Abstract
The infection of human fetal foreskin fibroblasts (HFFF2) with human cytomegalovirus (HCMV) resulted in the induction of autophagy. This was demonstrated by the increased lipidation of microtubule-associated protein 1 light chain 3 (LC3), a hallmark of autophagy, and by the visualization of characteristic vesicles within infected cells. The response was detected first at 2 h postinfection and persisted for at least 3 days. De novo protein synthesis was not required for the effect, since HCMV that was irradiated with UV light also elicited the response, and furthermore the continuous presence of cycloheximide did not prevent induction. Infection with herpes simplex virus type 1 (HSV-1) under conditions that inhibited viral gene expression provoked autophagy, whereas UV-irradiated respiratory syncytial virus did not. The induction of autophagy occurred when cells were infected with HCMV or HSV-1 that was gradient purified, but HCMV dense bodies and HSV-1 light particles, each of which lack nucleocapsids and genomes, were inactive. The depletion of regulatory proteins Atg5 and Atg7, which are required for autophagy, reduced LC3 modification in response to infection but did not result in any detectable difference in viral or cellular gene expression at early times after infection. The electroporation of DNA into HFFF2 cultures induced the lipidation of LC3 but double-stranded RNA did not, even though both agents stimulated an innate immune response. The results show a novel, early cellular response to the presence of the incoming virion and additionally demonstrate that autophagy can be induced by the presence of foreign DNA within cells.
INTRODUCTION
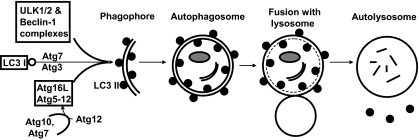
Autophagy is a process by which cellular organelles and abnormal proteins are degraded (recently reviewed in references 16, 43, and 75). It is an important mechanism for maintaining cell viability in times of starvation and stress, as it recycles cell components to provide nutrients. In addition, autophagy provides a means to eliminate toxic protein aggregates or damaged organelles from the cell. Three types of autophagy are recognized, named macroautophagy, microautophagy, and chaperone-mediated autophagy, and the term autophagy used here will refer only to macroautophagy. Studies of yeast first defined a number of proteins, given the prefix Atg, that control autophagy, and mammalian orthologs have been described for most of these. A simplified schematic of autophagy is shown in Fig. 1. The process is initiated by the formation of a phagophore, a double-membraned structure that can be visualized as crescent shaped by electron microscopy. The initiation of the phagophore requires the activities of two multiprotein complexes, the ULK1/2 and Beclin 1 complexes. The phagophore undergoes elongation, a stage that requires the modification of microtubule-associated protein 1 light chain 3 (LC3) by the conjugation of phosphatidylethanolamine, and the activity of a complex consisting of Atg5, Atg12, and Atg16L. The extended phagophore proceeds to engulf cell material or toxic proteins and closes to form a vesicle named the autophagosome. Lysosomes then fuse with autophagosomes to produce autolysosomes, which have a single membrane and contain enzymes that digest the cargo. Although autophagy is important for cell survival under starvation and other stress conditions, the process can result in cell death if allowed to continue unchecked. Thus, in some circumstances autophagy is an alternative to apoptosis as a cell death pathway.
Fig. 1.
Schematic diagram of autophagy. Shown is a simplified diagram that highlights the proteins and structures referred to in the text. The formation of the phagophore and elongation to produce the autophagosome are dependent upon the activities of three complexes, together with the lipidation of LC3. The ULK1/2 and Beclin-1 complexes each consist of at least five proteins, and these complexes are important for integrating signals that initiate autophagy. The small molecule Atg12 is covalently attached to Atg5 by the action of Atg7 (E1-like, using the nomenclature applied to ubiquitin conjugation) and Atg10 (E2-like). The Atg5-Atg12 fusion protein interacts with Atg16L to form a complex. Phosphatidylethanolamine is conjugated to LC3 form I (open circles) by the action of the enzymes Atg7 (E1-like) and Atg3 (E2-like). The Atg5-Atg12/16 complex and lipidated LC3 form II (filled circles) are required for the phagophore membrane to elongate and engulf cytoplasmic contents. LC3 II remains with the autophagosomal membrane. Fusion with lysosomes results in the formation of autolysosomes. The internal contents of the autolysosome, including LC3 II on the inner membrane, are digested. LC3 II on the outer membrane is released.
Since autophagy is important for the clearance of foreign or aggregated proteins, it is not surprising that the process affects the replication of intracellular parasites such as viruses in a number of ways (7, 15, 16, 18, 47). In some cases, viruses subvert autophagy to facilitate the progress of the infection cycle. The replication of poliovirus, the coronavirus mouse hepatitis virus, and hepatitis C virus is reduced when autophagy is inhibited. This is thought to occur because membranes derived from the initiation of the phagophore are important for the assembly of viral replication complexes (28, 50, 60, 71). In other examples, viruses possess mechanisms to prevent autophagy. The influenza A virus M2 protein blocks the formation of autolysosomes, although this property does not influence virus yields in tissue culture systems (20). HIV utilizes the autophagic machinery during early stages in replication, but in macrophages the viral nef protein inhibits the conversion of autophagosomes to autolysosomes, thereby preventing the loss of virus by proteolytic degradation (35). The engulfment of virus particles by autophagosomes and autolysosomes is named xenophagy.
Studies with herpesviruses have contributed to understanding the significance of autophagy during virus infection. The herpes simplex virus type 1 (HSV-1)-specific early protein ICP34.5 binds to Beclin 1, and this interaction requires ICP34.5 residues 68 to 87 (46). A virus mutant lacking this region (named Δ68-87) replicates normally in mouse embryo fibroblasts (MEFs), as does wild-type HSV-1 in Atg5−/− MEFs, demonstrating that autophagy does not affect virus replication in this tissue culture system (2). In mice, however, Δ68-87 is neuroattenuated in terms of both virus replication and the survival of the animals, showing that the inhibition of autophagy is important for the virulence of HSV-1 in vivo (46). The likely reason is the counteraction of xenophagy, since there were greater numbers of virions per autophagosome in MEFs, or cultured mouse neurons, at late times after infection with ICP34.5-null mutants than for wild-type HSV-1 (70). In addition, MEFs infected with an ICP34.5-null mutant exhibited a 2-fold increase in the number of autophagosomes per cell compared to that observed with wild-type virus, and a similar effect was observed in mouse neurons infected with Δ68-87 (2, 46). The HSV-1 ICP34.5 protein therefore contributes to virulence by preventing the xenophagic destruction of virions.
Investigations have been carried out on autophagy in cells infected with other herpesviruses. The infection of MRC-5 human fibroblasts with human cytomegalovirus (HCMV) resulted in the inhibition of autophagy, as shown by the failure of a green fluorescent protein-LC3 fusion protein to assume a characteristic punctate appearance in transfected cells at 24 h postinfection (hpi) and the absence of autophagosomes or autolysosomes at later times (8). Autophagy was induced upon infection with varicella-zoster virus at late times after infection, although viral DNA synthesis was not required for the effect (64). The Kaposi's sarcoma-associated herpesvirus (KSHV) induced autophagy during lytic reactivation, with the response being dependent on the activity of the viral RTA transcription activator (74). In addition, KSHV-encoded proteins vBcl-2 and vFLIP inhibited autophagy, suggesting roles in the survival of transformed cells (37, 49). In the case of Epstein-Barr virus, the latency-associated protein LMP1 stimulated autophagy in B cells, initiating a homeostatic interaction in which the consequent increased degradation of LMP1 prevented its overexpression and thereby optimized the survival of transformed cells (36).
Mammalian cells possess multiple mechanisms for recognizing infection by intracellular parasites and mounting innate immune responses (3, 24, 66). Toll-like receptors (TLRs) that detect the presence of foreign lipoproteins, lipopolysaccharide, RNA, or unmethylated DNA are found on the cell surface and in endosomes. Interaction with their respective ligands initiates signaling cascades that culminate in the production of cytokines, interferon-stimulated gene (ISG) products, and type I interferons, which make major contributions to innate immune responses (34). Signaling through TLRs results in the activation of NF-κB and interferon regulatory factor 3 (IRF3), a transcription factor that stimulates the expression of ISGs and alpha or beta interferon (10, 34, 59). The activation of IRF3 by phosphorylation can also occur through pathways initiated by TLR-independent sensors of foreign RNA or DNA (24, 26, 45, 61, 65). Double-stranded RNA (dsRNA) is recognized by the RNA helicases retinoic acid-inducible gene 1 (RIG-1) and melanoma-differentiated gene 5 (MDA5), which transmit signals via the adaptor molecule mitochondrial antiviral signaling protein (MAVS; also named IPS-1, VISA, and CARDIF) to result in the phosphorylation of IRF3 (33, 38, 76). Double-stranded DNA is detected by four known sensors (24). TLR9 is an endosomal protein that responds to the presence of unmethylated DNA but does not activate IRF3. RNA polymerase III can transcribe DNA to produce dsRNA, which then activates IRF3 through the MAVS/RIG-1-dependent pathway (1, 11, 26). The protein DNA-dependent activator of IRFs (DAI; also named ZBP1 and DLM-1) recognizes cytosolic DNA and mediates the induction of beta interferon through the activation of IRF3 (65, 73). The remaining known mechanism to sense DNA involves protein absent in melanoma 2 (AIM2). This factor activates the inflammasome, resulting in the production of proinflammatory cytokines but not the activation of IRF3 (3, 19, 23, 44).
Sensing infection with DNA viruses involves many of the mechanisms outlined above. The innate response to HSV-1 includes pathways controlled by TLR9, MAVS DAI, and AIM2 depending on the host cell type, indicating that the recognition of the viral genome is achieved through a variety of pathways (reviewed in reference 10). The activation of IRF3 and induction of beta interferon occurs after the infection of human fibroblasts with HCMV, and this complex response is due to multiple interactions of the virus particle with the host cell (4–6, 12–14, 21, 30, 45, 48, 51, 58). Recently, a role for DAI in stimulating beta interferon production via IRF3 has been demonstrated after the infection of cells with HCMV, and it is suggested that the viral genome is responsible for the activation of this pathway (12, 14).
Cells possess a variety of mechanisms to maintain viability and integrity after infection with viruses. These responses target many stages of infection, and the initial interaction of the virus with the host cell represents a point at which the probability of cell survival is optimal, provided a robust defense is mounted. The studies reported here demonstrate that the initial interactions of the HCMV or HSV-1 virion with the cell result in the induction of autophagy, and that this early event in infection is additional to innate immune responses mediated by IRF3. Evidence is presented that autophagy is triggered by the presence of foreign DNA, a novel finding that has widespread implications for understanding cell responses to intrusion of viral genomes.
MATERIALS AND METHODS
Cells and viruses.
Human fetal foreskin fibroblast (HFFF2) cells were propagated in Dulbecco's modified Eagle's medium supplemented with 5% (vol/vol) fetal calf serum, 5% (vol/vol) newborn calf serum, nonessential amino acids, and 100 U of penicillin plus 100 μg of streptomycin per milliliter (designated D5 + 5). For infection and other treatments for cells, Dulbecco's modified Eagle's medium supplemented with 2% (vol/vol) fetal calf serum, nonessential amino acids, and 100 U of penicillin plus 100 μg of streptomycin per milliliter (DF2) was used. HCMV, strain AD169, was propagated in HFFF2 cells, and stocks were prepared by centrifugation of medium harvested from infected monolayers, first at 1, 000 × g for 15 min to remove cellular debris and then 23,000 × g for 2 h. Pellets from the latter centrifugation were resuspended in D5 + 5 and purified further by negative viscosity-positive density gradient centrifugation (68). Bands identified as virions and dense bodies were removed from the gradient, pelleted by centrifugation at 23,000 × g for 2 h, and resuspended in D5 + 5. HCMV strain Merlin (17) was kindly supplied by D. Dargan (MRC-University of Glasgow Centre for Virus Research [CVR], Glasgow, United Kingdom). The HSV-1 mutant in1312 was constructed, propagated, and titrated as described previously (53). Virions and light particles of HSV-1 strain 17 were purified by Ficoll gradient centrifugation as described previously (62). Respiratory syncytial virus (RSV) was kindly provided by V. Cowton (CVR, Glasgow, United Kingdom). Numbers of virions, dense bodies, and light particles were estimated by electron microscopy. HCMV and RSV were UV irradiated in a Stratalinker (Stratagene) with a dose sufficient to reduce viral gene expression at 24 hpi to undetectable levels, as described previously (39). The induction of autophagy was achieved by the treatment of cells with DF2 containing methyl-beta-cyclodextrin (MβCD) (9).
Infection of cells.
HFFF2 monolayers, consisting of 1.5 × 105 cells, were routinely infected with 2 PFU of HCMV per cell or an equivalent amount of virus after UV irradiation. Cultures were infected with HSV-1 mutant in1312 at a multiplicity of 30 PFU per cell, representing 2 × 108 particles per monolayer, or with 10 PFU of RSV per cell or an equivalent amount of UV irradiated virus.
Electroporation of cells.
HFFF2 cultures were electroporated using an Amaxa nucleofector (Lonza) by following the manufacturer's instructions. For the introduction of short interfering RNA (siRNA), 2 × 106 cells were electroporated with 80 pmol of siRNA, and the cell suspension was used to seed 20 wells of 24-well plates. Cultures were incubated at 37°C for 3 days prior to infection. The sources of siRNAs were Atg5, Atg7, a control (no. 102655310, 102655373, and 1027280, respectively; Qiagen), and IRF3 (Dharmacon Thermo smartpool; no. M-006875-02). For the introduction of nucleic acids, 1.2 × 106 cells were electroporated with 0.5 μg of poly(dAT:dAT), 0.15 μg of poly(I:C), or 1.5 μg of HFFF2 DNA and used to seed 3 wells of 24-well plates. HFFF2 DNA and HCMV DNA from purified virus were prepared by use of a DNeasy tissue kit (Qiagen). HFFF2 DNA was sheared by repeated passage through a 21-gauge needle.
Protein blotting.
Cell extracts were prepared and analyzed by blotting as described previously (40). Actin was used as a loading control. Sources of primary antibodies were anti-actin, LC3, Atg5, and Atg7 (Sigma Aldrich); anti-ISG15 and IRF3 (Santa Cruz); anti-HCMV ICP36 and pp65 (Abcam); and anti-HCMV IE1/IE2 (AbD Serotec). Monoclonal antibodies specific for HCMV pp71 (32) and HSV-2 VP16 (42) were kindly provided by T. Shenk (Princeton University) and A. Minson (Cambridge University, United Kingdom), respectively.
Electron microscopy.
HFFF2 monolayers in 35-mm-diameter dishes were fixed in 2.5% glutaraldehyde, treated with 1% osmium tetroxide, and dehydrated through a series of increasing concentrations of ethanol. Cells were flat embedded in epoxy resin, and thin sections were stained with uranyl acetate and lead citrate prior to examination in a JEOL 1200 EXII electron microscope.
Analysis of protein degradation.
A culture of HFFF2 cells was radiolabeled by incubation for 24 h in D5 + 5 containing 35 μCi per milliliter of a mixture of [35S]methionine and [35S]cysteine (NEG772; Perkin-Elmer). The culture then was trypsinized and samples dispensed in D5 + 5 without radiolabeling mixture. After incubation for a further 12 h to allow short-lived proteins to decay, replicate monolayers were mock infected or infected with UV-HCMV. Samples of the medium were taken at various times and the radioactivity measured. Control experiments ascertained that the radioactivity was entirely soluble in 10% trichloroacetic acid. Total incorporation in the cell pellet was determined at the end of the experiment.
Immunofluorescence.
Cultures of 1 × 105 HFFF2 cells on coverslips were overlaid with DF2 or 5 mM MβCD in DF2 for 3 h at 37°C or were infected with UV-irradiated HCMV (UV-HCMV) and maintained at 37°C for 6 h. Cells were fixed by incubation for 15 min in 4% paraformaldehyde, washed with phosphate-buffered saline (PBS), and permeabilized by incubation in 100 μg of digitonin per milliliter for 10 min. After being washed with PBS, fixed cells were incubated with mouse anti-LC3 antibody (MBL) followed by fluorescein isothiocyanate (FITC)-conjugated sheep anti-mouse serum (Sigma Aldrich). Nuclei were stained by the incubation of cells in 1 μg of propidium iodide per milliliter for 1 min. Coverslips were mounted and analyzed on a Zeiss LSM510 confocal microscope with associated software.
RESULTS
LC3 lipidation occurs early after infection with HCMV.
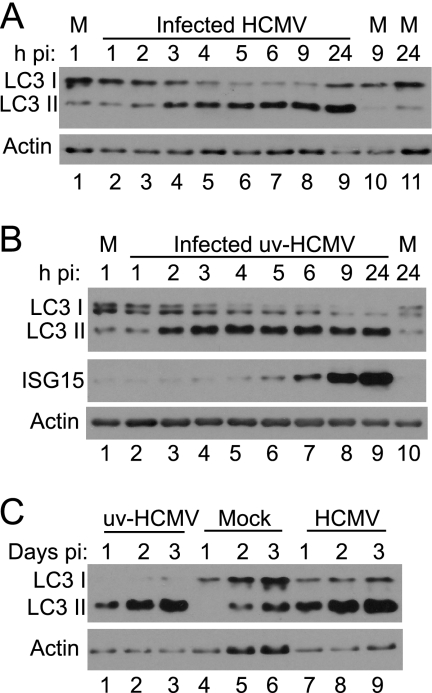
The modification of LC3 by lipidation, resulting in a protein that migrates more rapidly in SDS-polyacrylamide gels, is a hallmark of autophagy (31). The status of LC3 after the infection of HFFF2 cells with HCMV was investigated. An increase in the intensity of the characteristic faster-migrating form of LC3 (LC3 II) was detected by 2 hpi, and the extent of modification increased until it reached completion at approximately 5 hpi (Fig. 2A). The change in mobility was retained throughout infection up to 24 hpi. The apparent increase in the total amount of LC3 reflects the fact that most antibodies, including that used here, preferentially bind to the lipidated form of the protein. The LC3 I band often appeared as a doublet for reasons that are not clear to us. Since the change in LC3 occurred shortly after infection, the requirement for viral gene expression was investigated by infecting cultures with an HCMV sample that was irradiated with a dose of UV light sufficient to completely prevent the detectable expression of the major IE proteins. Infection with UV-HCMV induced the modification of LC3, with the effect again first detectable at 2 hpi and persisting until 24 hpi (Fig. 2B). As a measure of the rapidity of the response, the levels of interferon-stimulated gene 15-specific protein (ISG15) were assessed, since this protein is known to be induced after infection with UV-HCMV (13, 78, 79). Increased levels of ISG15 were first detected at 5 hpi, significantly later than the modification of LC3. To ascertain whether the effect persisted for even longer periods, samples were taken at 1, 2, and 3 days pi. The increased modification of LC3 compared to that in mock-infected cells was detected throughout this period in cultures infected with UV-HCMV or unirradiated virus (Fig. 2C).
Fig. 2.
Modification of LC3 after infection with HCMV. HFFF2 monolayers were mock infected (M), infected with HCMV, or infected with UV-HCMV. Cell lysates were analyzed for LC3 modification and ISG15 synthesis at various times pi. (A) Infection with HCMV AD169; (B) infection with UV-HCMV; (C) infection with UV-HCMV, mock infection, or infection with HCMV AD169 for 3 days.
Characterization of LC3 modification.
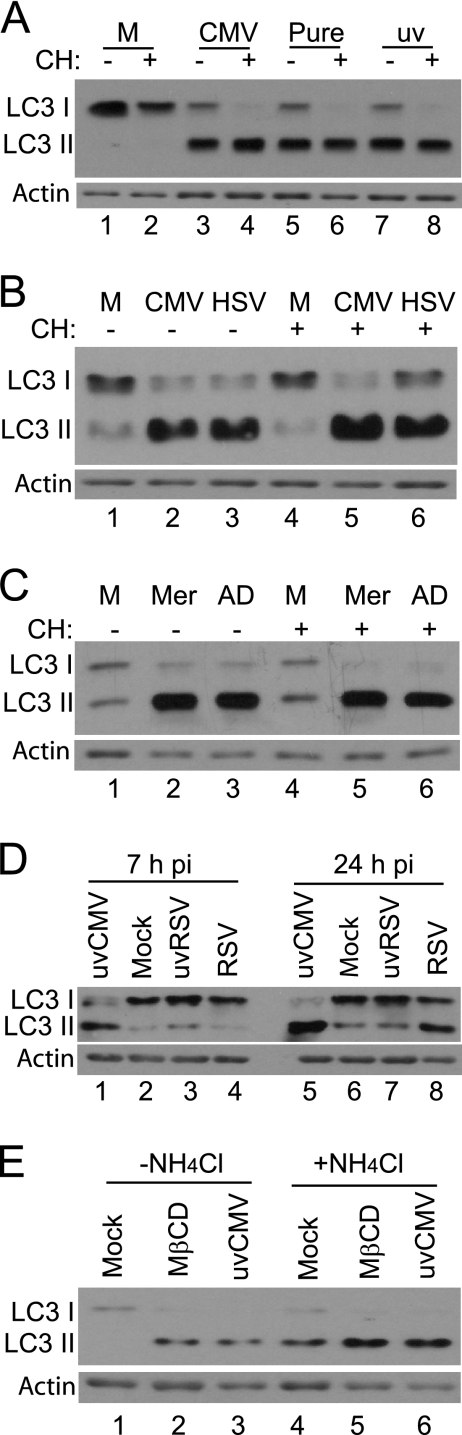
The HCMV stocks used routinely were partially purified by the centrifugation of medium taken from infected cells. To ensure that the virus particle truly was the agent responsible for inducing LC3 modification, virus was purified by gradient centrifugation and used to infect HFFF2 cultures. The requirement for protein synthesis also was tested by carrying out infection in the continuous presence of cycloheximide. The results presented in Fig. 3A demonstrate that purified HCMV was as effective as virus pelleted from the medium, whether or not it was UV irradiated (Fig. 3A). The data also demonstrate that ongoing protein synthesis was not required for LC3 modification, since the change in migration was observed when cycloheximide was present throughout infection (Fig. 3A). The modification of LC3 therefore appears to be a cellular response to the presence of the incoming virus particle that occurs even in the absence of de novo protein synthesis. To investigate whether the response was observed after infection with other enveloped viruses, HFFF2 cells were infected with the HSV-1 mutant in1312, which does not undergo detectable viral gene expression in the vast majority of cells due to mutations in VP16 and the immediate-early (IE) proteins ICP0 and ICP4 (53). Infection with this virus, which is unable to synthesize early and late gene products, including ICP34.5, the antagonist of autophagy (46), resulted in the induction of LC3 modification in both the absence and presence of cycloheximide (Fig. 3B). To investigate whether LC3 modification occurred with an alternative strain of HCMV, HFFF2 cells were infected with the clinical isolate Merlin, which is known to contain intact open reading frames of all viral genes, with the exception of mutations in RL13 and UL128 (17). Merlin induced LC3 modification as efficiently as strain AD169 (Fig. 3C). Infection with RSV, either UV irradiated sufficiently to prevent viral gene expression at 24 hpi or left unirradiated, did not elicit the response at 7 hpi (Fig. 3D, lanes 1 to 4). At 24 hpi, cells infected with unirradiated virus did show the modification of LC3 but UV-irradiated virus did not, suggesting that active RSV replication was responsible for this observation (Fig. 3D, lanes 5 to 8). Therefore, the early LC3 modification observed after infection with HCMV or HSV-1 was not due simply to the entry of an enveloped virus into the host cell.
Fig. 3.
Characterization of LC3 modification after infection. (A) HFFF2 monolayers were mock infected or infected with HCMV AD169 (CMV), gradient-purified HCMV AD169 (Pure), or UV-HCMV (uv) in the absence or continuous presence of 100 μg of cycloheximide (CH) per milliliter. Cell lysates were prepared at 6 hpi for the analysis of LC3. (B) HFFF2 monolayers were mock infected (M) or infected with HCMV AD169 (CMV) or HSV-1 in1312 (HSV) and incubated at 38.5°C in the absence or continuous presence of CH. Cell lysates were prepared for the analysis of LC3 at 6 hpi. (C) HFFF2 monolayers were mock infected (M) or infected with HCMV strain Merlin (Mer) or AD169 (AD) in the absence or continuous presence of CH. Cell lysates were prepared for the analysis of LC3 at 6 hpi. (D) HFFF2 monolayers were mock infected or infected with UV-HCMV (uvCMV), UV-irradiated RSV (uvRSV), or untreated RSV. Cell lysates were prepared for the analysis of LC3 at 7 hpi (lanes 1 to 4) or 24 hpi (lanes 5 to 8). (E) HFFF2 monolayers were mock treated for 3 h (M), treated with 3 mM MβCD for 2 h, or infected with UV-HCMV for 3 h in the absence (lanes 1 to 3) or presence (lanes 4 to 6) of 10 mM NH4Cl. Cell lysates were prepared for the analysis of LC3.
The detection of LC3 modification does not necessarily indicate an increase in the onset of autophagy, therefore a further test was carried out. To distinguish between an effect on the initiation of autophagy and a decrease in the turnover of LC3, the acidification of autolysosomes was inhibited by the inclusion of 10 mM NH4Cl in the culture medium. As a control for autophagy, cells were treated with the cholesterol-depleting agent MβCD, which is known to induce autophagy in human fibroblasts (9). The addition of either MβCD or UV-HCMV induced LC3 modification (Fig. 3E, lanes 1 to 3). The presence of NH4Cl resulted in the increased accumulation of modified LC3 in otherwise-untreated cultures, indicating that autophagy was routinely in progress in normal HFFF2 cells (Fig. 3E, lane 4). The amount of modified LC3 in MβCD- and UV-HCMV-treated cells was greater in NH4Cl-treated cells than in untreated cells (Fig. 3E, lanes 4 to 6), demonstrating that the effect of both MβCD and UV-HCMV was detectable even when processing within the autolysosome was inhibited. The results are consistent with an increase in the initiation of autophagy rather than a slowing of later stages.
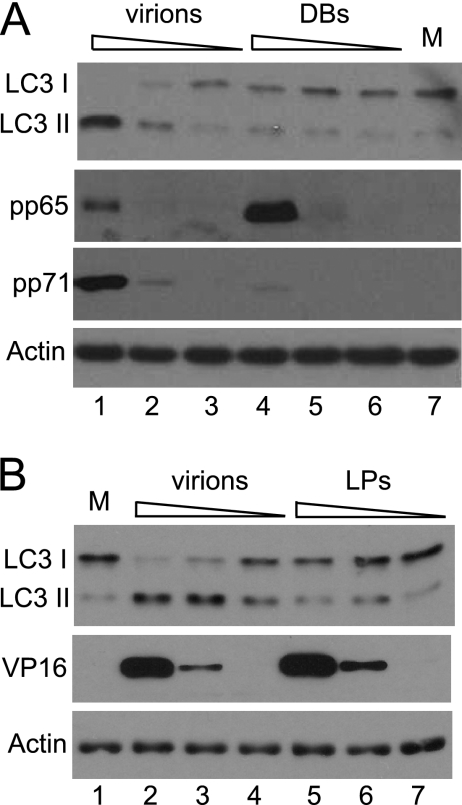
Preparations of HCMV and HSV-1 purified by centrifugation of medium from infected cells are known to contain noninfectious particles lacking genomes in addition to intact virions. The noninfectious particles, known as dense bodies or light particles (L-particles) for HCMV and HSV-1, respectively, are able to enter cells and deliver tegument proteins. Their abilities to stimulate LC3 modification were investigated (Fig. 4). Preparations containing equivalent numbers of HCMV virions and dense bodies were applied to HFFF2 monolayers in 10-fold dilutions. Purified virions stimulated LC3 modification when 6 × 107 and, to a lesser extent, 6 × 106 particles were applied to monolayers (Fig. 4A, lanes 1 to 3). Dense bodies, in contrast, did not elicit a response even when 6 × 107 particles were added (Fig. 4A, lanes 4 to 6). The uptake of the particles was confirmed by analyzing cells for the presence of pp65, which is highly enriched in dense bodies, and pp71, which is preferentially accumulated in virions (72). An analogous experiment was carried out with purified HSV-1 virions and L-particles (Fig. 4B). In this case, cycloheximide was present throughout to prevent the synthesis of ICP34.5. The increased modification of LC3 was observed after infection with 6 × 108 virions, and a partial effect was elicited by 6 × 107 virions (Fig. 4B, lanes 2 to 4). In contrast, even 6 × 108 L-particles failed to induce the detectable modification of LC3 (Fig. 4B, lanes 5 to 7). Equal uptake of virions and L-particles was confirmed by the demonstration that equivalent amounts of VP16 were present in cells at 2 hpi, since the same amount of this protein is present in the two types of particle (41).
Fig. 4.
Lipidation of LC3 after infection with virions, dense bodies, or L-particles. (A) HFFF2 monolayers were infected with purified HCMV virions (lanes 1 to 3) or dense bodies (DBs) (lanes 4 to 6) or were mock infected (M) (lane 7). Cell lysates were prepared at 6 hpi for the analysis of LC3 or at 2 hpi for the analysis of pp65 and pp71. The numbers of particles added to 1.5 × 105 cells were 6 × 107 (lanes 1 and 4), 6 × 106 (lanes 2 and 5), or 6 × 105 (lanes 3 and 6). (B) HFFF2 monolayers were mock infected (lane 1) or infected with HSV-1 virions (lanes 2 to 4) or L-particles (lanes 5 to 7), with 100 μg of cycloheximide per milliliter present throughout. Cell lysates were prepared at 6 hpi for the analysis of LC3 or 2 hpi for the analysis of VP16. The number of particles added to 1.5 × 105 cells was 6 × 108 (lanes 2 and 5), 6 × 107 (lanes 3 and 6), or 6 × 106 (lanes 4 and 7).
Further analysis of autophagy in HCMV-infected cells.
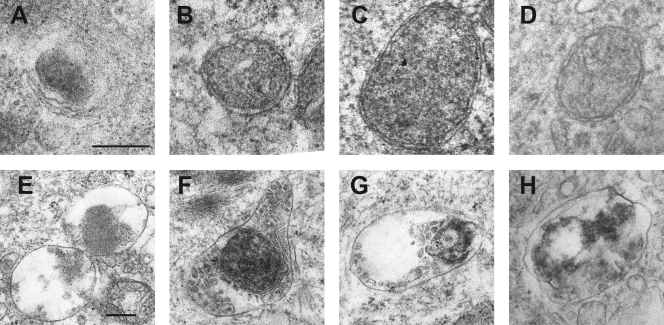
Additional evidence for the induction of autophagy by HCMV was obtained by the examination of cells infected with UV-HCMV or gradient-purified HCMV under the electron microscope. At 6 hpi, vesicles with the characteristic appearances of autophagosomes or autolysosomes were present, as shown in Fig. 5. Autophagosomes with double membranes were observed (Fig. 5B to D), as were autolysosomes containing cargo at various stages of digestion (Fig. 5E to H). Figure 5A may depict an early stage of engulfment by a phagophore membrane. Counts were made of autophagy-related vesicles in individual cell profiles from multiple fields and nonserial sections. In mock-infected cultures, 50 profiles exhibited a total of 22 vesicles classified as autophagosomes or autolysosomes, whereas 50 profiles from cultures infected with UV-HCMV or gradient-purified HCMV revealed 265 and 326 vesicles, respectively. Therefore, HCMV-infected cells contained increased numbers of vesicles, which is characteristic of autophagy.
Fig. 5.
Autophagy-related structures in HCMV-infected HFFF2 cells. Images from the electron microscopic examination of HFFF2 cells infected with gradient-purified HCMV or UV-HCMV for 6 h. (A) Possible engulfment by a phagophore structure; (B to D) double-membraned autophagosomes; (E to H) autolysosomes with cargo in different stages of digestion. The bar in panel A represents 200 nM for panels A to D, and the bar in E represents 200 nM for panels E to H.
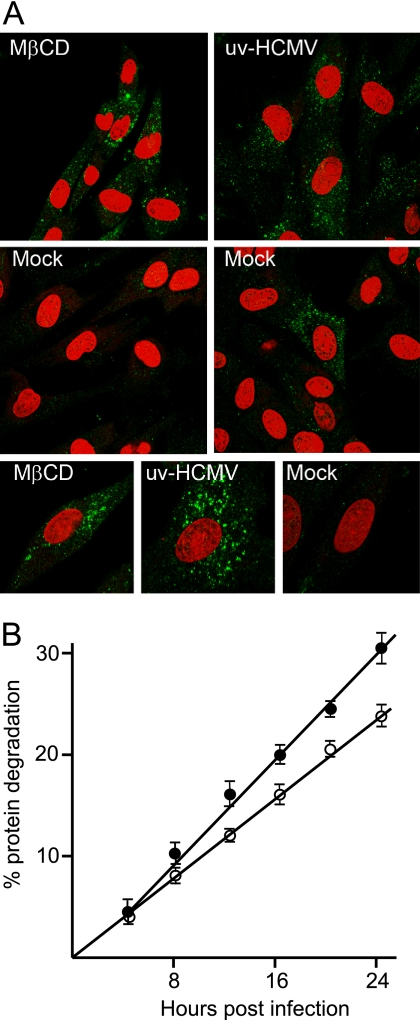
Immunofluorescence was carried out on cultures infected with UV-HCMV or treated with MβCD (Fig. 6A). Increased numbers of punctate vesicles recognized by anti-LC3 antibody per cell were observed after either treatment, in contrast to the small number present in most cells of mock-infected cultures. A few cells in mock-infected cultures also contained multiple LC3-positive vesicles, an example of which is shown. Taken together, the microscopy studies provide strong evidence that authentic autophagy occurs in response to infection with HCMV without the need for the de novo synthesis of viral or cellular proteins.
Fig. 6.
Characterization of HCMV-induced autophagy. (A) HFFF2 monolayers were mock treated for 3 h, treated with 5 mM MβCD for 3 h, or infected with UV-HCMV for 6 h. Cultures were fixed and analyzed for the presence of LC3 with propidium iodide counterstaining to identify nuclei. The mock infection panel in the second row on the right was selected to show the presence of a rare cell exhibiting obvious LC3 staining. The three lower panels show individual cells at higher magnification. (B) Radiolabeled HFFF2 monolayers were mock infected (open circles) or infected with UV-HCMV (filled circles) and incubated at 37°C. At various times, radioactivity released into the supernatant was measured. Six independent determinations were taken at each point, and the error bars show the standard deviations of the samples. The amount of released radioactivity was significantly greater in the infected cultures (P < 0.01) at all times except 4 hpi.
Autophagy results in the increased degradation of long-lived cell proteins. To investigate whether this occurred after HCMV infection, cell proteins were radiolabeled by the incubation of HFFF2 cells with [35S]methionine and [35S]cysteine, followed by a chase in normal medium for 12 h to permit the decay of short-lived proteins. The release of acid-soluble radioactivity into the cell supernatant, representing the degradation of long-lived cell proteins, was measured in mock-infected and UV-HCMV-infected cultures (Fig. 6B). An increase in the rate of protein degradation was observed from 4 hpi until at least 24 hpi, demonstrating that this feature of autophagy was induced by infection with UV-HCMV.
Depletion of Atg5 and Atg7 reduces HCMV-induced LC3 lipidation.
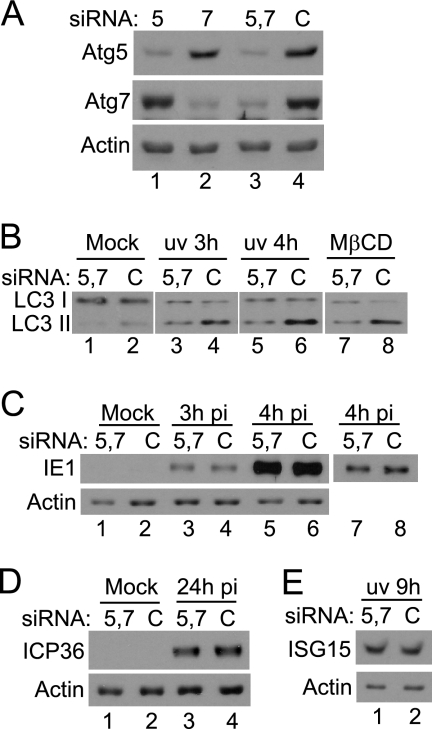
To investigate the significance of autophagy for HCMV infection, siRNAs specific for Atg5 and Atg7 or a control siRNA were electroporated into HFFF2 cells, and the response to infection with HCMV was analyzed. Treatment with the Atg-specific siRNAs reduced the levels of the target proteins (Fig. 7A). Cultures in which Atg5 or Atg7 was depleted individually exhibited detectable reduction in LC3 lipidation after infection with UV-HCMV or treatment with MβCD (results not shown), but a greater effect was observed when the siRNAs were used in combination (Fig. 7B). The extent of LC3 modification was reduced in cultures treated with siRNAs specific for Atg5 and Atg7, compared to that in cultures treated with control siRNA, after infection with UV-HCMV or the addition of MβCD (Fig. 7B). The response was not completely abolished, however, since the accumulation of the faster-migrating LC3 band was greater in UV-HCMV-infected cells than in mock-infected cells. To determine whether this compromised autophagy affected early events in infection, cultures electroporated with a mixture of Atg5- and Atg7-specific siRNAs were infected with HCMV and the production of IE1 protein was analyzed at 3 or 4 hpi (Fig. 7C). There were no differences in the levels of IE1 protein at 3 hpi, the earliest time at which it could be detected, or at 4 hpi. In addition, no difference in the production of the early protein ICP36 was detected at 24 hpi (Fig. 7D). The induction of ISG15 was examined at 9 h after infection with UV-HCMV, and this cellular response also was unaffected by the treatment of cells with the Atg-specific siRNAs (Fig. 7E). The reduction of LC3 modification upon the depletion of Atg5 and Atg7 further confirms that the response to HCMV truly represents autophagy. At the levels of depletion achieved, however, no effects on virus gene expression or the induction of ISG15 were detected. There also was no difference in virus yields from control and Atg-depleted cultures at 3 days postinfection (4.2 × 104 PFU/ml and 4.4 × 104 PFU/ml, respectively).
Fig. 7.
Effects of reducing Atg5 and Atg7 levels. (A) HFFF2 cultures were electroporated with siRNAs specific for Atg5 (lane 1), Atg7 (lane 2), both Atg5 and Atg7 (lane 3), or control siRNA (C) (lane 4). Extracts were analyzed for Atg5 or Atg7 at 3 days after electroporation. The band detected by the anti-Atg5 antibody is the Atg5-Atg12 conjugate. (B) HFFF2 cultures, electroporated with Atg5- and Atg7-specific siRNAs or control siRNA as described for panel A, were mock infected (lanes 1 and 2), infected with UV-HCMV for 3 (lanes 3 and 4) or 4 h (lanes 5 and 6), or treated with 3 mM MβCD for 2 h (lanes 7 and 8). Cell lysates were analyzed for the modification of LC3. (C) HFFF2 cultures, electroporated as described for panel A, were mock infected (lanes 1 and 2) or infected with HCMV for 3 (lanes 3 and 4) or 4 h (lanes 5 and 6). Lysates were analyzed for the presence of IE1 protein. Lanes 7 and 8 show a lower exposure of the signal in lanes 5 and 6. (D) HFFF2 cultures, electroporated as described for panel A, were mock infected (lanes 1 and 2) or infected with HCMV for 24 h (lanes 3 and 4). Cell lysates were analyzed for the presence of ICP36. (E) HFFF2 cultures, electroporated as described for panel A, were infected with UV-HCMV for 9 h. Cell lysates were analyzed for the presence of ISG15.
The role of innate immune responses in induction of autophagy.
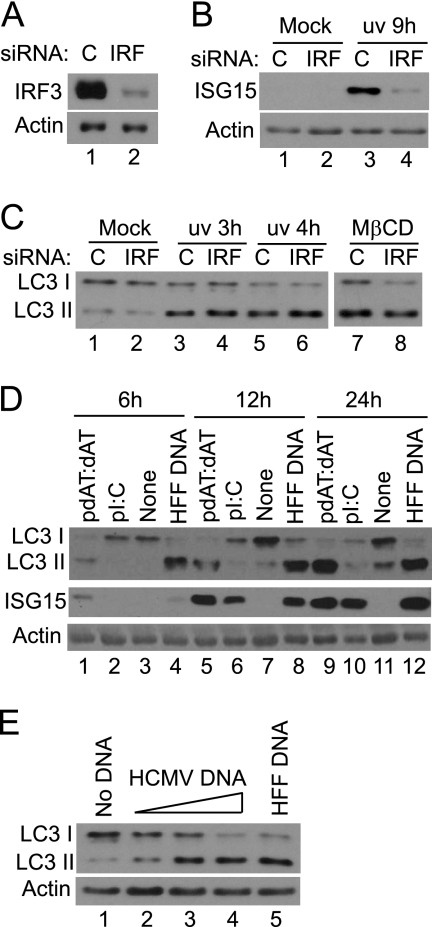
The activation of the interferon response is known to occur upon infection with UV-HCMV, therefore the possible requirement for the activation of IRF3 in eliciting LC3 modification after infection was investigated. A pool of four IRF3-specific siRNAs or a control siRNA was electroporated into HFFF2 cells, and the modification of LC3 upon infection was analyzed. The effective depletion of IRF3 was achieved by this approach (Fig. 8A), and as a consequence the induction of ISG15 after infection with UV-HCMV was inhibited (Fig. 8B). There was, however, no effect on the extent of LC3 modification after infection with UV-HCMV (Fig. 3C). This experiment shows that the activation of IRF3 by UV-HCMV does not contribute to the induction of LC3 modification. In considering alternative signaling pathways that might be involved in the response to UV-HCMV, the finding that HCMV and HSV-1, but not RSV, dense bodies or L-particles induced autophagy suggested a role for the viral nucleocapsid or genome. The presence of foreign DNA within cells is known to trigger innate immune responses (26, 27, 61), and a recent study has shown that immune-stimulatory DNA (ISD) activates IRF3 in human foreskin fibroblasts (14). We investigated whether introducing ISD into HFFF2 cells induces LC3 modification. Cultures were electroporated with poly(dAT:dAT), a potent inducer (14, 26), poly(I:C), a dsRNA mimic, or sheared HFFF2 DNA, and the modification of LC3 was monitored (Fig. 8D). The amounts of inducer used represented the greatest that HFFF2 cells tolerated without exhibiting cytopathology, as determined in preliminary experiments (results not shown). Both DNAs strongly induced the lipidation of LC3, whereas poly(I:C) did not elicit this response, even after 24 h, despite the strong activation of ISG15 synthesis by the homopolymer. In addition, purified HCMV DNA was as efficient as sheared HFFF2 DNA (Fig. 8E), giving support to the hypothesis that viral DNA is responsible for the induction of autophagy.
Fig. 8.
Involvement of cell immune responses in modification of LC3. (A) HFFF2 cultures were electroporated with a control siRNA (C) (lane 1) or IRF3-specific siRNAs (lane 2). After 3 days, cell lysates were analyzed for the presence of IRF3. (B) HFFF2 cultures, electroporated as described for panel A, were mock infected (lanes 1 and 2) or infected with UV-HCMV for 9 h (lanes 3 and 4). Cell lysates were analyzed for the presence of ISG15. (C) HFFF2 cultures, electroporated as described for panel A, were mock infected (lanes 1 and 2), infected with UV-HCMV for 3 (lanes 3 and 4) or 4 h (lanes 5 and 6), or treated with 3 mM MβCD for 3 h. Cell lysates were analyzed for the modification of LC3. (D) Poly(dAT:dAT), poly(I:C), no DNA, or sheared HFFF2 DNA (HFF DNA) was electroporated into HFFF2 cells, and cell lysates were analyzed for the modification of LC3 and ISG15 levels after 6 (lanes 1 to 4), 12 (lanes 5 to 8), or 24 h (lanes 9 to 12). (E) No DNA (lane 1), 0.15, 0.5, or 1.5 μg of HCMV DNA (lanes 2 to 4), or 1.5 μg of sheared HFFF2 DNA (lane 5) was electroporated into HFFF2 cells, and cell lysates were analyzed for LC3 modification after 12 h.
DISCUSSION
We report here that the induction of autophagy occurs very early after infection with HCMV or HSV-1, and that de novo protein synthesis is not required for the response. HCMV was the more efficient inducer based on the fact that fewer virus particles were required to cause the detectable lipidation of LC3, a hallmark of autophagy (31). We are not aware of any published reports that document such an early activation of autophagy after infection with either HCMV or HSV-1, and indeed there have been no studies that question the extent of viral gene expression required to elicit the response for these herpesviruses or, to our knowledge, any other viruses. Research to date has focused on the potential significance of xenophagy in affecting viral yields at late times of infection, in view of the seminal observation that the inhibition of autophagy by ICP34.5 is important for HSV-1 pathogenesis (46). An increase in numbers of autophagosomes after infection with ICP34.5-null HSV-1 has been recorded, but the viral stimulus for this effect was not investigated (2, 46). The only published study on HCMV examined the cells at late times of infection, when autophagy was inhibited relative to that of uninfected cells (8). Our observation of the continued modification of LC3 for 3 days appears incompatible with the conclusion of Chaumorcel et al. (8) that infection with HCMV inhibits autophagy, but it should be noted that different methods were used to assess the cellular changes in their study and ours, and further work is required to resolve the apparent discrepancies.
In addition to the lipidation of LC3, other features of autophagy were detected after infection with HCMV, and indeed the response was very similar to that elicited by MβCD (9). The increased modification of LC3 upon infection when the acidification of the autolysosome was blocked by NH4Cl argues that infection increases the rate of the initiation of autophagy rather than inhibiting the degradation of LC3 at a late stage in the process (43). Electron microscopic analysis of HCMV-infected cells revealed the presence of vesicles that could be identified as autophagosomes, with a characteristic double membrane, and autolysosomes containing cargo undergoing digestion, and the numbers of these per cell profile were more than 10-fold greater than those of mock-infected cultures. Structures with loosely apposed membranes also were observed, but we consider that additional characterization would be required before they could be classed as phagophores. Further evidence that our findings represent authentic autophagy is provided by the presence of LC3-positive puncta in virtually every cell of infected cultures, the observation of increased protein degradation after infection, and the finding that the depletion of Atg5 and Atg7 reduced the extent of LC3 modification in response to infection.
The prevailing hypothesis posits that, during infection with HSV-1, the activation of dsRNA-activated protein kinase (PKR) as a consequence of viral dsRNA synthesis is responsible for the induction of autophagy (69, 70). The observation that PKR−/− MEFs, in contrast to PKR+/+ cells, did not exhibit increased protein degradation upon infection with ICP34.5-null HSV-1 suggests that the activation of PKR has a role in the response (69). It is possible that infection with UV-HCMV or the HSV-1 mutant in1312 induced autophagy through the activation of PKR, but we consider this explanation unlikely for a number of reasons. First, the activation of PKR upon HSV-1 infection is a relatively late event, described as depending on viral DNA synthesis (63). Second, UV-HCMV has been exposed to a dose of irradiation sufficient to completely block the production of the major IE protein and therefore is unable to produce functional mRNAs. It is possible that small dsRNAs escape inactivation by irradiation, although the only such candidate product described to date is a single microRNA (22). Similarly, HSV-1 mutants with multiple defects in immediate-early gene expression, such as in1312, are severely impaired for the transcription of the viral genome and are unlikely to produce dsRNA (52, 56, 57) Third, poly(I:C) did not induce LC3 modification, even though it induced an immune response as shown by the production of ISG15.
The use of siRNA to reduce autophagy after infection with HCMV was only partially successful. Despite the obvious depletion of Atg5 and Atg7, the reduction in LC3 modification was modest and incomplete, as found by others (25). There was no corresponding change in the parameters of infection that we investigated, including the onset of immediate-early gene expression and the induction of ISG15, but more effective knockdown of Atg protein levels might reveal effects in cell culture. On the basis of the data presented here, it appears that autophagy does not have a major effect on the early stages of HCMV infection in human fibroblasts. It is possible that a role for the process will be revealed in a cell system that reproduces more closely the replication of the virus in vivo, as found in the case of HSV-1 (46).
Dense bodies and L-particles can bind to and enter cells, delivering tegument proteins, and therefore the failure of these particles to induce LC3 suggests that these early stages of infection are not required for the induction of autophagy. This appears to eliminate UV-HCMV-activated TLR2- and TLR4-mediated pathways, initiated by the binding of viruses to the cell surface, from involvement in the autophagy response (4, 30). Only virions induced LC3 modification, suggesting a requirement for components of the nucleocapsid or for the viral genome. Although we have not provided a definitive distinction between these two possibilities, the observation that immune-stimulatory DNA and HCMV DNA also induced the response points to the involvement of the viral genome. Since IRF3 activity is not required for the induction of LC3 modification, any of the four known DNA-sensing mechanisms could mediate the response, intersecting with autophagy-inducing pathways at a point upstream of IRF3 activation. Thus, TLR9 might transmit signals even though this receptor does not lead to IRF3 activation (34). A role for the RNA polymerase III pathway, leading to dsRNA, seems less likely, since poly(I:C) did not induce LC3 modification. The protein DAI recently has been shown to have a critical role in the induction of beta interferon after infection with HCMV (12, 14). DAI mediates this effect through the activation of IRF3, but it is possible that intermediate molecules in the cascade, such as the adaptor STING/MITA (27, 77), also activate autophagy. The activation of the inflammasome is another response to cytosolic DNA that might provoke autophagy, although it currently is not clear whether all components of the inflammasome are present in human fibroblasts. It is noteworthy that all of the known sensors are triggered by the presence of cytosolic DNA, whereas herpesvirus genomes are thought to be protected within the nucleocapsid until delivery to the cell nucleus. Possibly, sufficient viral DNA is released within the cytoplasm to activate the sensor(s); alternatively, either the known pathways or others yet to be identified are able to respond to nuclear foreign DNA.
It has been recognized recently that components of the autophagy pathway can affect cellular immune responses (54). The Atg5-Atg12 conjugate directly associates with MAVS and RIG-1, exerting a negative effect on beta interferon production after infection with vesicular stomatitis virus or stimulation with poly(I:C) (29, 67). More relevant to the results described here, stimulation with poly(dAT:dAT) causes STING to relocate to unidentified membrane-bound compartments, where it colocalizes with LC3 and Atg9a. The latter protein is essential for autophagy and has a negative effect on the poly(dAT:dAT)-mediated IRF3 activation of beta interferon production. The results suggest that Atg9a dampens the immune response through the translocation of STING to the unidentified cell compartments (54, 55). To our knowledge, however, the converse situation, in which autophagy is induced as a response to DNA, has not been reported previously. Our finding therefore suggests that autophagy is an additional cellular protection mechanism that guards against harmful effects of intracellular pathogens, possibly not aimed at herpesviruses but effective against other infectious agents.
The studies detailed here describe novel aspects of the interaction between infecting HCMV particles and the host cell. The induction of autophagy at such an early time, with the effect persisting for much of the viral replication cycle, is a surprisingly dramatic alteration to the cell milieu. The stimulation of LC3 lipidation in response to DNA adds to the spectrum of cell defenses. Future studies on the mechanism of the induction of autophagy by herpesviruses will reveal the viral and cellular molecules that mediate this response to infection.
ACKNOWLEDGMENTS
We thank Derrick Dargan, Vanessa Cowton, Tony Minson, and Tom Shenk for generous gifts of viruses and antibodies.
The study was supported by the Medical Research Council.
Footnotes
Published ahead of print on 16 February 2011.
REFERENCES
- 1. Ablasser A., et al. 2009. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed intermediate. Nat. Immunol. 10:1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alexander D. E., Ward S. L., Mizushima N., Levine B., Lieb D. A. 2007. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J. Virol. 81:12128–12134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alnemri E. S. 2010. Sensing cytoplasmic danger signals by the inflammasome. J. Clin. Immunol. 30:512–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boehme K. W., Guerrero M., Compton T. 2006. Human cytomegalovirus envelope glycoproteins B and H are necessary for TLR2 activation in permissive cells. J. Immunol. 177:7094–7102 [DOI] [PubMed] [Google Scholar]
- 5. Boehme K. W., Singh J., Perry S. T., Compton T. 2004. Human cytomegalovirus elicits a coordinated cellular antiviral response via envelope glycoprotein B. J. Virol. 78:1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Browne E. P., Wing B., Coleman D., Shenk T. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cavignac Y., Esclatine A. 2010. Herpesviruses and autophagy: catch me if you can. Viruses 2:314–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chaumorcel M., Souquere S., Pierron G., Codogno P., Esclatine A. 2008. Human cytomegalovirus controls a new autophagy-dependent cellular antiviral defense mechanism. Autophagy 4:46–53 [DOI] [PubMed] [Google Scholar]
- 9. Cheng J., Ohsaki Y., Tauchi-Sato K., Fujita A., Fujimoto T. 2006. Cholesterol depletion induces autophagy. Biochem. Biophys. Res. Commun. 351:246–252 [DOI] [PubMed] [Google Scholar]
- 10. Chew T., Taylor K. E., Mossman K. L. 2009. Innate and adaptive immune responses to herpes simplex virus. Viruses 1:979–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chiu Y. H., Macmillan J. B., Chen Z. J. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138:576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. DeFilippis V. R., Alvarado D., Sali T., Rothenburg S., Fruh K. J. 2010. Human cytomegalovirus induces the interferon response via the DNA sensor ZBP. J. Virol. 84:585–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. DeFilippis V. R., et al. 2006. Interferon regulatory factor 3 is necessary for induction of antiviral genes during human cytomegalovirus infection. J. Virol. 80:1032–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. DeFilippis V. R., et al. 2010. Activation of the interferon response by human cytomegalovirus occurs via cytoplasmic double-stranded DNA but not glycoprotein B. J. Virol. 84:8913–8925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deretic V. 2010. Autophagy in infection. Curr. Opin. Cell. Biol. 22:252–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deretic V., Levine B. 2009. Autophagy, immunity, and microbial adaptations. Cell Host Microbe 5:527–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dolan A., et al. 2004. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 85:1301–1312 [DOI] [PubMed] [Google Scholar]
- 18. Espert L., Codogno P., Biard-Piechaczyk M. 2007. Involvement of autophagy in virus infections: antiviral function and subversion by viruses. J. Mol. Med. 85:811–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fernandes-Alnemri T., Yu J. W., Datta P., Wu J., Alnemri E. S. 2009. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458:509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gannagé M., et al. 2009. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 6:367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gravel S.-P., Servant M. J. 2005. Roles of IkB kinase-related pathway in human cytomegalovirus-infected vascular smooth muscle cells. J. Biol. Chem. 280:7477–7486 [DOI] [PubMed] [Google Scholar]
- 22. Grey F., et al. 2005. Identification and characterization of human cytomegalovirus-encoded microRNAs. J. Virol. 79:12095–12099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hornung V., et al. 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458:514–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hornung V., Latz E. 2010. Intracellular DNA recognition. Nat. Rev. Immunol. 10:123–130 [DOI] [PubMed] [Google Scholar]
- 25. Hosokawa N., Hara Y., Mizushima N. 2006. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 580:2623–2629 [DOI] [PubMed] [Google Scholar]
- 26. Ishii K. J., et al. 2006. A toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat. Immunol. 7:40–48 [DOI] [PubMed] [Google Scholar]
- 27. Ishikawa H., Ma Z., Barber G. N. 2009. STING regulates intracellular DNA-mediated, type 1 interferon-dependent innate immunity. Nature 461:788–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jackson W. T., et al. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3:861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jounai N., et al. 2007. The Atg5-Atg12 conjugate associates with innate immune responses. Proc. Natl. Acad. Sci. U. S. A. 104:14050–14055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Juckem L. K., Boehme K. W., Feire A. L., Compton T. 2008. Differential initiation of innate immune responses induced by human cytomegalovirus entry into fibroblast cells. J. Immunol. 180:4965–4977 [DOI] [PubMed] [Google Scholar]
- 31. Kabeya Y., et al. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kalejta R. F., Bechtel J. T., Shenk T. 2003. Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors. Mol. Cell. Biol. 23:1885–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kang D. C., et al. 2002. Mda-5: an interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl. Acad. Sci. U. S. A. 99:637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kawai T., Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11:373–384 [DOI] [PubMed] [Google Scholar]
- 35. Kyei G. B., et al. 2009. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J. Cell Biol. 186:255–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee D. Y., Sugden B. 2008. The latent membrane protein 1 oncogene modifies B-cell physiology by regulating autophagy. Oncogene 27:2833–2842 [DOI] [PubMed] [Google Scholar]
- 37. Lee J. S., et al. 2009. FLIP-mediated autophagy regulation in cell death control. Nat. Cell Biol. 11:1355–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Loo Y. M., et al. 2008. Distinct RIG-1 and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82:335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lukashchuk V., McFarlane S., Everett R. D., Preston C. M. 2008. Human cytomegalovirus protein pp71 displaces the chromatin-associated factor ATRX from nuclear domain 10 at early stages of infection. J. Virol. 82:12543–12554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marshall K. R., et al. 2002. Activity and intracellular localization of the human cytomegalovirus protein pp71. J. Gen. Virol. 83:1601–1612 [DOI] [PubMed] [Google Scholar]
- 41. McLauchlan J. 1997. The abundance of the herpes simplex virus type 1 UL37 tegument protein in virus particles is closely controlled. J. Gen. Virol. 78:189–194 [DOI] [PubMed] [Google Scholar]
- 42. McLean C., et al. 1982. Monoclonal antibodies to three non-glycosylated antigens of herpes simplex virus type 2. J. Gen. Virol. 63:297–305 [DOI] [PubMed] [Google Scholar]
- 43. Mizushima N., Yoshimori T., Levine B. 2010. Methods in mammalian autophagy research. Cell 140:313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Muruve D. A., et al. 2008. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature 452:103–107 [DOI] [PubMed] [Google Scholar]
- 45. Noyce R. S., Collins S. E., Mossman K. L. 2009. Differential modification of interferon regulatory factor 3 following virus particle entry. J. Virol. 83:4013–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Orvedahl A., et al. 2007. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 1:23–35 [DOI] [PubMed] [Google Scholar]
- 47. Orvedahl A., Levine B. 2009. Eating the enemy within: autophagy in infectious diseases. Cell Death Differ. 16:57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Paladino P., Cummings D. T., Noyce R. S., Mossman K. L. 2006. The IFN-independent response to virus particle entry provides a first line of antiviral defense that is independent of TLRs and retinoic acid-inducible gene I. J. Immunol. 177:8008–8016 [DOI] [PubMed] [Google Scholar]
- 49. Pattingre S., et al. 2005. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122:927–939 [DOI] [PubMed] [Google Scholar]
- 50. Prentice E., Jerome W. G., Yoshimori T., Mizushima N., Denison M. R. 2004. Coronavirus replication complex formation utilises components of cellular autophagy. J. Biol. Chem. 279:10136–10141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Preston C. M., Harman A. N., Nicholl M. J. 2001. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J. Virol. 75:8909–8916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Preston C. M., Nicholl M. J. 1997. Repression of gene expression upon infection of cells with herpes simplex virus type 1 mutants impaired for immediate-early protein synthesis. J. Virol. 71:7807–7813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Preston C. M., Rinaldi A., Nicholl M. J. 1998. Herpes simplex virus type 1 immediate early gene expression is stimulated by inhibition of protein synthesis. J. Gen. Virol. 79:117–124 [DOI] [PubMed] [Google Scholar]
- 54. Saitoh T., Akira S. 2010. Regulation of innate immune responses by autophagy-related proteins. J. Cell Biol. 189:925–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Saitoh T., et al. 2009. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl. Acad. Sci. U. S. A. 106:20842–20846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Samaniego L. A., Neiderhiser L., DeLuca N. A. 1998. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J. Virol. 72:3307–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Samaniego L. A., Wu N., DeLuca N. A. 1997. The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected-cell survival in the absence of ICP4 and ICP27. J. Virol. 71:4614–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sato M., et al. 2000. Distinct and essential roles for transcription factors IRF-3 and IRF-7 in response to viruses for IFN-a/b gene induction. Immunity 13:539–548 [DOI] [PubMed] [Google Scholar]
- 59. Shayakhmetov D. M., Di Paulo N. C., Mossman K. L. 2010. Recognition of virus infection and innate host responses to viral gene therapy vectors. Mol. Ther. 18:1422–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sir D., Chen W. L., Wakita T., Yen T. S. B., Ou J. H. J. 2008. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology 48:1054–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stetson D. B., Medzhitov R. 2006. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 93:93–103 [DOI] [PubMed] [Google Scholar]
- 62. Szilagyi J. F., Cunningham C. 1991. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J. Gen. Virol. 72:661–668 [DOI] [PubMed] [Google Scholar]
- 63. Taddeo B., Luo T. R., Zhang W., Roizman B. 2003. Activation of NF-kB in cells productively infected with HSV-1 depends on activated protein kinase R and plays no apparent role in blocking apoptosis. Proc. Natl. Acad. Sci. U. S. A. 100:12408–12413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Takahashi M. N., et al. 2009. Varicella-zoster virus infection induces autophagy in both cultured cells and human skin vesicles. J. Virol. 83:5466–5476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Takaoka A., et al. 2007. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune responses. Nature 448:501–505 [DOI] [PubMed] [Google Scholar]
- 66. Takeuchi O., Akira S. 2009. Innate immunity to virus infection. Immunol. Rev. 227:75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tal M. C., et al. 2009. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl. Acad. Sci. U. S. A. 106:2770–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Talbot P., Almeida J. D. 1977. Human cytomegalovirus: purification of enveloped virions and dense bodies. J. Gen. Virol. 36:345–349 [DOI] [PubMed] [Google Scholar]
- 69. Talloczy Z., et al. 2002. Regulation of starvation- and virus-induced autophagy by the eIF2a kinase signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 99:190–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tallóczy Z., Virgin H. W., Levine B. 2006. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy 2:24–29 [DOI] [PubMed] [Google Scholar]
- 71. Tanida I., et al. 2009. Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy 5:937–945 [DOI] [PubMed] [Google Scholar]
- 72. Varnum S. M., et al. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang Z., et al. 2008. Regulation of innate immune sensing responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc. Natl. Acad. Sci. U. S. A. 105:5477–5482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wen H. J., Yang Z., Zhou Y., Wood C. 2010. Enhancement of autophagy during lytic replication by the Kaposi's sarcoma-associated herpesvirus replication and transcription activator. J. Virol. 84:7448–7458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yang Z., Klionsky D. J. 2010. Eaten alive: a history of autophagy. Nat. Cell Biol. 12:814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yoneyama M., et al. 2004. The RNA helicase RIG-1 has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737 [DOI] [PubMed] [Google Scholar]
- 77. Zhong B., et al. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29:538–550 [DOI] [PubMed] [Google Scholar]
- 78. Zhu H., Cong J. P., Mamtora T., Gingeras T., Shenk T. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. U. S. A. 95:14470–14475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhu H., Cong J. P., Shenk T. 1997. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. U. S. A. 94:13985–13990 [DOI] [PMC free article] [PubMed] [Google Scholar]