Abstract
The RNA genome of Seneca Valley virus (SVV), a recently identified picornavirus, contains an internal ribosome entry site (IRES) element which has structural and functional similarity to that from classical swine fever virus (CSFV) and hepatitis C virus, members of the Flaviviridae. The SVV IRES has an absolute requirement for the presence of a short region of virus-coding sequence to allow it to function either in cells or in rabbit reticulocyte lysate. The IRES activity does not require the translation initiation factor eIF4A or intact eIF4G. The predicted secondary structure indicates that the SVV IRES is more closely related to the CSFV IRES, including the presence of a bipartite IIId domain. Mutagenesis of the SVV IRES, coupled to functional assays, support the core elements of the IRES structure model, but surprisingly, deletion of the conserved IIId2 domain had no effect on IRES activity, including 40S and eIF3 binding. This is the first example of a picornavirus IRES that is most closely related to the CSFV IRES and suggests the possibility of multiple, independent recombination events between the genomes of the Picornaviridae and Flaviviridae to give rise to similar IRES elements.
INTRODUCTION
Seneca Valley virus (SVV) is a recently discovered member of the picornavirus family. It was found as a contaminant in PER.C6 cell cultures, and its natural host has not yet been definitively identified, but a number of closely related viruses have been isolated from pigs (16). The complete genome sequence of SVV-001 (16) and the crystal structure of the virus capsid (52) have now been determined. The virus is most closely related to the cardioviruses, but there are some significant differences (see below), and hence it has been recommended that the virus is classified as a new species within a new genus (Senecavirus) of the Picornaviridae.
SVV-001 and two of the related viruses (isolates 1278 and 66289) were inoculated into pigs; evidence of viral replication was obtained for all three viruses and for transmission of isolate 66289. However, in none of the experiments was any sign of illness observed (unpublished data and personal communication from J. Landgraf, USDA). An important feature of SVV is its ability to replicate selectively within human tumor cells. Owing to this novel activity and lack of observed pathogenicity in animals and humans, there is interest in using SVV as an oncolytic virus against neuroendocrine cancers (39), for which it is currently in clinical trials.
All picornaviruses have a positive-sense, single-stranded RNA genome that is infectious and has to act both as an mRNA and as a template for RNA replication (32). Picornavirus RNA includes a single large open reading frame (ORF), encoding a polyprotein, which is flanked by a long 5′ untranslated region (UTR) of approximately 600 to 1,300 nucleotides (nt) (depending on the virus) plus a shorter 3′ UTR (<100 nt) with a poly(A) tail. The viral RNA lacks the 5′ m7GpppN... cap structure found on all eukaryotic cytoplasmic mRNAs. Instead, a small virus-encoded peptide (VPg or 3B) is covalently attached to the 5′ terminus of the genomic RNA, although this is lost from the RNA within the cell. Recognition of cellular mRNAs by the cellular protein synthesis machinery is normally achieved through the binding of the 5′ cap by eIF4E. This protein is one component of the cap-binding complex, eIF4F, which also includes eIF4A (an RNA helicase) and eIF4G, a scaffold protein which makes numerous contacts with other cellular proteins [including eIF3 and the poly(A) binding protein (PABP)], and these interactions serve to bridge between the mRNA and the small ribosomal subunit (49). Translation initiation on picornavirus RNAs occurs by a different mechanism. The 5′ UTR of all picornavirus genomes contains an internal ribosome entry site (IRES) that directs cap-independent internal initiation of protein synthesis (reviewed in reference 11). To date, four distinct classes of picornavirus IRES element have been described (3, 17). The enteroviruses and rhinoviruses (e.g., poliovirus [PV]) share one type of element, while the cardioviruses and aphthoviruses (e.g., encephalomyocarditis virus [EMCV] and foot-and-mouth disease virus [FMDV], respectively) share a second type. Hepatitis A virus (HAV) has a third distinct class of IRES which is unique in requiring an intact form of eIF4F (1, 5). It has been found recently that certain picornaviruses contain a fourth class of IRES element which is closely related to the IRES found in hepatitis C virus (HCV; a Hepacivirus within the Flaviviridae). The pestiviruses (e.g., classical swine fever virus [CSFV]) are also members of the Flaviviridae and have an IRES that is similar to the HCV IRES, but there are distinct differences (12). For example, stem 1 of the pseudoknot is interrupted in CSFV, whereas in HCV it forms a single fully base-paired stem. Furthermore, the CSFV IRES has an extra subdomain termed IIId2 which is absent from the HCV IRES. This additional domain is thought to lie within the ribosome-binding interface formed by domains IIId, IIIe, and IIIf, which make direct contact with the small ribosomal subunit (50).
Picornavirus IRES elements related to the HCV IRES were initially identified in the porcine teschovirus-1 (PTV-1) genome (6, 22, 35) and subsequently have been demonstrated in a variety of other porcine, avian, and simian picornaviruses (namely porcine enterovirus-8 [PEV-8] [7], simian virus 2 [SV2] [7], avian encephalomyelitis virus [AEV] [2], and simian picornavirus 9 [SPV9] [8]). Furthermore, on the basis of sequence information and RNA structure prediction, it has been suggested that the duck hepatitis virus 1 (9), a seal picornavirus (24), SVV (17), and porcine kobuvirus (40) also contain this type of IRES.
The HCV-like IRES elements differ from those of the other picornaviruses in that they have no requirement for any part of the eIF4F complex; hence, they continue to function both when eIF4G is cleaved (e.g., induced by the PV 2A protease) and also in the presence of hippuristanol, a small molecule inhibitor of eIF4A (4). Hippuristanol provides a useful tool for identifying the HCV-like IRES elements since, by contrast, it strongly inhibits the PV and EMCV IRES elements.
The HCV and CSFV IRES elements each contain two major domains, termed domain II and domain III. The domain III contains a number of different stem-loop structures and a pseudoknot (domain IIIf) (10, 12, 29, 54). Domain II appears to facilitate 80S complex assembly (27), while domain III is required for direct interactions of the IRES with the 40S ribosomal subunit and eIF3 (reviewed in reference 14).
We now demonstrate that the SVV genome indeed contains a functional HCV-like IRES element despite the general similarity of SVV to the cardioviruses and furthermore show that this picornavirus IRES is novel in being more closely related, in terms of its secondary structure, to the CSFV IRES than to the HCV IRES. For the first time, we have probed the secondary structure and function of the additional IIId2 domain within CSFV-like IRES elements and suggest that it has no role in IRES function.
MATERIALS AND METHODS
Reporter plasmids.
DNA preparations and manipulations were performed using standard methods as described previously (45) or as stated in manufacturer's instructions. The reporter plasmids pGEM-CAT/EMCV/LUC containing the EMCV IRES cDNA and pGEM-CAT/LUC (lacking any IRES) have been described previously (44). These plasmids use a T7 promoter to express dicistronic mRNAs encoding chloramphenicol acetyltransferase (CAT) and firefly luciferase (fLUC).
SVV cDNA.
The plasmid pNTX-01 consists of SVV-001 sequences from nt 1 to 1074 in pGEM-4Z. Reporter plasmids containing the SVV 5′ UTR (nt 1 to 666; pGEM-CAT/SVVs/LUC) or longer sequences including 31, 55, or 79 nucleotides of coding sequence were prepared by PCR using pNTX-01 as the template with the SVV forward primer and the appropriate reverse primer, all designed to add a BamHI site to each end of the product (see Table 1). The PCR products were digested with BamHI, gel purified, and ligated between the two open reading frames of the similarly digested, dephosphorylated, dicistronic reporter plasmid pGEM-CAT/LUC (Fig. 1A). The constructs were confirmed by restriction enzyme digestion and sequencing of the inserts.
Table 1.
Primers used to prepare SVV sequences
| Primer | Sequencea |
|---|---|
| SVV For2 | ATATGGATCCTTTGAAATGGGGGGCTG |
| SVV utr Rev | ATGGATCCATTTGTATGTGCTAC |
| SVV +30 Rev | ATGGATCCTGTATCGAAAGAAA |
| SVV +55 Rev | ATATGGATCCTTACATCTTCAAAGGTGCCAG |
| SVV +75 Rev | ATATGGATCCTTCAACGATCTTGACTTTTGTTC |
| Mut1 For | CTGATAGGGCGACGGGGTAGTCGTGTCGGTTC |
| Mut1 Rev | GAACCGACACGACTACCCCGTCGCCCTATCAG |
| Mut1c For | CGGGGTAGTCGTGTCGGTTCTATACCTAGCACATAC |
| Mut1c Rev | GTATGTGCTAGGTATAGAACCGACACGACTACCCCG |
| Mut2 For | CGGGAACTGTAGCTACCCCTTAGCGTGCCTTG |
| Mut2 Rev | CAAGGCACGCTAAGGGGTAGCTACAGTTCCCG |
| Mut3 For | CTAGCATAGCGAGCTGGAGCGGGAACTGTAGC |
| Mut3 Rev | GCTACAGTTCCCGCTCCAGCTCGCTATGCTAG |
| Mut4 For | CTAGCATAGCGAGGGGGAGCGGGAACTGTAGCTAGGC |
| Mut4 Rev | GCCTAGCTACAGTTCCCGCTCCCCCTCGCTATGCTAG |
| Mut4c For | GCGAGGGGGAGCGGGAACTCCCCCTAGGCCTTAGCG |
| Mut4c Rev | CGTAAGGCCTAGGGGGAGTTCCCGCTCCCCCTCGC |
| Mut5 For | CTGTAGCTAGGCCT Δ GTGCCTTGGATACTGCCTG |
| Mut5 Rev | CAGGCAGTATCCAAGGCAC Δ AGGCCTAGCTACAG |
| Mut6 For | GCGAGCTACAGCCCCAACTGTAGCTAGGCCTTAGC |
| Mut6 Rev | GCTAAGGCCTAGCTACAGTTGGGGCTGTAGCTCGC |
| Mut7 Overlap For | ACTGTAGCTA Δ TTGGATACTGCCTGATAGGGCG |
| Mut7 Overlap Rev | CAGTATCCAA Δ TAGCTACAGTTCCCGCTGTAGC |
| Luc Rev | CATACTGTTGAGCAATTCAC |
| CAT For | TCACTGCATTCTAGTTGTGG |
BamHI site nucleotides are underlined, mutated nucleotides are in bold typeface, and Δ indicates that nucleotides have been deleted.
Fig. 1.
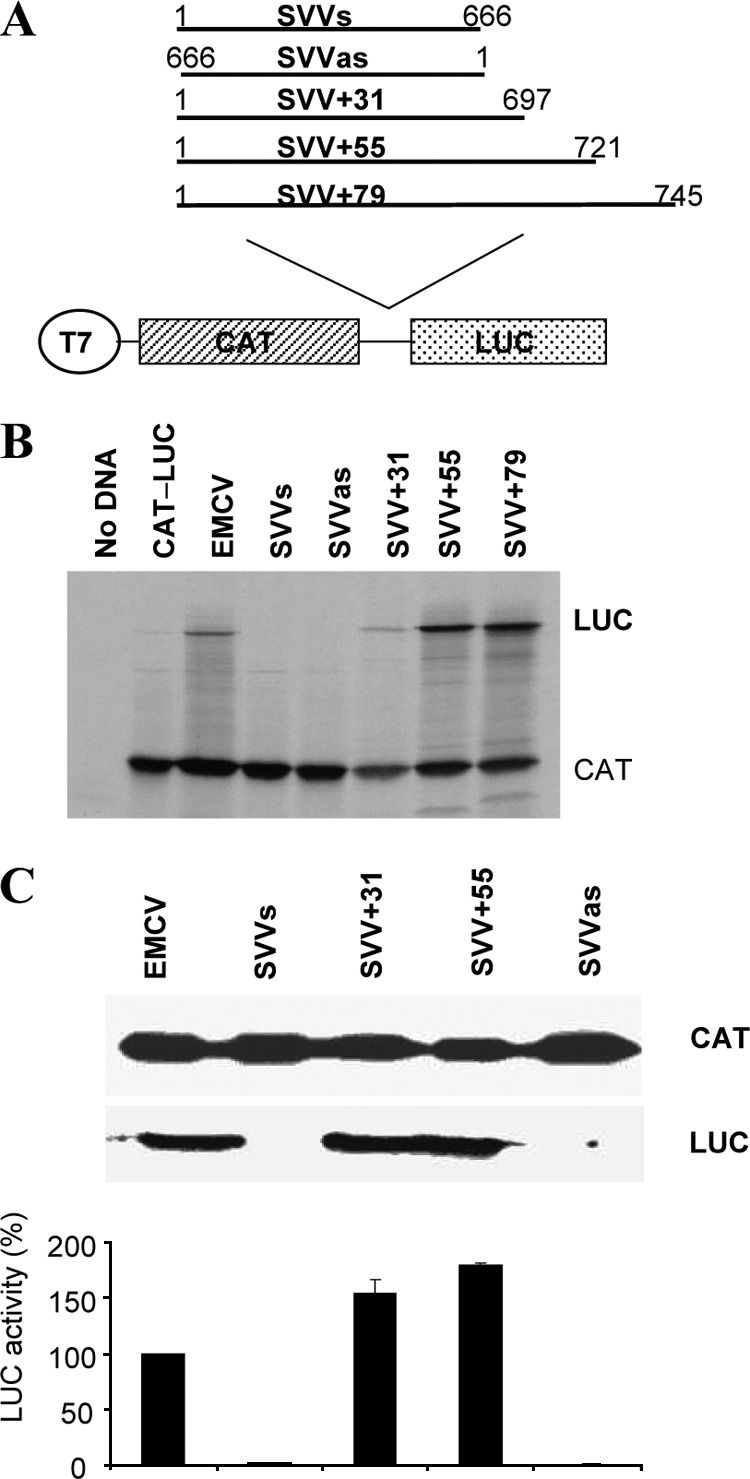
The SVV IRES extends into the viral coding sequence. (A) Structures of the dicistronic plasmids used in this study. The SVV 5′ UTR plus either 31, 55, or 79 nt of coding sequence were amplified by PCR, including added BamHI sites. These products were digested and inserted into a similarly digested plasmid containing the CAT and LUC ORFs, between the two ORFs at a unique BamHI site. (B) In vitro coupled transcription and translation reactions (RRL) containing [35S]methionine were programmed with the dicistronic plasmids. Reactions were analyzed by SDS-PAGE and autoradiography. The CAT and LUC protein products are indicated. (C) Transient expression assay in 293T cells. The dicistronic plasmids were transfected into 293T cells that had been previously infected with vTF7-3. After 20 h, cell lysates were prepared and analyzed for CAT and LUC expression as outlined in Materials and Methods. Extracts were also subjected to SDS-PAGE and immunoblotting. The LUC expression data were obtained from three separate transfections and the results standardized to the LUC expression value from the EMCV IRES, which was set at 100%. LUC activities were normalized against CAT expression values. Mean LUC values (plus standard errors of the means) are shown.
Mutagenesis of the SVV cDNA.
Mutated versions of the pGEM-CAT/SVV+55/LUC construct were prepared using a site-directed mutagenesis kit (QuikChange; Stratagene). Primary mutants (Mut1 to Mut6) were generated using pGEM-CAT/SVV+55/LUC as a template and the appropriate primer pairs (Table 1). Compensatory mutations (Mut1c and Mut4c) were prepared in the same way, except that the corresponding primary mutant was used as the template for the generation of the compensatory mutant.
Mutant 7 (in which the IIId2 stem-loop was deleted) was prepared by overlap PCR. The pGEM-CAT/SVV+55/LUC plasmid was used as the template for PCRs to generate fragments of 589 and 117 bp, one with each primer pair (SVV For and Mut7 Overlap Rev, and Mut7 Overlap For and SVV+55 Rev [Table 1]). After purification, the products were mixed and a further PCR was carried out using the SVV For and SVV+55 Rev primers. The resulting PCR product was digested with BamHI, gel purified, and ligated into pGEM-CAT/LUC as described before. The construct was verified by sequencing.
In vitro translation reactions.
The dicistronic reporter plasmids (1 μg) were assayed in the rabbit reticulocyte lysate (RRL) coupled transcription and translation (TNT) system (Promega) using [35S]methionine as described by the manufacturer. Products were analyzed by SDS-PAGE and autoradiography.
Transient expression assays.
The dicistronic reporter plasmids (2 μg) described above were assayed in human 293T cells (or HeLa or BHK cells when indicated) alone or with the pAΔ802 (0.2 μg) plasmid, which expresses the PV 2A protease as described previously (44). Briefly, the plasmids were transfected into cells (35-mm dishes) previously infected with the recombinant vaccinia virus vTF7-3 (15), which expresses T7 RNA polymerase, using Lipofectin (8 μl; Invitrogen) and Optimem (192 μl; Gibco BRL). Cell lysates were prepared 20 h after transfection and were analyzed by SDS-PAGE and immunoblotting to determine CAT and LUC expression. Detection was achieved with anti-CAT (Sigma) or anti-fLUC (Promega) antibodies and peroxidase-labeled anti-rabbit (Amersham) or anti-goat (Dako Cytomation) antibodies, respectively, using chemiluminescence reagents (Pierce). The fLUC expression was also quantified using a firefly luciferase assay kit (Promega) with a luminometer, while CAT protein was measured by enzyme-linked immunosorbent assay (ELISA) (Boehringer).
RNA secondary structure prediction.
The SVV-001 5′ UTR sequence (EMBL accession number DQ641257) was aligned with those from HCV (EMBL accession number AB016785) and CSFV (EMBL accession number J04358) using ClustalW and edited manually. Models of secondary structure elements (other than the pseudoknot) were generated using Mfold (56).
Translation assays in the presence of hippuristanol.
The requirement of the SVV IRES element for eIF4A was investigated both in vitro and in vivo using hippuristanol, a specific inhibitor of eIF4A (4). Dicistronic plasmid DNAs were assayed in the TNT RRL system (as described above) with or without hippuristanol (10 μM; kind gift from Jerry Pelletier, McGill University, Canada). The plasmids were also assayed in 293T cells, with or without the addition of 0.5 μM hippuristanol; cell lysates were prepared 20 h after transfection, and the inhibitor was added for the final 10 h.
Secondary structure probing.
The secondary structure of the SVV IRES was probed using dimethyl sulfate (DMS), N-cyclohexyl-N′-[N-methylmorpholino-ethyl]-carbodiimid-4-toluenesulfonate (CMCT), and RNase V1 as described previously (55). RNA (2 pmol) was resuspended in 20 μl of 20 mM Tris, 100 mM K acetate, 200 mM KCl, 2.5 mM MgCl2, and 1 mM dithiothreitol (DTT) (pH 7.5) (DMS), 20 mM Tris, 5 mM MgCl2, and 100 mM KCl (pH 7.5) (V1), or 50 mM borate-NaOH and 1 mM EDTA (pH 8.0) (CMCT), denatured for 1 min at 95°C and cooled on ice. DMS (0.395 M), CMCT (2, 4, or 10 mg/ml), or RNase V1 (0.01, 0.02, or 0.05 U) was added, and the mixture was incubated for 1, 5, or 10 min (DMS), 20 min (CMCT), or 5 min (RNase V1). The modified RNA was then immediately ethanol precipitated on dry ice in the presence of 0.3 M ammonium acetate, washed with 70% ethanol, and resuspended in 8 μl of water. Modifications were revealed by reverse transcriptase using 32P-labeled primer and avian myeloblastosis virus reverse transcriptase (Promega). To analyze the IIId2 domain, the primer 5′-TAGAACCGACACGACTAGGC-3′ was used. The products were resolved on a 7 M urea 6% polyacrylamide gel and revealed using a fluorescent screen and a Personal FX imager (Bio-Rad).
Binding of 40S subunits and eIF3 to the SVV IRES.
RNA transcripts were made in the presence of α-32P-UTP (3,000 mCi/mmol) using T7 RNA polymerase from PCR products containing the T7 promoter sequence and purified by size exclusion chromatography as previously described (46, 47). The initiation factor eIF3 and 40S ribosomal subunits were prepared following previously established procedures (37).
Radiolabeled RNA (50 fmol) in binding buffer (10 μl; 20 mM Tris [pH 7.6], 100 mM KCl, 2 mM DTT, 2 mM MgCl2) was denatured by heating to 85°C for 1 min and then slowly cooled to room temperature. Serial dilutions of eIF3 or 40S subunits were prepared, added to a 10-μl reaction, and incubated at 37°C for 15 min. Filter binding assays were accomplished essentially as previously described using two filters (28). Bound RNA was quantified using a Personal FX imager (Bio-Rad). To determine the apparent dissociation constant (Kd), the data were fitted to a Langmuir isotherm described by the equation θ = P/(P + Kd), where θ is the fraction of RNA bound and P is either the 40S subunit or eIF3 concentration. Reported values are the average of results from three repetitions with standard errors. All calculations were performed with GraphPad Prism 5.
RESULTS
The SVV IRES extends into the viral coding sequence.
The dicistronic reporter plasmid pGEM-CAT/SVVs/LUC containing the complete SVV 5′ UTR (nt 1 to 666) sequence inserted between the CAT and fLUC open reading frames (Fig. 1A), the negative-control plasmid lacking any IRES sequence (pGEM-CAT/LUC), and the dicistronic plasmid, pGEM-CAT/EMCV/LUC, containing the EMCV IRES as a positive control, were first assayed in coupled transcription and translation reactions (TNT) in rabbit reticulocyte lysates (RRL) reactions. SDS-PAGE and autoradiography for CAT and fLUC expression showed that all plasmids efficiently expressed CAT, as expected (Fig. 1B). Furthermore, the EMCV IRES directed efficient fLUC expression; however, the SVV 5′ UTR (nt 1 to 666) did not display any IRES activity, as indicated by lack of fLUC expression, in either the sense (s) or antisense (as) orientation.
Since some HCV-like IRES elements have been found, in some contexts, to require the presence of a viral coding sequence to display IRES activity (e.g., see references 41 and 13), three additional constructs were prepared containing 31, 55, and 79 nt of viral coding sequence, termed pGEM-CAT/SVV+31/LUC, pGEM-CAT/SVV+55/LUC, and pGEM-CAT/SVV+79/LUC. The inclusion of 31 nt of coding sequence resulted in the generation of a functional IRES. However, the addition of 55 nt from the coding sequence (as in pGEM-CAT/SVV+55/LUC) resulted in more efficient IRES activity from the SVV sequence (Fig. 1B), but the addition of 79 nt did not result in any further increase in IRES activity.
The SVV+31 and SVV+55 constructs were also assayed in a transient expression assay in human 293T cells infected with the vaccinia virus vTF7-3 (15) which expresses the T7 RNA polymerase. The plasmids were transfected into the cells, which were then harvested 20 h later; cell lysates were assayed for CAT and fLUC expression by SDS-PAGE and immunoblotting and in a quantitative manner by ELISA and enzymatic assay, respectively. All plasmids efficiently expressed CAT, as expected (Fig. 1C). Consistent with the data in Fig. 1B, fLUC activity assays indicated that the SVV 5′ UTR alone did not display IRES activity, but the addition of 31 or 55 nt of coding sequence resulted in efficient IRES activity, with the +55 construct again showing higher activity. The SVV IRES was almost twice as efficient as the EMCV IRES in these cells. Similar results were obtained when these plasmids were assayed in HeLa and BHK-21 cells (data not shown). Taken together, these results demonstrate that the SVV 5′ UTR, in conjunction with 55 nt of coding sequence, functions efficiently as an IRES element.
The SVV IRES does not require the eIF4F factors for function.
Having established that the SVV IRES element functions both in cells and in the cell-free RRL system in vitro, we next examined the requirement of this IRES for components of the eIF4F initiation factor complex. Cleavage of eIF4G can be induced by expression of the PV 2A protease or FMDV leader (L) protease and results in the inhibition of cap-dependent protein synthesis (44). However, most picornavirus IRES elements (with the exception of the HAV IRES) function efficiently under these conditions. Similarly, the HCV and related IRES elements from picornaviruses function efficiently in the presence of these proteases (reviewed in reference 3). We studied the effect of eIF4G cleavage on SVV IRES activity in cells to determine the degree of similarity to its effect on other viral IRES elements. The dicistronic reporter plasmids were transfected into 293T cells alone or with a plasmid (pAΔ802) (23) that expresses the PV 2A protease. After 20 h, cell lysates were prepared and efficient cleavage of eIF4G in the presence of PV 2Apro was confirmed by Western blot analysis as described before (2) (data not shown). Expression of CAT and fLUC was analyzed as above. All plasmids expressed CAT efficiently when assayed alone, but CAT expression was significantly reduced in the presence of the PV 2Apro (Fig. 2A), consistent with the loss of cap-dependent protein synthesis. As expected, the assays for fLUC expression demonstrated that EMCV IRES activity was unaffected by PV 2Apro-induced cleavage of eIF4G. The SVV IRES was able to direct efficient fLUC expression in the presence of 2Apro, although activity was reduced by about 50%. This modest inhibition of SVV IRES activity in the presence of cleaved eIF4G is comparable to results for some other HCV-like picornavirus IRES elements that have recently been described (see Discussion).
Fig. 2.
The SVV IRES functions in the presence of cleaved eIF4G and an eIF4A inhibitor. (A) Dicistronic plasmids (2 μg) of the form CAT/IRES/LUC were transfected into 293T cells in the absence (−) or presence (+) of the plasmid pAΔ802 (0.5 μg), which expresses the PV 2A protease. After 20 h, cell extracts were prepared and analyzed for CAT and LUC expression as described in Fig. 1. LUC and CAT assays were also performed on extracts from three separate experiments, and the mean LUC values are shown, standardized to the LUC expression values from the EMCV IRES-containing construct, which was set at 100%. LUC activities were normalized to CAT expression as described in the legend to Fig. 1. (B) The same dicistronic plasmids were transfected into 293T cells in the absence (−) or presence (+) of the eIF4A inhibitor, hippuristanol (Hipp.). Cells were harvested 20 h after transfection, and the hippuristanol (0.5 μM) was added for the last 10 h of the incubation. Cell extracts were analyzed for CAT and LUC expression as described in the legend to Fig. 1. The data shown are representative of results from two independent experiments.
We next assayed the requirement of the SVV IRES for another component of the eIF4F complex, the RNA helicase eIF4A. While cap-dependent translation and many picornavirus IRES elements (including those from PV and EMCV) need eIF4A for activity, the HCV and the HCV-like picornavirus IRES elements do not (33, 34, 51). We analyzed the effect of the eIF4A inhibitor, hippuristanol, on SVV IRES activity. The HCV and HCV-like IRES elements within picornavirus genomes have been shown to be resistant to this inhibitor (4, 7). The addition of hippuristanol to transfected 293T cells severely inhibited cap-dependent expression of CAT and also fLUC expression directed by the EMCV IRES, as expected (Fig. 2B). However, the SVV IRES activity displayed marked resistance to this inhibitor, as seen with the AEV and HCV IRES elements (2, 4). This indicates that the SVV IRES does not require eIF4A for function, emphasizing its similarity to the other HCV-like IRES elements.
The SVV IRES bears a striking resemblance to the CSFV IRES.
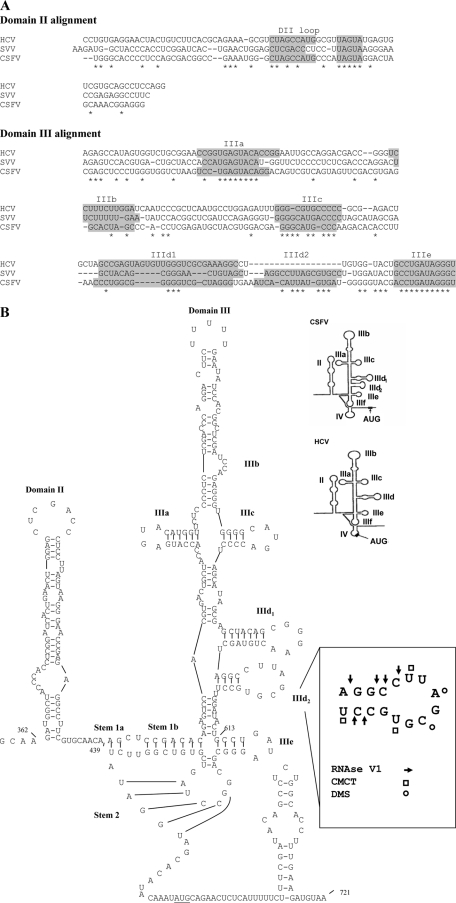
As described above, the functional properties of the SVV IRES resembled those of the recently discovered HCV-like picornavirus IRES elements. We therefore performed a sequence alignment of the SVV IRES to the HCV IRES sequence using ClustalW. We found that the SVV IRES closely resembles the HCV IRES (52% identity) and the CSFV IRES (47% identity). However, it should be noted that the high level of sequence identity between all three IRES elements occurs only within particular regions (see Fig. 3A), notably domain IIIe, domain IIIa, domain IIIc, and short motifs within domain II and domain IIId1 (including a GGG motif involved in 40S ribosome interaction) (20, 21, 25, 29, 30, 35, 36, 38, 42, 48). In between these stem-loop structures, the level of sequence identity between all three of them is rather low. We have derived a secondary structure prediction for the SVV IRES (Fig. 3B) which includes the 55 nt of the coding sequence. This structure resembles that of the HCV IRES but also reveals the presence of a bipartite stem 1 within the pseudoknot and an additional stem-loop structure adjacent to the domain IIId, which has been termed domain IIId2 in the CSFV IRES (12). This stem-loop is absent from the HCV IRES (19). These features, along with the requirement for about 55 nt of coding sequence for optimal SVV IRES activity, indicate that the SVV IRES is strikingly similar to the CSFV IRES. This is the first report of a CSFV-like IRES (a Pestivirus within the Flaviviridae) within a picornavirus genome and suggests the possibility of genetic exchange between several different members of these virus families.
Fig. 3.
The SVV IRES is an HCV-like picornavirus IRES. (A) Alignment of the SVV, CSFV, and HCV 5′ UTR sequences that comprise domain II and domain III. Sequences were aligned with ClustalW and edited manually; the nucleotides which are shared by all three IRES elements are marked with an asterisk. Gaps introduced to maximize alignment are indicated by dashes. Shaded areas highlight specific stem-loop structures. The individual SVV IRES domains are indicated above the sequence. The overall sequence identity between SVV and HCV IRES sequences is 52.4%, but some regions, e.g., domains IIIa and IIIe, are highly conserved among all three viruses. (B) Proposed secondary structure of the SVV 5′ UTR. Domains are labeled by analogy to the domains in the HCV IRES. The structure was derived from comparative sequence analyses and using Mfold (50) to predict the most thermodynamically favorable structures. Outlines of the HCV and CSFV IRES element structures are also shown for comparison. Note that in the SVV IRES (as in the CSFV IRES) there is an additional IIId2 stem-loop structure. In addition, a model of the 55-nt sequence from the coding sequence is included. Secondary structure probing of the IIId2 domain is also indicated. Nucleotides affected by RNase V1, CMCT, or DMS are noted by arrows, circles, and squares, respectively.
Mutational analysis of the domain III region of the SVV IRES.
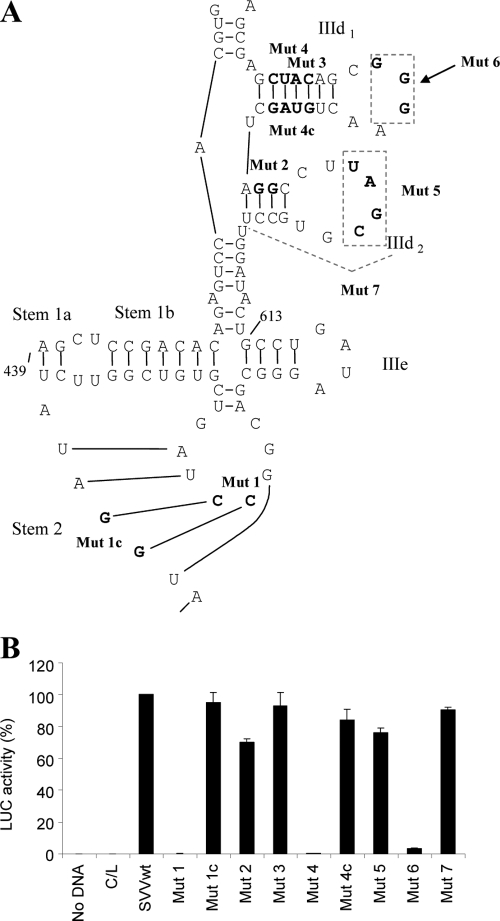
RNA structural elements within the domain III region of all the HCV-like IRES elements have been shown to be critical for function (3). We therefore tested our predicted secondary structure model of the SVV IRES by performing mutational analysis of this region. First, we tested the sequences involved in the putative pseudoknot. Modifications in the stem 2 region of the pseudoknot changed nt 630 and 631 from CC to GG; these changes were predicted to disrupt the base pair interaction with nt 651 and 652 (GG) (Fig. 4A). This mutant (Mut1) was analyzed using the dicistronic reporter construct in 293T cells as described above. SVV IRES activity was severely inhibited (<0.3% of wild-type [wt] activity [Fig. 4B]). Introduction of compensatory mutations at nt 651 and 652 to change GG to CC (Mut1c) restored SVV IRES activity to >80% that of the wild type. The inability to restore wt activity fully with these compensatory mutations has been observed previously (2, 6) and suggests that the primary sequences may be involved in other interactions (for example, with trans-acting factors) as well as forming part of the pseudoknot structure.
Fig. 4.
Mutational analysis of the domain III region of the SVV IRES. (A) Predicted secondary structure of the domain IIId, IIIe, and IIIf regions of the SVV IRES. The stems that form the pseudoknot (stem 1 and stem 2) and the IIId1 and IIId2 stem-loops are shown. The nucleotides that were modified in the mutational analysis experiments are shown in bold type. Suffix “c” indicates compensatory mutations. Areas indicated with dashed lines (as in Mut7) indicate that the whole IIId2 region was deleted. (B) Dicistronic plasmids of the form CAT/IRES/LUC, containing the indicated mutations within the domain III region of the SVV+55 construct, were transfected into 293T cells as described in the legend to Fig. 1. Cell extracts were analyzed for CAT and LUC expression as described in the legend to Fig. 1. The results are derived from three independent experiments, and the mean values plus standard errors are shown. LUC activities from the mutant IRES elements are expressed relative to the activity from the wt SVV+55 IRES, which was set at 100%. LUC activities were normalized against CAT expression as described in the legend to Fig. 1.
As the predicted secondary structure indicated the presence of a IIId2 stem-loop, as seen in the CSFV IRES structure, further mutational analysis focused on the IIId1 and IIId2 stem-loop regions. Mutation of nt 571 and 572 from AC to GG in stem IIId1 (Mut3) had only a modest effect on IRES activity (Fig. 4B), reducing activity to about 80% that of the wt IRES. However, mutation of four nucleotides in this region (nt 569 to 572) from CUAC to GGGG (Mut4), which would be expected to disrupt the stem, severely inhibited IRES activity. Furthermore, introduction of compensatory mutations at nt 583 to 586 that should restore base pairing in this stem (GUAG to CCCC; Mut4c) effectively restored IRES activity to over 80% of wt activity. The importance of the IIId1 stem-loop was also highlighted by mutational analysis of the loop region. Modification of the GGG motif (nt 576 to 578) to CCC as in Mut6 had a severe effect on IRES activity. These data support the predicted structure of the IIId1 loop of the SVV IRES and indicate that this structure is important for IRES activity as observed for the HCV and CSFV elements previously (12).
To assess the role of the predicted IIId2 stem-loop structure, we changed nt 590 and 591 (GG) to CC (Mut2) and assayed this modified IRES in 293T cells. This mutant retained about 70% of wt activity, suggesting that this stem structure is not critical for IRES activity. We also deleted nucleotides in the loop region of IIId2 (UAGC) (Mut5), and this also had little effect on IRES activity (this mutant retained about 80% of wt IRES activity). Finally, deletion of the entire IIId2 stem-loop (Mut7) had no significant effect on IRES activity; this mutant retained 90% of wt IRES activity. These data suggest that this predicted stem-loop structure is not important for IRES function.
These results are intriguing, as this additional domain lies between domains IIId1 and IIIe, which are both important for ribosome binding. Therefore, we investigated further whether the predicted IIId2 stem-loop structure was actually formed in solution as proposed by the secondary structure model we and others have predicted (Fig. 3) (17) and its role in factor recruitment. Thus, we performed enzymatic and chemical probing of the SVV IRES. Our results showed that domain IIId2 is indeed formed as predicted (Fig. 3B). This is the first time that the structure of this region has been defined for a CSFV-like IRES element and confirms its existence. Next we investigated the impact of domain IIId2 in the recruitment of 40S subunits and eIF3, the key factors mediating initiation of translation on HCV-like IRES. We performed filter binding assays of eIF3 and the 40S subunit to the SVV IRES in the presence or absence of domain IIId2. Both IRES elements were able to bind eIF3 and 40S subunits directly, as previously reported for the HCV IRES (25), and no significant differences were seen between the wt and mutant IRES lacking the IIId2 domain (Fig. 5). This confirmed that the IIId2 stem-loop has no role in assembly of initiation complexes on the SVV IRES and may suggest a role at another stage of the SVV replication cycle.
Fig. 5.
The SVV IRES directly binds to 40S subunits and eIF3. (A) Binding curves of 32P-labeled wild-type and IIId2 deletion mutant RNAs to purified 40S subunits. Labeled RNAs were incubated with 40S subunits and binding assessed by filter binding assay. (B) Binding curves of 32P-labeled wild-type and IIId2 deletion mutant RNAs to purified eIF3. Labeled RNAs were incubated with eIF3, and binding was assessed by filter binding assay. Reported values are the average of results from three repetitions with standard errors. All calculations were performed with GraphPad Prism 5.
DISCUSSION
The results presented here show that the 5′ UTR of the SVV genome, together with 31 to 55 nt of viral coding sequence, contains a functional IRES element. Based on its predicted structure and functional properties, the SVV IRES should be placed within the recently described HCV-like class of picornavirus IRES elements (see reference 3). Optimal IRES activity from the HCV IRES has been shown to require some 10 codons (30 nt) of the viral coding sequence (41), while the CSFV IRES requires between 12 and 17 codons of the coding sequence (36 to 51 nt) for optimal activity (13). However, it should be noted that under certain conditions, and with some reporter gene sequences, no absolute requirement for any coding sequence has been observed for the HCV and CSFV IRES elements (see reference 43). It therefore seems likely that the accessibility of the initiation codon in the different RNAs in some way determines this requirement. The picornavirus HCV-like IRES elements from PTV-1, PEV-8, and AEV do not require any coding sequence (2, 7, 22); however, the SV2 IRES is considerably more active when about 50 nt of coding sequence is present in the RNA (7). Thus, the SVV IRES is distinct from the other picornavirus IRES elements in that it showed an absolute requirement for the presence of over 30 nt of coding sequence, and in this respect it is most similar to the CSFV IRES. Our secondary structure prediction for the 55 nt downstream of the AUG codon suggests the presence of a stem-loop structure which is preceded by a pyrimidine-rich sequence. However, since the SVV+31 construct lacks the intact stem-loop structure and still functions efficiently, it is unlikely that this structure is critical to IRES function. It may be that the presence of the pyrimidine-rich sequence is important for maintaining this region as unstructured, as suggested for this region of the CSFV IRES (12).
The predicted secondary structure of the SVV IRES was also more similar to the CSFV structure than to the HCV IRES structure (Fig. 3B). The key distinguishing features are the presence of a bipartite stem 1 within the pseudoknot in CSFV and SVV, while a single stem is present in HCV (and PTV-1), and the CSFV and SVV IRES elements are each predicted to contain a IIId2 stem-loop structure (26). We suspected that this additional domain might have a functional role in translation initiation. Indeed, this domain lies between domains IIId1 and IIIe, both of which make direct contact with the ribosome, as seen in the structure of binary HCV IRES-40S subunit complexes (50), and significantly contribute to the high affinity of the IRES RNA for the ribosome (25, 31). While we confirmed that the IIId2 domain folds in solution into the predicted stem-loop structure, extensive modification of the IIId2 element in SVV did not have any significant effect on the activity of the IRES (Fig. 4) or on the ability of the IRES to bind eIF3 or 40S subunits (Fig. 5). These results, and the conservation of this motif in other HCV-like IRES elements (BDV, CSFV), suggest that this structure may have some other function in replication other than translation initiation; thus, our current studies are focusing on the role of this domain in virus replication.
The pseudoknot structure within domain III of the SVV IRES is critical for activity. Modification of stem 2 greatly reduced IRES activity, but restoration of the structure, through compensatory mutations, restored activity. These results are analogous to those obtained with the HCV, CSFV, PTV, and AEV IRES elements (2, 7, 51). The GGG motif in the loop of domain IIId is conserved across all the HCV-like IRES elements (20), and this sequence in the HCV IRES is protected from chemical modification by interaction with the 40S ribosomal subunit (25). Thus, it was not surprising that modification of this motif also had a significant effect on the activity of the SVV IRES. The domain IIIe region is known to interact with 40S ribosomal subunits (25, 30, 31), and this region is highly conserved between SVV and CSFV; 10 of 12 nucleotides are identical, and the two nucleotide differences still retain the base pairing required to maintain the conserved secondary structure.
The HCV-like IRES elements do not need any of the components of eIF4F for activity; however, there is some evidence that their activity can be enhanced in the presence of the fully functional factors. For example, the AEV IRES was shown to be inhibited by about 50% in the presence of cleaved eIF4G (2). Similarly, initiation complex formation on the related SPV9 IRES was enhanced by the addition of eIF4F (8). The results presented here with the SVV IRES also indicate that the activity is optimal when eIF4G is intact (Fig. 2A) and when eIF4A activity is not inhibited by hippuristanol (Fig. 2B). This differs slightly from the HCV IRES itself, which is apparently unaffected by the lack of eIF4G (34, 35). It seems therefore that the picornavirus HCV-like IRES elements may be stimulated in the presence of eIF4F through an unknown mechanism. As the SVV IRES functions very efficiently in RRL, it is clear that it does not need additional factors beyond those already present within the RRL, in contrast to the poliovirus or rhinovirus IRES elements that require supplementation with cell lysates (10).
The mechanisms by which SVV selectively infects and replicates in animals and human tumor cells is currently unknown, although interaction with specific cell membrane proteins appears to be at least one determinant (39). The HCV-like IRES elements are found in diverse picornaviruses from multiple genera with differing species and cellular tropisms (3, 17). However, SVV is the first example of a picornavirus having an IRES which is more closely related, at the level of the secondary structure, to the CSFV IRES rather than the HCV IRES. The fact that the likely host of SVV is the pig is consistent with this possibility, since CSFV can establish a relatively long-term persistent infection in these animals, offering ample opportunity for coinfection. It is of interest that another porcine picornavirus, the porcine kobuvirus (40), has recently been shown to contain an HCV-like IRES within its 5′ UTR; in this case, the IRES is most similar to the duck hepatitis virus and porcine teschovirus-1 HCV-like IRES elements. This suggests that not only may there have been multiple occasions when genetic exchange has occurred between members of the Picornaviridae and the Flaviviridae, but also there may have been exchange of HCV-like IRES elements within the Picornaviridae family.
Although SVV is not considered an important pathogen of animals, its importance lies in the fact that it has a unique potential for targeted cancer cell therapy. The virus exhibits oncolytic activity in certain tumor cells of neuroendocrine origin and is currently in a phase I/II clinical trial to test for safety and efficacy against small-cell lung carcinoma, with a view to developing an alternative treatment for this form of lung cancer. Cell lines derived from small-cell lung cancer, neuroblastoma, and neuroendocrine pediatric cancer have all been shown to be sensitive to SVV-001-mediated killing, while normal primary human cells have been shown to be resistant. Wadhwa et al. (53) have demonstrated a reduction in invasive disease and metastasis with oncolytic SVV-001 in a retinoblastoma xenograft mouse model. Therefore, a full understanding of what underlies this specific cell tropism is of medical importance, and the IRES could influence the ability of the virus to replicate in certain cell types.
ACKNOWLEDGMENTS
We thank Jerry Pelletier (McGill University, Montreal, Canada) for the kind gift of hippuristanol and Richard Jackson (University of Cambridge, United Kingdom) for the HCV IRES dicistronic reporter plasmid.
M.B. gratefully acknowledges receipt of a scholarship from the Ministry of Science, Research and Technology, and the Ministry of Jihad-Agriculture of Iran.
Footnotes
Present address: Poultry Vaccines Department, Razi Vaccine & Serum Research Institute, P.O. Box 31975/148, Karaj, Iran.
Published ahead of print on 16 February 2011.
REFERENCES
- 1. Ali I. K., McKendrick L., Morley S. J., Jackson R. J. 2001. Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J. Virol. 75:7854–7863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bakhshesh M., et al. 2008. The picornavirus avian encephalomyelitis virus possesses a hepatitis C virus-like internal ribosome entry site element. J. Virol. 82:1993–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belsham G. J. 2009. Divergent picornavirus IRES elements. Virus Res. 139:183–192 [DOI] [PubMed] [Google Scholar]
- 4. Bordeleau M.-E., et al. 2006. Functional characterization of IRESes by a novel inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2:213–220 [DOI] [PubMed] [Google Scholar]
- 5. Borman A. M., Kean K. M. 1997. Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology 237:129–136 [DOI] [PubMed] [Google Scholar]
- 6. Chard L. S., Kaku Y., Jones B., Nayak A., Belsham G. J. 2006. Functional analyses of RNA structures shared between the internal ribosome entry sites of hepatitis C virus and the picornavirus porcine teschovirus 1 Talfan. J. Virol. 80:1271–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chard L. S., Bordeleau M.-E., Pelletier J., Tanaka J., Belsham G. J. 2006. Hepatitis C virus-related internal ribosome entry sites are found in multiple genera of the Picornaviridae. J. Gen. Virol. 87:927–936 [DOI] [PubMed] [Google Scholar]
- 8. de Breyne S., Yu Y., Pestova T. V., Hellen C. U. T. 2008. Factor requirements for translation initiation on the simian picornavirus internal ribosome entry site. RNA 14:367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ding C., Zhang D. 2007. Molecular analysis of duck hepatitis virus type 1. Virology 361:9–17 [DOI] [PubMed] [Google Scholar]
- 10. Doudna J., Sarnow P. 2007. Translation initiation by viral internal ribosome entry sites, p. 129–153 In Mathews M. B., Sonenberg N., Hershey J. W. B. (ed.), Translational control in biology and medicine, 3rd ed. CSHL Press, Cold Spring Harbor, NY [Google Scholar]
- 11. Fitzgerald K. D., Semler B. L. 2009. Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochim. Biophys. Acta 1789:518–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fletcher S. P., Jackson R. J. 2002. Pestivirus internal ribosome entry site (IRES) structure and function: elements in the 5′ untranslated region important for IRES function. J. Virol. 76:5024–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fletcher S. P., Ali I. K., Kaminski A., Digard P., Jackson R. J. 2002. The influence of viral coding sequences on pestivirus IRES activity reveals further parallels with translation initiation in prokaryotes. RNA 8:1558–1571 [PMC free article] [PubMed] [Google Scholar]
- 14. Fraser C. C., Doudna J. A. 2007. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat. Rev. Microbiol. 5:29–38 [DOI] [PubMed] [Google Scholar]
- 15. Fuerst T. R., Niles E. G., Studier F. W., Moss B. 1986. Eukaryotic transient expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 83:8122–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hales L. M., et al. 2008. Complete genome sequence analysis of Seneca Valley virus-001, a novel oncolytic picornavirus. J. Gen. Virol. 89:1265–1275 [DOI] [PubMed] [Google Scholar]
- 17. Hellen C. U. T., de Breyne S. 2007. A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: evidence for modular exchange of functional noncoding RNA elements by recombination. J. Virol. 81:5850–5863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hellen C. U. 2009. IRES-induced conformational changes in the ribosome and the mechanism of translation initiation by internal ribosomal entry. Biochim. Biophys. Acta 1789:558–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Honda M., Beard M. R., Ping L. H., Lemon S. M. 1999. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 73:1165–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ji H., Fraser C. S., Yu Y., Leary J., Doudna J. A. 2004. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc. Natl. Acad. Sci. U. S. A. 101:16990–16995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jubin R., et al. 2000. Hepatitis C virus internal ribosome entry site (IRES) stem loop IIId contains a phylogentically conserved GGG triplet essential for translation and IRES folding. J. Virol. 74:10430–10437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaku Y., Chard L. S., Inoue T., Belsham G. J. 2002. Unique characteristics of a picornavirus internal ribosome entry site from the porcine teschovirus-1 Talfan. J. Virol. 76:11721–11728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaminski A., Howell M. T., Jackson R. J. 1990. Initiation of encephalomyocarditis virus RNA translation: the authentic initiation site is not selected by a scanning mechanism. EMBO J. 9:3753–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kapoor A., Victoria J., Simmonds P., Wang C., Shafer R. W., Nims R., Nielsen O., Delwart E. 2008. A highly divergent picornavirus in a marine mammal. J. Virol. 82:311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kieft J. S., Zhou K., Jubin R., Doudna J. A. 2001. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7:194–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kolupaeva V. G., Pestova T. V., Hellen C. U. 2000. Ribosomal binding to the internal ribosomal entry site of classical swine fever virus. RNA 6:1791–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Locker N., Easton L. E., Lukavsky P. J. 2007. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 26:795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Locker N., Chamond N., Sarguueil B. 2010. A conserved structure within the HIV gag open reading frame that controls translation initiation directly recruits the 40S subunit and eIF3. Nucleic Acids Res. [Epub ahead of print.] doi:10.1093/nar/gkq1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lukavsky P. J., Otto G. A., Lancaster A. M., Sarnow P., Puglisi J. D. 2000. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 7:1105–1110 [DOI] [PubMed] [Google Scholar]
- 30. Lytle J. R., Wu L., Robertson H. D. 2001. The ribosome binding site of hepatitis C virus mRNA. J. Virol. 75:7629–7636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Otto G. A., Puglisi J. D. 2004. The pathway of HCV IRES-mediated translation initiation. Cell 119:369–380 [DOI] [PubMed] [Google Scholar]
- 32. Paul A. V. 2002. Possible unifying mechanism of picornavirus genome replication, p. 227–246 In Semler B. L., Wimmer E. (ed.), Molecular biology of picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 33. Pause A., Méthot N., Svitkin Y., Merrick W. C., Sonenberg N. 1994. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13:1205–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pestova T. V., Shatsky I. N., Fletcher S. P., Jackson R. J., Hellen C. U. T. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pisarev A. V., et al. 2004. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J. Virol. 78:4487–4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pisarev A. V., Shirokikh N. E., Hellen C. U. T. 2005. Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites. C. R. Biol. 328:589–605 [DOI] [PubMed] [Google Scholar]
- 37. Pisarev A. V., Unbehaun A., Hellen C. U., Pestova T. V. 2007. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 30:147–177 [DOI] [PubMed] [Google Scholar]
- 38. Psaridi L., Georgopoulou U., Varaklioti A., Mavromara O. 1999. Mutational analysis of a conserved tetraloop in the 5′ untranslated region of hepatitis C virus identifies a novel RNA element essential for the internal ribosome entry site function. FEBS Lett. 453:49–53 [DOI] [PubMed] [Google Scholar]
- 39. Reddy P. S., et al. 2007. Seneca Valley virus, a systemically deliverable oncolytic picornavirus, and the treatment of neuroendocrine cancers. J. Natl. Cancer Inst. 99:1623–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reuter G., Boldizsár A., Pankovics P. 2009. Complete nucleotide and amino acid sequences and genetic organization of porcine kobuvirus, a member of a new species in the genus Kobuvirus, family Picornaviridae. Arch. Virol. 154:101–108 [DOI] [PubMed] [Google Scholar]
- 41. Reynolds J. E., et al. 1995. Unique features of internal initiation of hepatitis-C virus-RNA translation. EMBO J. 14:6010–6020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rijnbrand R., van der Straaten T., van Rijn P. A., Spaan W. J., Bredenbeek P. J. 1997. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J. Virol. 71:451–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rijnbrand R., et al. 2001. The influence of downstream protein-coding sequence on internal ribosome entry on hepatitis C virus and other flavivirus RNAs. RNA 7:585–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roberts L. O., Seamons R. A., Belsham G. J. 1998. Recognition of picornavirus internal ribosome entry sites within cells; influence of cellular and viral proteins. RNA 4:520–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sambrook J., MacCallum P., Russell D. 2006. The condensed protocols from molecular cloning: a laboratory manual. CSHL Press, Cold Spring Harbor, NY [Google Scholar]
- 46. Sargueil B., McKenna J., Burke J. M. 2000. Analysis of the functional role of a G.A sheared base pair by in vitro genetics. J. Biol. Chem. 275:32157–32166 [DOI] [PubMed] [Google Scholar]
- 47. Sargueil B., Hampel K. J., Lambert D., Burke J. M. 2003. In vitro selection of second site revertants analysis of the hairpin ribozyme active site. J. Biol. Chem. 278:52783–52791 [DOI] [PubMed] [Google Scholar]
- 48. Sizova D. V., Kolupaeva V. G., Pestova T. V., Shatsky I. N., Hellen C. U. T. 1998. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 72:4775–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sonenberg N., Hinnebusch A. G. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Spahn C. M. T., et al. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science 291:1959–1962 [DOI] [PubMed] [Google Scholar]
- 51. Svitkin Y. V., et al. 2001. The requirement for eukaryotic initiation factor 4A (eIF4A) in translation is directly proportional to the degree of mRNA 5′ secondary structure. RNA 7:382–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Venkataraman S., Reddy S. P., Loo J., Idarnakanti N., Hallenbeck P. L. 2008. Structure of Seneca Valley virus-001: an oncolytic picornavirus representing a new genus. Structure 16:1555–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wadhwa L., Hurwitz M. Y., Chevez-Barrios P., Hurwitz R. L. 2007. Treatment of invasive retinoblastoma in a murine model using an oncolytic picornavirus. Cancer Res. 67:10653–10656 [DOI] [PubMed] [Google Scholar]
- 54. Wang C., Le S. Y., Ali N., Siddiqui A. 1995. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA 1:526–537 [PMC free article] [PubMed] [Google Scholar]
- 55. Weill L., et al. 2010. A new type of IRES within gag coding region recruits three initiation complexes on HIV-2 genomic RNA. Nucleic Acids Res. 38:1367–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zuker M. 2003. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]