Abstract
Foamy viruses (FVs) synthesize the Pol precursor protein from a specific transcript. Thus, in contrast to what was found for orthoretroviruses, e.g., human immunodeficiency virus, no Gag-Pol precursor protein is synthesized. Foamy viral Pol consists of a protease (PR) domain, a reverse transcriptase domain, and an integrase domain and is processed into a mature protease-reverse transcriptase (PR-RT) fusion protein and the integrase. Protease activity has to be strictly regulated in order to avoid premature Gag and Pol processing before virus assembly. We have demonstrated recently that FV protease is an inactive monomer with a very weak dimerization tendency and postulated protease activation through dimerization. Here, we identify a specific protease-activating RNA motif (PARM) located in the pol region of viral RNA which stimulates PR activity in vitro and in vivo, revealing a novel and unique mechanism of retroviral protease activation. This mechanism is strikingly different to that of orthoretroviruses, where the protease can be activated even in the absence of viral RNA during the assembly of virus-like particles. Although it has been shown that the integrase domain is important for Pol uptake, activation of the foamy virus protease is integrase independent. We show that at least two foamy virus PR-RT molecules bind to the PARM and only RNAs containing the PARM result in significant activation of the protease. DNA harboring the PARM is not capable of protease activation. Structure determination of the PARM by selective 2′ hydroxyl acylation analyzed by primer extension (SHAPE) revealed a distinct RNA folding, important for protease activation and thus virus maturation.
INTRODUCTION
The replication of spumaretroviruses or foamy viruses (FVs) differs in several aspects from that of orthoretroviruses (for reviews, see references 20, 31, and 32). One important deviation from the replication strategy concerns the mode of pol expression. In orthoretroviruses, in addition to the Gag polypeptide a Gag-Pol fusion protein is synthesized at a ratio of approximately 1 Gag-Pol molecule to 20 Gag molecules. Both polypeptides are taken up into the virus particle by interaction of the Gag region with the viral RNA (reviewed in reference 7). In contrast, FV Pol is synthesized from a specific mRNA independently from Gag, leading to a discrete Pol precursor protein (3, 5, 16, 21, 22, 39). Consequently, the uptake mechanism for FV Pol must be different.
It has been suggested that the C terminus of FV Gag as well as parts of the FV genome contain determinants important for Pol encapsidation (15, 18, 28, 33). Once assembly of the viral components has taken place, the viral protease (PR) processes the Gag and Pol precursor proteins into mature structural and functional proteins. In FVs, only the viral integrase (IN) is cleaved off from Pol, whereas the PR remains covalently linked to the reverse transcriptase (RT) domain, thus leading to a mature protease-reverse transcriptase (PR-RT) protein (30). Retroviral PRs are active only as homodimers (26). In contrast to this, it has been shown previously that prototype FV (PFV) and macaque simian foamy virus (SFVmac) PR-RT and the SFVmac PR domain are monomeric (2, 11, 13). The free 10-kDa PR, lacking the RT domain, forms only transient dimers, which constitute a very small fraction (<5%) of PR molecules in solution (12).
Consequently, since they are predominantly monomeric proteins, FV PR and PR-RT lack proteolytic activity under physiologically relevant conditions in vitro (11). The regulatory mechanism for PR activation of retroviruses is only poorly understood. For the PR domain of human immunodeficiency virus type 1 (HIV-1), it was shown that transient dimerization events of two PR domains in the context of the Gag-Pol precursor protein lead to N-terminal autoprocessing and thus activation of the PR (35). However, in FVs separate Gag and Pol precursors are formed, with the PR domain being located at the N terminus of Pol. This indicates that FV PR activity needs to be regulated by means other than HIV-1 PR. So far, activation of FV PR in vitro either with the free 10-kDa PR domain or in the context of PR-RT could be achieved only at very high NaCl concentrations of 2 to 3 M (11–13). It is obvious that these conditions do not reflect the physiological situation in living cells but somehow create an environment that facilitates dimerization, probably by hydrophobic interaction of two monomers.
To identify factors important for PR stimulation in vivo, we postulated that RNA regions which are essential for virus replication and/or Pol uptake might also be important to stimulate PR dimerization and thus activation. In previous studies, two cis-acting sequences in the FV genomic RNA, CasI and CasII, were shown to be essential and sufficient for the transfer of FV vectors (6, 14, 15, 38), indicating a relevant role in virus assembly. CasI is located in the 5′ leader region and the 5′ gag region of the pregenomic RNA of PFV spanning nucleotides (nt) 1 to 645. Cas II is situated in the 3′ region of the pol gene spanning nt 3869 to 5884 (Fig. 1A).
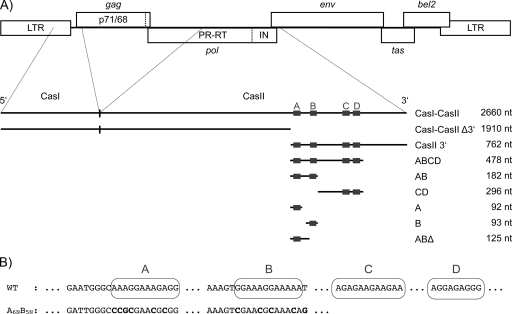
Fig. 1.
Localization and DNA sequences of the PFV purine-rich sequences used in this study. (A) Gene map of the PFV provirus (LTR, long terminal repeat; the open reading frames of the viral genes are shown by open boxes). The localization of cis-acting sequences CasI and CasII is indicated. CasI is localized in the 5′ and gag regions of the viral RNA (nt 1 to 645), and CasII is located in the 3′ region of the pol gene. The various RNAs used are displayed below the map. The relative positions of the purine-rich elements A to D and the lengths of the different RNAs used are indicated. (B) DNA sequences of the PFV purine-rich elements A, B, C, and D. WT, wild type.
It has been demonstrated that FVs harbor a second, so-called central polypurine tract (cPPT) in addition to the 3′ PPT (17, 27, 28, 36). FVs share this feature of two PPTs with lentiviruses, although they are members of different subfamilies. The 3′ PPT is located upstream of the 3′ long terminal repeat (LTR) and is required for plus-strand DNA synthesis. The cPPT is located in the pol open reading frame of the pregenomic viral RNA and thus is part of the CasII sequence. Within this region, FVs contain four motifs of purine-rich sequences (elements A, B, C, and D) (Fig. 1B) (27). The A and B elements appear to play a role in Pol protein encapsidation, whereas the C element is required for regulation of gene expression. Only the sequence of the D element is 100% identical to that of the 3′ PPT and is likely to constitute the actual cPPT. However, definite functions for either of the purine-rich elements, in particular of the A and B sequences, have not been identified so far (27).
To determine whether these cis-acting sequences play a role in FV PR activation, we expressed in vitro-transcribed RNAs containing CasI and CasII as well as several 5′- and 3′-truncated RNA versions thereof and tested the influence of these RNAs on the PR activity of PFV and SFVmac. We were able to identify a protease-activating RNA motif (PARM) which embraces a sequence that starts 5′ of the A element and ends 3′ of the B element of the viral cPPT region. Furthermore, although recent data from Lee et al. (19) suggest a contribution of the FV IN to PR activation, our results demonstrate clearly that PR activation occurs in the absence of IN.
MATERIALS AND METHODS
Protein purification.
SFVmac and PFV PR-RTs and the GB1 (immunoglobulin binding domain B1 of streptococcal protein G)-green fluorescent protein (GFP) PR substrate were purified as described previously (10, 11, 13).
RNA synthesis.
Synthesis of all RNAs used in this study was done with a T7 or T3 MEGAscript kit (Applied Biosystems, Austin, TX). To obtain 32P-labeled RNA, 0.74 MBq of α[32P]UTP (Hartmann Analytic, GmbH, Braunschweig, Germany) was included in the in vitro transcription assay. All RNAs were purified over MicroSpin columns (GE Healthcare, Munich, Germany). The integrity of the RNAs was analyzed by denaturing gel electrophoresis (5 to 10% polyacrylamide, 7 M urea). RNA folding was performed in the corresponding buffers used for the experiments by incubation for 2 min at 95°C, followed by 10 min at 65°C and slow cooling to 30°C within 45 to 60 min directly before use.
The investigated RNA fragments of PFV CasI-CasII (positions 1 to 2660) span the regions comprising positions 1 to 1910 (CasI-CasII Δ3′), 1898 to 2660 (CasII 3′), 1898 to 2376 (ABCD), 1898 to 2080 (AB), 2080 to 2376 (CD), 1898 to 1990 (A), 1987 to 2080 (B), and 1898 to 2023 (ABΔ) of plasmid pMD9 (14). The numbers of the RNAs refer to the transcriptional start site of the CasI-CasII RNA.
FV PR activity assays.
In vitro PR activity assays were essentially performed as described previously (13). In brief, 10 μM GB1-GFP substrate, harboring the SFVmac RT-IN cleavage site of the Pol polyprotein (ATQGSYVVH↓CNTTP) between GB1 and GFP, was incubated with 2.5 μM PR-RT or SFVmac PRshort and 0.5 μM DNA or RNA as indicated for 2 h at 37°C in 50 mM Na2HPO4-NaH2PO4 (pH 6.4), 100 mM NaCl, in a total volume of 20 μl. The cleavage products were separated by electrophoresis on 10% BisTris gels (Invitrogen, Karlsruhe, Germany) in 50 mM MES (morpholineethanesulfonic acid) buffer (pH 7.3), 50 mM Tris base, 0.1% SDS, 1 mM EDTA.
For in vivo FV PR assays, 5 × 105 HEK 293T cells were cotransfected with codon-optimized versions of PFV Gag (3 μg of pcoPG4) (34) and PFV Pol (0.1 μg of pcoPP) and 2 μg of the wild-type pMD9 vector genome (14) or the A6NB5N mutated form thereof (Fig. 1B) (27). To generate the codon-optimized version of the pol expression plasmid, a strategy analogous to that for the generation of the gag expression plasmid was applied (34). Gene synthesis and optimization of expression were done by Geneart (Regensburg, Germany). Plasmid pUC19 was used in control assays. Two days after transfection, cells were lysed and analyzed by Western blotting using monoclonal antibodies against Gag and IN (28) or a polyclonal antiserum against the cellular enzyme glycerol aldehyde phosphate dehydrogenase (GAPDH) (Sigma-Aldrich Chemie, GmbH, Taufkirchen, Germany).
HIV-1 PR activity assays.
HEK 293T cells (5 × 105) were transfected with either 2 μg of either pCMVsynGag or pCMVSynGagPol (8), which harbors a codon-optimized version of the HIV-1 gag or gag-pol gene, respectively, but lacks the uptake signal on the transcribed RNA. As a control, cells were transfected with the proviral HIV-1 clone pNL4-3 (1). Two days after transfection, supernatants were harvested and viruses (in the case of pNL4-3) or virus-like particles (VLPs) were concentrated by centrifugation at 40,000 rpm at 4°C using a TST-55-1 rotor. Concentrated particles and cellular lysates were analyzed by Western blotting using HIV-1 p21 capsid (CA)-specific antibodies or control antibodies specific for the cellular GAPDH.
Protein-protein cross-linking.
For cross-linking reactions, 2 μM PFV or SFVmac PR-RT was incubated with 0.1 mM bis-sulfosuccinimidyl suberate (BS3) (Sigma-Aldrich Chemie, GmbH, Taufkirchen, Germany) in the presence or absence of 0.5 μM RNA as indicated for 15 min at room temperature in 50 mM Na2HPO4-NaH2PO4 (pH 6.4), 100 mM NaCl in a total volume of 5 μl. Control reactions were performed without BS3. The reactions were stopped by the addition of an equal volume of 1 M Tris (pH 9). To hydrolyze the RNA, the mixture was then incubated for 15 min at 45°C. Reaction products were separated by electrophoresis on 10% BisTris gels (Invitrogen, Karlsruhe, Germany) in 50 mM MES buffer (pH 7.3), 50 mM Tris base, 0.1% SDS, 1 mM EDTA. Protein bands were photographed and quantitated by densitometry using the software program AIDA (version 4.15; raytest, Straubenhardt, Germany).
EMSAs.
For electrophoretic mobility shift assays (EMSAs), 0.5 μM sense or antisense [32P]-labeled AB RNA was mixed with 0 to 2.0 μM PFV or SFVmac PR-RT as indicated in 50 mM Na2HPO4-NaH2PO4 (pH 6.4), 100 mM NaCl, and 10% glycerol in a total volume of 10 μl. For displacement of the labeled AB RNA, unlabeled sense or antisense AB RNA was added as indicated. The mixtures were incubated at room temperature for 5 min. Half of the mixture was loaded on a 6% DNA retardation gel (Invitrogen, Karlsruhe, Germany). Electrophoresis was carried out in 0.5× TBE buffer (45 mM Tris-HCl [pH 8.0], 45 mM borate, and 10 mM EDTA) at 100 V for 3 h at 4°C. Gels were dried for 2 h at 80°C. RNA bands were visualized using a phosphorimaging device (FLA 3000; Raytest, Straubenhardt, Germany) and quantitated by densitometry using the software program AIDA.
5′-end labeling of the 3′ AB primer.
One hundred picomoles of 3′ AB primer (5′-GGTCTTCCTACTAGCAGTTTAGTTAAAAGTCGTTTTATATC) (IBA, Göttingen, Germany) was labeled with 3.7 MBq γ[32P]ATP (Hartmann Analytic, GmbH, Braunschweig, Germany) and 4 U T4 polynucleotide kinase (New England BioLabs, Frankfurt, Germany) in a total volume of 20 μl at 37°C for 1 h, followed by inactivation of the kinase for 20 min at 65°C. The labeled primer was finally purified from unincorporated nucleotides via a MicroSpin column (GE Healthcare, Munich, Germany).
SHAPE.
Selective 2′ hydroxyl acylation analyzed by primer extension (SHAPE) (24, 25, 37) was used to determine the secondary structure of AB RNA. In principle, the protocol developed by Wilkinson et al. (37) was used. A total of 2 pmol AB RNA in 12 μl 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA was heated to 95°C for 2 min and then put on ice for 2 min. Folding of the RNA was completed by the addition of 6 μl RNA folding mix (333 mM HEPES [pH 8.0], 20 mM MgCl2, and 333 mM NaCl) and incubation at 37°C for 20 min. Half of the mixture was removed and modified by adding 18 mM N-methylisatoic anhydride (NMIA) (Invitrogen, Karlsruhe, Germany) dissolved in 100% dimethyl sulfoxide (DMSO) and incubation for 45 min at 37°C. The other half served as a control and was treated with DMSO only. Both samples were ethanol precipitated, and the resulting pellet was dissolved in 10 μl 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA.
For primer annealing, ca. 100 nM 32P-labeled 3′ AB primer was incubated with RNA for 5 min at 65°C. The reaction mixture was then slowly cooled down to 35°C within 1 h. Reverse transcription was carried out in 50 mM Tris-HCl (pH 8.3), 50 mM KCl, 3 mM MgCl2, 5 mM dithiothreitol (DTT), and 500 μM each deoxynucleoside triphosphate (dNTP) for 10 min in a total volume of 20 μl. After preincubation of the mixture for 1 min at 52°C, the reaction was started by the addition of 200 U of Superscript III reverse transcriptase (Invitrogen, Karlsruhe, Germany) and further incubation at 52°C for 10 min. Subsequently, to degrade the RNA template, 1 μl of 4 M NaOH was added and the mixture was incubated for 5 min at 95°C. The reaction was stopped by an equal volume of 8 M urea in 1 M Tris-HCl (pH 8.0), 50 mM boric acid, and 50 mM EDTA, containing traces of bromophenol blue and xylene cyanol. Eight-microliter aliquots were analyzed by denaturing gel electrophoresis (10% polyacrylamide, 7 M urea). Gels were submerged in 10% acetic acid for 5 min and then dried for 2 h at 80°C. The reaction products were visualized and quantitated by densitometry using a phosphorimaging device (FLA 3000; Raytest, Straubenhardt, Germany) and the software program AIDA.
For calculation of relative SHAPE intensities, the difference between the integrated band intensities of the reactions with and without NMIA was divided by the highest measured value in the experiment. Relative SHAPE intensities higher than 0.35 were considered significant. The RNA secondary structure was predicted using RNAfold (9, 23). Nucleotides showing significant SHAPE intensities (for the exception, see Results) were considered unpaired.
Sequencing.
To assign the SHAPE reaction products, sequencing reactions were run in parallel using the same 32P-end-labeled 3′ AB primer and a vector harboring the CasI-CasII DNA sequences as a template. Sequencing reactions were performed with the Sequenase 7-deaza-dGTP sequencing kit (USB, Cleveland, OH) using the 32P-end-labeled 3′ AB primer.
RESULTS
Identification of a PR activation RNA motif.
To analyze the impact of viral RNA on PR activity, we constructed different RNAs containing either the complete CasI and CasII sequences or shorter versions thereof as well as mutant RNAs (Fig. 1). PR activity in vitro was observed using a model substrate and recombinant monomeric PR-RTs of PFV and of SFVmac purified from Escherichia coli (10, 11). The model substrate GB1-GFP contains the natural RT-IN PR cleavage site in the Pol precursor between the GB1 (immunoglobulin binding domain B1 of streptococcal protein G) and GFP domains (13). This allows the GB1-GFP substrate as well as the cleavage products thereof, GB1 and GFP, to be visualized by SDS-polyacrylamide gel electrophoresis (PAGE). We have shown previously that in the absence of other factors, PR could be activated only at high NaCl concentrations of 2 to 3 M (11, 13). Here, we decreased the NaCl concentration to 100 mM to achieve conditions that are biologically more relevant. Analysis of the cleavage products by SDS-PAGE obtained with PFV PR-RT revealed that indeed PFV CasI-CasII RNA could stimulate PR activity (Fig. 2A).
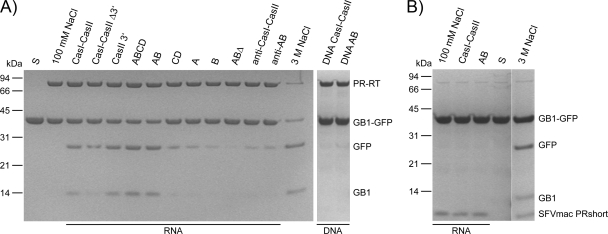
Fig. 2.
PR activity assays with PFV PR-RT (A) and SFVmac PRshort (B) in the presence of different nucleic acids. GB1-GFP substrate (10 μM) was incubated with 2.5 μM PR-RT and 0.5 μM RNA or DNA as indicated at 37°C for 2 h in 50 mM Na2HPO4-NaH2PO4 (pH 6.4), 100 mM NaCl. Reaction products were separated on 10% BisTris gels. The sizes and positions of the standard proteins are indicated on the left. S, unprocessed substrate.
To identify the RNA sequence(s) sufficient for PR stimulation, we created two shorter PFV RNA fragments, CasI-CasIIΔ3′, lacking the cPPT region, and CasII-3′, harboring the cPPT and the adjacent purine-rich elements (Fig. 1B). Since the stimulatory effect was much more pronounced with the use of CasII-3′ (Fig. 2B), we produced additional deletions in CasII-3′ to determine the minimal region necessary for PR stimulation. The in vitro-transcribed RNAs embraced sequences of the central purine-rich A-to-D elements of PFV (ABCD, AB, CD, A, B, and ABΔ, which contains a deletion of the 3′ region of the B element). In addition, antisense RNAs of CasI-CasII (anti-CasI-CasII) and of AB (anti-AB) were tested as controls. Figure 2A shows that in contrast to what was found for CD RNA, the cleavage activity of PFV PR-RT in the presence of AB RNA is comparable with that of ABCD or CasII 3′, whereas neither A nor B alone could stimulate PR in a similar manner. Additionally, truncation of AB at the 3′ end (ABΔ) by 57 nt as well as anti-CasI-CasII and anti-AB RNA resulted only in weak PR stimulation. Obviously, RNA in general has a weak stimulatory effect on PR activity; however, we could not observe any PR activation when the corresponding DNAs of CasI-CasII or AB were used (Fig. 2A).
Addition of 6 mM MgCl2 to the reaction mixture or refolding of the RNAs after in vitro transcription by heating and slow cooling did not change the results (data not shown). Moreover, the PR domain expressed and purified separately, i.e., lacking the RT domain, could not be stimulated by CasI-CasII or AB RNA (Fig. 2B), indicating strongly that the RT domain is required for RNA binding and thus dimerization of the PR domain.
We used these RNAs to investigate the PR activities of both PFV (Fig. 2) and SFVmac PR-RT (see Fig. S1 in the supplemental material) and obtained comparable results. PFV AB RNA could stimulate SFVmac PR-RT to a similar extent, although the sequence of the A element differs by two bases (Fig. S1A). We compared PFV and SFVmac PR-RTs for all the following in vitro experiments but never saw any significant differences.
Oligomerization of PR-RT upon RNA binding.
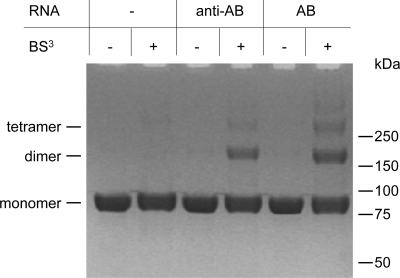
As dimerization of the PR domain is a prerequisite for PR activity, we analyzed whether PR-RT dimerizes by specifically binding to AB RNA or whether dimerization can also be achieved by binding to nonspecific anti-AB RNA. Therefore, PFV PR-RT was cross-linked with the cross-linking reagent bis-sulfosuccinimidyl suberate (BS3) in the presence or absence of AB RNA or anti-AB RNA, respectively. To exclude cross-linking of PR-RT to RNA, RNA hydrolysis under alkaline conditions was performed before the samples were analyzed by SDS-PAGE (Fig. 3). Clearly, dimers and, to a much lower extent, tetramers of PFV PR-RT could be detected in the presence of RNA. No dimers or multimers are visible in the control assays lacking RNA. Quantification of the cross-linking products showed that dimerization using AB RNA is more pronounced (34% cross-linked protein) than that using anti-AB RNA (24%), indicating a preference for AB RNA. The protein cross-links prove that FV PR-RTs dimerize or oligomerize when binding to RNA. Furthermore, with reference to Fig. 2, these data indicate that although anti-AB RNA can be bound by PR-RT, this does not necessarily lead to substrate cleavage. Obviously, binding of the correct RNA by the RT domain is required for PR activity.
Fig. 3.
Cross-linking reactions were performed with 2 μM PFV PR-RT and 0.1 mM BS3 in the presence or absence of 0.5 μM anti-AB or AB RNA as indicated for 15 min at room temperature in 50 mM Na2HPO4-NaH2PO4 (pH 6.4), 100 mM NaCl. After RNA hydrolysis, reaction products were separated on 10% BisTris gels. The proposed oligomerization state is indicated on the left, and the sizes and positions of the standard proteins are displayed at the right.
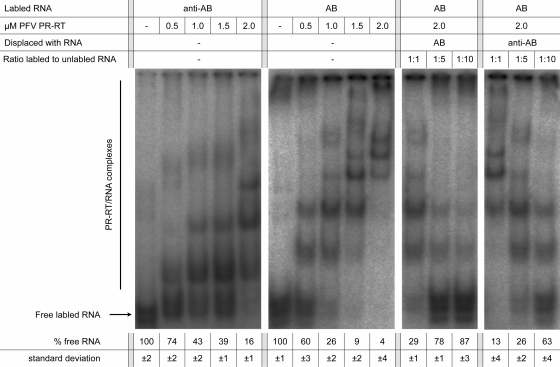
To further analyze RNA binding specificities, we performed electrophoretic mobility shift assays (EMSAs) with PFV PR-RT and AB RNA as well as anti-AB RNA (Fig. 4). Not surprisingly, since RTs are nucleic acid binding proteins, multiple shift bands indicated that dimers and higher multimers were formed with both RNAs. As the RNAs used are rather long (182 nt), this might be due to multiple binding sites with different binding affinities. However, quantification shows again that the affinity of PFV PR-RT to AB RNA is higher. This can be seen for all protein concentrations used. At the highest PR-RT concentration (2 μM), about 16% of unbound anti-AB RNA is still present whereas only 4% of AB RNA is not shifted. Displacement of bound AB RNA with either unlabeled AB RNA or anti-AB RNA confirms that anti-AB RNA is less effective in displacing bound AB RNA (Fig. 4). Even though we were not able to determine dissociation constants for the two RNA species, due to multiple shifted bands, these data indicate that FV PR-RT preferentially binds AB RNA.
Fig. 4.
Electrophoretic mobility shift assay. A radioactively labeled anti-AB or AB RNA (500 nM) was incubated with increasing concentrations of PFV PR-RT in 50 mM Na2HPO4-NaH2PO4 (pH 6.4), 100 mM NaCl, 10% glycerol at room temperature for 5 min. For displacement, increasing concentrations of unlabeled anti-AB or AB RNA were added to a mixture containing 500 nM radioactively labeled AB RNA and 2 μM PFV PR-RT as indicated above the gels. Complexes were separated on 6% DNA retardation gels at 4°C. Numbers below the gels show the remaining free RNA in percentages (including the standard deviations), as quantitated by densitometry.
Determination of the structure of the PR-activating RNA motif by SHAPE.
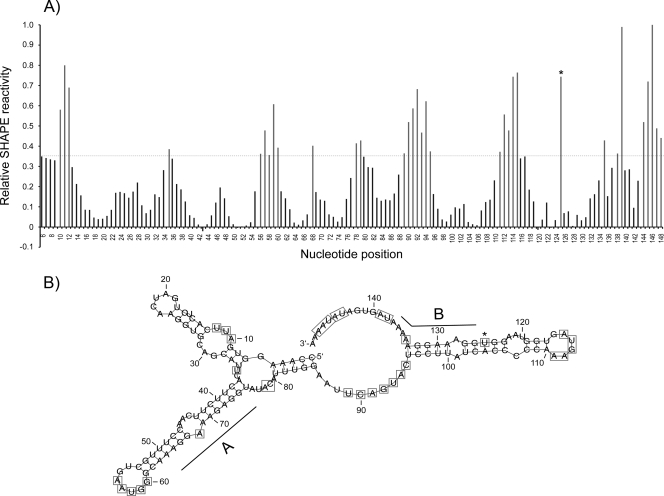
We assumed that the structure of the RNA might play an important role for PR-RT binding and thus PR activation. Secondary-structure prediction of the PFV cPPT AB RNA by use of the program RNAfold resulted in a molecule with several hairpin loop structures (see Fig. S2 in the supplemental material). To determine the actual structure of the PFV AB RNA, we performed selective 2′ hydroxyl acylation analyzed by primer extension (SHAPE) (24, 25, 37). N-Methylisatoic anhydride (NMIA) preferentially reacts with conformationally flexible nucleotides, e.g., nucleotides that are not base paired, at the 2′ OH of the ribose. Sequencing reactions of RNA treated with NMIA will stop at positions modified with NMIA. Thus, nucleotides showing significantly higher band intensities in sequencing reactions after treatment with NMIA correspond to single-stranded RNA regions.
We indentified several regions in the AB RNA sequence, which were unpaired (Fig. 5A). Calculations of the RNA secondary structure including constraints for the unpaired regions (Fig. 5B) confirmed the theoretically predicted RNA secondary structure (see Fig. S2 in the supplemental material) to a great extent. The RNA consists of three hairpin loop structures and a central as well as a 3′ unpaired region (nt 86 to 95 and 134 to 150). The A and B elements are each located in a stem-loop structure of approximately 15 nt in length. The 3′ side of the stem-loop structures almost exclusively consists of purines (Fig. 5B). Furthermore, the secondary structure analysis excludes the possibility that interactions of the unpaired RNA loops occur.
Fig. 5.
SHAPE analysis of AB RNA. (A) Relative SHAPE intensities as a function of nucleotide position. (B) RNA secondary-structure model of AB RNA using SHAPE constraints. Bases with relative SHAPE intensities higher than 0.35 are highlighted by boxes. Base 125 was not constrained (*; see Results).
The nucleotide at position 125 showed an unusual behavior. Only NMIA concentrations higher than 14 mM resulted in a strong signal, implying an unpaired nucleotide, whereas lower NMIA concentrations did not show this effect. Furthermore, setting nucleotide 125 unpaired in secondary structure calculations led to several unpaired nucleotides between positions 96 and 133, which were predicted to be paired. Thus, nt 125 was considered to be paired in our secondary structure calculation.
Since the predicted and the actual secondary structures of the AB element are almost identical, we only performed secondary-structure predictions by the program RNAfold to determine the structures of the following RNAs.
Structure and effect of mutant AB RNAs in vitro.
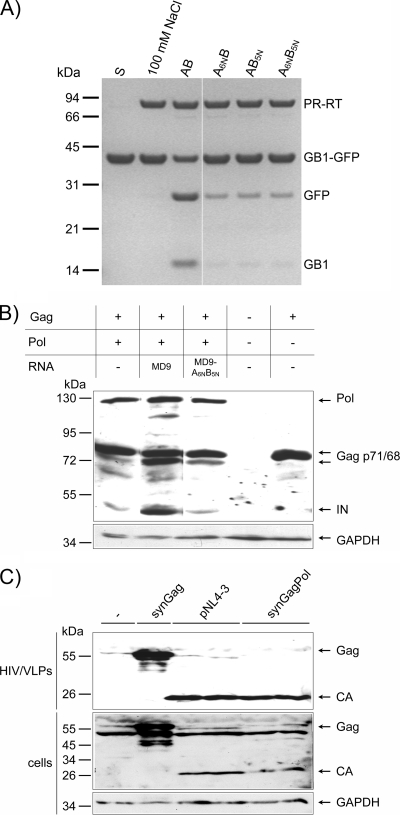
To investigate the influence of the AB RNA structure on PR activation, we exchanged several nucleotides within the A (6 nt; A6N) and B (5 nt; B5N) elements (Fig. 1B). These mutations were used by Peters et al. and restricted the incorporation of appreciable amounts of Pol protein into viral particles (27). We chose these multiple-nucleotide exchanges because secondary-structure predictions indicated that the original folding of the A or B element was destroyed, leading to significantly different stem-loop structures of A or B, respectively (see Fig. S2 in the supplemental material). Mutations with fewer nucleotide exchanges (27) did not result in complete structural disintegration in the secondary-structure predictions and thus might lead to ambiguous interpretation of the results. The importance of the integrity of the A and B elements is clearly demonstrated in the PR activity assay (Fig. 6A), where none of the mutated RNAs, A6NB, AB5N, and A6NB5N stimulated PR activity significantly, indicating that PR-RT recognizes the AB RNA structure.
Fig. 6.
PR activities of PFV PR-RT and of HIV-1. (A) In vitro PR activity of PFV PR-RT in the presence of wild-type and mutant AB RNAs. GB1-GFP substrate (10 μM) was incubated with 2.5 μM PFV PR-RT and 0.5 μM wild-type or mutant AB RNAs as indicated at 37°C for 2 h in 50 mM Na2HPO4-NaH2PO4 (pH 6.4), 100 mM NaCl. Reaction products were separated on 10% BisTris gels. The sizes and positions of the standard proteins are indicated on the left. S, uncleaved substrate. (B) In vivo PR activity of PFV PR-RT in the presence of wild-type and mutant AB RNAs. Western blot analysis of a cotransfection in HEK 293T cells of minimal amounts of codon adapted PFV Pol and Gag expression constructs and either a wild-type vector genome (pMD9) or an A6NB5N mutant thereof. Antisera against Gag and IN were used for Gag and Pol detection. (C) In vivo HIV-1 PR activity is independent of genomic RNA. Results are shown for Western blotting of concentrated HIV-1 VLPs/viruses and HEK 293T cells transfected with plasmids expressing synGag or synGagPol as indicated on top or with the proviral HIV-1 clone (pNL4-3). GAPDH amounts were visualized as a loading control.
Effects of wild-type and (A6NB5N) mutated RNAs in vivo.
To analyze the effects of the described PR activation motif in vivo, we cotransfected HEK 293T cells with codon-optimized PFV Pol (pcoPP) and Gag (pcoPG4) expression constructs (34) in order to monitor Gag processing as a readout for PR activity in the presence and absence of vector genomes harboring either wild-type CasI-CasII or mutated CasI-CasII(A6NB5N) sequences (Fig. 6B). Only minimal amounts of the pol-expressing plasmid were transfected to avoid pol overexpression and thus precursor processing in the cells before virus budding has occurred. Furthermore, a codon-optimized version of pol was used to exclude any influence on PR activation by the pol expression construct. The codon-optimized pol construct is not conserved in the region in question on the nucleotide level, i.e., it does not harbor the correct AB sequence and structure.
The activity of the PR domain was visualized in a Western blot using monoclonal antibodies against Gag and IN to identify Gag and Pol processing, respectively. Figure 6B shows that in the presence of the wild-type AB elements, Gag and Pol processing occurs and can be detected by partial processing of the 71-kDa Gag precursor into the 68-kDa product and by the presence of the cleaved-off IN. These results confirm that proteolytic activity of Pol depends on the presence of the wild-type AB elements, whereas neither cellular RNAs, which are ubiquitously present in the cell, nor the transfected mutated CasI-CasII(A6NB5N) sequences exhibited a significant stimulatory effect on PR activity in vivo.
These data are in strong contrast to those for HIV-1, where virus-like particles (VLPs) harboring processed Gag-Pol can be formed in the complete absence of genomic viral RNA. To demonstrate this difference, VLPs were isolated from cells expressing codon usage-adapted HIV-1 gag and gag-pol genes, which lack the uptake signal for the viral RNA. The purified VLPs were subjected to Western blot analysis using an HIV-1 capsid (CA)-specific antibody (Fig. 6C). Detection of the free CA protein proves that RNA-free HIV-1 VLPs are capable of processing the Gag precursor protein.
DISCUSSION
Orthoretroviral PRs investigated so far are present in the virion as rather stable dimers with catalytic activity (4, 26). Recently, a study showed that the HIV-1 PR domain can form transient dimers in the Gag-Pol precursor protein (35). Thus, premature activation of PR can be prevented before virus assembly. Packaging of the Pol proteins in HIV-1 is mediated by RNA binding of Gag within the Gag-Pol polyprotein (reviewed in reference 7). As FVs express Gag and Pol independently from each other, Pol packaging and PR regulation have to be different. FV Pol packaging has been shown to be dependent on a cis-acting sequence (CasII) located at the 3′ end of the pol gene in the viral RNA (27).
Here, we identified an RNA sequence needed for FV PR activation by truncating the FV CasI-CasII RNA at the 5′ and 3′ ends (Fig. 1). This PR-activating RNA motif (PARM) spans the A and B regions of the central purine-rich sequences of the PFV (pre)genomic RNA. PARM enables FV PR-RT to form a proteolytically active dimer in vitro as well as in vivo (Fig. 2A and 6B). Our data indicate that important interactions between Pol and PARM occur during virus assembly and maturation as suggested by Peters et al. (27). Therefore, RNA binding of Pol is required not only for Pol encapsidation but for PR activation as well. This leads to a model where Gag and Pol processing is inhibited until virus assembly takes place, since Pol interaction with viral RNA is necessary for PR activation.
The fact that DNA could not stimulate PR (Fig. 2A) has far-reaching implications on the viral life cycle. FV PR cleaves the Gag p71 once, resulting in Gag p68 (Fig. 1A and 6B). It has been suggested that an additional cleavage occurs in the Gag protein at amino acid positions 311/312 of Gag upon virus disassembly shortly after the virus has infected the host cell (29). However, since reverse transcription occurs late in the FV replication cycle and particles contain a DNA genome (40), the FV PR will presumably be inactive in the target cell. Thus, this cleavage is probably not catalyzed by the viral PR.
EMSA analyses imply sequence-independent binding of PR-RT to RNA (Fig. 4). In the presence of AB RNA as well as anti-AB, binding of several PR-RT molecules could be detected. A preference for AB RNA could be shown by displacement analyses. However, due to multiple binding events, it was not possible to determine dissociation constants by this method. Cross-linking experiments also revealed that in the presence of RNA, dimers or tetramers are formed (Fig. 3). Nevertheless, only AB RNA induced strong PR stimulation, indicating that nonspecific PR-RT binding to nucleic acid is not sufficient for efficient PR activation. Rather, in order to induce the formation of a correct and active PR homodimer, at least two molecules of PR-RT need to bind to PARM. Only then, the correct spatial orientation and proximity of the two PR domains allows for PR activation. Additionally, IN was recently suggested to be involved in Pol uptake and in PR activation (18, 19). However, since we can detect PR activation in the absence of IN, our data prove that IN is not necessary for PR activation.
Secondary-structure analysis by SHAPE showed that the A and B elements are part of hairpin loop structures (Fig. 5). The loop and the 3′ sequence of the stem are formed almost exclusively by purines. Disrupting the fold of either the A or the B element by the introduction of several mutations resulted in RNAs unable to stimulate PR activation in vitro as well as in vivo (Fig. 6). While Fig. 2 shows that both A and B have to be present for PR activation, Fig. 6 illustrates the importance of their structural and/or sequential integrity. This is in good agreement with the findings of Peters et al., where mutations in the A or B element completely abolished Pol encapsidation in vivo (27).
In conclusion, our data indicate that PR-RT dimerizes upon RNA binding. However, only the distinctly folded viral AB RNA leads to the formation of a correctly folded and functional dimer with high catalytic PR activity.
We propose that in the host cell the PR domain of FVs is inactive within the Pol polyprotein due to inefficient dimerization (12). Packaging of Pol and activation of PR are achieved through binding of the RT domain to the PARM of the pregenomic viral RNA (15, 27). Binding of PR-RT to PARM is essential for the formation of proteolytically active dimers that catalyze cleavage of Pol and Gag, finally resulting in mature virus particles.
This mechanism of PR regulation and activation by RNA is novel and unique and sets FV PRs apart from all other known retroviral PRs. It explains the necessity of a PR-RT fusion protein, since PR activity is dependent on binding of the RT domain to RNA. Furthermore, identification of PARM may contribute to the construction of improved FV vectors for gene transfer.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft DFG (Wo630/7-3, Bo3006/2-1, and SFB 479), the Graduate School in the Elite Network of Bavaria “Lead Structures of Cell Functions,” and the University of Bayreuth.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 16 February 2011.
REFERENCES
- 1. Adachi A., et al. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Benzair A. B., Rhodes-Feuillette A., Emanoil-Ravicovitch R., Peries J. 1982. Reverse transcriptase from simian foamy virus serotype 1: purification and characterization. J. Virol. 44:720–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bodem J., et al. 1996. Characterization of the spliced pol transcript of feline foamy virus: the splice acceptor site of the pol transcript is located in gag of foamy viruses. J. Virol. 70:9024–9027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dunn B. M., Goodenow M. M., Gustchina A., Wlodawer A. 2002. Retroviral proteases. Genome Biol. 3:1465–6914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Enssle J., Jordan I., Mauer B., Rethwilm A. 1996. Foamy virus reverse transcriptase is expressed independently from the Gag protein. Proc. Natl. Acad. Sci. U. S. A. 93:4137–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Erlwein O., Bieniasz P. D., McClure M. O. 1998. Sequences in pol are required for transfer of human foamy virus-based vectors. J. Virol. 72:5510–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goff S. P. 2007. Retroviridae: The retroviruses and their replication, p. 1999–2069 In Knipe D. M., Howley P. M. (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 8. Graf M., et al. 2000. Concerted action of multiple cis-acting sequences is required for Rev dependence of late human immunodeficiency virus type 1 gene expression. J. Virol. 74:10822–10826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gruber A. R., Lorenz R., Bernhart S. H., Neubock R., Hofacker I. L. 2008. The Vienna RNA websuite. Nucleic Acids Res. 36:W70–W74 doi:10.1093/nar/gkn188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hartl M. J., et al. 2008. AZT resistance of simian foamy virus reverse transcriptase is based on the excision of AZTMP in the presence of ATP. Nucleic Acids Res. 36:1009–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hartl M. J., Mayr F., Rethwilm A., Wöhrl B. M. 2010. Biophysical and enzymatic properties of the simian and prototype foamy virus reverse transcriptases. Retrovirology 7:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hartl M. J., et al. 2010. Formation of transient dimers by a retroviral protease. Biochem. J. 427:197–203 [DOI] [PubMed] [Google Scholar]
- 13. Hartl M. J., Wöhrl B. M., Rösch P., Schweimer K. 2008. The solution structure of the simian foamy virus protease reveals a monomeric protein. J. Mol. Biol. 381:141–149 [DOI] [PubMed] [Google Scholar]
- 14. Heinkelein M., et al. 2002. Improved primate foamy virus vectors and packaging constructs. J. Virol. 76:3774–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heinkelein M., et al. 2002. Pregenomic RNA is required for efficient incorporation of pol polyprotein into foamy virus capsids. J. Virol. 76:10069–10073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jordan I., Enssle J., Guttler E., Mauer B., Rethwilm A. 1996. Expression of human foamy virus reverse transcriptase involves a spliced pol mRNA. Virology 224:314–319 [DOI] [PubMed] [Google Scholar]
- 17. Kupiec J. J., et al. 1988. Evidence for a gapped linear duplex DNA intermediate in the replicative cycle of human and simian spumaviruses. Nucleic Acids Res. 16:9557–9565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee E.-G., Linial M. L. 2008. The C terminus of foamy retrovirus Gag contains determinants for encapsidation of Pol protein into virions. J. Virol. 82:10803–10810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee E.-G., et al. 2011. Foamy retroviral integrase contains a Pol dimerization domain required for protease activation. J. Virol. 85:1655–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Linial M. 2007. Foamy viruses, p. 2245–2262 In Knipe D. M., Howley P. M. (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 21. Linial M. L. 1999. Foamy viruses are unconventional retroviruses. J. Virol. 73:1747–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Löchelt M., Flügel R. M. 1996. The human foamy virus pol gene is expressed as a Pro-Pol polyprotein and not as a Gag-Pol fusion protein. J. Virol. 70:1033–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mathews D. H., Sabina J., Zuker M., Turner D. H. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911–940 [DOI] [PubMed] [Google Scholar]
- 24. Merino E. J., Wilkinson K. A., Coughlan J. L., Weeks K. M. 2005. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 127:4223–4231 [DOI] [PubMed] [Google Scholar]
- 25. Mortimer S. A., Weeks K. M. 2007. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 129:4144–4145 [DOI] [PubMed] [Google Scholar]
- 26. Pearl L. H., Taylor W. R. 1987. A structural model for the retroviral proteases. Nature 329:351–354 [DOI] [PubMed] [Google Scholar]
- 27. Peters K., Barg N., Gärtner K., Rethwilm A. 2008. Complex effects of foamy virus central purine-rich regions on viral replication. Virology 33:51–60 [DOI] [PubMed] [Google Scholar]
- 28. Peters K., Wiktorowicz T., Heinkelein M., Rethwilm A. 2005. RNA and protein requirements for incorporation of the Pol protein into foamy virus particles. J. Virol. 79:7005–7013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pfrepper K. I., et al. 1999. Molecular characterization of proteolytic processing of the Gag proteins of human spumavirus. J. Virol. 73:7907–7911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pfrepper K. I., et al. 1998. Molecular characterization of proteolytic processing of the Pol proteins of human foamy virus reveals novel features of the viral protease. J. Virol. 72:7648–7652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rethwilm A. 2005. Foamy viruses, p. 1304–1321 In ter Meulen V., Mahy B. W. J. (ed.), Topley & Wilson's microbiology and microbial infections: virology, vol. 2, 10th ed Hodder Arnold, London, United Kingdom [Google Scholar]
- 32. Rethwilm A. 2007. Foamy virus vectors: an awaited alternative to gammaretro- and lentiviral vectors. Curr. Gene Ther. 7:261–271 [DOI] [PubMed] [Google Scholar]
- 33. Stenbak C. R., Linial M. L. 2004. Role of the C terminus of foamy virus Gag in RNA packaging and Pol expression. J. Virol. 78:9423–9430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stirnnagel K., et al. 2010. Analysis of prototype foamy virus particle-host cell interaction with autofluorescent retroviral particles. Retrovirology 7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tang C., Louis J. M., Aniana A., Suh J. Y., Clore G. M. 2008. Visualizing transient events in amino-terminal autoprocessing of HIV-1 protease. Nature 455:693–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tobaly-Tapiero J., et al. 1991. Further characterization of the gapped DNA intermediates of human spumavirus: evidence for a dual initiation of plus-strand DNA synthesis. J. Gen. Virol. 72:605–608 [DOI] [PubMed] [Google Scholar]
- 37. Wilkinson K. A., Merino E. J., Weeks K. M. 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1:1610–1616 [DOI] [PubMed] [Google Scholar]
- 38. Wu M., Chari S., Yanchis T., Mergia A. 1998. cis-Acting sequences required for simian foamy virus type 1 vectors. J. Virol. 72:3451–3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu S. F., Baldwin D. N., Gwynn S. R., Yendapalli S., Linial M. L. 1996. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science 271:1579–1582 [DOI] [PubMed] [Google Scholar]
- 40. Yu S. F., Sullivan M. D., Linial M. L. 1999. Evidence that the human foamy virus genome is DNA. J. Virol. 73:1565–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.