Abstract
Maraviroc (MVC) inhibits the entry of human immunodeficiency virus type 1 (HIV-1) by binding to and modifying the conformation of the CCR5 extracellular loops (ECLs). Resistance to MVC results from alterations in the HIV-1 gp120 envelope glycoproteins (Env) enabling recognition of the drug-bound conformation of CCR5. To better understand the mechanisms underlying MVC resistance, we characterized the virus-cell interactions of gp120 from in vitro-generated MVC-resistant HIV-1 (MVC-Res Env), comparing them with those of gp120 from the sensitive parental virus (MVC-Sens Env). In the absence of the drug, MVC-Res Env maintains a highly efficient interaction with CCR5, similar to that of MVC-Sens Env, and displays a relatively modest increase in dependence on the CCR5 N terminus. However, in the presence of the drug, MVC-Res Env interacts much less efficiently with CCR5 and becomes critically dependent on the CCR5 N terminus and on positively charged elements of the drug-modified CCR5 ECL1 and ECL2 regions (His88 and His181, respectively). Structural analysis suggests that the Val323 resistance mutation in the gp120 V3 loop alters the secondary structure of the V3 loop and the buried surface area of the V3 loop–CCR5 N terminus interface. This altered mechanism of gp120-CCR5 engagement dramatically attenuates the entry of HIV-1 into monocyte-derived macrophages (MDM), cell-cell fusion activity in MDM, and viral replication capacity in MDM. In addition to confirming that HIV-1 escapes MVC by becoming heavily dependent on the CCR5 N terminus, our results reveal novel interactions with the drug-modified ECLs that are critical for the utilization of CCR5 by MVC-Res Env and provide additional insights into virus-cell interactions that modulate macrophage tropism.
INTRODUCTION
The entry of human immunodeficiency virus type 1 (HIV-1) is initiated by the interaction between the gp120 glycoproteins of the HIV-1 envelope (Env) and CD4 expressed on the target cell surface (reviewed in references 20 and 70). CD4 binding occurs with high affinity and triggers a conformational change in gp120 that exposes the binding site for a cellular coreceptor, either CCR5 or CXCR4 (reviewed in reference 70). Current models of gp120 binding to coreceptor suggest that the crown of the gp120 V3 loop interacts principally with the second extracellular loop (ECL2) region of the coreceptor, while the gp120 bridging sheet, which is formed between the C1, C2, and C4 domains of gp120 after CD4 binding, and the stem of the V3 loop interacts with the N terminus of the coreceptor (5, 7, 17, 29). While the coreceptor N terminus and ECL2 region appear to be important for gp120-coreceptor binding, the ECL1 and ECL3 regions may also influence coreceptor function (8, 9, 16). The interaction of CD4-bound gp120 with the coreceptor induces additional conformational changes in gp120, which lead to a structural rearrangement in gp41 that enables fusion and virus entry (reviewed in reference 70).
Maraviroc (MVC) is an inhibitor of HIV-1 entry that binds to a hydrophobic pocket in the transmembrane helices of CCR5, altering the conformation of the CCR5 extracellular loops (ECLs) such that they are no longer recognized by gp120 (reviewed in references 22 and 70). Thus, MVC and other CCR5 antagonists, such as vicriviroc (VVC) and aplaviroc (APL), are allosteric inhibitors of HIV-1 entry (10, 44, 45, 59, 72). MVC has been approved for use as an HIV-1 antiretroviral therapy (ART) for treatment-experienced and ART-naïve adults who have no evidence of CXCR4-using virus in plasma (22). As with other antiretrovirals, treatment with CCR5 antagonists can result in drug resistance, leading to virological rebound. Although treatment failure can arise from the emergence of CXCR4-using HIV-1 strains that were present at very low levels prior to the initiation of therapy (27, 38, 75), genuine resistance to CCR5 antagonists results from adaptive alterations in gp120 enabling recognition of the drug-bound conformation of CCR5 (3, 36, 49, 51, 55, 56, 69, 71).
Since CCR5 antagonists are allosteric inhibitors of virus entry, resistance to these drugs is evidenced by plateaus in virus inhibition curves that do not reach 100% inhibition rather than by increases in the concentration of the drug necessary to achieve complete inhibition (76). The magnitude of the reduction in the plateau height can be quantified as the maximal percentage of inhibition (MPI), which can differ from subject to subject (47). For example, for subjects who fail MVC therapy as a result of genuine resistance, the MPI can be high (>80%), signifying a relatively inefficient ability of gp120 to utilize the drug-bound conformation of CCR5, or low (<20%), signifying highly efficient utilization of drug-bound CCR5. Although MPIs can be influenced by differences in the level of CCR5 expression on the target cell populations (51, 56, 71), the results from the MOTIVATE clinical trials of MVC showed that the MPIs of most MVC-resistant viruses in subjects failing therapy fall within the range of 80 to 95% by the PhenoSense assay (47, 52).
Previous studies of HIV-1 resistance to CCR5 antagonists have underscored the remarkable flexibility of the interaction between gp120 and different functional domains of CCR5 (reviewed in reference 20). While viruses sensitive to CCR5 antagonists rely on interactions with both the N terminus and the ECL2 region of CCR5, studies using CCR5 mutants have shown that viruses engineered to have resistance to CCR5 antagonists and those that arise in patients failing therapy have an increased reliance on the N-terminal region (3, 48, 49, 51, 71). Furthermore, resistant viruses that show a predominant interaction with the CCR5 N terminus may have broad cross-resistance to different CCR5 antagonists, whereas those that exhibit increased dependence on the CCR5 N terminus but retain dependence on the drug-modified ECLs have a narrower cross-resistance profile (71). Sequence analysis has revealed that alterations in the V3 region of gp120 are predominantly responsible for the ability of CCR5 antagonist-resistant viruses to use the drug-bound receptor, although one study identified sequence alterations in gp41 that resulted in resistance to AD101, a preclinical precursor to VVC (2). Considering the broad range of MPIs that can be achieved by different HIV-1 strains with CCR5 antagonists resistance, these studies, taken together, suggest that in most instances, resistance arises from HIV-1 becoming able to use CCR5 differently, but not always more efficiently.
The consequences of altered coreceptor engagement by CCR5 antagonist-resistant viruses for HIV-1 pathogenesis have yet to be determined. Interestingly, a recent study showed that a viral variant with resistance to APL, which displayed relatively inefficient usage of drug-bound CCR5, had altered tropism on primary CD4+ T cells with relative sparing of the central memory CD4+ T-cell population (51), providing evidence that continued use of CCR5 antagonists even after the development of resistance could potentially be beneficial for some subjects. In addition to CD4+ T cells, tissue macrophages are a significant HIV-1 reservoir (30, 31). Furthermore, macrophage tropism (M tropism) is an important pathophysiological phenotype of CCR5-using (R5) viruses (11, 26, 42) and is essential for HIV-1 neurotropism and for the development of HIV-1-associated neurocognitive impairment (12–14, 21, 23, 50). However, not all R5 viruses are M tropic (reviewed in reference 20). Previous studies have shown that M tropism is associated with particular R5 viruses that have enhanced Env-mediated fusion activity (50, 64, 68) and altered recognition of CCR5, characterized by increased dependence on the CCR5 ECLs and reduced dependence on the CCR5 N terminus (66). Thus, diminishing the M-tropic and fusogenic properties of R5 HIV-1 strains could potentially attenuate HIV-1 pathogenicity.
Only one previous study has characterized the mechanism of CCR5 engagement of an MVC-resistant strain of HIV-1 (71), which exhibited unusually efficient utilization of drug-bound CCR5 compared to most MVC-resistant viruses from patients failing therapy (47, 52). Here we characterized the mechanisms underlying the resistance of an MVC-resistant HIV-1 strain that displayed relatively inefficient utilization of the drug-bound conformation of CCR5, which is more typical of the majority of MVC-resistant viruses arising in patients (47, 52), with a view to better understanding of how MVC resistance may alter HIV-1 tropism. We demonstrate an increased dependence on the CCR5 N terminus and an altered recognition of the drug-modified ECLs that is associated with escape from MVC and that dramatically attenuate M tropism. Structural analysis suggests that the Val323 resistance mutation in the gp120 V3 loop alters the secondary structure of the V3 loop and the buried surface area of the V3 loop–CCR5 N terminus interface, which may facilitate escape from MVC. In addition to illustrating novel virus-cell interactions associated with MVC resistance, our results provide further evidence that continued use of MVC even after resistance develops could potentially be beneficial in certain cases (for example, those viruses that exhibit resistance profiles with relatively high MPIs in CCR5-expressing cell lines) by promoting virus attenuation.
MATERIALS AND METHODS
Cells.
293T cells, JC53 cells (53), TZM-bl cells (74) and NP2-CD4/CCR5 cells (60) were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% (vol/vol) fetal calf serum (FCS) and 100 μg of penicillin and streptomycin per ml. U87-CD4 cells (4) were cultured in DMEM supplemented with 15% (vol/vol) FCS, 100 μg of penicillin and streptomycin per ml, and 0.5 mg of G418 per ml. The dually inducible 293-Affinofile cell line (32), in which expression of CD4 and CCR5 can be induced and regulated by the addition of minocycline or ponasterone A (ponA), respectively, was maintained in DMEM supplemented with 10% (vol/vol) FCS, 100 μg of penicillin and streptomycin per ml, 50 μg blasticidin per ml, and 200 μg G418 per ml. Peripheral blood mononuclear cells (PBMC) were purified from the blood of healthy HIV-1-negative donors by density gradient centrifugation. Monocytes were purified from PBMC by plastic adherence and were allowed to differentiate into monocyte-derived macrophages (MDM) by culturing for 5 days in Iscove modified Eagle medium (IMEM) supplemented with 10% (vol/vol) pooled AB+ human sera, 100 μg of penicillin and streptomycin per ml, and 12.5 ng of macrophage colony-stimulating factor per ml.
Env cloning and sequencing.
Infectious recombinant viruses containing the env gene of HIV-1 that had acquired resistance to MVC by serial passage in PBMC through increasing concentrations of MVC (termed MVC-Res), or containing the env gene from the parental HIV-1 strain similarly passaged without the drug (termed MVC-Sens), have been described previously (76). To characterize the the env genes from these recombinant viruses in isolation, the 2.1-kb KpnI-to-BamHI env fragment, corresponding to nucleotide positions 6348 to 8478 in HXB2, was amplified from the viral supernatants using reverse transcription-PCR and was cloned into the pSVIII-Env expression plasmid (18), as described previously (24, 25, 64). The MVC-Res and MVC-Sens Env clones were shown to be functional and to be able to support HIV-1 entry into JC53 cells when the Env clones were pseudotyped onto an Env-deficient luciferase reporter virus (data not shown). Envs were sequenced by BigDye Terminator sequencing and were analyzed using a model 3130 Genetic Analyzer (Applied Biosystems).
Production and quantitation of Env-pseudotyped luciferase reporter viruses.
Env-pseudotyped luciferase reporter viruses were produced by transfection of 293T cells with plasmids pCMVΔP1ΔenvpA, pHIV-1Luc, and pSVIII-Env or pcDNA3-Env using Lipofectamine 2000 (Invitrogen) at a ratio of 1:3:1, as described previously (24, 64, 79). Supernatants were harvested 48 h later, filtered through 0.45-μm-pore-size filters, and stored at −80°C. The 50% tissue culture infective doses (TCID50) of virus stocks were determined by titration in JC53 cells (53), as described previously (25, 64).
Production and quantitation of full-length, replication-competent NL4-3 HIV-1 variants carrying different env genes.
Full-length chimeric proviral HIV-1 plasmids were constructed by replacing the XhoI-to-EcoRI fragment of pNL4-3 (corresponding to amino acids 5743 to 8887 of pNL4-3), which encodes the NL4-3 HIV-1 provirus (1), with the corresponding fragments from the MVC-Res and MVC-Sens viruses that have been described previously (76). Viral stocks were produced by transfection of 293T cells with 5 μg of a chimeric HIV-1 plasmid by using the polyethyleneimine (PEI) transfection reagent (Polysciences Inc., Warrington, PA). Supernatants were collected 48 h posttransfection, filtered through 0.45-μm-pore-size filters, and stored at −80°C. The TCID50 of virus stocks were determined by titration in TZM-bl cells as described previously (74, 80).
Single-round HIV-1 entry assays.
For single-round entry assays using JC53 cells, NP2-CD4/CCR5 cells, U87-CD4 cells expressing wild-type (WT) or mutant CCR5 coreceptors, or 293-Affinofile cell populations, 2 × 104 cells cultured in 96-well plates were inoculated with 200 TCID50 of Env-pseudotyped luciferase reporter virus (corresponding to a multiplicity of infection [MOI] of 0.01) in a volume of 100 μl for 12 h at 37°C. The cells were washed twice with culture medium to remove the residual inoculum and were incubated for a further 60 h at 37°C. For single-round entry assays using MDM, cell monolayers that were approximately 90% confluent in 48-well tissue culture plates were inoculated with 1,500 TCID50 of Env-pseudotyped luciferase reporter virus in a volume of 300 μl for 12 h at 37°C. The MDM were then washed twice with culture medium to remove the residual inoculum and were incubated for a further 96 h at 37°C. For single-round entry assays in PBMC, 2 × 105 cells were inoculated with 2,000 TCID50 of Env-pseudotyped luciferase reporter virus in a volume of 100 μl for 12 h at 37°C. The cells were then washed twice with culture medium to remove the residual inoculum and were incubated for a further 72 h at 37°C. For all cell types, the level of HIV-1 entry was measured by luciferase activity in cell lysates (Promega) according to the manufacturer's protocol. Luminescence was measured using a FLUOStar microplate reader (BMG Labtech, GmbH, Germany). Negative controls included mock-infected cells that were incubated with culture medium instead of virus, as well as cells inoculated with a luciferase reporter virus pseudotyped with the nonfunctional ΔKS Env (15).
Drug sensitivity assays.
In experiments measuring the inhibition of HIV-1 entry by MVC, VVC, or T-20, target cells either were left untreated or were preincubated with dilutions of the drug (5-fold dilutions for MVC and T-20; 10-fold dilutions for VVC) for 30 min at 37°C prior to inoculation with an Env-pseudotyped luciferase reporter virus as described above. Drug concentrations ranged from 0.064 to 5,000 nM for MVC, from 0.01 to 1,000 nM for VVC, and from 0.032 to 100 μg per ml for T-20. These concentrations were maintained during virus inoculation and the subsequent 60-h culture period. The level of HIV-1 entry was measured as described above. After the background activity was subtracted, the amount of luciferase activity in cells treated with an inhibitor was expressed as a percentage of that in untreated cells. The percentage of inhibition was calculated by subtracting this number from 100. The data were fitted with a nonlinear function, and alterations in drug sensitivity were assessed by reductions in the MPI as described previously (76), or by changes in 50% inhibitory concentrations that were calculated by least squares regression analysis using Prism, version 4.0c (GraphPad Software, San Diego, CA) as described previously (64, 66).
Affinofile cell assays and quantitative vector analysis.
293-Affinofile cells were infected with Env-pseudotyped luciferase reporter viruses as described previously (32). Briefly, 48 populations of cells expressing different combinations of CD4 and CCR5 levels were generated by inducing the cells with 2-fold serial dilutions of minocycline (0.156 to 5.0 ng per ml, resulting in 6 induction levels of CD4 increasing linearly from approximately 1,500 to 160,000 CD4 molecules per cell) and ponasterone A (0.0156 to 2.0 μM, resulting in 8 induction levels of CCR5 increasing linearly from approximately 5,000 to 140,000 CCR5 molecules per cell). CD4 and CCR5 concentrations were determined by quantitative flow cytometry (qFACS) as described previously (32, 41). The induced cell populations were then either left untreated or treated with 10 μM MVC for 30 min at 37°C, after which they were inoculated with equivalent amounts of Env-pseudotyped reporter virus and were analyzed for levels of HIV-1 entry as described above. In experiments using MVC-treated cells, the MVC concentration was maintained during virus inoculation and the subsequent culture period. The relative level of virus entry achieved by each Env tested was expressed as a percentage of that achieved in 293-Affinofile cells expressing the highest concentrations of CD4 and CCR5. The relative dependence of Env-pseudotyped reporter viruses on CD4 and CCR5 expression levels was mathematically modeled using the VERSA computational platform (http://versa.biomath.ucla.edu), as described previously (32, 51). With this model, viral infectivity is quantified using a single vector. The vector magnitude reflects the efficiency of virus entry, and the vector angle represents the relative dependence on CD4 or CCR5. In theoretical extremes, viruses that have the greatest possible sensitivity to alterations in CD4 expression but are not affected by alterations in CCR5 expression have a vector angle of 0°, and conversely, viruses that have the greatest possible sensitivity to alterations in CCR5 expression but are not affected by alterations in CD4 expression have a vector angle of 90°.
HIV-1 replication kinetics in monocyte-derived macrophages.
MDM cultures that were approximately 90% confluent in 24-well tissue culture plates were either left untreated or treated with 1 μM MVC for 30 min at 37°C prior to inoculation with 3 × 104 TCID50 of chimeric NL4-3 virus or HIV-1 AD8 (67) in a volume of 500 μl for 16 h at 37°C. The virus inoculum was then removed; the cells were washed 3 times with phosphate-buffered saline (PBS); and then 1 ml fresh culture medium was added to the cells. Cells were cultured for 14 days at 37°C, with 50% medium changes and supernatant samples collected for assessment of virus production on days 1, 4, 7, 10, and 14 postinfection. For MVC-treated cells, drug concentrations were maintained during virus inoculation and the subsequent culture period. Virus production was assessed by measuring HIV-1 reverse transcriptase activity in supernatant samples as described previously (19). These infections were conducted in duplicate using MDM purified from 4 independent donors.
Macrophage fusion assays.
293T effector cells seeded in 6-well tissue culture plates were cotransfected with 3.4 μg of an Env-expressing plasmid and 0.6 μg pSVL-Tat by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. At 48 h posttransfection, 3 × 104 293T effector cells were added to 90% confluent monolayers of MDM cultured in 48-well tissue culture plates, and the cocultures were incubated at 37°C for 12 h in 300 μl of culture medium. The cocultures were then washed 3 times with PBS to remove all unfused 293T cells, after which the cells were fixed in 4% (wt/vol) paraformaldehyde, and syncytium formation was scored by light microscopy. The fusion activity mediated by MVC-Res and MVC-Sens Envs (i.e., syncytium formation) was compared to the levels attained in control fusion assays using the poorly fusogenic/poorly M tropic JRCSF and highly fusogenic/highly M tropic YU2 Envs. Fusion activity was scored accordingly as absent (−), scant (+/−), low (+), moderate (++), or high (+++).
gp120 structural modeling.
Three-dimensional protein structures of the MVC-Sens and MVC-Res gp120 sequences were prepared using the Discovery Studio suite, version 2.5 (Accelrys, San Diego, CA), as we have described recently (65, 66). The crystal structure of CD4-bound YU2 gp120 containing the V3 variable loop and docked with the nuclear magnetic resonance (NMR) structure of an N-terminal peptide of CCR5 (residues 2 to 15) (kindly provided by P. D. Kwong [28]) was used as a template to introduce the V3 loop sequences of the MVC-Sens and MVC-Res Envs by using the Mutate Protein protocol. Harmonic constraints were applied prior to optimization using the Steepest Descent protocol, which incorporates iterative cycles of conjugate-gradient energy minimization against a probability density function that includes spatial restraints derived from the template and residue specific properties (58). Similarities in 3-dimensional structure were measured by the root mean square deviation (RMSD) of the distances between main-chain atoms (N, Ca, C, and O atoms) from crystal and model structures after rigid body superposition, where an RMSD of <1 Å signifies a high level of homology of 3-dimensional structure between overlaid proteins. The overall quality of the geometry of gp120 models generated was verified using PROCHECK (39). Secondary structure was predicted by the method of Kabsch and Sander (33) using the Discovery Studio suite, version 2.5 (Accelrys). The interface between gp120 and the CCR52–15 sulfopeptide was mapped to atoms predicted to be within 4 Å of the ligand. The buried surface areas (BSA) of individual residues within the V3 loop and the CCR52–15 sulfopeptide were determined using the PISA (Protein Interfaces, Surfaces, and Assemblies) computational platform (37), and the change in BSA (ΔBSA) was calculated by subtracting the MVC-Res BSA from the MVC-Sens BSA at individual residues.
RESULTS
CCR5 antagonist resistance profiles of MVC-Res Env.
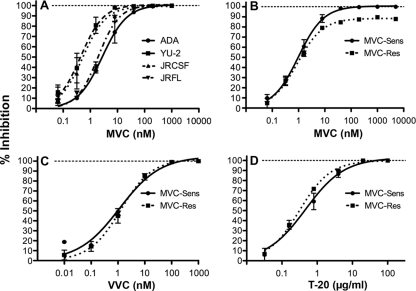
Our previous studies showed that recombinant HIV-1 carrying the MVC-Res env gene is capable of utilizing drug-bound CCR5 for entry (76). In order to more precisely dissect the virus-cell interactions contributing to escape from MVC, the MVC-Res Env, as well as the MVC-Sens Env isolated from the parental MVC-sensitive virus, was cloned into the pSVIII-Env expression vector (18). The CCR5 antagonist resistance profiles of luciferase reporter viruses pseudotyped with MVC-Res or MVC-Sens Env in JC53 cells, or with the R5 ADA, YU-2, JRCSF, or JRFL control Envs, are shown in Fig. 1. MVC completely inhibited the entry of R5 control Envs that exhibited differences in baseline sensitivity to MVC (Fig. 1A) and completely inhibited the entry of MVC-Sens Env (Fig. 1B). However, MVC-Res Env exhibited a plateau of incomplete inhibition at saturating drug concentrations, with MPIs of 88% ± 1.3%, suggesting relatively inefficient usage of drug-bound CCR5 for HIV-1 entry (Fig. 1B). The MPIs of MVC-Res Env were similar in TZM-bl cells (83% ± 3.4%) but considerably lower in NP2-CD4/CCR5 cells (55.3% ± 2.7%) (data not shown). Despite resistance to MVC, MVC-Res Env remained sensitive to VVC (Fig. 1C). As expected, the MVC-Res and MVC-Sens Envs remained equally sensitive to the fusion inhibitor T-20 (Fig. 1D). These results confirm that MVC resistance is associated with relatively inefficient utilization of drug-bound CCR5 by MVC-Res Env, which is typical of most MVC-resistant viruses in patients failing therapy (47, 52), and suggest a narrow profile of cross-resistance to other CCR5 antagonists, which has been reported recently (71).
Fig. 1.
Profiles of resistance to CCR5 inhibitors. Env-pseudotyped luciferase reporter viruses were used to infect JC53 cells in the presence of increasing concentrations of MVC (A and B), VVC (C), or T-20 (D) as described in Materials and Methods. Virus inhibition curves were constructed using Prism, version 4.0c (GraphPad Software), as described in Materials and Methods. The data shown are means of triplicates (error bars represent standard deviations) and are representative of 3 (A, C, and D) or 5 (B) independent experiments.
An increased and critical dependence on the CCR5 N terminus facilitates escape from MVC by MVC-Res Env.
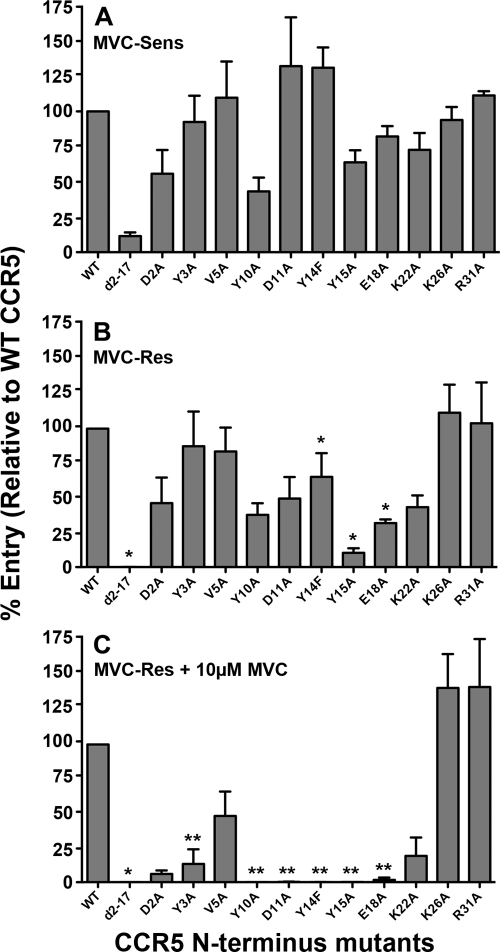
To elucidate alterations in the mechanism of CCR5 engagement associated with HIV-1 escape from MVC, single-round entry assays were conducted in U87-CD4 cells expressing either wild-type (WT) CCR5 or CCR5 containing various mutations in the N-terminal domain by using luciferase reporter viruses pseudotyped with MVC-Res or MVC-Sens Envs (Fig. 2). The levels of virus entry in cells expressing CCR5 mutants were expressed as percentages of that attained in cells expressing equivalent levels of WT CCR5, which was verified by flow cytometry as described previously (66) (data not shown). In the absence of drug, MVC-Res Env displays a relatively modest alteration in CCR5 engagement compared to that of MVC-Sens Env, characterized by increased reliance on Tyr14, Tyr15, and Glu18 in the CCR5 N terminus (Fig. 2A and B). However, in the presence of the drug, MVC-Res Env became critically reliant on these residues, as well as on Tyr3, Tyr10, and Asp11, in the CCR5 N terminus (Fig. 2C). These results indicate a critical reliance on the CCR5 N terminus for recognition of the drug-bound CCR5 complex by MVC-Res Env.
Fig. 2.
Increased dependence on the CCR5 N terminus by MVC-Res Env. Luciferase reporter viruses pseudotyped with MVC-Sens Env (A) or with MVC-Res Env (B and C) were used to infect U87-CD4 cells expressing equivalent levels of WT CCR5 or CCR5 with alternative mutations in the N terminus, as described in Materials and Methods, in the absence (A and B) or presence (C) of MVC. The CCR5-expressing cell populations were generated and characterized with regard to their CCR5 expression levels as described previously (66). The level of virus entry into cells expressing a particular CCR5 mutant was expressed as a percentage of entry into cells expressing WT CCR5. The data shown are means and standard errors from a compilation of 4 independent experiments. Asterisks indicate the significance (P < 0.05) of the increase in dependence on a particular residue over that of MVC-Sens Env (*) or over that of MVC-Res Env in the absence of the drug (**) (determined by an unpaired t test).
Altered recognition of the drug-modified ECLs is critical for escape from MVC by MVC-Res Env.
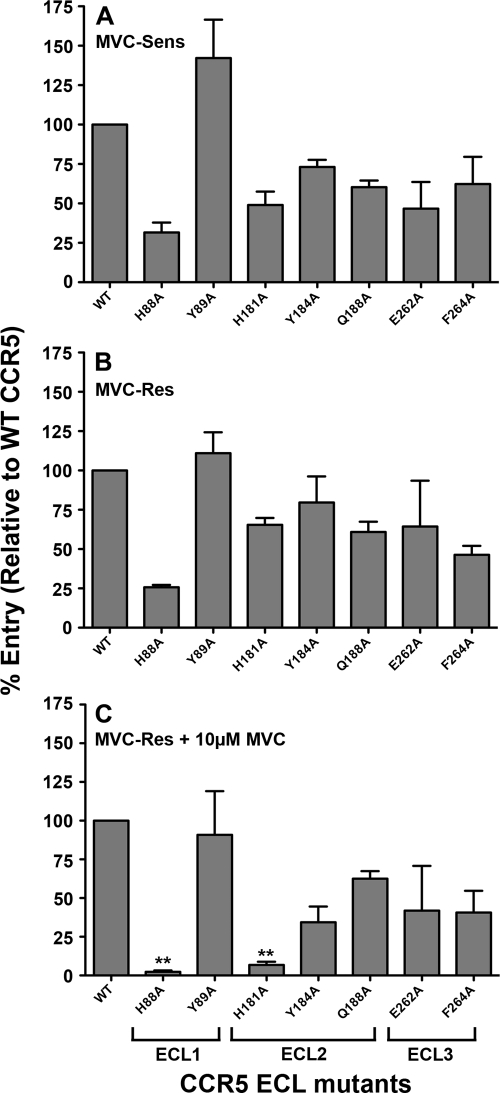
MVC-resistant HIV-1 with narrow cross-resistance to other CCR5 antagonists has been shown to remain dependent on the drug-modified ECLs for entry (71). To elucidate alterations in the way MVC-Res Env may recognize the drug-modified CCR5 ECLs, single-round entry assays were conducted in U87-CD4 cells expressing either WT CCR5 or CCR5 containing various mutations in the ECL1, ECL2, or ECL3 region by using luciferase reporter viruses pseudotyped with MVC-Res or MVC-Sens Envs (Fig. 3). HIV-1 entry levels were measured and expressed as described above. In the absence of the drug, MVC-Sens and MVC-Res Envs displayed similar profiles of dependence on these regions, at least with the CCR5 mutants tested. However, in the presence of the drug, MVC-Res Env became critically reliant on His88 and His181 in the CCR5 ECL1 and ECL2 regions, respectively. These results reveal novel interactions with charged elements of the CCR5 ECL1 and ECL2 regions that are critical for the recognition of drug-bound CCR5 by MVC-Res Env.
Fig. 3.
Altered recognition of the drug-modified ECLs by MVC-Res Env. Luciferase reporter viruses pseudotyped with MVC-Sens Env (A) or with MVC-Res Env (B and C) were used to infect U87-CD4 cells expressing equivalent levels of WT CCR5 or CCR5 with alternative mutations in the ECL1, ECL2, or ECL3 region, as described in Materials and Methods, in the absence (A and B) or presence (C) of MVC. The CCR5-expressing cell populations were generated and characterized with regard to their CCR5 expression levels as described previously (66). The level of virus entry into cells expressing a particular CCR5 mutant was expressed as a percentage of entry into cells expressing WT CCR5. The data shown are means and standard errors from a compilation of 3 independent experiments. Double asterisks indicate that the increase in dependence on a particular residue over that of MVC-Res Env in the absence of the drug is significant (P, <0.05 by an unpaired t test).
Escape from MVC by MVC-Res Env imparts a less-efficient interaction between gp120 and CCR5.
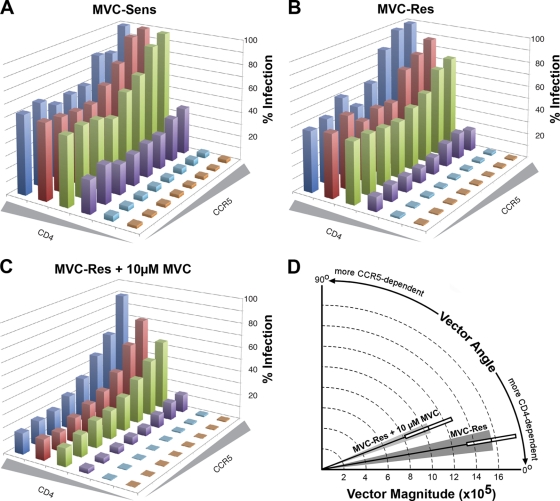
The results of the studies discussed above reveal a highly atypical interaction between gp120 and the N terminus and ECLs of CCR5 that allows MVC-Res Env to escape MVC. Moreover, the relatively high MPIs of MVC-Res Env (Fig. 1B), together with the observation that MVC-Res Env has only a modest alteration in dependence on the CCR5 N terminus in the absence of drug (Fig. 2B), implies a fitness cost associated with escape from MVC. Therefore, we next used the 293-Affinofile affinity-profiling system (32) to better understand the consequences of altered coreceptor engagement for gp120–CD4/CCR5 interactions. In this system, CD4 and CCR5 expression is controlled by separate inducible promoters, permitting independent variation of CD4 and CCR5 expression over a physiological concentration range (32). When 48 differentially induced cell populations are subjected to single-round entry assays with Env-pseudotyped luciferase reporter viruses, and data sets are analyzed quantitatively by mathematical modeling using the VERSA computational platform (32), vector metrics are generated; the vector angles measure the degrees of CD4 and CCR5 dependence, and the vector magnitude measures the efficiency of virus entry. These quantitative data can be used to dissect gp120–CD4/CCR5 interactions. For example, viruses displaying relatively low vector angles and relatively high vector magnitudes are typical of those that have a more efficient interaction with CCR5 (51, 69).
The results of single-round entry assays showed that in the absence of MVC, both MVC-Res and MVC-Sens Envs were highly sensitive to alterations in CD4 levels but much less sensitive to alterations in CCR5 levels (Fig. 4A and B). In fact, at the lowest level of CCR5 expression on the cell surface, these Envs achieved 50 to 60% of maximal entry when moderate to high levels of CD4 were present. In contrast, in the presence of the drug, MVC-Res Env was highly sensitive to alterations in both CD4 and CCR5 levels, requiring high levels of both receptors to achieve maximal levels of HIV-1 entry (Fig. 4C). In these cell populations, MVC completely inhibited the entry of MVC-sensitive Envs, including those shown previously to have highly efficient CCR5 usage, such as ADA, YU2, NB6-C3, and NB8-C2 Envs (64, 66) (data not shown). These results show that utilization of drug-bound CCR5 by MVC-Res Env increases CCR5 dependence, suggesting a reduction in the ability of gp120 to interact with and scavenge low levels of CCR5.
Fig. 4.
Increased CCR5 dependence of MVC-Res Env in the presence of MVC. (A to C) 293-Affinofile cells were treated with increasing concentrations of minocycline and ponasterone A to induce 48 cell populations expressing different combinations of cell surface CD4 and CCR5 levels, as described in Materials and Methods, and were inoculated with equivalent infectious units of a luciferase reporter virus pseudotyped with MVC-Sens (A) or MVC-Res (B and C) Envs in the absence (A and B) or presence (C) of MVC. In the absence of the drug (A and B), virus entry levels were normalized against that in cells expressing the highest levels of both CD4 and CCR5. In the presence of the drug (C), levels of entry by viruses with MVC-Res Env were expressed as percentages of that attained when no drug was present. The results shown are means from 4 independent experiments, each performed in duplicate. Shaded wedges along each axis represent increasing concentrations of CD4 or CCR5, as described in Materials and Methods. (D) Graphical representation of the VERSA vectors showing the efficiency of entry (vector magnitude) and relative dependence on CD4 and CCR5 levels (vector angle). The shaded wedges represent the standard errors of the means of the vector angles, and the boxes represent the standard errors of the means of the vector magnitudes. The difference between the vector angles shown was statistically significant (P, 0.0018 by an unpaired t test).
These results are reflected quantitatively by the VERSA vector metrics. In the absence of the drug, MVC-Sens and MVC-Res Envs had remarkably similar vector angles of 10.4 ± 2.0° and 10.5 ± 2.1°, respectively. These vector angles were well below the range of what we normally observe among subtype B HIV-1 Envs using this system (30 to 60° [32]), signifying highly efficient interactions with CCR5. In contrast, MVC-Res Env had significantly higher vector angles of 22.6 ± 2.7° for recognition of drug-bound CCR5 (P, 0.0018 by an unpaired t test) and a nonsignificant trend toward vector magnitudes lower than those in the absence of MVC (1.05 × 106 ± 0.45 × 106 compared to 1.56 × 106 ± 0.45 × 106; P, 0.16 by an unpaired t test) (Fig. 4D), confirming a less efficient interaction with CCR5. Thus, escape from MVC by MVC-Res Env compromises the efficiency of the interaction between gp120 and CCR5.
Potential structural basis for altered CCR5 engagement by MVC-Res Env.
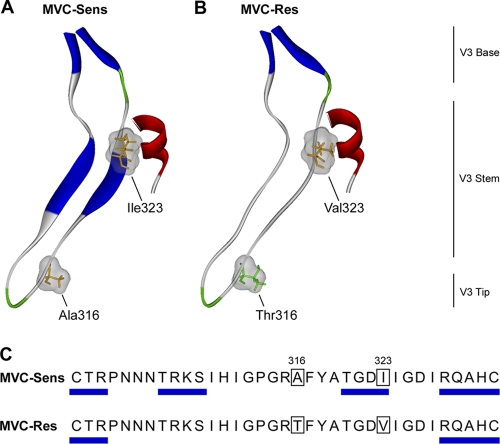
Our previous mutagenesis studies mapped the resistance mutations in MVC-Res Env to amino acids Thr316 and Val323 in the V3 region of gp120 (76). To identify potential structural changes associated with these mutations, we constructed molecular models of gp120 based on the crystal structure of CD4-bound YU2 gp120 (28). This structure is particularly useful because it is docked to a CCR5 N-terminal peptide (CCR52–15) and thus permits the investigation of the molecular interactions between the stem of the V3 loop and the CCR5 N terminus. The positions of the resistance mutations and their predicted effects on V3 loop structure are shown in Fig. 5. The Thr316 resistance mutation is near the tip of the V3 loop and presumably plays a role in binding to the ECLs, while the Val323 resistance mutation is at the V3 stem–CCR5 N terminus interface (Fig. 5B). The relative positions of these resistance mutations suggests that they may act separately yet in concert to permit recognition of the drug-modified CCR5, consistent with the mutagenesis studies which showed that both changes are necessary for complete resistance but that either change by itself confers partial resistance (76). Analysis of predicted secondary structures shows the loss of beta sheets in the MVC-Res V3 loop stem, resulting in a more disordered stem structure (Fig. 5A and B). Mapping of the predicted beta sheets on the amino acid sequence alignment of the MVC-Sens and MVC-Res V3 loops showed that this disruption in secondary structure overlaps with Val323 (Fig. 5C).
Fig. 5.
Structural analysis of MVC resistance mutations in the V3 loop. (A and B) Ribbon representations of the V3 loop regions of the MVC-Sens (A) and MVC-Res (B) gp120 proteins docked to the CCR5 N-terminal sulfopeptide. Secondary structure was predicted using the method of Kabsch and Sander (33) and was color coded as follows: blue, beta sheets; red, helices; green, coils; gray, disordered regions. Residues at positions 316 and 323 (HIVHXB2 numbering), shown by mutagenesis studies to confer MVC resistance on MVC-Res Env (76), are colored according to polarity (orange, nonpolar; green, polar), with their molecular surfaces shown in gray. (C) The predicted beta sheet structures are annotated by blue rectangles on the amino acid sequence alignment of the MVC-Sens and MVC-Res V3 loops.
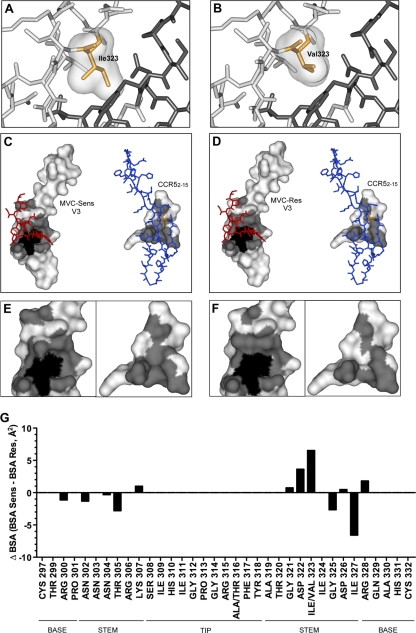
In addition to the possible effects of the Val323 resistance mutation on the secondary structure of the V3 loop, the difference between the space occupied by the bulky Ile side chain in the MVC-Sens V3 loop and the space occupied by the smaller Val side chain in the MVC-Res V3 loop may also affect the interactions between the V3 loop and the CCR5 N terminus (Fig. 6A and B). The smaller Val side chain may reduce steric interference and thus permit a closer interaction with residues of the N terminus of CCR5. To investigate this possibility further, the V3 stem-CCR52–15 interface was mapped for the MVC-Sens and MVC-Res V3 loops (Fig. 6C to F). The interface areas of both the V3 loop and CCR52–15 are reduced in the MVC-Res model (Fig. 6D and F), suggesting a more restricted binding surface that is more easily disrupted by mutations in CCR5. Furthermore, when the BSA was calculated for both the MVC-Sens and MVC-Res V3 models, the Val323 resistance mutation caused a decrease in the BSA at this position (as shown by a positive difference from the BSA for Ile323) and was associated with localized rearrangements in the BSA in the ascending and descending strands of the V3 loop stem (Fig. 6G). These areas of rearrangement overlapped, or were immediately adjacent to, disrupted beta sheet structures (see Fig. 5C).
Fig. 6.
Analysis of MVC resistance mutations in the context of the V3-CCR52–15 sulfopeptide protein interface. (A and B) Close-up views of the V3-CCR52–15 binding sites of the MVC-Sens (A) and MVC-Res (B) Envs, with V3 loops (light gray) and the CCR52–15 sulfopeptide (dark gray) shown as stick models. Ile323 of MVC-Sens V3 and Val323 of MVC-Res V3 are shown as orange stick models with molecular surfaces in gray. (C) The molecular surface of MVC-Sens V3 (left) in complex with the CCR52–15 sulfopeptide (red stick model) and the molecular surface of the CCR52–15 peptide (right) in complex with the V3 loop (blue stick model) are shown in white, with the interface area in gray. The molecular surfaces of buried residues (less than 10.00% solvent-accessible surface) are shaded black. (D) The molecular surfaces of MVC-Res V3 (left) and the CCR52–15 sulfopeptide (right) are colored as in panel C. Ile323 and Val323 in the stick models of the MVC-Sens and MVC-Res V3 loops are colored orange. (E and F) Close-up views of panels C and D, respectively, without the stick models. (G) The change in the buried surface area (ΔBSA, expressed in Å2) between the MVC-Sens and MVC-Res Envs at individual residue positions in the V3 loop was calculated as described in Materials and Methods and was plotted using Prism, version 5.0a (GraphPad Software). Residues corresponding to the V3 loop base, stem, and tip were identified according to the work of Xiang et al. (78).
Together, these structural predictions suggest that the Val323 resistance mutation in the MVC-Res V3 stem may lead to a more disordered or less rigid V3 loop structure with a modified CCR5 binding cavity, which may permit altered interactions between V3 and the N terminus of drug-bound CCR5.
Maraviroc abolishes the entry and replication capacity of HIV-1 carrying MVC-Res Env in monocyte-derived macrophages.
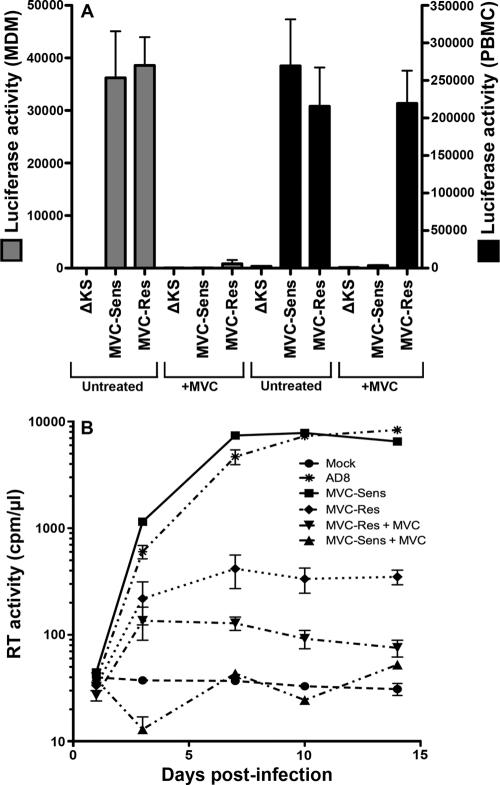
Efficient HIV-1 entry into MDM is associated with R5 Envs that have reduced dependence on the CCR5 N terminus (66). Since escape from MVC by MVC-Res Env results in increased dependence on the CCR5 N terminus (Fig. 2), we next reasoned that escape from MVC may attenuate M tropism. Single-round entry assays using luciferase reporter viruses pseudotyped with MVC-Sens or MVC-Res Env, or with the nonfunctional ΔKS Env as a control, were conducted in MDM in the presence or absence of MVC (Fig. 7A). Similar levels of HIV-1 entry into MDM were obtained with MVC-Sens and MVC-Res Envs in the absence of drug, consistent with the similar CD4/CCR5 dependence profiles in the differentially induced 293-Affinofile cells in the absence of MVC (Fig. 4A and B). Maraviroc completely inhibited the entry of MVC-Sens Env into MDM and inhibited the entry of MVC-Res Env by >95%. In contrast, MVC-Res Env remained strongly resistant to MVC in PBMC. Thus, MVC potently inhibits the entry of MVC-Res Env into MDM but not into PBMC.
Fig. 7.
MVC attenuates the entry and replication of MVC-Res Env in MDM. (A) Luciferase reporter viruses pseudotyped with MVC-Res or MVC-Sens Envs, or with the nonfunctional ΔKS Env, were used to infect cultures of MDM or PBMC in the presence or absence of 5 μM MVC as described in Materials and Methods. The level of entry is shown by luciferase activity in cell lysates. (B) Replication-competent HIV-1 strains carrying the MVC-Sens or MVC-Res env genes were constructed, titrated, and used to infect MDM cultures in the presence or absence of 1 μM MVC, as described in Materials and Methods. The replication of these viruses and the M-tropic R5 HIV-1 strain AD8 was monitored by measuring reverse transcriptase (RT) activity in culture supernatants. The data shown are means and standard deviations of duplicate (MDM) or triplicate (PBMC) cultures and are representative of 4 independent experiments conducted with cells obtained from different donors.
To further investigate the impact of MVC on the tropism of MVC-Res Env in macrophages, MDM were inoculated with equivalent infectious units of replication-competent HIV-1 carrying either MVC-Sens or MVC-Res env genes in the presence or absence of MVC, and the levels of virus replication were measured over 14 days (Fig. 7B). In the absence of the drug, MVC-Sens virus replicated to levels similar to those of the control HIV-1 M-tropic R5 strain AD8, and with similar kinetics, but in the presence of the drug, replication was completely inhibited. MVC-Res virus replicated to comparatively lower levels in the absence of MVC, and replication was almost completely inhibited in the presence of the drug, consistent with the very low levels of HIV-1 entry observed when MVC was present (Fig. 7A). Interestingly, MVC-Res virus replicated to significantly lower levels than MVC-Sens virus in MDM in the absence of the drug, despite similar levels of HIV-1 entry (Fig. 7A). Taking the virus entry and replication data together, the major conclusion is that MVC dramatically attenuates the M tropism of MVC-Res Env.
Reduced fusogenicity of MVC-Res Env is associated with the postentry restriction of HIV-1 replication in MDM.
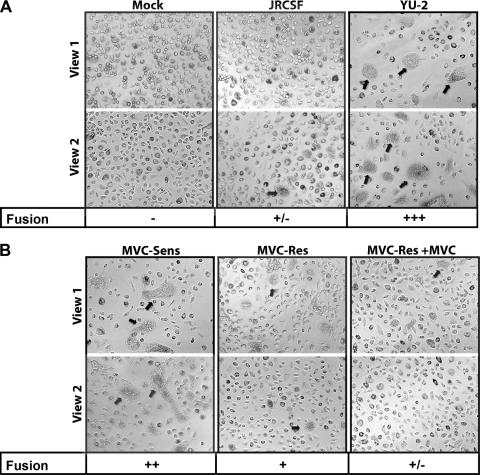
HIV-1 replication in MDM requires efficient cell-cell spread of virus, in addition to efficient entry of the virus into the cell. We therefore hypothesized that the discrepant levels of replication in MDM between viruses carrying MVC-Res and MVC-Sens Envs in the absence of drug, despite equivalent entry, may be due to reduced Env-mediated cell-cell fusion activity. We conducted cell-cell fusion assays using 293T cells expressing Env as effector cells and MDM as target cells (Fig. 8). Effector cells expressing the poorly fusogenic/poorly M tropic JRCSF Env or the highly fusogenic/highly M tropic YU-2 Env were included as controls for comparison (Fig. 8A). Compared to the controls, MVC-Sens Env induced moderately high levels of cell-cell fusion in the absence of MVC, as demonstrated by moderately high levels of syncytium formation (Fig. 8B), which was completely inhibited by MVC (data not shown). In contrast, MVC-Res Env induced comparatively low levels of cell-cell fusion, which was almost completely inhibited by MVC. Similar levels of JRCSF, YU-2, MVC-Sens, and MVC-Res Envs were detected on the surfaces of effector cells by flow cytometry (data not shown), which was conducted as described previously (64). These results suggest that the reduced cell-cell fusion activity of MVC-Res Env in MDM in the absence of drug, likely the result of the relatively modest increase in the dependence of this Env on the CCR5 N terminus in the absence of drug (Fig. 2B), may contribute to the observed postentry restriction of replication capacity in macrophages.
Fig. 8.
Env-mediated fusion activity influences the replication of MVC-Res Env in MDM. Cell-cell fusion assays using 293T effector cells expressing equivalent levels of JRCSF or YU2 Env (A) or MVC-Sens or MVC-Res Env (B) and MDM as target cells were conducted as described in Materials and Methods. Mock cultures were transfected with the nonfunctional ΔKS Env (A). Fusion levels were scored as described in Materials and Methods. Arrows point to representative fusion events. The experiments were conducted in duplicate, and the results shown are representative of 4 independent experiments conducted with MDM obtained from different donors.
DISCUSSION
Although treatment failure associated with the use of CCR5 antagonists can result from the outgrowth of preexisting CXCR4-using HIV-1 strains (27, 38, 75), genuine resistance to the CCR5 antagonists MVC, APL, VVC, AD101, and SCH-D occurs through adaptive alterations in gp120 that enable the utilization of the drug-bound form of CCR5 (3, 36, 49, 51, 55, 56, 69, 71). Maraviroc is the only CCR5 antagonist approved for the treatment of HIV-1 infection. Furthermore, the clinical use of MVC is predicted to escalate due to its recent approval for use in ART-naïve subjects in the United States (22) and the impending availability of alternatives to the Trofile assay that will offer more-rapid and -accessible HIV-1 tropism testing (57, 77), such as the Geno2Pheno tropism assay (46). A thorough knowledge of the mechanisms underlying HIV-1 resistance to MVC and its clinical consequences will be important for detecting, interpreting, and managing resistance given the likelihood of an increase in the use of MVC.
Only one study, by Tilton et al. (71), has characterized mechanisms contributing to the altered recognition of drug-bound CCR5 by an MVC-resistant HIV-1 strain. This study demonstrated very efficient usage of drug-bound CCR5 in a subject failing therapy, characterized by increased dependence on the CCR5 N terminus as well as retention of dependence on the drug-modified ECLs. In our study, we demonstrate a similar yet distinct mechanism of escape from MVC by MVC-Res Env, which has comparatively less efficient usage of drug-bound CCR5. Resistance to MVC may potentially be quantified by the MPI of virus inhibition curves in CCR5-expressing cell lines (47). Among subjects from the MOTIVATE clinical trials who developed MVC resistance, 9/15 (60%) had MPIs of 80 to 95%, 5/15 (33%) had MPIs of 20 to 80%, and 1/15 (7%) had an MPI of <20%, when their viruses were tested in U87-CD4/CCR5 cells (47). By comparison, the MVC-Res Env studied here had MPIs of 83 to 88% in JC53 and TZM-bl cells (Fig. 1 and data not shown). Not unexpectedly, the MPI of MVC-Res Env was substantially lower in NP2-CD4/CCR5 cells (approximately 55%), which express substantially higher levels of CCR5 (60, 61). These results suggest that MVC-Res Env has a lower level of MVC resistance than the MVC-resistant virus studied by Tilton et al. (71), which had MPIs as low as −25% in NP2-CD4/CCR5 cells. Thus, the MVC-resistant Envs characterized by Tilton et al. (71) probably represent unusually high level MVC resistance, whereas the MVC-Res Env studied here appears to be more representative of resistant strains that arise in most subjects with MVC resistance, at least in terms of the comparable MPIs. We acknowledge, however, that MVC-Res Env was generated in vitro and that further studies on primary MVC-resistant viruses with different MPIs are required to confirm the biological relevance of our results. Nonetheless, the characterization of MVC resistance by our study and that of Tilton et al. (71) synergizes the understanding of the molecular basis of this process and has the potential to identify conserved as well as Env-specific mechanistic insights into MVC resistance.
The relatively modest increase in the dependence of MVC-Res Env on the CCR5 N terminus in the absence of drug, but its critical dependence on this region for the recognition of drug-bound CCR5 (Fig. 2), implies that heavy reliance on the CCR5 N terminus may impart a replication disadvantage. In support of this view, we showed, using the 293-Affinofile assay (32), that while both MVC-Res and MVC-Sens Envs have highly efficient interactions with CCR5 in the absence of drug, escape from MVC results in significantly higher sensitivity of MVC-Res Env to alterations in CCR5 expression (Fig. 4). In support of our data, a recent study using the same MVC-Sens and MVC-Res Envs showed, using time-of-addition experiments with T-20, that MVC-Res Env has delayed kinetics of HIV-1 entry in the presence of MVC but not in the absence of MVC (40). Together, these data indicate that utilization of drug-bound CCR5 by MVC-Res Env is significantly less efficient than utilization of unmodified CCR5, a conclusion consistent with the results of a recent study of APL-resistant HIV-1 (51). In contrast, the MVC-resistant strain studied by Tilton et al. (71) maintained a highly efficient interaction with CCR5 even in the presence of the drug, which probably reflects the very high level of MVC resistance exhibited by this particular virus.
Further support for the view that altered interaction with drug-bound CCR5 by MVC-Res Env impairs replication is provided by its entry and replication in MDM (Fig. 7). We showed that while MVC-Res Env can enter MDM as efficiently as MVC-Sens Env in the absence of the drug, MVC almost completely inhibited entry. Furthermore, MVC almost completely inhibited the replication capacity of a full-length virus carrying the MVC-Res env gene and almost completely inhibited the fusion activity of MVC-Res Env in MDM (Fig. 8). Thus, MVC ostensibly abolishes the M-tropic and fusogenic properties of MVC-Res Env. Given the heavy reliance of MVC-Res Env on the CCR5 N terminus and the weaker interaction with drug-bound CCR5 imposed by MVC (Fig. 2 and 4), these results are consistent with those of our recent studies showing that M-tropic R5 Envs typically have reduced dependence on the CCR5 N terminus, increased dependence on the CCR5 ECL2 region, and an increased ability to scavenge low levels of CCR5 on the macrophage surface (66).
M tropism and fusogenicity are important viral phenotypes that contribute to HIV-1 pathogenicity. For example, enhanced fusogenicity and M tropism contribute to CD4+ T-cell apoptosis in vitro and CD4+ T-cell decline in subjects with R5 HIV-1 (26, 42, 64, 73), and M tropism is essential for HIV-1 neurotropism (12–14, 21, 23, 50). Furthermore, increased Env-mediated fusogenicity contributes to the pathogenicity of chimeric simian-HIV strains in vivo (6, 34, 35, 43, 62, 63) and is evident in humans as multinucleated giant cells in autopsied brain tissues of subjects with HIV-1 encephalitis (54). Finally, tissue macrophages are an important viral reservoir (30, 31). It is possible, then, that the continued presence of MVC may attenuate the M-tropic and fusogenic properties of viruses that have acquired a relatively low level of MVC resistance. The observation that MVC attenuates these pathophysiological phenotypes of MVC-Res Env is consistent with a recent study by Pfaff et al. (51), which showed that the presence of APL resulted in a degree of sparing of the central memory CD4+ T-cell subset by an HIV-1 variant that acquired relatively low level APL resistance. Our results raise the possibility that the continued use of MVC for some patients (for example, those for whom clinical testing confirms a relatively high MPI) could lead to virus attenuation, which may potentially have some clinical benefit. However, we cannot exclude the possibility that continuance of therapy may also lead to further lowering of the MPI and, therefore, to increased levels of resistance.
Not only does MVC-Res Env become heavily reliant on the CCR5 N terminus to escape MVC, it remains critically dependent on the drug-modified ECLs, in particular His88 and His181 in the ECL1 and ECL2 regions, respectively. This provides a likely explanation for the lack of cross-resistance to VVC, consistent with the results of recent studies (71). Alteration of either of these residues almost completely inhibited the ability of MVC-Res Env to recognize drug-bound CCR5. In contrast, these residues had only a moderate influence on the MVC-resistant virus characterized by Tilton et al. (71). His88 and His181 can affect the coreceptor activity of CCR5 (8, 16, 25, 66), but they do not normally exert such a potent influence on coreceptor function. These results suggest that the altered recognition of the CCR5 N terminus by MVC-Res Env is dependent on a highly atypical interaction between the gp120 V3 loop and charged elements within the drug-modified ECLs.
The molecular interactions associated with altered recognition of drug-bound CCR5 may be strain specific and may reflect the level of resistance to CCR5 antagonists. By modeling the gp120 V3 loop when docked to an N-terminal peptide of CCR5 (28), a previous study showed the potential for additional hydrogen bonding between Asn320 in the V3 loop and Asp11 in the CCR5 N terminus, associated with very high level of VVC resistance (49). Our results obtained with a similar modeling approach suggest that replacement of Ile323 with Val323 in the V3 stem of MVC-Res Env may contribute to a more specific interaction between V3 and the CCR5 N terminus that is highly sensitive to changes within the N terminus. This appears to occur by altering the secondary structure and the buried surface area of the CCR5 interaction domain of the V3 loop stem, potentially increasing the exposure of this domain. Since no structural information is available on the interaction between the V3 loop and the CCR5 ECLs, the molecular basis for the altered interaction between MVC-Res Env and the drug-modified CCR5 ECLs is unclear. However, replacement of the nonpolar Ala316 with the polar Thr316 at the V3 loop tip in MVC-Res Env could potentiate additional hydrogen bonding with His88 and His181 due to the hydroxyl group on the Thr side chain. Further studies are required in order to better understand the role of His88 and His181 in the CCR5 ECLs in facilitating escape from MVC by MVC-Res Env.
In conclusion, we show that altered recognition of CCR5 by an MVC-resistant variant that exhibits relatively low level resistance, characterized by critical reliance on the CCR5 N terminus and on charged elements of the drug-modified CCR5 ECLs, dramatically attenuates M tropism. These results add further support for the view, proposed initially by Pfaff et al. (51), that continuing therapy with coreceptor antagonists even after the development of resistance could potentially be beneficial in some cases. Our results suggest that these cases may be those that exhibit resistance profiles with relatively high MPIs in CCR5-expressing cell lines, whereby continuance of therapy may promote virus attenuation. Hence, our results highlight the need for further studies investigating the functional consequences of MVC-resistant viruses arising in patients that have various MPIs and underscore the potential utility of the MPI as a clinical tool for the management of patients receiving CCR5 antagonists.
ACKNOWLEDGMENTS
We thank J. Sodroski for providing the pSVIII-HXB2, pSVIII-YU2, pSVIII-ADA, and pSVIII-ΔKS Env plasmids and the pcDNA3-CD4, pcDNA3-CCR5, pCMVΔP1ΔenvpA, and pHIV-1Luc plasmids and CCR5 mutants. We also thank H. Gottlinger for providing the pSVL-Tat plasmid, D. Gabuzda for providing the pcDNA3-JRFL Env plasmid, D. Kabat for providing JC53 cells, R. Doms for providing CCR5 mutants, N. Shimizu and H. Hoshino for permission to use NP2-CD4/CCR5 cells, and D. Mosier and R. Nedellec for supplying the NP2-CD4/CCR5 cells. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: TZM-bl cells from John C. Kappes, Xiaoyun Wu, and Tranzyme, Inc.; U87-CD4 cells from HongKui Deng and Dan R. Littman; and T-20 fusion inhibitor from Roche.
This study was supported by a grant from the Australian Center for HIV and Hepatitis Virology Research (ACH2) to P.R.G. and M.J.C. and by a grant from NIH/NIAID to B.L. (R21 AI092218). M.R. is supported by a Monash University Postgraduate Research Scholarship. P.R.G. is the recipient of an Australian National Health and Medical Research Council (NHMRC) Level 2 Biomedical Career Development award. P.A.R. is the recipient of an NHMRC R. Douglas Wright Biomedical Career Development award. S.R.L. is the recipient of an NHMRC Practitioner Fellowship. We gratefully acknowledge the contribution to this work of the Victorian Operational Infrastructure Support Program, received by the Burnet Institute.
Footnotes
Published ahead of print on 23 February 2011.
REFERENCES
- 1. Adachi A., et al. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anastassopoulou C. G., Ketas T. J., Klasse P. J., Moore J. P. 2009. Resistance to CCR5 inhibitors caused by sequence changes in the fusion peptide of HIV-1 gp41. Proc. Natl. Acad. Sci. U. S. A. 106:5318–5323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berro R., Sanders R. W., Lu M., Klasse P. J., Moore J. P. 2009. Two HIV-1 variants resistant to small molecule CCR5 inhibitors differ in how they use CCR5 for entry. PLoS Pathog. 5:e1000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Björndal A., et al. 1997. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 71:7478–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brelot A., et al. 1999. Effect of mutations in the second extracellular loop of CXCR4 on its utilization by human and feline immunodeficiency viruses. J. Virol. 73:2576–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cayabyab M., et al. 1999. Changes in human immunodeficiency virus type 1 envelope glycoproteins responsible for the pathogenicity of a multiply passaged simian-human immunodeficiency virus (SHIV-HXBc2). J. Virol. 73:976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cormier E. G., Dragic T. 2002. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J. Virol. 76:8953–8957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doranz B. J., et al. 1997. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J. Virol. 71:6305–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Doranz B. J., et al. 1999. Identification of CXCR4 domains that support coreceptor and chemokine receptor functions. J. Virol. 73:2752–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dragic T., et al. 2000. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc. Natl. Acad. Sci. U. S. A. 97:5639–5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Duenas-Decamp M. J., Peters P. J., Burton D., Clapham P. R. 2009. Determinants flanking the CD4 binding loop modulate macrophage tropism of human immunodeficiency virus type 1 R5 envelopes. J. Virol. 83:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dunfee R., et al. 2006. Mechanisms of HIV-1 neurotropism. Curr. HIV Res. 4:267–278 [DOI] [PubMed] [Google Scholar]
- 13. Dunfee R. L., Thomas E. R., Gabuzda D. 2009. Enhanced macrophage tropism of HIV in brain and lymphoid tissues is associated with sensitivity to the broadly neutralizing CD4 binding site antibody b12. Retrovirology 6:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dunfee R. L., et al. 2006. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc. Natl. Acad. Sci. U. S. A. 103:15160–15165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Etemad-Moghadam B., et al. 2000. Envelope glycoprotein determinants of increased fusogenicity in a pathogenic simian-human immunodeficiency virus (SHIV-KB9) passaged in vivo. J. Virol. 74:4433–4440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farzan M., et al. 1998. A tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J. Virol. 72:1160–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Farzan M., et al. 1999. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96:667–676 [DOI] [PubMed] [Google Scholar]
- 18. Gao F., et al. 1996. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. J. Virol. 70:1651–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gorry P., Purcell D., Howard J., McPhee D. 1998. Restricted HIV-1 infection of human astrocytes: potential role of nef in the regulation of virus replication. J. Neurovirol. 4:377–386 [DOI] [PubMed] [Google Scholar]
- 20. Gorry P. R., Ancuta P. 2011. Coreceptors and HIV-1 pathogenesis. Curr. HIV/AIDS Rep. 8:45–53 [DOI] [PubMed] [Google Scholar]
- 21. Gorry P. R., et al. 2001. Macrophage tropism of human immunodeficiency virus type 1 isolates from brain and lymphoid tissues predicts neurotropism independent of coreceptor specificity. J. Virol. 75:10073–10089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gorry P. R., Ellett A., Lewin S. R. 2010. Maraviroc, p. 2869–2876 In Grayson M. L., Crowe S., McCarthy J., Mills J., Mouton J., Norrby S. R., Paterson D., Pfaller M. (ed.), Kucers' the use of antibiotics, 6th ed. Hodder Arnold, London, England [Google Scholar]
- 23. Gorry P. R., et al. 2002. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J. Virol. 76:6277–6292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gray L., et al. 2006. Genetic and functional analysis of R5X4 human immunodeficiency virus type 1 envelope glycoproteins derived from two individuals homozygous for the CCR5Δ32 allele. J. Virol. 80:3684–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gray L., et al. 2009. Tissue-specific sequence alterations in the human immunodeficiency virus type 1 envelope favoring CCR5 usage contribute to persistence of dual-tropic virus in the brain. J. Virol. 83:5430–5441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gray L., et al. 2005. Uncoupling coreceptor usage of human immunodeficiency virus type 1 (HIV-1) from macrophage tropism reveals biological properties of CCR5-restricted HIV-1 isolates from patients with acquired immunodeficiency syndrome. Virology 337:384–398 [DOI] [PubMed] [Google Scholar]
- 27. Gulick R. M., et al. 2007. Phase 2 study of the safety and efficacy of vicriviroc, a CCR5 inhibitor, in HIV-1-infected, treatment-experienced patients: AIDS clinical trials group 5211. J. Infect. Dis. 196:304–312 [DOI] [PubMed] [Google Scholar]
- 28. Huang C. C., et al. 2007. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317:1930–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang C. C., et al. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Igarashi T., et al. 2001. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): implications for HIV-1 infections of humans. Proc. Natl. Acad. Sci. U. S. A. 98:658–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Igarashi T., Imamichi H., Brown C. R., Hirsch V. M., Martin M. A. 2003. The emergence and characterization of macrophage-tropic SIV/HIV chimeric viruses (SHIVs) present in CD4+ T cell-depleted rhesus monkeys. J. Leukoc. Biol. 74:772–780 [DOI] [PubMed] [Google Scholar]
- 32. Johnston S. H., et al. 2009. A quantitative affinity-profiling system that reveals distinct CD4/CCR5 usage patterns among human immunodeficiency virus type 1 and simian immunodeficiency virus strains. J. Virol. 83:11016–11026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kabsch W., Sander C. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637 [DOI] [PubMed] [Google Scholar]
- 34. Karlsson G. B., et al. 1997. Characterization of molecularly cloned simian-human immunodeficiency viruses causing rapid CD4+ lymphocyte depletion in rhesus monkeys. J. Virol. 71:4218–4225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karlsson G. B., et al. 1998. The envelope glycoprotein ectodomains determine the efficiency of CD4+ T lymphocyte depletion in simian-human immunodeficiency virus-infected macaques. J. Exp. Med. 188:1159–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kitrinos K. M., Amrine-Madsen H., Irlbeck D. M., Word J. M., Demarest J. F. 2009. Virologic failure in therapy-naive subjects on aplaviroc plus lopinavir-ritonavir: detection of aplaviroc resistance requires clonal analysis of envelope. Antimicrob. Agents Chemother. 53:1124–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krissinel E., Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797 [DOI] [PubMed] [Google Scholar]
- 38. Lalezari J., et al. 2005. Antiviral activity and safety of 873140, a novel CCR5 antagonist, during short-term monotherapy in HIV-infected adults. AIDS 19:1443–1448 [DOI] [PubMed] [Google Scholar]
- 39. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. 1993. PROCHECK—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291 [Google Scholar]
- 40. Latinovic O., et al. 2011. CCR5 antibodies HGS004 and HGS101 preferentially inhibit drug-bound CCR5 infection and restore drug sensitivity of maraviroc-resistant HIV-1 in primary cells. Virology 411:32–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee B., Sharron M., Montaner L. J., Weissman D., Doms R. W. 1999. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc. Natl. Acad. Sci. U. S. A. 96:5215–5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li S., et al. 1999. Persistent CCR5 utilization and enhanced macrophage tropism by primary blood human immunodeficiency virus type 1 isolates from advanced stages of disease and comparison to tissue-derived isolates. J. Virol. 73:9741–9755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu Z. Q., et al. 1999. Derivation and biological characterization of a molecular clone of SHIV(KU-2) that causes AIDS, neurological disease, and renal disease in rhesus macaques. Virology 260:295–307 [DOI] [PubMed] [Google Scholar]
- 44. Maeda K., et al. 2006. Structural and molecular interactions of CCR5 inhibitors with CCR5. J. Biol. Chem. 281:12688–12698 [DOI] [PubMed] [Google Scholar]
- 45. Maeda K., et al. 2008. Involvement of the second extracellular loop and transmembrane residues of CCR5 in inhibitor binding and HIV-1 fusion: insights into the mechanism of allosteric inhibition. J. Mol. Biol. 381:956–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McGovern R. A., et al. 2010. Population-based V3 genotypic tropism assay: a retrospective analysis using screening samples from the A4001029 and MOTIVATE studies. AIDS 24:2517–2525 [DOI] [PubMed] [Google Scholar]
- 47. Mori J., et al. 2008. Characterization of maraviroc resistance in patients failing treatment with CCR5-tropic virus in MOTIVATE 1 and MOTIVATE 2 (24 week analysis), poster 51. Prog. Abstr. 6th Eur. Drug Resist. Workshop, Budapest, Hungary, 26 to 28 March 2008 [Google Scholar]
- 48. Ogert R. A., et al. 2009. Structure-function analysis of human immunodeficiency virus type 1 gp120 amino acid mutations associated with resistance to the CCR5 coreceptor antagonist vicriviroc. J. Virol. 83:12151–12163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ogert R. A., et al. 2010. Clinical resistance to vicriviroc through adaptive V3 loop mutations in HIV-1 subtype D gp120 that alter interactions with the N terminus and ECL2 of CCR5. Virology 400:145–155 [DOI] [PubMed] [Google Scholar]
- 50. Peters P. J., et al. 2004. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusigenicity for macrophages. J. Virol. 78:6915–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pfaff J. M., et al. 2010. HIV-1 resistance to CCR5 antagonists associated with highly efficient use of CCR5 and altered tropism on primary CD4+ T cells. J. Virol. 84:6505–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pfizer, Inc 2007. Maraviroc tablets NDA 22-128: Antiviral Drugs Advisory Committee (AVDAC) briefing document. Pfizer Inc., New York, NY [Google Scholar]
- 53. Platt E. J., Wehrly K., Kuhmann S. E., Chesebro B., Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Price R. W. 1996. Neurological complications of HIV infection. Lancet 348:445–452 [DOI] [PubMed] [Google Scholar]
- 55. Pugach P., et al. 2007. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology 361:212–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pugach P., et al. 2009. Inefficient entry of vicriviroc-resistant HIV-1 via the inhibitor-CCR5 complex at low cell surface CCR5 densities. Virology 387:296–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Reeves J. D., Coakley E., Petropoulos C. J., Whitcomb J. M. 2009. An enhanced-sensitivity Trofile HIV coreceptor tropism assay for selecting patients for therapy with entry inhibitors targeting CCR5: a review of analytical and clinical studies. J. Viral Entry 3:94–102 [Google Scholar]
- 58. Sali A., Blundell T. L. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815 [DOI] [PubMed] [Google Scholar]
- 59. Seibert C., et al. 2006. Interaction of small molecule inhibitors of HIV-1 entry with CCR5. Virology 349:41–54 [DOI] [PubMed] [Google Scholar]
- 60. Shimizu N., et al. 2009. Broad usage spectrum of G protein-coupled receptors as coreceptors by primary isolates of HIV. AIDS 23:761–769 [DOI] [PubMed] [Google Scholar]
- 61. Soda Y., et al. 1999. Establishment of a new system for determination of coreceptor usages of HIV based on the human glioma NP-2 cell line. Biochem. Biophys. Res. Commun. 258:313–321 [DOI] [PubMed] [Google Scholar]
- 62. Stephens E. B., et al. 1996. Initial characterization of viral sequences from a SHIV-inoculated pig-tailed macaque that developed AIDS. J. Med. Primatol. 25:175–185 [DOI] [PubMed] [Google Scholar]
- 63. Stephens E. B., et al. 1997. A cell-free stock of simian-human immunodeficiency virus that causes AIDS in pig-tailed macaques has a limited number of amino acid substitutions in both SIVmac and HIV-1 regions of the genome and has offered cytotropism. Virology 231:313–321 [DOI] [PubMed] [Google Scholar]
- 64. Sterjovski J., et al. 2007. Asn 362 in gp120 contributes to enhanced fusogenicity by CCR5-restricted HIV-1 envelope glycoprotein variants from patients with AIDS. Retrovirology 4:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sterjovski J., et al. 2011. CD4-binding site alterations in CCR5-using HIV-1 envelopes influencing gp120-CD4 interactions and fusogenicity. Virology 410:418–428 [DOI] [PubMed] [Google Scholar]
- 66. Sterjovski J., et al. 2010. An altered and more efficient mechanism of CCR5 engagement contributes to macrophage tropism of CCR5-using HIV-1 envelopes. Virology 404:269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Theodore T. S., et al. 1996. Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res. Hum. Retroviruses 12:191–194 [DOI] [PubMed] [Google Scholar]
- 68. Thomas E. R., et al. 2007. Macrophage entry mediated by HIV Envs from brain and lymphoid tissues is determined by the capacity to use low CD4 levels and overall efficiency of fusion. Virology 360:105–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tilton J. C., et al. 2010. HIV type 1 from a patient with baseline resistance to CCR5 antagonists uses drug-bound receptor for entry. AIDS Res. Hum. Retroviruses 26:13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tilton J. C., Doms R. W. 2010. Entry inhibitors in the treatment of HIV-1 infection. Antiviral Res. 85:91–100 [DOI] [PubMed] [Google Scholar]
- 71. Tilton J. C., et al. 2010. A maraviroc-resistant HIV-1 with narrow cross-resistance to other CCR5 antagonists depends on both N-terminal and extracellular loop domains of drug-bound CCR5. J. Virol. 84:10863–10876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tsamis F., et al. 2003. Analysis of the mechanism by which the small-molecule CCR5 antagonists SCH-351125 and SCH-350581 inhibit human immunodeficiency virus type 1 entry. J. Virol. 77:5201–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wade J., et al. 2010. Enhanced CD4+ cellular apoptosis by CCR5-restricted HIV-1 envelope glycoprotein variants from patients with progressive HIV-1 infection. Virology 396:246–255 [DOI] [PubMed] [Google Scholar]
- 74. Wei X., et al. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Westby M., et al. 2006. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J. Virol. 80:4909–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Westby M., et al. 2007. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J. Virol. 81:2359–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Whitcomb J. M., et al. 2007. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob. Agents Chemother. 51:566–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Xiang S. H., et al. 2010. A V3 loop-dependent gp120 element disrupted by CD4 binding stabilizes the human immunodeficiency virus envelope glycoprotein trimer. J. Virol. 84:3147–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yang X., Wyatt R., Sodroski J. 2001. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J. Virol. 75:1165–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yap S. H., et al. 2007. N348I in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance. PLoS Med. 4:e335. [DOI] [PMC free article] [PubMed] [Google Scholar]