Abstract
After binding to its cell surface receptor ganglioside GM1, simian virus 40 (SV40) is endocytosed by lipid raft-mediated endocytosis and slowly transported to the endoplasmic reticulum, where partial uncoating occurs. We analyzed the intracellular pathway taken by the virus in HeLa and CV-1 cells by using a targeted small interfering RNA (siRNA) silencing screen, electron microscopy, and live-cell imaging as well as by testing a variety of cellular inhibitors and other perturbants. We found that the virus entered early endosomes, late endosomes, and probably endolysosomes before reaching the endoplasmic reticulum and that this pathway was part of the infectious route. The virus was especially sensitive to a variety of perturbations that inhibited endosome acidification and maturation. Contrary to our previous models, which postulated the passage of the virus through caveolin-rich organelles that we called caveosomes, we conclude that SV40 depends on the classical endocytic pathway for infectious entry.
INTRODUCTION
To enter their host cells, the majority of animal viruses take advantage of endocytic mechanisms offered by the cell (23, 39). Penetration into the cytosol usually occurs from endosomes, often triggered by the low lumenal pH. However, there are viruses that deviate from this standard itinerary. These viruses include members of the polyomavirus family, such as mouse polyomavirus (mPy) and simian virus 40 (SV40). These viruses are nonenveloped DNA viruses that replicate in the nucleus. The interest and importance of this virus family are rapidly growing with the increasing number of human polyomaviruses identified. The most recently discovered human pathogens include KI polyomavirus (KIPyV), WU polyomavirus (WUPyV), and Merkel cell polyomavirus (MCPyV) (1, 17, 20). MCPyV is associated with the aggressive neuroendocrine skin cancer Merkel cell carcinoma.
Most polyomaviruses bind to gangliosides on the cell surface and are internalized into small tight-fitting vesicles devoid of a clathrin coat (26, 28, 30, 34, 55, 59). Instead of using endosomes for penetration, they travel to the lumen of the endoplasmic reticulum (ER), in which they are activated by lumenal thiol oxidoreductases and chaperones before penetrating into the cytosol or possibly directly into the nucleoplasm (28, 37, 46, 53).
In this study, we focus on SV40, a virus that binds to GM1 and is internalized via caveola/lipid raft-dependent endocytic mechanisms (2, 11, 45, 57, 59). Some virus particles are endocytosed via caveolae, and others enter through a parallel clathrin- and caveolin-independent mechanism. Uptake is slow and nonsynchronous, with transfer into the ER and penetration through the ER membrane occurring several hours after the initial endocytosis (53).
Exactly where SV40 spends the intervening hours is not clear. Unlike ER-targeted bacterial toxins such as cholera toxin and Shiga toxin, the virus particles are not observed in the trans-Golgi network or the cisternae of the Golgi complex. Some of them are seen in early endosomes (EEs) by confocal and electron microscopy (28, 42). We also reported the accumulation of incoming SV40 particles in so-called “caveosomes,” which we defined as large caveolin-1-positive, pH-neutral, endocytic organelles devoid of transferrin and other EE markers (44). Our observation of viruses in these organelles led us to propose a model whereby the caveosomal route allows incoming SV40 to bypass the classical endosomes during its transit from the plasma membrane to the ER (44). However, our most recent results indicate that caveosomes are not independent organelles but rather modified late endosomes (LEs) or endolysosomes in which caveolin-1 accumulates after overexpression or after interference with caveolar assembly (25).
In the light of these findings, the intracellular trafficking pathway followed by SV40 needs reevaluation. To analyze the transport of incoming viruses and the pathway to infection, in this study we have used a variety of techniques that allowed the visualization, tracking, and localization of viruses at different stages of entry into cells in which caveolar assembly was not interfered with. The experiments involved video microscopy of live cells, electron microscopy (EM), and a variety of perturbants in the form of small interfering RNAs (siRNAs) and pharmacological inhibitors. To avoid fixation artifacts that affect the visualization of late endosomes (25), many of the experiments were performed with live cells. The results provided a detailed picture of the intracellular route followed by the infectious virus and led to a thorough revision of our previous model of SV40 entry.
MATERIALS AND METHODS
Cell culture.
Human cervix carcinoma (HeLa CNX) cells were provided by Lucas Pelkmans (43), and African green monkey kidney cells (CV-1) were purchased from the American Type Culture Collection. Caveolin-1 knockout mouse lung fibroblast cells (caveolin-1 KO cells) were described previously (12). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS; Invitrogen).
Plasmids, antibodies, and other reagents.
Expression plasmids for Rab5a, Rab7, and Rab9, provided by Marino Zerial, were subcloned previously by Sonnichsen et al and Vonderheit and Helenius (56, 62). Rab5Q79L was described previously (42). LAMP1-enhanced green fluorescent protein (EGFP) was provided by Jean Gruenberg (University of Geneva), Hrs-EGFP was provided by Harald Stenmark (48), and rat DNM2(aa)-EGFP and rat DNM2(aa)K44A-EGFP were provided by Mark McNiven (8).
The mouse anti-LAMP1 antibody (sc-18821) was purchased from Santa Cruz Biotechnology; mouse anti-EEA1, anti-caveolin-1, and anti-DNM2 were obtained from BD Transduction Laboratories (catalog number BD-610457); and mouse anti-beta actin (A1978), anti-Rab4, anti-Rab5, and anti-NEDD4L were obtained from Sigma. Mouse anti-cofilin and anti-ezrin/villin antibodies were obtained from Cell Signaling Technologies. Mouse anti-T-antigen antibody 1605 was produced in-house, and the anti-mouse secondary antibodies labeled with Alexa Fluor 647 (AF647), AF594, and AF488 were obtained from Invitrogen. All chemicals were purchased from Sigma-Aldrich.
SV40 purification.
The protocol for virus purification was based on those described previously (10, 15). In brief, monkey kidney CV-1 cells were cultured in complete medium (DMEM [Gibco] supplemented with 10% FCS [LabForce, Nunningen, Switzerland] and 4 mM GlutaMAX) at 37°C in 5% CO2. Forty T175 flasks of subconfluent CV-1 cells were infected with SV40 at a multiplicity of infection (MOI) of 0.01 in DMEM without additives. Cells were cultured in complete medium for 14 days. To harvest virus, cells were put through three freeze-thaw cycles and then centrifuged at 10,000 × g for 10 min at 4°C. Twenty milliliters of virus-containing supernatant was loaded onto a 10-ml cushion of CsCl (1.4 g ml−1) in 10 mM HEPES (pH 7.4). Following centrifugation at 76,000 × g for 3 h at 4°C in an SW28 rotor (Beckman), the banded virus in the CsCl cushion was harvested. The density of the CsCl fraction containing SV40 was checked, and the fraction was adjusted to a CsCl density of 1.34 g/ml in 10 mM HEPES (pH 7.4). Following equilibrium centrifugation at 100,000 × g for 16 h at 4°C in a 70.1 Ti rotor (Beckman), the lower virus band was isolated and dialyzed against a solution containing 50 mM HEPES (pH 8.0), 150 mM NaCl, and 1 mM CaCl2 (virus buffer). The purified infectious virus was stored in aliquots at −80°C.
Fluorescent labeling of SV40.
The fluorescent labeling procedure was done as described previously (44). The modified SV40 virions were able to bind, enter, and infect CV-1 cells as described previously (44).
SV40 infection.
CV-1 cells in 6- or 12-well plates were infected with SV40 in inoculation medium (R-medium [Gibco] containing 50 mM HEPES buffer [Invitrogen] and 0.5% bovine serum albumin [BSA; Fluka] [pH 6.8]) at 37°C in 5% CO2 for 2 h at an MOI of 1, resulting in 20 to 30% infection, or at an MOI of 5, resulting in 30 to 50% infection. Subsequently, cells were washed with phosphate-buffered saline (PBS; pH 7.4) and maintained in DMEM with 10% FCS at 37°C in 5% CO2 for an additional 22 h until they were fixed with 4% formaldehyde in PBS for 20 min. Cells were permeabilized, labeled for SV40 T-antigen expression, and subjected to fluorescence-activated cell sorter (FACS) analysis.
For experiments with pharmacological inhibitors, cells were incubated with the respective drug (sample) or solvent alone (control sample) an hour before the addition of the virus and during infection. Drugs were used at the following concentrations: 0.2 mM genistein in dimethyl sulfoxide (DMSO) (Calbiochem), 5 μM nocodazole in DMSO (Sigma), 100 mM orthovanadate (OV) in H2O (Sigma), 0.5 μg/ml brefeldin A (BFA) in DMSO (Sigma), 80 μM dynasore in DMSO (Sigma), 100 nM bafilomycin A1 (Baf) in DMSO (Fluka), 10 μM monensin in H2O (Sigma), 20 mM NH4Cl in H2O (Sigma), and 100 nM okadaic acid (OA) in DMSO (Sigma). Culture medium (pH 7.4) was additionally buffered with 50 mM HEPES when NH4Cl was used (29).
FACS analysis.
For FACS analysis, cells were fixed with 4% formaldehyde in PBS, permeabilized (0.1% [wt/vol] saponin, 2% FCS, 20 mM EDTA, and 0.02% NaN3 in PBS), and stained with primary antibodies for 2 h at room temperature (RT) (mouse anti-T antigen [1:100]), followed by Alexa Fluor 647-labeled goat anti-mouse IgG (1:500; Invitrogen) for 45 min at RT. Cells were analyzed with a FACSCalibur cytometer using CellQuest 3.1 software (Becton Dickinson Immunocytometry Systems). At least 10,000 cells were analyzed for each sample.
Transfection, subcellular localization, immunostaining, and microscopy.
For localization studies, CV-1 cells were transiently transfected with 2 μg plasmid DNA encoding fluorescently tagged versions of the proteins using the Amaxa electroporation system and plated into 12-well dishes with coverslips or 8-well glass-bottom chambers. After 12 h in culture, fluorescently labeled SV40 (MOI of ∼10) was added to cells for 1 h on ice, unbound virus was washed away, and the cells expressing small amounts of the XFP-tagged fusion proteins were imaged live at 37°C at different time points postwarming with an inverted Zeiss 200 M spinning-disc microscope, an argon/krypton and helium/neon laser, a Hamamatsu C9100-13 electron-multiplying charge-coupled-device (EM-CCD) camera with a 100× 1.47-numerical-aperture (NA) objective, and a temperature-controlled incubation chamber using Metamorph (Molecular Devices) or with an inverted Zeiss laser scanning confocal microscope (model 510Meta; Carl Zeiss MicroImaging, Inc.) equipped with a heating device, a 100× Zeiss apochromat objective (1.4 NA), and an argon laser (458, 477, 488, and 514 nm) at 30 mW, an HeNe laser (543 nm) at 1 mW, or an HeNe laser (633 nm) at 5 mW.
For loading the cells with AF488-dextran, CV-1 cells were plated into 8-well glass-bottom chambers. After 12 h in culture, AF488-dextran (catalog number D-22910; Invitrogen) was added for 4 h at 37°C. The cells were washed and incubated for a further 20 h at 37°C before fluorescently labeled SV40-AF647 (MOI of ∼10) was added to the cells for 1 h on ice. Unbound virus was washed away, and cells were incubated for another 4 h before being imaged live at 37°C with the microscope settings described above.
For immunofluorescence detection of cellular proteins after incubation with fluorescent virus, cells were fixed with 4% formaldehyde solution in PBS and either permeabilized in PBS containing 3% BSA (wt/vol) and 0.05% (wt/vol) Triton X-100 (TX-100) for staining with anti-EEA1 (1:500) or permeabilized with PBS containing 0.1% (wt/vol) saponin and 3% BSA for staining with anti-LAMP1 (1:200) antibody, followed by secondary antibodies coupled to AF488, AF594, or AF647 (Invitrogen). Samples were mounted using ImmuMount (Thermo Shandon), and imaging was performed with a Zeiss LSM 510 Meta confocal microscope system. Images were processed by using Image J (National Institutes of Health) or Adobe Photoshop (Adobe Systems).
Quantification of colocalization in fixed and live cells.
Quantification of the colocalization of the virus in live CV-1 cells expressing XFP-tagged constructs was performed by taking single confocal slices with an inverted Zeiss 200 M spinning-disc microscope (described above) at the indicated time points and analyzing the images with a MATLAB-based colocalization program (Peter Horvath, Light Microscopy Center, ETH Zurich). The program considered a virus to colocalize with a given marker if at least 50% of the virus overlapped with the marker and if the marker signal exhibited a clear local maximum below the virus at the same time. Quantification of the colocalization of the virus with LAMP1 and EEA1 in fixed cells was done by using images taken with the CellR wide-field microscope system equipped with an inverted Olympus IX80 microscope using a 100× 1.47-NA oil UplanSApo objective. Samples were illuminated with a xenon lamp (MT20 CellR unit) and recorded with a Hamamatsu Orca ER camera. Five z stacks were taken for each cell, and the intensity was projected and analyzed with the MATLAB-based colocalization program.
All samples within one experiment were acquired with the same microscope settings using Metamorph software (Molecular Devices), the LSM 510 software package (Carl Zeiss MicroImaging, Inc.), or the CellR software version from the manufacturer.
Internalization assay.
SV40-AF488 (MOI of ∼10) was added to CV-1 cells, transfected with Rab7-monomeric RFP (mRFP) to visualize the cell boundaries, on ice for 1 h. Subsequently, cells were warmed to 37°C in the presence of 5% CO2. The fluorescent signal of the dye was recorded at 4 h postwarming for live cells at 37°C in 5% CO2 with a Zeiss 200 M spinning-disc confocal microscope equipped with a highly sensitive Hamamatsu C9100-13 EM-CCD camera. For the control samples, images were taken at room temperature 10 min after incubation on ice. A z stack, with each slice being 0.5 μm in thickness, of at least five cells was taken. The addition of trypan blue (0.4% [wt/vol]; Invitrogen) at a dilution of 1:50 immediately shifted the emission spectrum of particles exposed on the cell surface and led to a loss of detectable fluorescence in the 505- to 530-nm channel. Fluorescence images were exported as 12-bit tagged image file format (TIFF) files and merged, and the maximum intensity was projected with Image J (NIH) and quantified with a custom-written colocalization program implemented in MATLAB (Peter Horvath, Light Microscopy Center, ETH Zurich).
Electron microscopy.
For thin-section EM, CV-1 cells plated onto 12-mm coverslips were incubated with 100 nM Baf or 5 μM nocodazole or in the absence of drugs and with SV40 (MOI of 200) in R-medium at 37°C for 2 or 4 h before fixation with 2.5% glutaraldehyde (with 0.05 M sodium cacodylate [pH 7.2], 50 mM KCl, 1.25 mM MgCl2, and 1.25 mM CaCl2) for 60 min at RT, followed by 1.5 h of incubation in 2% OsO4 on ice. Dehydration, embedding, and thin sectioning were performed as previously described (28). For negative staining, 0.4-μm mesh copper grids were coated with a 4-nm carbon film. A 10-μl sample containing 0.5 mg/ml SV40 was added for 30 s, drained of excess liquid, and stained for an additional 30 s with 2% uranyl acetate in distilled water. After transmission electron microscopy (Zeiss EM 91 microscope), images were exported as 8-bit TIFF files.
RNAi screen.
An RNA interference (RNAi) screen of SV40 infection was performed with siRNAs against 108 selected genes encoding proteins that play a role in clathrin-dependent and -independent endocytosis, signaling, endocytic membrane trafficking, as well as cytoskeletal organization.
HeLa cells were transfected in 96-well black optical-bottom plates (Nunc) independently with three nonoverlapping targeting siRNAs per gene (Qiagen) (see Table S1 in the supplemental material), with each siRNA in duplicate and in two independent experiments. To monitor transfection efficiency, an siRNA causing cell death (Hs_KIF11_6; Qiagen) was used in each plate (66). AllStarsNegative siRNA was added as a nontargeting control in quadruplets per plate. To avoid inhomogeneous cell distribution in the outermost wells, only the inner 60 wells of a 96-well plate were used, while the outermost 36 wells were filled with medium.
Transfection was performed according to a reverse transfection protocol. Briefly, siRNA (20 nM final concentration) and Lipofectamine RNAiMAX (final dilution, 1:1,000; Invitrogen) were prediluted in Opti-MEM (Invitrogen), spotted onto 96-well plates, and incubated for at least 10 min at RT to allow siRNA-lipid complex formation. A total of 1,300 HeLa cells per well in DMEM supplemented with 10% FCS and GlutaMAX (Invitrogen) were added to the lipid-siRNA complex and incubated for 72 h at 37°C. Plates that showed an uneven cell distribution or insufficient cell death in AllStarsDeath positive-control wells were discarded.
Cells were washed once with PBS and inoculated with SV40 suspension in R-medium (RPMI medium, pH 6.8) at an MOI resulting in 10 to 30% infected cells for 120 min at 37°C. The virus inoculum was replaced by full medium (DMEM), and the cells were incubated for an additional 22 h at 37°C. After fixation with formaldehyde (4% final concentration), the cells were washed, permeabilized (PERM buffer [0.1% Triton X-100, 3% BSA, PBS] filtered through a 0.4-μm membrane to remove fibers), and incubated with a 1:1,000 dilution of primary antibody 1605 in PERM buffer against the small T antigen of SV40 for 120 min. Plates were washed with PBS, and a dilution of secondary antibody (1:1,000 dilution of goat anti-mouse antibody coupled to Alexa Fluor 488; Invitrogen) and Hoechst 33258 dye (1:10,000; Invitrogen) in PERM buffer was applied for 30 min.
Sixteen images per well for each channel (nucleus/Hoechst 33258 and T antigen/Alexa Fluor 488) were acquired on a Pathway 855 automated microscope station (Becton Dickinson) using a 10× objective (Olympus) and a laser-based autofocus every second image. Cell numbers (CN) and raw infection indices (rawII) for each well were determined using a MATLAB-based infection scoring procedure (Mathworks). Since cell density affects SV40 infection, the effect of cell density on the infection index was determined with an independent checkerboard. Briefly, various amounts of HeLa cells per well, transfected with AllStarsNegative siRNA, were infected with SV40 under screening conditions. From the obtained infection indices for each cell density, a polynomial regression function, normII = f(CN), was calculated. Density-corrected relative infection indices (corrRII) for each well of the screen were determined with the following equation:
CN and corrRII data were further processed by using the RNAi screen analysis software package cellHTS2 (7). Both features were independently normalized against the AllStarsNegative control within each plate, followed by the z-score calculation (zCN and zII, respectively) and standardization over all replicate experiments essentially as described in the cellHTS2 software manual (http://bioconductor.org/packages/2.5/bioc/html/cellHTS2.html). A cell number cutoff of zCN′target siRNA ≥ −3 (about 500 imaged cells per well) was applied to discard all siRNAs that strongly reduced the cell number. From the remaining siRNAs, those siRNAs whose z score of infection was outside a bandwidth of 5-fold the standard deviation of all AllStarsNegative control siRNA z scores [zII′target siRNA > |5 × SD(zII′AllStarsNegative)|] were considered “significant.” Genes affecting SV40 infection (“hits”) were defined by at least 2 significant targeting siRNAs decreasing virus infection. The change of infection for each hit as a percentage of the negative control (100%), shown in Table 1, was approximated from the mean zII of all significant siRNAs of this particular hit based on a logarithmic regression of the siRNA z scores of infection to their corresponding infection index averaged over all replicate experiments.
Table 1.
Thirty hits from the SV40 siRNA infection screena
| Gene | Description | Protein class | Function | Reduction of infection (%) |
|---|---|---|---|---|
| RAB5C | RAB5C, member of the RAS oncogene family | Small GTPase | Endosomal protein sorting | −69.9 |
| RAB4A | RAB4A, member of the RAS oncogene family | Small GTPase | Endosomal protein sorting | −68.1 |
| CFL1 | Cofilin 1 (nonmuscle) | Actin-binding protein | Cytoskeletal reorganization | −67.5 |
| CBLC | Cas-Br-M (murine) ecotropic retroviral transforming sequence c | E3 ubiquitin protein ligase | Endosomal protein (EGF) sorting | −64.4 |
| PAK2 | P21 protein (Cdc42/Rac)-activated kinase 2 | Serine/threonine protein kinase | Cytoskeletal reorganization | −63.7 |
| NCK2 | NCK adaptor protein 2 | Adaptor protein | EGF signaling/cytoskeletal reorganization | −63.3 |
| EHD1 | EH domain-containing 1 | Adaptor protein | EGF signaling/cytoskeletal reorganization | −63.3 |
| ARHGAP10 | Rho GTPase-activating protein 10 | GTPase-activating protein | Cytoskeletal reorganization | −62.4 |
| STX5A | Syntaxin 5 | SNARE protein | Vesicle fusion/endosome-to-TGN transport | −61.3 |
| RAB5A | RAB5A, member of the RAS oncogene family | Small GTPase | Endosomal protein sorting | −60.8 |
| CBL | Cas-Br-M (murine) ecotropic retroviral transforming sequence | E3 ubiquitin protein ligase | Endosomal protein (EGF) sorting | −60.4 |
| UBASH3A | Ubiquitin-associated and SH3 domain-containing A; TULA | Adaptor protein | Endosomal protein sorting | −58.3 |
| RAB5B | RAB5B, a member of the RAS oncogene family | Small GTPase | Endosomal protein sorting | −56.2 |
| PRKCG | Protein kinase C gamma | Serine/threonine protein kinase | Protein signaling | −55.5 |
| HGS | Hepatocyte growth factor-regulated tyrosine kinase substrate | Adaptor protein | Endosomal protein sorting | −55.5 |
| GRB2 | Growth factor receptor-bound protein 2 | Adaptor protein | EGF signaling | −55.4 |
| DNM2 | Dynamin 2 | Small GTPase | Vesicle scission | −55.4 |
| ARRB2 | Arrestin beta 2 | Adaptor protein | Protein sorting from Golgi complex to endosome | −54.9 |
| NEDD4L | Neural precursor cell expressed, developmentally downregulated 4-like | E3 ubiquitin protein ligase | Endosomal protein (EGF) sorting | −53.9 |
| GGA2 | Golgi-associated, gamma-adaptin ear-containing, Arf-binding protein 2 | Adaptor protein | Protein sorting/TGN-to-lysosome transport | −53.2 |
| PAK3 | P21 protein (Cdc42/Rac)-activated kinase 3 | Serine/threonine protein kinase | Cytoskeletal reorganization | −52.9 |
| PIK3C3 | Phosphoinositide-3-kinase, class 3 | Phosphoinositide-3-kinase | Endosomal protein sorting | −52.8 |
| NEDD4 | Neural precursor cell-expressed, developmentally downregulated 4 | E3 ubiquitin protein ligase | Endosomal protein (EGF) sorting | −52.2 |
| RAB1A | RAB1A, member of the RAS oncogene family | Small GTPase | Regulator of vesicle traffic from ER to Golgi complex | −51.5 |
| DAB2 | Disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila melanogaster) | Adaptor protein | Endosomal protein sorting | −51.3 |
| DYNC2H1 | Dynein, cytoplasmic 2, heavy chain 1 | Motor protein | Vesicle transport | −51.3 |
| RAB22A | RAB22A, member of the RAS oncogene family | Small GTPase | Endosomal protein sorting | −49.8 |
| PRKCZ | Protein kinase C zeta | Serine/threonine protein kinase | Protein signaling | −49.2 |
| SNX15 | Sorting nexin 15 | Sorting nexin | Endosomal protein sorting | −48.2 |
| VIL2 | Villin-2; ezrin | Actin-binding protein | Cytoskeletal reorganization | −47.6 |
See also Fig. S1 and S2 in the supplemental material. EGF, epidermal growth factor; TGN, trans-Golgi network; Arf, ADP-ribosylating factor.
RESULTS
SV40 infection depends on endosomal proteins.
To identify cellular factors involved in SV40 infection, we performed a targeted siRNA silencing screen with HeLa cells against a selection of 108 cellular proteins known to be involved in various aspects of endocytosis (see Fig. S1A in the supplemental material). Table S1 in the supplemental material shows the full list of proteins targeted and the siRNAs used. Materials and Methods provides a detailed description of the experimental procedure. Three nonoverlapping siRNAs were used against each gene. Seventy-two hours after siRNA transfection, cells were infected with SV40, and after a further 24 h, they were fixed and subjected to Hoechst staining to visualize all nuclei and indirect immunofluorescence using antibodies against the SV40 T antigen to identify the nuclei of infected cells (Fig. S1D and S1E) (43).
Proteins were defined as hits when at least two of the three siRNAs showed a significant decrease or increase in the number of infected cells, i.e., when the average z score of infection was at least five times the standard deviation of the z scores of all negative-control siRNAs (see Fig. S1C in the supplemental material). Typically, this meant a reduction in infected cells by 50% or more. Figure S1B in the supplemental material shows the range of effects that transfection with siRNAs had on the cell number and on the infection index in the full set of samples analyzed. Also shown is the effect of a nontargeting siRNA control (AllStarsNegative) and a transfection control (Hs_KIF11_6/EG5; Qiagen), which were present in every plate. Of the 324 siRNAs used, 26 were found to be toxic, judging by a dramatic drop in cell number. These were not considered in our analysis.
A total of 30 proteins were identified as hits (Table 1). They fell into many different protein classes. However, it was evident that a large number (18 in total) had functions associated with endosomes. The endosome-related hits included Rab5a, Rab5b, Rab5c, Rab4a, Rab22a, the GTPase dynamin-2 (DNM2), and hepatocyte-related substrate (Hrs or Hgs). The knockdown levels of some of the proteins were determined by a quantitative analysis of Western blots. For example, the expression of the proteins Rab4a, Rab5a, Hrs, and DNM2 was reduced to less than 17% for the best siRNAs (see Fig. S2 in the supplemental material).
Further studies are needed to validate the hits and to elucidate the roles that the 30 proteins play in infection. However, the finding that almost two-thirds of the hits were intimately associated with the regulation and function of classical endosomal compartments implied that endosomes play an important role in SV40 infection.
SV40 enters endosomes.
To analyze to what extent the incoming virus entered endosomes, indirect immunofluorescence microscopy, quantitative live-cell imaging, and electron microscopy (EM) were employed. Imaging of live cells was used for most of the colocalization and quantitation experiments because we recently found that many LEs and endolysosomes in CV-1 cells burst and lose their contents during fixation with the formaldehyde-based fixatives used for immunofluorescence (25). Moreover, the rounded shape of late endosomal compartments familiar from EM and live-cell imaging is often lost. Compare, for example, the stainings of LAMP1, a marker for LEs and lysosomes, in Fig. 2A (live) and Fig. 1F (fixed).
Fig. 2.
Colocalization of internalized SV40 with EE and LE markers (see also Movie S4 in the supplemental material). (A and B) SV40-AF647 was added to CV-1 cells transfected with Rab7-mRFP and LAMP1-EGFP or Rab9-EYFP and imaged with a spinning-disc confocal microscope at 153 min postwarming. (C) Fluorescently labeled SV40 internalized into CV-1 cells transfected with the constitutively active mutant Rab5Q79L-EGFP and imaged live with a confocal microscope at 160 min postwarming. (D) Fluorescently labeled dextran was added to CV-1 cells for 2 h, incubated overnight, and subjected to SV40-AF647 for binding on ice for 2 h. At 240 min after warming to 37°C, cells were imaged with a confocal microscope.
Fig. 1.
Internalized SV40 colocalizes with endosomal markers (see also Movies S1 to S3 in the supplemental material). (A) CV-1 cells were incubated with SV40-AF488 for 120 min at 37°C. Cells were mixed and immunostained with antibodies against the EE marker EEA1. A confocal section was imaged using the Zeiss LSM 510 confocal microscope system. (B to D) Fluorescently labeled SV40 was added to CV-1 cells transfected with different XFP-tagged endosomal markers and imaged live with a spinning-disc confocal microscope (except C, which was imaged with the Zeiss LSM 510 system) at the indicated time points. (E) Percentage of viruses colocalizing with the markers Rab7-EGFP and Rab5-mRFP at different times postwarming. It was calculated from images such as those shown in D. Error bars are standard errors of the means (SEM) of data from each time point for 5 to 15 cells from three different experiments, with an average of 50 to 100 particles per cell. (F) Same as A, but cells were incubated with SV40-AF594 and stained against the LE marker LAMP1.
The fixation artifacts make the visualization and quantitation of cargo such as viruses in late endocytic organelles by immunofluorescence difficult and irreproducible. This may, in fact, explain in part why in the past we have failed to observe viruses in LEs.
To avoid adverse effects of overexpression, we limited our observations to cells that expressed reporter constructs at very low levels. This was made possible by the use of a spinning-disc confocal microscope equipped with a highly sensitive EM-CCD camera. Colocalization was quantified by using a MATLAB-based algorithm. Viruses were detected by pixel size and intensity. When 50% of the virus pixels or more overlapped with the objects (endosomal vesicles) in the second channel, viruses were counted as colocalizing. For quantification, 50 to 100 viruses (per cell) in 5 to 15 cells from at least three different experiments were analyzed per time point.
The colocalization of fluorescently labeled SV40 with EE markers was reproducibly observed both by indirect immunofluorescence microscopy of fixed cells and by live-cell imaging of cells expressing XFP-tagged EE markers. Figure 1A shows a confocal section of a fixed cell in which the virus particles are green and the EEA1-containing organelles are immunostained in red. Many of the EEA1-positive endosomes contained virus particles. The green spots at the periphery of the cell represent viruses that have not been internalized. After 90 min, the fraction of SV40 cells colocalizing with endogenous EEA1 amounted to 7.6% ± 1.7% of the total cell-associated viruses.
Figure 1B shows a similar confocal section (although closer to the plasma membrane plane) with an extensive colocalization of red fluorescent virus with green Rab5-EGFP in a living, unfixed cell. Viruses could also be detected in Hrs-EGFP-positive organelles in live cells expressing this ESCRT 0 component (Fig. 1C).
The Rab5-EGFP-positive EEs containing SV40 were often small and peripheral, whereas EEs containing EEA1 and Hrs-EGFP were larger and located in the perinuclear region of the cytoplasm (Fig. 1A and C). In live cells, SV40 particles could be seen to move together with Rab5- and Rab7-mRFP-containing organelles in the cytoplasm, indicating that colocalization was not coincidental (Fig. 1D and see Movies S1 to S3 in the supplemental material). When quantified after 150 min, 16.8% ± 3.6% of the total cell-associated virus particles colocalized with Rab5-mRFP, in agreement with previously reported observations (42). The fraction of virus in EEs remained unchanged for at least another 2 h (Fig. 1E).
SV40 enters late endosomes.
Although the preservation of LEs in fixed cells was poor, as discussed above, virus particles (red) could be seen in organelles containing endogenous, immunostained LAMP1 (green) (Fig. 1F). In live cells expressing LAMP1-EGFP, Rab9-enhanced yellow fluorescent protein (EYFP), or Rab7-mRFP, the colocalization was much clearer and more extensive (Fig. 1D and 2A and B). The LEs and endolysosomes were round and displayed ring-like membrane fluorescence. The labeled SV40 could be seen to move together with the endosomes through the cytoplasm (see Movies S3 and S4 in the supplemental material).
In a previous publication, we had observed that infection is inhibited by the expression of constitutively active Rab5Q (Rab5Q79L) (42). It was recently shown that the overexpression of Rab5Q causes an efficient block in LE maturation at the level of the maturing endosome (65). To identify the intracellular location of SV40 in CV-1 cells overexpressing Rab5Q, we added SV40-AF647. At 2 h after the addition of virus, most virus particles localized to the sorting-defective, enlarged endosomes (Fig. 2C).
Quantitation after fixation and immunofluorescence showed that the colocalization of SV40 particles with endogenous LAMP1 peaked at 15.6% ± 1.9% at 90 min postwarming. However, in live cells where quantitation was not affected by the fixation-induced organelle disruption, the level of colocalization with LAMP1-EGFP and Rab7-EGFP was much higher. In Rab7-EGFP-expressing cells, it increased linearly with time and reached 49.9% ± 10.1% of the cell-associated virus after 4.5 h (Fig. 1E).
To confirm that SV40 transport to late endosomal compartments also occurred in untransfected cells, we preloaded the late endosomes and lysosomes by exposing CV-1 cells to fluorescein isothiocyanate (FITC)-dextran for 4 h, followed by a 20-h incubation in the absence of the fluorescent fluid-phase marker. SV40-AF647 was then allowed to enter the cells for 4 h. When the cells were viewed by confocal microscopy, we clearly observed the majority of viruses within FITC-dextran-containing late endosomes and lysosomes (Fig. 2D).
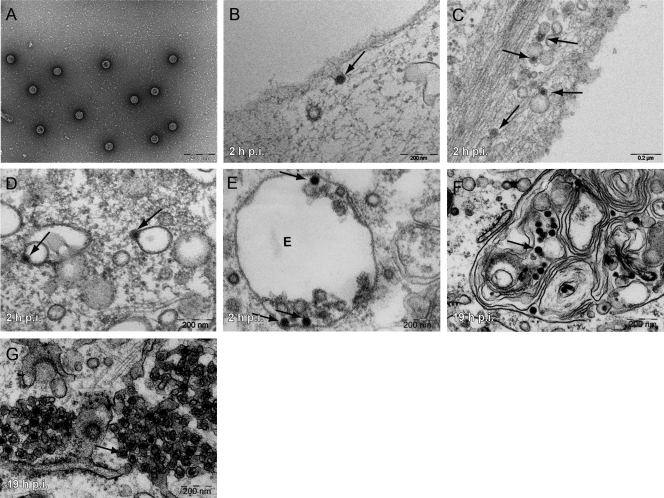
When EM was performed 2 h after the addition of virus to cells, SV40 particles (Fig. 3A) were seen in smooth-walled, close-fitting invaginations of the plasma membrane (Fig. 3B), in membrane-bounded vesicles in the peripheral cytoplasm (Fig. 3C), in endosome-like structures devoid of intralumenal vesicles (Fig. 3D) (26, 28), and in multivesicular body (MVB) late endosomes (Fig. 3E). At 19 h, they had reached smooth regions of the ER (Fig. 3G), but some were present in multilamellar bodies (MLBs) (Fig. 3F). At no time were virus particles seen in cisternae of the Golgi complex or trans-Golgi network.
Fig. 3.
SV40 enters endosomal structures and the ER. (A) Purified SV40 visualized by negative staining. (B to G) Purified viruses were incubated with CV-1 cells at different times postwarming, fixed, embedded in plastic, sectioned, and imaged. Arrows point toward single virus particles. Viruses are seen as single particles (A) and in invaginations of the plasma membrane (B), endosomal structures (C to F), and smooth regions of the ER (G). The letter E in E stands for “endosome.” Scale bars indicate 200 nm.
Taken together, the data indicated that the internalized SV40 entered peripheral and perinuclear EEs and also LEs, many of which were present as MVBs and MLBs. After 4 h, when the first virus particles enter the ER (53), more than 80% of the internalized viruses were associated with endosomal compartments. The reason that the EE-associated pool remained relatively small (less than 20% of the total cell-associated virus) was most likely the slow and asynchronous arrival of virus from the plasma membrane combined with a continuous movement of viruses from EEs to LEs. A similar progression from EEs to LEs was recently observed for mPy, which also transits via endosomes to the ER (46).
Elevation of vacuolar pH blocks SV40 infection.
The association of SV40 with endosomes raised the question of whether this trafficking route plays a role in productive infection. First, we analyzed the effects of inhibitors known to raise the lumenal pH of endosomes and lysosomes. In the presence of NH4Cl, the number of T-antigen-expressing cells was reduced to 15% of the control, as determined by flow cytometry. With monensin, a carboxylic ionophore, and bafilomycin A1 (Baf), an inhibitor of the vacuolar ATPase responsible for endosome acidification, infection was almost completely inhibited (Fig. 4A).
Fig. 4.
Acid dependence of internalization and infection. (A) CV-1 cells were pretreated for 1 h with 100 nM bafilomycin A1 (Baf), 20 mM NH4Cl, or 10 μM monensin (Mon) and were infected with SV40 for 24 h (MOI of 1) in the continued presence of the drugs. The infection level was determined by the immunostaining of the SV40 T antigen and flow cytometry and was normalized to drug-free controls. (B) Cells were infected with SV40 (MOI of 1), and Baf was added to the cells at different times. Infection was determined as described above for A. (C) Same as A but with mouse fibroblast caveolin-1 knockout cells and Baf. Data shown represent means ± SEM of data from three independent experiments, each with triplicate samples (A to C). (D) Percentages of SV40 particles internalized in the presence of NH4Cl, Baf, and Mon. Error bars indicate SEM of each time point for 5 to 15 cells from three different experiments, with an average of 50 to 100 particles per cell. (E to G) EM images of CV-1 cells incubated with SV40 particles in the presence of Mon (E and F) and Baf (G and H) at different times postwarming. Arrows point toward virus particles. Scale bars indicate 200 nm.
When Baf was added at different times after the addition of SV40, the sensitive step was found to occur with a midpoint of 90 min after cell warming. This fast-acting inhibitor thus affected an early step in SV40 infection coinciding approximately with virus internalization (Fig. 4B). Baf also inhibited SV40 infection in fibroblasts derived from caveolin-1 knockout mice, indicating that the caveolin-independent infection pathway also required acidification (Fig. 4C). The drugs did not affect cell viability. Overall, cell morphology was normal except for the swelling of endocytic vacuoles observed with NH4Cl and monensin (see below).
To analyze which steps in the infectious pathway were blocked, we employed a quantitative internalization assay based on a specific fluorophore, AF488, covalently attached to SV40 particles. AF488 can be quenched by the addition of the membrane-impermeable dye trypan blue (63) (Fig. 5).
Fig. 5.
Internalization assay. (A) CV-1 cells were transfected with Rab7-mRFP 20 h before the addition of fluorescently labeled SV40. Cells were incubated in the cold for 1 h, unbound virus was washed away, and cells were immediately transferred to the microscope. Five z sections of each cell were imaged with a spinning-disc confocal microscope before (10 min after washing) and after (15 min after washing) the addition of trypan blue to quench extracellular fluorescence. (B) Same as A, but cells were incubated at 37°C for 240 min before quenching (at 244 min). (C) Same as A, but cells were preincubated with Baf 1 h before and during incubation with the virus for 240 min at 37°C.
The results showed that in control cells, 80% of AF488-labeled virus particles were internalized within 4 h. When Baf, NH4Cl, or monensin was present, only 20% of the virus particles were internalized (Fig. 4D). The block in SV40 internalization caused by monensin was reported previously (54). EM confirmed that in monensin-treated cells at as late as 22 h postwarming, most SV40 particles remained trapped in plasma membrane invaginations (Fig. 4E). The minor populations of particles internalized were in MVBs and MLBs (Fig. 4F).
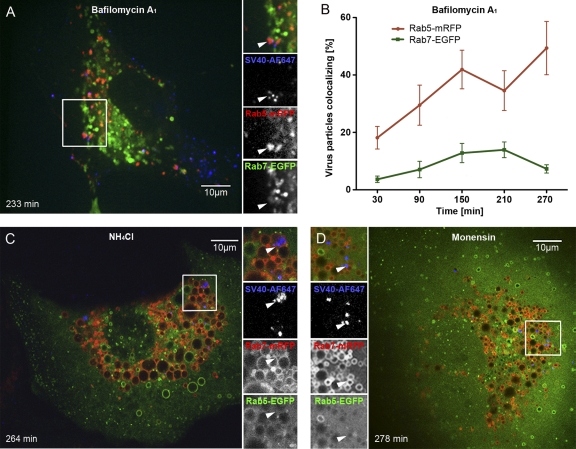
Also, in Baf-treated cells, the majority of SV40 particles remained on the plasma membrane (Fig. 4G), but in this case, the few particles that were internalized were seen in endosomal structures devoid of intralumenal membranes (Fig. 4H). The result could be confirmed by live-cell microscopy: the few internalized SV40-AF647 particles colocalized exclusively with Rab5-mRFP in Baf-treated cells, while in monensin- and in NH4Cl-treated cells, the viruses colocalized mainly with Rab7-mRFP (Fig. 6 and see Movies S5 and S6 in the supplemental material). Baf caused a loss of the round, ring-like Rab5 staining observed for control cells, whereas NH4Cl and monensin caused an expansion of both Rab5- and Rab7-positive vacuoles (Fig. 6A, C, and D).
Fig. 6.
SV40 trafficking is inhibited by an elevation of the vacuolar pH (see also Movies S5 and S6 in the supplemental material). (A, C, and D) SV40-AF647 was incubated with cells transfected with XFP-tagged proteins, and 100 nM bafilomycin A1 (Baf), 20 mM NH4Cl, or 10 μM monensin was added; cells were imaged by confocal microscopy at different time points postwarming. Note that NH4Cl and monensin cause a swelling of endosomes and lysosomes, and Baf leads to a loss of the round appearance of endosomes. (B) Percentage of viruses colocalizing with the markers Rab7-EGFP and Rab5-mRFP at different time points postwarming in the presence of 100 nM Baf, calculated from images such as those shown in A. Error bars are SEM of data from each time point for 5 to 15 cells from three independent experiments, with an average of 50 to 100 particles per cell.
Taken together, the data showed that infection was inhibited by all three pH perturbants. While the binding of viruses to the cell surface was unaffected, internalization was strongly reduced. NH4Cl and monensin allowed the few viruses that were internalized to enter LEs and MVBs. Baf seemed to interfere with endosome maturation at an earlier stage, with the virus arrested in modified EEs. These observations were consistent with the notion that SV40 uses the endosomal pathway for infection and that an elevation of the pH in acidic organelles can introduce multiple blocks in the infectious pathway (5, 6, 9, 61).
Molecular requirements for passage to the ER via LEs.
The movement of cargo from EEs to LEs relies on a complicated endosomal maturation process that involves a multitude of cellular factors (39). Among them, microtubules are important because they mediate the movement of the maturing endosomes from the periphery to the perinuclear cytoplasm and regulate fusion with lysosomes (3, 5, 6, 13). The disruption of microtubules by nocodazole efficiently inhibits SV40 infection but does not affect internalization (Fig. 7A and F). The virus particles do not reach the ER (44).
Fig. 7.
Molecular requirements for SV40 passage through the late endosomal pathway and to the ER. (A) Percentage of SV40 particles internalized in the presence of 5 μM nocodazole (Noc) compared to mock-treated controls. (B and C) EM images of SV40 particles added to CV-1 cells in the presence of 5 μM nocodazole for 4 h. Viruses are indicated by black arrows. (D) SV40-AF647 was incubated with CV-1 cells transfected with Rab7-mRFP and Rab5-EGFP in the continuous presence of 5 μM nocodazole. A confocal slice of a representative cell imaged after 230 min is shown. (E) Percentage of virus particles colocalizing with Rab7-mRFP and Rab5-EGFP at different time points postwarming in the presence of 5 μM nocodazole, as quantified from images such as those shown in D. Error bars are SEM of data from each time point for 5 to 15 cells from three different experiments, with an average of 50 to 100 particles per cell. (F) CV-1 cells incubated with 5 μM nocodazole were infected with SV40 at an MOI of 5 for 24 h. The drug was washed out at 8 h postwarming, or different drugs were added after the washout. The infection level was determined by the immunostaining of the SV40 T antigen and flow cytometry normalized to drug-free controls. Shown are means ± SEM of data from three independent experiments, each with triplicate samples. Abbreviations: Baf, bafilomycin A1; Mon, monensin; BFA, brefeldin A; OV, orthovanadate; Gen, genistein; OA, okadaic acid; CalC, calphostin C; Dyn, dynasore. (G) Percentage of SV40 particles internalized in the presence of 80 μM dynasore compared to mock-treated controls.
At 4 h postwarming, EM of nocodazole-treated cells showed viral particles in endosome-like organelles (Fig. 7B) and in MVBs (Fig. 7C). With live-cell microscopy, SV40-AF647 was present in vacuoles positive for both Rab5-EGFP and Rab7-mRFP (Fig. 7D). Over time, we could observe that the majority of internalized virus particles (Fig. 7A and E) accumulated in two types of organelles: Rab5-positive endosomes devoid of Rab7, and Rab5- and Rab7-positive maturing endosomes in which the viruses were located in the Rab7-positive domains (Fig. 7D). The latter organelles corresponded to previously described intermediates in LE maturation arrested by the inhibition of microtubule function (3, 5, 62).
Being reversible, the nocodazole effect provided a convenient method to divide the entry pathway into two phases: early events up to and including the maturing endosomes and late events during which the viruses moved to LEs and eventually to the ER. When CV-1 cells were incubated with nocodazole for 8 h after the addition of SV40 followed by a nocodazole washout, infection recovered to 75% of control levels (Fig. 7F) (53). When nocodazole was replaced at the time of washout with other inhibitors, it was possible to study the inhibitor sensitivity of steps in the entry pathway downstream of the nocodazole block (Fig. 7F).
Among the drugs that inhibited infection after the nocodazole washout were the acidification inhibitors monensin and Baf. The inhibitory effects of genistein (a general tyrosine kinase inhibitor), orthovanadate (OV) (an inhibitor of tyrosine phosphatases), and okadaic acid (OA) (an inhibitor of Ser/Thr phosphatases) pointed to a role for tyrosine and Ser/Thr phosphorylation. Judging by the strong inhibitory effect of brefeldin A (BFA), Arf proteins seemed to play a role both early and late in the pathway, consistent with previously reported data (41, 50). In addition to disrupting vesicle transport within the Golgi complex and from the Golgi complex to the ER, BFA is known to induce the tubulation of endosomes and inhibit cargo transport from EEs to endolysosomes (33, 58). The finding that calphostin C failed to inhibit infection after the nocodazole washout suggested that the role of protein kinase C (PKC) (two hits in the siRNA screen) was upstream of the nocodazole block.
One of the inhibitors that blocked infection in a postnocodazole step was dynasore, an inhibitor of DNM2 (36). At a concentration that did not cause cell toxicity, it caused more than an 80% reduction in infection (Fig. 7F). DNM2 was also a strong hit in the siRNA infection screen (Table 1). The overexpression of a dominant negative DNM2 construct was shown previously to reduce infection (45). However, the quantitative internalization assay showed that in the case of SV40, it was not the endocytic uptake that was inhibited by dynasore but rather a downstream event (Fig. 7G).
Taken together, the results indicated that the regulation of the latter part of the pathway of intracellular virus transport is complex and dependent on numerous cellular factors. It was apparent that some of the inhibitors such as BFA, the acidification inhibitors, and genistein affected processes both before and after the nocodazole block. The effects seen were consistent with the passage of the infectious virus through the LEs on its way to the ER.
DISCUSSION
The most important conclusion from our studies is that SV40 passes through a series of classical endocytic organelles during infectious entry into host cells. These include EEs, maturing hybrid endosomes, LEs with the properties of multivesicular and multilamellar bodies, and, most likely, endolysosomes. The pathway is schematically shown in Fig. 8. Passage occurs slowly and nonsynchronously, with the arrival of viruses in the ER several hours after the initial internalization (53).
Fig. 8.
Model of infectious SV40 entry into CV-1 cells. SV40 binds to its receptor (GM1), partitions into lipid rafts, and induces internalization from the plasma membrane either by a caveola-mediated or a caveolin-1-independent, lipid raft-dependent endocytosis mechanism. The virus is transported to Rab5-, EEA1-, and Hrs-positive EEs. When these endosomes acquire Rab7, SV40 associates with the Rab7-positive domains. Through endosome maturation, viruses become lumenal components of LAMP1-, Rab9-, and Rab7-positive LEs and eventually endolysosomes. The vacuolar ATPase (v-ATPase) is responsible for the acidification of endosomes and lysosomes. Acidification is required for SV40 internalization and subsequent transport steps. Virus transport to the ER occurs from the late compartments of the endocytic pathway by an unknown mechanism either directly or, less likely, via the Golgi complex. Early and late events in the entry pathway can be blocked by various inhibitors and other perturbants. References to the effects of different drugs reported in the literature are given.
Taken together, the observations failed to support our previous model in which SV40 was routed to the ER via caveolin-1-positive nonendosomal organelles, which we called caveosomes, bypassing the endosomes (44). The main reasons for abandoning the caveosome model at this time are 2-fold. First, the results presented here speak clearly for the endosome-based trafficking of the incoming virus and a role for endosomes in productive infection. Second, we have found in recent studies that caveosomes as originally described are likely to represent modified LEs or endolysosomes (25). We observed that caveolin-rich “caveosomes” arise when caveolar assembly is compromised by the overexpression of caveolin-1 constructs, by the reduction of cholesterol, and by the depletion of cavins. The unassembled caveolin-1 generated under these conditions is ubiquitinated and endocytosed, and it accumulates in intralumenal vesicles (ILVs) within LEs prior to lysosomal degradation. The original experiments that led to the caveosome model for SV40 entry were performed with cells overexpressing caveolin-1 (42, 44). The confusion was enhanced by the lack of appropriate LE markers and the difficulty that we now recognize in the fixation of late endosomal compartments in CV-1 cells with formaldehyde without a loss of contents, including virus particles (25).
A central role for endosomes in SV40 entry was underscored by a cluster of endosomal hits in our targeted siRNA silencing screen in HeLa cells. A decrease in infectivity was observed after depleting proteins such as Rab5a, Rab5b, Rab5c, Hrs, c-Cbl, Rab4a, Rab11b, SNX15, PIK3C3, DNM2, and arrestin beta-2. These proteins are known to play a variety of roles in cargo transport to endosomes, in endosome function, and in endosome regulation (24).
Light microscopy experiments confirmed that more than 80% of the internalized SV40 particles trafficked to Rab5-positive, EEA1-positive, and Hrs-positive EEs as well as to LAMP1-positive, Rab7-positive, Rab9-positive, and FITC-dextran-positive LEs and endolysosomes. EM substantiated the presence of viruses in endosomal structures.
Agents that raise the pH of endocytic organelles were found to inhibit infection. Their main inhibitory effect involved the internalization of bound virus. The mechanisms behind the endocytosis block are not clear, but the effect explains in part the high sensitivity of SV40 infection to pH perturbants. In addition, the fraction of viruses that was internalized (about 20%) failed to move beyond Rab5-positive EEs in the presence of Baf and beyond LEs in the presence of monensin. This was not unexpected, since a block in endosome maturation and cargo transport to lysosomes is one of the consequences often observed after interfering with the acidification of endosomes (5, 6, 9, 54, 61).
The inhibitory effect of monensin on the internalization and intracellular trafficking of SV40 was observed previously by others (54). In addition, it was recently reported that mPy and two human polyomaviruses, BK and JC viruses, require acidification for infection (4, 27, 32). However, in two previous reports, investigators failed to observe an inhibition of SV40 infection in the presence of acidification perturbants (4, 60). The difference may be technical: in our experiments we took into consideration that the entry process is slow, and we extended the drug treatment accordingly. We also made certain that the lysosomotropic weak base NH4Cl remained active throughout the experiment by including efficient buffers in the medium to prevent a drop in the medium pH and, thus, a decrease in the effectiveness of inhibition (29).
The finding that the sorting of SV40 deep into the degradative branch of the endocytic pathway is a requirement for infection was best illustrated by the effect of perturbations that interfered with endosome maturation. Previously (42), we had observed that infection is inhibited by the expression of constitutively active Rab5Q, which causes an efficient block in LE maturation at the level of the maturing endosome (18, 65). In addition, we observed that infection was inhibited when the formation of intralumenal vesicles was blocked by depleting Hrs and other components of the ubiquitin-dependent machinery necessary for LE formation (Table 1) (49). Furthermore, infection was inhibited when endosomal trafficking along microtubules was disturbed by nocodazole and when acidification inhibitors were used (54). SV40 shares its dependence on endosome maturation with a number of late-penetrating viruses such as influenza A virus, rhinovirus, mPy, bunyavirus, and parvovirus (19, 35, 39, 67).
Several inhibitors and perturbants were found to prevent infection after a nocodazole block (Fig. 7F). The Baf and monensin sensitivity demonstrated a requirement for acidification late in the pathway in addition to its role during early entry (5, 6, 9, 61). The inhibitory effect of BFA confirmed a late role for Arf proteins (41, 50, 64).
Kinases and phosphatases apparently play a role in both early and late stages of SV40 entry (43). OV, an inhibitor of tyrosine phosphatases that dramatically increases the internalization of SV40 (45), blocked infection late in the pathway, most likely by interfering with SV40 transport from endosomes to the ER. The finding that DNM2 was needed in a postnocodazole step was shown by the inhibitory effects of dynasore. In addition to its well-characterized role in primary endocytic processes, DNM2 was reported previously to support the transport of cholesterol and other cargo from endosomes to the ER and from endosomes to the Golgi complex (31, 40, 51).
The entry of SV40 shares many features with those of other polyomaviruses. The human BK virus, JC virus, and MCPyV as well as the mouse polyomavirus seem to require gangliosides for infectivity (14, 15, 21, 22, 30, 34, 46, 55, 59). While the entry pathways of the most recently discovered human polyomaviruses have not yet been investigated in detail, mPy, BK virus, and JC virus are known to enter endosomes and depend on acidification (4, 27, 32, 38, 46, 47). In the case of the well-studied mPy, a role for recycling endosomes and late endosomes was proposed (32, 38, 46). Like SV40, other polyomaviruses are transported to the ER lumen and seem to take advantage of the ER machinery for uncoating and membrane penetration (27, 37, 46).
Why do polyomaviruses use such a complicated entry pathway? One reason is that since their capsids are stabilized by a network of disulfide bonds, they require access to thiol oxidoreductases and chaperones in the ER lumen for initial uncoating (37, 53). Moreover, there is increasing evidence that the membrane penetration of the viruses depends on components of the ER-associated degradation pathway (27, 37, 53). An endosomal pathway to the ER is also used by bacterial toxins such as Shiga and cholera toxins. Like polyomaviruses, these have homopentameric binding subunits that associate with and cluster glycosphingolipid receptors (15, 16, 52). The similarities suggest that multimeric glycosphingolipid binding and association with cholesterol-containing lipid domains provide viruses and toxins an entry ticket not only to endosomes but also, as recently suggested by Qian et al., to an extension of the journey from endosomes to the ER (46).
Supplementary Material
ACKNOWLEDGMENTS
We thank the following colleagues for reagents: Jean Gruenberg, Marino Zerial, Harald Stenmark, and Mark McNiven. We thank Jason Mercer, Yohei Yamauchi, and Alicia Smith for critically reading the manuscript and Roger Geiger for experimental help. We are grateful for the support of the Light Microscopy Center (LMC), especially Peter Horvath, and the RNAi screening center (RISC) of the Department of Biology and the Electron Microscopy Center (EMEZ) of ETH Zurich.
The work was supported by funding from the SystemsX program through LipidX, ETH Zurich, and the ERC. S.E. was supported by a grant from the Boehringer-Ingelheim Fonds and the Roche Research Foundation.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 23 February 2011.
REFERENCES
- 1. Allander T., et al. 2007. Identification of a third human polyomavirus. J. Virol. 81:4130–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson H. A., Chen Y., Norkin L. C. 1996. Bound simian virus 40 translocates to caveolin-enriched membrane domains, and its entry is inhibited by drugs that selectively disrupt caveolae. Mol. Biol. Cell 7:1825–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aniento F., Emans N., Griffiths G., Gruenberg J. 1993. Cytoplasmic dynein-dependent vesicular transport from early to late endosomes. J. Cell Biol. 123:1373–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ashok A., Atwood W. J. 2003. Contrasting roles of endosomal pH and the cytoskeleton in infection of human glial cells by JC virus and simian virus 40. J. Virol. 77:1347–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baravalle G., et al. 2005. Transferrin recycling and dextran transport to lysosomes is differentially affected by bafilomycin, nocodazole, and low temperature. Cell Tissue Res. 320:99–113 [DOI] [PubMed] [Google Scholar]
- 6. Bayer N., et al. 1998. Effect of bafilomycin A1 and nocodazole on endocytic transport in HeLa cells: implications for viral uncoating and infection. J. Virol. 72:9645–9655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boutros M., Bras L. P., Huber W. 2006. Analysis of cell-based RNAi screens. Genome Biol. 7:R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cao H., Thompson H. M., Krueger E. W., McNiven M. A. 2000. Disruption of Golgi structure and function in mammalian cells expressing a mutant dynamin. J. Cell Sci. 113(Pt. 11):1993–2002 [DOI] [PubMed] [Google Scholar]
- 9. Clague M. J., Urbe S., Aniento F., Gruenberg J. 1994. Vacuolar ATPase activity is required for endosomal carrier vesicle formation. J. Biol. Chem. 269:21–24 [PubMed] [Google Scholar]
- 10. Cole C. N., Cozen S. D. 2001. Polyomaviridae: the viruses and their replication, p. 2141–2229 In Knipe D. M., et al. (ed.), Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 11. Damm E. M., et al. 2005. Clathrin- and caveolin-1-independent endocytosis: entry of simian virus 40 into cells devoid of caveolae. J. Cell Biol. 168:477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Drab M., et al. 2001. Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science 293:2449–2452 [DOI] [PubMed] [Google Scholar]
- 13. Driskell O. J., Mironov A., Allan V. J., Woodman P. G. 2007. Dynein is required for receptor sorting and the morphogenesis of early endosomes. Nat. Cell Biol. 9:113–120 [DOI] [PubMed] [Google Scholar]
- 14. Erickson K. D., Garcea R. L., Tsai B. 2009. Ganglioside GT1b is a putative host cell receptor for the Merkel cell polyomavirus. J. Virol. 83:10275–10279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ewers H., et al. 2010. GM1 structure determines SV40-induced membrane invagination and infection. Nat. Cell Biol. 12:11–18 [DOI] [PubMed] [Google Scholar]
- 16. Ewers H., et al. 2005. Single-particle tracking of murine polyoma virus-like particles on live cells and artificial membranes. Proc. Natl. Acad. Sci. U. S. A. 102:15110–15115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Feng H., Shuda M., Chang Y., Moore P. S. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319:1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Flinn R. J., Yan Y., Goswami S., Parker P. J., Backer J. M. 2010. The late endosome is essential for mTORC1 signaling. Mol. Biol. Cell 21:833–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fuchs R., Blaas D. 2008. Human rhinovirus cell entry and uncoating, p. 1–14 In Cheng R. H., Miyamura T. (ed.), Structure-based study of viral replication. World Scientific Publishing, Hackensack, NJ [Google Scholar]
- 20. Gaynor A. M., et al. 2007. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 3:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gilbert J., Benjamin T. 2004. Uptake pathway of polyomavirus via ganglioside GD1a. J. Virol. 78:12259–12267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gilbert J., et al. 2005. Ganglioside GD1a restores infectibility to mouse cells lacking functional receptors for polyomavirus. J. Virol. 79:615–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gruenberg J. 2009. Viruses and endosome membrane dynamics. Curr. Opin. Cell Biol. 21:582–588 [DOI] [PubMed] [Google Scholar]
- 24. Gruenberg J., Stenmark H. 2004. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 5:317–323 [DOI] [PubMed] [Google Scholar]
- 25. Hayer A., et al. 2010. Caveolin-1 is ubiquitinated and targeted to intralumenal vesicles in endolysosomes for degradation. J. Cell Biol. 191:615–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hummeler K., Tomassini N., Sokol F. 1970. Morphological aspects of the uptake of simian virus 40 by permissive cells. J. Virol. 6:87–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang M., Abend J. R., Tsai B., Imperiale M. J. 2009. Early events during BK virus entry and disassembly. J. Virol. 83:1350–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kartenbeck J., Stukenbrok H., Helenius A. 1989. Endocytosis of simian virus 40 into the endoplasmic reticulum. J. Cell Biol. 109:2721–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kielian M. C., Marsh M., Helenius A. 1986. Kinetics of endosome acidification detected by mutant and wild-type Semliki Forest virus. EMBO J. 5:3103–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Komagome R., et al. 2002. Oligosaccharides as receptors for JC virus. J. Virol. 76:12992–13000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lauvrak S. U., Torgersen M. L., Sandvig K. 2004. Efficient endosome-to-Golgi transport of Shiga toxin is dependent on dynamin and clathrin. J. Cell Sci. 117:2321–2331 [DOI] [PubMed] [Google Scholar]
- 32. Liebl D., et al. 2006. Mouse polyomavirus enters early endosomes, requires their acidic pH for productive infection, and meets transferrin cargo in Rab11-positive endosomes. J. Virol. 80:4610–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lippincott-Schwartz J., et al. 1991. Forskolin inhibits and reverses the effects of brefeldin A on Golgi morphology by a cAMP-independent mechanism. J. Cell Biol. 112:567–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Low J. A., Magnuson B., Tsai B., Imperiale M. J. 2006. Identification of gangliosides GD1b and GT1b as receptors for BK virus. J. Virol. 80:1361–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lozach P. Y., et al. 2010. Entry of bunyaviruses into mammalian cells. Cell Host Microbe 7:488–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Macia E., et al. 2006. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell 10:839–850 [DOI] [PubMed] [Google Scholar]
- 37. Magnuson B., et al. 2005. ERp29 triggers a conformational change in polyomavirus to stimulate membrane binding. Mol. Cell 20:289–300 [DOI] [PubMed] [Google Scholar]
- 38. Mannova P., Forstova J. 2003. Mouse polyomavirus utilizes recycling endosomes for a traffic pathway independent of COPI vesicle transport. J. Virol. 77:1672–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mercer J., Schelhaas M., Helenius A. 2010. Virus entry by endocytosis. Annu. Rev. Biochem. 79:803–833 [DOI] [PubMed] [Google Scholar]
- 40. Nicoziani P., et al. 2000. Role for dynamin in late endosome dynamics and trafficking of the cation-independent mannose 6-phosphate receptor. Mol. Biol. Cell 11:481–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Norkin L. C., Anderson H. A., Wolfrom S. A., Oppenheim A. 2002. Caveolar endocytosis of simian virus 40 is followed by brefeldin A-sensitive transport to the endoplasmic reticulum, where the virus disassembles. J. Virol. 76:5156–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pelkmans L., Burli T., Zerial M., Helenius A. 2004. Caveolin-stabilized membrane domains as multifunctional transport and sorting devices in endocytic membrane traffic. Cell 118:767–780 [DOI] [PubMed] [Google Scholar]
- 43. Pelkmans L., et al. 2005. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature 436:78–86 [DOI] [PubMed] [Google Scholar]
- 44. Pelkmans L., Kartenbeck J., Helenius A. 2001. Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat. Cell Biol. 3:473–483 [DOI] [PubMed] [Google Scholar]
- 45. Pelkmans L., Puntener D., Helenius A. 2002. Local actin polymerization and dynamin recruitment in SV40-induced internalization of caveolae. Science 296:535–539 [DOI] [PubMed] [Google Scholar]
- 46. Qian M., Cai D., Verhey K. J., Tsai B. 2009. A lipid receptor sorts polyomavirus from the endolysosome to the endoplasmic reticulum to cause infection. PLoS Pathog. 5:e1000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Querbes W., O'Hara B. A., Williams G., Atwood W. J. 2006. Invasion of host cells by JC virus identifies a novel role for caveolae in endosomal sorting of noncaveolar ligands. J. Virol. 80:9402–9413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Raiborg C., Bache K. G., Mehlum A., Stang E., Stenmark H. 2001. Hrs recruits clathrin to early endosomes. EMBO J. 20:5008–5021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Raiborg C., Malerod L., Pedersen N. M., Stenmark H. 2008. Differential functions of Hrs and ESCRT proteins in endocytic membrane trafficking. Exp. Cell Res. 314:801–813 [DOI] [PubMed] [Google Scholar]
- 50. Richards A. A., Stang E., Pepperkok R., Parton R. G. 2002. Inhibitors of COP-mediated transport and cholera toxin action inhibit simian virus 40 infection. Mol. Biol. Cell 13:1750–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Robinet P., et al. 2006. Dynamin is involved in endolysosomal cholesterol delivery to the endoplasmic reticulum: role in cholesterol homeostasis. Traffic 7:811–823 [DOI] [PubMed] [Google Scholar]
- 52. Romer W., et al. 2007. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature 450:670–675 [DOI] [PubMed] [Google Scholar]
- 53. Schelhaas M., et al. 2007. Simian virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell 131:516–529 [DOI] [PubMed] [Google Scholar]
- 54. Shimura H., Umeno Y., Kimura G. 1987. Effects of inhibitors of the cytoplasmic structures and functions on the early phase of infection of cultured cells with simian virus 40. Virology 158:34–43 [DOI] [PubMed] [Google Scholar]
- 55. Smith A. E., Lilie H., Helenius A. 2003. Ganglioside-dependent cell attachment and endocytosis of murine polyomavirus-like particles. FEBS Lett. 555:199–203 [DOI] [PubMed] [Google Scholar]
- 56. Sonnichsen B., De Renzis S., Nielsen E., Rietdorf J., Zerial M. 2000. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J. Cell Biol. 149:901–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stang E., Kartenbeck J., Parton R. G. 1997. Major histocompatibility complex class I molecules mediate association of SV40 with caveolae. Mol. Biol. Cell 8:47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tooze J., Hollinshead M. 1992. In AtT20 and HeLa cells brefeldin A induces the fusion of tubular endosomes and changes their distribution and some of their endocytic properties. J. Cell Biol. 118:813–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tsai B., et al. 2003. Gangliosides are receptors for murine polyoma virus and SV40. EMBO J. 22:4346–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Upcroft P. 1987. Simian virus 40 infection is not mediated by lysosomal activation. J. Gen. Virol. 678:2477–2480 [DOI] [PubMed] [Google Scholar]
- 61. van Weert A. W., Dunn K. W., Gueze H. J., Maxfield F. R., Stoorvogel W. 1995. Transport from late endosomes to lysosomes, but not sorting of integral membrane proteins in endosomes, depends on the vacuolar proton pump. J. Cell Biol. 130:821–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vonderheit A., Helenius A. 2005. Rab7 associates with early endosomes to mediate sorting and transport of Semliki forest virus to late endosomes. PLoS Biol. 3:e233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wan C. P., Park C. S., Lau B. H. 1993. A rapid and simple microfluorometric phagocytosis assay. J. Immunol. Methods 162:1–7 [DOI] [PubMed] [Google Scholar]
- 64. Ward T. H., Polishchuk R. S., Caplan S., Hirschberg K., Lippincott-Schwartz J. 2001. Maintenance of Golgi structure and function depends on the integrity of ER export. J. Cell Biol. 155:557–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wegner C. S., et al. 2010. Ultrastructural characterization of giant endosomes induced by GTPase-deficient Rab5. Histochem. Cell Biol. 133:41–55 [DOI] [PubMed] [Google Scholar]
- 66. Weil D., et al. 2002. Targeting the kinesin Eg5 to monitor siRNA transfection in mammalian cells. Biotechniques 33:1244–1248 [DOI] [PubMed] [Google Scholar]
- 67. Whittaker G. R., Digard P. 2006. Entry and intracellular transport of influenza virus, p. 37–64 In Kawaoka Y. (ed.), Influenza virology: current topics. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.