav 1.8 and potassium channel TASK1, interact with S100A10/p11 (13, 20), and binding of S100A10/p11 to a cytosolic domain of these integral plasma membrane proteins was necessary and sufficient for presentation on the cell surface (reviewed in reference 21). BTV evidently usurps this pathway, among others such as the ESCRT pathway (31), in mammalian cells.
As an arthropod-borne virus, BTV also replicates in insect cells, where, compared to mammalian cells, less is known about virus replication, trafficking, and release. Release of BTV from infected insect cells is predominantly nonlytic with little or no cytopathic effect (18). Accordingly, the NS3 mutants used for an analysis of function in BSR cells were also used to assess replication in KC (Culicoides-derived) cells. In contrast to growth in mammalian cells, all mutant viruses grew in KC cells but with variable degrees of attenuation. Notably, attenuation was apparent with BTVM1, which exhibited a WT phenotype in BSR cells, suggesting from the outset that the pathways of assembly and release differ significantly between the two hosts. The attenuation shown by BTVM1 virus suggests that the second methionine in NS3, which initiates the expression of NS3A, is essential in the insect cell background. Further, infection of insect cells with BTVR8E, which retained NS3A, exhibited higher virus release than that with BTVM1, which lacked the NS3A. A comparison between total titer and virus titer in the supernatant suggested that the primary point of attenuation was in virus release. In support of this, ultrathin sections of KC cells infected with selected NS3 mutants revealed that while virus release from WT BTV-infected cells was associated with virus entrapment in membrane-bound vesicle-like structures, no such ultrastructure was apparent for any of the NS3 mutants tested; rather, particles were scattered throughout the cytosol. BTV association with vesicles in insect cells has not been previously reported. However, other nonenveloped viruses have been shown to be vesicle associated; Peruvian horse sickness virus, another member of the genus Orbivirus, associates with vesicles in mosquito-derived C6/36 cells (1), and for both the parvovirus minute virus of mice and the picornavirus poliovirus, egress has also been shown to be mediated by vesicles (24). These data suggest that while the N terminus of NS3 is not essential for virus growth in insect cells (as shown by growth of BTVM14), it is nonetheless involved in the trafficking of virus to exit the cell. It is possible that the N-terminal domain interacts with an as-yet-unidentified insect cell factor involved in egress, that the attenuation is mediated due to misfolding of the NS3A protein induced by residue changes in the N terminus, or that both NS3 and NS3A are required for optimum replication. Further analysis of the pathways of virus release in insect cells will be required to resolve this point.
In summary, our study has demonstrated that NS3 and NS3A, the smallest of the BTV-encoded proteins, play important roles in virus assembly or virus release in both vector and mammalian cells. NS3 appears to engage with various cellular factors, including cellular exocytic pathway proteins, to enable efficient virus release, the exact mechanism of which will require further study. The different requirements for NS3 and NS3A in the two cell types offer a possible explanation for the conservation of the second initiation codon in the S10 gene of members of the orbiviruses.
ACKNOWLEDGMENTS
We thank Maria McCrossan (LSHTM) for technical help with electron microscopy experiments.
This work was funded by the Biotechnology and Biological Sciences Research Council (BBSRC), United Kingdom.
Footnotes
Published ahead of print on 16 March 2011.
REFERENCES
- 1. Attoui H., et al. 2009. Peruvian horse sickness virus and Yunnan orbivirus, isolated from vertebrates and mosquitoes in Peru and Australia. Virology 394:298–310 [DOI] [PubMed] [Google Scholar]
- 2. Balasuriya U. B., et al. 2008. The NS3 proteins of global strains of bluetongue virus evolve into regional topotypes through negative (purifying) selection. Vet. Microbiol. 126:91–100 [DOI] [PubMed] [Google Scholar]
- 3. Beaton A. R., Rodriguez J., Reddy Y. K., Roy P. 2002. The membrane trafficking protein calpactin forms a complex with bluetongue virus protein NS3 and mediates virus release. Proc. Natl. Acad. Sci. U. S. A. 99:13154–13159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bhattacharya B., Roy P. 2008. Bluetongue virus outer capsid protein VP5 interacts with membrane lipid rafts via a SNARE domain. J. Virol. 82:10600–10612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boyce M., Celma C. P., Roy P. 2008. Development of a reverse genetics system for bluetongue virus: recovery of infectious virus from synthetic RNA transcripts. J. Virol. 82:8339–8348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Broome A. M., Ryan D., Eckert R. L. 2003. S100 protein subcellular localization during epidermal differentiation and psoriasis. J. Histochem. Cytochem. 51:675–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Celma C. C., Roy P. 2009. A viral nonstructural protein regulates bluetongue virus trafficking and release. J. Virol. 83:6806–6816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clayson E. T., Brando L. V., Compans R. W. 1989. Release of simian virus 40 virions from epithelial cells is polarized and occurs without cell lysis. J. Virol. 63:2278–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Demirov D. G., Freed E. O. 2004. Retrovirus budding. Virus Res. 106:87–102 [DOI] [PubMed] [Google Scholar]
- 10. Freed E. O. 2002. Viral late domains. J. Virol. 76:4679–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. French T. J., Inumaru S., Roy P. 1989. Expression of two related nonstructural proteins of bluetongue virus (BTV) type 10 in insect cells by a recombinant baculovirus: production of polyclonal ascitic fluid and characterization of the gene product in BTV-infected BHK cells. J. Virol. 63:3270–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garrus J. E., et al. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55–65 [DOI] [PubMed] [Google Scholar]
- 13. Girard C., et al. 2002. p11, an annexin II subunit, an auxiliary protein associated with the background K+ channel, TASK-1. EMBO J. 21:4439–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guirakhoo F., Catalan J. A., Monath T. P. 1995. Adaptation of bluetongue virus in mosquito cells results in overexpression of NS3 proteins and release of virus particles. Arch. Virol. 140:967–974 [DOI] [PubMed] [Google Scholar]
- 15. Homan E. J., Yunker C. E. 1988. Growth of bluetongue and epizootic hemorrhagic disease of deer viruses in poikilothermic cell systems. Vet. Microbiol. 16:15–24 [DOI] [PubMed] [Google Scholar]
- 16. Hyatt A. D., Eaton B. T., Brookes S. M. 1989. The release of bluetongue virus from infected cells and their superinfection by progeny virus. Virology 173:21–34 [DOI] [PubMed] [Google Scholar]
- 17. Hyatt A. D., Zhao Y., Roy P. 1993. Release of bluetongue virus-like particles from insect cells is mediated by BTV nonstructural protein NS3/NS3A. Virology 193:592–603 [DOI] [PubMed] [Google Scholar]
- 18. Jennings M., Boorman J. 1979. The susceptibility of cell lines of Aedes aegypti (Linn.), Aedes albopictus (Skuse) and Aedes pseudoscutellaris (Therobald) to infection with bluetongue virus. Arch. Virol. 59:121–126 [DOI] [PubMed] [Google Scholar]
- 19. Martin-Serrano J., Zang T., Bieniasz P. D. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313–1319 [DOI] [PubMed] [Google Scholar]
- 20. Okuse K., et al. 2002. Annexin II light chain regulates sensory neuron-specific sodium channel expression. Nature 417:653–656 [DOI] [PubMed] [Google Scholar]
- 21. Rescher U., Gerke V. 2008. S100A10/p11: family, friends and functions. Pflugers Arch. 455:575–582 [DOI] [PubMed] [Google Scholar]
- 22. Reynolds E. S. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roy P. 2008. Functional mapping of bluetongue virus proteins and their interactions with host proteins during virus replication. Cell. Biochem. Biophys. 50:143–157 [DOI] [PubMed] [Google Scholar]
- 24. Taylor M. P., Burgon T. B., Kirkegaard K., Jackson W. T. 2009. Role of microtubules in extracellular release of poliovirus. J. Virol. 83:6599–6609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tucker S. P., Thornton C. L., Wimmer E., Compans R. W. 1993. Vectorial release of poliovirus from polarized human intestinal epithelial cells. J. Virol. 67:4274–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Niekerk M., et al. 2003. Variation in the NS3 gene and protein in South African isolates of bluetongue and equine encephalosis viruses. J. Gen. Virol. 84:581–590 [DOI] [PubMed] [Google Scholar]
- 27. Verwoerd D. W., Els H. J., De Villiers E. M., Huismans H. 1972. Structure of the bluetongue virus capsid. J. Virol. 10:783–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wechsler S. J., McHolland L. E. 1988. Susceptibilities of 14 cell lines to bluetongue virus infection. J. Clin. Microbiol. 26:2324–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wechsler S. J., McHolland L. E., Tabachnick W. J. 1989. Cell lines from Culicoides variipennis (Diptera: Ceratopogonidae) support replication of bluetongue virus. J. Invertebr Pathol. 54:385–393 [DOI] [PubMed] [Google Scholar]
- 30. Weiner M. P., et al. 1994. Site directed mutagenesis of double stranded DNA by the polymerase chain reaction. Gene 151:119–123 [DOI] [PubMed] [Google Scholar]
- 31. Wirblich C., Bhattacharya B., Roy P. 2006. Nonstructural protein 3 of bluetongue virus assists virus release by recruiting ESCRT-I protein Tsg101. J. Virol. 80:460–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu X., Chen S. Y., Iwata H., Compans R. W., Roy P. 1992. Multiple glycoproteins synthesized by the smallest RNA segment (S10) of bluetongue virus. J. Virol. 66:7104–7112 [DOI] [PMC free article] [PubMed] [Google Scholar]