Abstract
Infection of mice with pneumonia virus of mice (PVM) provides a convenient experimental pathogenesis model in a natural host for a human respiratory syncytial virus-related virus. Extending our previous work showing that the PVM nonstructural (NS) proteins were pathogenicity factors in mice, we identify both the NS1 and NS2 proteins as antagonists of alpha/beta interferon (IFN-α/β) and IFN-λ by use of recombinant PVM (rPVM) with single and combined deletions of the NS proteins (ΔNS1, ΔNS2, and ΔNS1 ΔNS2). Wild-type and NS deletion PVMs were evaluated for growth and pathogenesis by infecting knockout mice that lack functional receptors to IFN-α/β, IFN-λ, or both. The absence of the receptor to IFN-α/β (IFNAR) or IFN-λ (interleukin-28 receptor α chain [IL-28Rα]) individually did not reverse the attenuated virulence of the NS deletion viruses although loss of IFNAR partially restored replication efficiency. When both receptors were deleted, replication and virulence were largely rescued for rPVM ΔNS1 and were significantly but not completely rescued for rPVM ΔNS2. As for rPVM ΔNS1 ΔNS2, the effect was mostly limited to partial enhancement of replication. This indicates that both IFN-α/β and IFN-λ contributed to restricting the NS deletion viruses, with the former playing the greater role. Interestingly, the replication and virulence of wild-type PVM were completely unaffected by the presence or absence of functional receptors to IFN-α/β and IFN-λ, indicating that both systems are strongly suppressed during infection. However, pretreatment of mice with IFN-α/β was protective against lethal rPVM challenge, whereas pretreatment with IFN-λ delayed but did not prevent disease and, in some cases, reduced mortality. The fact that virulence of rPVM lacking NS2 was not recovered completely when both interferon receptors were deleted suggests that NS2 may have further functions outside the IFN system.
INTRODUCTION
Interferons (IFNs) play a crucial role in the immune response against viruses by mounting an early antiviral innate immune response and by modulating innate and subsequent adaptive immune responses (for a detailed review, see reference 26). IFNs are divided into three classes, type I, type II, and type III IFNs, that are characterized by recognition of distinct and specific receptors. Type I IFNs comprise IFN-α and IFN-β, as well as a number of less-characterized subtypes, e.g., IFN-ε, -δ, -τ, -κ, and -ω, some of which are expressed in a species-dependent fashion (24). Whereas humans and mice encode a single IFN-β gene, there are 13 and 14 IFN-α subtypes expressed in human and murine genomes, respectively, that share a high degree of amino acid identity within each species (37). Synthesis of IFN-α/β is induced by pattern recognition receptor-mediated recognition of viral nucleic acid or proteins and, as a key event, involves activation and nuclear translocation of the transcription factors IFN regulatory factor 3 (IRF-3) and -7 and NF-κB. Upon secretion, IFN-α/β acts by binding in an autocrine and paracrine fashion to the ubiquitously expressed heterodimeric IFN-α/β receptor (IFNAR) consisting of IFNAR1 and IFNAR2 chains (26). Binding of IFN-α/β to IFNAR activates the JAK-STAT signal transduction pathway, resulting in transcription of more than 100 IFN-stimulated gene (ISG) products that have direct or indirect (i.e., modulatory) antiviral activity, thus establishing an antiviral state.
Type II IFN has IFN-γ as its sole member. It is not directly induced by viral infection and activates natural killer (NK) cells and macrophages.
In 2003, the IFN-λs were identified as a new class of IFN, the type III IFNs, by in silico methods (13, 31). They comprise IFN-λ1, IFN-λ2, and IFN-λ3, also called interleukin-28A (IL-28A), IL-28B, and IL-29, that belong to the IL-10 superfamily of cytokines. Whereas all three type III IFNs are expressed in humans, IFN-λ1 is a pseudogene in the mouse (16). Type III IFNs resemble type I IFNs with respect to induction pathways and stimulation of ISGs (8, 19, 40). However, they act via a different heterodimeric receptor consisting of IL-10 receptor β (IL-10Rβ) and IL-28Rα chains (13, 31). Since the IL-10Rβ subunit is shared with IL-10 and other members of the IL-10 superfamily, it is the IL-28Rα subunit that confers specificity for IFN-λ. In contrast to the ubiquitously expressed IFN-αβ receptor, the expression of the IL-28Rα subunit and, thus, the function of IFN-λ are predominantly restricted to cells of epithelial origin (8, 22, 32). Reflecting this distribution, type III IFNs are thought to be important in epithelial tissues at high risk for infections, e.g., the lung (22, 32).
Human and bovine respiratory syncytial virus (HRSV and BRSV, respectively), which are important pathogens of men and cattle, respectively, and their murine homologue, pneumonia virus of mice (PVM), are members of the genus Pneumovirus, family Paramyxoviridae. The pneumoviruses are distinguished from all other members of the family Paramyxoviridae by two nonstructural (NS) proteins, NS1 and NS2, that are encoded by the first two genes proximal to the 3′ end of the negative-sense genomic RNA. For HRSV and BRSV, the NS1 and NS2 proteins have been identified as potent multifunctional antagonists of the IFN response that act upon IFN induction (4, 28, 29, 33, 34, 36) as well as on the IFN signaling pathways (9, 18, 25). Interestingly, although both proteins appear to cooperate in their inhibitory functions, there are species-specific differences, such that the NS1 protein of HRSV is a more potent IFN antagonist than NS2 (33), whereas in BRSV the roles are reversed (36). NS1 and NS2 of both RSVs have been shown to inhibit activation and translocation of the transcription factor IRF-3 into the nucleus (4, 33, 34). For HRSV, it was shown that this inhibition may be mediated by direct interaction of NS2 with RIG-I (17). In addition, both HRSV NS proteins appear to mediate degradation of intracellular STAT2 levels, with a more efficient role for NS2 (9, 18, 25). The NS1 protein appears to function as an E3 ubiquitin ligase, causing polyubiquitination and proteasomal degradation of STAT2 (9). Other studies suggest an inhibitory effect of the HRSV NS proteins on apoptosis that appears to be independent of IFN (3). However, the evaluation of the roles of the NS proteins and the effects of IFN antagonism for pneumoviruses in vivo have been limited to date to gross attenuation phenotypes. Thus, HRSV lacking either or both NS proteins is attenuated in chimpanzees and humans (35, 39), and the corresponding BRSV mutants are attenuated in cattle (36).
PVM is the murine counterpart of the RSVs and appears to model important features of HRSV disease in humans (10, 27). Although characterization of the PVM proteins is less complete than that of HRSV and BRSV proteins, 11 of the 12 encoded proteins appear to be functional homologues of their RSV and BRSV counterparts (14). Thus, PVM provides a convenient small-animal model to study pneumoviral pathogenesis in a natural host. Recently, a reverse genetic system became available, permitting generation of virulent recombinant PVM (rPVM) (15), which we used in previous work to identify the NS proteins as pathogenicity factors in mice by generating rPVM with deletions of the NS1 and NS2 proteins individually or in combination (5). In this previous study, the NS2 protein was identified as the major antagonist of the IFN-α/β response, whereas the participation of the NS1 protein was unclear.
In the present study, we have extended this work by showing that both proteins act as antagonists of both type I and type III IFNs. The use of mouse strains lacking the receptors to type I, type III, or both types of IFNs showed that both types contribute to restricting replication and virulence of the NS deletion viruses although type I IFN played the larger role. Interestingly, however, replication and virulence of wild-type rPVM were not detectably affected by the presence or absence of these receptors, indicating that the wild-type virus strongly suppresses both IFN systems during infection in vivo. However, pretreatment of mice with type I IFN provided protection against disease and death, whereas pretreatment with type III IFN delayed but did not prevent disease, and, in some cases, reduced mortality. Virulence of rPVM lacking NS2 was not recovered completely when both interferon receptors were deleted, suggesting that NS2 may have further functions outside the IFN system.
MATERIALS AND METHODS
Mice.
Six-to-eight-week-old C57BL/6 (B6) mice were obtained from Charles River Laboratories. B6 IFNAR-deficient (B6.IFNAR0/0), B6.IL28Rα0/0, and B6.IFNAR0/0.IL28Rα0/0 mice, all bred on a B6.Mx− or B6.Mx+ background, were described previously (1, 21, 23). Both backgrounds were used in this study after confirming that results were completely independent from presence or absence of Mx (data not shown).
For infection with virus, mice were anesthetized either by intraperitoneal (i.p.) injection of 100 mg of ketamine and 5 mg of xylazine per kg of body weight or by isoflurane inhalation. Mice were infected intranasally with the indicated doses of viruses diluted with phosphate-buffered saline (PBS) to a final volume of 80 μl. Mice were closely observed for signs of illness such as ruffled fur, hunching, and listless behavior and were weighed daily as a quantitative measure of disease. Mice were sacrificed if the weight loss exceeded 20% for more than 2 days or if the animals were obviously in extremis. For analyzing virus replication, mice were sacrificed at the time points indicated on the figures and lungs were removed, homogenized in 3 ml of Eagle's minimal essential medium (MEM) supplemented with 50 mM HEPES, 100 mM MgSO4, and 10% fetal calf serum (FCS), and stored at −80°C until use. Viral titers were determined by plaque assays as described previously (15).
For treatment with IFN, mice that had been anesthetized as described above were treated intranasally with 106 units of a mixture containing recombinant human IFN-α/β/ω (huIFN-α/β/ω; Novartis, Basle, Switzerland) or 5 μg of recombinant murine IFN-λ2 (muIFN-λ2; R&D Systems) in a volume of 100 μl of PBS. Control animals received just PBS. Eight hours later, the treated mice were infected with 5,000 PFU of rPVM as described above. All procedures were approved by the local animal care committee.
Viruses.
Generation, propagation, and titration of recombinant pneumonia virus of mice (rPVM) and of rPVM in which the NS1 and NS2 genes were deleted individually and in combination have been described previously (5, 15). Briefly, a fragment of pPVM flanked by a unique XmaI site immediately upstream of the T7 promoter on the vector side and a naturally occurring unique Acc65I site in the N gene (accession number AY729016; nucleotide [nt] 1654) was excised and transferred to a pGEM vector. In consequence, this fragment contained the T7 promoter, the PVM leader sequence, the complete NS1 and NS2 genes, and part of the N gene. The NS1 and NS2 genes were deleted individually or in combination by PCR amplification of the whole plasmid, with exception of the respective genes, according to the method of Byrappa et al. (6) (primer sequences are available upon request). The resulting XmaI-Acc65I fragments lacking one or both NS genes were confirmed in their sequence, excised, and reintroduced into the pPVM and the pPVM-green fluorescent protein (GFP) backbone (15), resulting in, respectively, plasmids pPVM ΔNS1 and pPVM-GFP ΔNS1, pPVM ΔNS2 and pPVM-GFP ΔNS2, pPVM ΔNS1ΔNS2, and pPVM-GFP ΔNS1ΔNS2. These plasmids were used to recover the corresponding rPVM. In the case of the ΔNS2 mutant, the sequence of the NS1-N intergenic region differed from that previously described in that it was identical to the natural sequence of the NS2-N intergenic region.
Cells.
BHK-21 and Vero cells were maintained in Glasgow MEM and Eagle's MEM, respectively, all supplemented with 10% FCS. The RAW 264.7 macrophage cell line was maintained in RPMI 1640 medium supplemented with 20% FCS. Mouse embryonic fibroblasts (MEF) were prepared from 14-day-old embryos by removing brain and liver, mincing the residual tissue with scissors, and digestion with trypsin-EDTA solution. Following inactivation of trypsin with FCS, digested tissue was filtered through sterile gauze to remove tissue debris. The flowthrough was concentrated by low-speed centrifugation and seeded into cell culture dishes. MEF were propagated in Dulbecco's modified Eagle high-glucose medium (DMEM high glucose) supplemented with 10% FCS. All MEF used for experiments were below passage 10. Mouse peritoneal macrophages were obtained by lavage of the peritoneal cavity with 10 ml of RPMI medium (supplemented with 20% FCS, 10 U/ml penicillin, 10 μg/ml streptomycin, and 2 mM l-glutamine) 4 days following intraperitoneal injection of sterile 4% thioglycolate broth. Peritoneal macrophages were maintained in RPMI 1640 medium supplemented with 20% FCS.
Growth curves.
Replication kinetics in BHK-21 and MEF cells were performed as described previously (15). Briefly, replicate cultures were infected at a multiplicity of infection (MOI) of 0.01 PFU/cell, and, at the indicated time points, cells and supernatants were harvested and subjected to freeze-thaw cycles to release cell-bound virus particles. In the case of mouse peritoneal macrophages and the RAW 264.7 macrophage cell line, the overlaying medium was collected and replaced with fresh medium at the time points indicated on the figures. The collected medium supernatants were flash-frozen and stored at −80°C until titration. Virus titers were determined by plaque assay as described previously (15).
Determination of IFN induction.
For analyzing IFN mRNA levels in cell cultures, MEF, peritoneal macrophages, or RAW 264.7 cells were infected at an MOI of 1 PFU per cell of rPVM, rPVM ΔNS1, rPVM ΔNS2, or rPVM ΔNS1 ΔNS2. Total cellular RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer's instructions. One microgram of total RNA was reverse transcribed using random hexamers and SuperScript II reverse transcriptase (Invitrogen). Subsequently, 2 μl of the reverse transcription (RT) reaction products were subjected to 40 PCR cycles using GoTaq polymerase (Promega). All polymerases were used according to the manufacturer's instructions. The following specific primer pairs and PCR conditions were used: for IFN-α5, primer and conditions according to (32); for IFN-β (accession number NM_010510) forward primer 5′-ATGAACAACAGGTGGATCCTCC-3′, reverse primer 5′-TTCAAGTGGAGAGCAGTTGAG-3′, and annealing temperature of 55°C; and for IFN-λ2/3 (accession number AY86969), forward primer 5′-CCGCAGTGCTGACAAGAACC-3′, reverse primer 5′-ACTGGCCACACACTTGAGG-3′, and annealing temperature of 55°C; for PVM N gene (nucleotide positions 1548 to 2056 of the PVM genome [14]; accession number AY720016), forward primer 5′-CCAGATTGAGGTGTGATAGTTC-3′, reverse primer 5′-TGACCCTATGATGCCTAAAC-3′, and annealing temperature of 53°C; for total actin (accession number X03672), forward primer 5′-GTTGAGGTGTTGAGGCAGCCAGGGCT-3′, reverse primer 5′-TGGCGCTTTTGACTCAGGAT-3′, and annealing temperature of 55°C.
For determining IFN expression levels by quantitative reverse transcription-PCR (qRT-PCR), RNA samples were digested with RNase-free DNase I (Qiagen), and 1 μg of RNA was reverse transcribed using SuperScript III (Invitrogen) in a 25-μl mixture using random primers. The reverse transcription product was diluted 4-fold, and 2 μl of the diluted cDNA mix was used in each quantitative TaqMan PCR using the Universal PCR Master Mix (Applied Biosystems, Foster City, CA) for quantification of mRNAs of IFN-β (Applied Biosystems; Mm00439552_s1), IFN-α5 (Mm00833976_s1), and IFN-λ (Mm00663660_g1). For quantification of PVM RNA, the following primers and probes were used: NPVM forward (5′-GGACACTCGGCATGTTCTTACTT-3′), NPVM reverse (5′-GAGCCCTATTTCTGCCACTTCTT-3′), and NPVM probe (5′-CTGCCTTCAACCGTTG-3′); FPVM forward (5′-TCCAATGCCTTGAAGTCCAAA-3′), FPVM reverse (5′-GCAGCTCCGAGACCAAGAAT-3′), and FPVM probe (5′-AAGAAGAGGTTCCTCGGTT-3′). Results were analyzed using the comparative threshold cycle (ΔΔCT) method, normalized to 18S rRNA. rPVM ΔNS2 in C57BL/6 infected cells was used as a calibrator. Results are expressed as fold change over mock infection levels (C57BL/6) for IFN or fold change over rPVM infection levels (C57BL/6) for the viral RNA.
For analyzing IFN induction in the lung, C57BL/6 mice were infected with 105 PFU of the respective virus, and the lungs were taken at 24 h postinfection (p.i.) and homogenized in 2 ml of phosphate-buffered saline (PBS), and tissue debris was removed by low-speed centrifugation. Total celllular RNA was isolated from 200 μl of each sample and subjected to IFN-specific RT-PCR as described above. The residual supernatants from the lung homogenates were subjected to IFN-α- and -β-specific enzyme-linked immunosorbent assay (ELISA; PBL Biomedical Laboratories) according to the manufacturer's instructions.
Cytometric bead array.
Bronchoalveolar lavage (BAL) was performed with 0.8 ml of phosphate-buffered saline (PBS). Cells and debris were removed by low-speed centrifugation, and the supernatants were stored at −20°C until use. Fifty microliters of the supernatants was analyzed for the following secreted cytokines: interleukin-6 (IL-6), interleukin-10 (IL-10), monocyte chemoattractant protein 1 (MCP-1), IFN-γ, tumor necrosis factor alpha (TNF-α), and IL-12p70 using a cytometric bead array (CBA) mouse inflammation kit (BD Pharmingen) according to the manufacturer's instructions. The data were analyzed by using CBA software (BD Pharmingen).
Histopathology.
For evaluating histopathology, mice were sacrificed on day 8 after infection with 500 PFU of rPVM or 50,000 PFU of the NS deletion mutants. The lungs were intratracheally instilled with 0.5 ml of 4% formalin, removed, immersed in 4% formalin overnight, and embedded in paraffin. Sections of 4 μm were stained with hematoxylin and eosin (H&E) and scored in a blinded fashion by a single experienced observer. The samples were examined for the extent and cellular composition of inflammatory infiltrates.
RESULTS
The NS1 and NS2 proteins of PVM interfere with the type I and type III interferon responses.
The replication efficiencies of recombinant wild-type PVM (rPVM) and rPVM in which the NS1 and NS2 genes were deleted individually and in combination were compared in IFN-competent and IFN-incompetent cells. Cell monolayers were infected at a multiplicity of infection (MOI) of 0.01 PFU/cell, and viral titers in the supernatants over a period of 7 days were determined by plaque assay.
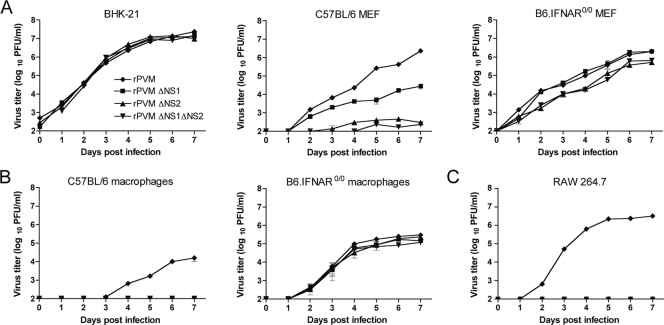
All four viruses, rPVM and the ΔNS1, ΔNS2, and ΔNS1 ΔNS2 mutants, replicated with identical kinetics and reached titers of approximately 107 PFU/ml by day 5 in BHK-21 cells, which have been reported to contain a defect in the type I IFN system (20) (Fig. 1A, left panel). In IFN-competent mouse embryonic fibroblasts (MEF) isolated from C57BL/6 (B6) mice, the replication of all three ΔNS mutants was significantly restricted compared with that of wild-type rPVM, with a larger effect on rPVM ΔNS2 and rPVM ΔNS1 ΔNS2 than on rPVM ΔNS1 (Fig. 1A, middle panel). Although the titer of rPVM ΔNS1 increased steadily over time, the increase was significantly reduced compared to that of rPVM, with a maximum difference of 100-fold at the last time point. Following infection with the ΔNS2 or ΔNS1 ΔNS2 mutant, no virus was detected up to day 2, and virus titers did not exceed 4 × 102 PFU/ml (Fig. 1A, middle panel). In contrast, this restriction of replication on the ΔNS mutants was largely lost when they were used to infect MEF lacking the IFN-α/β receptor (Fig. 1A, right panel).
Fig. 1.
Multicycle growth kinetics of wild-type rPVM and rPVM mutants lacking one or both NS proteins in IFN-competent and incompetent cell cultures. (A) Replicate cultures of BHK-21 cells or MEF from C57BL/6 or B6.IFNAR0/0 mice were infected with rPVM or the ΔNS1, ΔNS2 or ΔNS1ΔNS2 mutants at an MOI of 0.01 PFU per cell. At the indicated time points, cells and medium were harvested, clarified, and flash frozen. In addition, peritoneal macrophages from C57BL/6 or B6.IFNAR0/0 mice (B) or RAW 264.7 cells (C) were infected as described for panel A. At the indicated time points, the medium supernatants were taken and flash frozen, and fresh medium was added. Virus titers were determined by plaque assay. For all experiments, each time point for each virus is represented by duplicate cultures. Mean values of two independent experiments are shown, with standard error of the means indicated.
The restriction of the ΔNS1, ΔNS2, and ΔNS1 ΔNS2 mutants in IFN-competent cells was even more prominent when the replication kinetics were analyzed in peritoneal macrophages isolated from C57BL/6 mice (Fig. 1B, left panel) or in the macrophage cell line RAW 267.4 (Fig. 1C): in the supernatants of both of these IFN-competent cell types, productive replication was detectable only for rPVM and not for the ΔNS mutants. The restriction of the ΔNS1, ΔNS2, and ΔNS1 ΔNS2 mutants was completely lost when peritoneal macrophages from B6.IFNAR0/0 mice were used (Fig. 1B, right panel), indicating that the restriction of replication was mediated by type I IFN.
Next, we investigated the induction of IFN-α, IFN-β, and of the type III IFN-λ after infection with rPVM and the three ΔNS mutants in MEF and macrophages. MEF, peritoneal macrophages of C57BL/6 mice, or RAW 267.4 cells were infected at an MOI of 1 PFU/cell with the panel of viruses. Total cellular RNA was isolated at the time points indicated on Fig. 2 and investigated by RT-PCR for the presence of mRNA for the rPVM N gene, total actin, IFN-α5, IFN-β, and IFN-λ2/3.
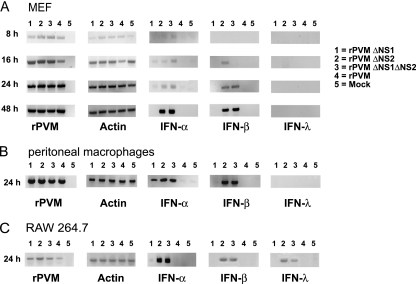
Fig. 2.
Induction of IFN-α, IFN-β, and IFN-λ in infected cell cultures, as detected by RT-PCR. MEF (A), peritoneal macrophages of C57BL/6 mice (B), or the RAW 264.7 macrophage cell line (C) were either mock infected or infected with rPVM or the ΔNS1, ΔNS2, or ΔNS1ΔNS2 mutant at an input MOI of 1 PFU per cell. At the indicated time points, total cellular RNA was extracted, and 1 μg of total RNA was subjected to RT-PCR to amplify mRNA specific for rPVM N, total actin, IFN-α5, IFN-β, or IFN-λ2/3. The RT-PCR products were subjected to electrophoresis on agarose gels and visualized with ethidium bromide.
In MEF, IFN-β-specific mRNA was detected starting at 16 and 24 h after infection with the ΔNS2 and ΔNS1 ΔNS2 mutants, respectively, and continued until 48 h p.i., the last time point measured (Fig. 2A). In comparison, rPVM and rPVM ΔNS1 did not detectably induce IFN-β at any time point. The same expression pattern for IFN-β was observed following infection of peritoneal macrophages (Fig. 2B) or RAW 264.7 cells (Fig. 2C). Infection with the ΔNS2 and ΔNS1 ΔNS2 mutants also induced the transcription of IFN-α5 in all cells tested (Fig. 2A, B, and C). Interestingly, we also observed a less abundant band specific for IFN-α5 mRNA after infection of MEF or peritoneal macrophages with rPVM ΔNS1 (Fig. 2A and B). As observed for IFN-β, rPVM did not induce synthesis of IFN-α5 at any time point in any of the tested cells.
Since the NS proteins have been reported to interfere with the induction of type III IFN (33), we also investigated transcription of IFN-λ2/3. No induction of IFN-λ was detected in MEF or peritoneal macrophages after infection with rPVM or any of the NS deletion mutants. However, transcription of IFN-λ mRNA was detected 24 h after infection of RAW 264.7 cells with the ΔNS2 or ΔNS1 ΔNS2 mutant (Fig. 2C), indicating that the PVM-NS2 protein antagonizes the induction of this cytokine.
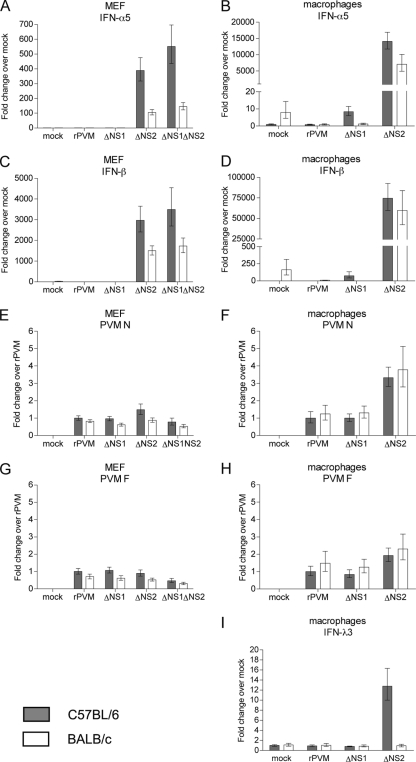
To compare IFN induction in cells derived from different mouse strains, MEF and peritoneal macrophages were isolated from C57BL/6 and BALB/c mice. Cells were infected with rPVM or the NS mutants, and, 24 h later, total RNA was isolated as described above. Induction of IFN-α5, IFN-β, and IFN-λ3 was determined by quantitative RT-PCR and analyzed relative to C57BL/6-derived mock-infected cells. For comparison of virus replication, viral RNA levels for PVM N and F proteins were measured by quantitative RT-PCR of the same RNA preparations.
As observed earlier using conventional RT-PCR (Fig. 2), IFN-α5 and -β were strongly induced in all cell types infected with the ΔNS2 and ΔNS1 ΔNS2 mutants (Fig. 3A to D) but not in rPVM-infected cells. In addition, we detected low levels of IFN-λ3 (12.8-fold over mock) in macrophages derived from C57BL/6 mice after infection with rPVM ΔNS2 (Fig. 3I). IFN-λ3 expression was not detected in any other samples (data not shown). With regard to the ΔNS1 mutant, IFN-α5 appeared to be induced at low levels (8.4-fold over mock) in C57BL/6-derived macrophages, compared to the corresponding mock control (Fig. 3B), which is similar to results shown in Fig. 2B. In addition, infection with rPVM ΔNS1 appeared to induce some IFN-β mRNA (75-fold over mock) in C57BL/6 macrophages. However, neither IFN-α5 nor IFN-β was significantly detectable in macrophages from BALB/c mice infected with the same mutant or in MEF from either mouse strain. We noted that uninfected BALB/c cells, particularly macrophages, appeared to express IFN-α5- and IFN-β-specific mRNA at levels similar to the level induced by the ΔNS1 mutant in C57BL/6 macrophages (Fig. 3B and D). These background expression levels were reduced by infection with rPVM and the ΔNS1 mutant. Overall, despite the background expression, both type I IFNs appeared to be expressed to somewhat lower levels in BALB/c cells than in C57BL/6 cells infected by the ΔNS2 mutants. Thus, moderate differences in the magnitude of the IFN response of different mouse strains may account for the differences in IFN induction observed for ΔNS1 and IFN-λ.
Fig. 3.
Induction of IFN-α5, IFN-β, and IFN-λ3 in infected primary cell cultures detected by qRT-PCR. MEF or peritoneal macrophages from C57BL/6 mice (gray bars) or BALB/c mice (white bars) were either mock infected or infected with rPVM or the ΔNS1, ΔNS2 or ΔNS1ΔNS2 mutant as indicated. Cells were harvested 24 h later, total cellular RNA was extracted, and 1 μg of RNA was subjected to qRT-PCR with primers and probes specific for IFN-α5, IFN-β, IFN-λ3, PVM N, or PVM F. Results are normalized to 18S rRNA and show fold increase of cytokine mRNA relative to mock infection (C57BL/6) (A to D and I) and levels of viral RNA relative to rPVM (C57BL/6) (E to H). Shown are mean values derived from a single experiment performed in triplicates for MEF or duplicates for macrophages. Error bars represent upper and lower range of values. Of note, the ranges of the y axes were adjusted to account for the variation of expression levels.
Induction of type I and type III interferons in the lungs of infected mice.
To analyze the IFN-inducing capacity of wild-type rPVM and the ΔNS mutants in mouse lungs, C57BL/6 mice were mock infected or infected with 1 × 105 PFU of each virus. Mice were sacrificed 24 h later, and the lungs were removed and homogenized; one aliquot of homogenate was analyzed by ELISA for IFN-β or IFN-α (Fig. 4A), and a second aliquot was subjected to total cellular RNA extraction and RT-PCR specific for IFN-β, -α5, and -λ2/3 (Fig. 4B).
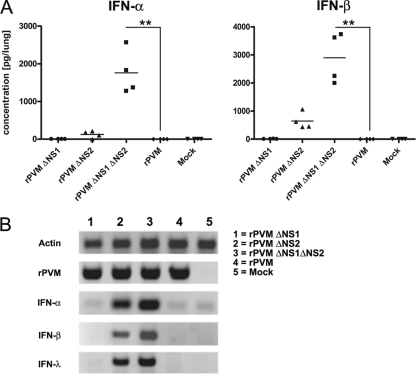
Fig. 4.
Induction of IFN-α, IFN-β, and IFN-λ in the lungs of infected mice. C57BL/6 mice in groups of four were mock infected or infected intranasally with 50,000 PFU of rPVM or the ΔNS1, ΔNS2, or ΔNS1ΔNS2 mutant. The animals were sacrificed after 24 h, and the lungs were removed, homogenized, and aliquoted. (A) One aliquot was used to determine IFN-α and -β protein levels by ELISA. Statistical significance was calculated by analysis of variance, followed by Tukey-Kramer posthoc test analysis. Columns marked with asterisks differ significantly (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001) from values for rPVM. Data from one of two independent experiments providing comparable results are shown. (B) Total RNA was extracted from a second aliquot of lung homogenate and IFN-α5-, -β-, and -λ2/3-specific mRNA, as well as PVM- and actin-specific mRNA, was amplified by RT-PCR and visualized by gel electrophoresis and ethidium bromide staining. Shown are exemplary samples of one mouse per infection group.
Similar to our results in vitro, no IFN-α or -β was detected in the lungs of rPVM-infected mice by either ELISA (Fig. 4A) or RT-PCR (Fig. 4B). However, we also did not detect IFN-α or -β in the lungs of mice infected with rPVM ΔNS1 although it should be noted that in a single animal an IFN-β-specific band was detectable by RT-PCR, and a very small amount of IFN-β protein (5 pg/ml) was detectable by ELISA (data not shown), indicating that a weak induction of IFN might occur that is close to the detection limit. As expected, IFN-α and IFN-β were induced following infection with the ΔNS2 and ΔNS1 ΔNS2 mutants, with the latter virus resulting in significant protein levels of 880 pg/lung and 1,267 pg/lung, respectively. Whereas type I IFN-specific RT-PCR products were clearly detectable from infected-cell samples for rPVM ΔNS2, only small amounts of protein (mean IFN-α, 62 pg/lung; mean IFN-β, 320 pg/lung) were detected, which were not significantly above background.
Since a commercially available IFN-λ ELISA (1) resulted in high background signals in mock-infected lung homogenates (B. Heinze and C. D. Krempl, unpublished observation), only RT-PCR was available for detection of IFN-λ. No IFN-λ-specific mRNA was amplified following infection with recombinant wild-type virus or rPVM ΔNS1. Comparable to the data in vitro, transcription of IFN-λ2/3 genes was observed in lungs of mice infected with the ΔNS2 and ΔNS1 ΔNS2 mutants, thus identifying the PVM-NS2 protein as an antagonist of IFN-λ induction in vivo.
Replication and virulence of rPVM with deletion of the NS proteins in mice lacking a functional receptor for type I, type III, or both types of IFN.
To evaluate the contributions of IFN-α/β and IFN-λ to the pathogenesis of PVM, wild-type C57BL/6 mice or derivatives lacking functional receptors for either or both IFNs (i.e., B6.IFNAR0/0, B6.IL28Rα0/0 [1], and B6.IFNAR0/0.IL28Rα0/0[21]) were infected intranasally with 5,000 PFU per mouse of rPVM or the respective ΔNS mutant in an inoculum of 80 μl. Mice were observed closely and weighed daily as a measure of disease. For analyzing replication efficiency, mice were sacrificed on days 3 and 7 postinfection, respectively; lungs were removed and homogenized, and viral titers were determined by plaque assay.
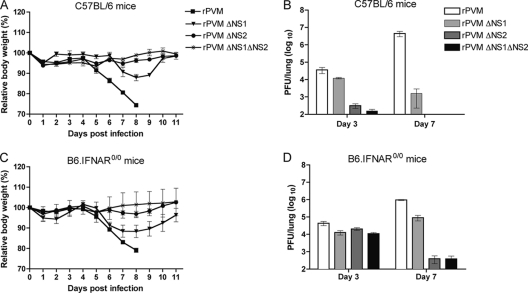
Since 5,000 PFU of wild-type PVM per C57BL/6 mouse is a highly lethal dose, all mice infected with rPVM rapidly lost weight beginning on day 4 and died or were sacrificed in extremis by day 8, consistent with previous results (10) (Fig. 5A). In comparison, all of the wild-type mice infected with any of the three ΔNS mutants survived, which corresponds to our previous data using BALB/c mice (5). For the ΔNS mutants, the only substantial weight loss observed was for mice infected with rPVM ΔNS1, involving a transient weight loss of approximately 10% (Fig. 5A). This was indicative of residual virulence for the ΔNS1 mutant. These differences in the extent of virulence were reflected in the ability of the viruses to replicate in the lungs of mice. In C57BL/6 mice infected with rPVM, the virus replicated to a mean titer of 3 × 104 PFU/lung on day 3 and increased to 4 × 106 PFU/lung on day 7 (Fig. 5B). In comparison, the virus load in mice infected with rPVM ΔNS1 reached a mean titer of only 104 PFU/lung on day 3 and decreased to 103 PFU/lung on day 7. The ΔNS2 and ΔNS1 ΔNS2 mutants were severely attenuated in the lower respiratory tract of C57BL/6 mice, such that virus was detectable in only 80% and 60% of the mice, respectively, with titers close to the detection limit on day 3 and no detectable virus in any of the mouse lungs at day 7 (Fig. 5B).
Fig. 5.
Virulence and replication of rPVM and NS deletion mutants in the respiratory tract of C57BL/6 mice and B6.IFNAR0/0 mice. C57BL/6 (A and B) and B6.IFNAR0/0 (C and D) mice were infected intranasally with 5,000 PFU of rPVM or the ΔNS1, ΔNS2, or ΔNS1ΔNS2 mutant in an volume of 80 μl. Mice were observed closely, and body weight was monitored daily (A and C). For analyzing replication (B and D), mice were sacrificed on day 3 and day 7 after infection, and the lungs were removed, homogenized, flash frozen, and stored until use. Viral titers were determined by plaque assay. Results represent mean values and standard errors of the mean derived from a minimum of five animals per group.
In knockout mice lacking the type I IFN receptor (B6.IFNAR0/0) (Fig. 5C and D), the type III IFN receptor (B6.IL28Rα0/0) (Fig. 6A and B), or both receptors (B6.IFNAR0/0.IL28Rα0/0) (Fig. 6C and D), the replication and virulence of wild-type rPVM were essentially indistinguishable from what was observed in C57BL/6 mice (Fig. 5A and B). In the wild-type mice and all three transgenic mouse strains, weight loss started from day 4 to 5, with 100% mortality by day 8, and virus load was 3 × 104 to 4 × 104 PFU/lung and 3 × 106 to 4 × 106 PFU/lung on days 3 and 7, respectively. The lack of difference in virus replication and virulence between C57BL/6 mice and knockout strains indicates that wild-type rPVM very efficiently counteracts the induction/action of type I IFNs such that intact type I and type III IFN systems did not restrict replication detectably.
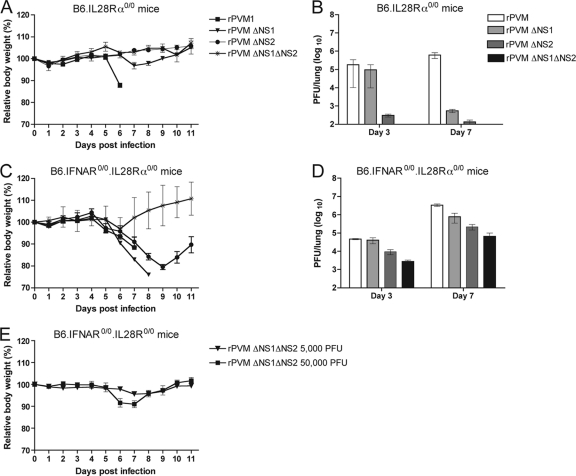
Fig. 6.
Virulence and replication of rPVM and the NS deletion mutants in the respiratory tract of B6.IL28Rα0/0 and B6.IFNAR0/0.IL28Rα0/0 mice. (A and B) B6.IL28Rα0/0 and (C and D) B6.IFNAR0/0.IL28Rα0/0 mice were infected intranasally with 5,000 PFU of rPVM or the ΔNS1, ΔNS2, or ΔNS1ΔNS2 mutant in a volume of 80 μl. As described in the legend to Fig. 5, body weight was monitored daily (A and C); mice were sacrificed on days 3 and 7, and lung homogenates were assayed for virus titer (B and D). (E) In addition, B6.IFNAR0/0.IL28Rα0/0 mice were infected intranasally with 5,000 or 50,000 PFU of rPVM ΔNS1ΔNS2, and body weight was monitored as described above. Results represent mean values and standard errors of the mean derived from a minimum of five animals per group.
However, in contrast to observations in tissue culture, a defect in type I IFN signaling did not reverse the attenuated phenotypes of the NS deletion mutants in vivo. In B6.IFNAR0/0 mice, replication and virulence of all three NS deletion mutants exhibited a spectrum of attenuation. Following infection with rPVM ΔNS1, the virus titers in the lungs of B6.IFNAR0/0 mice and wild-type mice were identical on day 3. However, on day 7, the titers of rPVM ΔNS1 increased in the B6.IFNAR0/0 mice, reaching 8 × 104 PFU/lung on day 7 (Fig. 5B and D), whereas the titers decreased in wild-type mice. However, in both mouse strains, the titer of the ΔNS1 virus was reduced compared to that of wild-type rPVM. The somewhat increased permissiveness of B6.IFNAR0/0 mice to replication of the ΔNS1 mutant was also reflected in a more pronounced weight loss, although that barely exceeded 10% (Fig. 5C), and no fatalities were observed. Following infection of B6.IFNAR0/0 mice with the ΔNS2 and ΔNS1 ΔNS2 mutants, the pulmonary virus titers were similar to the titer of rPVM ΔNS1 on day 3 but decreased until day 7 such that the titers were close to the detection limit in one-third of the animals and below the detection limit in the other two-thirds. Correspondingly, no significant weight loss was observed for B6.IFNAR0/0 mice infected with the ΔNS2 or ΔNS1 ΔNS2 mutant. Overall, mice lacking the type I IFN receptor were somewhat more susceptible to rPVM ΔNS1 with regard to replication and virulence and supported increased titers of the ΔNS2 or ΔNS1 ΔNS2 virus but only on day 3.
Replication and virulence of all four viruses in B6.IL28Rα0/0 mice, lacking the receptor for type III IFN, were not significantly different from earlier results obtained with wild-type mice (Fig. 5A and B and 6A and B). This showed that type I IFN accounts for the attenuation observed for the ΔNS viruses.
Mice lacking the receptors to both type I and type III IFNs (B6.IFNAR0/0.IL28Rα0/0) were generally more susceptible to infection with the ΔNS1, ΔNS2, and ΔNS1 ΔNS2 mutants than mice nonresponsive to either IFN alone. Infection with rPVM and rPVM ΔNS1 at 5,000 PFU was uniformly lethal for the double-knockout mice although the endpoint was delayed 1 day for the latter virus (Fig. 6C). This disease pattern was reflected by similar replication efficiencies such that the titers of wild-type rPVM and the ΔNS1 mutant were identical on day 3, whereas the titer of rPVM was somewhat higher on day 7 (Fig. 6D). In comparison, infection with 5,000 PFU of rPVM ΔNS2 did not cause any lethality but resulted in significant weight loss starting on day 5 and reaching a maximum of approximately 20% on day 9. The mean virus load increased from 9 × 103 PFU/lung on day 3 to 2 × 105 PFU/lung on day 7. However, the titer of the ΔNS2 mutant was still approximately 5-fold lower than that of rPVM (Fig. 6D), thus reflecting the attenuated phenotype.
Interestingly, infection with 5,000 PFU of rPVM ΔNS1 ΔNS2 was almost nonpathogenic for B6.IFNAR0/0.IL28Rα0/0 mice (Fig. 6C). The replication efficiency of the ΔNS1 ΔNS2 virus was significantly increased in the double-knockout mice compared to the other mouse strains, but nonetheless the virus titers of 2 × 103 PFU/lung on day 3 and 6 × 104 PFU/lung on day 7 were the lowest of any of the viruses tested. Infection of B6.IFNAR0/0.IL28Rα0/0 mice with a 10-fold higher dose of the ΔNS1 ΔNS2 virus resulted in moderate weight loss of approximately 10%, indicating that additional deletion of NS1 to NS2 increases attenuation more than 10-fold (Fig. 6E). The observation that the virulence and replication of PVM lacking the major IFN antagonist NS2 were not completely rescued in B6.IFNAR0/0.IL28Rα0/0 mice is suggestive of further protective immune mechanisms or further functions involving the NS proteins.
Histopathological analysis and cytokine profiles of C57BL/6 and B6.IFNAR0/0.IL28Rα0/0 mice infected with rPVM or the ΔNS mutants.
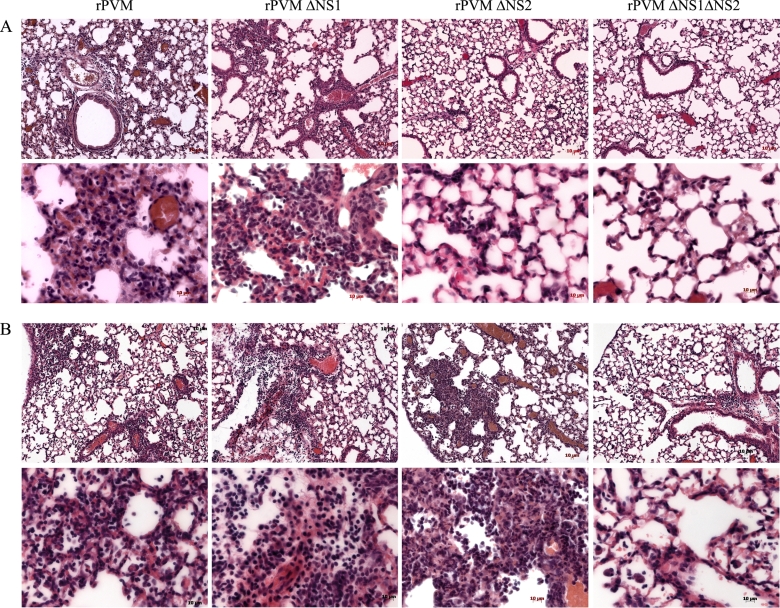
To further investigate the observed pathology of rPVM and the ΔNS mutants in IFN-α/β/λ-nonresponsive mice, wild-type and B6.IFNAR0/0.IL28Rα0/0 mice were infected with rPVM or the NS deletion mutants. The following doses per mouse were used in order to account for the different virulence levels of the viruses: 250 PFU of rPVM, 5,000 PFU of rPVM ΔNS1, and 50,000 PFU of rPVM ΔNS2 or rPVM ΔNS1 ΔNS2. Mice were sacrificed on day 8 after infection, and the lungs were removed and processed for histological analysis.
The lungs of C57BL/6 mice infected with rPVM or the ΔNS1 mutant showed a heterogeneous interstitial pneumonia with lymphoplasmatic infiltrates and signs of hyperemia (Fig. 7A). In comparison, lungs of wild-type mice infected with rPVM ΔNS2 or rPVM ΔNS1 ΔNS2 showed only minimal, noncharacteristic infiltrates or no signs of inflammation at all (Fig. 7A).
Fig. 7.
Histopathology of C57BL/6 and B6.IFNAR0/0.IL28Rα0/0 mice after infection with rPVM or the NS deletion mutants. Histopathological analysis of lung sections (H&E staining) from C57BL/6 (A) and B6.IFNAR0/0.IL28Rα0/0 (B) mice that had been infected with 250 PFU of rPVM, 5,000 PFU of rPVM ΔNS1, or 50,000 PFU of the ΔNS2 and ΔNS1ΔNS2 mutants 8 days previously. The panels show representative sections from histological analysis of two to four mice per group. Scale bars are indicated in the panels.
When B6.IFNAR0/0.IL28Rα0/0 mice were infected with rPVM, there were no obvious differences in pathological severity in the lungs compared to lungs of C57BL/6 wild-type mice (Fig. 7A and B). However, in addition to lymphocytes and plasma cells, scattered neutrophils were present in the infiltrates of rPVM-infected B6.IFNAR0/0.IL28Rα0/0 mice (Fig. 7B). The lungs of B6.IFNAR0/0.IL28Rα0/0 mice infected with the ΔNS1 mutant exhibited interstitial pneumonia and peribronchiolar chronic inflammation (ca. 45% of tissue affected) with only few granulocytes. In contrast to lungs of wild-type mice, the lungs of B6.IFNAR0/0.IL28Rα0/0 mice after infection with the ΔNS2 mutant displayed the pathology of an interstitial pneumonia with patchy tissue involvement, with infiltrates consisting predominantly of neutrophils and lymphocytes. Following infection with the ΔNS1 ΔNS2 mutant, the pathological changes in B6.IFNAR0/0.IL28Rα0/0 mice were less severe than for the other viruses. They were characterized by rather mild perivascular and peribronchiolar lymphocytic infiltrates (Fig. 7B, far right panel).
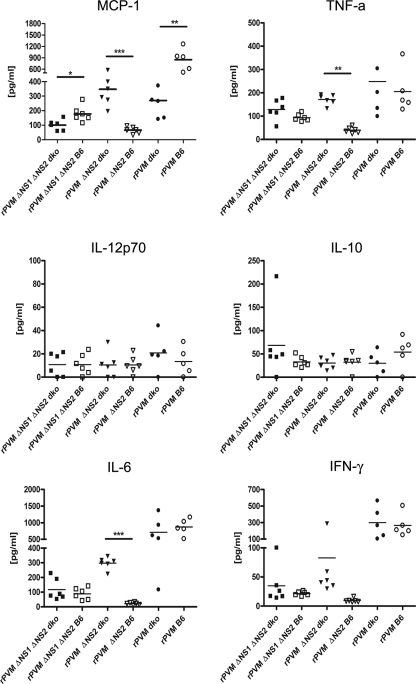
In addition, we measured the concentration of several inflammatory cytokines and of the anti-inflammatory IL-10 in the BAL fluid 8 days after infection of C57BL/6 or B6.IFNAR0/0.IL28Rα0/0 mice with 500 PFU of rPVM or 50,000 PFU of rPVM ΔNS2 or rPVM ΔNS1 ΔNS2 (Fig. 8).
Fig. 8.
Cytokine levels in bronchoalveolar lavage (BAL) fluid of C57BL/6 (B6) and B6.IFNAR0/0.IL28Rα0/0 (dko) mice infected with rPVM or the ΔNS mutants. Mice were intranasally infected with 500 PFU of rPVM or 50,000 PFU of rPVM ΔNS2 or rPVM ΔNS1ΔNS2 in a volume of 80 μl. On day 8 after infection, the mice were sacrificed, and a BAL with PBS was performed. Following clarification by low-speed centrifugation, cytokine levels in the BAL fluids were determined by cytometric bead array. The cytokine levels (pg/ml) from five to six mice per group are shown. Statistical significance was calculated by analysis of variance, followed by Tukey-Kramer posthoc test analysis. Groups marked with asterisks differ significantly (**, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
Infection of C57BL/6 mice with wild-type rPVM induced substantial amounts of MCP-1, TNF-α, IL-6, and IFN-γ but only negligible amounts of IL-12p70 and IL-10. There were no significant differences between the levels of cytokines in rPVM-infected wild-type and B6.IFNAR0/0.IL28Rα0/0 mice except in the case of MCP-1, which was expressed at a higher level in the parent mouse strain. However, this observation is in accordance with previously reported IFN-α/β dependency of MCP-1 synthesis after infection with murine cytomegalovirus (MCMV) (11).
In contrast, expression of all six cytokines in response to the ΔNS2 virus was low in C57BL/6 mice but was increased dramatically in the double-knockout mice for MCP-1, TNF-α, and IL-6 and was noticeably but not significantly increased for IFN-γ. Thus, the increase in the expression of these cytokines in ΔNS2-infected B6.IFNAR0/0.IL28Rα0/0 mice reflected the increase in virus replication and pathogenicity. In comparison, infection of either mouse strain with the ΔNS1 ΔNS2 virus induced only low levels of any of the six cytokines although the expression of MCP-1 was modestly but significantly higher in wild-type mice. Thus, unlike the case with the ΔNS2 mutant, the increased replication of the ΔNS1 ΔNS2 mutant in B6.IFNAR0/0.IL28Rα0/0 versus C57BL/6 mice was not accompanied by an increase in cytokine expression. Since the ΔNS1 ΔNS2 mutant replicated less efficiently under all conditions than rPVM and the ΔNS2 mutant, its replication may have been insufficient to trigger cytokine induction. Thus, pathological changes and cytokine synthesis reflect replication efficiency and virulence of the rPVM or the ΔNS mutants in the respective mouse strain.
Pretreatment with type I IFN, but not with type III IFN, protects mice against a lethal PVM infection.
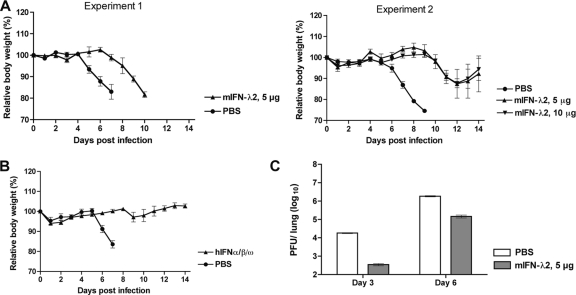
Treatment with IFN-α is associated with significant side effects, likely due to its systemic activity. Therefore, with regard to its organ-specific activity, it is convenient to speculate about using IFN-λ for topical treatment to control respiratory virus infections. To evaluate the protective capacity of the respective IFNs against infection with wild-type rPVM, C57BL/6 mice were treated intranasally with 5 or 10 μg of muIFN-λ2 (Fig. 9A and C), 106 units of a mixture of huIFN-α/β/ω (Fig. 9B), or an equal volume of PBS (controls) 8 h prior to infection with 5,000 PFU of rPVM. The amount of 5 μg of IFN-λ2 has previously been shown to be active and protective in the mouse system (1, 2).
Fig. 9.
Protective capacity of exogenous type I and type III IFN in a lethal PVM infection. C57BL/6 mice in groups of five were treated intranasally with 5 or 10 μg of murine IFN-λ2 (A), 106 units of a mixture of IFN-α/β/ω (B), or PBS (A and B) with a volume of 100 μl. Eight hours later, the mice were infected intranasally with 5,000 PFU of wild-type PVM in a volume of 80 μl. The mice were observed closely, and body weight was monitored daily. (C) To determine the virus load of mice pretreated with IFN-λ2, C57BL/6 mice in groups of 10 were treated as described for panel A; five mice of each group were sacrificed on day 3 and day 6 after infection, the lungs were removed and homogenized, and viral titers were determined by plaque assay.
In all experiments, PBS-treated mice succumbed to PVM infection in about the same time pattern observed in all previous experiments (Fig. 9A and B). In contrast, mice treated with IFN-α/β/ω did not develop any significant symptoms of disease (Fig. 9B). Surprisingly, IFN-λ2 treatment did not protect mice from disease and in several cases did not even ameliorate the outcome: in an initial experiment, treatment with 5 μg of IFN-λ2 merely delayed the onset of disease for 2 days (Fig. 9A, experiment 1). Thereafter, mice started to loose weight after day 6, and all mice died or were sacrificed in extremis by day 10, compared to day 4 and day 7, respectively, for the PBS-treated animals. Doubling the IFN-λ dose did not improve the course of disease and outcome compared to the 5-μg dose (Fig. 9A, experiment 2). However, in this experiment some IFN-treated animals survived infection; i.e., two out of four of the 5-μg group and three out of five of the 10-μg group survived. We also noted that the overall course of the disease in all groups of the second experiment was somewhat delayed compared to the course of experiment 1, indicating some variation in the infection efficiency between the two experiments. Delayed disease in the IFN-λ2-treated animals correlated with significantly delayed virus replication at day 3 (Fig. 9C). Thus, in the case of PVM, pretreatment with IFN-λ2 was not sufficient to prevent infection, disease, or mortality although some restriction of virus replication was observed at early time points during infection, as well as a reduction of mortality.
DISCUSSION
The NS1 and NS2 proteins of human and bovine RSV have been identified as potent inhibitors of the type I and type III IFN response (4, 28, 29, 33, 34, 36). Whereas the multifunctionality of these proteins and their effects on the IFN response have been well defined, investigation of their role as pathogenicity factors in vivo has been limited by the lack of a permissive small-animal model. In recent years, PVM infection of mice has been increasingly used by us and by other investigators as a pathogenicity model in a natural host for pneumoviral infections. The availability of a reverse genetic system permitting generation of virulent recombinant PVM greatly facilitates evaluation of viral and cellular pathogenicity factors in vivo, e.g., by using a variety of transgenic mouse strains. Previously, we identified the PVM NS proteins as pathogenicity factors in inbred mouse strains and showed that the NS2 protein is a potent inhibitor of the induction of IFN-α/β. This was done by the use of rPVM derivatives with single and combined deletions of the NS1 and NS2 genes (5). In the present study, we deepened and broadened the approach by not only confirming a dominant IFN-antagonizing role of the PVM NS2 protein but also clearly identifying the PVM NS1 protein as an antagonist of the IFN response. Replication of PVM with deletion of the NS1 protein was moderately impaired in MEF and highly attenuated in primary peritoneal macrophages, both from C57BL/6 mice and in the IFN-competent RAW 264.7 cell line. This attenuation was almost completely rescued in corresponding cells unresponsive to IFN-α/β (i.e., from IFNAR0/0 mice) (Fig. 1), thus linking the attenuation of rPVM ΔNS1 in wild-type cells to the integrity of the type I IFN system. In addition, attenuation of rPVM ΔNS1 in IFN-competent cells was accompanied by some production of IFN-α and -β mRNA (Fig. 2 and 3), although at very low levels compared to rPVM ΔNS2 or rPVM ΔNS1 ΔNS2. Furthermore, restriction of rPVM ΔNS1 in mice was at least partially dependent on a functional IFN response (Fig. 5 and 6C and D), thus further substantiating the function of NS1 as an IFN antagonist.
This conclusion extends our previous identification of PVM NS1 as a pathogenicity factor in wild-type mice (5). However, our current results with respect to NS1 appear somewhat contradictory to our previous observations that deletion of NS1 did not reveal any effect on virus replication and IFN induction in MEF. However, this appeared to be due to the use of different mouse strains and cell types in the two studies, i.e., CF3 (MEF) and BALB/c mice (previous study) versus C57BL/6 mice (present study) or MEF versus macrophages. In the present study, we detected low levels of IFN induction by rPVM ΔNS1 in macrophages derived from C57BL/6 mice but not from BALB/c mice (Fig. 2 and 3). In addition, this ΔNS1-dependent expression pattern appears to be cell type dependent since it was observed in peritoneal macrophages but not in MEF (Fig. 3). We also noted that upon virus infection, expression of type I IFNs appeared to be somewhat more efficient in C57BL/6 cells than in BALB/c cells (Fig. 3). Thus, these moderate differences in the IFN response to PVM may account for the differences with respect to ΔNS1 or IFN-λ, particularly, if the inhibition of IFN induction is close to the detection limit. In conclusion, the IFN-antagonistic activity of NS1 is low and seems to be below the detection limit of certain cell and mouse systems. Also, the exact IFN-antagonizing mechanism of NS1 remains elusive, whereas NS2 seems to interfere with the induction of IFN-α/β. Further work is needed to determine the detailed mechanisms of the NS proteins.
In the present study, we further show that PVM efficiently antagonizes the type III IFN response in addition to that of type I IFN. IFN-λ2/3 was induced in macrophages and in the lungs of mice following infection with PVM lacking NS2 (i.e., rPVM ΔNS2 and rPVM ΔNS1 ΔNS2) (Fig. 2C, 3I, and 4B) but not following infection with rPVM or rPVM ΔNS1. Consequently, the NS2 protein appears to be the main antagonist of type III IFN, as was the case with type I IFN. However, the pathogenicity of rPVM ΔNS1 was fully rescued only in mice not responsive to both type I and III IFNs but not in mice not responsive to type I or type III IFN alone (Fig. 5 and 6) (see below). Thus, as with the type I IFN response, NS1 also appears to contribute to inhibition of the type III IFN response.
The ability of cells to synthesize IFN-λ appears to be a general property, but its ability to act is restricted to epithelial tissue by the limited expression pattern of its receptor (22, 32). Because of a high content of epithelial tissue, organs such as the respiratory tract, skin, and gastrointestinal tract may be particularly responsive to IFN-λ, which in turn would serve as a defense mechanism against pathogens with tropism for such organs. In fact, functional defects in the receptors for IFN-α/β and IFN-λ render mice significantly more susceptible than single IFN receptor deficiency to several human respiratory viruses, including HRSV (21, 22), thus substantiating a protective role of type III IFN in the respiratory tract. In human bronchial epithelial cells from asthmatic patients and in human volunteers, rhinovirus replication or asthma exacerbations were inversely correlated with levels of IFN-λ and -β (7), which indicates their importance in controlling natural infections. However, experimental studies in species-conforming in vivo models have been lacking so far. For HRSV in particular, the potent IFN-α/β and -λ antagonists that are expressed by the pathogen to prevent early IFN induction in the natural human host are not fully functional in the mouse.
Our studies using PVM in B6.IFNAR0/0.IL28Rα0/0 mice account for the specificity. Our results indicate that IFN-λ has the capability to confer protection in PVM infection in vivo since replication and pathogenicity were significantly increased for all of the ΔNS mutants in B6.IFNAR0/0.IL28Rα0/0 double-knockout mice compared to B6.IFNAR0/0 mice. Furthermore, as discussed above, rPVM ΔNS2 and rPVM ΔNS1 ΔNS2 are able to induce production of IFN-λ in the lungs of mice.
However, this effect is limited to a situation where IFN-α/β and/or -λ is actually induced, i.e., in the absence of the functional IFN antagonists NS1 and NS2. In the case of the wild-type virus, replication and pathogenicity were completely unaffected by the presence or absence of functional type I and type III IFN receptors. In the IFN-competent wild-type and all IFN-incompetent transgenic mouse strains, infection-induced weight loss started on day 4 to 5, leading to 100% mortality by day 8. In addition, no significant differences in virus load were observed on days 3 and 7. Thus, in a natural pneumovirus infection, the protective activity of the IFN-α/β and IFN-λ response appears to be completely prevented by the NS proteins.
Of note, in B6.IFNAR0/0.IL28Rα0/0 mice replication and pathogenicity were recovered in the case of rPVM ΔNS1 but not in the case of PVM lacking the most dominant IFN antagonist, NS2 (i.e., rPVM ΔNS2 and rPVM ΔNS1 ΔNS2). Infection with a dose of rPVM ΔNS2 that otherwise was highly lethal when used for infections with wild-type virus caused severe weight loss but no lethality. This correlated with an overall increased virus load that, however, was still about 5-fold reduced compared to wild-type PVM at the peak of replication (day 7). The additional deletion of NS1 added to the attenuation since the identical dose of the ΔNS1 ΔNS2 mutant did not cause any symptoms, and a 10-fold-higher dose caused only minor weight loss of a maximal 10%. Since deletion of NS2 did not affect virus replication in IFN-incompetent cells (i.e., BHK-21, B6.IFNAR0/0 MEF, and macrophages), we exclude direct involvement of NS2 in the viral replication process and instead assume further inhibitory functions outside the IFN system. These functions appear to be secondary in importance to the IFN-antagonistic activity since the induction of the IFN type I/III systems clearly achieved a higher attenuation of the ΔNS mutants in wild-type and IFNAR0/0 mice. Interestingly, the NS proteins of HRSV have been reported to antagonize premature apoptosis by an IFN-independent mechanism (3). In the case of PVM, preliminary experiments are pointing toward a comparable antiapoptotic effect in IFN-incompetent BHK-21 cells that appears to be mainly mediated by NS2 (data not shown). If confirmed, premature apoptosis of pulmonary cells in mice infected with the ΔNS2 or ΔNS1 ΔNS2 mutant might account for the restriction of viral replication in B6.IFNAR0/0.IL28Rα0/0 mice. Further detailed experiments are required to characterize the additional functions of the NS proteins.
It still remains tempting to speculate about the use of IFN-λ for therapeutic purposes in respiratory infections. However, we were surprised to find that pretreatment with IFN-λ did not prevent disease and, in several cases, did not prevent a lethal course of PVM infection. This is in contrast to symptomatic infection of mice with influenza virus, which was completely prevented by pretreatment with IFN-λ (21), thus pointing toward a significantly higher resistance of PVM to IFN-λ. PVM infects a variety of pulmonary cell types, including ciliated epithelial cell, Clara cells, type I and type II pneumocytes, and macrophages (5; also B. Heinze and C. D. Krempl, unpublished), which represents a cell tropism similar to that found in postmortem autopsies of HRSV-infected patients (12, 38). Interestingly, in a comparison of autopsy tissues from fatal cases of HRSV and influenza virus, antigen of influenza virus was detected primarily in epithelial cells of larger airways but not in alveolar tissue (38). This difference in cell tropism may explain differences in the effectiveness of treatment since it has been found previously that the receptor to IFN-λ is present on epithelia of the larger airways but not on alveolar cells in mice (22). Pretreatment with IFN-λ would induce an antiviral state in cells that are targeted by influenza virus, thus preventing infection. In comparison, the pneumoviruses HRSV and PVM additionally infect cells in small airways and alveoli that are insensitive to IFN-λ. In consequence, infection would be reduced to these cells but not prevented, and the course of disease would be delayed. Thus, the significance of IFN-λ for respiratory viruses depends on the effectiveness of their IFN antagonists and their cell tropism.
ACKNOWLEDGMENTS
We thank ZymoGenetics Inc. for providing breeding pairs of IL28Rα0/0 mice. We thank Thomas McCarty for performing the quantitative PCRs and Kristina Prenko for excellent technical assistance. Also we thank Stephan Schulz, Institute of Pathology, TU Munich, for support during preparation of the manuscript.
This work has been supported by grants KR2964/1-2 and EH145/4-2 from the German Research Foundation to C.D.K. and S.E. P.L.C. and U.J.B. were supported by the NIAID Intramural Program.
Footnotes
Published ahead of print on 9 February 2011.
REFERENCES
- 1. Ank N., et al. 2008. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J. Immunol. 180:2474–2485 [DOI] [PubMed] [Google Scholar]
- 2. Ank N., et al. 2006. Lambda interferon (IFN-λ), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J. Virol. 80:4501–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bitko V., et al. 2007. Nonstructural proteins of respiratory syncytial virus suppress premature apoptosis by an NF-κB-dependent, interferon-independent mechanism and facilitate virus growth. J. Virol. 81:1786–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bossert B., Marozin S., Conzelmann K.-K. 2003. Nonstructural proteins NS1 and NS2 of bovine respiratory syncytial virus block activation of interferon regulatory factor 3. J. Virol. 77:8661–8668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buchholz U. J., et al. 2009. Deletion of nonstructural proteins NS1 and NS2 from pneumonia virus of mice attenuates viral replication and reduces pulmonary cytokine expression and disease. J. Virol. 83:1969–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Byrappa S., Gavin D. K., Gupta K. C. 1995. A highly efficient procedure for site-specific mutagenesis of full-length plasmids using Vent DNA polymerase. Genome Res. 5:404–407 [DOI] [PubMed] [Google Scholar]
- 7. Contoli M., et al. 2006. Role of deficient type III interferon-lambda production in asthma exacerbations. Nat. Med. 12:1023–1026 [DOI] [PubMed] [Google Scholar]
- 8. Doyle S. E., et al. 2006. Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology 44:896–906 [DOI] [PubMed] [Google Scholar]
- 9. Elliott J., et al. 2007. Respiratory syncytial virus NS1 protein degrades STAT2 by using the elongin-cullin E3 ligase. J. Virol. 81:3428–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frey S., Krempl C. D., Schmitt-Gräff A., Ehl S. 2008. Role of T cells in virus control and disease after infection with pneumonia virus of mice. J. Virol. 82:11619–11627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hokeness K. L., Kuziel W. A., Biron C. A., Salazar-Mather T. P. 2005. Monocyte chemoattractant protein-1 and CCR2 interactions are required for IFN-alpha/beta-induced inflammatory responses and antiviral defense in liver. J. Immunol. 174:1549–1556 [DOI] [PubMed] [Google Scholar]
- 12. Johnson J. E., Gonzales R. A., Olson S. J., Wright P. F., Graham B. S. 2007. The histopathology of fatal untreated human respiratory syncytial virus infection. Mod. Pathol. 20:108–119 [DOI] [PubMed] [Google Scholar]
- 13. Kotenko S. V., et al. 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 4:69–77 [DOI] [PubMed] [Google Scholar]
- 14. Krempl C. D., Lamirande E. W., Collins P. L. 2005. Complete sequence of the RNA genome of pneumonia virus of mice (PVM). Virus Genes 30:237–249 [DOI] [PubMed] [Google Scholar]
- 15. Krempl C. D., et al. 2007. Identification of a novel virulence factor in recombinant pneumonia virus of mice. J. Virol. 81:9490–9501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lasfar A., et al. 2006. Characterization of the mouse IFN-lambda ligand-receptor system: IFN-lambdas exhibit antitumor activity against B16 melanoma. Cancer Res. 66:4468–4477 [DOI] [PubMed] [Google Scholar]
- 17. Ling Z., Tran K. C., Teng M. N. 2009. Human respiratory syncytial virus nonstructural protein NS2 antagonizes the activation of beta interferon transcription by interacting with RIG-I. J. Virol. 83:3734–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lo M. S., Brazas R. M., Holtzman M. J. 2005. Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness. J. Virol. 79:9315–9319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marcello T., et al. 2006. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology 131:1887–1898 [DOI] [PubMed] [Google Scholar]
- 20. Mifune K., Desmyter J., Rawls W. E. 1970. Effect of exogenous interferon on rubella virus production in carrier cultures of cells defective in interferon production. Infect. Immun. 2:132–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mordstein M., et al. 2008. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 4:e1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mordstein M., et al. 2010. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J. Virol. 84:5670–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Muller U., et al. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918–1921 [DOI] [PubMed] [Google Scholar]
- 24. Pestka S., Krause C. D., Walter M. R. 2004. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 202:8–32 [DOI] [PubMed] [Google Scholar]
- 25. Ramaswamy M., et al. 2006. Respiratory syncytial virus nonstructural protein 2 specifically inhibits type I interferon signal transduction. Virology 344:328–339 [DOI] [PubMed] [Google Scholar]
- 26. Randall R. E., Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1–47 [DOI] [PubMed] [Google Scholar]
- 27. Rosenberg H. F., Domachowske J. B. 2008. Pneumonia virus of mice: severe respiratory infection in a natural host. Immunol. Lett. 118:6–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schlender J., Bossert B., Buchholz U., Conzelmann K. K. 2000. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J. Virol. 74:8234–8242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schlender J., Walliser G., Fricke J., Conzelmann K. K. 2002. Respiratory syncytial virus fusion protein mediates inhibition of mitogen-induced T-cell proliferation by contact. J. Virol. 76:1163–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reference deleted.
- 31. Sheppard P., et al. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4:63–68 [DOI] [PubMed] [Google Scholar]
- 32. Sommereyns C., Paul S., Staeheli P., Michiels T. 2008. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 4:e1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Spann K. M., Tran K.-C., Chi B., Rabin R. L., Collins P. L. 2004. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages [corrected]. J. Virol. 78:4363–4369 (Erratum, 78:6705.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Spann K. M., Tran K. C., Collins P. L. 2005. Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-κB, and proinflammatory cytokines. J. Virol. 79:5353–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Teng M. N., et al. 2000. Recombinant respiratory syncytial virus that does not express the NS1 or M2-2 protein is highly attenuated and immunogenic in chimpanzees. J. Virol. 74:9317–9321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Valarcher J.-F., et al. 2003. Role of alpha/beta interferons in the attenuation and immunogenicity of recombinant bovine respiratory syncytial viruses lacking NS proteins. J. Virol. 77:8426–8439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van Pesch V., Lanaya H., Renauld J. C., Michiels T. 2004. Characterization of the murine alpha interferon gene family. J. Virol. 78:8219–8228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Welliver T. P., et al. 2007. Severe human lower respiratory tract illness caused by respiratory syncytial virus and influenza virus is characterized by the absence of pulmonary cytotoxic lymphocyte responses. J. Infect. Dis. 195:1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wright P. F., et al. 2006. The interferon antagonist NS2 protein of respiratory syncytial virus is an important virulence determinant for humans. J. Infect. Dis. 193:573–581 [DOI] [PubMed] [Google Scholar]
- 40. Zhou Z., et al. 2007. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the JAK-STAT pathway and the mitogen-activated protein kinases. J. Virol. 81:7749–7758 [DOI] [PMC free article] [PubMed] [Google Scholar]