Abstract
Neutralizing antibodies have a role in controlling hepatitis C virus (HCV) infection. A successful vaccine will need to elicit potently neutralizing antibodies that are capable of preventing the infection of genetically diverse viral isolates. However, the specificity of the neutralizing antibody response in natural HCV infection still is poorly understood. To address this, we examined the reactivity of polyclonal antibodies isolated from chronic HCV infection to the diverse patient-isolated HCV envelope glycoproteins E1 and E2 (E1E2), and we also examined the potential to neutralize the entry of pseudoparticles bearing these diverse E1E2 proteins. The genetic type of the infection was found to determine the pattern of the antibody recognition of these E1E2 proteins, with the greatest reactivity to homologous E1E2 proteins. This relationship was strongest when the component of the antibody response directed only to linear epitopes was analyzed. In contrast, the neutralization serotype did not correlate with genotype. Instead, serum-derived antibodies displayed a range of neutralization breadth and potency, while different E1E2 glycoproteins displayed different sensitivities to neutralization, such that these could be divided broadly into neutralization-sensitive and -resistant phenotypes. An important additional observation was that entry mediated by some E1E2 proteins was enhanced in the presence of some of the polyclonal antibody fractions isolated during chronic infection. These data highlight the need to use diverse E1E2 isolates, which represent extremes of neutralization sensitivity, when screening antibodies for therapeutic potential and for testing antibodies generated following immunization as part of vaccine development.
INTRODUCTION
Hepatitis C virus (HCV) is a major cause of chronic hepatitis, hepatocellular carcinoma, and liver cirrhosis (1). Current treatments are inadequate, and an effective vaccine has yet to be developed. This is due in part to the great genetic diversity exhibited by the virus, which is a consequence of the error-prone replication of the RNA genome combined with high viral replication rates (64). As a result, individual HCV isolates can differ by up to 30% of their nucleotide sequence (55). HCV currently is classified into at least six distinct genotypes, which differ in their geographic distribution and phenotype (54).
The E1 and E2 (E1E2) glycoproteins mediate the entry of the virus into host cells (3, 14, 45) and are important targets for the host neutralizing antibody response (23, 24, 30, 44, 46). Several lines of evidence suggest that neutralizing antibodies have a protective role in vivo. The production of potently neutralizing antibodies in acute infections has been shown to correlate with clearance in a single-source outbreak cohort (47). In vaccinated chimpanzees, a sustained antibody response to E1 and E2 correlates with reduced viral load (63), while the passive administration of neutralizing monoclonal antibodies (MAbs) in a uPA-SCID chimeric mouse model of infection was able to protect against challenge with an HCV quasispecies inoculum (27).
The majority of neutralizing antibodies described to date are directed against linear or conformation-sensitive epitopes located within or proximal to the CD81 binding regions (23, 27, 35, 43, 44, 46) or to epitopes located in the first hypervariable region of E2. Antibodies targeting the latter region presumably mediate their neutralizing effect by blocking the interaction between E2 and scavenger receptor class B type I (SR-BI) (5, 49).
It is unclear to what degree genotypic variability will need to be taken into consideration in the development of antibody-based vaccines and, in particular, whether or not antibodies induced by exposure to one genotype will have any neutralizing effect against strains within the same or different genotypes. Cross-reactive neutralizing antibodies have been described in the sera of both acutely and chronically infected patients, but neutralization potencies were tested against only a limited number of strains (2, 30). We have demonstrated previously that antibodies directed to the E1E2 glycoproteins preferentially recognize homologous proteins in genotype 1 and 3 infections (22). Here, we have used our panel of well-characterized E1E2 glycoprotein clones (26, 27, 43, 44, 46) to test the cross-reactivity and neutralization potency of a panel of homologous and heterologous human serum IgG from genotype 1, 2, and 3 infections. Using immobilized native and denatured E1E2, we demonstrate that cross-reactive antibodies generally target conformational epitopes. Antibodies recognizing linear epitopes are more genotype restricted. In general, antigenic serotype, but not neutralization serotype, corresponded with genotype. Importantly, our data demonstrate that E1E2 glycoproteins differ in their sensitivity to neutralization, with patient-isolated E1E2 being more resistant to neutralization than reference molecular clone H77c. We also demonstrate that some HCV-infected sera harbor antibodies capable of enhancing infection of particular strains of HCV.
MATERIALS AND METHODS
HCV E1E2 glycoproteins, sera, and monoclonal antibodies.
A panel of HCV glycoproteins representing functional, primary patient isolates of genotypes 1 to 6 were isolated previously from chronic HCV infections (26, 43). We selected a panel of proteins consisting of at least two independently isolated functional E1E2 genes that represented genotypes 1 to 3 and, because of its widespread use in other studies, E1E2 from the molecular clone H77c (sample H77.20) (45). All clones were generated in the pcDNA3.1 mammalian expression vector. To verify the genotype and designations, the E1E2 panel sequences, together with representative sequences obtained from the Los Alamos HCV database, were subjected to maximum-likelihood phylogenetic analysis under a GTR+G+I model using PAUP, version 10.
E1 and E2 proteins were expressed in cis in HEK 293T cells, as described previously (57). Proteins were separated using nonreducing 9% SDS-PAGE and analyzed by Western blotting with the broadly reactive anti-E2 MAbs AP33 and ALP98 (42). Confirmation that the expressed proteins were correctly folded was achieved by CD81 pulldown assay essentially as described previously (10). Briefly, E1E2 proteins were captured to a glutathione S-transferase (GST)-CD81 large extracellular loop (LEL) fusion construct immobilized to glutathione-agarose beads (Sigma). After the complex was washed, binding was demonstrated by boiling the protein complex and analyzing the input and bound protein by Western blotting. GST was used as a negative control in these experiments. Precipitated E2 protein was quantified using densitometry (Alphaease v4.0).
Glycoprotein conformation also was assessed by reactivity to a panel of conformation-sensitive antibodies. Monoclonal antibodies CBH2, CBH5, CBH7, A8, AR3A, AR3b, AR3c, 1:7, L1, AR1A, AR1B, AR2A, CBH4B, CBH4G, CBH4D, and CBH8 were used in this study and have been described previously (21, 23, 24, 27). Antibody binding was performed as described previously (23, 44).
A panel of 35 HCV-infected human sera was selected from the Trent HCV cohort (37). These represented chronically infected individuals who tested positive in second-generation commercial antibody tests (Innogenetics). All patients were diagnosed as HIV negative. Where possible, the duration of infection, estimated by the date of their first risk behavior, was matched between samples. The infecting genotype was determined using Inno-LiPA (version 2x). Sera were diluted in 20 mM sodium phosphate buffer and mixed with protein G-agarose beads (Sigma) for 1 h. Unbound material was removed by five washes with phosphate buffer, and IgG was recovered by incubation for 10 min with IgG elution buffer (Pierce). Samples were neutralized with 1 M Tris (pH 8.8).
Serum reactivity assays.
Each serum sample was tested for reactivity to the panel of E1E2 glycoproteins immobilized on Galanthus nivalis agglutinin (GNA)-coated wells (9). To ensure the equivalent capture of different E1E2 protein constructs, initial experiments were performed using a range of GNA concentrations to capture serial dilutions of cell lysates. Bound E1E2 was detected by cross-reactive MAbs AP33 and ALP98. From these experiments it was determined that lysates captured onto GNA coated at 150 ng ml−1 resulted in saturating binding. GNA diluted in carbonate-bicarbonate buffer (Sigma) was coated onto the wells of an assay plate (Maxisorp; Nunc), and nonspecific binding sites were blocked by incubation for 2 h with PBS–0.05% Tween 20 (PBST) containing 5% low-fat milk. Saturating amounts of HEK 293T cell lysates were captured for 4 h at room temperature. After being washed, sera were diluted 1/100 in PBST and incubated for 2 h, followed by the addition of an anti-human IgG antibody conjugated to alkaline phosphatase (Sigma). To assay reactivity to conformation-insensitive epitopes, E1E2 proteins first were denatured as described previously (40, 58) before appropriate dilution in PBST and addition to GNA-coated enzyme-linked immunosorbent assay (ELISA) wells. Identical amounts of E1E2 protein were assayed in both native and denatured forms.
Neutralization of HCVpp entry by purified polyclonal antibody isolated from HCV-infected sera.
Immunoglobulin G fractions were isolated by affinity chromatography using protein G columns. IgG purified from a pool of normal human serum was used as a negative control. These IgG preparations were used to neutralize the entry of HCV pseudoparticles (HCVpp) into Huh7 cells essentially as described previously (2). Briefly, HCVpp preparations possessing E1E2 from genotypes 1, 2, and 3 were expressed. HCVpp preparations containing equivalent amounts of murine leukemia virus (MLV) core protein were mixed with purified polyclonal antibodies either at a standard concentration of 25 μg ml−1 or serially diluted for dose-dependent experiments. These mixtures were added to Huh7 cells, and luciferase activity was measured after 72 h using a BMG Labtech Fluorostar Optima.
Neutralization of replication of cell culture infectious clones of HCV (HCVcc) by purified polyclonal antibody isolated from HCV-infected sera.
Plasmids containing JFH-1 or JFH-1GND genome cDNA (29, 61) were linearized using XbaIFD (Fermentas) and used as the template for in vitro HCV genomic RNA transcript generation using a MEGAscript T7 high-yield transcription kit (Ambion). Genomic transcripts were cleaned up using an RNeasy minikit (Qiagen). For each sample, 10 μg HCV RNA was electroporated into 7 × 106 Huh7.5 (6) cells using a GenePulser Xcell electroporator (Bio-Rad). Medium harvested after 48 h was filtered through a 0.45-μm membrane, and virus particles were quantified by serially diluting cell supernatants in replicates and infecting Huh7.5 cells. Fixed cells were permeabilized with 0.5% Triton X-100 and stained with anti-NS5A MAb 9E10 (29), followed by the addition of horseradish peroxidase (HRP)-conjugated polyclonal rabbit anti-mouse IgG (DakoCytomation). HRP activity was detected using the NovaRED (Vector) substrate kit. The 50% tissue culture infective doses (TCID50) were calculated using the Reed and Muench method (48). Neutralization assays were performed by mixing 100 focus-forming units (FFU) of virus with purified polyclonal IgG for 1 h before addition to Huh7.5 cells. Infection was detected as described above, and the percentage of infection was determined by comparison to the number of cells infected in the presence of IgG isolated from a pool of donors not infected with HCV.
Statistical analysis.
Calculated reactivity and neutralization data were analyzed using SPSS, version 14.1 (SPSS Inc.). To control for potential differences in the titer of HCV-specific antibodies in each human serum IgG preparation, a normalization method similar to that described by Moore et al. (39) was adopted. For each serum or IgG sample analyzed, the highest signal was assigned 100% reactivity. From this, the relative percentage of reactivity to each of the other proteins was calculated. Proteins, serum IgG reactivities, and neutralization profiles then were compared by agglomerative clustering with unweighted pair-group mean distances of squared Euclidian distance measurements. The resulting matrices were visualized by two-way hierarchical tree building using J Express 2009 (Molmine). The robustness of the two-way reactivity/neutralization data was determined by bootstrap analysis and significance testing using the program Heatmap (as implemented in the Los Alamos National Laboratory HIV database; http://www.hiv.lanl.gov).
Differences in median reactivity data were compared using the Kruskal-Wallis test with Dunn's correction for multiple comparisons, as implemented in GraphPad Prism 4.0.
RESULTS
Phylogenetic relationships between E1E2 antigens.
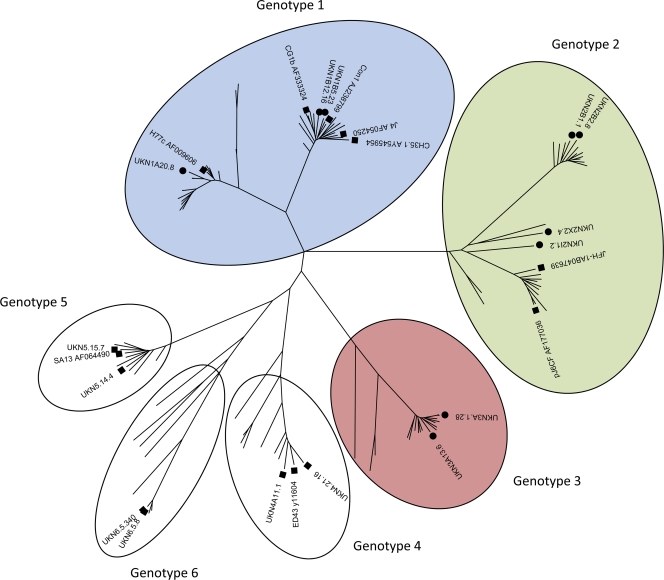
Primary isolates of the E1E2 genes were recovered from chronically infected patients using high-fidelity PCR, as described previously (57) (Table 1). These proteins had been demonstrated previously to permit the entry of HCV pseudoparticles. To determine the evolutionary relationships between the nucleotide sequences encoding these proteins, we performed high-resolution phylogenetic analysis on the entire E1 and E2 genes (Fig. 1). To adequately address intergenotypic seroreactivity and neutralization profiles, we selected isolates from genotypes 1, 2, and 3, the most prevalent genotypes in our patient cohort.
Table 1.
Details of study E1E2 isolatesa
| HCV genotype | Patient code | E1E2 clone name | Accession no. |
|---|---|---|---|
| 1a | H | H77.20 | AF011751 |
| 1a | UKN1A20 | UKN1A20.8 | EU155192 |
| 1b | UKN1B5 | UKN1B5.23 | AY734976 |
| 1b | UKN1B12 | UKN1B12.16 | AY734974 |
| 2i | UKN2A1 | UKN2A1.2 | AY734977 |
| 2x | UKN2A2 | UKN2A2.4 | AY934979 |
| 2b | UKN2B1 | UKN2B1.1 | AY734982 |
| 2b | UKN2B2 | UKN2B2.8 | AY734983 |
| 3a | UKN3A1 | UKN3A1.28 | AY734984 |
| 3a | UKN3A13 | UKN3A13.6 | AY894683 |
The consensus clone H77.20 has been described previously (45). All other samples were primary E1E2 isolates obtained from sera isolated from patients recruited into the UK Trent HCV cohort (38). Each sample is coded by genotype. The ability of the E1E2 clones to mediate the entry of retroviral pseudoparticles has been reported previously (27, 43, 44, 58).
Fig. 1.
Phylogenetic analysis of E1 and E2 genes isolated from patient isolates. Patient-derived genes were aligned with the E1/E2 coding sequences from genotype reference strains (53), and clones were analyzed in previous studies of functional E1E2 (23, 25, 33, 34, 43). Symbols: ●, sequence from patient isolated and analyzed in this study; ■, other functional E1/E2 sequences reported in the available literature. Genotypes are color coded for clarity of presentation in subsequent figures. Blue corresponds to genotype 1 samples, green to genotype 2, and red to genotype 3.
Expression of monomeric, correctly folded E1E2.
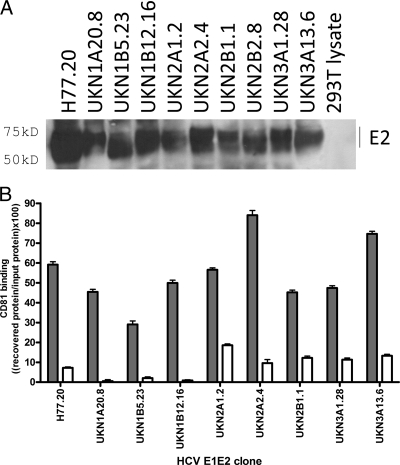
To validate the E1E2 patient clones selected for study, E1E2 proteins present in cell lysates of retroviral pseudoparticle (HCVpp)-producing HEK 293T cells (3) were analyzed by nonreducing Western blotting with a mixture of two broadly reactive anti-E2 monoclonal antibodies (Fig. 2). In agreement with previous studies (11), all of the clones generated detectable monomeric E2 protein with a molecular mass of approximately 60 kDa. Higher-molecular-mass aggregates, corresponding to incorrectly folded proteins, also were observed; importantly, densitometry indicated that the relative proportion of monomer to aggregated protein was similar for all proteins included in the study (data not shown). To determine if the expressed proteins were correctly folded, they were captured onto GNA, and their reactivity to a panel of conformation-sensitive human monoclonal antibodies was assessed (Table 2). Each of the proteins reacted with at least seven of these antibodies, demonstrating the presence of correctly folded E1E2 protein. These proteins form noncovalent heterodimers, as demonstrated by radiolabeled immunoprecipitations (data no shown). A pulldown assay using a GST-CD81 large extracellular loop (LEL) fusion protein also was performed (Fig. 2B). This interaction is dependent on the correct conformation of E2 (17). CD81-LEL was able to pull down E2 in all cell lysates tested, further demonstrating the correct conformation of the different patient-isolated E1E2 glycoproteins.
Fig. 2.
Analysis of protein expression of genetically diverse functional E1E2 proteins. (A) E1E2 constructs were expressed in HEK 293T cells and analyzed by nonreducing Western blotting, using a mixture of the broadly reactive monoclonal antibodies ALP98 and AP33 for detection. Monomeric E2 protein was observed as a 60-kDa monomer for all samples. (B) Binding of glycoproteins to CD81 was analyzed by pulldown with a GST-CD81 fusion protein. Proteins were revealed using Western blotting with a mixture of the antibodies AP33 and ALP98, and relative quantities of E2 protein were assessed by densitometry. Data are presented as a proportion of the recovery of input protein. All E1E2 proteins, independently of genotype, interacted with GST-CD81 LEL (gray bars), unlike the control GST (white bars). The recovery of E2 varied between 30 and 85% of input protein. Sample UKN2A2.8 was not included in this analysis.
Table 2.
Reactivity of well-defined human monoclonal antibodies to patient-isolated E1E2 proteins expressed in HEK 293T cellsa
| MAb | E1E2 glycoprotein sample |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| H77.20 | UKN |
|||||||||
| 1A20.8 | 1B12.16 | 1B5.23 | 2A2.4 | 2A1.2 | 2B1.1 | 2B2.8 | 3A1.28 | 3A13.6 | ||
| CBH2 | − | − | +++ | +++ | ++ | ++ | +++ | ++ | − | − |
| CBH5 | ++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | + | ++ |
| CBH7 | +++ | +++ | +++ | +++ | − | +++ | +++ | +++ | ++ | + |
| A8 | ++ | ++ | +++ | +++ | + | + | ++ | + | ++ | ++ |
| AR3A | +++ | +++ | +++ | +++ | + | +++ | +++ | ++ | ++ | + |
| AR3B | +++ | +++ | +++ | +++ | ++ | +++ | +++ | +++ | ++ | + |
| AR3C | +++ | +++ | +++ | +++ | ++ | ++ | +++ | + | − | + |
| 1:7 | ++ | +++ | ++ | + | + | ++ | ++ | + | ++ | + |
| L1 | − | + | + | − | − | − | + | − | − | − |
| AR1A | +++ | +++ | − | − | − | − | − | − | − | − |
| AR1B | +++ | +++ | + | + | − | − | ++ | − | − | − |
| AR2A | +++ | − | − | − | − | + | + | − | − | − |
| CBH4B | + | + | +++ | +++ | ++ | + | +++ | +++ | − | +++ |
| CBH4D | − | − | + | +++ | − | + | +++ | − | − | ++ |
| CBH4G | − | + | +++ | +++ | +++ | +++ | +++ | +++ | + | +++ |
| CBH8 | − | − | +++ | +++ | − | + | ++ | ++ | − | + |
Each of the patient isolates reacted with at least seven of these antibodies, demonstrating that these isolates are correctly folded and have accessible epitopes for anti-E1/E2 antibodies.
Human serum antibodies exhibit genotype-restricted binding to E1E2 glycoproteins.
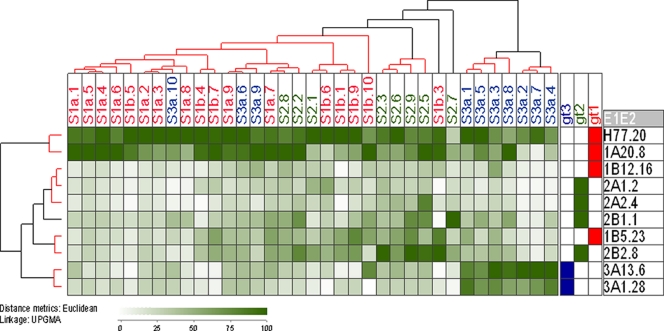
The reactivity of a panel of human sera from individuals chronically infected with HCV genotype 1, 2, or 3 to the E1E2 glycoprotein panel was assessed by GNA capture enzyme immunoassay (EIA). To ensure that valid comparisons between proteins could be made, saturating amounts of target glycoprotein were used in the assays. This was achieved by performing serial dilutions of the GNA capture molecule and dose-binding curves of the E1E2 cell lysates, detecting bound E1E2 with cross-reactive MAbs. The saturation of E1E2 capture occurred at 150 ng ml−1 GNA (data not shown). To take into account different titers of anti-E1E2 antibodies in each of the patients' sera, the binding reactivity of the sera to each protein was calculated as a binding percentage by calculating the binding percentage for each sample as (OD/highest sample OD for that serum) × 100, where OD is optical density. A two-way comparison of proteins and serum binding then was performed by using the percent reactivity as input data for a Euclidean distance analysis. Heat plots and hierarchical trees were generated from the resulting Euclidean distance matrices (Fig. 3). To confirm that serum correctly represented the binding properties of the IgG component of sera in these assays, preliminary assays were performed with selected sera and purified polyclonal antibodies in parallel. Equivalent patterns of reactivity were observed with both sample types (data not shown).
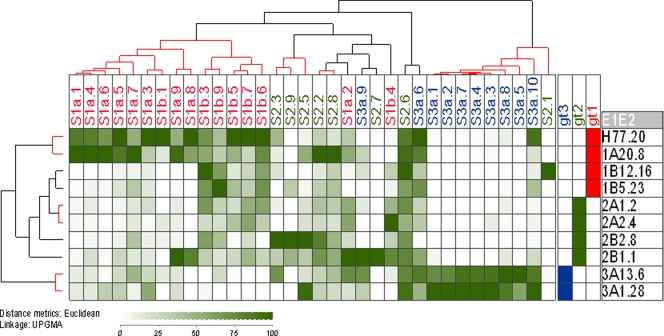
Fig. 3.
Cross-reactivity of the human polyclonal antibody response to E1E2 proteins representing genotypes 1 to 3. E1E2 proteins expressed in human cells were captured on GNA-coated ELISA plate wells and probed with human serum samples. Patient sera are listed across the top of the matrix, with E1E2 clones labeled down the side of the matrix along with their genotype designation. Blue corresponds to genotype 1 samples, green to genotype 2, and red to genotype 3. Normalized binding data were analyzed using JExpress (Molmine). Increasing reactivity to each protein is indicated by the increasing darkness of green color. The serum reactivity clusters (indicated as a hierarchical cluster above the matrix) were analyzed by calculating Euclidean distances for the data set and then calculating the unweighted pair-group mean averages (UPGMA) between groups. The confidence of clustering was determined with the program Heatmap (Los Alamos National Laboratory). Clusters of samples determined to have greater than 90% probability of multiscale bootstrap resampling are indicated by red highlighted dendrograms. Reactivity to homologous genotype protein was generally at the highest level, although cross-reactivity also was observed for proteins of other genotypes. This demonstrated that the specificity of the antibody response is restricted by the subtype of infection. The antigenic similarity of E1E2 proteins was plotted as a dendrogram (indicated on the left side of the reactivity matrix). These reactivity data broadly defined that the antigenic properties are shared between samples isolated from the same genotype. An important exception to this was the genotype 1b samples, which did not cluster with genotype 1a but clustered with genotype 2 samples based on their antibody reactivity.
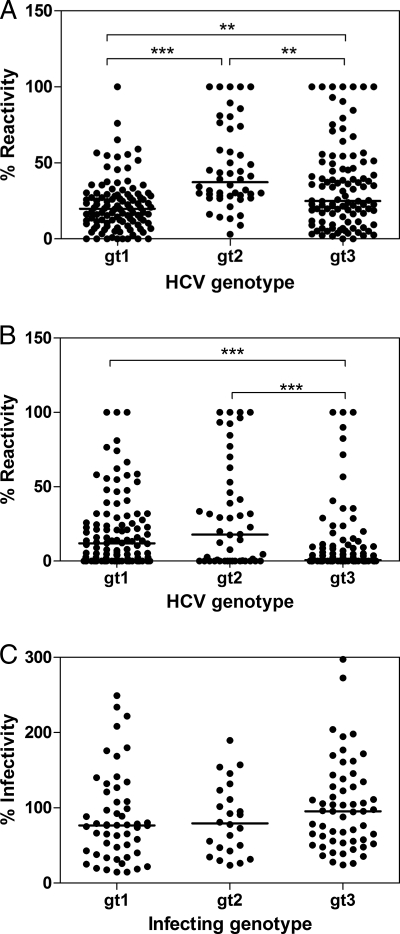
The highest reactivity generally was observed between serum and E1E2 obtained from the homologous genotype, suggesting that the majority of the serum IgG samples were targeting genotype-restricted epitopes. However, reactivity to heterologous E1E2 also was observed, indicating the presence of antibodies recognizing cross-reactive epitopes. Hierarchical cluster analysis of the data revealed that many of the sera isolated from infections with the same genotype clustered together. There were two antibody clusters exclusively from genotype 3 infections, one cluster containing mainly genotype 2, and one large cluster representing the majority of sera derived from the genotype 1 infections. However, there were some notable exceptions to this trend. For example, three sera obtained from individuals infected with genotype 3 (S3a.6, S3a.9, and S3a.10) showed the greatest antibody reactivity to genotype 1 E1E2 and lower reactivity to homologous genotype 3 and therefore clustered with genotype 1 sera. Some sera contained more broadly cross-reactive antibodies; this was most evident for those sera obtained from individuals with genotype 2 infection as well as two sera (S1b.3 and S1b.10) derived from individuals with genotype 1 infection. The analysis of the median heterologous reactivity of the sera (Fig. 4A) showed decreasing levels of cross-reactivity in the order genotype 2 > genotype 3 > genotype 1. Taken together, these data indicated that the antibody response to the E1 and E2 proteins elicited in different genotypes is determined, in general, by the genotype of the infecting virus, but that some cross-reactive antibodies are produced in all infections.
Fig. 4.
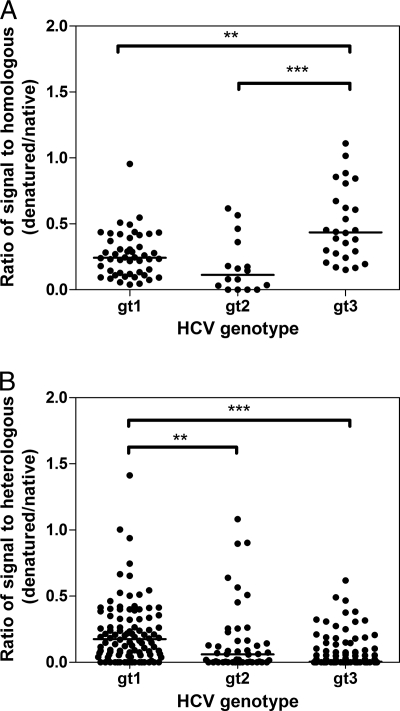
Cross-genotype reactivity and neutralization by antibodies isolated from patient sera. The normalized reactivities of antibodies, present in the sera isolated from individuals with genotype 1, 2, and 3 infection, to native (A) and denatured (B) heterologous (cross-genotype) HCV envelope glycoproteins were plotted. (C) Similarly, the percentage of the infectivity of HCVpp supplemented with genotype 1 to 3 E1E2 in the presence of 25 μg ml−1 IgG obtained from patients with a heterologous cross-genotype also were plotted. Differences in the median values were assessed using the Kruskal-Wallis test with Dunn's correction for multiple comparisons. **, P < 0.01; ***, P < 0.001. Each data point represents the normalized reactivity of one serum sample with one specific E1E2 sample of heterologous genotype. Each combination of serum IgG and E1E2 proteins was assessed. Genotype 2 infections resulted in significantly greater cross-genotype reactivity to heterologous proteins than either genotype 1 or 3. However, this did not result in the broader neutralization of HCV pseudoparticles bearing heterologous glycoproteins by antibodies generated in genotype 2 infections.
We also considered the antigenic relationships between the different E1E2 proteins, as defined by their serum reactivity profiles. These relationships are represented as a dendrogram to the left of the matrix in Fig. 3. Proteins generally clustered according to subtype. However, the two genotype 1b proteins did not form a distinct cluster based on their antigenic properties. UKN1B12.16 showed similar patterns of reactivity to genotype 2a proteins, while UKN1B5.23 was more closely related to genotype 2b. The statistical analysis of these antigenic similarities revealed four discrete groups, with genotype 1a and genotype 3a forming distinct antigenic groups and two clusters containing UKN1B5.23/UKN2B2.8 and UKN1B12.16/UKN2A1.2/UKN2A2.4, respectively. Thus, these data suggest that the viral subtype generally corresponds to the antigenic serotype, although some proteins possess epitopes shared across genotypes, which leads to their clustering with heterologous proteins.
Human polyclonal IgG demonstrates greater type specificity for denatured E1E2 proteins.
Euclidian distance analyses showed that serum reactivity to denatured proteins was far more restricted (see Fig. 6). The majority of sera clustered according to infecting genotype, with many having little reactivity to heterologous denatured protein. Genotype 1 and 2 infections generally yielded antibodies with greater cross-reactivity than genotype 3 infections, and this was statistically significant in a comparative analysis of the median intergenotypic reactivity (Fig. 4b). In addition, some genotype 1 and genotype 3 infections resulted in antibodies with some degree of cross-reactivity (for example, S1a.2, S1b.3, S3a.6, and S3a.10). One sample, S2.6, retained broad cross-reactivity to all glycoproteins tested, demonstrating that conserved linear epitope(s) are recognized by this polyclonal antibody.
Fig. 6.
Effect of denaturation on reactivity of serum IgG to homologous and heterologous E1E2 proteins. Sera, obtained from individuals infected with different HCV genotypes, were used to detect homologous (same genotype) (A) and heterologous (cross-genotype) (B) E1E2 proteins representing genotypes 1 to 3. The ratio of absorbance values obtained for each serum when detecting native and denatured E1E2 was plotted, and differences in the median values were compared using the Kruskal-Wallis test with Dunn's correction for multiple comparisons. **, P < 0.01; ***, P < 0.001. Each data point represents the normalized reactivity of one serum sample with one specific E1E2 sample of either homologous genotype (A) or heterologous genotype (B). Each combination of serum and E1E2 proteins was plotted. The median reactivity was reduced for all samples when the target E1E2 was denatured. Genotype 3 infections retained a larger amount of their homologous reactivity while losing a larger amount of their heterologous reactivity.
Denatured E1E2 proteins, clustered based on their recognition by the different sera, fell into subtype-specific groups (Fig. 5), which were more obvious than those in the analysis of native protein. While proteins from the same subtype clustered together (e.g., subtypes 1a, 2a, and 3a), proteins of the same genotype did not always cluster together. This is best illustrated by the subtype 1b proteins, which form a distinct group within the genotype 2 grouping. Thus, in general, conserved epitopes within E1E2 are conformation sensitive, while linear epitopes have more restricted recognition.
Fig. 5.
Cross-reactivity of the human polyclonal antibody response to denatured E1E2 proteins representing genotypes 1 to 3. E1E2 proteins expressed in human cells were denatured as previously described (58), captured on GNA-coated ELISA plate wells, and probed with human serum samples. Patient sera are listed across the top of the matrix with E1E2 clones labeled down the side of the matrix, along with their genotype designation. Blue corresponds to genotype 1 samples, green to genotype 2, and red to genotype 3. Reactivity was analyzed as described in the legend for Fig. 3. Reactivity generally was subtype restricted, suggesting that linear epitopes recognized by the polyclonal antibody response vary between subtypes. Some notable exceptions were sera S2.6 and S3a.6, which displayed greater cross-reactivity. Sample S1b.10 had no reactivity to any protein in this assay, and as such it was omitted from this analysis. The clustering of the antigenic properties of the denatured E1E2 proteins was performed similarly to that for the patient antibody samples. Each of the genotypes and subtypes clustered, demonstrating that common antigenic properties are shared by E1E2 isolates of the same genotype.
The loss of antibody reactivity following denaturation provides insight into the relative contribution of antibodies recognizing linear and conformational epitopes to the overall reactivity. The denaturation of E1E2 frequently reduced serum reactivity, and this was more noticeable for the heterologous (Fig. 6B) than the homologous (Fig. 6A) antibody response. This suggested that a large proportion of antibodies targeted conformation-sensitive epitopes, especially those conserved across genotypes. Also, the reduction in antibody reactivity differed between genotypes. Sera from genotype 1 infections maintained more of their heterologous and homologous reactivity following the denaturation of the target proteins than sera obtained from genotype 2. Sera from genotype 3 infections lost most of their reactivity to heterologous proteins following denaturation, demonstrating that a greater proportion of their cross-reactive antibodies targeted conformational epitopes. However, a minority of sera retained most of their reactivity to heterologous denatured E1E2, suggesting that the majority of cross-reactive antibodies in these samples recognized conserved linear epitopes.
Genotype does not correspond to neutralization serotype.
To investigate if the genotype-restricted antibody reactivity to E1 and E2 resulted in restricted neutralization serotypes, a retroviral pseudotype model of infection was used to determine the neutralization potential of purified patient antibodies. As a control, MLV pseudotyped with vesicular stomatitis virus (VSV) G glycoproteins was included. To perform these analyses, IgG was purified from sera using protein G affinity chromatography. IgG preparations that demonstrated any inhibitory effect on these control pseudoparticles were omitted from these analyses (data not shown).
In contrast to the reactivity experiments, the association between the genotype of infection and neutralization serotype was minimal (Fig. 7). The majority of IgG preparations neutralized HCVpp incorporating heterologous E1E2. These IgG samples were grouped into three clusters that were supported by greater than 90% confidence values. The largest group possessed representatives of all three genotypes and neutralized some of the E1E2 isolates. A second cluster of IgG samples (S1a.7, S1a.1, and S1a.4) potently neutralized the entry of diverse E1E2 isolates. These broadly reactive IgG samples achieved greater than 90% inhibition of some HCVpp and neutralized all patient pseudoparticles to greater than 50% at a concentration of 25 μg ml−1. One of the most striking features of this analysis was the presence of a third group of antibodies that had little neutralizing activity and that enhanced the entry of some patient-derived HCVpp. This group included samples S1a.6, S1a.9, S3a.4, S3a.5, and S3a.7. The enhancement of infection observed by these IgG samples typically ranged from 110 to 200%. However, one IgG sample (S3a.9) had very little neutralizing potency and enhanced the infection of some of the HCVpp by almost 3-fold. In essence, a minority of patient IgG samples were capable of broad heterologous neutralization, a larger group was capable of more restricted neutralization, and a third minor group was poorly neutralizing and often enhanced infection mediated by some patient isolates.
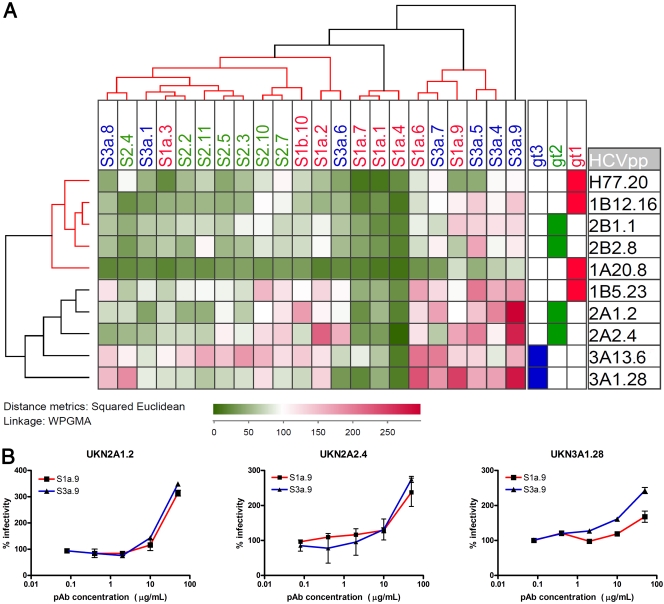
Fig. 7.
Effect of patient-isolated polyclonal antibodies on the entry of pseudoparticles bearing diverse HCV E1E2 glycoproteins. (A) Neutralization was performed on each of the retroviral pseudoparticle preparations, using 25 μg ml−1 IgG purified from patients' sera. Patient IgG are listed across the top of the matrix, with E1E2 clones labeled down the side of the matrix along with their genotype designation. Within the matrix, the increasing darkness of green color represents increasing neutralization of entry, while red indicates enhancement. White indicates no effect on pseudoparticle entry (100% infectivity). Baseline values in the presence of IgG purified from a pool of HCV-negative donors ranged from 1,626 to 28,780 relative light units (RLU) dependent on the HCVpp clone, with a mean value of 326 RLU for pseudoparticles possessing no glycoprotein. The IgG neutralization clusters for each serum IgG and the neutralization phenotype of each E1E2 (indicated as a hierarchical cluster above and to the left of the matrix, respectively) were analyzed by calculating Euclidean distances for the data set and then calculating the unweighted pair-group mean averages between groups. Confidence of clustering was determined with the program Heatmap (Los Alamos National Laboratory). Three significant groups of antibodies were defined. These clusters are highlighted as red branches of the dendrogram. The first group neutralized a subset of the diverse pseudoparticles, a second group potently neutralized diverse pseudoparticles representing all three genotypes, and a third group showed minimal neutralization and frequent enhancement of infection. Two discrete groups of E1E2 proteins were apparent: a neutralization-sensitive group and a second group that exhibited more limited neutralization. (B) Dose-dependent enhancement of infection by polyclonal antibody preparations purified from chronic HCV infections was demonstrated. Antibody preparations S1a.9 (■) and S3a9 (▴) were selected as representatives of the group of enhancing pAb. Increasing concentrations of purified IgG preparations were incubated with HCV pseudoparticles bearing patient-isolated E1E2 glycoproteins before infecting Huh7 cells. Data are expressed as a proportion of the infectivity observed in the presence of polyclonal IgG isolated from a pool of HCV-negative donors. In each case, dose-dependent enhancement was observed with these combinations of purified IgG and HCV pseudoparticles.
The cluster analysis of the E1E2 proteins based on their neutralization phenotype also was performed. Two main groups emerged. One group, containing isolates H77.20, UKN1A20.8, UKN1B12.16, UKN2B1.1, and UKN2B2.8, was neutralization sensitive. A second group, containing isolates UKN2A1.2, UKN2A2.4, UKN3A1.28, UKN3A13.6, and UKN1B5.23, was more refractive to neutralization and even enhanced by some IgG preparations. The statistical analysis of these groups revealed that they had 94 and 89% clustering probability, respectively.
Neutralization and enhancement of HCVcc infection.
To further support the results obtained with diverse HCVpp, the JFH1 HCVcc clone was treated with selected purified antibody preparations isolated from genotype 1a and 3a infections. The results broadly confirmed the findings with HCVpp (Fig. 8). One of the antibody preparations that enhanced HCVpp (S1a.9) showed a dose-dependent enhancement of JFH1 infection, while another (S3a.9) had no effect on infectivity. Two samples that neither neutralized nor enhanced HCVpp entry (S1a.2 and S1a.3) also had no effect on JFH1 infection, while two of the broadly neutralizing polyclonal antibodies (S1a.4 and S1a.7) both neutralized JFH1 infectivity in a dose-dependent manner.
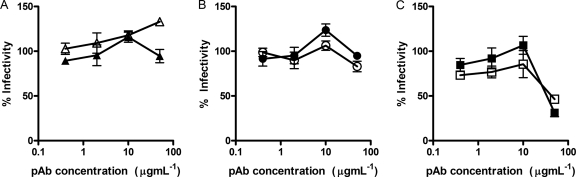
Fig. 8.
Effect of purified polyclonal antibodies on replication of HCVcc strain JFH1. Two antibody preparations were selected to represent each of the three different HCVpp neutralization phenotypes identified for patient-isolated IgG. (A) HCVpp enhancing antibodies; (B) antibodies with restricted neutralizing activity; (C) broadly neutralizing antibodies. Purified antibody preparations were incubated at the indicated concentrations with 100 FFU of JFH1 virus before infecting Huh7.5 cells. Infection was determined after 48 h by staining for the presence of core protein in infected cells. Data are presented as a percentage of an uninhibited control. All reactions were performed in triplicate. Some evidence of dose-dependent enhancement was observed for IgG isolated from patient S1a.9 (Δ) but not from patient S3a.9 (▴). Both preparations of restricted neutralizing, nonenhancing IgG, Sa1.2 (●) and Sa1.3 (○), had little effect on JFH1 infectivity. In contrast, potent, broadly neutralizing antibodies S1a.4 (□) and S1a.7 (■) both neutralized infectivity in a dose-dependent manner.
Taken together, these data suggested that the neutralization serotype of E1E2 proteins is not strictly defined by their genotype, and different patient-isolated E1E2 proteins demonstrate markedly different sensitivities to neutralization by IgG isolated from patients' sera.
DISCUSSION
This study characterized the cross-reactivity and the breadth of neutralization of polyclonal antibodies produced in natural HCV infections, together with the neutralization sensitivity and antigenic structure of a panel of independently isolated functional E1E2 clones representing diverse genotypes of HCV.
An important requirement for studies of antibody recognition of the HCV glycoproteins is a robust model of the target antigen. Previous studies of antibody reactivity have often utilized HCV E1E2 clones that were not checked for correct folding. We selected E1E2 patient-isolated proteins that had been shown previously to confer entry in the HCVpp model of entry. To ensure that these functional clones were presented in their correct native conformation in our assays, we assessed binding to a large panel of human antibodies recognizing conformation-sensitive epitopes as well as the conformation-sensitive binding to the large extracellular loop of human CD81 (17). Immunoprecipitation also demonstrated that heterodimers formed for these expressed proteins (data not shown), which is consistent with previous reports of functional glycoproteins (41). These analyses confirmed that the global conformation of the proteins was correct in these assays.
The comparative analysis of the cross-reactivity of serum samples is complicated by the fact that HCV-specific antibodies comprise only a fraction of the total antibody response and titers of the HCV-specific antibodies could, therefore, differ between samples. To compensate for this, we utilized a method of normalization first applied to the comparative analysis of HIV-1 serum antibody reactivity, where differences in antibody titer are negated by expressing each binding signal as a percentage of the highest value observed for a particular serum sample (39). It also was important to ensure that equal amounts of E1E2 glycoprotein were used in these assays. All of the glycoproteins used in the assays first were shown to produce similar ratios of correctly folded monomeric to aggregated E2, and saturating amounts of these proteins were captured using GNA. The HCVpp neutralization assay was used to determine the neutralizing potency of each serum-derived polyclonal IgG (2). A variety of intergenotypic cell culture infectious clones of HCV (HCVcc) are now available (8, 18, 19); however, many of these clones possess mutations in the envelope glycoproteins acquired during the process of adaptation to culture (12, 62, 65), and there are data emerging that some of these mutations can affect neutralization sensitivity (20). The HCVpp assay permits better assessment of neutralizing antibody responses against uncultured, nonadapted E1E2, and a larger number of native functional E1E2 clones are available for these studies. We also have demonstrated previously that chimeric HCVcc bearing a patient-isolated genotype 2 E1E2 protein is more easily neutralized by human monoclonal antibodies than the equivalent HCVpp (44). Therefore, the HCVpp assay provides a stringent assessment of neutralization in the context of ex vivo-derived E1E2. Having chosen to utilize the HCVpp to assay for cross-neutralization, it was necessary to ensure that different HCVpp preparations contained similar amounts of viral particles. To address this, we ensured that HCVpp preparations contained equivalent amounts of MLV core protein. Although still an approximation of the relative number of HCVpp, this approach has been widely used to normalize amounts of HCVpp (26, 33), and therefore it ensures that our data are comparable to previously published data.
Our data highlighted an association between antigenic serotype and genotype but not between neutralization serotype and genotype. While antigenic serotype correlated with infecting subtype, this association was more marked when these sera were recognizing denatured E1E2. In addition, the overall reactivity to denatured protein was reduced compared to that of native protein. Taken together, these findings suggested that a major component of the antibody response is directed toward conformational epitopes and that linear epitopes are, in general, genotype restricted. Consistently with previous analyses, genotype 1 and 3 infections yielded antibodies that preferentially recognized homologous E1E2 (22). However, in genotype 2 infections this association was less clear. This suggests that in genotype 2, E1E2 type-restricted linear epitopes are less well exposed or are less immunogenic than those present in genotype 1 and 3 E1E2. This was supported by the finding that the denaturation of homologous E1E2 resulted in the greatest loss of reactivity for genotype 2 infections. While it cannot be discounted that some patients might have been previously infected with multiple genotypes of HCV, which led to a broader antibody repertoire, the detection of multiple infections is reported to be rare (28). Similarly, the restricted recognition of denatured E1E2 would argue against this possibility.
Interestingly, the retained homologous reactivity to denatured E1E2 protein was greatest for genotype 3 infections. We have described previously a genotype-restricted variable region in the E2 protein that displays length polymorphisms around amino acids 570 to 580 in the HCV polyprotein (7). This region is under positive selection in genotype 3 viruses, and it is plausible that this region is an immunodominant linear epitope in genotype 3 viruses. Genotype-restricted linear epitopes previously have been reported for studies on the serum antibody response to the HIV-1 gp120 glycoprotein (40).
Sera that harbored IgG with the broadest neutralization failed to recognize heterologous denatured E1E2, indicating that the breadth of neutralization most likely was attributable to antibodies targeting conformation-sensitive epitopes. For example, serum S1a.1 and S1a.4, two of the most potent and broadly neutralizing IgG used in this study, failed to recognize many of the denatured proteins, e.g., UKN2B2.8 and UKN1B12.16, which both were highly sensitive to neutralization by the same IgG preparations. Again, the lack of cross-reactivity to heterologous denatured proteins shown by these sera also suggests that the broad neutralization observed is unlikely to be due to infection by multiple genotypes, as one might expect that infection by multiple genotypes would elicit multiple antibody specificities that would recognize linear epitopes present on more than one genotype.
Few naturally occurring antibodies to E1 have been described (35). Instead, the major targets for naturally occurring neutralizing antibodies reside in E2. The first to be described was the first hypervariable region of E2 (16) (31, 52, 66). However, this region tolerates extensive genetic variability, and therefore antibodies targeting HVR1 tend to be highly restricted (15). The second major target for neutralizing antibodies is the CD81 binding site. Broadly conserved, linear neutralization epitopes, for example, those recognized by MAb AP33 and MAb 3/11, have been described (43, 58), but antibodies targeting these epitopes are rare in natural infection (56). The majority of neutralizing antibodies targeting the CD81 binding site recognize conformation-sensitive epitopes located on discontinuous regions of E2 (23, 27, 44, 46). It remains to be demonstrated if the broadly neutralizing antibodies described here recognize epitopes similar to those previously described for CD81 binding site-targeted MAbs; it is unlikely that they recognize the same epitope as MAb AP33, as none reacted to a peptide recognized by MAb AP33 (58 and data not shown). When comparing the breadth of neutralization with cross-reactivity, some serum antibody samples, such as S1a.7, cross-reacted with native and denatured E1E2 and also exhibited broad neutralization. Studies are under way to determine if this serum harbors IgGs that recognize novel conserved linear neutralizing epitopes.
The observed disparity in the neutralization sensitivity of E1E2 proteins is an important issue. It has been reported previously that heterologous neutralizing antibodies are frequently isolated in chronic HCV infection (30, 34), yet of the 22 sera we analyzed, only 3 showed potent neutralization against all of the E1E2 proteins tested. This apparent discrepancy can be explained, at least in part, by the fact that previous studies have tended to focus on the ability of sera to neutralize relatively small numbers of heterologous E1E2 and often have relied on molecular clones such as H77c. Our data show that some E1E2s, such as H77c, are particularly susceptible to neutralization. This highlights the importance of utilizing panels that contain E1E2 proteins representing the extremes of neutralization sensitivity in studies testing the likely efficacy of therapeutic antibodies or antibody-based vaccines. In this context it will also be important to assess the neutralizing phenotype of E1E2 cell culture-adapted infectious clones of HCV. Differences in the neutralization properties of primary viral isolates and cell culture-adapted isolates are well documented in HIV-1, where laboratory-adapted strains have a greater sensitivity to antibody neutralization (32, 38). Furthermore, recent studies have demonstrated that a point mutation in the E2 coding region (G451R) of an adapted strain of JFH-1 renders it more sensitive to neutralization by MAbs targeting the CD81 binding site and patient sera (20).
Serum-associated factors, particularly high-density lipoprotein, have been shown to enhance infection and interfere with neutralizing antibodies (4, 13, 59, 60). For this reason, immunoglobulins were purified from sera for the neutralization studies. Using purified IgG, we found that some sera harbored antibodies that enhanced the infection of specific HCVpp. The observed enhancement may be facilitated by neonatal FcR (FcRn) on the surface of Huh7 cells (50). FcR-mediated enhancement of HCV infection has been proposed previously (36), and it is possible that this mechanism will enhance infectivity into hepatocytes. Alternatively, FcR-independent enhancement might arise through the antibody-mediated dimerization of viral glycoproteins, as has been reported for HIV-1 gp120 (51). The mechanism of enhancement of HCV entry remains to be elucidated, as do the specific epitopes that are targeted by these enhancing antibodies. It will be important to ensure that future antibody-based vaccines avoid eliciting any infection-enhancing antibody responses.
In conclusion, these studies have discovered that the genetic background of the envelope glycoproteins of HCV defines their immunogenicity and antigenicity. However, the neutralization serotype is not defined by the viral genotype. This has implications for the design of broadly acting antibody therapeutics and vaccines. In addition, diverse HCV glycoproteins differed in their relative susceptibility to antibody-mediated neutralization, and some sera harbored antibodies capable of enhancing infection. Clearly, the neutralizing potential of a particular serum is defined by a complex interplay between the titer and specificity of the neutralizing response balanced against the relative sensitivity of the virus isolate and the contribution of enhancing serum factors and antibodies. Certainly, cross-reactive antibodies were present in some individuals with chronic infection, but these were far less frequent than previously reported in the literature (2, 30). Further studies of the polyclonal antibody response in acute and chronic infection are needed to better understand determinants associated with broad neutralization and the enhancement of infection. It will be essential for future studies to use patient-derived glycoproteins to ensure the accurate assessment of neutralization potency.
ACKNOWLEDGMENTS
We thank Francois-Loic Cosset for the provision of the HCV pseudoparticle system, Charles Rice for the provision of Huh7.5 cells and MAb 9E10, Takaji Wakita for the JFH1 HCVcc strain, and Mats Persson, Steven Foung, Dennis Burton, and Arvind Patel for the provision of anti-E2 monoclonal antibodies.
This work was supported by the MRC (G0801169), the University of Nottingham Biomedical Research Committee, and EU contract MRTN-CT-2006-035599.
Footnotes
Published ahead of print on 16 February 2011.
REFERENCES
- 1. Alter H. J., Seeff L. B. 2000. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin. Liver Dis. 20:17–35 [DOI] [PubMed] [Google Scholar]
- 2. Bartosch B., et al. 2003. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc. Natl. Acad. Sci. U. S. A. 100:14199–14204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartosch B., Dubuisson J., Cosset F. L. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartosch B., et al. 2005. An interplay between hypervariable region 1 of the hepatitis C virus E2 glycoprotein, the scavenger receptor BI, and high-density lipoprotein promotes both enhancement of infection and protection against neutralizing antibodies. J. Virol. 79:8217–8229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartosch B., et al. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624–41630 [DOI] [PubMed] [Google Scholar]
- 6. Blight K. J., McKeating J. A., Rice C. M. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brown R. J., et al. 2007. Cross-genotype characterization of genetic diversity and molecular adaptation in hepatitis C virus envelope glycoprotein genes. J. Gen. Virol. 88:458–469 [DOI] [PubMed] [Google Scholar]
- 8. Bungyoku Y., et al. 2009. Efficient production of infectious hepatitis C virus with adaptive mutations in cultured hepatoma cells. J. Gen. Virol. 90:1681–1691 [DOI] [PubMed] [Google Scholar]
- 9. Clayton R. F., et al. 2002. Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles. J. Virol. 76:7672–7682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cocquerel L., Kuo C. C., Dubuisson J., Levy S. 2003. CD81-dependent binding of hepatitis C virus E1E2 heterodimers. J. Virol. 77:10677–10683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cocquerel L., et al. 2003. Recognition of native hepatitis C virus E1E2 heterodimers by a human monoclonal antibody. J. Virol. 77:1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Delgrange D., et al. 2007. Robust production of infectious viral particles in Huh-7 cells by introducing mutations in hepatitis C virus structural proteins. J. Gen. Virol. 88:2495–2503 [DOI] [PubMed] [Google Scholar]
- 13. Dreux M., et al. 2006. High density lipoprotein inhibits hepatitis C virus-neutralizing antibodies by stimulating cell entry via activation of the scavenger receptor BI. J. Biol. Chem. 281:18285–18295 [DOI] [PubMed] [Google Scholar]
- 14. Drummer H. E., Boo I., Maerz A. L., Poumbourios P. 2006. A conserved Gly436-Trp-Leu-Ala-Gly-Leu-Phe-Tyr motif in hepatitis C virus glycoprotein E2 is a determinant of CD81 binding and viral entry. J. Virol. 80:7844–7853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Esumi M., et al. 1999. Experimental vaccine activities of recombinant E1 and E2 glycoproteins and hypervariable region 1 peptides of hepatitis C virus in chimpanzees. Arch. Virol. 144:973–980 [DOI] [PubMed] [Google Scholar]
- 16. Farci P., et al. 1996. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc. Natl. Acad. Sci. U. S. A. 93:15394–15399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Flint M., et al. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235–6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gottwein J. M., et al. 2007. Robust hepatitis C genotype 3a cell culture releasing adapted intergenotypic 3a/2a (S52/JFH1) viruses. Gastroenterology 133:1614–1626 [DOI] [PubMed] [Google Scholar]
- 19. Gottwein J. M., et al. 2009. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology 49:364–377 [DOI] [PubMed] [Google Scholar]
- 20. Grove J., et al. 2008. Identification of a residue in hepatitis C virus E2 glycoprotein that determines scavenger receptor BI and CD81 receptor dependency and sensitivity to neutralizing antibodies. J. Virol. 82:12020–12029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hadlock K. G., et al. 2000. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J. Virol. 74:10407–10416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hamed M. R., et al. 2008. Association of antibodies to hepatitis C virus glycoproteins 1 and 2 (anti-E1E2) with HCV disease. J. Viral Hepat. 15:339–345 [DOI] [PubMed] [Google Scholar]
- 23. Johansson D. X., et al. 2007. Human combinatorial libraries yield rare antibodies that broadly neutralize hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 104:16269–16274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keck Z. Y., et al. 2004. Hepatitis C virus E2 has three immunogenic domains containing conformational epitopes with distinct properties and biological functions. J. Virol. 78:9224–9232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lagging L. M., et al. 2002. Neutralization of pseudotyped vesicular stomatitis virus expressing hepatitis C virus envelope glycoprotein 1 or 2 by serum from patients. J. Infect. Dis. 185:1165–1169 [DOI] [PubMed] [Google Scholar]
- 26. Lavillette D., et al. 2005. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology 41:265–274 [DOI] [PubMed] [Google Scholar]
- 27. Law M., et al. 2008. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat. Med. 14:25–27 [DOI] [PubMed] [Google Scholar]
- 28. Lee J.-H., Roth W. K., Zeuzem S. 1997. Evaluation and comparison of different hepatitis C virus genotyping and serotyping assays. J. Hepatol. 26:1001–1009 [DOI] [PubMed] [Google Scholar]
- 29. Lindenbach B. D., et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 30. Logvinoff C., et al. 2004. Neutralizing antibody response during acute and chronic hepatitis C virus infection. Proc. Natl. Acad. Sci. U. S. A. 101:10149–10154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Majid A., et al. 1999. Ontogeny of hepatitis C virus (HCV) hypervariable region 1 (HVR1) heterogeneity and HVR1 antibody responses over a 3 year period in a patient infected with HCV type 2b. J. Gen. Virol. 80:317–325 [DOI] [PubMed] [Google Scholar]
- 32. Mascola J. R., et al. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. The National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. J. Infect. Dis. 173:340–348 [DOI] [PubMed] [Google Scholar]
- 33. McKeating J. A., et al. 2004. Diverse hepatitis C virus glycoproteins mediate viral infection in a CD81-dependent manner. J. Virol. 78:8496–8505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meunier J. C., et al. 2005. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc. Natl. Acad. Sci. U. S. A. 102:4560–4565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meunier J. C., et al. 2008. Isolation and characterization of broadly neutralizing human monoclonal antibodies to the e1 glycoprotein of hepatitis C virus. J. Virol. 82:966–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meyer K., Ait-Goughoulte M., Keck Z. Y., Foung S., Ray R. 2008. Antibody-dependent enhancement of hepatitis C virus infection. J. Virol. 82:2140–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mohsen A. H. 2001. The epidemiology of hepatitis C in a UK health regional population of 5.12 million. Gut 48:707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Montefiori D. C., et al. 1998. Neutralizing antibodies in sera from macaques infected with chimeric simian-human immunodeficiency virus containing the envelope glycoproteins of either a laboratory-adapted variant or a primary isolate of human immunodeficiency virus type 1. J. Virol. 72:3427–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moore J. P., et al. 1996. Inter- and intraclade neutralization of human immunodeficiency virus type 1: genetic clades do not correspond to neutralization serotypes but partially correspond to gp120 antigenic serotypes. J. Virol. 70:427–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moore J. P., Ho D. D. 1993. Antibodies to discontinuous or conformationally sensitive epitopes on the gp120 glycoprotein of human immunodeficiency virus type 1 are highly prevalent in sera of infected humans. J. Virol. 67:863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Op De Beeck A. O., et al. 2004. Characterization of functional hepatitis C virus envelope glycoproteins. J. Virol. 78:2994–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Owsianka A., Clayton R. F., Loomis-Price L. D., McKeating J. A., Patel A. H. 2001. Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J. Gen. Virol. 82:1877–1883 [DOI] [PubMed] [Google Scholar]
- 43. Owsianka A., et al. 2005. Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J. Virol. 79:11095–11104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Owsianka A. M., et al. 2008. Broadly neutralizing human monoclonal antibodies to the hepatitis C virus E2 glycoprotein. J. Gen. Virol. 89:653–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Owsianka A. M., et al. 2006. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J. Virol. 80:8695–8704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Perotti M., et al. 2007. Identification of a broadly cross-reacting and neutralizing human monoclonal antibody directed against the Hcv\E2 protein. J. Virol. 82:1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pestka J. M., et al. 2007. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc. Natl. Acad. Sci. U. S. A. 104:6025–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reed L. J., Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 24:493–497 [Google Scholar]
- 49. Scarselli E., et al. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schilling R., et al. 2003. Endocytosis of hepatitis B immune globulin into hepatocytes inhibits the secretion of hepatitis B virus surface antigen and virions. J. Virol. 77:8882–8892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schutten M., Andeweg A. C., Rimmelzwaan G. F., Osterhaus A. D. 1997. Modulation of primary human immunodeficiency virus type 1 envelope glycoprotein-mediated entry by human antibodies. J. Gen. Virol. 78:999–1006 [DOI] [PubMed] [Google Scholar]
- 52. Shang D., Zhai W., Allain J. P. 1999. Broadly cross-reactive, high-affinity antibody to hypervariable region 1 of the hepatitis C virus in rabbits. Virology 258:396–405 [DOI] [PubMed] [Google Scholar]
- 53. Simmonds P., et al. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962–973 [DOI] [PubMed] [Google Scholar]
- 54. Simmonds P., et al. 1993. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J. Gen. Virol. 74:2391–2399 [DOI] [PubMed] [Google Scholar]
- 55. Simmonds P., et al. 1994. Identification of genotypes of hepatitis C virus by sequence comparisons in the core, E1 and NS-5 regions. J. Gen. Virol. 75:1053–1061 [DOI] [PubMed] [Google Scholar]
- 56. Tarr A. W., et al. 2007. Determination of the human antibody response to the epitope defined by the hepatitis C virus-neutralizing monoclonal antibody AP33. J. Gen. Virol. 88:2991–3001 [DOI] [PubMed] [Google Scholar]
- 57. Tarr A. W., Owsianka A. M., Szwejk A., Ball J. K., Patel A. H. 2007. Cloning, expression, and functional analysis of patient-derived hepatitis C virus glycoproteins. Methods Mol. Biol. 379:177–197 [DOI] [PubMed] [Google Scholar]
- 58. Tarr A. W., et al. 2006. Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology 43:592–601 [DOI] [PubMed] [Google Scholar]
- 59. Voisset C., et al. 2005. High density lipoproteins facilitate hepatitis C virus entry through the scavenger receptor class B type I. J. Biol. Chem. 280:7793–7799 [DOI] [PubMed] [Google Scholar]
- 60. Voisset C., et al. 2006. High-density lipoproteins reduce the neutralizing effect of hepatitis C virus (HCV)-infected patient antibodies by promoting HCV entry. J. Gen. Virol. 87:2577–2581 [DOI] [PubMed] [Google Scholar]
- 61. Wakita T., et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yi M., Ma Y., Yates J., Lemon S. M. 2007. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J. Virol. 81:629–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Youn J. W., et al. 2005. Sustained E2 antibody response correlates with reduced peak viremia after hepatitis C virus infection in the chimpanzee. Hepatology 42:1429–1436 [DOI] [PubMed] [Google Scholar]
- 64. Zeuzem S. 2001. The kinetics of hepatitis C virus infection. Clin. Liver Dis. 5:917–930 [DOI] [PubMed] [Google Scholar]
- 65. Zhong J., et al. 2006. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J. Virol. 80:11082–11093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zibert A., Kraas W., Meisel H., Jung G., Roggendorf M. 1997. Epitope mapping of antibodies directed against hypervariable region 1 in acute self-limiting and chronic infections due to hepatitis C virus. J. Virol. 71:4123–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]