Abstract
The fusion of enveloped viruses with cellular membranes is mediated by proteins that are anchored in the lipid bilayer of the virus and capable of triggered conformational changes necessary for driving fusion. The flavivirus envelope protein E is the only known viral fusion protein with a double membrane anchor, consisting of two antiparallel transmembrane helices (TM1 and TM2). TM1 functions as a stop-transfer sequence and TM2 as an internal signal sequence for the first nonstructural protein during polyprotein processing. The possible role of this peculiar C-terminal helical hairpin in membrane fusion has not been investigated so far. We addressed this question by studying TM mutants of tick-borne encephalitis virus (TBEV) recombinant subviral particles (RSPs), an established model system for flavivirus membrane fusion. The engineered mutations included the deletion of TM2, the replacement of both TM domains (TMDs) by those of the related Japanese encephalitis virus (JEV), and the use of chimeric TBEV-JEV membrane anchors. Using these mutant RSPs, we provide evidence that TM2 is not just a remnant of polyprotein processing but, together with TM1, plays an active role in fusion. None of the TM mutations, including the deletion of TM2, affected early steps of the fusion process, but TM interactions apparently contribute to the stability of the postfusion E trimer and the completion of the merger of the membranes. Our data provide evidence for both intratrimer and intertrimer interactions mediated by the TMDs of E and thus extend the existing models of flavivirus membrane fusion.
INTRODUCTION
Membrane fusion is a crucial step during the cell entry of enveloped viruses and is mediated by specific membrane-anchored viral surface proteins (fusion proteins) (11, 24). According to their molecular architecture, these have been assigned to three different structural classes (classes I, II, and III) (11, 41). They all drive fusion by conformational changes that are triggered by interactions with the host cell (such as receptor binding or exposure to acidic pH) and presumably involve protein-protein interactions at the fusion site (41). Classes I and III, and the class II fusion proteins of alphaviruses, possess a single transmembrane (TM) domain that functions as a membrane anchor and is followed by a cytoplasmic tail of varying length (41). In contrast, the flavivirus class II viral fusion protein E is unique in possessing a hairpin-like double-membrane-spanning carboxy terminus, derived from a special combination of stop-transfer and internal signal sequences, required for the intracellular sorting and processing of the flaviviral polyprotein (21) (Fig. 1A).
Fig. 1.
Representations of the flavivirus E protein (A), membrane fusion (B), modifications engineered into the transmembrane (TM) region of E (C), and sequence alignment of TM regions of E (D). (A) Membrane topologies of E and NS1 during polyprotein processing at the ER membrane. TM2 functions as an internal signal sequence for NS1. The red arrow indicates host signalase cleavage at the E-NS1 junction. Color code for E: domain I, red; domain II, yellow; domain III, blue; stem, purple; anchor, gray; FP, green. (B) Flavivirus membrane fusion. 1, side view of an E dimer at the surface of a flavivirus particle at neutral pH; 2, low-pH-induced E dimer dissociation, extension of the stem, and interaction of the FPs with the target membrane; 3, E trimerization and initiation of hairpin formation (DIII relocates to the side of the molecule, and the stem zippers along the trimeric core); 4, formation of a stalk-like hemifusion intermediate with only the apposing leaflets merged; 5, formation of the hairpin-like postfusion structure and the opening of a fusion pore. The color code for E domains is as in panel A. Color code for membranes: viral membrane, orange; host cell membrane, gray. (C) Diagram showing C-terminal modifications of E in the constructs used for producing mutant RSPs. The numbers indicate amino acid (aa) positions in TBLV E. (D) Alignment of the amino acid sequences of the membrane anchor regions of several flavivirus E proteins (TBE numbering): TBEV (Genbank accession number U27495), Powassan virus (POWV) (Genbank accession number L06436), dengue virus (DENV) type 1 (Genbank accession number FJ687432), DENV type 2 (Genbank accession number NC_001474), DENV type 3 (Genbank accession number FJ850055), DENV type 4 (Genbank accession number AY618990), JEV (Genbank accession number EF571853), St. Louis encephalitis virus (SLV) (Genbank accession number M16614), West Nile virus (WNV) (Genbank accession number DQ211652), and yellow fever virus (YFV) (Genbank accession number AY640589). Identical amino acid residues are highlighted in yellow. The predicted TM1 and TM2 helices are indicated at the bottom.
Flaviviruses are members of the genus Flavivirus (family Flaviviridae) and comprise a number of important human pathogens, including the dengue viruses, Japanese encephalitis virus (JEV), yellow fever virus, West Nile virus, and tick-borne encephalitis virus (TBEV) (10, 40). They are small, enveloped, positive-strand RNA viruses that are assembled in the endoplasmic reticulum (ER) in an immature noninfectious form containing three structural proteins designated C (capsid), prM (precursor of M [membrane]), and E (envelope). During exocytosis, the prM protein is proteolytically cleaved by furin in the trans-Golgi network, and the C-terminal membrane-bound cleavage product of prM (M) remains associated with mature secreted virions (43, 44). As revealed by cryo-electron microscopy (cryo-EM), immature and mature viruses display different envelope protein organizations, but both have icosahedral symmetry, with a specific arrangement of 60 trimers of prM-E heterodimers and 90 E dimers, respectively (14). E functions as both a receptor binding and a fusion protein, and the atomic structures of C-terminally truncated soluble forms of E (sE) have been determined in their pre- and postfusion conformations for several flaviviruses (35). sE is composed of three distinct domains (DI, DII, and DIII), and an S-S bridge stabilized loop at the tip of DII functions as the fusion peptide (FP) (Fig. 1A). The organization of the C-terminal part of the molecule—not present in the X-ray structures—was elucidated by cryo-EM of mature dengue virions (45). It is composed of two mostly amphipathic alpha helices that are half buried in the outer leaflet of the viral membrane (designated the “stem”) and connect the E ectodomain to the C-terminal transmembrane elements (TM1 and TM2 helices) (Fig. 1A). These are arranged in an antiparallel coiled-coil hairpin structure linked by four to six mostly polar amino acids (45) and display only a low degree of sequence conservation among flaviviruses (Fig. 1D). Both have important functions during biosynthesis and processing of the viral polyprotein, with the TM1 element serving as a stop-transfer sequence for E and the TM2 element as an internal signal sequence for the translocation of the first nonstructural protein (NS1) into the lumen of the ER (21) (Fig. 1A).
The existing models of flavivirus fusion are based on the pre- and postfusion structures of sE, as well as on biochemical studies (Fig. 1B). After virus uptake by receptor-mediated endocytosis, the low endosomal pH causes the exposure of the FPs and their insertion into the target membrane. Further changes involve trimerization of E and the formation of its hairpin-like postfusion structure with the FPs and the TM domains (TMDs) on the same side of the molecule. These molecular rearrangements and interactions finally result in the opening of a fusion pore (reviewed in references 11, 14, and 36) (Fig. 1B). In the current flavivirus fusion models, the TMDs have not been ascribed a specific functional role, except for anchoring E in the viral membrane.
Since the presence of a double membrane anchor is a unique structural feature not found in other viral fusion proteins, it was the objective of our work to study the contributions of these sequence elements to the different stages of flavivirus membrane fusion. We were specifically interested in whether the TM2 helix is merely a functional remnant of polyprotein processing or whether it has an additional role in the fusion process. As an experimental system, we chose noninfectious capsid-less recombinant subviral particles (RSPs) of TBE virus, because they have fusion characteristics similar to those of whole virions (6) and modifications of the double membrane anchor are uncoupled from the TM2 signal sequence function that would be required for virus replication. We modified the membrane anchor of E by deleting the TM2 helix, replacing both TMDs with those of a heterologous flavivirus (JEV), and shuffling the TMDs between TBEV and JEV. Functional analyses of these mutant RSPs demonstrate that the TM2 helix is completely dispensable for the early steps of membrane fusion (including FP exposure, interaction with target membranes, and E trimerization) but contributes to E trimer stability and is essential for later fusion steps, including the stage of hemifusion. Our data not only provide evidence for a role of the TMDs in intratrimer interactions, but also indicate the participation of TMDs in intertrimer interactions that are both required for efficient fusion.
MATERIALS AND METHODS
Virus sequences.
The different constructs used in this study were based on Western subtype TBEV strain Neudoerfl (GenBank accession number U27495) and JEV strain Nakayama (GenBank accession number EF571853).
RSP plasmids and cloning procedures (Fig. 1C).
SV-PE (wild type ([WT]) was the parent plasmid for mutant construction and WT controls (1). The plasmid contains nucleotides 388 to 2550 from TBEV and includes the last 31 codons of the C gene, the entire prM and E genes, and the first 30 codons of the NS1 gene under the control of the simian virus 40 early promoter. Construction of the ΔTM2 deletion mutant, in which a TAG stop codon replaces the codon for the amino acid at position 473 of the E gene of the SV-PE WT plasmid (3), was reported previously. Constructs containing cytoplasmic tails (CT) instead of the TM2 region and shuffled or heterologous anchors were generated by using unique restriction sites to introduce chemically synthesized DNA fragments (GeneArt) into the parent plasmid. The plasmids SV-PE CT-Marburg and SV-PE CT-Synthetic corresponded to SV-PE WT up to the codon of amino acid residue 472 of E, followed by the nucleotide sequences ATCTTTACTAAATATATCGGATAG (Marburg) and AGAAGAAGAAGATAG (Synthetic). These encode the cytoplasmic tail of the Marburg virus fusion protein (RIFTKYIG) and a synthetic oligo(R) sequence. In plasmid SV-PE JE1-JE2, the complete double-membrane-spanning anchor, including the short loop connecting the two helices of TBEV, was replaced by the corresponding JEV sequences, together with the first 30 codons of the NS1 gene. In SV-PE TBE1-JE2, the TBE TM2 domain plus the following NS1 sequence, and in SV-PE JE1-TBE2, the TBE TM1 domain, were replaced. In both constructs, the short loop between the two TM regions was derived from TBEV.
Production and quality controls of RSPs.
For the production of RSPs, COS-1 cells were electroporated with the corresponding recombinant plasmids as described previously (33). Particles secreted into cell culture supernatants were harvested 48 h after electroporation by ultracentrifugation for 2 h at 44,000 rpm. The pelleted particles were resuspended in TAN buffer (50 mM triethanolamine [TEA], 100 mM NaCl, pH 8.0). For coflotation and lipid-mixing experiments, particles were subsequently purified by sucrose gradient centrifugation (2, 33). For lipid-mixing assays, the particles were metabolically labeled with 1-pyrenehexadecanoic acid (Molecular Probes; Invitrogen) as described previously (2, 6). RSPs secreted from transfected cells were quantified in a four-layer enzyme-linked immunosorbent assay (ELISA) after solubilization with 0.4% SDS at 65°C for 30 min (12). The hemagglutination (HA) activity of RSPs was measured at pH 6.4 by the method of Clarke and Casals, using goose erythrocytes (5). The structural integrity of the conformation of E on the particle surface was confirmed by epitope mapping with 22 E-specific monoclonal antibodies (MAbs) in ELISA (2, 16). To confirm that the mutations did not affect the intracellular maturation process, the presence of prM was analyzed in mutant RSPs and compared to the contents of WT particles by Western blotting as described previously (8).
Virus plasmids and cloning procedures.
Plasmid pTNd/c contains a full-length genomic cDNA insert of TBEV strain Neudoerfl cloned into the vector pBR322 under the control of a T7 transcription promoter (23). The membrane anchor region of TBEV E in the pTNd/c vector was replaced by chemically synthesized DNA fragments (GeneArt) containing the heterologous membrane anchor of JEV (TBEV JE1-JE2) or shuffled membrane anchors (TBEV JE1-TBE2 and TBEV TBE1-JE2) using unique restriction sites. In contrast to RSP mutants JE1-JE2 and TBE-JE2, the NS1 sequence was not replaced by that of JEV in the corresponding TBEV mutants. Full-length DNA templates for in vitro transcription of TBEV plasmids were generated as described previously (17).
RNA transfection and quantification.
In vitro transcription and transfection of BHK-21 cells by electroporation were performed as described previously (39). Briefly, RNA was synthesized from full-length cDNA clones using the T7 Megascript kit (Ambion) according to the manufacturer's instructions. The template DNA was digested with DNase I, and the quality of the RNA was checked by electrophoresis in a 1% agarose gel containing 6% formalin. RNA was purified with an RNeasy minikit (Qiagen) and quantified spectrophotometrically, and equimolar amounts of the corresponding RNAs were used for the transfection of BHK cells. The number of particles containing genomic RNA (RNA equivalents) in the cell culture supernatant was determined 48 h posttransfection by quantitative PCR (qPCR) after reverse transcription of the viral RNA, as described previously (18).
Liposomes.
Large unilamellar liposomes were prepared as described previously (37). Phosphatidylcholine, phosphatidylethanolamine (Avanti Polar Lipids), and cholesterol (Sigma-Aldrich) were mixed at a molar ratio of 1:1:2 in chloroform. The mixture was dried to a homogeneous film in high vacuum and hydrated in liposome buffer (10 mM TEA, 140 mM NaCl, pH 8.0) by 5 freeze-thaw cycles. Prior to use, liposomes were subjected to 21 extrusion cycles through polycarbonate membranes (200-nm pore size) using a LiposoFast-Basic extruder (Avestin).
Fusion assay.
Fusion of pyrene-labeled RSPs with liposomes was measured by monitoring the decrease in pyrene excimer fluorescence caused by the dilution of pyrene-labeled phospholipids in the RSP membrane in the unlabeled liposome membrane (8). Briefly, pyrene-labeled RSPs were mixed with liposomes (total lipid, 0.3 mM) in a continuously stirred fluorimeter cuvette at 37°C. Lipid mixing was induced by adding 300 mM morpholinoethanesulfonic acid (MES) to yield a pH of 5.4. Fluorescence was continuously monitored by a Perkin-Elmer LS-50B fluorescence spectrophotometer. The initial excimer fluorescence after mixing was defined as 0% fusion. To determine the residual excimer fluorescence at infinite dilution of the probe (defined as 100% fusion for calculating the fusion extents), the detergent n-octa(ethylene glycol)n-dodecyl monoether (C12E8) was added to a final concentration of 10 mM to disperse the viral and liposomal membranes.
FP exposure assay.
Microtiter plates were coated with polyclonal anti-TBEV immunoglobulin G to capture native RSPs (0.5 μg/ml E) in phosphate-buffered saline, pH 7.4, containing 2% lamb serum as described previously (34). The exposure of the FP was measured by the addition of biotinylated MAb A1 (FP-specific MAb) in MES buffer (50 mM MES, 100 mM NaCl) adjusted to the appropriate pH by titration with 1 N NaOH. After incubation for 1 h at 37°C, the bound MAb A1 was detected by using streptavidin-peroxidase (Sigma-Aldrich) (8).
Liposome coflotation assay.
RSPs were mixed with liposomes (1 μg E and 200 nmol lipids), acidified with 300 mM MES to yield a pH of 5.4, and incubated for 15 min at 37°C. The acidified samples were back neutralized to pH 8.0 by the addition of 150 mM TEA and adjusted to 0.6 ml 20% sucrose (wt/wt) in TAN buffer, pH 8.0 (50 mM triethanolamine, 100 mM NaCl). These samples were layered onto a 1-ml 50% sucrose cushion and overlaid with 1.4 ml of 15% sucrose and 1 ml 5% sucrose as described previously (8). Centrifugation was carried out for 2 h at 50,000 rpm at 4°C in a Beckman SW55 rotor. Fractions of 0.2 ml were collected by upward displacement using a Piston Gradient Fractionator (BioComp Instruments Inc.), and the amount of E protein in each fraction was determined by a quantitative four-layer ELISA (12).
E trimerization and trimer stability assays.
The oligomeric state of E, after incubation of RSPs at acidic or neutral pH, was determined by sedimentation analysis as described previously (2, 8). RSPs (3 μg) in TAN buffer, pH 8.0, were incubated for 10 min at pH 5.4 or 8.0 by the addition of 300 mM MES or TAN buffer, pH 8.0, respectively. Acidified samples were back neutralized with 150 mM TEA, and all samples were solubilized with 1% Triton X-100 for 1 h at room temperature. For thermostability experiments, RSPs were acidified and solubilized as described above before being incubated for 10 min at 37°C or 70°C (8). All samples were applied on top of 7 to 20% (wt/wt) continuous sucrose gradients containing 0.1% Triton X-100. Centrifugation was carried out at 38,000 rpm and 15°C for 20 h in a Beckmann SW40 rotor. Fractions of 0.6 ml were collected by upward displacement using a Piston Gradient Fractionator (BioComp Instruments Inc.). The amount of E in the fractions was determined by a quantitative four-layer ELISA (12).
Immunofluorescence.
RNA-transfected or virus-infected BHK-21 cells were seeded in 24-well tissue culture plates containing microscope coverslips and incubated for 24 h at 37°C. The cells were fixed and permeabilized with acetone-methanol (1:1) as described previously (39). Staining was performed by successive incubations with a rabbit polyclonal anti-TBEV serum recognizing the structural proteins and a fluorescein-isothiocyanate-conjugated anti-rabbit antibody (Jackson Immune Research Laboratory).
Focus formation assay.
Supernatants from transfected BHK-21 cells were harvested 2 days postelectroporation and added to confluent cell monolayers in serial 10-fold dilutions. After 3 h of incubation at 37°C, the cells were covered with a 3% carboxymethyl cellulose overlay. Two days after infection, the cells were fixed and permeabilized with acetone-methanol (1:1) for 10 min at −20°C and incubated with a polyclonal rabbit anti-TBEV serum. Antibody-labeled cells were detected with goat anti-rabbit immunoglobulin G-alkaline phosphatase (Sigma-Aldrich).
RESULTS
RSP mutagenesis.
We used the TBEV RSP system to investigate the effects of TMD modifications on the different steps of the flavivirus membrane fusion process. For this purpose, we generated mutant RSPs with modifications of the TM hairpin as follows (Fig. 1C): (i) deletion of the TM2 helix (ΔTM2); (ii) replacement of the TM2 helix by the carboxy-terminal tail of the Marburg virus fusion protein (CT-Marburg) or an oligo(R) tail (CT-Synthetic); (iii) replacement of the complete TM hairpin by the heterologous TM hairpin of JEV, a distantly related flavivirus (JE1-JE2); (iv) generation of a chimeric hairpin in which the TM2 domain was replaced by that of JEV (TBE1-JE2); or (v) generation of a chimeric hairpin in which the TM1 domain was replaced by that of JEV (JE1-TBE2).
Since the modification of the membrane anchor could potentially interfere with processes unrelated to fusion, we performed a series of quality control experiments to ensure WT-like folding and oligomerization of E, as well as particle formation and maturation of RSPs. The data from these analyses are summarized in Table 1. None of the introduced mutations affected the proper folding of E, as shown by reactivity with conformation-sensitive monoclonal antibodies (see Materials and Methods). The dimeric state of E was confirmed by sedimentation analysis after solubilization of RSPs with Triton X-100 (see Materials and Methods). The particulate organization of the mutants was verified by ultracentrifugation. Western blots using prM-specific polyclonal sera revealed that the maturation state of the mutants was identical to that of the WT control.
Table 1.
Quality controls of TBEV WT and mutant RSPsa
| RSP | Reactivity with monoclonal antibodies | Oligomeric state of E | Hemagglutination activity | Particulate nature (ultracentrifugation) | Maturation cleavage |
|---|---|---|---|---|---|
| WT | + | Dimer | + | + | + |
| ΔTM2 | + | Dimer | + | + | + |
| CT-Marburg | + | Dimer | + | + | + |
| CT-Synthetic | + | Dimer | + | + | + |
| JE1-JE2 | + | Dimer | + | + | + |
| TBE1-JE2 | + | Dimer | + | + | + |
| JE1-TBE2 | + | Dimer | + | + | + |
+, WT-like behavior.
The TM hairpin is required for efficient membrane fusion.
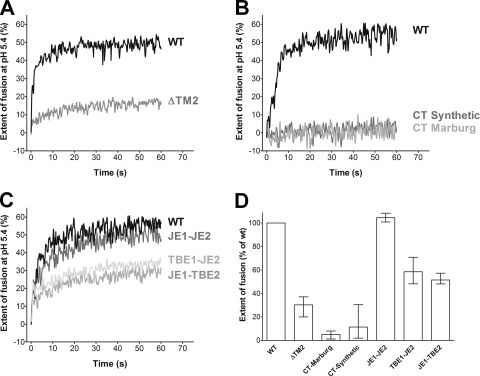
To assess the requirement for the TM2 domain in the overall fusion process, we used an in vitro lipid-mixing assay with pyrene-labeled particles and liposomes (see Materials and Methods). Deletion of the TM2 helix resulted in strong impairment of the rate and extent of acidic-pH-induced fusion compared to the WT control (Fig. 2A). The addition of carboxy-terminal tails to the TM1 helix as a replacement for the TM2 helix (making E more similar to other viral fusion proteins) did not exert a compensating effect on fusion (Fig. 2B). RSPs containing E with either the cytoplasmic tail of the Marburg virus fusion protein or a synthetic poly(R) tail were even more impaired in membrane fusion than RSP ΔTM2. Further information about the contribution of the TM hairpin to membrane fusion was obtained by analyzing RSPs with a completely heterologous TM hairpin (JE1-JE2) and chimeric hairpins (TBE1-JE2 and JE1-TBE2) in E. RSP JE1-JE2 displayed virtually the same fusion characteristics as the WT with respect to the rate as well as the extent of fusion (Fig. 2C). In contrast, chimerization of the hairpin resulted in impairment, but not loss, of fusion activity (Fig. 2C). The quantitative evaluation of the effects of all TMD modifications on the extent of fusion relative to the WT are displayed in Fig. 2D.
Fig. 2.
Fusion activities of pyrene-labeled WT and mutant RSPs. (A) Fusion kinetics of RSP WT (black line) and ΔTM2 (gray line) at pH 5.4. (B) Fusion kinetics of RSP WT (black line) and RSPs in which the second TMD of E was replaced by a cytoplasmic tail (gray lines) at pH 5.4. (C) Fusion kinetics of RSP WT (black line) and RSPs with heterologous or shuffled E membrane anchors (gray lines) at pH 5.4. (D) Extent of fusion of mutant RSPs relative to RSP WT (= 100%) at pH 5.4 after 60 s. The data represent the means of at least two independent experiments, and the error bars indicate the observed range.
From these experiments, we conclude that (i) the TM hairpin of E represents a functional element in the fusion process, (ii) the deletion of the TM2 helix cannot be compensated for by cytoplasmic tails, (iii) a heterologous flavivirus membrane anchor can completely substitute for the functions of the homologous TM hairpin in fusion, and (iv) mutants with chimeric anchors display fusion activities that are intermediate between the WT and ΔTM2. Since the highest fusion extents were achieved by RSPs with TM anchors containing matching helices (RSP WT and RSP JE1-JE2), it can be assumed that interactions between the TM1 and TM2 helices contribute to efficient fusion.
The TM hairpin is not required for early steps of membrane fusion.
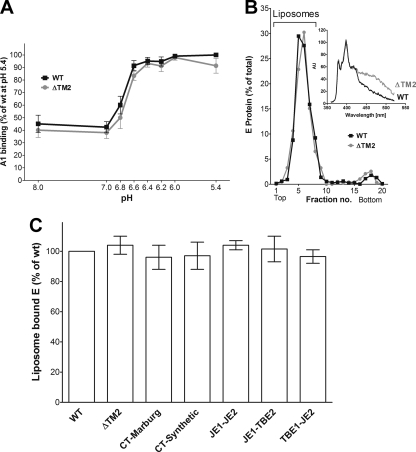
The fusion experiments revealed an important role of the TM2 helix and TM1-TM2 helix interactions in the overall process. However, it remained unclear for which stage of fusion these interactions were required. We therefore employed an experimental design that allowed us to dissect the fusion process and to specifically study the stages of fusion initiation, i.e., acidic-pH-induced FP exposure by the use of an FP-specific monoclonal antibody (MAb A1) in ELISA and interactions with target membranes by liposome coflotation. As shown in Fig. 3A, the deletion of TM2 had no measurable effect on the pH threshold and the extent of FP exposure. Also, the patterns of acidic-pH-induced liposome coflotation were virtually superimposable for RSP WT and RSP ΔTM2 (Fig. 3B). Consistent with the fusion assays, the use of pyrene-labeled RSPs in coflotation experiments confirmed that ΔTM2 RSPs were mostly attached to, but not fused with, liposomes (Fig. 3B, inset). The same results were also obtained with the other TM mutant RSPs, none of which were affected in their association with liposomes at acidic pH (Fig. 3C).
Fig. 3.
Interactions of WT and mutant RSPs with target membranes. (A) Acidic-pH-induced FP exposure measured by binding of MAb A1. The results are expressed as percentages of the maximal absorbance of A1 obtained with the WT at pH 5.4. The data are the means of four independent experiments performed in duplicate, and the error bars indicate the standard errors of the means. WT, squares; ΔTM2, circles. (B) Acidic-pH-induced coflotation of WT and ΔTM2 RSPs with liposomes. The top fractions containing RSPs cofloated with the liposomes are indicated by a bracket. (Inset) Fluorescence spectrum of cofloated pyrene-labeled mutant (gray) and WT (black) RSPs. AU, arbitrary units. (C) Extent of acidic-pH-induced coflotation with liposomes obtained with mutant RSPs relative to the WT (= 100%). The data are the means of two independent experiments, and the error bars indicate the observed range.
These data indicate that the second TM helix is fully dispensable for the early stages of membrane fusion and that the membrane anchorage of E by the TM1 helix alone is sufficient for the exposure of the FP, as well as its insertion into target membranes.
Reduced thermostability of trimers lacking the TM2 helix.
Since the deletion of the TM2 helix did not affect the initiation of fusion, the block observed with this mutant in overall fusion must occur at later stages of the multistep fusion process. Trimerization and the conformational switch from the prefusion into the postfusion conformation of E represent crucial events in the fusion process and are believed to provide the energy for merging the two membranes. We therefore analyzed the effects of TMD modifications on trimer formation (Fig. 4) and trimer stability (Fig. 5).
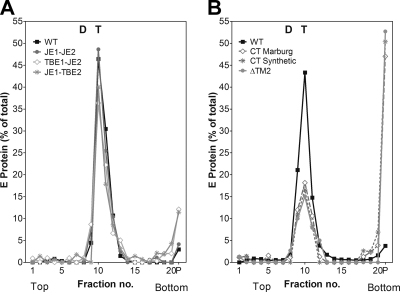
Fig. 4.
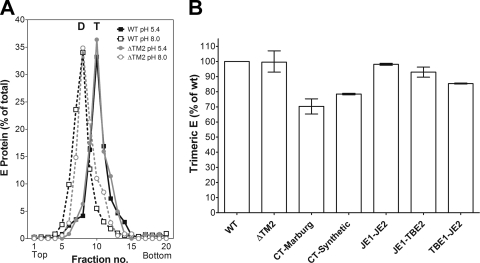
E trimer formation of WT and mutant RSPs. Analysis of acidic-pH-induced trimer formation by sucrose gradient centrifugation, as described in Materials and Methods, after Triton X-100 solubilization of acidic-pH-pretreated and untreated RSPs. (A) Sedimentation patterns of E from RSP ΔTM2 (gray lines) and RSP WT (black lines). Dotted lines, untreated RSPs; solid lines, acidic-pH-treated RSPs. The sedimentation direction is from left to right, and the positions of E dimers (D) and trimers (T) are indicated. (B) Extent of E trimer formation obtained with mutant RSPs compared to the WT (= 100%). The results are expressed as percentages of E found in the trimer peak fractions relative to the total amount of E in the gradient. The data represent the means of at least two independent experiments, and the error bars indicate the observed range.
Fig. 5.
Thermostability of E trimers of WT and mutant RSPs. Acidic-pH-induced E trimers of WT and mutant RSPs were exposed to 70°C and subjected to rate zonal sucrose density gradient centrifugation. The sedimentation direction is from left to right, and the positions of E dimers (D) and trimers (T) are indicated. The pellet (P) was resuspended in 0.6 ml, corresponding to the volume of a single fraction. (A) JE1-JE2, TBE1-JE2, and JE1-TBE2 mutant trimers, displaying trimer stability similar to that of the WT. (B) ΔTM2, CT-Marburg, and CT-Synthetic mutant trimers displaying strongly reduced thermostability compared to the WT. The data are representative examples from two or more independent experiments.
As revealed by rate zonal centrifugation of low-pH-pretreated and solubilized RSPs, all mutant E proteins were able to undergo the dimer-to-trimer transition similarly to the WT (Fig. 4A and B). A slight reduction in the efficiency of trimerization was observed with the constructs carrying carboxy-terminal tails, indicating that the added sequence elements somehow interfered with the formation of trimers (Fig. 4B).
The stability of the mutant E trimers was investigated in thermal denaturation experiments. After low-pH pretreatment and solubilization of RSPs, the samples were heated, and the effect on the oligomeric state of the solubilized E trimers was analyzed by sedimentation in sucrose gradients. As shown in Fig. 5A and B, the WT trimers were stable up to 70°C but started to denature and aggregate at about 75°C (data not shown). Mutant trimers containing a double membrane anchor (both heterologous and chimeric anchors) displayed stability similar to that of the WT and were found to sediment in the corresponding fractions in sucrose gradients (Fig. 5A). In contrast, the deletion of the TM2 helix or its replacement by cytoplasmic tails resulted in a reduction of thermostability, as revealed by a strong aggregation of E trimers at 70°C and sedimentation of most of the material into the pellet at the bottom of the gradients (Fig. 5B).
In conclusion, these data show that the TM2 helix is dispensable for trimer formation but contributes to interactions that stabilize the postfusion trimer.
Heterologous and chimeric anchors strongly reduce the production of virus particles.
We attempted to assess the significance of the observations made with RSP TM mutants also in the context of infectious virions and therefore engineered heterologous and chimeric membrane anchors (JE1-JE2, TBE1-JE2, and JE1-TBE2) into an infectious cDNA clone of TBEV. Since the TM2 element is indispensable for polyprotein translation and thus for the generation of infectious virions (Fig. 1A), a ΔTM2 mutant could not be analyzed in this part of the study.
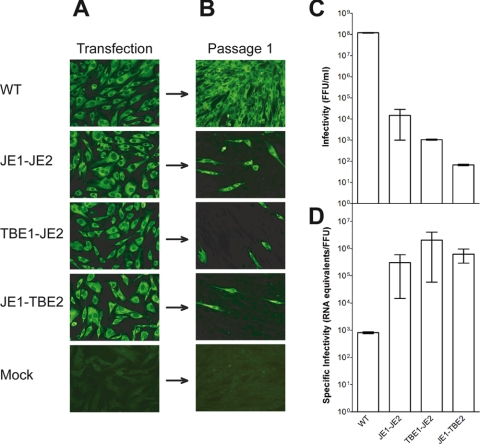
Immunofluorescence staining of transfected cells did not reveal any differences between WT and mutant viruses (Fig. 6A). To determine whether the transfected cells not only produced viral proteins but also released infectious virions, the cell culture supernatants of the experiments displayed in Fig. 6A were transferred to fresh cells, which were stained after 24 h. As can be seen from Fig. 6B, only a few positive cells were detected in the case of the mutants in contrast to the WT control. Quantification of the primary cell culture supernatants (Fig. 6A) by focus formation assays showed that the infectious titers of the mutant viruses were at least ∼10,000-fold lower than that of the WT control (Fig. 6C). A comparative analysis of RNA equivalents in these supernatants revealed that the specific infectivities of the mutants were also dramatically reduced (Fig. 6D).
Fig. 6.
Infectivity of TMD virus mutants. (A) Immunofluorescence staining of BHK cells transfected with mutant or WT viral RNAs, as indicated on the left. (B) Immunofluorescence staining of cells infected with the supernatants from panel A harvested 48 h after transfection. Staining was carried out using a polyclonal serum recognizing the structural proteins of TBEV. The data are representative examples from two or more independent experiments. (C) Quantification of virus in the cell culture supernatants of panel A harvested 48 h after transfection by focus formation assays. FFU, focus-forming units. (D) Determination of the specific infectivities (RNA equivalents per infectious unit) of virus in the cell culture supernatants of panel A harvested 48 h after transfection. Two independent experiments were carried out in duplicate. The data represent the means of these two experiments, and the error bars indicate the observed range.
Due to these low yields of mutant viruses, it was not possible to produce the amounts of virus necessary for the biochemical analyses conducted with RSPs.
DISCUSSION
The double-membrane-spanning C-terminal anchor (TM1 and TM2) of the envelope protein E (consisting of two antiparallel helices) is a unique distinguishing structural feature of the flavivirus fusion protein E. The TM2 helix is a remnant of flavivirus polyprotein processing and has an essential function, serving as an internal signal sequence for the synthesis of the first nonstructural protein, NS1 (Fig. 1A) (21). All other viral fusion proteins analyzed so far have single TM anchors that are C-terminally extended by intracytoplasmic tails of various lengths (41). With class I and III fusion proteins, it was shown in several studies that TMD modifications affected late stages of membrane fusion, such as the hemifusion state and/or the opening of the fusion pore (reviewed in references 20 and 41). We demonstrate in this work, using virus-like particles of TBEV as a model system, that the flavivirus TM2 helix is apparently multifunctional and not only essential for viral polyprotein processing, but also indispensable for efficient membrane fusion.
It is an important finding of our study that all of the initial steps of flavivirus fusion (i.e., acidic-pH-induced dissociation of E, FP exposure, and its interaction with target membranes) can proceed completely unimpaired in the absence of the TM2 helix, whereas later steps leading to hemifusion and full fusion are blocked. In the current models (11, 14, 36), these later steps include the trimerization of E and its conversion into an energetically more stable postfusion conformation, as well as possible further protein-protein interactions. Together, these structural changes and interactions are believed to provide the energy necessary for merging the two membranes and the formation of a fusion pore (Fig. 1B). Since the replacement of both TBEV TMDs with those from the distantly related flavivirus JEV did not impair fusion activity, it can be assumed that intra- and/or interprotein interactions between the TM helices are essential for flavivirus fusion. Such interactions clearly contribute to trimer stability, because all trimers lacking the TM2 domain were significantly less thermostable than trimers with both TM helices. WT-like trimer stability, however, is apparently not dependent on a homologous match between the two TM helices, because the mutants with chimeric TM hairpins, derived from TBEV and JEV, displayed a trimer stability phenotype similar to that of the WT. It is therefore justified to assume that interactions between homologous TM2 helices contribute to the stability of the postfusion trimer. Despite this unimpaired trimer stability, the overall fusion activity of RSPs with chimeric TM hairpins was reduced compared to those with homologous TM hairpins, suggesting an additional functional role of the TM helices in fusion. One likely possibility is that they also contribute to intertrimer interactions involved in the formation of higher-order structures at the site of fusion pore formation. Such ring-like clusters of interacting fusion proteins have been proposed to form during the process of alphavirus fusion (9), i.e., for another family of icosahedral enveloped viruses with class II fusion proteins, like flaviviruses.
It must be kept in mind that the structural details of the interactions of the TM1 and TM2 helices within the anchor and with other parts of E are not known for the postfusion conformation of E. The crystal structures of truncated sE trimers of TBEV and dengue virus, however, indicate a juxtaposition of the TM hairpin and the FPs on the same side of the molecule (4, 26, 29). Therefore, interactions not only between the two helices within the anchor, but also of the TM anchor with the FPs, might contribute to fusion (38). The short loop connecting the two TM helices is unlikely to be important for these interactions, because the TM1-TM2 chimeras, which contained either a TM1 loop or a TM2 loop match, behaved identically in all our analyses.
Given the structural similarity of alphavirus and flavivirus fusion proteins (15, 32), their difference with respect to the membrane anchors—single TM and double TM anchors, respectively—is noteworthy, especially when the essential role of the TM2 helix for flavivirus fusion is considered. Inspection of the alphavirus particle, however, reveals that there are other TMDs that could serve functions similar to that of the flavivirus TM2 helix and provide interactions that contribute to efficient fusion. First, the TM segments of the two envelope proteins E1 and E2 are strongly intertwined (22, 28), and it has been suggested that these interactions exert a cooperative effect at an as yet undefined step of alphavirus membrane fusion (42). This is in strong contrast to the TM regions of the two envelope proteins of flaviviruses (prM/M and E), which were shown by cryo-EM to be separate and not to interact (45). Second, alphavirus particles contain the 6K protein, a remnant of polyprotein processing, which appears to contribute significantly to the efficiency of alphavirus fusion (25). Experiments with recombinant viruses lacking 6K showed that they were strongly impaired in fusion activity (25), very much like the ΔTM2 mutants in our work. It can therefore be speculated that E1-6K interactions during alphavirus fusion are related to those of the TM1 and TM2 helices in the case of flaviviruses.
The engineering of recombinant TBEV with completely heterologous TM anchors or chimeric TM anchors resulted in a dramatic reduction of specific infectivities (RNA equivalents to focus-forming units) and a severe impairment of virus growth (Fig. 6C and D), prohibiting the production of sufficient amounts of mutant viruses that would have been necessary for conducting fusion experiments similar to those presented for RSPs. There are different possible, not mutually exclusive explanations for these findings. In the case of the chimeric TM hairpin mutants, reduced fusion activities, as observed with the RSPs, could contribute to the low infectivities. However, the specific infectivity of the JE1-JE2 mutants was also strongly reduced and led to ∼10,000-times-lower yields than the WT, although the same mutation had no effect on fusion activity in the RSP system. It therefore must be assumed that modifications of the TM helices in the virus also affected other steps of virus replication, independent of membrane fusion. Particle assembly would be a primary candidate for such effects, involving mechanisms that contribute to virion, but not to subviral particle, formation. RSPs can be formed through lateral interactions between prM-E heterodimers only, and they are smaller than whole virions (33). In mature RSPs, the E homodimers are arranged in a regular T = 1 icosahedral lattice (7), whereas on the virus surface, the E proteins are more tightly packed in a herringbone-like arrangement (19, 27). It is possible that the more complex organization of the viral envelope requires interactions of the TMDs of prM-E heterodimers during assembly and/or budding that are dispensable for RSP formation. Such an interpretation would be consistent with other studies that had revealed that alterations in the membrane anchor of E could severely impair the production of infectious viruses (13, 30, 31).
Irrespective of the possible involvement of the TM helices of E in virus assembly, our data provide evidence that they play an important role in the late stages of flavivirus membrane fusion and provide both intra- and intermolecular E trimer contacts that are necessary to drive the fusion process to completion. This is an extension of existing models of flavivirus fusion that have focused on the relocation of DIII and the zippering of the stem to serve as primary energy sources for this process (11).
ACKNOWLEDGMENTS
This work was supported by the Austrian Science Fund (FWF) (P19843-B13 and P20533-B03).
We thank Cornelia Stöckl for excellent technical assistance and Harald Rouha for help with the quantitative-PCR experiments.
Footnotes
Published ahead of print on 16 February 2011.
REFERENCES
- 1. Allison S. L., Mandl C. W., Kunz C., Heinz F. X. 1994. Expression of cloned envelope protein genes from the flavivirus tick-borne encephalitis virus in mammalian cells and random mutagenesis by PCR. Virus Genes 8:187–198 [DOI] [PubMed] [Google Scholar]
- 2. Allison S. L., Schalich J., Stiasny K., Mandl C. W., Heinz F. X. 2001. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J. Virol. 75:4268–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allison S. L., Stiasny K., Stadler K., Mandl C. W., Heinz F. X. 1999. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J. Virol. 73:5605–5612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bressanelli S., et al. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23:728–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clarke D. H., Casals J. 1958. Techniques for hemagglutination and hemagglutination-inhibition with arthropod-borne viruses. Am. J. Trop. Med. Hyg. 7:561–573 [DOI] [PubMed] [Google Scholar]
- 6. Corver J., et al. 2000. Membrane fusion activity of tick-borne encephalitis virus and recombinant subviral particles in a liposomal model system. Virology 269:37–46 [DOI] [PubMed] [Google Scholar]
- 7. Ferlenghi I., et al. 2001. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell 7:593–602 [DOI] [PubMed] [Google Scholar]
- 8. Fritz R., Stiasny K., Heinz F. X. 2008. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J. Cell Biol. 183:353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gibbons D. L., et al. 2004. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427:320–325 [DOI] [PubMed] [Google Scholar]
- 10. Gubler D., Kuno G., Markhoff L. 2007. Flaviviruses, p. 1153–1252 In Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E. (ed.), Fields virology, 5th ed. Lippincott. Williams & Wilkins., Philadelphia, PA [Google Scholar]
- 11. Harrison S. C. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heinz F. X., et al. 1994. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology 198:109–117 [DOI] [PubMed] [Google Scholar]
- 13. Hsieh S. C., Tsai W. Y., Wang W. K. 2010. The length of and nonhydrophobic residues in the transmembrane domain of dengue virus envelope protein are critical for its retention and assembly in the endoplasmic reticulum. J. Virol. 84:4782–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaufmann B., Rossmann M. G. 2010. Molecular mechanisms involved in the early steps of flavivirus cell entry. Microbes Infect. 13:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kielian M., Rey F. A. 2006. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 4:67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kiermayr S., Stiasny K., Heinz F. X. 2009. Impact of quaternary organization on the antigenic structure of the tick-borne encephalitis virus envelope glycoprotein E. J. Virol. 83:8482–8491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kofler R. M., Heinz F. X., Mandl C. W. 2002. Capsid protein C of tick-borne encephalitis virus tolerates large internal deletions and is a favorable target for attenuation of virulence. J. Virol. 76:3534–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kofler R. M., Hoenninger V. M., Thurner C., Mandl C. W. 2006. Functional analysis of the tick-borne encephalitis virus cyclization elements indicates major differences between mosquito-borne and tick-borne flaviviruses. J. Virol. 80:4099–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuhn R. J., et al. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Langosch D., Hofmann M., Ungermann C. 2007. The role of transmembrane domains in membrane fusion. Cell. Mol. Life Sci. 64:850–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lindenbach B. D., Thiel H. J., Rice C. M. 2007. Flaviviridae: the viruses and their replication, p. 1101–1152 In Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E. (ed.), Fields virology, 5th ed. Lippincott, Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 22. Mancini E. J., Clarke M., Gowen B. E., Rutten T., Fuller S. D. 2000. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol. Cell 5:255–266 [DOI] [PubMed] [Google Scholar]
- 23. Mandl C. W., Ecker M., Holzmann H., Kunz C., Heinz F. X. 1997. Infectious cDNA clones of tick-borne encephalitis virus European subtype prototypic strain Neudoerfl and high virulence strain Hypr. J. Gen. Virol. 78:1049–1057 [DOI] [PubMed] [Google Scholar]
- 24. Martens S., McMahon H. T. 2008. Mechanisms of membrane fusion: disparate players and common principles. Nat. Rev. Mol. Cell Biol. 9:543–556 [DOI] [PubMed] [Google Scholar]
- 25. McInerney G. M., Smit J. M., Liljestrom P., Wilschut J. 2004. Semliki Forest virus produced in the absence of the 6K protein has an altered spike structure as revealed by decreased membrane fusion capacity. Virology 325:200–206 [DOI] [PubMed] [Google Scholar]
- 26. Modis Y., Ogata S., Clements D., Harrison S. C. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313–319 [DOI] [PubMed] [Google Scholar]
- 27. Mukhopadhyay S., Kim B. S., Chipman P. R., Rossmann M. G., Kuhn R. J. 2003. Structure of West Nile virus. Science 302:248. [DOI] [PubMed] [Google Scholar]
- 28. Mukhopadhyay S., et al. 2006. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure 14:63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nayak V., et al. 2009. Crystal structure of dengue virus type 1 envelope protein in the postfusion conformation and its implications for membrane fusion. J. Virol. 83:4338–4344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Op De Beeck A., et al. 2003. Role of the transmembrane domains of prM and E proteins in the formation of yellow fever virus envelope. J. Virol. 77:813–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Orlinger K. K., Hoenninger V. M., Kofler R. M., Mandl C. W. 2006. Construction and mutagenesis of an artificial bicistronic tick-borne encephalitis virus genome reveals an essential function of the second transmembrane region of protein E in flavivirus assembly. J. Virol. 80:12197–12208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sánchez-San Martín C., Liu C. Y., Kielian M. 2009. Dealing with low pH: entry and exit of alphaviruses and flaviviruses. Trends Microbiol. 17:514–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schalich J., et al. 1996. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J. Virol. 70:4549–4557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stiasny K., Brandler S., Kossl C., Heinz F. X. 2007. Probing the flavivirus membrane fusion mechanism by using monoclonal antibodies. J. Virol. 81:11526–11531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stiasny K., Fritz R., Pangerl K., Heinz F. X. 2009. Molecular mechanisms of flavivirus membrane fusion. Amino Acids. doi:10.1007/s00726-009-0370-4 [DOI] [PubMed] [Google Scholar]
- 36. Stiasny K., Heinz F. X. 2006. Flavivirus membrane fusion. J. Gen. Virol. 87:2755–2766 [DOI] [PubMed] [Google Scholar]
- 37. Stiasny K., Koessl C., Heinz F. X. 2003. Involvement of lipids in different steps of the flavivirus fusion mechanism. J. Virol. 77:7856–7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sultana H., et al. 2009. Fusion loop peptide of the West Nile virus envelope protein is essential for pathogenesis and is recognized by a therapeutic cross-reactive human monoclonal antibody. J. Immunol. 183:650–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taucher C., Berger A., Mandl C. W. 2010. A trans-complementing recombination trap demonstrates a low propensity of flaviviruses for intermolecular recombination. J. Virol. 84:599–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thiel H. J., et al. 2005. Family Flaviviridae, p. 981–998 In Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA [Google Scholar]
- 41. White J. M., Delos S. E., Brecher M., Schornberg K. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43:189–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu S. R., et al. 2007. The dynamic envelope of a fusion class II virus. Prefusion stages of semliki forest virus revealed by electron cryomicroscopy. J. Biol. Chem. 282:6752–6762 [DOI] [PubMed] [Google Scholar]
- 43. Yu I. M., et al. 2009. Association of the pr peptides with dengue virus at acidic pH blocks membrane fusion. J. Virol. 83:12101–12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yu I. M., et al. 2008. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319:1834–1837 [DOI] [PubMed] [Google Scholar]
- 45. Zhang W., et al. 2003. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 10:907–912 [DOI] [PMC free article] [PubMed] [Google Scholar]