Abstract
A chimeric porcine circovirus (PCV1-2) with the capsid gene of pathogenic PCV2 cloned into the genomic backbone of nonpathogenic PCV1 is attenuated in pigs but elicits protective immunity against PCV2. In this study, short epitope tags were inserted into the C terminus of the capsid protein of the chimeric PCV1-2 vaccine virus, resulting in a tractable marker virus that is infectious both in vitro and in vivo. Pigs experimentally infected with the epitope-tagged PCV1-2 vaccine viruses produced tag-specific antibodies, as well as anti-PCV2 neutralizing antibodies, indicating that the epitope-tagged viruses could potentially serve as a positive-marker modified live-attenuated vaccine.
Porcine circovirus (PCV) is a small nonenveloped virus with a single-stranded circular DNA genome of approximately 1.7 kb in the family Circoviridae (11, 28). Type 1 PCV (PCV1) was discovered as a contaminant of PK-15 cells and is nonpathogenic in pigs (26, 27). Type 2 PCV (PCV2) was discovered in piglets with postweaning multisystemic wasting syndrome and is the causative agent of PCV-associated disease (1, 2, 7, 14, 17). At least two subtypes of PCV2, PCV2a and PCV2b, have been recognized worldwide (4). The genome contains two open reading frames (ORF); ORF1 encodes the replication proteins (Rep and Rep′), and ORF2 encodes the capsid protein (Cap) (5, 6, 16, 20). We have previously demonstrated that a chimeric PCV (PCV1-2) with the capsid gene of PCV2 inserted into the backbone of PCV1 is infectious but attenuated in pigs (9, 10), and an inactivated commercial vaccine based on chimeric PCV1-2 is currently on the market (9, 10, 13, 23). Since PCV2 infection is mostly subclinical, it is important to design a new vaccine that can track the virus's spread and herd level immunity. Immunogenic epitopes have been expressed on surface-exposed domains of viral proteins in other viruses, resulting in specific immune responses (12, 18, 21, 22, 25). In part due to its small genome size, the ability of the PCV genome to tolerate insertion and display foreign epitopes has not been explored. In this study, we aimed to identify genomic locations that can tolerate small insertions of epitope tags and to produce an epitope-tagged vaccine virus for use as a potential tractable modified live-attenuated vaccine (MLV).
Identification of locations within the PCV genome that tolerate the insertion of small epitope tags.
The PCV2 infectious clone constructed in previous studies (4) was used as the genomic backbone for the constructions of four mutants each containing an influenza virus hemagglutinin (HA) tag (YPYDVPDYA) inserted in frame at the amino (N′) and carboxy (C′) termini of ORF1 and ORF2 (Table 1). Insertions were introduced into the infectious clone by site-directed mutagenesis, followed by the assembly of two overlapping PCR products by overlap extension PCR (Table 1) and subcloning as described previously (3). Each clone was completely sequenced to verify the introduced tag and confirm that no undesired mutations were introduced. Infectious virus stocks were generated by transfection of PK-15 cells with each of the concatemerized full-length clones, and infectivity titers of the mutant viruses in PK-15 cells were determined as previously described (8, 10). The HA tag was visualized by an immunofluorescence assay (IFA) using a fluorescein isothiocyanate (FITC)-labeled anti-HA monoclonal antibody (MAb; Sigma, St. Louis, MO). No infectious virus was detectable from cells transfected with N′-HA or C′-HA ORF1 mutants (Table 1), indicating that insertions at the termini of ORF1 directly interfered with Rep/Rep′ function and prevented virus replication. However, N′-HA and C′-HA capsid insertion mutants were infectious in PK-15 cells, with infectious titers of 103.5 and 105.0 50% tissue culture infective doses (TCID50)/ml, respectively (Table 1). The N-terminal domain of the PCV capsid is thought to interact with DNA on the interior of the virion, which may explain the lower detection level of the HA tag for the N′-HA mutant (15). The C terminus of the PCV2 capsid is a type-specific immunoreactive epitope that is believed to be displayed on the surface of the virion (15, 24).
Table 1.
Epitope tag insertion mutants of PCVs
| Strain ID | Parent strain | Insert location | Insert tag | Insert size (aa) | Primersa | Titer (TCID50/ml) |
|---|---|---|---|---|---|---|
| PCV2b | NAb | NA | NA | NA | 1 + 2 | 104.5 |
| PCV1-2 | NA | NA | NA | NA | 11 + 12 | 103.7 |
| 2-ORF1N-HA | PCV2 | N′-ORF1 | HA | 9 | 1 + 3, 2 + 4 | |
| 2-ORF1C-HA | PCV2 | C′-ORF1 | HA | 9 | 1 + 5, 2 + 6 | |
| 2-ORF2N-HA | PCV2 | N′-ORF2 | HA | 9 | 1 + 7, 2 + 8 | 103.5 |
| 2-ORF2C-HA | PCV2 | C′-ORF2 | HA | 9 | 1 + 9, 2 + 10 | 105.0 |
| PCV1-2-HA1 | PCV1-2 | C′-ORF2 | 1× HA | 9 | 12 + 13, 11 + 14 | 103.7 |
| PCV1-2-HA2 | PCV1-2 | C′-ORF2 | 2× HA | 18 | 12 + 15, 11 + 16 | 103.7 |
| PCV1-2-HA3 | PCV1-2 | C′-ORF2 | 3× HA | 27 | 12 + 17, 11 + 18 | 103.5 |
| PCV1-2-GLU | PCV1-2 | C′-ORF2 | GLU | 10 | 12 + 19, 11 + 20 | 103.5 |
| PCV1-2-KT3 | PCV1-2 | C′-ORF2 | KT3 | 11 | 12 + 21, 11 + 22 | 103.7 |
Primer sequences (5 prime to 3 prime): primer 1, AGCCCGCGGAAATTTCTGACAAACGTTAC; primer 2, TTTCCGCGGGCTGGCTGAACTTTTGAAAG; primer 3, ACATGTACCCATACGATGTTCCAGATTACGCTCCCAGCAAGAAGA; primer 4, GGAGCGTAATCTGGAACATCGTATGGGTACATGTTGCTGCTGAGG; primer 5, ACTACCCATACGATGTTCCAGATTACGCTTGAGTCTTTTTTATCAC; primer 6, ACTCAAGCGTAATCTGGAACATCGTATGGGTAGTAATTTATTTCA; primer 7, GTAGCGTAATCTGGAACATCGTATGGGTACATAGCTGAAAACGAAA; primer 8, CTATGTACCCATACGATGTTCCAGATTACGCTACGTATCCAAGGAG; primer 9, CATTAAGCGTAATCTGGAACATCGTATGGGTAAGGGTTAAGTGGGG; primer 10, CTTACCCATACGATGTTCCAGATTACGCTTAATGAATAATAAAAAC; primer 11, GGAGGTACCCGAAGGCCGATTTGAAGCAG; primer 12, TCGGGTACCTCCGTGGATTGTTCTCCAGC; primer 13, ATTTAAGCGTAATCTGGAACATCGTATGGGTAGGGTTTAAGTGGGG; primer 14, AACCCTACCCATACGATGTTCCAGATTACGCTTAAATGAATAAAAATAA; primer 15, ATTTAAGCGTAATCTGGAACATCGTATGGGTACGCATAGTCCGGGACGTCATACGGATAGGGTTTAAGTGGGG; primer 16, AACCCTATCCGTATGACGTCCCGGACTATGCGTACCCATACGATGTTCCAGATTACGCT TAAATGAATAAAAATAA; primer 17, ACGGATAAGCGTAATCTGGAACATCGTATGGGTACGCATAGTCCGGGACGTCATACGGATAGGGTTTAAGTGGGG; primer 18, TATGCGTACCCATACGATGTTCCAGATTACGCTTATCCGTATGACGTCCCGGACTATGCGTAAATGAATAAAAATAA; primer 19, ATTTATTCCATCGGCATATATTCTTCTTCTTCGCAGGGTTTAAGTGGGG; primer 20, AACCCTGCGAAGAAGAAGAATATATGCCGATGGAATAAATGAATAAAAATAA; primer 21, ATTTAGGTTTCCGGTTCCGGCGGCGGGGTCGGCGGTTTGGGTTTAAGTGGGG; primer 22, AACCCAAACCGCCGACCCCGCCGCCGGAACCGGAAACCTAAATGAATAAAAATAA.
NA, not applicable.
Effects of insertion sizes and different epitope tags in the C terminus of the capsid on PCV1-2 virus infectivity in vitro.
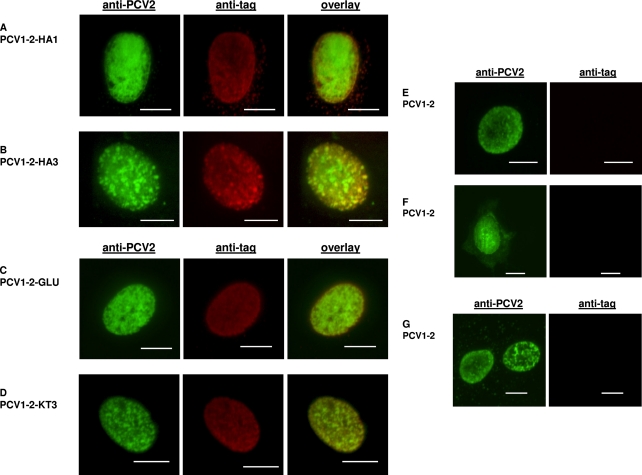
We further investigated the effects of insertion sizes of the HA tag and two additional epitope tags on the infectivity of chimeric PCV1-2 vaccine virus (10). Five PCV1-2 mutants were constructed and tested in vitro for infectivity, with each mutant containing a different tag inserted in frame in the C terminus of the capsid: a single HA tag (HA1), an HA tag dimer (HA2), an HA tag trimer (HA3), a glu-glu tag (GLU) from mouse polyomavirus medium T antigen (CEEEEYMPME), and a KT3 tag (KT3) from simian virus 40 large T antigen (KPPTPPPEPET) (Table 1). The results showed that each of the 5 mutants (PCV1-2-HA1, PCV1-2-HA2, PCV1-2-HA3, PCV1-2-GLU, and PCV1-2-KT3) was infectious in vitro, as lysates from transfected PK-15 cells had infectious titers of 103.5 to 103.7 TCID50/ml (Table 1). Confocal microscopy examination revealed that each of the inserted epitope tags was properly expressed on the surface of virions, resulting in double labeling of infected cell nuclei with both anti-PCV2 capsid and anti-epitope tag antibodies (Fig. 1). The HA, KT3, and GLU tags were visualized using polyclonal rabbit anti-HA (eBioscience, San Diego, CA), anti-KT3, and anti-GLU (GenScript, Piscataway, NJ) antibodies, respectively, followed by staining with an Alexa Fluor 647-labeled goat anti-rabbit antibody (Invitrogen, Carlsbad, CA). The PCV2 capsid was stained with a mouse anti-PCV2 capsid MAb (RTI, Brookings, SD), followed by an FITC-labeled goat anti-mouse antibody (KPL, Gaithersburg, MD). The stability of the epitope-tagged PCV1-2-KT3 and PCV1-2-GLU viruses was verified by 10 successful serial passages of the mutant viruses in PK-15 cells, followed by IFA and DNA sequencing confirmation of the passaged viruses (data not shown). The results showed that the C terminus of the capsid tolerates epitope tag insertions as large as 27 amino acid (aa) residues, as mutants with the HA tag dimer and trimer were both infectious in vitro (Fig. 1 and Table 1).
Fig. 1.
Confocal microscopy of double immunofluorescent staining of epitope tags and PCV2 capsid antigen in PK-15 cells infected with chimeric PCV1-2 containing different inserted epitope tags. PK-15 cells infected with different insertion mutants were dually labeled with respective rabbit anti-tag and mouse anti-PCV2 capsid antibodies (Rural Technologies, Inc., Brookings, SD) and then stained with a mixture of Alexa Fluor 647-labeled goat anti-rabbit (Invitrogen, San Diego, CA) and FITC-labeled goat anti-mouse (KPL, Gaithersburg, MD) antibodies: (A) PCV1-2-HA1 (a single HA tag), (B) PCV1-2-HA3 (HA tag trimer), (C) PCV1-2-GLU (a single GLU tag), and (D) PCV1-2-KT3 (a single KT3 tag). Cells infected with chimeric PCV1-2 vaccine virus (control) were dually labeled with mouse anti-PCV2 capsid and rabbit anti-HA (E), rabbit anti-GLU (F), and rabbit anti-KT3 (G) antibodies and then stained as described above. Infected cells were visualized at ×1,000 to ×1,500 magnification using a Nikon TE2000-E confocal microscope at 488 nm (525/50 emission filter) to detect the PCV2 capsid and at 647 nm (710/50 emission filter) to detect the epitope tags, and images were captured using a Cascade II 512 camera (Roper Scientific/Photometrics, Tucson, AZ). Scale bars all represent 5 μm.
Chimeric PCV1-2 vaccine viruses with epitope tags inserted in the C terminus of the capsid are infectious in specific-pathogen-free pigs, inducing both anti-epitope tag and anti-PCV2 neutralizing antibodies.
To determine whether the tagged chimeric PCV1-2 viruses are infectious and immunogenic, a total of 12 pigs, 7 weeks old, were randomly assigned to four groups of 3 each. Each group was inoculated intramuscularly with 2 × 103.5 TCID50 of PCV1-2, PCV1-2-KT3, PCV1-2-GLU, or phosphate-buffered saline (PBS). The HA-tagged viruses were not tested in vivo, as they are not considered viable MLV candidates in pigs due to potential antibody cross-reactivity in pigs naturally infected with swine influenza viruses. The pigs were each bled prior to inoculation and weekly thereafter until necropsy at 42 days postinoculation (dpi). A nested PCR (first-round primers 5′-TGGAGAAGAAGTTGTTGT-3′ [forward] and 5′-ATGACGTATCCAAGGAGGCGTTACCGCAGAAGAAGACACCGCCCCCGCAG-3′ [reverse] and second-round primers 5′-GGAGGTACCCGAAGGCCGATTTGAAGCAG-3′ [forward] and 5′-CCCTTTGAATACTACAGA-3′ [reverse]) was used to detect viremia in the sera of all pigs. An enzyme-linked immunosorbent assay (ELISA) was used to detect anti-PCV2 antibodies in the weekly sera (19). Synthetic-peptide-based ELISAs were used to detect anti-GLU and anti-KT3 tag antibodies in the sera. ELISA plates were coated with bovine serum albumin-conjugated GLU or KT3 synthetic peptides (GenScript, Piscataway, NJ). Diluted pig serum was added to each well and incubated 1 h at 37°C, followed by incubation with 1:2,000-diluted, horseradish peroxidase-labeled goat anti-swine IgG antibody (KPL, Gaithersburg, MD). The cutoffs for both the KT3 and GLU ELISAs were determined as the mean optical density at 450 nm (OD450) of all negative pig sera from 0 to 42 dpi plus 3 standard deviations. To determine if the anti-PCV2 antibodies in the serum of each pig at 42 dpi were neutralizing, an in vitro serum virus neutralization assay was performed. Serial 2-fold dilutions of each serum were mixed with an equal volume of PCV2a or PCV2b virus stock and incubated at 37°C for 1 h. PK-15 cells at 50% confluence in 96-well plates were infected in duplicate with 50 μl of each reaction mixture. After incubation at 37°C for 72 h, the infected cells were visualized by IFA (8, 10) and the percentage of virus neutralization was calculated.
Viremia was detected by nested PCR in 8/9 inoculated pigs, thus confirming infection by each chimeric virus (Table 2). PCV1-2-GLU had a noticeably lower frequency and duration of viremia than PCV1-2 and PCV1-2-KT3, which were similar. Sequencing and sequence analysis of all positive PCR products confirmed the authenticity and in vivo stability of each epitope-tagged virus (data not shown).
Table 2.
Detection by nested PCR of viremia of tagged and wild-type chimeric PCV1-2 in the sera of infected specific-pathogen-free pigs
| Inoculum | No. of pigs with detectable viremia/total no. of pigs at dpi: |
||||||
|---|---|---|---|---|---|---|---|
| 0 | 7 | 14 | 21 | 28 | 35 | 42 | |
| PBS | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 |
| PCV1-2 | 0/3 | 3/3 | 2/3 | 1/3 | 3/3 | 1/3 | 0/3 |
| PCV1-2-GLU | 0/3 | 2/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 |
| PCV1-2-KT3 | 0/3 | 3/3 | 2/3 | 2/3 | 1/3 | 1/3 | 1/3 |
Anti-PCV2 capsid antibodies were detected in all infected pigs but not in PBS control pigs (Fig. 2A). All pigs infected with PCV1-2 and PCV1-2-KT3 seroconverted to PCV2 antibodies by 28 dpi. One pig in the PCV1-2-GLU group seroconverted by 28 dpi, while the other two had a delayed seroconversion at 35 dpi. Anti-KT3 tag antibodies were detected in all PCV1-2-KT3-infected pigs but not in other groups (Fig. 2B). Anti-KT3 tag antibodies were detected in all three pigs by 14 dpi, which remained seropositive at 42 dpi. Similarly, anti-GLU tag antibodies were detected only in PCV1-2-GLU-infected pigs (Fig. 2C): two pigs seroconverted at 14 and 28 dpi, while one pig had detectable anti-GLU antibodies only at 28 dpi. Anti-PCV2 neutralizing antibodies were detected in all infected pigs at 42 dpi (Fig. 2D). The levels of neutralizing antibodies in the PCV1-2 and PCV1-2-KT3 pigs were similar, while the two PCV1-2-GLU pigs with delayed seroconversion had noticeably lower levels of neutralizing antibodies (Fig. 2D). Therefore, the results indicated that PCV1-2-KT3 induced an anti-PCV2 antibody response that was comparable to that induced by PCV1-2 but stronger than that induced by PCV1-2-GLU. The small animal numbers and pig-to-pig variation may play a role in the observed difference in antibody response among groups. Also, insertion of tags may affect the ability of the tagged viruses to replicate in pigs, although the viremia pattern of PCV1-2-KT3 was similar to that of wild-type PCV1-2 (Table 2). Importantly, both PCV1-2-KT3 and PCV1-2-GLU elicited anti-PCV2 neutralizing antibodies and anti-epitope tag antibodies.
Fig. 2.
Detection of specific anti-GLU and anti-KT3 tag antibodies, as well as anti-PCV2 neutralizing antibodies, in specific-pathogen-free pigs infected with chimeric PCV1-2 containing the inserted KT3 or GLU epitope. Specific antibody responses were examined in the sera of pigs infected with PCV1-2 (■), PCV1-2-KT3 (●), PCV1-2-GLU (▵), or PBS (○) using three different ELISAs, (A) a PCV2 capsid-specific ELISA, (B) a KT3 tag-specific ELISA, and (C) a GLU tag-specific ELISA. The mean OD450 ± the standard error of the mean is plotted for each treatment group throughout the experiment, with an asterisk indicating significant differences on that day. Treatments with different letters have statistically significant differences on that day. Statistical comparison was performed using repeated-measures analysis of variance, with the slice option of the Glimmix procedure, followed by Tukey's procedure for multiple comparisons. Statistical significance was set to alpha = 0.05. All analyses were performed using commercially available software (SAS version 9.2; SAS, Cary, NC). The dotted horizontal line indicates the cutoff of each assay. (D) PCV2-specific neutralizing antibodies (Ab) detected in sera of infected pigs at 42 dpi. Serum samples from each pig were tested for specific anti-PCV2a and anti-PCV2b neutralizing activity. Black bars represent PCV1-2, open bars represent PCV1-2-KT3, and shaded gray bars represent PCV1-2-GLU. The mean percentage of virus neutralization for each treatment group ± the standard deviation is plotted for five different serum dilutions.
In summary, we demonstrated that the C terminus of PCV2 capsid protein tolerates insertions of at least 27 aa, whereas short terminal insertions in rep rendered the virus nonviable. We further demonstrated that chimeric PCV1-2 vaccine viruses containing inserted epitopes in the C terminus of the Cap protein are infectious in vitro and in vivo and elicit both anti-epitope tag antibodies and anti-PCV2 neutralizing antibodies. These epitope-tagged PCV1-2 vaccine viruses will allow the serologic differentiation of vaccinated pigs from naturally infected ones and thus could potentially serve as a tractable MLV such as a compliance marker vaccine.
Acknowledgments
We thank Stephen Werre for assistance with statistical analysis and Scott Kenney, Barbara Dryman, Kylie Harrall, Brent Sanford, Shannon Viers, Pete Jobst, and Dustin Lucas for their technical assistance.
This study was supported by a Various Fund account of Virginia Tech.
Footnotes
Published ahead of print on 9 February 2011.
REFERENCES
- 1. Allan G. M., et al. 2004. PMWS: experimental model and co-infections. Vet. Microbiol. 98:165–168 [DOI] [PubMed] [Google Scholar]
- 2. Allan G. M., et al. 1998. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the U. S. A. and Europe. J. Vet. Diagn. Invest. 10:3–10 [DOI] [PubMed] [Google Scholar]
- 3. Beach N. M., Juhan N. M., Cordoba L., Meng X. J. 2010. Replacement of the replication factors of porcine circovirus (PCV) type 2 with those of PCV type 1 greatly enhances viral replication in vitro. J. Virol. 84:8986–8989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beach N. M., Ramamoorthy S., Opriessnig T., Wu S. Q., Meng X. J. 2010. Novel chimeric porcine circovirus (PCV) with the capsid gene of the emerging PCV2b subtype cloned in the genomic backbone of the non-pathogenic PCV1 is attenuated in vivo and induces protective and cross-protective immunity against PCV2b and PCV2a subtypes in pigs. Vaccine 29:221–232 [DOI] [PubMed] [Google Scholar]
- 5. Cheung A. K. 2003. The essential and nonessential transcription units for viral protein synthesis and DNA replication of porcine circovirus type 2. Virology 313:452–459 [DOI] [PubMed] [Google Scholar]
- 6. Cheung A. K. 2003. Transcriptional analysis of porcine circovirus type 2. Virology 305:168–180 [DOI] [PubMed] [Google Scholar]
- 7. Ellis J., et al. 1998. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can. Vet. J. 39:44–51 [PMC free article] [PubMed] [Google Scholar]
- 8. Fenaux M., et al. 2002. Cloned genomic DNA of type 2 porcine circovirus is infectious when injected directly into the liver and lymph nodes of pigs: characterization of clinical disease, virus distribution, and pathologic lesions. J. Virol. 76:541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fenaux M., Opriessnig T., Halbur P. G., Elvinger F., Meng X. J. 2004. A chimeric porcine circovirus (PCV) with the immunogenic capsid gene of the pathogenic PCV type 2 (PCV2) cloned into the genomic backbone of the nonpathogenic PCV1 induces protective immunity against PCV2 infection in pigs. J. Virol. 78:6297–6303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fenaux M., Opriessnig T., Halbur P. G., Meng X. J. 2003. Immunogenicity and pathogenicity of chimeric infectious DNA clones of pathogenic porcine circovirus type 2 (PCV2) and nonpathogenic PCV1 in weanling pigs. J. Virol. 77:11232–11243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Finsterbusch T., Mankertz A. 2009. Porcine circoviruses—small but powerful. Virus Res. 143:177–183 [DOI] [PubMed] [Google Scholar]
- 12. Gedvilaite A., et al. 2000. Formation of immunogenic virus-like particles by inserting epitopes into surface-exposed regions of hamster polyomavirus major capsid protein. Virology 273:21–35 [DOI] [PubMed] [Google Scholar]
- 13. Gillespie J., et al. 2008. A genetically engineered chimeric vaccine against porcine circovirus type 2 (PCV2) is genetically stable in vitro and in vivo. Vaccine 26:4231–4236 [DOI] [PubMed] [Google Scholar]
- 14. Grau-Roma L., Fraile L., Segales J. 2011. Recent advances in the epidemiology, diagnosis and control of diseases caused by porcine circovirus type 2. Vet. J. 187:23–32 [DOI] [PubMed] [Google Scholar]
- 15. Lekcharoensuk P., et al. 2004. Epitope mapping of the major capsid protein of type 2 porcine circovirus (PCV2) by using chimeric PCV1 and PCV2. J. Virol. 78:8135–8145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mankertz A., Hillenbrand B. 2001. Replication of porcine circovirus type 1 requires two proteins encoded by the viral rep gene. Virology 279:429–438 [DOI] [PubMed] [Google Scholar]
- 17. Meehan B. M., et al. 1998. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J. Gen. Virol. 79(Pt. 9):2171–2179 [DOI] [PubMed] [Google Scholar]
- 18. Murray K., Shiau A. L. 1999. The core antigen of hepatitis B virus as a carrier for immunogenic peptides. Biol. Chem. 380:277–283 [DOI] [PubMed] [Google Scholar]
- 19. Nawagitgul P., et al. 2002. Modified indirect porcine circovirus (PCV) type 2-based and recombinant capsid protein (ORF2)-based enzyme-linked immunosorbent assays for detection of antibodies to PCV. Clin. Diagn. Lab. Immunol. 9:33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nawagitgul P., et al. 2000. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 81:2281–2287 [DOI] [PubMed] [Google Scholar]
- 21. Neugebauer M., et al. 2006. Development of a vaccine marker technology: display of B cell epitopes on the surface of recombinant polyomavirus-like pentamers and capsoids induces peptide-specific antibodies in piglets after vaccination. Biotechnol. J. 1:1435–1446 [DOI] [PubMed] [Google Scholar]
- 22. Niikura M., et al. 2002. Chimeric recombinant hepatitis E virus-like particles as an oral vaccine vehicle presenting foreign epitopes. Virology 293:273–280 [DOI] [PubMed] [Google Scholar]
- 23. Segalés J., et al. 2009. A genetically engineered chimeric vaccine against porcine circovirus type 2 (PCV2) improves clinical, pathological and virological outcomes in postweaning multisystemic wasting syndrome affected farms. Vaccine 27:7313–7321 [DOI] [PubMed] [Google Scholar]
- 24. Shang S. B., et al. 2009. Fine mapping of antigenic epitopes on capsid proteins of porcine circovirus, and antigenic phenotype of porcine circovirus type 2. Mol. Immunol. 46:327–334 [DOI] [PubMed] [Google Scholar]
- 25. Slupetzky K., et al. 2001. Chimeric papillomavirus-like particles expressing a foreign epitope on capsid surface loops. J. Gen. Virol. 82:2799–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tischer I., Gelderblom H., Vettermann W., Koch M. A. 1982. A very small porcine virus with circular single-stranded DNA. Nature 295:64–66 [DOI] [PubMed] [Google Scholar]
- 27. Tischer I., Mields W., Wolff D., Vagt M., Griem W. 1986. Studies on epidemiology and pathogenicity of porcine circovirus. Arch. Virol. 91:271–276 [DOI] [PubMed] [Google Scholar]
- 28. Todd D., et al. 2005. Circoviridae, p. 327–334 In Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (ed.), Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA [Google Scholar]