Abstract
The Epstein-Barr virus (EBV) BRRF1 lytic gene product (Na) is encoded within the same immediate-early region as the BZLF1 (Z) and BRLF1(R) gene products, but its role during EBV infection has not been well defined. We previously showed that Na cooperates with the R protein to induce lytic gene expression in latently infected EBV-positive 293 cells, and in some EBV-negative cell lines it can activate the Z promoter in reporter gene assays. Here we show that overexpression of Na alone is sufficient to induce lytic gene expression in several different latently infected epithelial cell lines (Hone-Akata, CNE2-Akata, and AGS-Akata), while knockdown of endogenous Na expression reduces lytic gene expression. Consistent with its ability to interact with tumor necrosis factor receptor-associated factor 2 (TRAF2) in a yeast two-hybrid assay, we demonstrate that Na interacts with TRAF2 in cells. Furthermore, we show that TRAF2 is required for Na induction of lytic gene expression, that Na induces Jun N-terminal protein kinase (JNK) activation in a TRAF2-dependent manner, and that a JNK inhibitor abolishes the ability of Na to disrupt viral latency. Additionally, we show that Na and the tumor suppressor protein p53 cooperate to induce lytic gene expression in epithelial cells (including the C666-1 nasopharyngeal carcinoma cell line), although Na does not appear to affect p53 function. Together these data suggest that Na plays an important role in regulating the switch between latent and lytic infection in epithelial cells and that this effect requires both the TRAF2 and p53 cellular proteins.
INTRODUCTION
Epstein-Barr virus (EBV) is a gammaherpesvirus that causes infectious mononucleosis and infects a large percentage of the human population (51, 66). EBV is associated with several epithelial cell cancers, such as nasopharyngeal carcinoma (NPC) and gastric carcinoma, and B-cell cancers, including Burkitt lymphoma and Hodgkin lymphoma (51, 66). EBV normally establishes a life-long latent infection in the memory B cells of the host but is reactivated periodically to the lytic form of infection. Lytic reactivation in B cells occurs following B-cell receptor engagement and/or plasma cell differentiation (40). Although EBV infection of normal oral epithelial cells commonly results in lytic infection (51), EBV-positive epithelial tumors are composed primarily of cells with the latent form of viral infection (37, 51).
The switch between latent and lytic infection is induced by the EBV immediate-early (IE) proteins BZLF1 (Z; also known as Zta, Zebra, or EB1) and BRLF1 (R) (13, 15, 37, 61, 65). Z and R serve as transcriptional activators and function to transactivate one another's promoters and to activate early lytic gene expression (3, 13, 14, 16, 19, 22, 29, 32, 35, 43, 50, 52, 61, 65). The cellular and viral proteins that regulate transcription of the Z and R gene products thus play a key role in determining if EBV infection is latent or lytic.
The Z promoter (Zp) is regulated by a number of cellular factors. In latently infected cells, several repressors have been shown to inhibit Zp activity, including the ZEB1, ZEB2, and MEF2D cellular proteins (21, 30, 39, 45, 57). During lytic infection, a variety of cellular transcription factors, including CREB, ATF-1, ATF-2, c-Jun, and XBP-1, bind to the Zp ZII motif (a CRE binding site), leading to Zp activation (1, 6, 44, 47, 58, 63). Phosphorylation of the c-Jun transcription factor by the c-Jun N-terminal protein kinase (JNK) greatly enhances its activity (17), and activation of JNK activity has been shown to be important for lytic EBV induction mediated by a variety of different agents (9, 23, 24).
The EBV BRRF1 open reading frame, which encodes the Na protein, is contained within the same locus as the Z and R IE genes. Our lab previously showed that Na enhances R-mediated disruption of viral latency in 293 cells that are stably infected with an EBV mutant that does not express either R or Na (293RKO cells) (33). Likewise, the murine gammaherpesvirus 68 (MHV68) Na homologue (ORF49) has been reported to enhance the ability of the R homologue (Rta) to induce lytic viral gene expression (41). Furthermore, we showed that Na activates a Zp-driven reporter gene in EBV-negative HeLa cells through the Zp ZII motif (a c-Jun binding site), and that Na induces c-Jun phosphorylation and c-Jun transcriptional activity (33). However, the mechanism by which Na induces c-Jun activation and the role that Na plays during the switch between latent and lytic infection in epithelial cell lines remains unclear.
The Na protein was previously demonstrated to interact with the cellular protein tumor necrosis factor (TNF) receptor-associated factor 2 (TRAF2) in a yeast two-hybrid analysis (10), although the possible functions(s) of this interaction and whether it occurs in vivo remain unknown. TRAF2 is a signal transducer for the TNF receptor superfamily members, including TNFR1/2, CD40, and IRE1 (4, 8). TRAF2 signaling mediates several major signaling pathways following receptor activation, including those of mitogen-activated protein kinase, classical and noncanonical NF-κB, and JNK (4, 8). In EBV-infected B cells, TRAF2 interacts with latent membrane protein 1 (LMP1), and this interaction is thought to be important for LMP1 activation of NF-κB (18, 34, 56). Although the role of TRAF2 during latent infection of B cells has been well studied, the role of TRAF2 during EBV lytic infection remains unclear.
In this study, we demonstrate that overexpression of the Na protein induces lytic gene expression in several different latently infected epithelial cell lines and that, conversely, knockdown of endogenous Na expression inhibits lytic gene expression. We confirm that Na interacts with TRAF2 in vivo and show that TRAF2 expression is required for Na disruption of latency. We also demonstrate that Na induces JNK phosphorylation in a TRAF2-dependent manner and that JNK activation is required for Na-induced lytic gene expression. Furthermore, we demonstrate that the ability of Na to disrupt viral latency also requires the p53 cellular protein, consistent with a recent report showing that p53 is required for histone deacetylase (HDAC) inhibitor (HDACi)-induced EBV lytic infection in a latently infected NPC cell line (12). Together these results suggest that Na plays an important role in promoting EBV lytic reactivation in epithelial cells, and the results also reveal unexpected roles for TRAF2, as well as for p53, in Na-induced lytic reactivation.
MATERIALS AND METHODS
EBV cell lines.
Hone-Akata (a gift from Lawrence Young) and CNE2-Akata (a gift from K. W. Lo at The Chinese University of Hong Kong [received via Diane Hayward]) are NPC epithelial cell lines superinfected with the Akata strain of EBV (25, 46), and they were maintained in RPMI medium supplemented with 10% fetal bovine serum (FBS), penicillin-streptomycin, and G418 (400 μg/ml). CNE2-Akata cells contain an arginine-to-threonine mutation at codon 280 of p53 (20) as well as wild-type p53 (11). C666-1 cells, an NPC line, were maintained in RPMI medium supplemented with 10% FBS and penicillin-streptomycin and were grown on plates treated with fibronectin (Sigma). C666-1 cells (a gift from Dolly Huang) contain a deletion at codon 249 of p53 (64). 293 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FBS and penicillin-streptomycin. AGS gastric carcinoma cells were maintained in F-12 medium supplemented with 10% FBS and penicillin-streptomycin. AGS-Akata cells are gastric carcinoma cells superinfected with the Akata strain of EBV and were maintained in F-12 medium supplemented with 10% FBS, penicillin-streptomycin, and G418 (400 μg/ml). Raji cells (ATCC) were maintained in RPMI 1640 medium supplemented with 10% FBS and penicillin-streptomycin. MutuI cells (a gift from Jeff Sample) are an EBV-positive Burkitt lymphoma cell line and were maintained in RPMI medium supplemented with 10% FBS and penicillin-streptomycin.
Plasmids.
Plasmid DNA was purified using Qiagen maxi-prep columns according to the manufacturer's protocol. pSG5 and pCDNA3.1 were obtained from Stratagene and Invitrogen, respectively. pSG5-FLAGNa was generated by PCR amplification of the Na open reading frame from EBV B95.8 viral DNA with addition of a FLAG tag to the amino terminus of Na with oligonucleotides BRRF1-EcoRI-FLAG (5′-GCCCGAATTCACCATGGACATCAAGGACGACGATGACAAGGCTAGTAGTAACAGAGG-3′) and BRRF1-BamHI-Rev (5′-GCCCCGGATCCGACGCAGGTAAGAG-3′). The PCR product was then ligated into pSG5 digested with EcoRI and BamHI restriction enzymes. pSG5-R, a gift from Diane Hayward, contains the genomic sequence encoding R beginning from the second exon but lacks the Na-encoding first exon (32). pCMV-FLAGNa was a gift from Henri Gruffat and was previously described (55). The pOriP plasmid contains the EBV origin of replication P site and the cytomegalovirus (CMV) promoter (a gift from Bill Sugden). pOriP-FLAGNa was cloned by digesting pCMV-FLAGNa with HindIII and XbaI and ligating the FLAG-Na fragment into pOriP digested with HindIII and NheI. pHA-TRAF2 is a pCDNA3 vector containing a hemagglutinin (HA) epitope fused to the human TRAF2 open reading frame and was a gift from Bill Sugden (53). pNa-YFP was generated by PCR amplification of FLAG-Na by using pSG5-FLAGNa as template with oligonucleotides SacI-FLAG-FOR (5′-GGCGAGCTCATGGACTACAAGGACGACG-3′) and KpnI-BRRF1-REV (5′-GCCGGTACCTTTGTATTGCATGGCAGAACAG-3′). The PCR product was then ligated into pCMX-YFP digested with KpnI and SacI restriction enzymes. pTRAF2-YFP was generated by PCR amplification of HA-TRAF2 by using pHA-TRAF2 as a template with oligonucleotides KpnI-TRAF2-FOR (5′-GCCGGTACCATGGCTGCAGCTAGCGTG-3′) and KpnI-TRAF2-Rev (5′-GGCGGTACCGAGCCCTGTCAGGTCC-3′). The resulting PCR product was then ligated into pCMX-YFP digested with KpnI restriction enzyme and screened for correct orientation. pLKO.1 was obtained from Open Biosystems. shTRAF2 and shp53 were obtained from Open Biosystems and are mixtures of pLKO.1 vectors expressing short hairpin RNAs against various TRAF2 and p53 sequences. pG13-CAT (a gift from Bert Vogelstein) (36) contains 13 copies of the p53 consensus sequence upstream of the polyomavirus early promoter driving the chloramphenicol acetyltransferase (CAT) reporter gene. The p53 expression vector encodes p53 driven by the CMV promoter (pC53-SN3; a gift from Bert Vogelstein) (5). Mutant p53 constructs p53-175 and p53-248 were also a gift from Bert Vogelstein.
DNA transfection.
DNA was transfected into 293, Hone-Akata, CNE2-Akata, and C666-1 cells using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol.
siRNA transfections.
Small interfering RNAs (siRNAs) were transfected into cells using either Xtreme gene (Roche), Lipofectamine (Invitrogen), or Lipofectamine RNAimax (Invitrogen) according to the corresponding manufacturer's protocol. Control 2 siRNA was obtained from Ambion and targets a nonspecific sequence. TRAF2 siRNA (Santa Cruz Biotechnology) contains a mixture of TRAF2 siRNA sequences. Duplexed siRNA targeting Na (5′-AAUCUUUCCUCCAUGAGCUUU-3′) was obtained from Integrated DNA Technologies.
Chemical reagents.
The JNK inhibitor SP600125 (Sigma) was used at a final concentration of 10 μM and left on the cells for 2 days posttransfection. TNF-α (Sigma) was used at a final concentration of 10 ng/ml for the indicated times.
Virus titration assay.
Virus titration assays were performed as previously described (33). CNE2-Akata cells were transfected with control or FLAG-Na expression vectors. Medium was changed 2 days posttransfection, and supernatant was harvested and filtered through a 0.8-μm-pore-size filter 3 days posttransfection. Raji cells (2 × 105 cells/infection) were infected with various amounts of virus and incubated at 37°C. Phorbol-12-myristate-13-acetate (TPA; 20 ng/ml) and sodium butyrate (3 mM, final concentration) were added 24 h after infection. Green fluorescent protein (GFP)-positive Raji cells were counted 48 h postinfection to determine viral titers.
Immunoblot analysis.
Immunoblotting was performed as previously described (2, 7). Cell lysates were harvested in SUMO buffer containing protease inhibitor cocktail (Roche) and quantified in a SUMO protein assay (Bio-Rad). Equivalent amounts of protein were separated in sodium dodecyl sulfate–10% polyacrylamide gels and transferred to membranes. Membranes were blocked in phosphate-buffered saline (PBS) containing 5% milk and 0.1% Tween 20 solution and incubated with primary antibody. Immunoblots were probed with the following antibodies: anti-FLAG (1:2,000; Sigma), anti-R (1:250; Argene), anti-Z (1:250; BZ-1; Santa Cruz Biotechnology), anti-EAD (1:250; BMRF1; Vector), anti-β-actin (1:5,000; Sigma), anti-HA (1:500; Santa Cruz Biotechnology), anti-TRAF2 (1:750; Santa Cruz, Biotechnology), anti-phospho-JNK (1:1,000; Cell Signaling), anti-JNK (1:1,000; Cell Signaling), anti-p53 (1:250; Santa Cruz Biotechnolgoy), anti-IκBα (1:250; Santa Cruz Biotechnology), anti-p52 (1:1,000; Millipore), anti-Na (1:500; a kind gift from Henri Gruffat [55]), anti-tubulin (1:2,000; Sigma), and anti-lamin A/C (1:500; Santa Cruz Biotechnology).
Immunoprecipitation analysis.
293 cells were transfected with the indicated expression plasmids, and cell lysates were harvested in NP-40 lysis buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 0.5% NP-40, 0.75% IPEGAL) containing protease inhibitor cocktail (Roche) 48 h posttransfection. Protein levels were quantified by using the Bradford assay. Aliquots of 150 μg of protein were incubated with 1 μg FLAG or HA antibodies for 2 h. Protein-antibody complexes were then incubated with 25 μl protein A/G-agarose beads (Santa Cruz Biotechnology). Beads were washed three times with NP-40 lysis buffer, and samples were eluted by boiling the beads in twice sample buffer. Samples were resolved on an SDS-PAGE gel and probed for the indicated proteins as described above.
REAP cell fractionation.
Cell fractionation was performed using the rapid, efficient, and practical (REAP) method as previously described (60). CNE2-Akata cells were transfected with various expression vectors and harvested 48 h posttransfection. Cells were washed with PBS, scraped off the dishes, and pelleted. Cell pellets were resuspended in PBS containing 0.1% NP-40, and a portion was saved for whole-cell lysates. The remaining sample was pelleted, and the supernatant was saved as the cytosolic fraction. The pellet was washed in PBS–0.1%NP-40 and pelleted, and the supernatant was discarded. The remaining pellet was then resuspended in sample buffer and saved as the nuclear fraction.
Fluorescent microscopy.
Hone-Akata cells plated on glass coverslips were transfected with various yellow fluorescent protein (YFP) expression plasmids and fixed 24 h after transfection with 0.4% paraformaldehyde in PBS. Cell nuclei were stained with Hoechst 33342 (Sigma), and coverslips were mounted onto slides by using Vectashield hard set. Images were visualized using a Nikon A1R confocal microscope.
CAT reporter assay.
CAT assays were performed as previously described (27). CNE2-Akata cells were transfected with the indicated expression vectors and harvested in 0.25 M Tris (pH 7.5) and SUMO buffers 48 h after transfection. Tris buffer lysates were incubated with acetyl coenzyme A (CoA) and [14C]chloramphenicol, and acetyltransferase activity was determined following thin-layer chromatography. Activity was quantified on a Storm 840 PhosphorImager (Molecular Dynamics). Western blot analysis was performed on the SUMO buffer lysates to determine protein expression.
RESULTS
Na induces lytic gene expression in EBV-infected epithelial cells.
Although the Na protein was previously shown to induce c-Jun phosphorylation and activate the Z promoter in EBV-negative HeLa cells (33), overexpression of Na has not previously been shown to induce lytic EBV gene expression in the context of the intact viral genome. To examine this further, we transfected Hone-Akata cells (a nasopharyngeal carcinoma line superinfected with the Akata strain of EBV) with two different Na expression vectors (FLAG tagged), or the appropriate control vectors, and examined the expression levels of various lytic EBV proteins 48 h posttransfection. As shown in Fig. 1A, overexpression of the Na protein clearly induced expression of the EBV lytic proteins BMRF1 and Z. These results indicate that activation of Na expression is sufficient to disrupt viral latency in the Hone-Akata cell line.
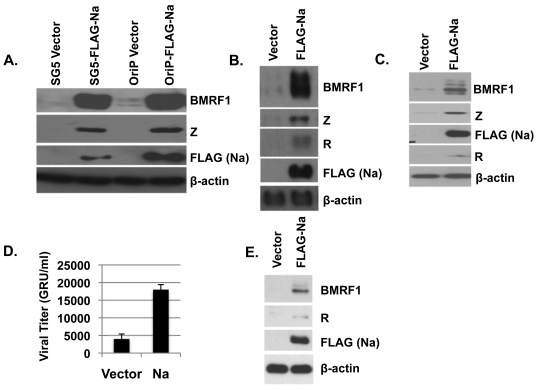
Fig. 1.
Overexpression of Na induces lytic infection. (A to C) Hone-Akata (A), CNE2-Akata (B), or AGS-Akata (C) cells were transfected with various expression plasmids encoding control vector, FLAG-Na, or R as indicated, and cell lysates were harvested 2 days later. Immunoblot analysis was performed using antibodies against the lytic proteins BMRF1, Z, and R. FLAG antibody was used to detect the transfected Na protein. β-Actin was included as an internal loading control. Na expression had similar effects when expressed in either an SG5-based vector (SG5-FLAG-Na) or an OriP-based vector (OriP-FLAG-Na). (D) CNE2-Akata cells were transfected with control or Na expression plasmids, and the titers of infectious viral particles released into the supernatant were determined 3 days posttransfection using the green Raji cell assay to determine the number of green Raji units (GRU). (E) The CNE2-Akata cells used in the experiment shown in panel D were harvested 3 days posttransfection, and the levels of BMRF1, R, FLAG (Na), and β-actin protein expression were examined by immunoblotting.
To ensure Na disruption of viral latency was not specific to the Hone-Akata line, the effect of Na overexpression was also examined in two additional EBV superinfected epithelial cell lines, CNE2-Akata (an NPC line) (Fig. 1B) and AGS-Akata (a gastric carcinoma cell line) (Fig. 1C). Similar to the Na effect in Hone-Akata cells, Na also induced expression of lytic EBV proteins (BMRF1, Z, and R) in CNE2-Akata and AGS-Akata cells. Together, these results indicate that overexpression of Na alone is sufficient to induce lytic gene expression in at least a subset of EBV-infected epithelial cell lines.
To determine if Na expression also results in enhanced lytic viral replication, CNE2-Akata cells were transfected with control or Na expression vectors, and the amount (titer) of infectious viral particles released into the supernatant was determined by using the green Raji cell assay 3 days posttransfection (Fig. 1D). Consistent with the ability of Na to induce early lytic protein expression (Fig. 1E), CNE2-Akata cells transfected with Na also had increased virus production in comparison to cells transfected with the control vector (Fig. 1D).
Knockdown of endogenous Na expression inhibits lytic reactivation in EBV-positive epithelial cells.
Given that overexpression of Na can induce lytic gene expression in some EBV-infected epithelial cell lines, we next examined the effect of inhibiting Na expression derived from the endogenous viral genome by transfecting cells with control siRNA, or an siRNA directed against Na, into Hone-Akata cells. As shown in Fig. 2A, knockdown of endogenous Na expression not only greatly inhibited the low-level constitutive lytic viral protein expression that sometimes occurs in this cell line but also attenuated the ability of the lysis-inducing agents TPA and sodium butyrate (NaBut) to activate lytic protein expression (Fig. 2A). These results demonstrate that Na derived from the endogenous viral genome contributes to both constitutive and chemically induced lytic viral protein expression in Hone-Akata cells.
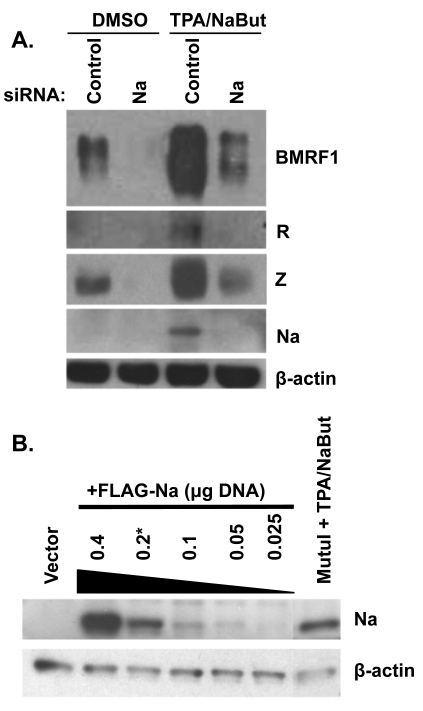
Fig. 2.
Knockdown of Na inhibits lytic protein expression. (A) CNE-2 Akata cells were transfected with siRNAs targeting control or Na sequences. Cells were then treated with either dimethyl sulfoxide (DMSO) or TPA (20 ng/ml) plus sodium butyrate (3 mM; NaBut). Cell lysates were harvested 48 h after induction, and immunoblot analysis was performed using antibodies against BMRF1, Na, R, Z, and β-actin. (B) CNE2-Akata cells were transfected with various amounts of the FLAG-Na vector as indicated (in a 12-well plate). At 48 h posttransfection, an immunoblot assay was performed (using an anti-Na antibody) to compare the level of Na expressed in the transfected CNE-Akata cells versus that expressed from the endogenous viral genome in MutuI cells treated with TPA/NaBut. β-Actin levels were examined as a protein loading control.
To determine if the level of Na protein obtained following Na plasmid transfection was physiologic with regard to the level of endogenous Na protein expressed during normal lytic infection, we transfected CNE2-Akata cells with different amounts of the FLAG-Na construct and used an anti-Na antibody to compare the level of transfected Na versus that expressed in TPA-sodium butyrate-treated MutuI cells (Fig. 2B). The amount of Na expressed in the lytically induced MutuI Burkitt lymphoma cells was comparable to the amount of expressed in the CNE2-Akata cells transfected with 0.2 μg Na expression vector (in a 12-well plate). Since most of the transfection experiments presented in this study used 0.2 μg (or less) of the Na vector, we conclude that the amount of Na expressed is physiologic.
Na induction of JNK activity is required for Na disruption of viral latency.
Our lab previously showed that Na induces c-Jun phosphorylation and enhances c-Jun transactivator function (33). Furthermore, expression of the Kaposi's sarcoma-associated herpesvirus (KSHV) homologue of Na, Orf49, in 293 and CV-1 cells induces phosphorylation of both JNK and c-Jun (26). To determine if Na likewise induces JNK phosphorylation, Hone-Akata cells were transfected with Na or control expression plasmids, and the level of JNK activation was examined by immunoblot analysis using antibodies directed against phosphorylated (p-JNK) versus total JNK. As shown in Fig. 3A, p-JNK levels increased in the presence of Na, while the total JNK level was not affected. Similar results were obtained in CNE2-Akata cells (Fig. 3B). These results suggest that Na, like the KSHV Orf49 protein, induces c-Jun activation by promoting JNK phosphorylation.
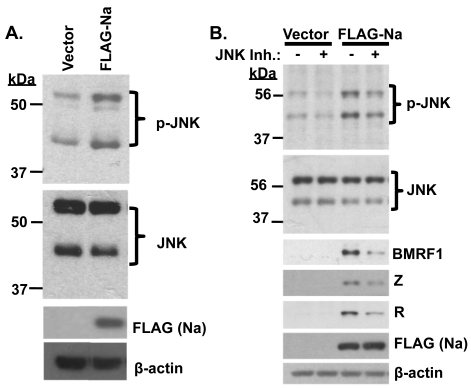
Fig. 3.
Na induction of JNK is required for Na to activate lytic protein expression. (A) Hone-Akata cells were transfected with control vector or FLAG-Na expression plasmids, and immunoblot analysis was performed using antibodies against phosphorylated JNK (p-JNK), total JNK, FLAG (to detect transfected Na), and β-actin. (B) CNE2-Akata cells were transfected with control vector or FLAG-Na expression plasmids in the presence or absence of the JNK inhibitor SP600125 (10 μM). Cell lysates were harvested, and immunoblot analysis was performed to examine p-JNK, total JNK, BMRF1, Z, R, FLAG (Na), and β-actin expression levels.
To determine if JNK activation is required for Na-mediated disruption of viral latency, CNE2-Akata cells were transfected with control or Na expression plasmids in the presence or absence of the JNK inhibitor SP600125 (10 μM). The ability of Na to induce expression of the lytic EBV proteins BMRF1, Z, and R was reduced in the presence of the JNK inhibitor (Fig. 3B). These results suggest that Na activation of JNK plays an important role in Na-induced disruption of viral latency.
Na interacts with the cellular protein TRAF2 in vivo.
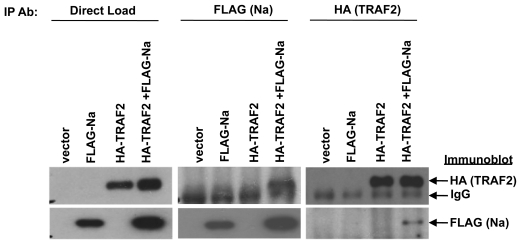
Since Na has been reported to interact directly with the cellular protein TRAF2 by yeast two-hybrid analysis (10) and TRAF2 is involved in activating JNK during certain forms of signaling, these observation suggest that the ability of Na to activate JNK may involve a direct interaction with the TRAF2 protein. Nevertheless, the TRAF2 protein is not thought to enter the nucleus in most cell types, while Na has been reported to be a nuclear protein (55). To determine whether the Na and TRAF2 proteins can interact in vivo, 293 cells were transfected with FLAG-Na and HA-TRAF2 expression plasmids, immunoprecipitation assays were performed using antibodies directed against the FLAG or HA tags, and then immunoblot analyses was conducted to detect coprecipitated FLAG-Na and HA-TRAF2 proteins. As shown in Fig. 4, an interaction between Na and TRAF2 was detected following immunoprecipitation with either FLAG or HA antibodies. These data demonstrate that Na and TRAF2 can interact when expressed in a cellular environment.
Fig. 4.
Na interacts with TRAF2. 293 cells were cotransfected with the indicated combinations of vector control, FLAG-Na, or HA-TRAF2. Cell lysates were harvested in NP-40 buffer 48 h posttransfection. Immunoprecipitation analysis was performed using antibodies against the FLAG or HA tags of transfected FLAG-Na and HA-TRAF2. Immunoblot analysis was then performed using antibodies against FLAG or HA to detect the immunoprecipitated proteins. Direct load samples were also used for immunoblot analysis as a control.
Na is present in both the nucleus and cytoplasm.
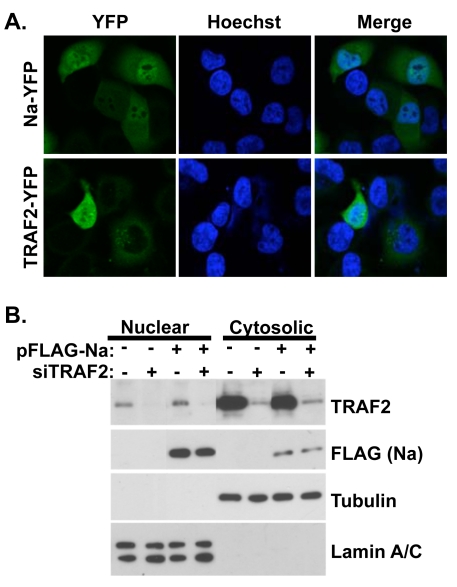
The finding that Na interacts with TRAF2 in cells suggests that a portion of Na may be located within the cytoplasm, similar to what has been described for the KSHV Orf49 homologue (26). To investigate the subcellular localization of both Na and TRAF2, we constructed plasmids in which the Na or TRAF2 proteins were fused to a C-terminal YFP tag and examined the cellular localization of each protein by confocal fluorescent microscopy in transfected Hone-Akata cells. As shown in Fig. 5A, the Na-YFP protein, similar to the Orf49 protein, can be detected both in the nucleus and the cytoplasm of cells. TRAF2-YFP localized primarily in the cytoplasm but was also detected in the nucleus of some Hone-Akata cells. Although TRAF2 is predominantly localized within the cell cytoplasm, other reports have also detected TRAF2 in the nucleus of the cell (38).
Fig. 5.
Na and TRAF2 are localized to both the cytoplasm and nucleus. (A) Hone-Akata cells were transfected with YFP-tagged Na or TRAF2 expression plasmids. At 24 h posttransfection, the cells were fixed with paraformaldehyde and nuclei were stained with Hoechst 33342. Confocal fluorescence microscopy was used to visualize the YFP-tagged proteins and cell nuclei. (B). CNE2-Akata cells were transfected with vector control or FLAG-Na expression plasmids, in the presence or absence of cotransfected control siRNA or TRAF2-directed siRNA. At 48 h posttransfection, cell lysates were fractionated into nuclear and cytosolic fractions. Immunoblot analysis was performed using antibodies against TRAF2 and FLAG-Na. Tubulin and lamin A/C were included as controls for proper fractionation.
To further examine Na and TRAF2 cellular localization, CNE2-Akata cells were pretreated with either control siRNA or TRAF2 siRNA and then transfected with control or FLAG-Na expression plasmids, and biochemical fractionation was performed on the cell lysates to separate the nuclear and cytosolic fractions (Fig. 5B). Immunoblot analysis showed that Na was present in both fractions of the cell; TRAF2 was also detected in both fractions but was predominantly in the cytosolic fraction. Control and TRAF2-directed siRNAs were included in this experiment to confirm that the TRAF2 band detected in the immunoblot assay of the nuclear fraction was indeed TRAF2. Similar results were obtained when EBV-negative CNE cells were transfected with Na and TRAF2 and fractionated into Triton X-100-soluble and -insoluble fractions (data not shown). These results suggest that Na and TRAF2 can potentially interact in either the cytoplasm and/or nucleus of EBV-infected cells. Additionally, it does not appear that Na and TRAF2 alter one another's cellular localization.
TRAF2 is required for Na-induced lytic EBV protein expression.
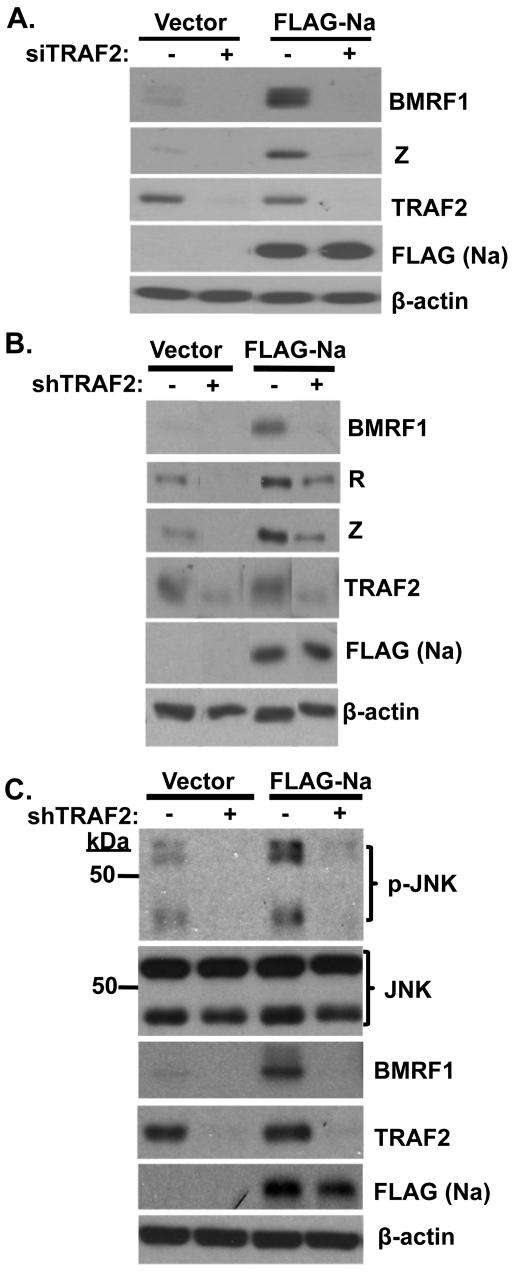
To determine if TRAF2 contributes to Na disruption of viral latency, Hone-Akata cells were pretreated with either control siRNA or TRAF2 siRNA and then transfected with control or Na expression vectors. As expected, Na induced BMRF1 and Z expression in cells transfected with control siRNA (Fig. 6A). However, following knockdown of TRAF2 expression, Na lost the ability to induce lytic viral protein expression. In addition, the low level of constitutive lytic protein expression in Hone-Akata cells was also reduced by loss of TRAF2 expression. Similar results were obtained when lentivirus vectors expressing control or short hairpin sequences against TRAF2 were used to reduce TRAF2 expression in CNE2-Akata cells (Fig. 6B). Together these data indicate that TRAF2 is required for Na-mediated disruption of EBV latency and suggest that the interaction between Na and TRAF2 is important for induction of lytic protein expression.
Fig. 6.
TRAF2 is required for Na-mediated induction of lytic protein expression. (A) Hone-Akata cells were transfected with control or TRAF2 siRNAs. At 48 h after transfection, the cells were retransfected with the control or TRAF2 siRNAs along with control or FLAG-Na expression plasmids. Immunoblot analysis was performed 2 days later using antibodies against BMRF1 and Z to examine lytic protein expression, or with TRAF2 and FLAG to detect TRAF2 and Na, respectively. β-Actin was used as a loading control. (B) CNE2-Akata cells were transfected with lentiviruses expressing shRNAs against either a control sequence or TRAF2. At 48 h posttransfection, cells were retransfected with the shRNA vectors along with either control or FLAG-Na expression plasmids. Cell lysates were harvested, and immunoblot analyses were performed using antibodies against TRAF2, BMRF1, FLAG (Na), R, Z, and β-actin. The data shown represent samples run on the same gel; irrelevant lanes were cropped from the figure. (C) Stable selected Hone-Akata cell lines transduced with lentiviruses expressing shRNAs against a nonspecific control or TRAF2 were transfected with either vector control or FLAG-Na expression vectors as described above. Immunoblot analysis was performed using antibodies against phosphorylated JNK (p-JNK), total JNK, BMRF1, TRAF2, FLAG (Na), and β-actin.
TRAF2 is required for Na-induced JNK activation.
To determine if TRAF2 is also involved in Na-mediated JNK activation, Hone-Akata cells were transduced with lentiviruses expressing TRAF2 shRNAs or control shRNAs, stably selected, and then transfected with control or Na expression plasmids. Na induced JNK phosphorylation as expected in cells expressing the control shRNAs (Fig. 6C). However, knockdown of TRAF2 expression greatly inhibited the ability of Na to induce JNK phosphorylation, as well as decreasing the constitutive level of p-JNK expression in cells transfected with control vector. As expected, TRAF2 knockdown also reduced the ability of Na to increase lytic BMRF1 protein expression. Together, these data suggest that TRAF2 is required both for the ability of Na to induce JNK phosphorylation and for its ability to initiate lytic viral protein expression.
Na induces constitutive p100/p52 processing but does not affect TNF-α signaling.
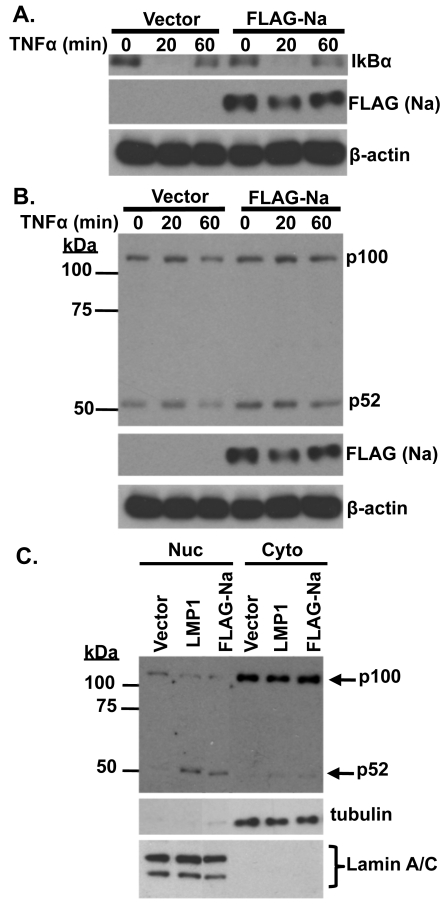
Since TRAF2 is involved in multiple different aspects of TNF-α-induced signaling, including activation of the classical NF-κB pathways, as well as JNK, and inhibition of the alternative NF-κB pathway, we also examined whether Na affects TNF-α-induced signaling. CNE2-Akata cells were transfected with control or Na expression vectors and treated with TNF-α (10 ng/ml) for 20 or 60 min prior to harvesting (Fig. 7A). Cell lysates were examined for IκBα, which becomes phosphorylated and rapidly degraded through classical NF-κB activation, as well as for the level of p100/p52 processing (which is increased during activation of the alternative NF-κB pathway) (Fig. 7B). Overexpression of Na did not affect the ability of TNF-α to decrease expression of IκBα (Fig. 7A) or to increase the level of JNK activation (data not shown). Interestingly, overexpression of Na consistently increased the level of constitutive p100/p52 processing (Fig. 7B). Since the CNE2-Akata cells have some level of constitutive p100/52 processing even in the absence of Na, the effects of Na on p100 processing were also examined in the EBV-negative AGS cell line, which has a lower baseline level of constitutive p100/p52 processing. Again, overexpression of Na led to an increase in p52 accumulation in the nucleus, as did transfected LMP1 protein (which is known to induce p100/p52 processing [49] and served as a positive control). These results suggest Na does not affect TNF-α signaling through the classical NF-κB pathway or the JNK pathway, but that it increases the level of p100/p52 processing in the alternative NF-κB pathway, even in the absence of TNF-α signaling.
Fig. 7.
Na induces p100/p52 processing. CNE2-Akata cells were transfected with control or FLAG-Na expression plasmids and treated with TNF-α (10 ng/ml) for 20 or 60 min prior to harvesting. (A) Immunoblot analysis was performed using antibodies against IκBα, FLAG (Na), and β-actin. (B) Immunoblot analysis was performed using antibodies against p52, FLAG (Na), and β-actin. (C) AGS cells were transfected with control, LMP1, or FLAG-Na expression plasmids and harvested 48 h posttransfection. Biochemical fractionation was used to obtain nuclear (Nuc) and cytosolic (Cyto) extracts. Immunoblot analysis was performed to examine p100 processing by using a p52 antibody. Tubulin and lamin A/C antibodies were used as controls for fractionation.
Na does not affect p53 function.
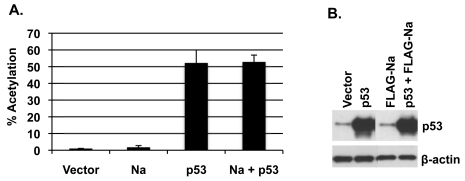
Since a recent study suggested that p53 is important for HDACi-mediated lytic reactivation in EBV-positive epithelial cell lines (12), and our results here indicate that Na is also important for viral reactivation by HDAC inhibitors (Fig. 2), we hypothesized that Na may enhance p53 transcriptional activity. To examine this, CNE2-Akata cells were transfected with a CAT reporter construct driven by 13 p53-responsive promoter elements upstream of the polyomavirus early promoter (pG13CAT), in the presence or absence of cotransfected Na and/or p53 vectors, and the level of CAT activity was measured. As shown in Fig. 8, p53 alone activated the pG13CAT construct as expected. However, cotransfected Na did not increase the ability of p53 to enhance CAT activity. Similar results were obtained in C666-1 cells (data not shown). These results suggest that Na does not affect p53 function.
Fig. 8.
Na does not alter p53 function. CNE2-Akata cells were cotransfected with vector control, FLAG-Na, or p53 expression plasmids along with a pG13-CAT reporter construct that contained 13 p53-responsive elements. (A) CAT assays were performed on the cell lysates 48 h after transfection and quantified by phosphorimaging analysis. Results were normalized to vector-alone results, which were set at 1. (B) Immunoblot analysis was performed on the cell lysates by using antibody against p53 to demonstrate that equal levels of p53 were transfected into the cells. β-Actin was included as a loading control. The data shown represent samples run on the same gel; irrelevant lanes were cropped from the figure.
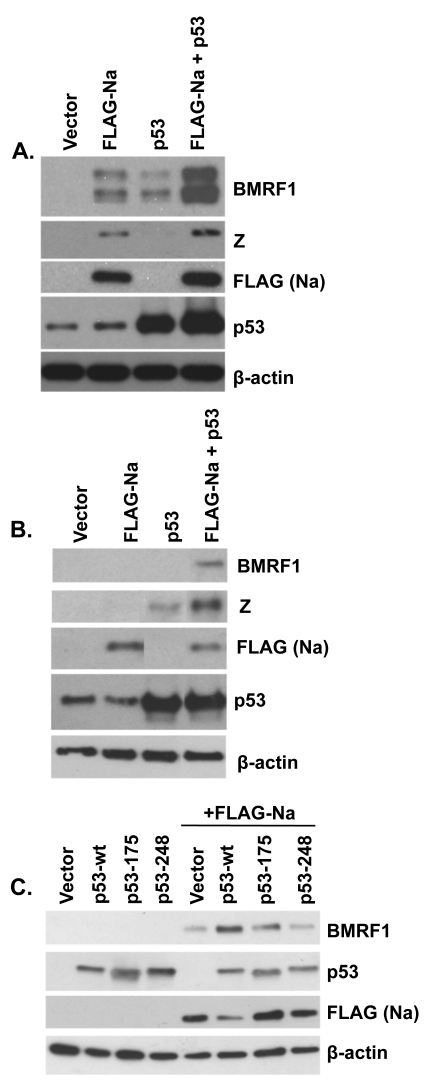
Na and p53 synergistically induce lytic protein expression.
Given the finding above that Na does not activate p53 transcriptional function, another potential explanation for why p53 alone in a previous paper was reported to be unable to activate lytic EBV gene expression except in conjunction with HDAC inhibitors might be that the HDAC inhibitors serve to induce endogenous Na expression and that the combination of Na and p53 then synergistically activates lytic gene expression. To examine this possibility, Hone-Akata cells were transfected with Na or p53 expression plasmids (alone or in combination), and an immunoblot analysis was performed to examine the level of lytic induction (Fig. 9A). Interestingly, we found that either Na or p53 expression alone was sufficient to induce BMRF1 expression in this cell type, but that the combination of p53 and Na expression together was clearly more effective than either protein alone. Furthermore, in the tightly latent C666-1 NPC cell line (the only NPC cell line which has remained persistently EBV positive in culture without selection), we found that only the combination of both p53 and Na was sufficient to induce expression of the early lytic BMRF1 protein (Fig. 9B). Interestingly, however, p53 alone was able to induce Z, but not BMRF1, expression in C666-1 cells. These results suggest that p53 and Na work synergistically to induce lytic protein expression in latently infected, EBV-positive epithelial cell lines.
Fig. 9.
Na and p53 synergistically induce lytic protein expression. Hone-Akata (A) or C666-1 (B) cells were cotransfected with control, FLAG-Na, or p53 expression plasmids; immunoblot analysis was performed 2 days later with antibodies against BMRF1 and Z to examine lytic gene expression and with antibodies against FLAG (Na) and p53 to examine levels of transfected Na and p53. (C) CNE2-Akata cells were cotransfected with combinations of control vector, FLAG-Na, and wild-type (wt) or mutant p53 (p53-175 and p53-248) expression vectors as indicated. Immunoblot analysis was performed 48 h posttransfection to examine BMRF1, FLAG (Na), and p53. β-Actin was used as an internal loading control.
Since p53 is mutated in many carcinoma cell lines, we next tested the ability of two different p53 mutant constructs to synergistically induce lytic protein expression with cotransfected Na in CNE2-Akata cells. As shown in Fig. 9C, while the p53-wt construct cooperated with Na to induce lytic protein expression, neither a p53 mutant with an altered residue 175 (which affects p53 conformation) nor a p53 mutant with an altered residue 248 (which inhibits p53 DNA binding) enhanced the effect of the FLAG-Na expression vector. These results indicate that wild-type p53 function is important for enhancing Na-induced lytic protein expression.
p53 is required for Na-mediated disruption of viral latency.
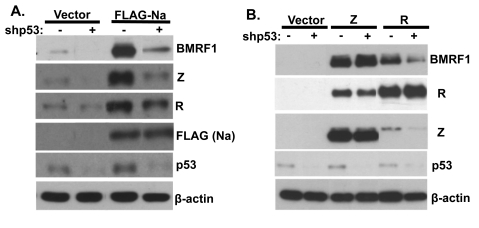
Finally, to examine whether endogenous p53 expression is required for the ability of Na to induce lytic viral protein expression, CNE2-Akata cells were transfected with lentivirus vectors expressing control shRNA or p53-directed shRNA (shp53) and then transfected with control or Na expression vectors. As shown in Fig. 10A, knockdown of endogenous p53 expression greatly reduced the ability of transfected Na to induce lytic EBV protein expression. In addition, knockdown of endogenous p53 expression reduced the low level of constitutive lytic EBV protein expression found in CNE2-Akata cells (Fig. 10A) and reduced the ability of transfected R protein to activate lytic EBV protein expression (Fig. 10B). In contrast, knockdown of p53 expression did not affect the ability of transfected Z protein to induce lytic gene expression (Fig. 10B). These results suggest that endogenous p53 activity is required for the ability of both Na and R (but not Z) to induce lytic EBV gene expression, and the results furthermore suggest that a key aspect of the p53 effect may be to help the Z/R proteins to activate Zp in the context of the intact viral genome.
Fig. 10.
p53 is required for Na activation of lytic protein expression. CNE2-Akata cells were transfected with lentivirus vectors expressing shRNAs against either control or p53 sequences. At 48 h posttransfection, the cells were cotransfected with the same shRNAs and control, FLAG-Na (A), or Z or R (B) expression plasmids. Immunoblot analysis was performed 2 days later using antibodies against BMRF1, Z, and R to examine EBV lytic gene expression and using antibody against FLAG or p53 to detect transfected Na and endogenous p53 expression. β-Actin was included as an internal loading control.
DISCUSSION
The switch between latent and lytic EBV infection is known to be activated by expression of either the BZLF1 (Z) and/or BRLF1 (R) IE proteins (13, 15, 37, 61, 65). Z and R activate one another's promoters (Zp and Rp) and cooperatively activate the early lytic viral promoters (3, 13, 14, 16, 19, 22, 29, 32, 35, 43, 50, 52, 65). Although the early lytic viral protein Na (the BRRF1 gene product) was previously reported to activate the Z promoter in HeLa cells and to cooperate with R to activate lytic gene expression in EBV-infected 293 cells, overexpression of Na by itself was not found to activate lytic gene transcription in 293 cells (33). Here we show that overexpression of Na is sufficient to induce lytic gene expression in a number of EBV-infected epithelial cell lines (Fig. 1) and that, conversely, knockdown of endogenous Na expression inhibits low-level constitutive lytic protein expression (Fig. 2).
We have demonstrated that Na interacts directly with TRAF2 in cells (Fig. 4) and that TRAF2 expression is required for the lysis-inducing effect of Na (Fig. 6). Furthermore, similar to a recent report indicating that HDAC inhibitors require wild-type p53 to activate lytic viral protein expression in NPC cell lines (12), we likewise found that wild-type p53 (but not mutant p53) enhanced both Na-induced as well as constitutive lytic viral protein expression in EBV-infected epithelial cell lines (Fig. 9 and10). Together these results suggest that Na plays an important regulatory role in promoting the switch between latent and lytic EBV infection in epithelial cells, and cellular factors that activate Na expression may in fact be sufficient to induce the whole lytic viral protein cascade expression in some cell lines.
Our results suggest that the ability of Na to induce lytic EBV protein expression is at least partially mediated through activation of the JNK pathway. We have shown that Na, like the KSHV homologue Orf49, induces JNK activation in cells, and that furthermore JNK activation is required for the ability of Na to activate lytic viral protein expression (Fig. 3). These results are consistent with our previous findings that Na induces c-Jun phosphorylation and activates c-Jun transcriptional function (33). Since c-Jun is a known activator of the Z promoter (binding to the Zp ZII motif), our results support a model in which Na expression leads to lytic EBV expression via its ability to activate the Z promoter in certain cell lines.
Although the Na and TRAF2 proteins were previously shown by yeast two-hybrid assays to directly interact (10), the potential significance of this interaction has been unclear, particularly since TRAF2 is thought to be a cytoplasmic protein, whereas Na has been reported to be a nuclear protein (55). However, since TRAF2 is known to be essential for activation of the JNK pathway following TNFR activation (42) and overexpression of TRAF2 alone has been reported to activate JNK in certain cell lines (31), these previous reports suggested the possibility that one or more Na effects may be mediated through TRAF2. We have demonstrated that Na interacts with TRAF2 in cells (Fig. 4), and we have shown that Na (similar to KSHV Orf49) is located within both the nucleus and cytoplasm (Fig. 5). Most importantly, we have demonstrated that constitutive cellular TRAF2 expression is required not only for Na-induced JNK activation but also for Na-induced lytic viral protein expression (Fig. 6).
Together, these results strongly suggest that the Na effect is at least partially mediated through its direct interaction with TRAF2 and that this Na-TRAF2 interaction mediates JNK activation. Nevertheless, the proof of this hypothesis will ultimately require the identification of a Na mutant that is specifically deficient in the ability to interact with TRAF2. Unfortunately, to date we have found that Na mutants unable to interact with TRAF2 are also unstable (data not shown). Additionally, due to the limitations of interaction studies using overexpressed proteins, the interaction between endogenous Na and TRAF2 should be confirmed. Also unclear at present is whether Na affects any other TRAF2 functions, for example, the ability of TRAF2 to regulate NF-κB signaling in response to the TNF-α ligand. While we did not observe any affect of Na on IκBα expression following TNF-α treatment of CNE2-Akata cells (Fig. 7A), suggesting that Na does not alter the canonical NF-κB signaling pathway, we routinely observed an increase in the level of constitutive p100/p52 processing in cells expressing Na (Fig. 7B and C). Since TRAF2 knockout mouse embryonic fibroblast cells have been reported to have an increased level of constitutive p100/52 processing (which is thought to reflect the loss of TRAF2-mediated degradation of NIK) (28, 62), the effect of Na on p100/52 processing may reflect its ability to inhibit this particular TRAF2 function. Although the mechanism(s) by which Na induces p100/52 processing remains to be explored, since this Na effect was observed in both EBV-negative as well as EBV-positive cell types, it does not seem to require any other EBV-encoded proteins, such as LMP1. Whether Na-induced p100/52 processing enhances or inhibits its ability to induce lytic viral gene expression will be an important focus for future studies.
In this study we have also demonstrated that p53 contributes to the ability of Na to induce lytic EBV protein expression. We show that coexpression of wild-type p53, but not mutant p53, enhances the ability of Na to induce lytic EBV protein expression in CNE2-Akata and Hone-Akata cells (Fig. 9A and C) and that Na cannot induce lytic gene expression in the tightly latent C666-1 NPC cell line unless it is coexpressed with a p53 expression vector (Fig. 9B). Furthermore, we have shown that knockdown of endogenous p53 expression in CNE2-Akata cells diminishes the ability of both Na and R to induce lytic gene expression (Fig. 10) while having no effect on the ability of transfected Z protein to disrupt viral latency (Fig. 10B). These results suggest that the effect of p53 is mediated at the level of Z promoter activation (and thereby effectively bypassed when Z is expressed under the control of a strong heterologous promoter). Similar to our results here, loss of p53 expression was recently shown to inhibit HDACi-induced lytic reactivation (12), while overexpression of p53 was reported to enhance early EBV lytic infection (54). Our finding that endogenous p53 expression appears to cooperate with Na to induce lytic EBV expression in CNE2-Akata and Hone-Akata cells, but not in C666-1 cells, is possibly explained by the fact that C666-1 cells contain only a mutant form of p53 (64), whereas the CNE2-Akata and Hone-Akata lines, although originally described as NPC lines, are at least partially derived from HeLa cells (11) and thus may express some wild-type p53 as well as mutant p53. Alternatively, the p53 mutant found in CNE2-Akata and Hone-Akata cells (p53 Thr280) retains the ability to cooperate with Na to disrupt viral latency.
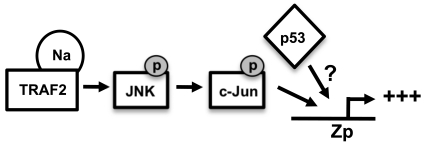
Together, the results presented here suggest a model in which Na interacts with TRAF2 to activate JNK signaling, leading to transactivation of Zp, and wild-type p53 enhances Na-mediated activation of Zp (Fig. 11). Although we demonstrated that wild-type p53 enhances Na induction of lytic protein expression (Fig. 9), the mechanism by which p53 and Na synergistically activate lytic protein expression remains to be determined. Since we did not find that Na activates p53 transcriptional function (Fig. 8) or consistently affects endogenous p53 expression, we speculate that Na and p53 enhance BZLF1 transcription in the context of the intact viral genome through independent mechanisms (Fig. 11). To date, we have been unable to demonstrate this synergistic activation of the BZLF1 promoter by Na and p53 using Zp-driven reporter gene assays (data not shown). This result suggests that the synergistic effect of the Na/p53 combination may not be observed in the absence of a proper chromatin structure, and/or that the p53-responsive element is not contained within our Zp construct. Another potential role for p53 is to either enhance transcription factors or inhibit transcriptional repressors involved in regulation of Zp. Additionally, since p53 is important during HDACi-induced lytic infection (12), p53 may play a role in regulating histone modifications and thereby affect regulation of lytic gene transcription. Alternatively, since a number of recent studies have suggested that p53 plays an important role in the regulation of microRNAs (miRNAs) (59), it is possible that the effect of p53 on lytic reactivation is at least partially mediated through this mechanism. For example, the miRNAs 200b and 429, which reduce the level of the Zp cellular repressor proteins ZEB-1 and ZEB-2, have been shown to enhance lytic gene expression (21). Interestingly, although p53 function appears to promote activation of Z transcription during the initial steps of viral reactivation, the Z protein subsequently acts to inhibit p53 function through multiple mechanisms (including targeting p53 for ubiquitin-mediated degradation, as well as inhibiting its transcriptional function) (48, 54a). Thus, the role of p53 during lytic EBV reactivation is complex and may be quite different during the early versus late stages of infection. Our results here suggest that the p53 status of different cell lines likely plays an important factor in determining if Na expression results in viral reactivation.
Fig. 11.
Hypothesized model for Na transactivation of the Z promoter. A potential model for Na-induced lytic protein expression is shown. Na interacts with TRAF2 to activate the JNK signaling cascade, leading to phosphorylation of c-Jun and activation of Zp. p53 independently induces activation of Zp and has a synergistic effect when coexpressed with Na. The mechanism by which p53 enhances Zp activity is currently unknown.
Although we have shown here that Na overexpression in certain EBV-infected epithelial cell lines is sufficient to induce lytic protein expression (particularly in conjunction with p53), we nevertheless have consistently found that the Z and R proteins induce lytic protein expression more efficiently than the Na protein in all cell lines tested. Thus, activation of the Z and/or R promoters by cellular transcription factors likely plays a more important role than activation of the Na promoter in promoting lytic infection in virally infected epithelial cells. Since the EBV-infected epithelial cell lines that respond to the lytic-inducing effect of Na alone can constitutively express lytic EBV proteins at a very low level, it remains possible that an important aspect of the Na lytic induction effect is to enhance R-mediated and/or Z-mediated transcriptional effects. In any event, our finding that knockdown of Na expression derived from the endogenous viral genome in Hone-Akata cells not only decreased chemically induced lytic protein expression (Fig. 2) but also shut down constitutive Z expression strongly suggests that Na has an important and essential role in promoting lytic viral protein expression during normal EBV infection of epithelial cells.
ACKNOWLEDGMENTS
This work was supported by grants T32 CA009681, T32 AI078985, R01-CA58853, R01-CA66519, P01-CA022443, and R01-H6064851 from the National Institutes of Health.
We thank Henri Gruffat for providing antibody against Na. We also thank Lance Rodenkirch at the W. M. Keck Laboratory for Biological Imaging, University of Wisconsin, for helping with the confocal microscopy.
Footnotes
Published ahead of print on 16 February 2011.
REFERENCES
- 1. Adamson A. L., et al. 2000. Epstein-Barr virus immediate-early proteins BZLF1 and BRLF1 activate the ATF2 transcription factor by increasing the levels of phosphorylated p38 and c-Jun N-terminal kinases. J. Virol. 74:1224–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adamson A. L., Kenney S. 2001. Epstein-barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J. Virol. 75:2388–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adamson A. L., Kenney S. C. 1998. Rescue of the Epstein-Barr virus BZLF1 mutant, Z(S186A), early gene activation defect by the BRLF1 gene product. Virology 251:187–197 [DOI] [PubMed] [Google Scholar]
- 4. Au P. Y., Yeh W. C. 2007. Physiological roles and mechanisms of signaling by TRAF2 and TRAF5. Adv. Exp. Med. Biol. 597:32–47 [DOI] [PubMed] [Google Scholar]
- 5. Baker S. J., Markowitz S., Fearon E. R., Willson J. K., Vogelstein B. 1990. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science 249:912–915 [DOI] [PubMed] [Google Scholar]
- 6. Bhende P. M., Dickerson S. J., Sun X., Feng W. H., Kenney S. C. 2007. X-box-binding protein 1 activates lytic Epstein-Barr virus gene expression in combination with protein kinase D. J. Virol. 81:7363–7370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bhende P. M., Seaman W. T., Delecluse H. J., Kenney S. C. 2005. BZLF1 activation of the methylated form of the BRLF1 immediate-early promoter is regulated by BZLF1 residue 186. J. Virol. 79:7338–7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bradley J. R., Pober J. S. 2001. Tumor necrosis factor receptor-associated factors (TRAFs). Oncogene 20:6482–6491 [DOI] [PubMed] [Google Scholar]
- 9. Bryant H., Farrell P. J. 2002. Signal transduction and transcription factor modification during reactivation of Epstein-Barr virus from latency. J. Virol. 76:10290–10298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Calderwood M. A., et al. 2007. Epstein-Barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. U. S. A. 104:7606–7611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chan S. Y., et al. 2008. Authentication of nasopharyngeal carcinoma tumor lines. Int. J. Cancer 122:2169–2171 [DOI] [PubMed] [Google Scholar]
- 12. Chang S. S., et al. 2008. Critical role of p53 in histone deacetylase inhibitor-induced Epstein-Barr virus Zta expression. J. Virol. 82:7745–7751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chevallier-Greco A., et al. 1986. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 5:3243–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Countryman J., Jenson H., Seibl R., Wolf H., Miller G. 1987. Polymorphic proteins encoded within BZLF1 of defective and standard Epstein-Barr viruses disrupt latency. J. Virol. 61:3672–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Countryman J., Miller G. 1985. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc. Natl. Acad. Sci. U. S. A. 82:4085–4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Darr C. D., Mauser A., Kenney S. 2001. Epstein-Barr virus immediate-early protein BRLF1 induces the lytic form of viral replication through a mechanism involving phosphatidylinositol-3 kinase activation. J. Virol. 75:6135–6142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Derijard B., et al. 1994. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76:1025–1037 [DOI] [PubMed] [Google Scholar]
- 18. Devergne O., et al. 1996. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-κB activation. Mol. Cell. Biol. 16:7098–7108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dickerson S. J., et al. 2009. Methylation-dependent binding of the Epstein-Barr virus BZLF1 protein to viral promoters. PLoS Pathog. 5:e1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Effert P., et al. 1992. Alterations of the p53 gene in nasopharyngeal carcinoma. J. Virol. 66:3768–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ellis A. L., Wang Z., Yu X., Mertz J. E. 2010. Either ZEB1 or ZEB2/SIP1 can play a central role in regulating the Epstein-Barr virus latent-lytic switch in a cell-type-specific manner. J. Virol. 84:6139–6152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Feederle R., et al. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19:3080–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Feng W. H., Hong G., Delecluse H. J., Kenney S. C. 2004. Lytic induction therapy for Epstein-Barr virus-positive B-cell lymphomas. J. Virol. 78:1893–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Feng W. H., et al. 2007. ZEB1 and c-Jun levels contribute to the establishment of highly lytic Epstein-Barr virus infection in gastric AGS cells. J. Virol. 81:10113–10122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Glaser R., et al. 1989. Two epithelial tumor cell lines (HNE-1 and HONE-1) latently infected with Epstein-Barr virus that were derived from nasopharyngeal carcinomas. Proc. Natl. Acad. Sci. U. S. A. 86:9524–9528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gonzalez C. M., et al. 2006. Identification and characterization of the Orf49 protein of Kaposi's sarcoma-associated herpesvirus. J. Virol. 80:3062–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gorman C. M., Moffat L. F., Howard B. H. 1982. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol. Cell. Biol. 2:1044–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grech A. P., et al. 2004. TRAF2 differentially regulates the canonical and noncanonical pathways of NF-κB activation in mature B cells. Immunity 21:629–642 [DOI] [PubMed] [Google Scholar]
- 29. Gruffat H., Manet E., Rigolet A., Sergeant A. 1990. The enhancer factor R of Epstein-Barr virus (EBV) is a sequence-specific DNA binding protein. Nucleic Acids Res. 18:6835–6843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gruffat H., Manet E., Sergeant A. 2002. MEF2-mediated recruitment of class II HDAC at the EBV immediate early gene BZLF1 links latency and chromatin remodeling. EMBO Rep. 3:141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Habelhah H., et al. 2004. Ubiquitination and translocation of TRAF2 is required for activation of JNK but not of p38 or NF-κB. EMBO J. 23:322–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hardwick J. M., Lieberman P. M., Hayward S. D. 1988. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J. Virol. 62:2274–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hong G. K., et al. 2004. The BRRF1 early gene of Epstein-Barr virus encodes a transcription factor that enhances induction of lytic infection by BRLF1. J. Virol. 78:4983–4992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kaye K. M., et al. 1996. Tumor necrosis factor receptor associated factor 2 is a mediator of NF-kappa B activation by latent infection membrane protein 1, the Epstein-Barr virus transforming protein. Proc. Natl. Acad. Sci. U. S. A. 93:11085–11090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kenney S., et al. 1989. The Epstein-Barr virus (EBV) BZLF1 immediate-early gene product differentially affects latent versus productive EBV promoters. J. Virol. 63:1729–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kern S. E., et al. 1992. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science 256:827–830 [DOI] [PubMed] [Google Scholar]
- 37. Kieff E., Rickinson A. B. 2007. Epstein-Barr virus and its replication, p. 2603–2654 In Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E. (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 38. Kim W. J., Back S. H., Kim V., Ryu I., Jang S. K. 2005. Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol. Cell. Biol. 25:2450–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kraus R. J., Perrigoue J. G., Mertz J. E. 2003. ZEB negatively regulates the lytic-switch BZLF1 gene promoter of Epstein-Barr virus. J. Virol. 77:199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Laichalk L. L., Thorley-Lawson D. A. 2005. Terminal differentiation into plasma cells initiates the replicative cycle of Epstein-Barr virus in vivo. J. Virol. 79:1296–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee S., et al. 2007. The ORF49 protein of murine gammaherpesvirus 68 cooperates with RTA in regulating virus replication. J. Virol. 81:9870–9877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee S. Y., et al. 1997. TRAF2 is essential for JNK but not NF-κB activation and regulates lymphocyte proliferation and survival. Immunity 7:703–713 [DOI] [PubMed] [Google Scholar]
- 43. Lieberman P. M., Hardwick J. M., Sample J., Hayward G. S., Hayward S. D. 1990. The zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J. Virol. 64:1143–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu P., Liu S., Speck S. H. 1998. Identification of a negative cis element within the ZII domain of the Epstein-Barr virus lytic switch BZLF1 gene promoter. J. Virol. 72:8230–8239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu S., Liu P., Borras A., Chatila T., Speck S. H. 1997. Cyclosporin A-sensitive induction of the Epstein-Barr virus lytic switch is mediated via a novel pathway involving a MEF2 family member. EMBO J. 16:143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lo A. K., et al. 2006. Epstein-Barr virus infection alters cellular signal cascades in human nasopharyngeal epithelial cells. Neoplasia 8:173–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. MacCallum P., Karimi L., Nicholson L. J. 1999. Definition of the transcription factors which bind the differentiation responsive element of the Epstein-Barr virus BZLF1 Z promoter in human epithelial cells. J. Gen. Virol. 80:1501–1512 [DOI] [PubMed] [Google Scholar]
- 48. Mauser A., et al. 2002. The Epstein-Barr virus immediate-early protein BZLF1 regulates p53 function through multiple mechanisms. J. Virol. 76:12503–12512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Paine E., Scheinman R. I., Baldwin A. S., Jr., Raab-Traub N. 1995. Expression of LMP1 in epithelial cells leads to the activation of a select subset of NF-kappa B/Rel family proteins. J. Virol. 69:4572–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ragoczy T., Heston L., Miller G. 1998. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol. 72:7978–7984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rickenson A. B., Kieff E. 2007. Epstein-Barr virus, p. 2655–2700 In Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E. (ed.), Fields virology, 5th ed. Lippincott Williams & Williams, Philadelphia, PA [Google Scholar]
- 52. Rooney C. M., Rowe D. T., Ragot T., Farrell P. J. 1989. The spliced BZLF1 gene of Epstein-Barr virus (EBV) transactivates an early EBV promoter and induces the virus productive cycle. J. Virol. 63:3109–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sandberg M., Hammerschmidt W., Sugden B. 1997. Characterization of LMP-1's association with TRAF1, TRAF2, and TRAF3. J. Virol. 71:4649–4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sato Y., et al. 2010. Transient increases in p53-responsible gene expression at early stages of Epstein-Barr virus productive replication. Cell Cycle 9:807–814 [DOI] [PubMed] [Google Scholar]
- 54a. Sato Y., et al. 2009. Expression of Epstein-Barr virus BZLF1 immediate-early protein induces p53 degradation independent of MDM2, leading to repression of p53-mediated transcription. Virology 388:204–211 [DOI] [PubMed] [Google Scholar]
- 55. Segouffin-Cariou C., Farjot G., Sergeant A., Gruffat H. 2000. Characterization of the Epstein-Barr virus BRRF1 gene, located between early genes BZLF1 and BRLF1. J. Gen. Virol. 81:1791–1799 [DOI] [PubMed] [Google Scholar]
- 56. Soni V., Cahir-McFarland E., Kieff E. 2007. LMP1 TRAFficking activates growth and survival pathways. Adv. Exp. Med. Biol. 597:173–187 [DOI] [PubMed] [Google Scholar]
- 57. Speck S. H., Chatila T., Flemington E. 1997. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 5:399–405 [DOI] [PubMed] [Google Scholar]
- 58. Sun C. C., Thorley-Lawson D. A. 2007. Plasma cell-specific transcription factor XBP-1s binds to and transactivates the Epstein-Barr virus BZLF1 promoter. J. Virol. 81:13566–13577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Suzuki H. I., Miyazono K. 2010. Dynamics of microRNA biogenesis: crosstalk between p53 network and microRNA processing pathway. J. Mol. Med. 88:1085–1094 [DOI] [PubMed] [Google Scholar]
- 60. Suzuki K., Bose P., Leong-Quong R. Y., Fujita D. J., Riabowol K. 2010. REAP: a two minute cell fractionation method. BMC Res. Notes 3:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Takada K., Shimizu N., Sakuma S., Ono Y. 1986. trans activation of the latent Epstein-Barr virus (EBV) genome after transfection of the EBV DNA fragment. J. Virol. 57:1016–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vallabhapurapu S., et al. 2008. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-κB signaling. Nat. Immunol. 9:1364–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang Y. C., Huang J. M., Montalvo E. A. 1997. Characterization of proteins binding to the ZII element in the Epstein-Barr virus BZLF1 promoter: transactivation by ATF1. Virology 227:323–330 [DOI] [PubMed] [Google Scholar]
- 64. Weinrib L., Li J. H., Donovan J., Huang D., Liu F. F. 2001. Cisplatin chemotherapy plus adenoviral p53 gene therapy in EBV-positive and -negative nasopharyngeal carcinoma. Cancer Gene Ther. 8:352–360 [DOI] [PubMed] [Google Scholar]
- 65. Zalani S., Holley-Guthrie E., Kenney S. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. U. S. A. 93:9194–9199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. zur Hausen H., et al. 1970. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature 228:1056–1058 [DOI] [PubMed] [Google Scholar]