Abstract
Noroviruses (NoVs) are one of the leading causes of gastroenteritis in children and adults. For the last 2 decades, genogroup II genotype 4 (GII.4) NoVs have been circulating worldwide. GII.4 NoVs can be divided into variants, and since 2002 they have circulated in the population before being replaced every 2 or 3 years, which raises questions about the role of their histo-blood group antigen (HBGA) ligands in their evolution. To shed light on these questions, we performed an analysis of the interaction between representative GII.4 variants and HBGAs, and we determined the role of selected amino acids in the binding profiles. By mutagenesis, we showed that there was a strict structural requirement for the amino acids, directly implicated in interactions with HBGAs. However, the ablation of the threonine residue at position 395 (ΔT395), an epidemiological feature of the post-2002 variants, was not deleterious to the binding of the virus-like particle (VLP) to the H antigen, while binding to A and B antigens was severely hampered. Nevertheless, the ΔT395 VLPs gained the capacity to bind to the Lewis x and sialyl-Lewis x antigens in comparison with the wild-type VLP, demonstrating that amino acid residues outside the HBGA binding site can modify the binding properties of NoVs. We also analyzed the attachment of baculovirus-expressed VLPs from six variants (Bristol, US95/96, Hunter, Yerseke, Den Haag, and Osaka) that were isolated from 1987 to 2007 to phenotyped saliva samples and synthetic HBGAs. We showed that the six variants could all attach to saliva of secretors irrespective of the ABO phenotype and to oligosaccharides characteristic of the secretor phenotype. Interestingly, Den Haag and Osaka variants additionally bound to carbohydrates present in the saliva of Lewis-positive nonsecretors. The carbohydrate binding profile and the genetic and mutagenesis analysis suggested that GII.4 binding to Lewis x and sialyl-Lewis x antigens might be a by-product of the genetic variation of the amino acids located in the vicinity of the binding site. Analysis of the binding properties for the six variants by surface plasmon resonance showed that only post-2002 variants (i.e., Hunter, Yerseke, Den Haag, and Osaka) presented strong binding to A and B antigens, suggesting that the GII.4 evolution could be related to an increased affinity for HBGAs for the post-2002 variants. The combination of increased affinity for ABH antigens and of a newly acquired ability to recognize glycans from Lewis-positive nonsecretors could have contributed to the epidemiological importance of strains such as the Den Haag GII.4 subtype.
INTRODUCTION
Noroviruses (NoVs) constitute one of the five genera of the Caliciviridae. They are divided into five genogroups (GI to GV) (14), and human NoVs belong to genogroups I, II, and IV. Genogroups I and II are divided into 8 (GI.1 to GI.8) and 17 (GII.1 to GII.17, excluding GII.11) genotypes, respectively (51). Norovirus outbreaks have been reported worldwide, in people of all ages, and in all ethnic groups. With the improvement in the molecular tools used for the detection of NoVs, laboratory networks set up in Europe and North America showed that genogroup II NoVs belonging to genotype 4 (GII.4) are by far the most predominant NoV genotype. The phylogenetic analysis of the circulating GII.4 isolates for the last 20 years showed that the GII.4 genogroup can be divided into distinct subgroups or variants. Moreover, several authors noticed that the evolutionary rate of the GII.4 strains has accelerated during the last 10 years, and the occurrence of new variants has increased since the appearance of the Farmington variants in 2002. The main feature of these variants is an additional amino acid residue inserted after residue 394, which is located in the hypervariable region of VP1 (12). Epidemiological data and a molecular survey of the NoVs circulating throughout the world showed that until now the insertion, a threonine for most of the variants, was conserved among the GII.4 NoVs (2). Before 2000, the US95/96 variants, which succeeded the Bristol variants and the archival CHDC-like strains (where CHDC refers to Children's Hospital National Medical Center, Washington, DC), were the first NoVs to be classified as pandemic (5). The US95/96 variants were then replaced by the Farmington variants. Since the emergence of the Farmington strain, new variants have been detected worldwide (i.e., Hunter, Yerseke, Den Haag, and, most recently, Apeldoorn) or locally (i.e., Chiba and Osaka), and some of these became predominant for a period of 2 to 3 years before being replaced by a new predominant variant.
The virion is nonenveloped, and the 34-nm capsid is composed of 180 monomeric protein units, VP1, which are organized into 90 dimers. Each VP1 protein can be divided into three domains: N, S, and P. N and S domains are internal and contribute to the architecture of the viral particle; they also interact with viral genomic RNA (35). The external P domain is subdivided into two domains, P1 and P2. In the absence of cell culture for human NoVs, the baculovirus-expressed VP1 protein and the bacterially expressed P domain, which can form virus-like particles (VLP) and P particles, respectively (19, 48), have provided useful molecular tools for the study of the attachment of the NoV to human blood group antigen (HBGA) ligands (A, B, H, and Lewis antigen) (16, 25, 28, 44, 45), which are present in body fluids (e.g., saliva) and epithelial cells, like those in the epithelial tract (27). The structural analysis by X-ray crystallography of the P particles from the GI.1 Norwalk strain and GII.4 VA387 strains bound to A and B trisaccharides revealed the presence of two symmetrical binding pockets located at the interface between the two monomers that compose the dimer unit of the capsid for GII NoV, while for GI NoV each binding pocket lies entirely on the monomer (8, 10). Other mutagenesis studies, which were based upon the results of the structural analysis, confirmed the importance of the amino acid residues implicated in the carbohydrate binding site or its surrounding on ligand recognition (49). Using saliva binding assays, GI and GII human NoVs could be divided into five ABH-related (saliva from secretor individuals) and three Lewis-related (saliva from nonsecretor individuals) binding profiles, as reviewed previously (49).
Human challenge studies showed there was a direct link between the presence of HBGAs in the gut of secretor individuals and the NoV infection (18, 24). Histological studies of human intestine biopsy specimens clearly showed binding of Norwalk virus VLPs to H antigen expressed at the surface of epithelial cells. Additionally, transfection experiments showed that the binding of VLPs was dependent of an active FUT2 gene, which encodes the α1,2-fucosyltransferase in the small intestine and defines the secretor status (28). For 20% of the Caucasian population defined as nonsecretors, the presence of a nonsense mutation in the FUT2 gene (G428A) completely abrogates expression of FUT2-dependent HBGAs (i.e., A, B, H, Lewis b, and Lewis y antigens) (22). Of note, for nonsecretor individuals, Lewis a and x antigens might be detected in saliva or intestinal cells, provided that the FUT3 and/or FUT5 gene is functional.
It has been suggested that a broader binding profile to HBGAs and a higher evolution rate might explain why GII.4 NoVs now predominate over NoVs from other genogroups (6, 46). However, little is known about the mechanisms that cause GII.4 variants to circulate for a couple of years before disappearing from the community. It has been hypothesized that genetic drift of the capsid allows GII.4 NoVs to escape herd immunity and modulates their binding capabilities (13).
We performed a qualitative analysis of the binding capabilities of the VLPs derived from six GII.4 variants that were present over a 20-year period. The analysis was based on binding experiments on phenotyped saliva and a panel of synthetic carbohydrates. The study includes the analysis of amino acid residues from the binding pocket that contribute to the recognition of HBGAs. In addition, using modified VLPs, we evaluated the biological importance of the newly inserted threonine residue, which is a key epidemiological and genetic feature of the new GII.4 NoVs. Finally, we analyzed the quantitative aspect of the attachment of the panel of VLPs to A, B, and H antigens using surface plasmon resonance (SPR) technology.
MATERIALS AND METHODS
NoV isolates.
The NoV strains used for this study were isolated from patients suffering from gastroenteritis in France (i.e., Dijon171 and E1057 strains were US95/96 and Hunter GII.4 variants, respectively), Egypt (i.e., Cairo7, Cairo1, and Cairo4 isolates belonged to the Yerseke, Den Haag, and Osaka GII.4 variants, respectively), and the United States (i.e., the MD145 strain was representative of the Bristol variant), as documented previously (15, 20, 21, 34) (Table 1).
Table 1.
List of the GII.4 isolates used in this study
| Isolate name (reference) | GenBank accession no. | Setting (year) | Varianta |
|---|---|---|---|
| MD145 (15) | AY032605 | Nursing home (1987) | Bristol |
| Dijon171 (34) | AF472623 | Community (1996) | US95/96 (1996) |
| E1057 (20) | EU876890 | Retirement home (2005) | Hunter (2004) |
| Cairo7 (20) | EU876887 | Children's hospital (2006) | Yerseke (2006a) |
| Cairo1 (20) | EU876892 | Children's hospital (2006) | Den Haag (2006b) |
| Cairo4 (20) | EU876884 | Children's hospital (2006) | Osaka (2007) |
Cloning strategy and recombinant baculovirus.
The primers used during the study were obtained from Eurogentec (Seraing, Belgium). Primers FW2, RT2, and RT5 have been described previously (20). For the Hunter variant and the Egyptian isolates, the cloning of the complete open reading frame 2 (ORF2) into the plasmid pGEM-T Easy has been documented previously (20). The ORF2 fragment was amplified using ORF2 pGEM-based constructs as the template with the forward primer 5′-GAAGATCTATGAAGATGGCGTCGAATGACGC-3′, containing a BglII restriction site at the 5′ end (underlined), and the reverse primer RT5 corresponding to the 5′ end of ORF3, described previously (20). The amplified fragment was gel purified and cloned into pGEM-T Easy vector prior to digestion with BglII and NotI restriction enzymes. The insert with an engineered BglII site was then gel purified and ligated into transfer vector pVL1392 (Pharmingen). The ligation mixture was used to transform JM109 competent cells (Promega). Positive clones were screened by PCR with primers FW2 and RT2 that were located in the conserved part of ORF2 (20). For the selected positive clones, the nucleotide sequence of the entire ORF2 was determined for the detection of any inadvertent mutation.
The recombinant baculovirus was obtained by cotransfection of the pVL transfer vector and the linearized baculovirus genome into Sf9 cells following the manufacturer's recommendations (Pharmingen). The recombinant baculovirus was plaque purified, and baculovirus clones expressing large amounts of VLPs were selected by using an immunoassay from RD-Biopharm (Saint-Didier au Mont d'Or, France). Titers of the recombinant baculovirus seeds were determined by plaque assay, and a high-titer viral stock was produced by infecting Sf9 cells at a low multiplicity of infection ([MOI] i.e., below 0.2).
The Dijon171 recombinant baculovirus has been documented previously (34). The MD145 baculovirus was a kind gift of Kim Green (NIAID, NIH, Bethesda, MD). Here, the recombinant VLPs of the GII.4 NoVs will be named after the variants to which the isolates belong (Table 1).
Mutagenesis of the Hunter transfer vector.
The transfer vector corresponding to ORF2 of the Hunter variant (E1057 isolate) was used as the template for site-directed mutagenesis. Site-directed mutagenesis of the Hunter construct was performed with a QuikChange mutagenesis kit (Stratagene) to replace (mutagenized Hunter VLPs: S343A, T344A R345Q, D373N, S442A, G443A, and Y444F) and delete [mutagenized Hunter VLPs: ΔS343, Δ(T344A R345Q), ΔD373, ΔS442, ΔG443, and ΔY444] the amino acid residues that are involved in the binding of the α-fucose ring of the HBGAs (Fig. 1A). The threonine residue at position 395 corresponding to the inserted amino acid of the post-2002 GII.4 isolates was also deleted to produce the recombinant ΔT395-Hunter baculovirus. The ORF2-mutagenized pVL constructs used in this study and the primers used for the mutagenesis are summarized in Table 2. Presence of the mutation was confirmed by sequencing of the complete ORF2 before production of the recombinant baculovirus as described above.
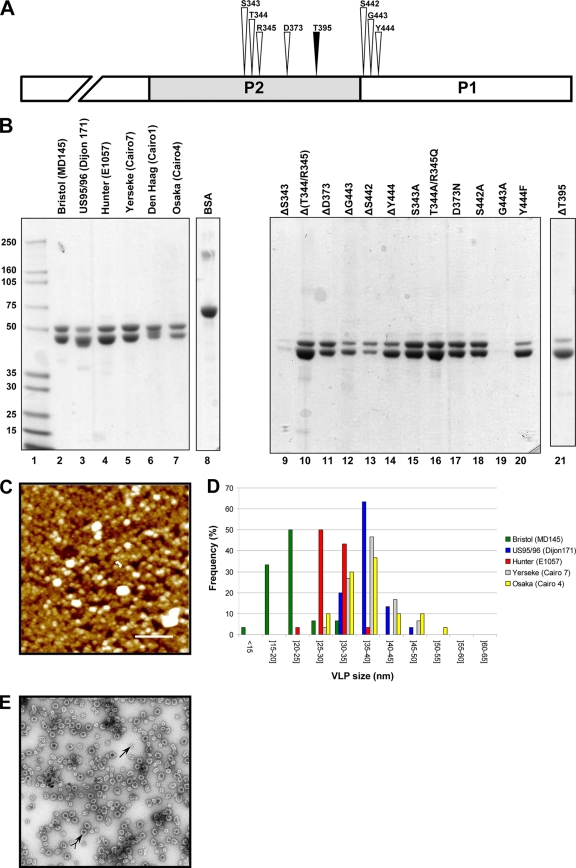
Fig. 1.
Characterization of the purified VLPs of the GII.4 variants. (A) Drawing to scale of the amino acid residues (S343, T344, R345, D373, S442, G443, and Y444) from the α-fucose binding site (white arrowhead) (8, 49) and the inserted threonine residue, T395 (black arrowhead). The NH2 terminus of the P1 domain is truncated. (B) SDS-PAGE analysis of the VLPs from the GII.4 variants (lanes 2 to 7) and the mutagenized Hunter VLPs (lanes 9 through 21). The name of the NoV strain is indicated in parentheses for each variant. The mutagenized VLPs originated from the Hunter variant (strain E1057), and the location of the amino acid residues is based upon the ORF2 amino acid sequence of the E1057 strain (GenBank accession number EU876890). Two micrograms of bovine serum albumin (Pierce) was added as a control for the protein estimation (lane 8). Lane 1 contains the protein molecular size markers that are indicated on the right side of the gel. (C) Atomic force microscopy imaging of the Hunter purified VLPs. The aggregates are shown in white. One of the VLPs is indicated by a bracket. Scale bar, 250 nm. (D) Size distribution of the VLPs as determined from atomic force microscopy imaging. The variant VLPs are color coded according to the legend on the right side of the graph. (E) Electron micrograph of the CsCl-purified Hunter VLPs after negative staining. Complete and subunit VLPs are indicated by arrows with and without a tail, respectively.
Table 2.
Oligonucleotides used for the site-directed mutagenesis of the E1057 pVL vector (Hunter variant)
| Mutation | Oligonucleotide sequencea |
|---|---|
| S343A | 5′-GAGGATGCGCGACCCGTGTCAC-3′ |
| ΔS343 | 5′-GAGGATGC(TCG)ACCCGTGTCAC-3′ |
| T344A R345Q | 5′-GAGGATGCTCGCCCAAGTCACAAAGCCAC-3′ |
| Δ(T344 R345) | 5′-GAGGATGCTCG(ACCCGT)GTCACAAAGCCAC-3′ |
| D373N | 5′-CTGATACAAGCAATAACTTTGAAACTGCC-3′ |
| ΔD373 | 5′-CTGATACAAGCAAT(GAC)TTTGAAACTGCC-3′ |
| ΔT395 | 5′-CAGATGTAGTACC(ACC)CACCAAAATGAACCC-3′ |
| S442A | 5′-CTATGCCCGCTGCGCGGTATCCCAACATG-3′ |
| ΔS442 | 5′-CTATGCCCGCTGC(AGC)GGTATCCCAACATG-3′ |
| G443A | 5′-GCCCGATGCAGCGCGTATCCCAACTG-3′ |
| ΔG443 | 5′-GCCCGATGCAGC(GG)TATCCCAACTG-3′ |
| Y444F | 5′-GATGCAGCGGTTTCCCAACATGAATTT-3′ |
| ΔY444 | 5′-GATGCAGCGG(TAT)CCCAACATGAATTT-3′ |
Modified codons are underlined. Deleted codons are indicated in parentheses.
Production and purification of the virus-like particles.
The VLPs were produced by infecting High-Five cells (Invitrogen) at a high MOI in serum-free Express Five medium (Invitrogen). The VLPs were concentrated and purified from the cell medium at the sixth day postinfection as described previously (3). The purified VLPs were resuspended overnight at 4°C in TNC buffer (10 mM Tris, 140 mM NaCl, 10 mM CaCl2, pH 7.4) containing 20 μg/ml leupeptin (Sigma, Saint-Quentin Fallavier, France). The purified VLPs were quantified using a bicinchoninic acid protein assay kit from Pierce (Perbio Science, Bezons, France), diluted to 1 mg/ml to prevent precipitation, and stored at 4°C or divided into aliquots in 10-μl vials and stored at −40°C for short- or long-term storage, respectively (Fig. 1B). Up to 4 μg of protein mixture was resolved by SDS-PAGE in a 4 to 12% Bis-Tris polyacrylamide (PAA) gel (Invitrogen). Proteins were visualized by staining with Coomassie blue (Pierce).
Saliva typing for carbohydrates and genotyping for the secretor phenotype.
Forty-six samples of saliva and swabs of buccal epithelial cells were collected from healthy individuals. The study was approved by the Nantes University Hospital Review Board (study number BRD02/2-P), and informed consent was obtained from all the donors. The saliva samples were typed by enzyme-linked immunosorbent assay (ELISA) for the presence of A, B, and O blood group antigens and Lewis antigens as described previously (26). Genetic material was extracted from swab samples; FUT2 genotyping (e.g., secretor status) was performed by PCR as described previously (26). ABO phenotyping was performed by classical hemagglutination and confirmed by ELISA on saliva samples from secretor individuals.
Binding assay of the VLPs on saliva.
For the saliva analysis, an ELISA was performed as described previously except that 1,000-fold dilutions of saliva and 500 ng of purified VLPs per assay were used (28). Additionally, the GII.4-bound VLPs were detected by a GII.4-specific in-house monoclonal antibody incubated for 1 h at room temperature. Peroxidase-conjugated anti-mouse antibodies (Vector/ABCYS, Paris, France) were incubated for 30 min at 37°C. Peroxidase activity was detected with 3,3′,5,5′-tetramethyl benzidine (KPL/Eurobio, Courtaboeuf, France). The reaction was stopped after an 8-min incubation at room temperature with 2N HCl prior to reading the absorbance at 450 nm. The background was arbitrarily fixed at an optical density (OD) of 0.2 and was subtracted from each OD value.
A panel of saliva samples from secretor and nonsecretor individuals was treated with 10 mM NaIO4. The ELISA plates were first coated with 1,000-fold dilutions of saliva boiled in carbonate/bicarbonate buffer, pH 9.6, and left overnight at 37°C. After the plates were washed three times with phosphate-buffered aline (PBS), pH 7.4, 100 μl of 10 mM NaIO4 diluted in 50 mM CH3COONa, pH 5, was added to each well and incubated for 30 min at room temperature. The wells were washed three times with PBS, and the ELISA was continued as described above.
Binding assay of the VLPs on synthetic carbohydrates.
The human serum albumin (HSA)- and bovine serum albumin (BSA)-conjugated carbohydrates were purchased from Isosep AB (Uppsala, Sweden) and Dextra laboratories (Reading, United Kingdom), respectively. The HSA and BSA carbohydrates were provided with an average of 17 and 20 carbohydrate moieties per protein, respectively (Table 3). The glycoconjugates that were used during the study are listed in Table 3. The ELISA was performed as described previously. Briefly, glycoconjugates were diluted in pH 9.6 carbonate/bicarbonate buffer at a concentration of 1 μg/well (unless otherwise indicated), coated onto Immulon MaxiSorp plates (Nunc), and left overnight at 37°C. The plates were washed three times with PBS–0.05% Tween prior to incubating 500 ng/well of purified VLPs at 37°C (i.e., HSA or BSA glycoconjugates) for 4 h. The method used to detect the bound VLPs by the monoclonal antibody was similar to that described above.
Table 3.
Neoglycoconjugates used for the study
| Carbohydrate moiety | Epitope | Carrier | Avg no. of sugar residues per carriera |
|---|---|---|---|
| Lacto-N-fucopentaose I | H type 1 | HSA | 25 |
| BSA | 20 | ||
| 2′-Fucosyllactosamine | H type 2 | HSA | 19 |
| A heptasaccharide | A heptasaccharide | HSA | 18 |
| A trisaccharide | A trisaccharide | HSA | 19 |
| BSA | 19 | ||
| B trisaccharide | B trisaccharide | HSA | 19 |
| BSA | 21 | ||
| Lacto-N-difucohexaose I | Lewis b | HSA | 8.5 |
| Lacto-N-fucopentaose III | Lewis x | HSA | 12 |
| Lewis y-tetrasaccharide | Lewis y | HSA | 15 |
| Lacto-N-fucopentaose II | Lewis a | HSA | 23 |
| Sialyl-Lewis x-hexaose | Si-Lewis x | HSA | 13 |
| Sialyl(mono), monofucosyllacto-N-tetraose | Si-Lewis a | HSA | 12 |
| Sialyl-lacto-N-fucopentaose | Si-LNFb | HSA | 19 |
| Sialyl-lacto-N-tetraose | Si-LNTb | HSA | N/A |
| Lacto-N-tetraose | LNTb | HSA | N/A |
| Sialyl-lacto-N-neotetraose | Si-LNnTc | HSA | N/A |
HSA and BSA linkers were acetyl phenylenediamine and six-atom spacer, respectively. N/A: not applicable.
Possesses type 1 precursor motif.
Possesses type 2 precursor motif.
Analysis of the VLP binding on carbohydrates by SPR.
The binding of purified VLPs to lacto-N-fucopentaose I (LNFP-I) and to A and B trisaccharides conjugated to BSA (Dextra Laboratories) (Table 3) was analyzed by surface plasmon resonance at 25°C with a Biacore 2000 instrument (GE Healthcare). The homemade gold chips for SPR were designed and fabricated as described previously (4). The chemical functionalization was obtained by using a mixture of 11-mercapto-1-undecanol (11-MUOH) and 16-mercapto-1-hexadecanoic acid (16-MHA) (Sigma-Aldrich). The mixture of 11-MUOH–16-MHA (97/3, by mole) at 1 mM in absolute ethanol was sonicated for 10 min using an Elma sonicator (power, 90W; frequency, 50/60 Hz). The chip was rinsed with ultrapure water. Then, the carboxyl groups were activated using N-hydroxysuccinimide (NHS) at 50 mM and N-ethyl-N′-(3-dimethyl aminopropyl)-carbodiimide hydrochloride (EDC) at 240 mM (Amine Coupling Kit; Biacore AB, Uppsala, Sweden) directly with the help of the Biacore's fluidic cartridge at 2 μl/min for 14 min. This procedure prepares the chips for the immobilization step. In this way, covalent immobilization of the three glycoconjugates was obtained in three separate lanes on the same chip, allowing the simultaneous analysis of the same VLP preparation on three different synthetic carbohydrates, with the fourth line being reserved as a control surface. For each glycoconjugate, 300 to 370 resonance units (RU) corresponding to 86 to 92.5 fmole/mm2 were linked on the sensor chip. VLPs were diluted in running HBS buffer (0.01 M HEPES, 0.15 M NaCl, 3 mM EDTA, and 0.005% surfactant P20 at pH 7.4) provided by the company and were injected for 120 s at a flow rate of 10 μl/min and a concentration of 2 ng/μl. The injection was stopped, and the dissociation was observed in running buffer for 120 s. The chip was recycled by the injection of 5 μl at 10 μl/min of 10 mM Gly solution at pH 2.5 prior to new analysis. The experimental chips were stored in a humid atmosphere at 4°C after use.
AFM.
For atomic force microscopy (AFM) imaging of the VLPs, the different samples were used at around 1 mg/ml. Twenty-five microliters of the sample was adsorbed on freshly cleaved mica for 30 min and washed three times with HBS buffer to remove nonadsorbed VLPs. The AFM experiment was carried out with a Nanoscope III instrument from Veeco (Santa Barbara, CA). Imaging was performed in contact and oscillating contact modes (TappingMode) using NP-S (for nitride probe, sharpened) oxide-sharpened silicon nitride probes (Veeco) with spring constants of 0.32 N/m or 0.58 N/m and resonance frequencies ranging from 8.5 to 9.5 kHz. For the feedback controls, typical values of set points for imaging were between 0.5 and 1.5 V, depending on the scan size and drive amplitude in the oscillating contact mode. The oscillation amplitude was generally maintained at 5 to 10 nm away from the surface. For each variant, the mean diameter of the VLP was determined from the diameter of 30 globular VLPs.
MST of the ORF2 amino acid sequences.
To study evolution between strains, a minimum spanning tree (MST) was constructed using the default setting from the Bionumerics package (Applied Maths BVBA, Sint-Martems-Latem, Belgium) as described previously (20). The MST was based upon 496 complete available ORF2 amino acid sequences of the GII.4 from GenBank corresponding to Bristol, US95/96, Farmington, Hunter, Chiba, Yerseke, Den Haag, Apeldoorn, and Osaka variants.
Statistical analysis.
The statistical analyses were performed with StataCorp statistical software (StataCorp LP, College Station, TX). For the saliva testing, a Kruskal-Wallis test was used to determine whether VLP binding to typed saliva samples was different for each variant, and US95/96 VLP was used as the standard for comparison. An analysis of variance (ANOVA) test was used for paired comparison of nonsecretor saliva. To study the relative affinity of the VLPs for the A, B, and H antigens, fractional polynomials were used to model the VLP-ligand interaction by linear regression. The same statistical model was also used to determine whether the changes observed in the binding profile of the Y444F and ΔT395 VLPs for the ABO antigens were statistically significant compared with binding of the wild-type (wt) Hunter VLPs.
RESULTS
Production and purification of the virus-like particles.
The VLPs were purified from 4 × 108 to 6 × 108 infected cells and yielded 1 to 10 mg of purified VLPs, depending on the recombinant baculovirus. The purified VLPs yielded a distinct band with a buoyant density of 1.31 g/ml in cesium chloride. The Y444F and T395 Hunter mutants were rather fragile in the cesium chloride gradient, and a discontinuous sucrose gradient was preferred for the purification step, as described previously (3). Following centrifugation, a wide band was observed at the interface between the 30 and 40% sucrose fractions. The protein analysis in polyacrylamide gel showed that VLPs were purified to greater than 90% homogeneity. The purified VLP fraction was composed of the VP1 protein at 57 kDa and a cleaved version at 54 kDa, as described previously (3) (Fig. 1B). The NoV origin of the doublet was confirmed by Western blotting using polyclonal serum raised against MD145 antigen (provided by Kim Green) (data not shown). The poor yield observed for the ΔS343 and G443A mutagenized VLPs suggests that the mutations of these amino acid residues made the recombinant VLPs unstable. Interestingly, unlike ΔS343, the S343A mutation was not detrimental to VLP production. In contrast, G443A hampered VLP production, whereas the deletion of G443 did not.
To control the integrity of the purified material, the size of each VLP was analyzed by AFM (Fig. 1C). The majority of the VLP preparations presented globular particles with an average diameter ranging between 30 and 38 nm (US95/96, 37.2 ± 3.7 nm; Hunter, 29.6 ± 3.2 nm; Den Haag, 37.6 ± 4.8 nm; Osaka, 36.8 ± 6.2 nm) and corresponded to complete VLPs (also called VP1180) as described previously (Fig. 1D) (37). For the Bristol isolate, the VLPs were smaller, with diameters averaging 21.3 ± 4.7 nm, which suggests that they might have been subunit particles and might have been composed of 60 monomers (also called VP160) (3, 37). The observation by electron microscopy of the cesium chloride-purified VLPs from the Hunter variant showed that the preparation was essentially composed of full and empty particles of 35 nm in diameter (Fig. 1E). Of note, the Yerseke VLPs did not adsorb on the mica surface using HBS buffer. Because we used low-concentration protein solutions for the AFM experiments, the salt concentration might have been too high and may have inhibited protein adsorption onto the mica, as described previously (11).
Mutagenesis of the amino acids participating in the HBGA binding site.
In previous studies, the amino acid residues participating in the attachment of the HBGA to the P1/P2 domains were characterized by crystallography, which showed that S343, T344, R345, D374, S441, G442, and Y443 residues of the GII.4 VA387 NoV strain (US95/96 variant) interacted directly with the carbohydrate antigens (Fig. 1A). Of note, these residues are conserved for all the GII.4 variants, including the recently described Osaka and Apeldoorn variants (data not shown). Even though it has been demonstrated that a bacterially expressed P domain had a blood group antigen binding profile similar to that of the entire VLP (43), we used mutagenized VLPs to study the HBGA binding site because deletions or modifications of these VLPs might also be detrimental to the structure of the entire VLP and to their binding to some antigens. Moreover, it has been reported that VLP-HBGA complexes are usually more stable than the interaction between the P domain dimer and HBGA (8). We first deleted each amino acid and showed that most of the VLPs could be recovered after isopycnic centrifugation into a cesium chloride gradient, suggesting that modified VP1 protein could still assemble into VLPs. Nevertheless, the deletion of any residue completely abolished binding to the HBGA associated with either HSA or polyacrylamide (PAA) (data not shown). Our results confirmed that each residue was involved in the attachment of a wide spectrum of carbohydrates. We also performed site-directed mutagenesis of each of these amino acid residues, which were replaced by homologous amino acids (Table 2). All but the Y444F mutation completely abolished attachment of the HBGA glycoconjugates. A previous study showed that the aromatic ring of the Tyr residue interacted with the α-fucose ring of the H antigen through van der Waals interactions (8). Using HSA glycoconjugates, binding of the Y444F VLPs was observed for Lewis b and y antigens and for glycoconjugates carrying the B trisaccharides, H type 1, and A tri-, hexa-, and heptasaccharides (Fig. 2A). For the other antigens, binding was absent or residual (i.e., OD at 450 nm [OD450] of <0.25). To determine whether the binding to A, B, and H blood group antigens was modified, wt and mutagenized Hunter VLPs were tested on serially diluted A trisaccharides, B trisaccharides, and LNFP-I–BSA glycoconjugates (Fig. 2B). The Hunter VLP was used as the standard for comparison for the statistical analysis using a fractional polynomial model. Given the similar numbers of carbohydrates that were attached to the BSA, it was possible to compare the VLPs' attachment patterns (Table 3). Binding of the mutagenized VLPs to A and B glycoconjugates was markedly lower than that observed for the wt Hunter VLPs (P = 0.001 for the A antigen). For the B antigen, the lower binding was not significant for Y444F VLPs (P = 0.057). For the H antigen, although the binding we observed was slightly lower for the mutant VLPs, the difference was not significant (P = 0.28). In conclusion, our analysis confirms the importance of amino acids from the GII.4 binding site previously deduced from alanine mutagenesis of P domains from other strains. Moreover, our data clearly showed that replacing the tyrosine at position 444 by a phenylalanine did not alter recognition of the H type 1 epitope, confirming the lack of involvement of the tyrosine hydroxyl group in binding to this motif, although this mild structural modification was sufficient to modify the attachment profile of the Hunter isolate.
Fig. 2.
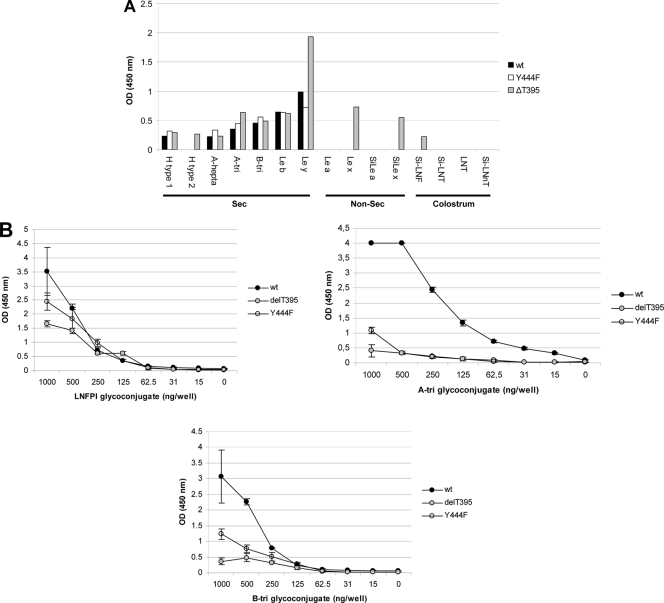
Comparison of the binding of the mutagenized VLPs Y444F and ΔT395 with the wt Hunter VLPs. (A) The VLPs were tested in duplicate on a panel of HSA glycoconjugates, and the mean values are plotted on the graph. Ordinate and abscissa indicate the optical density at 450 nm (OD450) and the nature of the glycoconjugate, respectively. The legend is at the right of the graph. (B) Relative binding of the Y444F, ΔT395, and wt Hunter VLPs for the LNFP-I (H type 1 pentasaccharide) and A and B BSA conjugates. LNFP-I and A and B trisaccharide conjugates were diluted 2-fold in carbonate/bicarbonate buffer, pH 9.6, from 103 to 0.15 μg per well. The amount of coated neoglycoconjugate is indicated in nanograms per well (abscissa). For each variant and neoglycoconjugate, the mean results of triplicate binding experiments and standard deviations are shown (vertical bars). Binding values are given by the absorbance at 450 nm (ordinate). The glycoconjugate used for each assay is indicated on the graph (abscissa). A-tri, A trisaccharide; B-tri, B trisaccharide; Le, Lewis; Sec, secretor; Non-sec, nonsecretor.
Role of the threonine residue at position 395.
Since 2002, the new GII.4 NoV strains have been characterized by conserved mutations in their capsids and the insertion of one amino acid at position 395 in the hypervariable P2 domain of the capsid proteins (12). This additional amino acid, which is mostly a threonine residue, has been conserved for all the GII.4 strains ever since the emergence of the Farmington variants. Our goal was to determine whether the additional threonine residue had a biological function in the attachment to carbohydrates. The threonine residue was deleted from the Hunter VLPs. The purified VLPs were screened using the panel of HSA-linked glycans. The attachment profile of the mutated VLPs was similar to that observed for the wt Hunter VLPs except that recognition of the Lewis y antigen was significantly higher than for the wt VLPs (Fig. 2A). In addition, we observed that, unlike wt Hunter VLPs, ΔT395 VLPs could attach to the H type 2, Lewis x, and sialyl-Lewis x motifs. Of note, the ΔT395 VLPs were also assayed with PAA glycoconjugates, where a marked binding of the VLPs was also observed for Lewis b and y and also to Lewis x-PAA conjugate (data not shown).
The binding of ΔT395 mutant VLPs to A and B glycoconjugates was markedly lower than that observed for the wt Hunter VLPs (P = 0.001 and P = 0.003 for the A and B antigens, respectively) but not for the H antigen (P = 0.08). Overall, our data suggest that deletion of the inserted threonine at position 395 induced qualitative (binding to Lewis x and sialyl-Lewis x antigens) and quantitative (decrease of binding to A and B epitopes) changes in the binding profile to HBGAs. Thus, deletion of the threonine residue suggests that the mutagenesis of amino acids that are not directly involved in the HBGA binding site can modify the attachment of NoVs to these carbohydrate ligands, as previously predicted by modeling the Farmington capsid (25).
Binding profile of the GII.4 variants. (i) Saliva binding assays.
The mutational analysis showed that certain amino acids might trigger the suppression of VLP attachment to HBGAs and/or the modification of the binding profile. We then extended our investigation to a panel of six GII.4 variants that were first tested on phenotyped saliva from secretor (n = 34) and nonsecretor (n = 12) individuals representative of the ABO and Lewis phenotypes (Fig. 3). We observed binding of the six variants to the ABO saliva samples, irrespective of the presence of the Lewis antigens. However, the magnitude of the binding significantly varied according to the nature of the VLPs and the antigen present in the saliva (P = 0.00001, two-way ANOVA). For Bristol, US95/96, Hunter, and Yerseke, no binding was observed for nonsecretor saliva. Conversely, we observed strong binding to nonsecretor saliva for Den Haag and Osaka variants. For the latter, the binding correlated with the presence of Lewis antigens (P = 0.0001, Kruskal-Wallis). The binding to Lewis-positive nonsecretor saliva was significantly higher for Den Haag than for Osaka (P = 0.0104, Kruskal-Wallis). This observation indicates that Den Haag and Osaka GII.4 variants are able to bind nonsecretor saliva through antigens synthesized by the FUT3 enzyme. Of note, the binding to nonsecretor saliva of the Den Haag and Osaka VLPs was abolished after sodium periodate treatment, showing the involvement of carbohydrates (data not shown).
Fig. 3.
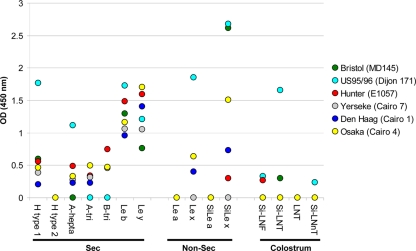
Saliva binding assays of the GII.4 variants. The binding experiments were performed in duplicate for each sample, and the mean values are given on the graph (OD450; ordinate). The Lewis status is indicated on the abscissa by minus (absence of Lewis antigen) and plus (presence of Lewis antigen) signs. The nonsecretor (non sec) and HBGAs are also indicated on the abscissa and are separated by dashed lines. The type of VLP is indicated on the right side of the graph.
(ii) Synthetic glycoconjugate binding assays.
To determine precisely which antigens might be recognized by the NoV GII.4 VLPs, binding assays with the GII.4 variants were performed by ELISA using 15 HSA glycoconjugates (Table 3 and Fig. 4). Binding to H type 1 was observed for all the variants using HSA glycoconjugates. Inversely, we observed no binding to H type 2. For the A antigen, we used hepta- and trisaccharides conjugated to BSA. Weak binding was observed for the Hunter, Yerseke, Den Haag, and Osaka variants, using A tri- or heptasaccharide conjugates. No binding was observed for the Bristol variant, and US95/96 VLPs bound only to the heptasaccharide conjugate. Our data suggest that the position of the ligand on the carrier is essential for the efficient binding of VLPs. Binding of the Hunter, Yerseke, Den Haag, and Osaka VLPs to the B trisaccharide was observed, while there was no attachment of the Bristol and US95/96 variants. The fact that all variants efficiently bound to the H type 1 antigen and additionally that Bristol and US95/96 variants bound poorly to A and B trisaccharides was later confirmed by using trisaccharides linked to BSA (Fig. 5). For the Lewis-related carbohydrates, we observed efficient binding of the variants to Lewis b and Lewis y antigens, which can be found in the secretor population. Inversely, no binding was observed to the Lewis a antigen, which is found in large amounts in Lewis-positive nonsecretor individuals. The data for Lewis x antigen were different from those for Lewis a antigen. We observed that US95/96, Den Haag, and Osaka VLPs could efficiently bind the Lewis x antigen. It has recently been reported that sialyl-Lewis x antigen could attach the US95/96 variant (36). Despite the fact that the biological meaning of this finding remains unknown and the fact that sialyl-Lewis antigens are usually absent from the gut of healthy individuals (9, 23, 31), we further investigated the binding of the GII.4 variant on sialylated neoglycoconjugates (i.e., sialyl-Lewis x and sialyl-Lewis a). The sialyl-Lewis x antigen was strongly recognized by the Bristol and US95/96 VLPs and less well by the Osaka, Den Haag, and Hunter VLPs. These results indicate that the binding to sialylated structures is variant dependent, but they do not show a relationship with the ability of the Den Haag and Osaka VLPs to bind to Lewis-positive, nonsecretor saliva, suggesting that binding to salivary FUT3-dependent structures of nonsecretors requires recognition of more complex motifs than those represented by the neoglycoconjugates that were tested. Thus, we cannot exclude the possibility that the Lewis a antigen is involved in binding to nonsecretor saliva because binding to synthetic Lewis a carbohydrate was negative.
Fig. 4.
Binding of the GII.4 variant VLPs to a panel of neoglycoconjugates. The binding experiments were performed in duplicate for each HSA glycoconjugate, and the mean values are plotted on the graph. Ordinate and abscissa indicate the OD at 450 nm and the nature and the origin of the oligosaccharide moiety, respectively. Details about the neoglycoconjugates are shown in Table 3. Variants are identified by the legend at right, with the name of the isolate in parentheses.
Fig. 5.
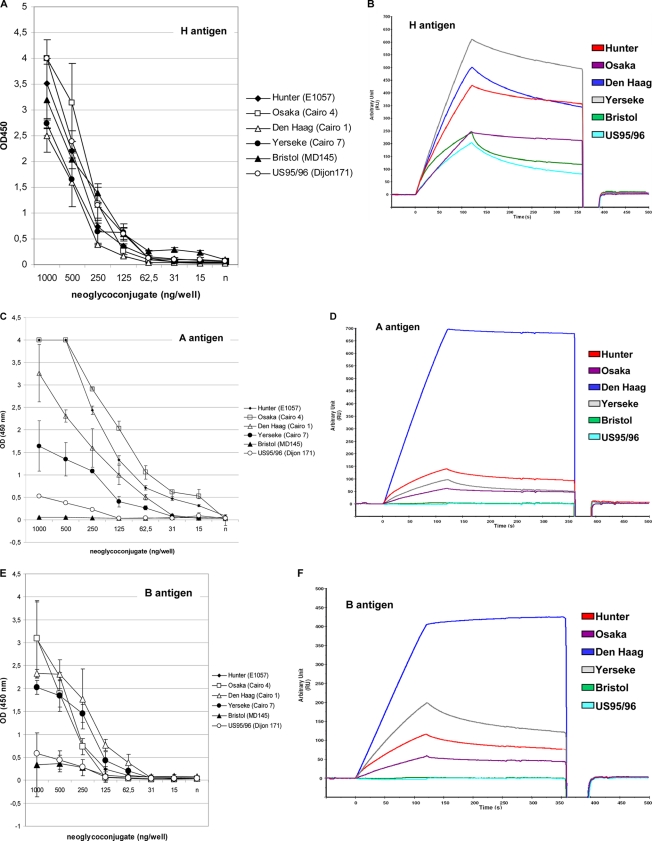
Relative binding affinity of the GII.4 NoV variants for the LNFP-I (H type 1 pentasaccharide) and A and B BSA conjugates by ELISA (A, C, and E) and SPR (B, D, and F). The H antigen (A and B), A antigen (C and D), and B antigen (E and F) BSA conjugates used for the ELISA and SPR analysis originated from the same stock. The neoglycoconjugates are indicated above each graph. The ELISA binding assay was similar to that described for the Hunter mutated VLPs (Fig. 2B). For each variant and neoglycoconjugate, mean results of triplicate binding experiments and standard deviations are shown (vertical bars). Binding values are given by the absorbance at 450 nm (ordinate). For the SPR binding assay, the same VLP and neoglycoconjugate preparations were used as for the ELISA. The response (ordinate) is given in resonance units (RU). The sensorgrams are color coded as indicated on the graphs.
It was recently shown that ganglioside-linked sialic acid might act as a murine norovirus (MNV) docking molecule (50). However, in our study, no binding of the GII.4 variants was observed using synthetic sphingolipid (globoside) and the GM1 pentasaccharide (ganglioside containing one sialic acid residue) (data not shown). This preliminary data suggest that the receptor of human NoVs might be somewhat different from the MNV receptor. Lacto-N-tetraose (LNT), sialyl-lacto-N-tetraose (Si-LNT), and sialyl-lacto-N-neotetraose (Si-LNnT) as well as lacto-N-fucopentaose type I (LNFP-I; presenting the H determinant) are free oligosaccharides that are found in human milk and may function as decoy receptors of human bacterial pathogens (33). Lacto-N-tetraose, Si-LNT, and sialyl-lacto-N-fucopentaose (Si-LNF) present a disaccharide motif, which is similar to type 1 precursor (Galβ1,3GlcNAcβ1). The Si-LNnT possesses a disaccharide motif, which is similar to type 2 precursor (Galβ1,4GlcNAcβ1). Weak binding was observed for Hunter and Bristol VLPs on Si-LNF and Si-LNT glycoconjugates, respectively. US95/96 VLPs could attach Si-LNF, Si-LNT, and Si-LNnT; strong binding was observed for Si-LNT. These data suggest that type 1 and 2 precursor antigens are poorly recognized by most of the GII.4 NoVs (except for the US95/96 variants). The role of the sialic acid moiety in the binding of VLPs remains to be determined.
In summary, our data showed that all the GII.4 variants could attach secretor-related antigens (i.e., A, B, H, Lewis b, and Lewis y antigens). We also observed that Lewis x and sialyl-Lewis x antigens could be recognized in a strain-dependent manner unrelated to the saliva binding patterns.
Relative affinity of GII.4 norovirus variants for A, B, and H blood group antigens.
Two methods were used to determine whether the relative affinity to glycans was variant dependent. We analyzed VLP binding to ABO blood group antigens by enzyme immunoassay (EIA) and SPR. Our previous experiment showed that VLP binding to synthetic carbohydrates was lower than that to saliva. Because binding to A and B HSA glycoconjugates was somewhat lower than that observed for H type 1, Lewis b, and Lewis y synthetic antigens (Fig. 4), we used trisaccharidic A and B antigens linked to a six-atom spacer and BSA to study the relative affinity of the GII.4 variants. Preliminary experiments showed that binding was more efficient and suitable for SPR analysis using these glycoconjugates (data not shown). To avoid any discrepancy between glycoconjugates, the same VLP preparation was used throughout the experiment using the three antigens, and the binding assays were performed in triplicate.
Overall, 95%, 77.6%, and 79.7% of the data fit the model for the H, A, and B antigens, respectively, using fractional polynomials. All of the GII.4 variants bound to the H antigen BSA glycoconjugates with similar dilution curves, and the detection limit was 62.5 ng/well of glycoconjugate. No statistical difference was observed with a P value above 0.05 for each type of VLP and the Hunter VLP used as the standard for comparison (Fig. 5A). Hunter and Osaka VLPs showed the highest binding for A antigen, followed by the Den Haag VLP, with a detection limit below 15 ng/well of glycoconjugate (Fig. 5C). No statistical difference was observed for the three variants. Conversely, the detection rate was lower for Yerseke, US95/96, and Bristol VLPs. The affinity of these three GII.4 variants for A antigen was statistically significantly lower than that of the Hunter VLP (P ≤ 0.001). For B antigen, all but US95/96 and Bristol VLPs showed similar binding affinities (P ≥ 0.05) (Fig. 5E). For US95/96 and Bristol variants, the signals were markedly lower, suggesting a lower relative affinity for B antigen. The difference was statistically significant (P = 0.03).
Surface plasmon resonance analysis.
Because the SPR signal is proportional to the refractive index at the surface of the chip (in the first 150 nm from the surface), we empirically estimated the coverage of the chip by the BSA glycoconjugates, as described previously, with 1,000 response units, which corresponded to 1 ng/mm2 of protein (41). For relevant comparison between glycoconjugates, equivalent amounts of glycoconjugates were coated on the chip. Seeing that the signal of the VLP-glycoconjugate interaction is mostly proportional to the mass of the VLP and that the signal intensity of the interaction is magnified by the large mass of the VLP, the dissociation constants (Kon and Koff) could not be accurately calculated since one VLP possesses 180 putative binding sites. Additionally, it was unclear how many HBGA molecules participated in the interaction. However, a relative binding affinity, based upon visual examination of the binding curves as reported previously, was estimated (32). Preliminary binding experiments by ELISA using Hunter VLPs and PBS and SPR-specific HBS buffer clearly showed that VLP binding with HBS buffer was as specific as that observed with PBS buffer (data not shown). Therefore, the HBS buffer was used for the SPR experiments. Preliminary assays using Hunter and Osaka VLPs suggested a concentration-dependent increase in the SPR response. Following treatment of the chip with NaIO4, no binding was observed, indicating that the carbohydrate moiety of the glycoconjugate was involved in the VLP binding (data not shown). The sensorgram showed an increasing relative affinity for the H glycoconjugate in the following order: Yerseke > Den Haag = Hunter > Osaka > Bristol and US95/96 variants (Fig. 5B). The sensorgram showed an increasing relative affinity for the A and B glycoconjugates in the following order: Den Haag ≫ Hunter > Yerseke = Osaka and Den Haag ≫ Yerseke > Hunter > Osaka (Fig. 5D and F, respectively). It is worth mentioning that no attachment was observed for US95/96 and Bristol variants, suggesting that neither of these two variants had detectable affinity for A and B carbohydrates although weak binding was detected by ELISA. For the dissociation curve, we observed that Den Haag and Osaka VLPs presented a marked stability for the three glycoconjugates, regardless of the VLP concentration during the injection. In contrast, the US95/96 and Bristol variants presented the lowest interaction with H antigen.
Minimum spanning tree of the ORF2 amino acid sequences.
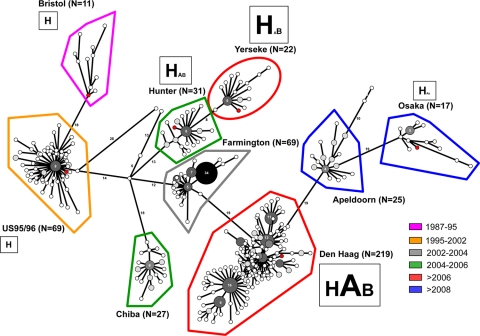
To determine the minimum number of amino acid changes between strains, an MST was constructed based upon the 496 complete amino acid sequences of ORF2 available in the GenBank (Fig. 6). The MST was built with 872 amino acid changes. The lineages and variants found for the MST of this study matched those observed for neighbor-joining and Bayesian trees as described previously (2). The MST suggested that Yerseke and Den Haag variants might be directly related to the former Hunter and Farmington variants, respectively, as previously described (39). The tree showed that Hunter, Farmington, and Chiba variants might all be related to the US95/96 variants via the Lanzhou isolate. The Den Haag variants are directly related to the Farmington variants. The large number of Den Haag isolates (n = 219) revealed the clonal diversification within this group, which resulted in the emergence of the Apeldoorn and Osaka variants. In our previous study, the data suggested that Osaka variants might be equally distant from the US95/96 and the Chiba variants (n = 23) (20). This change in the tree topology might be explained by the lower number of Osaka sequences included for the construction of the MST from the earlier study and the absence, at the time, of sequences corresponding to the Apeldoorn variant.
Fig. 6.
MST of the GII.4 variants. The number of sequences for each group of variants is indicated in parentheses. Identical sequences are represented by gray-shaded circles, which are scaled according to member count. The white and light gray circles represent one and two sequences, respectively. For dark gray and black circles, the number of identical sequences is indicated inside each circle. The six GII.4 isolates, which were analyzed during the study, are indicated by red circles. Each group of variants is color coded according to the date of circulation, as described previously (2). For the six variants that were analyzed, the binding profile to secretor and nonsecretor HBGAs is summarized in Table 4. For the A, B, and H antigens, the size of the letter is scaled according to the numbers of RU that were determined during the SPR analysis in the experiment shown in Fig. 5.
DISCUSSION
The genetic survey of NoVs from outbreaks of gastroenteritis showed the epochal evolution of GII.4 NoVs over the last 2 decades to become the predominant strain (25, 39). However, since 2002 and the spread of the Farmington variants, laboratory networks have observed an increasing pace in the genetic drift, with the emergence of new GII.4 variants every 2 or 3 years (40). Thorough analysis of the attachment of NoVs belonging to several genotypes allowed the characterization of eight binding profiles (16, 17), which can be classified into the ABO and Lewis binding groups. GII.4 NoVs present the widest binding spectrum since they can attach efficiently to saliva from secretor ABO individuals (80% of the Caucasian population), as reviewed previously (47).
Because the biological meaning and the mechanisms by which new GII.4 variants have emerged every 2 or 3 years for the last decade are still unclear, we performed an analysis of six representative GII.4 variants to determine whether epidemiological observations correlated with specific biological properties during the attachment of GII.4 NoV particles to HBGA ligands. We first performed a qualitative analysis of the variants by evaluating their attachment profiles on phenotyped saliva and a large panel of synthetic glycoconjugates (Table 4). It has been hypothesized that the occurrence of new variants was driven by herd immunity and was accompanied by new binding properties (7, 13). At first glance, our data clearly showed that the occurrence of new variants did not correlate with new and expanded binding profiles. Indeed, the variants that we analyzed could bind A, B, H type 1 (and not H type 2), Lewis b, and Lewis y synthetic antigens, which are all representative of the secretor phenotype. These data are consistent with those obtained on saliva, which showed that the six variants could bind secretor saliva regardless of the ABO phenotype, given that the level of binding of the VLPs was variant dependent. Nevertheless, the variations that we observed for the saliva binding assays between variants for a given antigen (i.e., A, B, and H antigen) might be explained by the expression level of the HBGAs in the saliva. Indeed, it has been shown that NoV binding was dependent on ligand density (26). Further studies will also be required to determine whether there is a synergistic association between Lewis and ABH antigens, which might explain differences in the binding. For nonsecretor individuals, we tested 12 saliva samples, and for six of these, which were Lewis positive, we demonstrated strong binding with Den Haag and Osaka VLPs. Our data supported the hypothesis that these variants might potentially infect all secretors and a major part of nonsecretors. Only Lewis-negative nonsecretor individuals would not be recognized by these two recent strains. In all major human populations, individuals with this phenotype represent 1 to 3% of the population (30) while Lewis-positive nonsecretors represent almost 20% of the population. Thus, the Den Haag and Osaka strains would have acquired an increased ability to circulate worldwide. Based on their newly acquired recognition of Lewis-related structures (FUT3-dependent) in nonsecretors, they might potentially infect over 95% of the population. Binding experiments using synthetic carbohydrates failed to detect which precise carbohydrate motif is recognized by these two strains in Lewis-positive nonsecretors, suggesting that more complex structural features are involved (e.g., branched structures or internal fucosylation), requiring further study. The sequence alignment of the GII.4 variants (CHDC, Bristol, US95/96, Farmington, Hunter, Yerseke, Chiba, Den Haag, Osaka, and Apeldoorn) showed that amino acids directly involved in the HBGA binding sites are all conserved (data not shown) (2). This observation was further supported by our mutagenesis analysis of the amino acid residues of the binding pocket that interact with the α-fucose ring, which is part of the A, B, and H oligosaccharides (8). The deletion or mutation of these residues totally abolished binding to the synthetic HBGAs and saliva samples. Conversely, as exemplified by the ΔT395 Hunter VLPs, amino acids that are outside the binding sites might vary under the influence of the genetic drift and the immune system. It has recently been shown that T395 is under positive selection (38). This amino acid is part of a triad involved in monoclonal antibody recognition, and other studies predicted that it should be involved in HBGA binding specificity (1, 25). Our data suggest that T395 is indeed involved in binding specificity. The deletion of a T395 insertion for the post-2002 Hunter strain allowed binding to Lewis x and sialyl-Lewis x antigens while no binding was observed for wt Hunter VLPs. Additionally, binding to sialyl-Lewis x was lowered or abrogated for the post-2002 variants. However, the biological relevance of the recognition of Lewis x and/or sialyl-Lewis x remains to be determined. Further mutagenesis studies will be required to verify which amino acid residues might influence the attachment of these antigens by the virion and to determine its biological relevance.
Table 4.
Summary of the binding properties of the GII.4 variants
| Isolate | Binding on nonsecretor saliva by antigena |
Neoglycoconjugateb |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Secretor |
Nonsecretor |
Colostrum |
|||||||||||||
| Le+ | Le− | H (RU) | A (RU) | B (RU) | Lewis b | Lewis y | Lewis a | Lewis x | Si-Lewis a | Si-Lewis x | Si-LNF | Si-LNT | LNT | Si-LNnT | |
| Bristol | Neg | Neg | 227 | 3 | 2 | +++ | ++ | − | − | − | ++++ | − | + | − | − |
| US95/96 | Neg | Neg | 191 | 1 | 1 | ++++ | +++ | − | ++++ | − | ++++ | + | ++++ | − | + |
| Hunter | Neg | Neg | 425 | 134 | 114 | +++ | ++++ | − | − | − | + | + | − | − | − |
| Yerseke | Neg | Neg | 606 | 88 | 195 | +++ | +++ | − | − | − | − | − | − | − | − |
| Den Haag | Pos | Neg | 490 | 694 | 407 | ++ | +++ | − | + | − | ++ | − | − | − | − |
| Osaka | Pos | Neg | 243 | 59 | 54 | +++ | ++++ | − | ++ | − | +++ | − | − | − | − |
VLP binding assay by ELISA. Le+, Lewis antigen positive; Le−, Lewis antigen negative. Pos, positive; Neg, negative.
VLP binding to ABH antigens was determined by SPR analysis. RU, resonance units. The results from ELISAs using neoglycoconjugates are indicated as follows: +, OD < 0.5; ++, 0.5 < OD < 1; +++, 1 < OD < 1.5; ++++, OD > 1.5.
It is noteworthy that the genetic shifts observed for the GII.4 variants might not be entirely driven by immune pressure. In the literature, the attachment of VLPs to carbohydrates is mostly analyzed by using ELISA techniques. Despite the fact that the VLP-ligand interaction was not stoichiometric, SPR technology has been a valuable tool to study the attachment of VLPs to and their dissociation from carbohydrates. The short incubation time of the VLPs in the presence of ligands (2 min) and the constant flow of buffer during the binding assay put the attachment of VLPs to HBGAs under more stringent conditions in SPR than in ELISA, where the incubation time is longer (2 h). Additionally, the slope and RU suggested that the relative affinity to a given glycoconjugate was HBGA and variant dependent. Our data indicated that “old” GII.4 variants (i.e., Bristol and US95/96) bound strongly only to antigen H while new GII.4 variants additionally exhibited strong binding to A and B antigens, with the Den Haag variant showing the highest relative affinity for A, B, and H antigens (Fig. 6). We might hypothesize that there is a relationship between the higher binding of the Den Haag variants to ABH antigens and the epidemiological predominance of these variants over the Yerseke variants which circulated at the same period (2). This is the first report to show that binding to secretor antigens might be chronologically based, with the most recent and predominant variants showing the highest relative affinity for HBGAs.
It should be acknowledged that our data were mainly obtained from in vitro experiments. Future studies should be undertaken to determine the binding pattern of GII.4 variants on intestinal tissues from secretor and nonsecretor individuals. In the event of Den Haag-related gastroenteritis cases, the analysis should include the determination of the secretor status to verify whether nonsecretor individuals can indeed be infected by GII.4 NoVs. Other factors should also be taken into account. For example, heparan sulfate molecules might play a role in NoV binding, as suggested previously (42). It has also been suggested that increased pathogenicity for GII.4 NoV might be related to an augmented activity of the viral polymerase (6).
In summary, as illustrated by the deletion of T395, changes in amino acids that are not directly involved in HBGA attachment might contribute to the creation/establishment of new binding profiles as predicted previously (25). The binding capability to secretor and nonsecretor populations might have contributed to the success of strains belonging to the Den Haag variant. However, the future evolution of these strains is difficult to predict since they can hardly expand their host range, which already represents over 95% of the population. One might hypothesize that the immune pressure would lead to the disappearance of the Den Haag variants to the benefit of new emerging strains, which will evade the immune system. Presently, it is difficult to predict whether the host range of emerging variants would be restricted to the secretor population. Additionally, our data highlighted the fact that the evolution of GII.4 resulted in an increase in the relative affinity to A, B, and H antigens, which was most evident in the most successful strains. The combination of a high relative affinity for ABH antigens expressed in secretors and the ability to recognize the majority of the nonsecretor population may have contributed to the dominance of Den Haag strains since their appearance in 2006. The routine analysis of binding to nonsecretor saliva might contribute to predicting the pathogenicity of emerging GII.4 variants in the population. In addition, our data suggest that it would be pertinent to determine NoV relative affinity toward HBGAs to determine whether certain emerging GII.4 NoV variants will become predominant worldwide.
ACKNOWLEDGMENTS
This work was supported by the National Reference Center for Enteric Viruses and the Public Hospital of Dijon (France). The biochip part of the project was financially supported by a fellowship in the Proteomic Platform program from the University of Franche-Comté and was conducted with the help of the Clinical and Innovation Proteomic Platform and the technologic platform MIMENTO (Besançon, France).
The 162 GII.4 NoV sequences from the study of Motomura et al. were kindly provided by Hironori Sato (29). We thank Kim Y. Green for sending recombinant MD145 baculovirus and Jean Lepeault for the electron micrograph. We thank Katia Ambert-Balay and Philip Bastable for technical and editorial assistance.
We declare that we have no conflicts of interest with regard to carrying out the study and writing the manuscript.
Footnotes
Published ahead of print on 23 February 2011.
REFERENCES
- 1. Allen D. J., et al. 2009. Characterisation of a GII-4 norovirus variant-specific surface-exposed site involved in antibody binding. Virol. J. 6:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Belliot G., Kamel A. H., Estienney M., Ambert-Balay K., Pothier P. 2010. Evidence of emergence of new GGII.4 norovirus variants from gastroenteritis outbreak survey in France during the 2007-to-2008 and 2008-to-2009 winter seasons. J. Clin. Microbiol. 48:994–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belliot G., et al. 2001. Characterization of capsid genes, expressed in the baculovirus system, of three new genetically distinct strains of “Norwalk-like viruses.” J. Clin. Microbiol. 39:4288–4295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boireau W., Rouleau A., Lucchi G., Ducoroy P. 2009. Revisited BIA-MS combination: entire “on-a-chip” processing leading to the proteins identification at low femtomole to sub-femtomole levels. Biosens. Bioelectron. 24:1121–1127 [DOI] [PubMed] [Google Scholar]
- 5. Bok K., et al. 2009. Evolutionary dynamics of GII.4 noroviruses over a 34-year period. J. Virol. 83:11890–11901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bull R. A., Eden J. S., Rawlinson W. D., White P. A. 2010. Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog. 6:e1000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cannon J. L., et al. 2009. Herd immunity to GII.4 noroviruses is supported by outbreak patient sera. J. Virol. 83:5363–5374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cao S., et al. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 81:5949–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carneiro F., Amado M., David L., Clausen H., Sobrinho-Simoes M. 1994. Glycosylation features of gastric carcinoma initiation and progression. A review with emphasis on simple mucin-type carbohydrates and histo-blood group antigens of the Lewis system. Eur. J. Cancer Prev. 3(Suppl. 2):39–46 [DOI] [PubMed] [Google Scholar]
- 10. Choi J. M., Hutson A. M., Estes M. K., Prasad B. V. 2008. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc. Natl. Acad. Sci. U. S. A. 105:9175–9180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Czajkowsky D. M., Shao Z. 2003. Inhibition of protein adsorption to muscovite mica by monovalent cations. J. Microsc. 211:1–7 [DOI] [PubMed] [Google Scholar]
- 12. Dingle K. E. 2004. Mutation in a Lordsdale norovirus epidemic strain as a potential indicator of transmission routes. J. Clin. Microbiol. 42:3950–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Donaldson E. F., Lindesmith L. C., Lobue A. D., Baric R. S. 2008. Norovirus pathogenesis: mechanisms of persistence and immune evasion in human populations. Immunol. Rev. 225:190–211 [DOI] [PubMed] [Google Scholar]
- 14. Green K. Y. 2007. Caliciviridae: the noroviruses, p. 949–980 In Knipe D. M., et al. (ed.), Fields virology, 5 ed., vol. 1 Lippincott, Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 15. Green K. Y., et al. 2002. A predominant role for Norwalk-like viruses as agents of epidemic gastroenteritis in Maryland nursing homes for the elderly. J. Infect. Dis. 185:133–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang P., et al. 2003. Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: identification of 4 distinct strain-specific patterns. J. Infect. Dis. 188:19–31 [DOI] [PubMed] [Google Scholar]
- 17. Huang P., et al. 2005. Norovirus and histo-blood group antigens: demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J. Virol. 79:6714–6722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hutson A. M., Airaud F., LePendu J., Estes M. K., Atmar R. L. 2005. Norwalk virus infection associates with secretor status genotyped from sera. J. Med. Virol. 77:116–120 [DOI] [PubMed] [Google Scholar]
- 19. Jiang X., Wang M., Graham D. Y., Estes M. K. 1992. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66:6527–6532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kamel A. H., et al. 2009. Predominance and circulation of enteric viruses in the region of Greater Cairo, Egypt. J. Clin. Microbiol. 47:1037–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kapikian A. Z., et al. 1972. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J. Virol. 10:1075–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kelly R. J., Rouquier S., Giorgi D., Lennon G. G., Lowe J. B. 1995. Sequence and expression of a candidate for the human secretor blood group alpha(1,2)fucosyltransferase gene (FUT2). Homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the non-secretor phenotype. J. Biol. Chem. 270:4640–4649 [DOI] [PubMed] [Google Scholar]
- 23. Kobayashi K., et al. 1993. Lewis blood group-related antigen expression in normal gastric epithelium, intestinal metaplasia, gastric adenoma, and gastric carcinoma. Am. J. Gastroenterol. 88:919–924 [PubMed] [Google Scholar]
- 24. Lindesmith L., et al. 2003. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 9:548–553 [DOI] [PubMed] [Google Scholar]
- 25. Lindesmith L. C., et al. 2008. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marionneau S., Airaud F., Bovin N. V., Le Pendu J., Ruvoen-Clouet N. 2005. Influence of the combined ABO, FUT2, and FUT3 polymorphism on susceptibility to Norwalk virus attachment. J. Infect. Dis. 192:1071–1077 [DOI] [PubMed] [Google Scholar]
- 27. Marionneau S., et al. 2001. ABH and Lewis histo-blood group antigens, a model for the meaning of oligosaccharide diversity in the face of a changing world. Biochimie 83:565–573 [DOI] [PubMed] [Google Scholar]
- 28. Marionneau S., et al. 2002. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 122:1967–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Motomura K., et al. 2010. Divergent evolution of norovirus GII/4 by genome recombination from May 2006 to February 2009 in Japan. J. Virol. 84:8085–8097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mourant A. 1983. Blood relations: blood groups and anthropology, 2nd ed. Oxford University Press, Oxford, England [Google Scholar]
- 31. Murata K., et al. 1992. Expression of blood group-related antigens, ABH, Lewis(a), Lewis(b), Lewis(x), Lewis(y), CA19-9, and CSLEX1 in early cancer, intestinal metaplasia, and uninvolved mucosa of the stomach. Am. J. Clin. Pathol. 98:67–75 [DOI] [PubMed] [Google Scholar]
- 32. Nam H. J., et al. 2006. Identification of the sialic acid structures recognized by minute virus of mice and the role of binding affinity in virulence adaptation. J. Biol. Chem. 281:25670–25677 [DOI] [PubMed] [Google Scholar]
- 33. Newburg D. S. 2009. Neonatal protection by an innate immune system of human milk consisting of oligosaccharides and glycans. J. Anim. Sci. 87:26–34 [DOI] [PubMed] [Google Scholar]
- 34. Nicollier-Jamot B., Pico V., Pothier P., Kohli E. 2003. Molecular cloning, expression, self-assembly, antigenicity, and seroepidemiology of a genogroup II norovirus isolated in France. J. Clin. Microbiol. 41:3901–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Prasad B. V., et al. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287–290 [DOI] [PubMed] [Google Scholar]
- 36. Rydell G. E., et al. 2009. Human noroviruses recognize sialyl Lewis x neoglycoprotein. Glycobiology 19:309–320 [DOI] [PubMed] [Google Scholar]
- 37. Shoemaker G. K., et al. 2010. Norwalk virus assembly and stability monitored by mass spectrometry. Mol. Cell. Proteomics 9:1742–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Siebenga J. J., et al. 2010. Phylodynamic reconstruction reveals norovirus GII.4 epidemic expansions and their molecular determinants. PLoS Pathog. 6:e1000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Siebenga J. J., et al. 2007. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J. Virol. 81:9932–9941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Siebenga J. J., et al. 2009. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001–2007. J. Infect. Dis. 200:802–812 [DOI] [PubMed] [Google Scholar]
- 41. Stenberg E., Persson B., Roos H., Urbaniczky C. 1991. Quantitative determination of surface concentration of protein with surface plasmon resonance using radiolabeled proteins. J. Colloid Interface Sci. 143:513–526 [Google Scholar]
- 42. Tamura M., Natori K., Kobayashi M., Miyamura T., Takeda N. 2004. Genogroup II noroviruses efficiently bind to heparan sulfate proteoglycan associated with the cellular membrane. J. Virol. 78:3817–3826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tan M., et al. 2008. Noroviral P particle: structure, function and applications in virus-host interaction. Virology 382:115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tan M., Hegde R. S., Jiang X. 2004. The P domain of norovirus capsid protein forms dimer and binds to histo-blood group antigen receptors. J. Virol. 78:6233–6242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tan M., et al. 2003. Mutations within the P2 domain of norovirus capsid affect binding to human histo-blood group antigens: evidence for a binding pocket. J. Virol. 77:12562–12571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tan M., Jiang X. 2005. Norovirus and its histo-blood group antigen receptors: an answer to a historical puzzle. Trends Microbiol. 13:285–293 [DOI] [PubMed] [Google Scholar]
- 47. Tan M., Jiang X. 2010. Norovirus gastroenteritis, carbohydrate receptors, and animal models. PLoS Pathog. 6:.e1000983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tan M., Jiang X. 2005. The P domain of norovirus capsid protein forms a subviral particle that binds to histo-blood group antigen receptors. J. Virol. 79:14017–14030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tan M., et al. 2008. Elucidation of strain-specific interaction of a GII-4 norovirus with HBGA receptors by site-directed mutagenesis study. Virology 379:324–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Taube S., et al. 2009. Ganglioside-linked terminal sialic acid moieties on murine macrophages function as attachment receptors for murine noroviruses. J. Virol. 83:4092–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zheng D. P., et al. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312–323 [DOI] [PubMed] [Google Scholar]
- 52. Zheng D. P., Widdowson M. A., Glass R. I., Vinje J. 2010. Molecular epidemiology of genogroup II-genotype 4 noroviruses in the United States between 1994 and 2006. J. Clin. Microbiol. 48:168–177 [DOI] [PMC free article] [PubMed] [Google Scholar]