Abstract
Varicella-zoster virus (VZV) is the alphaherpesvirus that causes chicken pox (varicella) and shingles (zoster). The two VZV glycoproteins gE and gI form a heterodimer that mediates efficient cell-to-cell spread. Deletion of gI yields a small-plaque-phenotype virus, ΔgI virus, which is avirulent in human skin using the xenograft model of VZV pathogenesis. In the present study, 10 mutant viruses were generated to determine which residues were required for the typical function of gI. Three phosphorylation sites in the cytoplasmic domain of gI were not required for VZV virulence in vivo. Two deletion mutants mapped a gE binding region in gI to residues 105 to 125. A glycosylation site, N116, in this region did not affect virulence. Substitution of four cysteine residues highly conserved in the Alphaherpesvirinae established that C95 is required for gE/gI heterodimer formation. The C95A and Δ105–125 (with residues 105 to 125 deleted) viruses had small-plaque phenotypes with reduced replication kinetics in vitro similar to those of the ΔgI virus. The Δ105–125 virus was avirulent for human skin in vivo. In contrast, the C95A mutant replicated in vivo but with significantly reduced kinetics compared to those of the wild-type virus. In addition to abolished gE/gI heterodimer formation, gI from the C95A or the Δ105–125 mutant was not recognized by monoclonal antibodies that detect the canonical conformation of gI, demonstrating structural disruption of gI in these viruses. This alteration prevented gI incorporation into virus particles. Thus, residues C95 and 105 to 125 are critical for gI structure required for gE/gI heterodimer formation, virion incorporation, and ultimately, effective viral spread in human skin.
INTRODUCTION
Varicella-zoster virus (VZV), an alphaherpesvirus, causes two forms of disease: chicken pox (varicella), a disseminated exanthema in primary infection, and shingles (zoster), typified by extensive lesions that emanate from peripheral nerve axons upon recrudescence of latent VZV (16). Although VZV infection is considered benign in normal patients, immunocompromised patients or nonimmune neonates are at risk of very severe disease (55). Children are particularly vulnerable if they are deficient in cell-mediated immunity following malignancy or chemotherapy. Importantly, live attenuated vaccines are available, but these are not recommended for the severely immunocompromised (48).
The 125-kbp double-stranded DNA (dsDNA) genome of VZV is the smallest of the alphaherpesviruses and is comprised of a unique long (UL) and unique short (US) region flanked by internal and terminal repeats (16). In common with all herpesviruses, VZV requires glycoproteins on the virion surface for cell entry and cell-to-cell spread. Nine glycoproteins are encoded by the VZV genome. Two glycoproteins critical to VZV, gE and gI, are encoded by open reading frame 68 (ORF68) and ORF67, respectively, located in the US region of the genome. Deletion of ORF68 (gE) is lethal to VZV (36, 41). In contrast, deletion of ORF67 (gI) yields a virus with a small-plaque phenotype in vitro, but the ΔgI virus cannot be recovered from inoculated human skin xenografts (36). The noncovalent heterodimer that forms between gE and gI can be disrupted by the deletion of the first cysteine-rich region in gE, severely impairing replication in human skin tissue (8, 47, 57). Thus, the gE/gI heterodimer is an important complex for targeted mutagenesis studies to define its role in VZV pathogenesis.
VZV has been phylogenetically classified in the α2 subgroup (varicella viruses), which is distinct from the α1 subgroup (simplex viruses) of the Alphaherpesvirinae (39). Despite their genetic diversity, the gE/gI heterodimer is conserved throughout the alphaherpesviruses and the family Herpesviridae. Studies using herpes simplex virus type 1 (HSV-1) and HSV-2, plus pseudorabies virus (PRV), which are closely related to VZV, have demonstrated that both gE and gI are dispensable for replication but are required for pathogenesis in their respective models of disease (18, 27, 63). Importantly, the gE/gI heterodimer is a neurovirulence factor for HSV and PRV in animal models of disease (12, 19, 50, 52). The role of the gE/gI heterodimer is unclear, but it appears to have multiple functions crucial for virus replication. gE/gI can function as an Fc receptor for HSV and VZV, which has been shown to contribute to HSV immune evasion (6, 32, 44). HSV gE/gI has been implicated in the secondary envelopment of virus particles at the trans-Golgi network (TGN), suggesting a role in virion assembly (21). In agreement with this hypothesis, VZV gI was reported to be required for virus particle envelopment in the TGN (54). The VZV gE/gI heterodimer is not required for the infection of human dorsal root ganglia in the SCIDhu xenograft model, as demonstrated by inoculation with the gE ΔCys mutant, which abrogates heterodimer formation (60, 62). However, this gE mutant failed to establish viral persistence in neurons. Despite similarities of the VZV gE/gI heterodimer to that of other herpesviruses, subtle differences in function remain.
VZV ORF67 generates a transcript of 1.65 kbp that encodes gI, previously known as gpIV, a type I transmembrane protein 354 amino acids (aa) in length (31, 51). gI can be divided into four predicted regions: the signal peptide (aa 1 to 17), the ectodomain (aa 18 to 277), the transmembrane region (aa 278 to 296), and the cytoplasmic domain (aa 297 to 354). In transfection studies, the immature form of gI is expressed as a 36-kDa protein that undergoes N-linked and O-linked glycosylation to a fully mature 64-kDa form (57). gI is phosphorylated, but this was reported to occur at a single residue, S343 (56, 59). In addition, residue T338 was shown to be involved in the TGN localization of gI (53). These studies imply that serine and threonine residues in the cytoplasmic domain of gI might be required for the function of gI and, potentially, the formation of the gE/gI heterodimer.
VZV gI has a low amino acid identity (15 to 16%) to the HSV-1 and HSV-2 homologs, but we noted that three cysteines in VZV gI, at residues 83, 95, and 106 (and, to a lesser extent, residue 200), are conserved across the alphaherpesviruses. Deletion of the N-terminal 209 amino acids of gI, which contains the four conserved cysteines, leads to a small-plaque phenotype and significantly reduces syncytium formation, a hallmark of wild-type VZV (36). The reason for this effect is unknown, and specifically, whether the cysteine residues within this region affect the structural stability of gI has not been investigated. In addition, formation of the gE/gI heterodimer was shown to occur in the endoplasmic reticulum by coexpression studies using a vaccinia virus-based system (57). Currently, the mechanism of heterodimer formation remains unclear and might be dependent on the structural conformation of gI.
Mutagenesis studies to analyze the VZV gE/gI heterodimer have focused predominantly on residues in gE required for effective dimerization and VZV virulence (3, 8, 9, 30). However, studies to date have not investigated the residues in gI required for the formation of the gE/gI heterodimer in the context of the VZV genome or their effects on pathogenicity. The present study was undertaken to investigate predicted phosphorylation sites in the cytoplasmic domain and residues in the ectodomain of gI to elucidate their function and role in pathogenicity. This is the first study to perform site-directed mutagenesis of single amino acids in VZV gI using cosmid-based mutagenesis and to identify a region in gI required to generate the gE/gI heterodimer, thereby providing an insight into its formation and function.
MATERIALS AND METHODS
Cell lines.
Melanoma cells were propagated in culture medium (minimal essential Eagle medium [MEM] supplemented with 10% fetal bovine serum [Gemini Bio-Products, Woodland, CA], nonessential amino acids [100 μM; Omega Scientific, Inc., Tarzana, CA], penicillin G [100 units/ml; Omega Scientific, Inc., Tarzana, CA], streptomycin [100 units/ml; Omega Scientific, Inc., Tarzana, CA], and amphotericin [0.5 mg/ml; Omega Scientific, Inc., Tarzana, CA]). Human embryonic lung fibroblasts (HELFs) were propagated in culture medium without nonessential amino acids. HEK-293 cells were propagated in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum and penicillin G (100 units/ml)-streptomycin (100 units/ml).
VZV-specific antibodies.
The v67 polyclonal rabbit antibody to gI was a kind gift from Saul Silverstein and was generated to amino acids 192 to 266 of gI (35). The 6B5 monoclonal antibody (MAb) to gI was a kind gift from Charles Grose (56). The following antibodies were commercially available. MAb SG4 to gI was originally generated from mice vaccinated with the Ellen strain of VZV (Meridian Life Science Inc., Saco, ME). gI from the Ellen strain shares 100% amino acid identity to that from vaccine Oka (vOka). MAb 8612 is specific for VZV gE (Millipore Biosciences, Temecula, CA). Rabbit polyclonal antibodies generated to gE and ORF23 have been described previously (14, 25).
Prediction of gI phosphorylation sites and secondary structure.
NetPhos was used to predict serine and threonine phosphorylation sites in the cytoplasmic domain of gI (10). The secondary structure of gI was predicted using the Dompred server (http://bioinf.cs.ucl.ac.uk) of the UCL Department of Computer Sciences Bioinformatics Group (11). The Dompred server was also used to predict the domains of gI and identify putative structurally similar proteins.
Generation of VZV rOka gI mutants.
The generation of cosmids for the complete genome from a vOka isolate of VZV has been reported previously (36). Initial studies were performed using vOka cosmids prior to the generation of parent Oka (pOka) cosmids. For continuity, all mutants were constructed using vOka cosmids. The gE and gI proteins of vOka and pOka are identical. Virus generated from these cosmids is referred to as recombinant Oka (rOka) throughout the paper. ORF67, which encodes gI, is located in cosmid pvSpe21 (Fig. 1). ORF67 spans the region between nucleotides (nt) 114497 and 115558. All restriction endonucleases were obtained from New England BioLabs, Inc., Ipswich, MA. The pSac6A vector, containing a 6-kbp SacI fragment of pvSpe21 subcloned into the pBluescript KS vector (Stratagene, La Jolla, CA), was used to construct the gI mutants as described previously (36).
Fig. 1.
Construction of VZV recombinants with mutations in ORF67, which encodes gI. (A) Schematic of the VZV genome. UL, unique long; US, unique short; TRL, terminal repeat long; IRL, internal repeat long; IRS, internal repeat short; TRS, terminal repeat short. (B) Schematic of the four cosmids (genome fragments in nucleotides), pvFsp4 (nt 1 to 33128), pvSpe5 (nt 21796 to 61868), pvPme19 (nt 53756 to 96035), and pvSpe21 (nt 94055 to 125123), used to generate recombinant rOka with mutations in gI. (C) Location of ORF67, which encodes gI, in the pvSpe21 cosmid. (D) Location of mutations in gI (ORF67). The shaded box, representing amino acids 1 to 17, shows the location of the signal peptide. Amino acids 18 to 277 in the ectodomain are represented by the first open box. The hatched box represents the transmembrane domain from amino acids 278 to 292, with the remaining residues of the cytoplasmic domain shown as the final open box in wild-type (WT) gI. Markers with filled circles indicate the locations of the four substituted cysteines C83, C95, C106, and C200. The marker with an open circle indicates the location of the substituted asparagine N116. Markers with stars indicate serine residues 296, 343, and 347, and markers with triangles indicate threonine residues 312 and 338, which were substituted with alanine. The two deletion mutants are indicated with a line connecting the boxes in the ectodomain of gI. (E) Predicted secondary structure of gI. The white bar indicates the carbon backbone, hatched boxes represent alpha helices, and gray rectangles represent the β-sheet. All figure panels are drawn to scale. The dashed lines indicate subsequent increases in scale from the previous figure panel.
Construction of gI phosphorylation mutants.
Alanine substitutions were performed at 5 amino acid residues in the cytoplasmic domain of gI to generate the three rOka mutant viruses P789 (S296, T312, and T338), P10 (S343), and P11 (S347). To generate each alanine substitution, two sequential PCRs were performed as described previously (4). Briefly, the first round of PCR yielded two products, one for each primer pair, which overlapped by 20 nucleotides and spanned the mutation site. These two fragments were gel purified with a QIAquick PCR purification kit (Qiagen, Inc., Valencia, CA), pooled, and used as a template for the second-round PCR, using external primers. The resulting PCR products of 1.8 kb were cloned into pCR4-TOPO vector (Invitrogen, Carlsbad, CA) and sequenced to verify the mutation. The 1.8-kb fragment was digested with PmlI and BsiWI and cloned into the pSac6A plasmid. For the P789 mutant, the single substitution was used as a template for the addition of a second substitution and the resulting double substitution for the third substitution in gI. Transfer of the newly generated gI mutants from pSac6A was performed by cloning the SgrAI/AvrII fragment into the pvSpe21ΔAvrII cosmid.
Construction of internal deletions in the gI N terminus.
The Δ105–125 and Δ37–167 deletion mutants (with residues 105 to 125 and residues 37 to 167, respectively, deleted) were generated using PCR and blunt-end ligation. Primer pairs Sac6A6036/Sac6A3831 and Sac6A3748/Sac6A2202 were used to generate the Δ105–125 mutant, and primer pairs gI37-167R2/gI37-167R1 and gI37-167L2/gI37-167L1 were used to generate the Δ37–167 mutant. The PCR products were gel purified, digested with BglII or AvrII, reisolated, ligated into pSac6A, and then transferred into the pvSpe21ΔAvrII cosmid.
Construction of gI cysteine mutants and the N116A mutant.
gI cysteine mutants and the N116A mutant were constructed using the same PCR triple ligation technique used for the N-terminal deletion mutants. Briefly, the gI upper and gI lower primers were paired with the appropriate mutagenesis primers (Table 1). The resulting PCR products were gel purified, digested with either BglII or KpnI, reisolated, ligated into the pSac6A vector, and then transferred into the pvSpe21ΔAvrII cosmid.
Table 1.
PCR primers used to generate rOka gI mutantsa
| Primer name | Mutation | Primer sequenceb (5′–3′) | Genome location (nt)c |
|---|---|---|---|
| Sac6A.Fe2452+ | NA | CAAGATCCTCCTGTGCAGACATTTGTGTG | 116263–116291 |
| Sac6A.Re4254− | NA | CACGGGATCATTTTCTTAACATTGTAGTAG | 114489–114518 |
| pSac6A.Fi3247 | S296A | CGCTTAACGGCTATTACAATAACAATAACC | 115467–115496 |
| pSac6A.Ri3264 | S296A | TGTAATAGCCGTTAAGCGACGTAGAATTAA | 115479–115508 |
| pSac6A.Fi3199 | S312A | CTTGTTTTTGCATTTGGGCGATAAATTGG | 115516–115544 |
| pSac6A.Ri3217 | S312A | GCCCAAATGCAAAAACAAGAAGGG | 115526–115549 |
| pSac6A.Fi3122 | T338A | CGCGAATCGCTGCTAGTTGTGCAATGGC | 115594–115621 |
| pSac6A.Ri3140 | T338A | CAACTAGCAGCGATTCGCGAAGAATCCC | 115603–115630 |
| pSac6A.Fi3109 | S343A | GGGGGGGCTTCTTCGCGAATCGTTGCT | 115608–115634 |
| pSac6A.Ri3124 | S343A | GCGAAGAAGCCCCCCCACATTCCGTT | 115619–115644 |
| pSac6A.Fi3093 | S347A | GTTTACAACGGCATGTGGGGGGGATTCTTC | 115621–115650 |
| pSac6A.Ri3112 | S347A | CCCCACATGCCGTTGTAAACCCGTTTGTT | 115631–115659 |
| gI37-167R2 | Δ37–167 | GCGTGAGCATTGAG | 112428–112445 |
| gI37-167R1d | Δ37–167 | GAGACTGCTGTTAACTTGC | 114690–114708 |
| gI37-167L2d | Δ37–167 | GCGGGCTCGCATCACAA | 115102–115118 |
| gI37-167L1 | Δ37–167 | TTTCGGCCAACTGATCC | 116638–116654 |
| Sac6A6036 | Δ105–125 | CCACCGCTTCCATCAGTAAG | 112688–112707 |
| Sac6A3831d | Δ105–125 | AATAAAAGCGCTCGTTCTAA | 114893–114912 |
| Sac6A3748d | Δ105–125 | GATGCTGGTGTAATGTTGAA | 114976–114995 |
| Sac6A2202 | Δ105–125 | CCCGTACAGGTTAATGAC | 116524–116541 |
| gI upper | NA | GGTTGGTCCGCTTCCTTCTTA | 116674–116694 |
| gI lowere | NA | TTACCGCCTTTGAGTGAGCTGA | NA |
| ORF67[228–246]d | C83A | AAACGCCACCGTATCCGCGTAT | 114825–114846 |
| ORF67[247–269]d | C83A | GCTTTCCGGTCAGTACAAGTAAT | 114847–114869 |
| ORF67[262–282]d | C95A | TCCGTCGTATCTTATTACTTG | 114862–114882 |
| ORF67[283–303]d | C95A | GCTCCCCGGATTAGAACGAGC | 114883–114903 |
| ORF67[293–315]d | C106A | CGAAATAAAAGCGCTCGTTCTAA | 114893–114915 |
| ORF67[316–337]d | C106A | GCTAGGTACAAACATTCGTGGC | 114916–114937 |
| ORF67[324–345]d | N116A | ACCATAATGCCACGAATGTTTG | 114924–114945 |
| ORF67[346–368]d | N116A | GCCTCAACGGATCGGATATCAAC | 114946–114968 |
| ORF67[575–597]d | C200A | CAAACGAGCTTGTTGAAAAAGGG | 115175–115197 |
| ORF67[598–618]d | C200A | GCTGATTTACCCGCGACACCC | 115198–115218 |
NA, not applicable.
Bold underlined letters indicate the sites of substitution in ORF67.
Locations of the primers within the VZV rOka genome (GenBank accession number AB097933).
Primer had a phosphate modification at the 5′ end.
Anneals in the pBluescript KS vector of the pSac6A construct.
All mutant cosmids were sequenced across and within the ligation sites to verify that unexpected mutations were not present. Mutant viruses were generated from these cosmids as previously described (36).
Expression constructs.
The complete ORF67 (nt 115601 to 115671) of rOka was amplified by PCR with the two primers HindIII_5ORF67 (TTAAAGCTTGCCACCATGTTTTTAATCCAATGTTTGATATCG) and 3ORF67_XhoI (TTACTCGAGTCACTATTTAACAAACGGGTTTACAACG), using Accuprime (Invitrogen, Carlsbad, CA). PCR products were gel purified (Qiagen, Inc., Valencia, CA), digested with either HindIII or XhoI, and then ligated into pCDNA3.1(+) to construct pCDNA-gI, which contained ORF67 (gI) under the cytomegalovirus (CMV) promoter. The dual expression vector pBudCE4.1 (Invitrogen) was used for the simultaneous expression of wild-type gE and wild-type gI or gI mutants. The pBud-gE/gI expression construct was generated using a two-step cloning procedure. The pCDNA-gE expression construct was described previously (7). This was digested with HindIII and pBudCE4.1 with KpnI. The overhangs of the digested DNA were filled with Klenow fragment (New England BioLabs, Inc., Ipswich, MA) and then purified with a nucleotide removal kit (Qiagen, Inc., Valencia, CA), followed by subsequent digestion with XhoI. The ORF68 (gE) excised from pCDNA-gE was then ligated into the EF-1α promoter site of pBudCE4.1 to generate the construct pBudE-gE with ORF68 (gE) under the EF-1α promoter. pCDNA-gI, pBudCE4.1, and pBudE-gE were digested with HindII and XbaI and gel purified, and ORF67 (gI) was ligated into the CMV promoter site of pBudCE4.1 to generate the construct pBudC-gI with ORF67 (gI) under the CMV promoter or into pBudE-gE to generate the construct pBud-gE/gI with ORF67 (gI) under the CMV promoter and ORF68 (gE) under the EF-1α promoter. All gI mutants were cloned into the pBud-gE/gI construct.
Immunoprecipitation of gI from transfected or infected cells.
Cells were lysed with a glycoprotein extraction buffer (0.1 M Tris-base [pH 8.8], 0.1 M NaCl, 5 mM KCl, 1 mM CaCl2, 0.5 mM MgCl2, 1% sodium deoxycholate, 1% NP-40, plus an EDTA-free protease inhibitor cocktail [Roche, CA]) as described previously (23). Monoclonal antibodies (MAbs) to gI (6B5 or SG4) or gE (MAb 8612) were cross-linked to immobilized protein A on agarose beads (Thermo Scientific, Rockford, IL) as previously described (24), or no primary antibody was used. Lysates were incubated overnight with bound antibodies, washed extensively, and then eluted into SDS sample buffer by incubation of the agarose beads at 100°C for 3 min. The precipitated proteins were resolved by SDS-PAGE, and Western blotting was performed as described previously (45). gI was detected using v67 rabbit polyclonal antiserum, gE was detected using the MAb 8612, and insulin-degrading enzyme (IDE) was detected using a rabbit serum (Covance, Princeton, NJ) by Western blotting.
Mass spectrometry identification of phosphorylation sites in the cytoplasmic domain of gI.
Melanoma cells (105 cells/cm2) were inoculated with rOka (3 log10 PFU/cm2) and harvested 48 h postinfection, and the gE/gI heterodimer was immunoprecipitated as described above. Samples were separated by SDS-PAGE, and proteins were visualized by silver staining. Bands were excised and digested with trypsin, and the proteins were identified using an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific, West Palm Beach, FL), with mass spectrometry performed by the Vincent Coates Foundation Mass Spectrometry Laboratory, Stanford University Mass Spectrometry (http://mass-spec.stanford.edu).
PNGase treatment of viral proteins and identification of gI multimers.
Melanoma cells infected with rOka or the gI mutants were lysed 48 h postinfection with glycoprotein extraction buffer. To identify gI multimers, cell monolayers were washed with phosphate-buffered saline (PBS) containing 20 mM N-ethylmaleimide (NEM; Sigma-Aldrich, St. Louis, MO) and lysed in glycoprotein extraction buffer containing 20 nM NEM. This was done to alkylate free sulfhydryl groups. Proteins were precipitated using methanol and chloroform, dried under vacuum, and then resuspended in 0.5% SDS. N-linked glycosylation sites were removed using PNGase (New England BioLabs, Inc., Ipswich, MA) and incubated at 37°C overnight. The proteins were resolved by SDS-PAGE under reducing or nonreducing conditions using Laemmli sample buffer with or without 5% β-mercaptoethanol, followed by Western blotting for gE and gI.
Confocal microscopy of transfected and infected cells.
Melanoma cells transfected with gE and gI expression constructs were fixed 48 h posttransfection with 4% formaldehyde for 10 min. The anti-gI rabbit serum v67 and the anti-gE MAb 8612 were used to stain the fixed cells. Primary antibodies were detected using donkey anti-rabbit Alexa Fluor 488 (Invitrogen, Carlsbad, CA) and donkey anti-mouse Alexa Fluor 555 (Invitrogen, Carlsbad, CA), and nuclei were stained with Hoechst 33342 (Invitrogen, Carlsbad, CA). Confocal microscopy of rOka- and gI mutant-infected melanoma cells was performed as described previously (7) using the primary antibodies to gE, gI, and the trans-Golgi network (TGN46, sheep polyclonal antibody; AbD Serotec, Oxford, United Kingdom), which were detected using anti-mouse, anti-rabbit, or donkey anti-sheep Alexa Fluor 647 (Invitrogen, Carlsbad, CA) secondary antibodies and staining for nuclei. To stain skin xenografts for confocal microscopy, high-temperature antigen retrieval was performed on paraffin-embedded, paraformaldehyde (PFA)-fixed sections by using vector antigen unmasking solutions following the manufacturer's instructions (Vector Laboratories, Inc., Burlingame, CA). Staining for gE, gI, ORF23, the TGN, and nuclei was performed with VZV protein-specific antibodies as described above. Images were captured using a Zeiss LSM510 confocal microscope equipped with two-photon excitation.
Replication kinetics and plaque sizes of the rOka gI mutant viruses in melanoma cells.
The replication kinetics of rOka and gI mutant viruses were determined as described previously (45). Briefly, 106 melanoma cells were inoculated with 3.0 log10 PFU of either rOka or one of the gI mutants. Infected cells were harvested at 24-h intervals and titrated on melanoma cells. Titer plates fixed at 4 days postinoculation with 4% paraformaldehyde were stained by immunohistochemistry using a high-titer human VZV antiserum. Images of the plaques were captured, and then the outline of each stained plaque was traced and the area (mm2) was calculated using ImageJ (1). Statistical analyses were performed using GraphPad Prism.
Immunoelectron microscopy of HELFs infected with rOka gI mutant viruses.
Samples were fixed in 4% PFA in PBS for 24 h, followed by fixation in 0.1% glutaraldehyde in PBS for 24 h. After dehydration in a graded ethanol series, samples were embedded in LR-white resin (SPI Supplies, Chester, PA). Ultrathin sections (70 nm) were blocked with digoxigenin (DIG) blocking solution (Roche Inc., Indianapolis, IN) and then incubated with rabbit polyclonal anti-gI (v67) and anti-gE (MAb 8612) antibodies. The primary antibodies were detected using gold-conjugated anti-mouse (15-nm gold) and anti-rabbit (5-nm gold) antibodies (BB International, Cardiff, United Kingdom). The immunolabeled sections were stained with 3.5% aqueous uranylacetate followed by 0.2% lead citrate. Sections were analyzed using a JEOL 1230 transmission electron microscope (TEM) at 80 kV, and digital photographs were taken with a Gatan Multiscan 701 digital camera.
Replication of the rOka gI mutant viruses in human skin xenografts.
SCID mice (CB-17scid/scid) were implanted bilaterally with fetal human skin at least 5 weeks before inoculation of viruses as described previously (43). The Stanford University Administrative Panel on Laboratory Animal Care approved all animal protocols. Human tissues were provided by Advanced Bioscience Resources (ABR, Alameda, CA) and were obtained in accordance with state and federal regulations. Virus-infected HELFs were used to inoculate xenografts in the SCID mice. Inocula were titrated on melanoma cells as described previously to determine virus titer. At 7, 10, and 21 days postinoculation, the implants were removed and homogenized for virus titration plus DNA. Quantitative PCR (qPCR) was performed using probes to ORF63 (IE63) and ORF31 (gB) to determine VZV genome copy numbers in infected tissues as previously described (61).
RESULTS
Characterization of gI from the rOka isolate of VZV.
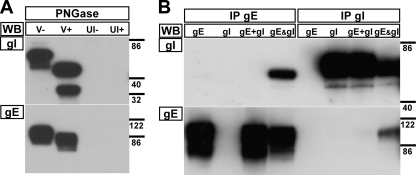
The properties of gI for the rOka isolate of VZV had not been characterized previously and were determined in the present study to allow comparisons with mutant viruses. gI from rOka-infected melanoma cells migrated as two major species of 65 and 53 kDa as determined by Western blotting (Fig. 2A, lane V−). N-linked glycosylation of rOka gI was demonstrated by treating lysates with PNGase. There was a decrease in molecular mass from the 65- and 53-kDa species of gI to 50 and 34 kDa, respectively, representing the polypeptide backbone and, presumably, an O-glycosylated form of gI (Fig. 2A, lane V+). To demonstrate that the rOka gE/gI heterodimer formed only when both of the proteins were expressed simultaneously, immunoprecipitations of gE or gI from lysates of transiently transfected HEK-293 cells were performed (Fig. 2B). When lysates that expressed gE or gI only were combined, they failed to produce a heterodimer during immunoprecipitation (Fig. 2B, lanes gE+gI). In contrast, cotransfection of pCDNA-gE and pCDNA-gI generated stable gE/gI heterodimers (Fig. 2B, lanes gE&gI). Thus, the gE/gI heterodimer must form during protein folding when the two proteins are in close proximity with each other.
Fig. 2.
rOka gI is N glycosylated and forms a heterodimer with gE only upon coexpression. (A) Proteins from lysates of infected (V) or uninfected (UI) melanoma cells were mock (−) or PNGase (+) treated to remove N-linked glycosylation sites. (B) Immunoprecipitation (IP) with MAb to gE (MAb 8612) or gI (6B5) and Western blotting (WB) of gE (MAb 8612) and gI (rabbit v67) from HEK-293 cells transfected with pCDNA-gE, pCDNA-gI, or both expression vectors (gE&gI). gE+gI denotes that lysates from HEK-293 cells transfected with pCDNA-gE or pCDNA-gI only were mixed in equal quantities. The molecular mass markers (in kilodaltons) are given to the right of the blots.
Phosphorylation sites in the cytoplasmic domain do not affect gI function, but residues 105 to 125 are essential for VZV virulence.
Phosphorylation sites that were potentially important for viral replication were identified using a combination of predictive bioinformatics and mass spectrometry analysis of the gE/gI heterodimer, the functional unit required for effective virus replication. Yao and Grose (56) had previously shown that gI is phosphorylated at a single residue, S343, but NetPhos predicted that gI was phosphorylated at residues S296, T312, T338, S343, and S347. Peptides generated by trypsin digest and detected by Orbitrap mass spectrometry confirmed that S343 was extensively phosphorylated (317GIQNATPESDVMLEAAIAQLATIREEpS343PPHSVVNPFVK354, n = 36 peptides; 341EEpS343PPHSVVNPFVK354, n = 9) but also demonstrated that residues T338 and S347 were phosphorylated, though the peptides were less abundant (317GIQNATPESDVMLEAAIAQLApT338IREESPPHSVVNPFVK354, n = 19; 317GIQNATPESDVMLEAAIAQLATIREESPPHpS347VVNPFVK354, n = 1). The phosphorylation status of residues S296 and T312 could not be determined, as peptides spanning this region were not detected in the mass spectrometry data. Therefore, alanine substitutions were incorporated at each of the predicted phosphorylation sites to generate three mutant viruses: P789 (S296A, T312A, and T338A), P10 (S343A), and P11 (S347A). None of the gI mutant viruses, P789, P10, or P11, were defective for replication in human skin xenografts. Each of the mutant viruses had titers similar to that of rOka at day 10 (rOka, 4.0 ± 0.2 log10 PFU; P10, 4.3 ± 0.3 log10 PFU; P11, 4.1 ± 0.2 log10 PFU) and day 21 (rOka, 4.6 ± 0.5 log10 PFU; P10, 5.1 ± 0.1 log10 PFU; P11, 4.5 ± 0.4 log10 PFU; [independent experiment] rOka, 3.2 ± 1.3 log10 PFU; P789, 3.4 ± 0.9 log10 PFU). Thus, these phosphorylation sites in gI do not have a role in virulence and, due to the lack of a phenotype in human skin, were not investigated further.
To map residues in the ectodomain required for gI function, two mutant viruses, the Δ37-167 and Δ105-125 viruses, were generated to determine their effect on VZV virulence. In contrast to the gI phosphorylation mutants, neither the Δ105-125 nor the Δ37-167 mutant could be recovered from human skin xenografts. Thus, further studies were performed to determine the role that residues within this region of gI could have in gE/gI heterodimer formation and VZV virulence.
Mutagenesis of gI identifies C95 as a critical residue for gE/gI heterodimer formation.
Prediction of the gI structure using the Dompred server indicated that VZV gI had a fold similar to that of HSV gE, having an immunoglobulin-type topology. From the initial studies with the Δ105–125 virus, two residues that could affect gI function were identified within the region from residues 105 to 125: a cysteine at residue 106, which is conserved in the Alphaherpesvirinae, and a predicted N-linked glycosylation site at asparagine residue 116. Both of these residues fall within a predicted β-sheet region of gI (Fig. 1E). Therefore, individual alanine substitutions of these two residues, plus three other conserved cysteines in the ectodomain of gI, were generated to determine the effect on gE trafficking and gE/gI heterodimer formation.
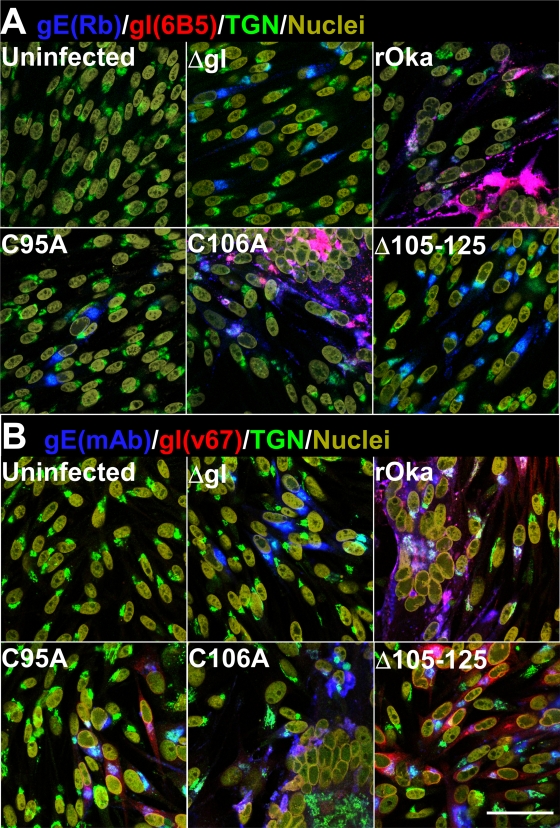
Expression of gE in the absence of gI resulted in a predominantly perinuclear staining pattern in transiently transfected melanoma cells (Fig. 3). In contrast, expression of gI in the absence of gE leads to diffuse gI staining in the cytoplasm and localization at the plasma membrane. The typical localization of gE on the plasma membrane and perinuclear localization were seen in melanoma cells transfected with a dual expression vector that coexpressed wild-type gE and gI. In comparison to wild-type rOka gE, there was aberrant localization of gE in melanoma cells transfected with the dual expression vectors expressing gE and the gI C95A, Δ105–125, and Δ37–167 mutants (Fig. 3). Very little gE was on the plasma membrane, and gE localization was predominantly perinuclear, compared to gE coexpression with wild-type gI. In contrast, none of the remaining gI substitutions had any observable effect on gE localization in melanoma cells. In addition, the gE/gI heterodimer was not detected for the C95A, Δ105–125, and Δ37–167 mutants by immunoprecipitation of gE and gI from HEK-293 cells transiently transfected with dual expression vectors (Fig. 4). The N116 residue was shown to be a glycosylation site by a reduction in relative molecular mass of fully mature gI from 65 kDa for rOka gI to 60 kDa for the N116A mutant (Fig. 4, lanes WT and N116, respectively). These transfection studies suggested that residues 95 and 105 to 125 might be required for gE/gI heterodimer formation plus efficient trafficking and localization of gE to the plasma membrane but that N-linked glycosylation at N116 does not have a role in these functions of gI. Based on these observations, gI mutagenesis experiments were conducted in the context of the viral genome.
Fig. 3.
The localization of gE was aberrant when gE was expressed alone or coexpressed with the gI C95A, Δ105–125, and Δ37–167 mutants. Confocal microscopy of melanoma cells transiently transfected with the pBud dual expression vector containing wild-type gE only (WT-gE), wild-type gI only (WT-gI), wild-type gE and gI (WT-gE/gI), or wild-type gE and the gI C83A, C95A, C106A, N116A, C200A, Δ105–125, or Δ37–167 mutant. The figure shows a merged image and, for clarity, individual channels for gE and gI staining. Green, gE (MAb 8612); red, gI (rabbit polyclonal antibody v67); blue, nuclei (Hoechst 33342). Bar, 50 μm.
Fig. 4.
gE did not form a heterodimer with the gI C95A, Δ105–125, or Δ37–167 mutant. Immunoprecipitation of the VZV gE/gI heterodimer from HEK-293 cells transiently transfected with the pBud dual expression vector containing gE and gI or the gI C83A (C83), C95A (C95), C106A (C106), N116A (N116), C200A (C200), Δ105–125 (Δ105), or Δ37–167 (Δ37) mutant. IP, immunoprecipitation with MAb to gE (MAb 8612) or gI (6B5). WB, Western blotting for gE (MAb 8612) or gI (rabbit v67). The arrowhead in the bottom panel indicates a nonspecific protein. The molecular mass markers (in kilodaltons) are given to the right of the blots. Blots for each IP were stripped and reprobed for consistency. We have noted that the C83A lane in this blot suggests reduced expression, but this was not representative.
Characterization of the spread and replication of rOka viruses carrying mutations in gI.
To determine whether the four cysteine residues 83, 95, 106, and 200 or the N-linked glycosylation site at N116 in gI played a role in VZV replication, the alanine substitutions were transferred into the rOka genome by using cosmid-based mutagenesis. Plaque sizes for the C83A, C106A, N116A, and C200A mutants were comparable to that of rOka (1.0 mm2 [±0.4]), ranging from 1.2 to 1.5 mm2 (±0.3 to 0.5) (Fig. 5A). In contrast, the C95A mutant had a significantly reduced plaque size (0.2 mm2 [±0.1]), which was comparable to that of the ΔgI, Δ105–125, and Δ37–167 viruses (0.1 to 0.2 mm2 [±0.1]). The replication kinetics for the C95A, Δ105–125, and Δ37–167 viruses during the logarithmic phase (days 1 to 3) were significantly reduced at each time point (P < 0.01) in melanoma cells compared to those for rOka, whereas those for the C83A, C106A, N116A, and C200A mutants were similar to those for the wild-type virus (Fig. 5B and C).
Fig. 5.
Plaque sizes were reduced and replication kinetics delayed for rOka C95A, Δ105–125, and Δ37–167 mutants. (A) Box and whisker plot of mean plaque sizes, with standard errors of the means, calculated from 30 individual plaques for each virus. (B and C) Replication of rOka and the gI mutants, with kinetics disparate from (B) or similar to (C) those for the rOka virus in melanoma cells. Standard errors of the means are shown.
Infection of melanoma cells causes extensive syncytium formation at 48 h postinfection. This is characterized by rosettes of nuclei and the accumulation of viral proteins at the periphery of the syncytium, while components of the TGN localize to its center. Similar to the plaque size data, syncytium formation was not affected at 48 h postinfection by the C83A, C106A, N116A, and C200A substitutions (Fig. 6A [only C106A shown]). Rosettes of nuclei were evident, and these were outlined with gE and gI, while TGN staining localized at the center of the syncytia, as determined by confocal microscopy. Individually infected cells at the periphery of the syncytia, indicating more recently infected cells, showed that gE and gI were localized to cell membranes and the TGN. In contrast, the C95A, Δ105–125, and Δ37–167 mutants, similar to the ΔgI virus, failed to induce syncytia at 48 h postinfection, with gE localization for these mutants predominantly in the TGN as demonstrated by confocal microscopy (Fig. 6A). In addition, gI was not detected using the conformation-dependent anti-gI MAb 6B5. To determine that gI could be detected in the virus mutants, confocal microscopy was performed using the v67 rabbit polyclonal anti-gI serum. The v67 serum detected gI in the C95A, Δ105–125, and Δ37–167 mutants at levels similar to that for wild-type rOka (Fig. 6B). This striking observation suggested that C95 and residues 105 to 125 are critical for the canonical structural conformation of VZV gI.
Fig. 6.
The canonical structural conformation of gI was altered for the C95A and Δ105–125 mutants. Confocal microscopy of melanoma cells infected with rOka and the ΔgI, C95A, C106A, and Δ105–125 mutants. (A) Blue, gE (rabbit [Rb]); red, gI (6B5); green, TGN (TGN46); yellow, nuclei (Hoechst 33342). (B) Blue, gE (MAb 8612); red, gI (rabbit v67); green, TGN (TGN46); yellow, nuclei (Hoechst 33342). Bar, 50 μm.
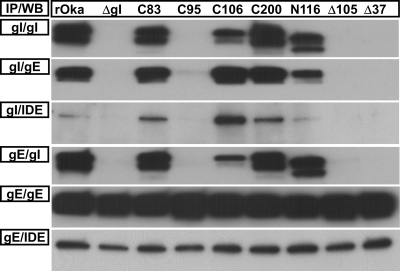
Effects of gI mutagenesis on VZV gE/gI heterodimer formation.
C95 and residues 105 to 125 were required for the formation of the gE/gI heterodimer in the transfection studies, but whether viral factors could contribute to the heterodimer formation was unclear. Immunoprecipitation of the gE/gI heterodimer from infected melanoma cells showed that the C95A, Δ105–125, and Δ37–167 gI mutants did not coimmunoprecipitate with gE (Fig. 7). As seen in the transfection experiments, the N116 residue of gI was confirmed to be a site for posttranslational modification in the context of virus replication. Mature gI from the N116A mutant had a reduced relative molecular mass of 60 kDa, compared to 65 kDa from the wild-type virus. Importantly, this substitution did not affect gE/gI heterodimer formation.
Fig. 7.
The gE/gI heterodimer formation was absent for the C95A, Δ105–125 (Δ105), and Δ37–167 (Δ37) mutants. Coimmunoprecipitation (IP) of the gE/gI heterodimer from the gI C83A (C83), C95A (C95), C106A (C106), C200A (C200), N116A (N116), Δ105–125 (Δ105), and Δ37–167 (Δ37) mutants using anti-gE (MAb 8612) or anti-gI (6B5) and Western blotting (WB) for gE (MAb 8612), gI (rabbit v67), or IDE (rabbit; Covance). The membranes for each IP were stripped and reprobed for consistency.
To ensure that coimmunoprecipitation was achievable for the C95A, Δ105–125, and Δ37–167 mutants, the presence of IDE, a protein known to interact strongly with VZV gE (8, 30), was used as a coimmunoprecipitation control. IDE was detected only when the gE/gI heterodimer was coimmunoprecipitated using the anti-gI MAb with lysates from the wild type plus the C83A, C106A, N116A, and C200A mutants (Fig. 7). As expected, IDE was not coimmunoprecipitated using the anti-gI MAb from the C95A, Δ105–125, and Δ37–167 mutants. In contrast, IDE was successfully coimmunoprecipitated from all of the mutant viruses when the anti-gE MAb was used, demonstrating that the lack of gI binding for the C95A, Δ105–125, and Δ37–167 mutants was not a consequence of unfavorable coimmunoprecipitation conditions. Thus, the lack of gE/gI heterodimer formation added weight to the hypothesis that residues C95 and 105 to 125 are required for the canonical structure of gI.
The presence of free cysteines in a molecule can result in the formation of protein multimers. To determine whether any of the substitutions had this effect on gI in the context of virus, protein lysates were prepared in the presence of the alkylating agent N-ethylmaleimide and digested with PNGase to remove N-linked glycosylation sites to improve protein resolution under reducing and nonreducing SDS-PAGE. Under these conditions, three proteins that were separated by both nonreducing and reducing SDS-PAGE were detected by Western blotting, at approximately 38, 51, and 56 kDa for rOka (Fig. 8). These are equivalent to the protein backbone (approximately 38 kDa) and posttranslationally modified forms of gI but lacking N-linked glycosylation (approximately 51 and 56 kDa). A series of multimers (approximately 105, 94, and 83 kDa) that approximately correlated with the relative molecular masses for dimers produced from the monomers (approximately 38, 51, and 59 kDa) detected under reducing conditions were detected for the C83A mutant. This suggested that an unpaired cysteine was contributing to intermolecular bond formation with C83A gI. In addition, the C83A and C200A mutants showed slight alterations in posttranslational modification compared to that for wild-type gI. The relative molecular mass of the mature monomeric forms of gI for both the C83A and the C200A mutant was approximately 59 kDa, 3 kDa higher than that for wild-type gI. The C95A mutant resulted in complex, higher-order multimer formation, as at least seven gI-specific proteins, ranging from approximately 90 to 152 kDa, were identified under nonreducing conditions. In contrast, a single, posttranslationally modified protein (approximately 51 kDa) was detected under reducing conditions, suggesting that this monomer could form posttranslationally modified dimers and trimers. Few multimers were seen for the remaining virus mutants, with the exception of the Δ105–125 mutant (Fig. 8, lane Δ105). Similar to the C95A mutant, the Δ105-125 mutant produced a single gI-specific protein (approximately 48 kDa) as observed under reducing conditions, but a pattern of multimers similar to that for the C95A mutant was formed, with the exception of those between approximately 117 and 152 kDa, likely due to the loss of the C106 residue in the Δ105-125 mutant (Fig. 8, lanes C95 and Δ105). These data support the hypothesis that significant changes occurred within the structures of gI for the C95A and Δ105-125 mutants, contributing to a lack of gE/gI heterodimer formation and subsequent effects on replication and plaque size in vitro.
Fig. 8.
Unpaired cysteine residues cause gI to form disulfide-linked multimers. Western blots of lysates from C83A (C83), C95A (C95), C106A (C106), C200A (C200), N116A (N116), or Δ105–125 (Δ105) mutant-infected melanoma cells that were treated with the sulfhydryl alkylating agent N-ethylmaleimide and PNGase to remove N-linked glycosylation. Proteins were resolved by SDS-PAGE under reducing (β-mercaptoethanol) and nonreducing conditions to identify disulfide-linked multimers in the gI mutants. The upper two panels are short (A) and long (B) exposures of blots to show more clearly the presence and abundance of multimers. (C) Blot of the stripped membranes for gE (MAb 8612), to show the levels of protein. The molecular mass markers (in kilodaltons) are given in the center.
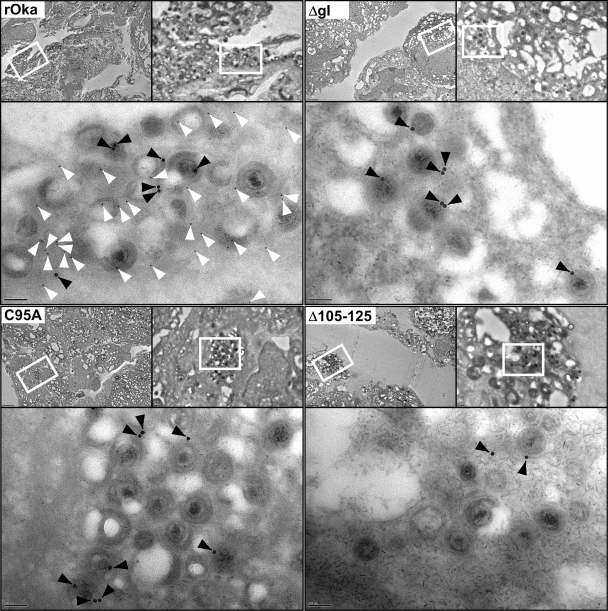
Loss of gE/gI heterodimer formation prevents gI incorporation into virus particles.
To determine whether the loss of heterodimer formation between gE and gI adversely affects incorporation of gI into virus particles, immunoelectron microscopy studies were performed. Both gE and gI were readily detected on rOka virus particles in infected HELFs by using immunogold labeling of gE (15 nm) and gI (5 nm) on negatively stained ultrathin sections by electron microscopy (Fig. 9). In contrast, gE but not gI was detected on virus particles in the C95A and Δ105–125 mutant-infected HELFs. As expected, gI was not detected for the ΔgI virus. This suggested that the loss of the gI interaction with gE prevented the incorporation of gI into virus particles.
Fig. 9.
gI was not incorporated into virus particles of the gI C95A and Δ105–125 mutants. Immunoelectron microscopy of HELFs infected with rOka or the ΔgI, C95A, or Δ105–125 mutant. gE (MAb 8612) was detected using 15-nm gold particles (black arrowheads), and gI (rabbit v67) was detected with 5-nm gold particles (white arrowheads). Each of the four panels is divided into three magnifications; upper left, ×1,000 (bar, 2 μm); upper right, ×5,000 (bar, 200 nm); bottom, ×20,000 (bar, 100 nm). The white rectangles outline the areas where the subsequent higher-magnification images were taken.
Effects of gI mutagenesis on VZV pathogenesis in human skin.
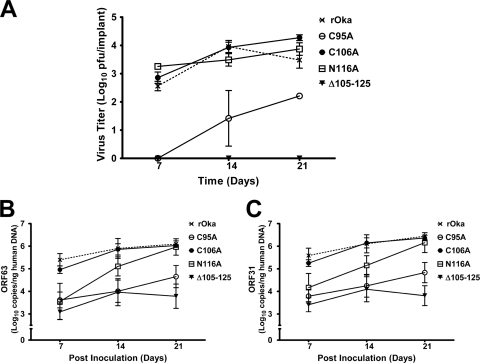
Based on the initial experiments showing a lack of Δ105–125 virus replication in the SCIDhu model of skin pathogenesis, the C95A, C106A, and N116A mutants were tested for replication and viral spread in human skin. As demonstrated in the preliminary study, the Δ105–125 virus could not be recovered from inoculated skin xenografts at any time point (Fig. 10A). In contrast, the C106A and N116A mutants were recovered from xenografts at titers similar to that of wild-type rOka at 7, 14, and 21 days postinoculation. Comparable to what was observed in vitro, the C95A mutant replicated poorly in human skin. Virus was not recovered at 7 days postinoculation and was recovered from only 2 of 6 xenografts on both 14 and 21 days postinoculation (Table 2), reaching a titer of 2.2 ± 0.1 log10 PFU by day 21, which was significantly lower (P < 0.01) than that for rOka (Fig. 10A). The differences in titers were not a result of inoculum titers, as these were not significantly different between the wild-type and mutant viruses (Table 2).
Fig. 10.
Virulence of the C95A mutant was significantly diminished in human skin xenografts. (A) Titers of rOka and the C95A, C106A, N116A, and Δ105–125 mutant viruses in human skin xenografts at 7, 14, and 21 days postinoculation. (B and C) Genome copies of rOka and the C95A, C106A, N116A, and Δ105–125 mutant viruses extracted from human skin xenografts, determined using qPCR of ORF63 and ORF31. Standard errors of the means are shown.
Table 2.
Frequency of skin implants from which virus was recovered at days 7, 14, and 21 postinoculation
| Virus | Inoculum titera, log10 PFU/ml (SEM) | No. of implants from which virus was recovered at day: |
||
|---|---|---|---|---|
| 7 | 14 | 21 | ||
| rOka | 6.1 (0.055) | 5/6 | 5/6 | 6/6 |
| C95A | 6.0 (0.113)* | 0/6 | 2/6 | 2/6 |
| C106A | 6.0 (0.040)* | 6/6 | 5/6 | 6/6 |
| N116A | 6.0 (0.028)* | 3/6 | 5/6 | 5/6 |
| Δ105–125 | 5.9 (0.078)* | 0/6 | 0/6 | 0/6 |
*, P > 0.05 when compared to rOka, as determined by analysis of variance.
qPCR was performed to establish whether viral DNA could be detected in the skin xenografts from which infectious virus was not recovered. It was also noted that genome copy number increased over the duration of the experiment for all of the viruses that could be recovered from human skin xenografts (Fig. 10B and C). rOka and the C106A mutant had similar increases in genome copy number from day 7 to day 21, which correlated well with virus titer. In contrast, the N116A mutant had genome copies at day 7, but these were significantly lower than those for rOka (P < 0.01 for ORF63 and P < 0.05 for ORF31) and closer to the range for the C95A and Δ105–125 mutants, where virus could not be recovered. This correlated with the recovery of virus from fewer implants (Table 2). The increase in C95A genome copies was consistent with the ability to recover virus from the skin xenografts. The Δ105–125 mutant showed some limited replication by qPCR, as indicated by an approximately 1 log10 increase in genome copy from day 7 to day 14, but this did not translate into recoverable virus.
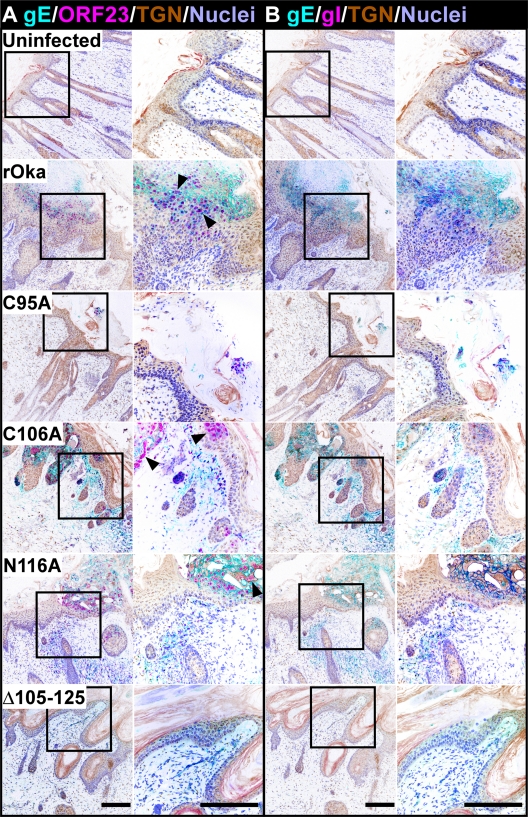
To determine the extent of lesion formation by the mutant viruses in human skin xenografts, confocal microscopy was performed on serial sections that were stained, in conjunction with a TGN marker, for either ORF23 and gE or gE and gI. Serial sections were stained to allow comparisons of virus-infected cells that were stained with the ORF23/gE and gE/gI antibodies. rOka formed typical lesions in the human skin xenografts that showed extensive replication in the epidermis and traversed across the basal layer into the dermis (Fig. 11). As expected, staining for the ORF23 capsid protein was localized to the nucleus, while the gE and gI staining localized to the plasma membranes of infected cells. Polykaryon formation, a signature for VZV replication in the skin, was evident in the stained sections. Staining patterns similar to that for the wild type were seen for both the C106A and the N116A mutant. In contrast to the wild-type lesions, small lesions that were localized to the epidermis were detected for the C95A mutant. The lesions of the C95A mutant did not penetrate across the basal layer into the dermis. VZV-specific staining was not detected for the Δ105–125 mutant or, as expected, in uninfected control xenografts. These data provide additional strong evidence that the gE/gI heterodimer of VZV is required for skin pathogenesis. Moreover, based on the minimal replication of the C95A mutant, which lacks heterodimer formation, these observations indicate that gI has functions in virulence distinct from heterodimer formation with gE.
Fig. 11.
The gI C95A mutant failed to penetrate into the dermal layer of skin xenografts. Confocal microscopy of skin implants infected with rOka and the gI C95A, C106A, N116A, and Δ105–125 mutants. (A) Cyan, gE (MAb 8612); pink, ORF23 (rabbit v67); brown, TGN (sheep; Serotec); lilac, nuclei (Hoechst 33342). (B) Cyan, gE (MAb 8612); pink, gI (rabbit v67); brown, TGN (sheep; Serotec); lilac, nuclei (Hoechst 33342). The staining was conducted on serial skin sections, allowing comparisons of VZV-infected cells to be made. The colors for the images were inverted to emphasize staining for viral proteins among the complex architecture of the skin and allow a clear differentiation between the infected and uninfected areas. Syncytia, the hallmark of VZV infection, are highlighted by the black arrowheads. Bar, 200 μm.
DISCUSSION
Mutagenesis and deletion studies of gE and gI have previously shown that the gE/gI heterodimer is a virulence determinant for the alphaherpesviruses (5, 9, 12, 18, 27, 60). However, studies with VZV have focused predominantly on the role that gE has in virulence in human tissues (7–9, 60). The present study clearly demonstrated that N-terminal residues C95 and 105 to 125 in gI are essential for gE/gI heterodimer formation and necessary for effective virulence. Substitution of C95 with alanine led to a significant replication defect in human skin xenografts comparable to that for the gE ΔCys mutant of VZV, which also fails to form gE/gI heterodimers (8). In contrast, the gI Δ105–125 mutant was avirulent in human skin, the same phenotype as that of the ΔgI virus (42). In addition to significantly reduced virulence, all of these viruses had small-plaque phenotypes, which indicated a reduction in their efficiency for cell-cell spread. Cell-cell spread of HSV, the human virus most closely related to VZV, is partially governed by the gE/gI heterodimer, but this heterodimer is unnecessary for virion attachment or fusion of the viral envelope, which are both requirements for virus entry (18, 19). Replication of the ΔgI, C95A, and Δ105–125 mutants demonstrated that, similar to HSV, functional gI is not required for VZV entry. IDE is a cellular molecule that contributes to cell-cell spread of VZV, a notion supported by the ΔY51-P187 avirulent gE mutant, which binds IDE poorly (3, 9, 29, 30). In the present study, none of the gI mutant viruses, including the C95A and Δ105–125 viruses, inhibited the binding of gE to IDE. Therefore, it is very unlikely that defective cell-cell spread or entry would be a consequence of aberrant gE/IDE interactions. Combined, these data strengthen the notion that the gE/gI heterodimer has a pivotal role in VZV virulence and that gI has additional functions independent of gE.
It is clear from this study that C95 is required for canonical gI structure, which is critical for its function. The loss of gE/gI heterodimer formation for both the C95A and the Δ105–125 virus, the lack of MAb 6B5 binding detected by confocal and immunoprecipitation studies, and the detection of higher-order disulfide-linked multimers suggest that neither of these gI mutants could undergo the canonical protein folding of wild-type gI. C95 is very likely to form a disulfide bond with residue C106, based on a previous study with feline herpesvirus (FHV) gI (40). The four cysteine residues C83, C95, C106, and C200 of VZV gI are analogous to C79, C91, C102, and C223 in FHV gI. Extrapolation of the FHV data, which were based on transfection studies using a truncated form of gI, suggested that two disulfide bonds form: one between C83 and C200 and a second between C95 and C106. In the present study, gI multimers were formed when C106 (in the C95A mutant) and C200 (in the C83A mutant) were free to form disulfide bonds. These data provide an insight into gI topology that can be inferred from glycoproteins of other alphaherpesviruses.
McGeoch (38) proposed that both gD and gI arose in the genomes of alphaherpesviruses via gene duplication. VZV lacks a gD homolog, which is an entry determinant for HSV (17, 22). However, the predicted structure of VZV gI suggests an immunoglobulin-like (Ig-like) fold similar to that demonstrated for HSV gD (13). The disulfide linkages within HSV gD are similar to those proposed for VZV gI, supporting the idea that gI could form an Ig-like domain (34, 38). The gE/gI heterodimer of HSV acts as an Fc receptor, and similar to gD, HSV gE exhibits an Ig-like domain, but the structure of HSV gI has not been resolved (6, 49). In the present study, VZV gI was predicted to have a topology similar to that of HSV gE. Therefore, the nature of the disulfide linkages in HSV gD can be used to predict the basis of the catastrophic effects that the C95A substitution had on VZV gI function without an available crystal structure for gI. In HSV gD, the disulfide bond formed between C118 and C127, which are a comparable distance to VZV gI C95 and C106, stabilizes a turn immediately after a β-sheet configuration, leading to additional interactions within the protein that are likely to form a subsequent complex fold. A serine substitution in HSV gD at C118, analogous to gI C95, prevented binding of conformation-dependent antibodies, and the mutant was unable to complement a gD-null virus (33). Because it is necessary for entry, gD is more critical to HSV replication than gI is for VZV, but the data indicate that the disulfide bond requirements for the two proteins are similar.
The data from the present study strongly suggest that both residues C83 and C95 are obscured by the topology of gI. The disulfide bond between C95 and C106 determines a protein conformation that stabilizes gI. However, the nature of this stabilizing bond is essential in a single direction only, C95 to C106, as the free cysteine C95 in the C106A mutant does not overtly affect gI conformation. This was clearly demonstrated in the present study, as the C106A mutant was not defective in gE/gI heterodimer formation, viral replication, or virulence in the skin. C106 is likely to be located close to the surface of gI, leaving C95 buried within the topology, but remains important for the stability of gI. Intriguingly, the stabilized fold of gI appears to play an essential role in the gE/gI heterodimer formation. The recent resolution of protein structures for the gH/gL heterodimer demonstrates that heterodimers of other herpesvirus glycoproteins fold simultaneously during coexpression (15). The requirement of gE/gI coexpression for heterodimer formation strongly supports this hypothesis. Simultaneous folding of gE and gI would lead to stabilization of the gI structure and prevent the free cysteine at C95 in the C106A mutant from forming the multimers that were observed when this residue was substituted.
The second disulfide bond between C83 and C200 might have a role in gI stability, but this bond was not essential for heterodimer formation. Importantly, substitution of FHV gI C79, a conserved cysteine analogous to C83 of VZV gI, was also heterodimer competent but did not affect replication in vitro (40). The remaining cysteine homologs in FHV gI were not investigated in the context of infectious virus. The similar relative molecular masses of the mature VZV C83 and C200 mutant proteins under reducing conditions, which differed from that of wild-type gI by 3 kDa, suggested that both mutants undergo similar maturation processes. The prominent dimers formed by the C83A mutant under nonreducing conditions indicated that C83 is also obscured by the topology of gI. In the C83A mutant, the free cysteine of C200 was available to form a disulfide bond, unlike the free cysteine in the C200A mutant, as the C83 could be hidden by cofolding with gE.
In contrast to the point mutants, the 21-amino-acid deletion in the Δ105-125 mutant is very likely to have severely disrupted the β-sheet configuration of gI, leading to a dramatic conformational change. This was demonstrated clearly by the loss of gE/gI heterodimer formation and MAb 6B5 binding in the present study. It might be anticipated that the severe structural alteration in Δ105-125 gI would cause extensive dimer formation. However, the presence of fewer multimers in the Δ105-125 mutant than in the C95A mutant is not surprising, as the deletion of C106 from the Δ105-125 mutant would not allow disulfide bond formation, similar to the result seen for the C95A mutant.
A previous study using linker insertion mutagenesis close to the signal peptide of baculovirus-derived gI showed a similar failure of gE/gI heterodimer formation (28). This disrupted region falls within the predicted β-sheet structure at the N terminus of gI and endorses the predictions made for the folded protein in the present study. In addition, the required coexpression of VZV gE with gI needed for heterodimer formation ensures trafficking of the heterodimer to the cell surface, which becomes endocytosed and trafficked back to the TGN for incorporation into virus particles during secondary envelopment (2, 37, 47). A previous study has shown that gI plays a role in the secondary envelopment of VZV particles (54). However, immunoelectron microscopy analyses in the present study clearly showed that the lack of gI incorporation into virus particles did not affect virus particle morphology or gE incorporation into the ΔgI, C95A, or Δ105-125 mutant. This demonstrates that VZV particle envelopment is possible in the absence of functional gI. Moreover, because the C95A and Δ105-125 mutants had independent phenotypes in human skin, gI must have a function in cells independent of gE and the gE/gI heterodimer.
The VZV gE/gI heterodimer has receptor properties, including phosphorylation of the cytoplasmic domains of gE and gI (32, 46, 58, 59). The present study also demonstrated for the first time that gI phosphorylation in the cytoplasmic domain does not affect VZV skin virulence. Previously, it was thought that VZV gI was phosphorylated at a single residue in the cytoplasmic domain of gI (56). However, mass spectrometry clearly identified that residues T338 and S347, in addition to S343, were phosphorylated. Moreover, whether these phosphorylation sites were required for VZV virulence was not known previously, but their substitution with alanine in this study showed that they were not required for VZV replication in vitro or skin virulence in vivo. The discrepancy in the numbers of residues phosphorylated between this and previous studies can be attributed to the superior sensitivity of new mass spectrometry tools compared to that of previous mutagenesis studies using radiolabeled proteins from transiently expressed gI (56). Another fundamental difference was the immunoprecipitation of the gE/gI heterodimer from VZV-infected rather than transfected cells. It is plausible that more-extensive phosphorylation might occur during viral infection than during transient transfection. The serine threonine kinases of VZV, ORF47 and ORF66, could play a role in the phosphorylation of gI at T338 and S347, but cellular kinases, such as casein kinase II, which leads to gE phosphorylation, might also be involved (20, 26). Nevertheless, mutation of these residues did not generate an observable phenotype in vitro or in vivo, which demonstrated that, in the context of functions related to the phosphorylation of the gI cytoplasmic domain, viral or cellular kinases are not relevant for VZV virulence in skin.
In conclusion, C95 in VZV gI has been identified as a critical residue required for the structure of gI. This is very likely conserved among the alphaherpesviruses. These experiments demonstrate that gI, like gE, has a multifunctional role in the pathogenicity of VZV.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AI20459 and AI053846.
We thank Chris Adams for providing assistance with mass spectrometry at the Vincent Coates Foundation Mass Spectrometry Laboratory, Stanford University. We thank Susan Vleck for valuable discussion and critical review of the manuscript.
Footnotes
Published ahead of print on 23 February 2011.
REFERENCES
- 1. Abramoff M. D., Magelhaes P. J., Ram S. J. 2004. Image processing with ImageJ. Biophotonics Int. 11:36–42 [Google Scholar]
- 2. Alconada A., Bauer U., Hoflack B. 1996. A tyrosine-based motif and a casein kinase II phosphorylation site regulate the intracellular trafficking of the varicella-zoster virus glycoprotein I, a protein localized in the trans-Golgi network. EMBO J. 15:6096–6110 [PMC free article] [PubMed] [Google Scholar]
- 3. Ali M. A., Li Q., Fischer E. R., Cohen J. I. 2009. The insulin degrading enzyme binding domain of varicella-zoster virus (VZV) glycoprotein E is important for cell-to-cell spread and VZV infectivity, while a glycoprotein I binding domain is essential for infection. Virology 386:270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baiker A., et al. 2004. The immediate-early 63 protein of varicella-zoster virus: analysis of functional domains required for replication in vitro and for T-cell and skin tropism in the SCIDhu model in vivo. J. Virol. 78:1181–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banfield B. W., Yap G. S., Knapp A. C., Enquist L. W. 1998. A chicken embryo eye model for the analysis of alphaherpesvirus neuronal spread and virulence. J. Virol. 72:4580–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baucke R. B., Spear P. G. 1979. Membrane proteins specified by herpes simplex viruses. V. Identification of an Fc-binding glycoprotein. J. Virol. 32:779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Berarducci B., et al. 2006. Essential functions of the unique N-terminal region of the varicella-zoster virus glycoprotein E ectodomain in viral replication and in the pathogenesis of skin infection. J. Virol. 80:9481–9496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berarducci B., et al. 2009. Deletion of the first cysteine-rich region of the varicella-zoster virus glycoprotein E ectodomain abolishes the gE and gI interaction and differentially affects cell-cell spread and viral entry. J. Virol. 83:228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Berarducci B., et al. 2010. Functions of the unique N-terminal region of glycoprotein E in the pathogenesis of varicella-zoster virus infection. Proc. Natl. Acad. Sci. U. S. A. 107:282–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blom N., Gammeltoft S., Brunak S. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294:1351–1362 [DOI] [PubMed] [Google Scholar]
- 11. Bryson K., et al. 2005. Protein structure prediction servers at University College London. Nucleic Acids Res. 33:W36–W38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Card J. P., Whealy M. E., Robbins A. K., Enquist L. W. 1992. Pseudorabies virus envelope glycoprotein gI influences both neurotropism and virulence during infection of the rat visual system. J. Virol. 66:3032–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carfi A., et al. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169–179 [DOI] [PubMed] [Google Scholar]
- 14. Chaudhuri V., Sommer M., Rajamani J., Zerboni L., Arvin A. M. 2008. Functions of varicella-zoster virus ORF23 capsid protein in viral replication and the pathogenesis of skin infection. J. Virol. 82:10231–10246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chowdary T. K., et al. 2010. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat. Struct. Mol. Biol. 17:882–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cohen J. I., Straus S. E., Arvin A. 2007. Varicella-zoster virus replication, pathogenesis and management, p. 3091 In Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E. (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 17. Davison A. J., Scott J. E. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759–1816 [DOI] [PubMed] [Google Scholar]
- 18. Dingwell K. S., et al. 1994. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J. Virol. 68:834–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dingwell K. S., Doering L. C., Johnson D. C. 1995. Glycoproteins E and I facilitate neuron-to-neuron spread of herpes simplex virus. J. Virol. 69:7087–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Erazo A., Kinchington P. R. 2010. Varicella-zoster virus open reading frame 66 protein kinase and its relationship to alphaherpesvirus US3 kinases. Curr. Top. Microbiol. Immunol. 342:79–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Farnsworth A., Goldsmith K., Johnson D. C. 2003. Herpes simplex virus glycoproteins gD and gE/gI serve essential but redundant functions during acquisition of the virion envelope in the cytoplasm. J. Virol. 77:8481–8494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fuller A. O., Spear P. G. 1987. Anti-glycoprotein D antibodies that permit adsorption but block infection by herpes simplex virus 1 prevent virion-cell fusion at the cell surface. Proc. Natl. Acad. Sci. U. S. A. 84:5454–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grose C., Edwards D. P., Weigle K. A., Friedrichs W. E., McGuire W. L. 1984. Varicella-zoster virus-specific gp140: a highly immunogenic and disulfide-linked structural glycoprotein. Virology 132:138–146 [DOI] [PubMed] [Google Scholar]
- 24. Harlow E., Lane D. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 25. Ito H., et al. 2005. Role of the varicella-zoster virus gene product encoded by open reading frame 35 in viral replication in vitro and in differentiated human skin and T cells in vivo. J. Virol. 79:4819–4827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kenyon T. K., Grose C. 2010. VZV ORF47 serine protein kinase and its viral substrates. Curr. Top. Microbiol. Immunol. 342:99–111 [DOI] [PubMed] [Google Scholar]
- 27. Kimman T. G., et al. 1992. Contribution of single genes within the unique short region of Aujeszky's disease virus (suid herpesvirus type 1) to virulence, pathogenesis and immunogenicity. J. Gen. Virol. 73:243–251 [DOI] [PubMed] [Google Scholar]
- 28. Kimura H., Straus S. E., Williams R. K. 1997. Varicella-zoster virus glycoproteins E and I expressed in insect cells form a heterodimer that requires the N-terminal domain of glycoprotein I. Virology 233:382–391 [DOI] [PubMed] [Google Scholar]
- 29. Li Q., Ali M. A., Cohen J. I. 2006. Insulin degrading enzyme is a cellular receptor mediating varicella-zoster virus infection and cell-to-cell spread. Cell 127:305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li Q., Krogmann T., Ali M. A., Tang W. J., Cohen J. I. 2007. The amino terminus of varicella-zoster virus (VZV) glycoprotein E is required for binding to insulin-degrading enzyme, a VZV receptor. J. Virol. 81:8525–8532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ling P., Kinchington P. R., Sadeghi-Zadeh M., Ruyechan W. T., Hay J. 1992. Transcription from varicella-zoster virus gene 67 (glycoprotein IV). J. Virol. 66:3690–3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Litwin V., Jackson W., Grose C. 1992. Receptor properties of two varicella-zoster virus glycoproteins, gpI and gpIV, homologous to herpes simplex virus gE and gI. J. Virol. 66:3643–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Long D., Cohen G. H., Muggeridge M. I., Eisenberg R. J. 1990. Cysteine mutants of herpes simplex virus type 1 glycoprotein D exhibit temperature-sensitive properties in structure and function. J. Virol. 64:5542–5552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Long D., Wilcox W. C., Abrams W. R., Cohen G. H., Eisenberg R. J. 1992. Disulfide bond structure of glycoprotein D of herpes simplex virus types 1 and 2. J. Virol. 66:6668–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lungu O., Panagiotidis C. A., Annunziato P. W., Gershon A. A., Silverstein S. J. 1998. Aberrant intracellular localization of varicella-zoster virus regulatory proteins during latency. Proc. Natl. Acad. Sci. U. S. A. 95:7080–7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mallory S., Sommer M., Arvin A. M. 1997. Mutational analysis of the role of glycoprotein I in varicella-zoster virus replication and its effects on glycoprotein E conformation and trafficking. J. Virol. 71:8279–8288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maresova L., Pasieka T. J., Homan E., Gerday E., Grose C. 2005. Incorporation of three endocytosed varicella-zoster virus glycoproteins, gE, gH, and gB, into the virion envelope. J. Virol. 79:997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McGeoch D. J. 1990. Evolutionary relationships of virion glycoprotein genes in the S regions of alphaherpesvirus genomes. J. Gen. Virol. 71:2361–2367 [DOI] [PubMed] [Google Scholar]
- 39. McGeoch D. J., Dolan A., Ralph A. C. 2000. Toward a comprehensive phylogeny for mammalian and avian herpesviruses. J. Virol. 74:10401–10406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mijnes J. D., et al. 1998. The disulfide-bonded structure of feline herpesvirus glycoprotein I. J. Virol. 72:7245–7254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mo C., Lee J., Sommer M., Grose C., Arvin A. M. 2002. The requirement of varicella zoster virus glycoprotein E (gE) for viral replication and effects of glycoprotein I on gE in melanoma cells. Virology 304:176–186 [DOI] [PubMed] [Google Scholar]
- 42. Moffat J., Ito H., Sommer M., Taylor S., Arvin A. M. 2002. Glycoprotein I of varicella-zoster virus is required for viral replication in skin and T cells. J. Virol. 76:8468–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moffat J. F., Stein M. D., Kaneshima H., Arvin A. M. 1995. Tropism of varicella-zoster virus for human CD4+ and CD8+ T lymphocytes and epidermal cells in SCID-hu mice. J. Virol. 69:5236–5242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nagashunmugam T., et al. 1998. In vivo immune evasion mediated by the herpes simplex virus type 1 immunoglobulin G Fc receptor. J. Virol. 72:5351–5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oliver S. L., Zerboni L., Sommer M., Rajamani J., Arvin A. M. 2008. Development of recombinant varicella-zoster viruses expressing luciferase fusion proteins for live in vivo imaging in human skin and dorsal root ganglia xenografts. J. Virol. Methods 154:182–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Olson J. K., Bishop G. A., Grose C. 1997. Varicella-zoster virus Fc receptor gE glycoprotein: serine/threonine and tyrosine phosphorylation of monomeric and dimeric forms. J. Virol. 71:110–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Olson J. K., Grose C. 1998. Complex formation facilitates endocytosis of the varicella-zoster virus gE:gI Fc receptor. J. Virol. 72:1542–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pickering L. K., et al. 2009. Immunization programs for infants, children, adolescents, and adults: clinical practice guidelines by the Infectious Diseases Society of America. Clin. Infect. Dis. 49:817–840 [DOI] [PubMed] [Google Scholar]
- 49. Sprague E. R., Wang C., Baker D., Bjorkman P. J. 2006. Crystal structure of the HSV-1 Fc receptor bound to Fc reveals a mechanism for antibody bipolar bridging. PLoS Biol. 4:e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tirabassi R. S., Enquist L. W. 2000. Role of the pseudorabies virus gI cytoplasmic domain in neuroinvasion, virulence, and posttranslational N-linked glycosylation. J. Virol. 74:3505–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vafai A., Wellish M., Wroblewska Z., Cisco M., Gilden D. 1987. Induction of antibody against in vitro translation products encoded by varicella-zoster virus glycoprotein genes. Virus Res. 7:325–333 [DOI] [PubMed] [Google Scholar]
- 52. Wang F., et al. 2005. Herpes simplex virus type 1 glycoprotein e is required for axonal localization of capsid, tegument, and membrane glycoproteins. J. Virol. 79:13362–13372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang Z. H., Gershon M. D., Lungu O., Zhu Z., Gershon A. A. 2000. Trafficking of varicella-zoster virus glycoprotein gI: T(338)-dependent retention in the trans-Golgi network, secretion, and mannose 6-phosphate-inhibitable uptake of the ectodomain. J. Virol. 74:6600–6613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang Z. H., et al. 2001. Essential role played by the C-terminal domain of glycoprotein I in envelopment of varicella-zoster virus in the trans-Golgi network: interactions of glycoproteins with tegument. J. Virol. 75:323–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Weller T. H. 1996. Varicella: historical perspective and clinical overview. J. Infect. Dis. 174(Suppl. 3):S306–S309 [DOI] [PubMed] [Google Scholar]
- 56. Yao Z., Grose C. 1994. Unusual phosphorylation sequence in the gpIV (gI) component of the varicella-zoster virus gpI-gpIV glycoprotein complex (VZV gE-gI complex). J. Virol. 68:4204–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yao Z., Jackson W., Forghani B., Grose C. 1993. Varicella-zoster virus glycoprotein gpI/gpIV receptor: expression, complex formation, and antigenicity within the vaccinia virus-T7 RNA polymerase transfection system. J. Virol. 67:305–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yao Z., Jackson W., Grose C. 1993. Identification of the phosphorylation sequence in the cytoplasmic tail of the varicella-zoster virus Fc receptor glycoprotein gpI. J. Virol. 67:4464–4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ye M., Duus K. M., Peng J., Price D. H., Grose C. 1999. Varicella-zoster virus Fc receptor component gI is phosphorylated on its endodomain by a cyclin-dependent kinase. J. Virol. 73:1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zerboni L., et al. 2011. Varicella-zoster virus glycoprotein E is a critical determinant of virulence in the SCID mouse-human model of neuropathogenesis. J. Virol. 85:98–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zerboni L., Ku C. C., Jones C. D., Zehnder J. L., Arvin A. M. 2005. Varicella-zoster virus infection of human dorsal root ganglia in vivo. Proc. Natl. Acad. Sci. U. S. A. 102:6490–6495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zerboni L., et al. 2007. Aberrant infection and persistence of varicella-zoster virus in human dorsal root ganglia in vivo in the absence of glycoprotein I. Proc. Natl. Acad. Sci. U. S. A. 104:14086–14091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zuckermann F. A., Mettenleiter T. C., Schreurs C., Sugg N., Ben-Porat T. 1988. Complex between glycoproteins gI and gp63 of pseudorabies virus: its effect on virus replication. J. Virol. 62:4622–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]