Abstract
Incoming capsids of herpes simplex virus type 1 (HSV-1) enter the cytosol by fusion of the viral envelopes with host cell membranes and use microtubules and microtubule motors for transport to the nucleus. Upon docking to the nuclear pores, capsids release their genomes into the nucleoplasm. Progeny genomes are replicated in the nucleoplasm and subsequently packaged into newly assembled capsids. The minor capsid protein pUL25 of alphaherpesviruses is required for capsid stabilization after genome packaging and for nuclear targeting of incoming genomes. Here, we show that HSV-1 pUL25 bound to mature capsids within the nucleus and remained capsid associated during assembly and nuclear targeting. Furthermore, we tested potential interactions between parental pUL25 bound to incoming HSV-1 capsids and host factors by competing for such interactions with an experimental excess of cytosolic pUL25. Overexpression of pUL25, GFPUL25, or UL25GFP prior to infection reduced gene expression of HSV-1. Electron microscopy and in situ hybridization studies revealed that an excess of GFPUL25 or UL25GFP prevented efficient nuclear import and/or transcription of parental HSV-1 genomes, but not nuclear targeting of capsids or the uncoating of the incoming genomes at the nuclear pore. Thus, the uncoating of HSV-1 genomes could be uncoupled from their nuclear import and gene expression. Most likely, surplus pUL25 competed with important interactions between the parental capsids, and possibly between authentic capsid-associated pUL25, and cytosolic or nuclear host factors required for functional interaction of the incoming genomes with the nuclear machinery.
INTRODUCTION
Herpes simplex virus type I (HSV-1) is the most thoroughly studied pathogen among the alphaherpesviruses and among the eight human-pathogenic herpesviruses. Its double-stranded DNA genome of 152 kb is packaged into a capsid that is surrounded by 33 capsid-associated and tegument proteins, which in turn are wrapped by a viral envelope (43, 51, 68, 73, 93). HSV-1 assembly commences in the nucleus with the formation of spherical, scaffold-containing procapsids that develop into three types of angularized icosahedral capsids characterized by different sedimentation coefficients, morphologies, protein compositions, and mechanical properties (A, B, and C capsids) (3, 28, 75). The C capsids, which are the heaviest, enclose the viral genome; the B capsids still contain scaffold remnants but no DNA; and the A capsids are empty and lack both scaffold and DNA. After degradation of the scaffold proteins VP21 and VP22a by viral proteases, the procapsids package the viral DNA and mature into C capsids. Predominantly, C capsids leave the nucleus by primary envelopment and de-envelopment at nuclear membranes and acquire their final envelope by secondary envelopment in the cytoplasm (51, 52, 92, 93). B and A capsids most likely result from a failure to initiate or to complete DNA packaging properly and are considered to be dead-end products that are formed during assembly (3, 85, 92).
The major capsid protein VP5 assembles into the 150 hexons of the capsid faces and edges, as well as the pentons located at 11 vertices of the icosahedrons. A dodecameric ring of pUL6 builds the 12th vertex. It provides the portal through which the viral genome is packaged during assembly and which the parental genomes most likely use to leave the capsids during cell entry (28, 55, 57, 90). In addition to UL6, the six HSV-1 genes UL15, UL17, UL25, UL28, UL32, and UL33 are required for HSV-1 DNA cleavage and stable packaging (3, 8, 95). Of their products, pUL6, pUL17, and pUL25 become structural proteins of the virions (68, 88, 92).
Quantitative immunoblot and immunoelectron microscopy studies suggest that pUL25 molecules are added to capsids as genome packaging proceeds and that each vertex accommodates up to 5 copies of pUL25 on the C capsid (13, 56, 75, 87). It remains capsid associated even at high salt concentrations (68, 96). Cryoelectron tomography studies suggest that an elongated C-capsid-specific component (CCSC) at unique vertex-adjacent sites that spans the two penton-adjacent triplexes is actually a heterodimer of pUL17 and pUL25 (13, 92). pUL25 can bind to the major capsid proteins VP5, VP19C, and VP23; the minor capsid proteins pUL17 and pUL6; and the inner tegument protein pUL36 (10, 12, 13, 59, 63, 68, 80, 87, 96). Structural analysis of an N-terminally truncated protein with amino acid residues 135 to 580 revealed a stable, almost brick-shaped core of multiple α-helices with a distinct electrostatic surface distribution and from which many flexible loops emanate that could potentially interact with other viral or host proteins (7).
The UL25 gene of HSV-1 encodes a protein of 63 kDa that is highly conserved among alphaherpesviruses. pUL77 of human cytomegalovirus (HCMV), a betaherpesvirus, has a sequence identity of 23% to HSV-1 pUL25. It is also a structural protein of extracellular virions and is located in the nuclei of infected cells (24, 94). Open reading frame 19 (ORF19) of Kaposi's sarcoma-associated herpesvirus (KSHV), a gammaherpesvirus, encodes a structural protein with a sequence identity of 25% to HSV-1 pUL25 (98). Herpesviruses share many similarities with double-stranded DNA (dsDNA) bacteriophages of the family Caudovirales, for example, the assembly of spherical procapsids that mature into angularized capsids (3, 4, 28, 70). Upon DNA packaging, many bacteriophages add scaffolding proteins to the capsid surface to retain the genome stably within the capsid. pUL25 and its homologues fulfill a similar function in herpesviruses, and pUL25 contributes to sealing the capsid after genome packaging (2, 50, 59, 86). The formation of C capsids in alphaherpesvirus mutants lacking UL25 indicates that pUL25 is not essential for encapsidation of viral DNA but increases the efficiency of stable genome packaging (36, 40, 61, 86). The CCSC heterodimer of pUL25 and pUL17 might also generate a “head-full” signal on the capsid surfaces to foster nuclear egress (10, 36, 40, 50, 59, 67, 86, 92).
HSV-1 capsids enter the cytosol after fusion of the viral envelopes with host cell membranes, and the microtubule-motor dynein transports those incoming capsids to the nucleus (20, 47, 48, 84). The subcellular fate of most structural proteins of herpesviruses upon cell entry is unknown. However, electron and fluorescence microscopy studies suggest that most tegument proteins, also called outer tegument proteins, remain bound to the viral envelope or are released into the cytosol, while only a few inner tegument proteins remain capsid associated during transport to the nucleus (12, 25, 46, 49, 68, 84). Biochemical data have shown that capsids covered with inner tegument proteins, but neither untegumented nor fully tegumented capsids, recruit the microtubule motors dynein, kinesin 1, and kinesin 2 and move along microtubules in vitro (68, 96).
Inner tegument proteins, as well as outer capsid proteins, such as pUL36, pUL37, or pUL25, could contribute to microtubule-mediated nuclear targeting, genome uncoating at the nuclear pore complexes (NPCs), and nuclear import of the viral genomes (68, 96). It has been suggested that the pUL25 proteins of HSV-1 and pseudorabies virus (PrV), an alphaherpesvirus of swine, associate with microtubules (27, 34). Furthermore, certain HSV-1 UL25 and HSV-1 UL36 mutants are unable to induce early viral-gene expression, and incoming parental HSV-1 pUL36, the inner tegument protein interacting with pUL25, needs to be cleaved to ensure early viral-gene expression (1, 5, 33, 61, 63, 67, 72). The host nuclear import factor importin β, as well as the NPC proteins Nup358/RanBP2 and Nup214/CAN, which both face the cytosol, are involved in capsid docking to the nuclear pores (14, 15, 62, 63). Furthermore, immunoprecipitates of cytoplasmic HSV-1 capsids from cells transfected with plasmids encoding nucleoporins contain Nup214/CAN, but not other nucleoporins, and pUL25 can interact with Nup214/CAN and hCG1, suggesting that incoming capsids might dock to NPCs via an interaction of capsid-associated pUL25 with Nup214/CAN (63).
Here, we tested potential interactions between parental pUL25 bound to incoming HSV-1 capsids and host factors by competing for such interactions with an experimental excess of cytosolic pUL25. Excess HSV-1 pUL25 mainly remained in the cytosol, with a minor enrichment on the nuclear envelope, but not on microtubules. Surplus HSV-1 pUL25 reduced HSV-1 gene expression, but not nuclear targeting of incoming capsids. Electron microscopy and in situ hybridization studies indicated that the HSV-1 genomes had left the capsids and were thus uncoated but were either not efficiently imported into the nucleoplasm or were maintained in a conformation that hampered transcription. Our data suggest that incoming capsid-associated pUL25 interacts either with nuclear import factors or with NPC components, such as Nup214/CAN or hCG1, to ensure proper nuclear import of functional HSV-1 genomes and gene expression.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Virus amplifications, purifications, and titrations of HSV-1 strain F (ATCC VR-733) or strain 17+ (provided by John Subak-Sharpe, MRC Virology Unit, Glasgow, United Kingdom), as well as culture of PtK2 (ATCC CCL-56), HeLa (ATCC CCL-2), and Vero cells (ATCC CCL-81), were performed as described previously (19, 20, 52, 84). All virus inocula used in this study had genome/PFU ratios of 20 to 30, indicative of a small amount of noninfectious particles (19). Vaccinia virus strain WR (78) and human adenovirus serotype 5 expressing β-galactosidase (79) were provided by Jacomine Krijnse-Locker (University of Heidelberg, Heidelberg, Germany) and Florian Kreppel (University of Ulm, Ulm, Germany), respectively. We used rabbit polyclonal antisera raised against the C-terminal 239 residues of pUL25 (R8-3 [2]), full-length glutathione S-transferase (GST)-pUL25 (polyclonal antibody [PAb] ID1 [38]), VP5 (NC-1 [11]), β-galactosidase (5-Prime–3 Prime, Inc., Boulder, CO), or the c-myc epitope (23). Furthermore, we used mouse monoclonal antibodies specific for VP5 of mature hexons (monoclonal antibody [MAb] 5C10 [91]), immature VP5 (MAb LP12 [66]), pUL6 (MAb IC9 [58]), ICP0 (MAb 11060 [22]), ICP8 (ATCC HB-8180; Rockville, MD [83]), vaccinia virus protein p35 (MAb H5R [76]), tubulin (MAb DM1A; Sigma-Aldrich, Schnelldorf, Germany), vimentin (MAb 7A3 [39]), actin (MAb 1501; Chemicon-Millipore, Schwalbach, Germany), or green fluorescent protein (GFP) (MAb JL-8; Boehringer, Ingelheim, Germany). We stained filamentous actin using tetramethyl rhodamine isocyanate (TRITC)-phalloidin (0.2 μg/ml in phosphate-buffered saline [PBS]; Sigma-Aldrich). The goat-derived secondary antibodies were conjugated with either lissamine-rhodamine B sulfonyl chloride (LRSC) or fluorescein-isothiocyanate (FITC) and highly preabsorbed against cross-reactivity to species other than the intended one (rabbit or mouse; Dianova, Hamburg, Germany) for fluorescence microscopy or conjugated with alkaline phosphatase or horseradish peroxidase (Thermo Scientific, Bonn, Germany) for immunoblotting.
Virus infection.
Cells were inoculated with HSV-1 diluted in RPMI 1640 (Cytogen, Sinn, Germany) supplemented with 0.1% (wt/vol) bovine serum albumin (BSA)-20 mM HEPES, pH 7.0, or in CO2-independent medium (Invitrogen, Paisley, United Kingdom) containing 0.1% (wt/vol) BSA for 2 h on ice, or with vaccinia virus in serum-free Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) for 30 min at room temperature (RT). After unbound HSV-1 or vaccinia virus was removed, the cells were shifted to regular growth medium at 37°C and 5% CO2. Adenovirus was added to cells in regular growth medium at 37°C and 5% CO2 for 12 h. We used 10 PFU/cell to study HSV-1 assembly, 50 to 200 PFU/cell to analyze HSV-1 cell entry by fluorescence microscopy or immunoblotting, 500 PFU/cell to analyze HSV-1 cell entry by electron microscopy, and 2 to 10 PFU/cell to measure HSV-1 gene expression (19, 20, 47, 84). Furthermore, 60 PFU/cell was added to monitor vaccinia virus gene expression (78) or 100 PFU/cell to study adenovirus gene expression (79). In cell entry experiments, the media contained 0.5 mM cycloheximide to prevent synthesis of progeny virions (84).
Immunoblotting.
Cells from a 10-cm dish were harvested in hot sample buffer (1% [wt/vol] SDS, 50 mM Tris-HCl, pH 6.8, 5% [wt/vol] glycerol, 1% [vol/vol] 2-mercaptoethanol, bromophenol blue) and approximately 3 × 105 to 4 × 105 cells were loaded per lane. After SDS-PAGE using linear 10 to 18% gradient gels, the proteins were transferred onto nitrocellulose membranes in 48 mM Tris, 380 mM glycine, 0.1% (wt/vol) SDS, 10% (vol/vol) methanol. The membranes were incubated with 5% (wt/vol) skim milk and 0.1% (vol/vol) Tween 20 in PBS for 1 h and incubated with primary antibodies for 2 h and with secondary antibodies coupled to alkaline phosphatase or peroxidase for 1 h. After being washed in PBS-Tween, the membranes were equilibrated with 10-fold-diluted SuperSignal West Femto substrate (Thermo Scientific) to detect peroxidase or with TSM buffer (100 mM Tris-HCl, pH 9.5, 100 mM NaCl, 5 mM MgCl2) and incubated with 0.2 mM nitroblue tetrazolium chloride and 0.8 mM 5-bromo-4-chloro-3-indolyl phosphate to detect alkaline phosphatase. Digital images were recorded using a flatbed scanner (Hewlett-Packard, Böblingen, Germany) or an LAS-3000 documentation system (Fuji Film, Düsseldorf, Germany).
Plasmids.
The HSV-1 UL25 gene was amplified from HSV-1 cosmid 28 (16) using the primers 5′-CGC GGA TCC GCC GCC ACC ATG GAC CCG TAC TGC CCA T-3′ and 5′-ATA GAA TTC CTA AAC CGC CGA CAG GTA C-3′ (restriction sites are underlined). The PCR product was cleaved with BamHI and EcoRI and ligated into pcDNA3.1(+) (Invitrogen, Karlsruhe, Germany) to generate pcDNA3-UL25 or ligated into pEGFP-C1 (Invitrogen) to generate pEGFPUL25. The HSV-1 UL25 ORF without the stop codon was amplified using primers 5′-CGC GGA TCC GCC GCC ACC ATG GAC CCG TAC TGC CCA T-3′ and 5′-CCG CTC GAG TAT ACC GCC GAC AGG TAC TGT-3′, cleaved with BamHI and XhoI, and cloned into BglII- and SalI-cut pEGFP-N1 (Invitrogen) to generate pUL25EGFP. HCMV-UL77 was amplified from plasmid 6KMfe-UL77HA (provided by Martin Messerle, Hannover Medical School) using primers 5′-GCC GAA TTC ATG AGT CTG TTG CAC ACC TTT-3′ and 5′-GCC GGT ACC TGC AAC ACC GCC ACG CTC GGA A-3′. The PCR product was cleaved with EcoRI and KpnI and ligated into pEGFP-N1 to generate pHCMV-UL77GFP. KSHV ORF19 was amplified from bacterial artificial chromosome 36 (BAC36) (provided by Thomas Schulz, Hannover Medical School) using 5′-GCG GCC GCG CCG CCA CCA TGC TGA CAT CAG AAA GGT CCT-3′ and 5′-CGC TCT AGA CTA CAG ATC TTC TTC AGA AAT AAG TTT TTG TTC AAC GAC CGC GAG-3′ to generate KSHV-ORF19myc with a C-terminal myc epitope. KSHV BAC36 was generated by homologous recombination of BAC vector sequences with the KSHV genome in BCBL1 cells (97). The PCR product was cleaved with NotI and XbaI prior to ligation into a NotI- and XbaI-cut pcDNA3.1(+) vector to generate pcDNA3-KSHV-ORF19myc. pUL25, pGFPUL25, pUL25GFP, pHCMV-UL77GFP, and pKSHV-ORF19myc were expressed under the control of the cytomegalovirus immediate-early promoter. All constructs were verified by sequence analysis.
Transfection.
The transfection reagents GeneJuice (Merck, Darmstadt, Germany) and Fugene (Roche, Mannheim, Germany) were used according to the manufacturers' protocols. Cells were seeded at a density of 3 × 104 (Vero) or 4 × 104 (PtK2) cells per well of 24-well plates containing glass coverslips or at 6 × 104 (Vero) or 8 × 104 (PtK2) cells per well of Lab-Tek II CC2 chamber slides (Nunc, Naperville, IL; 2-well slides). After 24 h, the cells were transfected using calcium phosphate with 1.4 (Vero) or 1.1 (PtK2) μg DNA per 24 wells (20, 77). At 2 h posttransfection, this mixture was replaced with 1 ml/well growth medium for 24 h. For transfection of cells grown in Lab-Tek chamber slides, twice as much plasmid DNA and medium per well was used.
Immunofluorescence microscopy.
For most experiments, the cells were fixed with 3% (wt/vol) paraformaldehyde (PFA) in PBS for 20 min and permeabilized with 0.1% Triton X-100 in PBS for 5 min at RT (20, 84). In some experiments, cells were simultaneously fixed and permeabilized using 100% methanol at −20°C for 4 min or using Phemo-fix (3.7% [wt/vol] PFA, 0.05% [wt/vol] glutaraldehyde, 0.5% [vol/vol] Triton X-100 in Phemo buffer [68 mM PIPES {piperazine-N,N′-bis(2-ethanesulfonic acid)}, 25 mM HEPES, pH 6.9, 15 mM EGTA, 3 mM MgCl2, 10% dimethyl sulfoxide {DMSO}]) for 10 min at 37°C (20, 47). Alternatively, cells were permeabilized using 0.5% Triton X-100 in 80 mM PIPES, pH 6.8, containing 2 mM MgSO4 and 10 μM taxol for 3 to 10 s at 37°C prior to methanol fixation (84). Furthermore, cells prefixed with 3% PFA/PBS were extracted with either 100% acetone or 100% methanol at −20°C for 4 min. To inactivate any residual PFA, the cells were treated with 50 mM NH4Cl in PBS for 10 min. For in situ hybridization, cells were fixed with a mixture of 95% ethanol and 5% glacial acetic acid at −20°C for 5 min. To block nonspecific protein binding, the cells were incubated prior to being immunolabeled with 0.5% (wt/vol) BSA in PBS, with 1% (vol/vol) fetal calf serum (FCS) in PBS, or with PBS containing both 0.5% (wt/vol) BSA and 10% (vol/vol) human serum from a healthy HSV-1-seronegative volunteer. Immunolabelings were performed as described previously, and the cells were embedded in Mowiol containing 2.5% (wt/vol) 1,4-diazabicyclo-[2.2.2]octane (19, 20, 84). The samples were examined using inverted fluorescence microscopes (DM IRB/E [Leica, Wetzlar, Germany]; Axiovert 200 and AxioObserver [Zeiss, Göttingen, Germany]) equipped with plan-apochromatic 40×, 63×, or 100× oil immersion objective lenses; appropriate filter sets; and mercury or 150 W xenon (Polychrome IV; T.I.L.L Photonics, Gräfeling, Germany) lamps. Images were recorded using digital interline charge-coupled device (CCD) cameras (Leica MicroMax-5MHz-782Y [Princeton Instruments Inc., Princeton, NJ] controlled by MetaMorph software 5.05 [Universal Imaging Corporation, West Chester, PA], Zeiss Axiovert 200 Imago-QE CCD camera controlled by TillVision software 4.0 [T.I.L.L Photonics], or AxioCam HRm controlled by Axiovision software 4.6.3.0 SP1 [Zeiss AxioObserver]). Confocal sections were recorded with an LSM 510 Meta (Zeiss), a plan-apochromat 63×/1.40 oil objective, and argon (Argon 2, 458 nm, 477 nm, 488 nm, and 514 nm) or helium-neon (HeNe1, 543 nm; HeNe2, 633 nm) lasers controlled by LSM 510 software. Images were processed using MetaMorph, Image J (Wayne Rasband, NIH; http://rsb.info.nih.gov/ij/), LSM Image Browser (Zeiss), or Adobe Photoshop CS (Adobe Systems, San Jose, CA). We imaged randomly sampled transfected or control cells by phase-contrast to determine the cell margins and the positions of the nuclei. To analyze HSV-1 gene expression, the average labeling intensities for the early nuclear HSV-1 proteins ICP0 and ICP8 were measured using MetaMorph or ImageJ 1.36b. Statistical tests were performed using a Mann-Whitney test and GraphPad software (version 5.02; GraphPad Software, Inc.).
Fluorescence-activated cell sorting and electron microscopy.
Vero cells in a 10-cm dish overexpressing GFP, UL25GFP, or GFPUL25 were infected with 500 PFU/cell of HSV-1. After 2 h, they were trypsinized, resuspended in 3 ml growth medium, and fixed for 20 min at RT by adding 3 ml of 2× fixative containing 8% (wt/vol) PFA and 0.2% (wt/vol) glutaraldehyde in 400 mM sodium cacodylate, pH 7.4. The cells were sedimented at 1,000 × g for 10 min at RT and resuspended in 500 μl PBS containing 7.5% (wt/vol) BSA. GFP-positive cells were collected by fluorescence-activated cell sorting (FACS) (MoFlo high-speed sorter; Cytomation, Fort Collins, CO) and sedimented at 10,000 × g for 5 min. The cell pellets were further fixed with 1% (wt/vol) glutaraldehyde in 200 mM cacodylate, pH 7.4, for 1 h; washed three times with cacodylate buffer; and rinsed three times with water. The cells were contrasted with 1% (wt/vol) OsO4 and 1.5% (wt/vol) K3Fe(III)(CN)6 for 1 h at RT and with 0.5% (wt/vol) uranyl acetate in 50% (vol/vol) ethanol at RT overnight and dehydrated in a graded ethanol series and propylenoxid prior to being embedded in Epon. Ultrathin sections 50 to 70 nm thick were further stained using lead citrate (71) and documented with a 10 CR electron microscope (Zeiss, Göttingen, Germany) or Tecnai (FEI, Eindhoven, Netherlands). The negatives were scanned with a flatbed scanner (AGFA Duo Scan; AGFA, Cologne, Germany) at 1,600 dots per inch (dpi) and further processed using Adobe Photoshop. For quantification, we sampled cells from several ultrathin sections derived from different Epon blocks. For the first quantification, the cytoplasm and the plasma membranes, as well as the nuclei, of 20 different, randomly selected cells were documented at ×12,500 magnification, resulting in 40 images (see Table 1, experiment 1). For the second quantification, 40 randomly selected cells that contained capsids were documented (experiment 2). We classified the capsids as filled or empty depending on the presence or absence of an electron-dense core and into cytosolic capsids and capsids at the NPC with a distance of 125 nm or less to the nuclear envelope.
Table 1.
HSV-1 nuclear targeting and genome uncoating despite surplus pUL25
| Experimental parameter |
Value |
|||||
|---|---|---|---|---|---|---|
| Expt 1 |
Expt 2 |
|||||
| Surplus protein | GFP | UL25GFP | GFPUL25 | GFP | UL25GFP | GFPUL25 |
| No. of cells | 22 | 23 | 21 | 46 | 32 | 40 |
| No. of capsids | 65 | 92 | 61 | 71 | 67 | 78 |
| Cytosolic, % filled | 34 | 60 | 28 | 21 | 40 | 25 |
| NPC, % filled | 12 | 5 | 3 | 7 | 3 | 5 |
| NPC, % empty | 52 | 31 | 66 | 71 | 54 | 69 |
| Cytosolic, % empty | 2 | 4 | 3 | 1 | 3 | 1 |
| Filled, total % | 46 | 65 | 31 | 28 | 43 | 30 |
| Cytosol, total % | 34 | 64 | 31 | 22 | 43 | 26 |
| NPC, total % | 64 | 36 | 69 | 78 | 57 | 74 |
| Empty, total % | 54 | 35 | 69 | 72 | 57 | 70 |
In situ hybridization.
HSV-1 probe synthesis and hybridization were performed as described previously (21). Cells were prehybridized for 30 min at 37°C in hybridization buffer (50% [vol/vol] formamide, 10% [vol/vol] dextran sulfate in 4× SSC [0.6 M NaCl, 0.06 M sodium citrate]). The coverslips were blotted dry, incubated at 42°C for 20 min, and then treated with 10 ng/μl HSV-1 Cy3-labeled probe in hybridization buffer. A 20- by 20-mm glass coverslip was placed on the cells and fixed onto a cover slide using fixogum rubber cement (Marabu, Tamm, Germany). These ensembles were heated for 4 min to 95°C using a PCR thermocycler (Cyclone 96 in situ; PeqLab, Erlangen, Germany) and then incubated overnight at 37°C in a humidified chamber. The cells were washed with 2× SSC at 60°C, with 2× SSC at RT, and with PBS containing 1% (vol/vol) FCS or 0.5% (wt/vol) BSA and labeled with antibodies as described above. The cell margins and nuclei were identified by phase-contrast microscopy. The average gray values of the nuclear hybridization signals were determined using the measuring tool of Image J. We subtracted the background signals of uninfected cell nuclei to obtain the relative fluorescence intensity correlated with the level of the hybridized Cy3-labeled probe and performed Mann-Whitney statistical tests using GraphPad software.
RESULTS
pUL25 expression and subcellular localization during HSV-1 infection.
To determine the expression kinetics of UL25, we infected Vero cells with HSV-1 at 10 PFU/cell (see Fig. S1A in the supplemental material). pUL25 and pUL6 expression was detected around 6 to 7 h postinfection (p.i.) and increased until 15 h p.i., as expected for late proteins with weak promoters (69). Next, we analyzed the subcellular localization of pUL25 during assembly in Vero cells also infected with 10 PFU/cell. The cells were labeled with antibodies against pUL25 and either against mature VP5 epitopes on capsid hexons (MAb 5C10) (19, 91) or against immature VP5 epitopes (MAb LP12) (19, 66). As described previously (52), HSV-1 capsids identified with antibodies against mature VP5 epitopes had already assembled at 5 h p.i. in the nucleus and had been exported to the cytoplasm by 9 h p.i. (see Fig. S1B to D in the supplemental material). Such capsids also contained pUL25, both in the nucleus and in the cytoplasm (Fig. S1B to D, white arrowheads). In contrast, antibodies against immature VP5 epitopes revealed diffuse nuclear labeling. For the most part, pUL25 did not colocalize with nuclear structures labeled by MAb LP12 (Fig. S1F), and cytoplasmic structures, detected by anti-pUL25 and thus most likely the progeny capsids, did not react with MAb LP12. These data indicate that mature nuclear capsids recruited pUL25 and that pUL25 remained capsid associated during nuclear egress and passage through the cytoplasm.
To determine the subcellular fate of pUL25 during cell entry, we infected Vero cells with 200 PFU/cell in the presence of cycloheximide to prevent synthesis of progeny viral proteins. The amount of pUL25 or VP5 did not decrease, and there was no indication of proteolytic cleavage of pUL25 (see Fig. S2A in the supplemental material). To analyze the subcellular localization of parental pUL25, Vero cells were infected with 70 PFU/cell in the presence of cycloheximide (see Fig. S2B to G in the supplemental material). As described previously (20, 84), incoming capsids labeled for VP5 were distributed throughout the cytoplasm at 1 h p.i. and often colocalized with pUL25. At 3 h p.i., pUL25 had remained associated with these incoming capsids at the nuclear rim. Our data show that pUL25 bound to nuclear capsids that had formed mature VP5 epitopes. It remained capsid associated during transport of incoming capsids from the cell periphery to the nucleus.
Subcellular localization of HSV-1 pUL25.
We never detected pUL25 on microtubules or other components of the host cytoskeleton during the entire HSV-1 life cycle either during assembly or during cell entry. However, Kaelin et al. (34) and Guo et al. (27) reported that after transfection, pUL25 proteins of PrV and HSV-1 accumulated on microtubules (27, 34). Therefore, we generated HSV-1 UL25 plasmids for eukaryotic transient expression to test whether HSV-1 pUL25 bound to the cytoskeleton when expressed alone. Immunoblot analysis showed an expected molecular mass of around 97 kDa for the fusion proteins, as well as the absence of any degradation products (Fig. 1A). Thus, we could use the GFP signal to infer the subcellular localization of GFPUL25 and UL25GFP. Our goal was then to identify a fixation method that would preserve the in vivo localization of the GFP-tagged UL25 proteins.
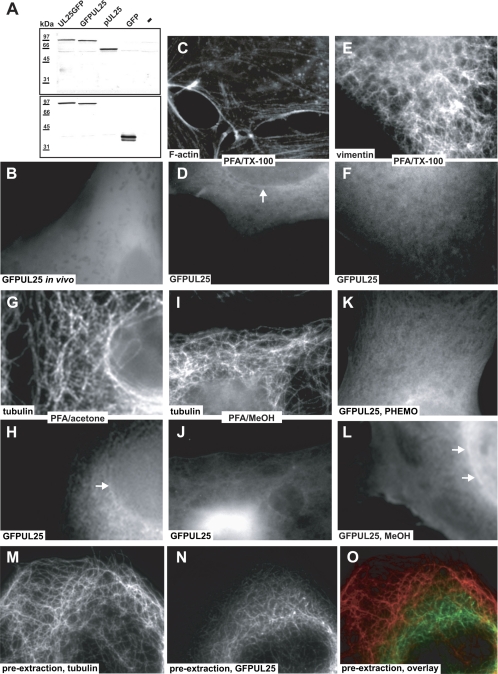
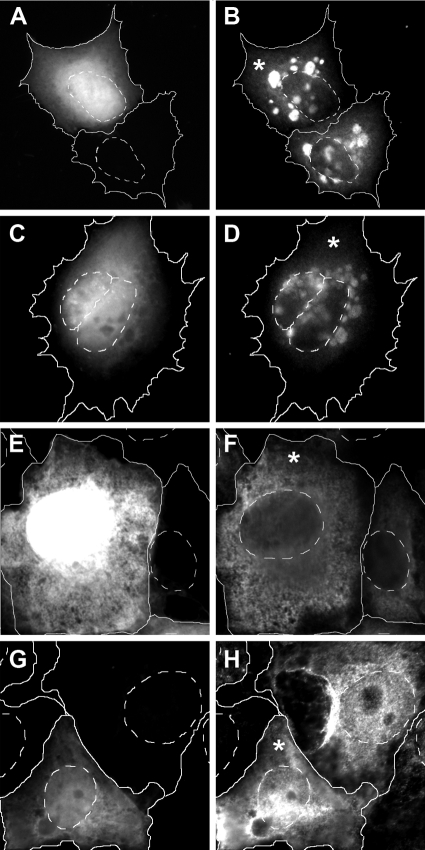
Fig. 1.
pUL25, GFPUL25, and UL25GFP do not colocalize with the cytoskeleton. (A) Immunoblot. Vero cells transfected with plasmids encoding UL25GFP, GFPUL25, UL25, or GFP or mock (−) treated were analyzed for pUL25 (PAb R8.3) and GFP (MAb JL-8). (B to O) Fluorescence microscopy. GFPUL25-transfected PtK2 cells were analyzed in vivo (B) or treated with 3% PFA and 0.1% TX-100 (C to F), 3% PFA and 100% acetone (G to H), 3% PFA and 100% methanol (I to J), Phemo-fix (K), 100% methanol at −20°C (L), or TX-100 in the presence of taxol and 100% methanol at −20°C (M to O). The samples were analyzed for filamentous actin (TRITC-phalloidin) (C), for vimentin (MAb 7A3) (E), or for tubulin (MAb DM1A) (G, I, and M) and for GFP (D, F, H, J, L, and N). GFPUL25 remained in the cytosol when the cells had been fixed prior to permeabilization (D, F, H, J, K, and L), although a portion of pUL25 accumulated in the nucleus after stronger extraction (J and L). Actin (C), vimentin (E), or microtubules (G and I) did not colocalize with GFPUL25. When the cells had been extracted with TX-100 prior to fixation, there was some overlap (O) of the tubulin labeling (M) with a filamentous GFPUL25 signal (N). Occasionally, GFPUL25 was enriched on the nuclear envelope (arrows).
In living cells, GFPUL25 (Fig. 1B) and UL25GFP (not shown) were exclusively localized in the cytosol. The cytoplasmic organelles and the nuclei appeared dark, whereas the GFP fluorescence highlighted the apparent cytosolic volume. After fixation and permeabilization, all proteins remained mostly located in the cytoplasm. In several cells, HSV-1 pUL25 was enriched on the nuclear envelope (arrow in Fig. 1D), but we did not detect any obvious colocalization of GFPUL25 with the actin cytoskeleton (Fig. 1C and D), the intermediate filaments (Fig. 1E and F), or the microtubule network (not shown) using a common protocol of PFA fixation followed by permeabilization with Triton X-100. Even after the large pool of cytosolic proteins was extracted by using PFA fixation followed by extraction with acetone (Fig. 1G and H) or with methanol (Fig. 1I and J) in order to reveal a potential minor population associated with microtubules, there was no apparent colocalization of microtubules (Fig. 1G and I) with GFPUL25 (Fig. 1H and J) or UL25GFP (data not shown). Furthermore, a combined fixation/permeabilization protocol (Phemo) that preserves microtubules very well (20, 47) also maintained the cytosolic distribution of GFPUL25 (Fig. 1K). After methanol fixation at −20°C, GFPUL25 partially redistributed to the nucleus but still did not colocalize with microtubules (Fig. 1L). However, again, pUL25 occasionally accumulated at the nuclear envelopes (arrows in Fig. 1H and L).
Only after a short preextraction using the detergent Triton X-100 in a microtubule-stabilizing buffer prior to fixation with methanol did we notice a filamentous pattern of GFPUL25 (Fig. 1N), but not of pUL25 or UL25GFP (not shown), that overlapped with microtubules only to a minor extent (Fig. 1M; yellow in overlay in panel O). However, this localization resembled neither the overall architecture of the microtubule network nor the in vivo localization of GFPUL25 (cf. Fig. 1B and N). Based on these observations, we concluded that wild-type HSV-1 pUL25 and HSV-1 pUL25 tagged with GFP at the N or C terminus predominantly localize to the cytosol with some enrichment on the nuclear envelope and, after preextraction, on cytoskeletal filaments.
Surplus HSV-1 pUL25 does not impair nuclear targeting of HSV-1 capsids.
If HSV-1 pUL25 functions in microtubule-mediated transport, an excess of pUL25 might behave as a dominant-negative inhibitor and compete with nuclear capsid targeting. Therefore, we investigated the subcellular localization of incoming capsids in pUL25-expressing cells. Cells expressing either GFP (Fig. 2A and C), UL25GFP (Fig. 2B and D), GFPUL25 (not shown), or pUL25 (not shown) were infected with HSV-1 in the presence of cycloheximide. At 1 h p.i., incoming capsids were localized throughout the cytoplasm, as in GFP (Fig. 2A and B) or mock-transfected cells (not shown). Irrespective of the overexpressed protein, capsids accumulated at the nucleus at 3 h p.i. (Fig. 2C and D). Thus, an excess of pUL25 during the early phase of the HSV-1 life cycle had no effect on capsid transport to the nucleus.
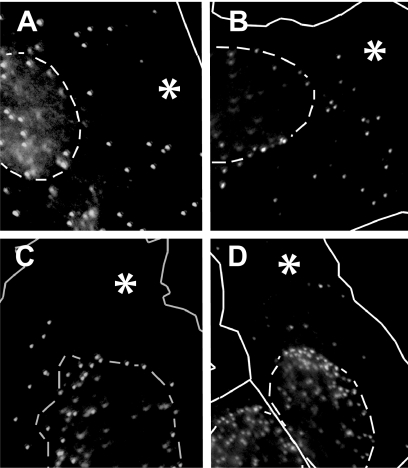
Fig. 2.
Surplus pUL25 does not affect nuclear targeting of incoming HSV-1 capsids. Cells overexpressing GFP (asterisks in panels A and C) or UL25GFP (asterisks in panels B and D) were infected with 70 PFU/cell of HSV-1 strain F in the presence of cycloheximide, fixed with 3% PFA, permeabilized with 0.1% TX-100, and processed for immunofluorescence microscopy. At 1 h p.i. (A and B), incoming capsids labeled for VP5 (MAb 5C10) were distributed throughout the cytosol. Almost all capsids accumulated at the nuclear rim at 3 h p.i. (C and D), while only a few capsids remained in the cytosol, irrespective of the transfected protein. The dashed lines indicate the positions of the nuclei as identified by phase-contrast microscopy.
Surplus HSV-1 pUL25 or its homologous proteins of HCMV and KSHV reduce HSV-1 gene expression.
To test whether overexpressed pUL25 competed with other important pUL25-host interactions during the HSV-1 life cycle, cells expressing a surplus of UL25 were infected for 2 (Fig. 3A and C) or 3 (Fig. 3B and D) h, fixed, and labeled with antibodies against ICP0, an immediate-early HSV-1 protein. In mock-transfected or GFP-expressing HSV-1-infected cells, ICP0 had a typical nuclear localization (Fig. 3A and B), as described previously (22), while only a few cells expressing UL25GFP (Fig. 3C and D), GFPUL25 (not shown), or UL25 (not shown) had synthesized ICP0. In addition, GFP also had no effect on the expression of ICP8, an early HSV-1 protein (Fig. 3E), whereas overexpression of GFPUL25 (Fig. 3F) and UL25GFP (Fig. 3G) inhibited it, suggesting that HSV-1 gene expression in general was inhibited.
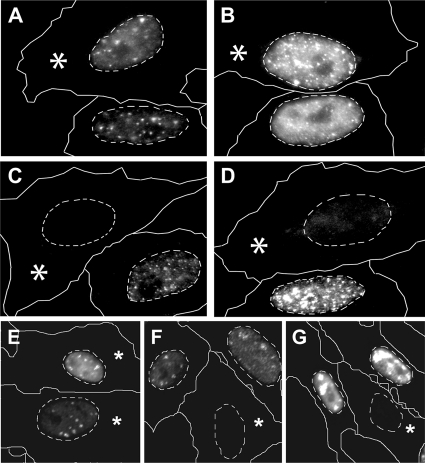
Fig. 3.
Surplus pUL25 reduces HSV-1 gene expression. Vero cells were transfected (asterisks) with plasmids coding for GFP (A, B, and E), GFPUL25 (F), or UL25GFP (C, D, and G). The cells were infected for 2 (A and C) or 3 (B and D to G) h with 2 (A to D) or 5 (E to G) PFU/cell of HSV-1 strain F, fixed with 3% PFA, permeabilized with 0.1% TX-100, and labeled for ICP0 (A to D) or ICP8 (E to G) for analysis by wide-field immunofluorescence microscopy. The dashed lines indicate the positions of the nuclei as identified by Hoechst staining. GFP-transfected (asterisks in panels A and B) or untransfected (without label) cells had synthesized more ICP0 than UL25GFP-expressing cells (asterisks in panels C and D). Cells overexpressing GFPUL25 (asterisk in panel F) or UL25GFP (asterisk in panel G) expressed less ICP8 than cells expressing GFP (asterisks in panel E) or untransfected cells (without label in panels E to G).
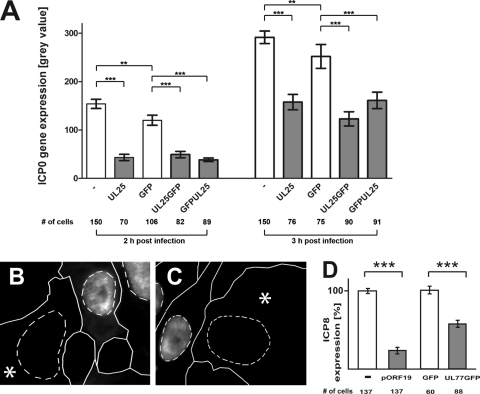
For quantification, we measured the ICP0 or ICP8 expression levels in randomly selected transfected or mock-transfected cells. Excess pUL25 reduced ICP0 expression by about 70% at 2 h and by 50% at 3 h postinfection compared to mock-transfected cells, and UL25GFP or GFPUL25 reduced ICP0 expression by about 60% at 2 h and 50% at 3 h p.i. compared to GFP-expressing cells (Fig. 4A). As in our previous study (20), transfection of GFP alone also reduced HSV-1 gene expression compared to cells that were not transfected. Furthermore, we tested KSHV-pORF19myc and HCMV pUL77GFP, which are homologues of HSV-1 pUL25, in this assay. KSHV-pORF19myc (Fig. 4B) and HCMV pUL77GFP (Fig. 4C) reduced HSV-1 ICP8 expression (Fig. 4D). Thus, an excess of HSV-1 pUL25, as well as its KSHV and HCMV homologues, impaired HSV-1 gene expression.
Fig. 4.
Surplus pUL25 or its homologous HCMV and KSHV proteins reduce HSV-1 gene expression. (A) Quantification. Vero cells were transfected and infected as described for Fig. 3A to D, and the average gray values of nuclear ICP0 fluorescence intensities (A) were measured as a means to quantify HSV-1 gene expression. The error bars represent standard errors of the mean (SEM). (B to D) Immunofluorescence microscopy and quantification. Vero cells were transfected with plasmids coding for pORF19-myc (B and D) or HCMV-pUL77GFP (C and D). The cells were infected for 4.5 h with 10 PFU/cell of HSV-1 strain F, fixed with 3% PFA, permeabilized with 0.1% TX-100, and labeled for ICP8 for analysis by wide-field immunofluorescence microscopy. The dashed lines indicate the positions of the nuclei as identified by Hoechst staining. Cells overexpressing KSHV-pORF19myc (anti-myc) (asterisk in panel B) or HCMV-pUL77GFP (asterisk in panel C) expressed less HSV-1 ICP8 than control cells (without label in panels B and C). The average gray values of nuclear ICP8 fluorescence intensities (D) were analyzed as a means to quantify HSV-1 gene expression. The error bars represent SEM. (A and D) ***, P value of <0.001; **, P value between 0.001 and 0.01.
Surplus HSV-1 pUL25 blocks neither vaccinia virus nor adenovirus gene expression.
To address whether pUL25 blocked gene expression or protein synthesis in general, we infected GFP- or UL25GFP-expressing HeLa cells with vaccinia virus, a double-stranded DNA virus that replicates in the cytoplasm (81). Cytoplasmic vaccinia virus replication compartments had formed at 3 h p.i. and contained similar amounts of the vaccinia virus protein p35 irrespective of whether GFP (Fig. 5A) or UL25GFP (Fig. 5C) had been ectopically expressed (Fig. 5B and D). Adenovirus capsids release their parental genomes into the nucleoplasm for viral-DNA transcription and replication (47, 89), similar to HSV-1. We therefore infected GFP-expressing (Fig. 5E) or UL25GFP-expressing (Fig. 5G) HeLa cells with an adenovirus encoding β-galactosidase (79). By 12 h p.i., the infected cells had synthesized β-galactosidase irrespective of the ectopically expressed protein (Fig. 5F and H). Therefore, entry, endocytosis, microtubule-mediated nuclear targeting of incoming capsids, uncoating, and gene expression of adenovirus proceeded normally in pUL25-overexpressing cells. Immunofluorescence microscopy experiments showed that overexpression of UL25 did not alter the subcellular localization of the nuclear import factor importin β or the transcriptional activator HSV-1 VP16 (data not shown). Thus, surplus UL25 proteins do not impair nuclear import and export, transcription, or protein synthesis in general but specifically block the gene expression of HSV-1.
Fig. 5.
pUL25 blocks neither vaccinia virus nor adenovirus gene expression. GFP-expressing (A, B, E, and F) or UL25GFP-expressing (C, D, G, and H) (asterisks) HeLa cells were infected with 60 PFU/cell of vaccinia virus for 3 h p.i., fixed with 3% PFA, permeabilized with 0.1% TX-100, and labeled for vaccinia virus protein p35/H5R (B and D) or infected with 100 PFU/cell of adenovirus encoding β-galactosidase for 12 h p.i. and labeled for β-galactosidase (F and H). The dashed lines indicate the positions of the nuclei as identified by phase-contrast microscopy.
Surplus HSV-1 pUL25 does not impair genome uncoating.
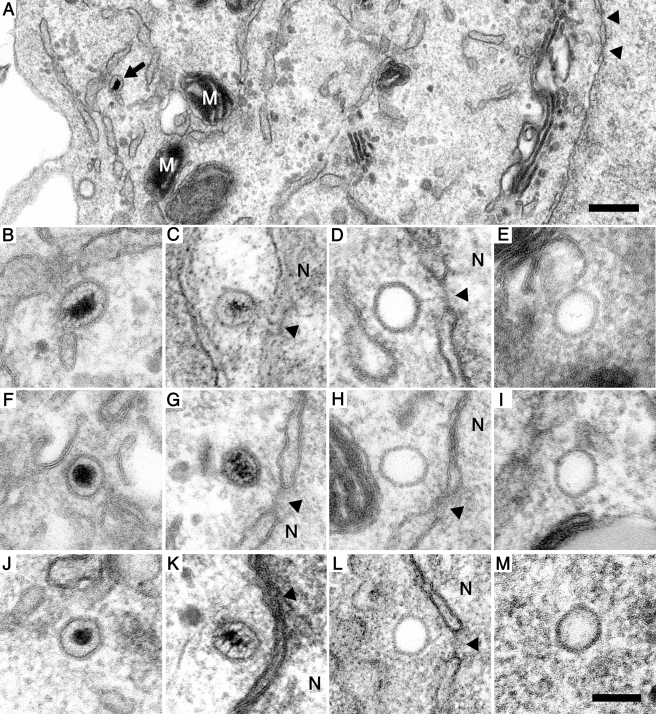
To address whether excess pUL25 impaired uncoating of parental genomes at the NPCs, we infected cells expressing GFP, GFPUL25, or UL25GFP with HSV-1 at a multiplicity of infection (MOI) of 500 PFU/cell for 2 h and analyzed them by electron microscopy. The mild prefixation, FACS sorting for GFP-expressing cells, postfixation, and processing for electron microscopy preserved the morphology of subcellular organelles, such as mitochondria or the nuclear membranes, well (Fig. 6A). We could distinguish DNA-containing, filled capsids (Fig. 6A [black arrow], B, C, F, G, J, and K) from empty capsids (Fig. 6D, E, H, I, L, and M), since the latter lacked an electron-dense core but maintained the typical hexagonal morphology of herpesvirus capsids, as described previously (5, 6, 26, 62, 84). Filled cytosolic capsids (Fig. 6B, F, and J), filled capsids at nuclear membranes (Fig. 6C, G, and K), empty capsids at nuclear membranes (Fig. 6D, H, and L), and empty cytosolic capsids (Fig. 6E, I, and M) were present under each condition. Occasionally, there were linear electron densities connecting one corner of the capsids with the center of the associated NPC (Fig. 6D and H; see also Fig. 1H in Sodeik et al. [84]). These resembled those emerging from T4 phages adsorbed to E. coli cells (see Fig. 17 in reference 82), those emerging from capsids of the gammaherpesvirus murine gammaherpesvirus 68 (MHV-68) docked at NPCs (see Fig. 3 in reference 64), and those emerging from HSV-1 capsids that had extruded their DNA in vitro (55, 64). We therefore presume that these densities represent HSV-1 DNA genomes that emerged from incoming capsids and entered the nucleoplasm via the NPCs.
Fig. 6.
HSV-1 nuclear targeting and genome uncoating in GFP-, UL25GFP-, or GFPUL25-expressing cells. GFP-, UL25GFP-, or GFPUL25-overexpressing cells were infected with HSV-1 strain F at 500 PFU/cell, initially fixed with PFA, sorted by FACS for GFP expression, fixed again with glutaraldehyde, and processed for electron microscopy. (A) The combined FACS-electron microscopy protocol maintained the GFP autofluorescence, as well as the cellular ultrastructure. GFP-expressing (A to E), UL25GFP-expressing (F to I), or GFPUL25-expressing (J to M) Vero cells were analyzed for cytosolic-DNA-filled capsids (B, F, and J), DNA-filled capsids at the nuclear membranes or NPCs (C, G, and K), empty capsids at NPCs (D, H, and L), and cytosolic empty capsids (E, I, and M). Note the presence of electron-dense structures connecting capsid corners with the centers of nuclear pores (arrowheads in panels D and H). N, nucleus; M, mitochondrion; arrow, DNA containing capsid; arrowheads, NPC. Scale bars: 500 nm (A) and 100 nm (B to M).
Consistent with our previous studies (62, 84), there were numerous cytosolic, DNA-containing capsids and many capsids at the NPCs that lacked an electron-dense core and appeared empty in GFP-expressing cells. For quantification, we randomly documented cells until we had collected at least 130 capsids for each sample. Cells transfected with GFPUL25 or UL25GFP prior to infection also contained many DNA-filled capsids in the cytosol, as well as empty capsids at the nuclear pores (Table 1). Under all conditions, numerous empty capsids had accumulated at the nucleus, indicative of nuclear targeting and genome uncoating. If an excess of pUL25 had indeed prevented genome release from the capsid, but not binding to the NPC, there would have been more DNA-filled capsids at the NPCs. Cells expressing UL25GFP contained more filled cytosolic capsids than cells expressing GFP or GFPUL25, while there were fewer empty capsids at the nucleus (Table 1). However, these differences seem to be minor, as our immunofluorescence microscopy studies had not revealed any gross change in the efficiency of nuclear capsid targeting in the presence of GFP, UL25GFP, or GFPUL25. In summary, a surplus of GFP, GFPUL25, or UL25GFP does not seem to impair HSV-1 early gene expression by preventing nuclear capsid targeting and viral genome uncoating per se.
Surplus HSV-1 pUL25 interferes with transcription of HSV-1 genomes.
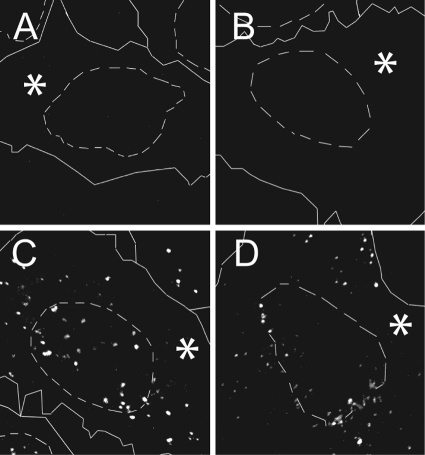
A reduction in HSV-1 gene expression might be the result of defective nuclear import or reduced transcription of viral genomes, or both. To analyze the amount and subcellular localization of parental viral genomes and transcripts, we used immunoFISH, a combination of fluorescence in situ hybridization (FISH) using a Cy3-labeled HSV-1 DNA probe (21) with immunolabeling for the transfected proteins. The harsh fixation and extraction conditions that are necessary for FISH did not preserve the autofluorescence of GFP and the epitopes of pUL25. GFP- or GFPUL25-expressing cells, therefore, identified by an anti-GFP antibody, had little background after mock infection and hybridization with an HSV-1 DNA probe (Fig. 7A and B). Two hours after inoculation with HSV-1 at an MOI of 50 PFU/cell in the presence of cycloheximide, cells expressing GFP revealed cytoplasmic and nuclear punctate genome labeling that was indistinguishable from that of mock-transfected neighboring cells (Fig. 7C). In contrast, the nuclei of inoculated cells expressing GFPUL25 contained fewer nuclear genomes and transcripts, since the nuclear labeling was less intense than in mock-transfected cells. However, there were parental genomes in the cytoplasm and at the nuclear rims (Fig. 7D). To obtain the relative fluorescence intensity derived from viral genomes and transcripts, we determined the average nuclear gray value of uninfected cells and subtracted this from the average nuclear gray values derived from the hybridized HSV-1 DNA probe. The nuclei of cells expressing GFPUL25 contained about 25% less probe than GFP-expressing cells. A statistical Mann-Whitney test showed that the reduction of nuclear genomes and/or transcripts by a surplus of GFPUL25 was significant (Table 2).
Fig. 7.
Surplus pUL25 reduces the amount of HSV-1 nuclear genomes and transcripts. Cells were infected with HSV-1 strain F at 50 PFU/cell, fixed after 2 h postinfection, and labeled by in situ hybridization using a Cy3-coupled HSV-1 DNA probe followed by immunolabeling for GFP (MAb JL-8), and analyzed by confocal fluorescence microscopy. The HSV-1 DNA probe did not label uninfected cells (A and B). After expression of GFPUL25 (D), but not of GFP (C), there were fewer viral genomes and/or transcripts at the nuclear rim and within the nucleus. Transfected cells are marked by asterisks; the solid and dashed lines indicate the cell boundaries and nuclei, respectively, as identified by phase-contrast microscopy.
Table 2.
GFPUL25 reduces the amounts of nuclear HSV1 genomes and transcripts
| Protein | No. of cells | Nuclear viral genomes and transcriptsa |
||
|---|---|---|---|---|
| Gray value (10−5 pixel−2) (%) | SD (%) | SEM (%) | ||
| GFP | 17 | 12.5 (100) | 8.2 (66) | 1.2 (10) |
| GFPUL25 | 29 | 9.2 (74) | 7.3 (59) | 0.9 (7) |
Mann-Whitney test, P = 0.0295.
DISCUSSION
The capsid protein pUL25 may interact with the host's intracellular microtubule transport system and the NPCs (27, 34, 63). We therefore tested whether an excess of pUL25 had any effect on the early stages of HSV-1 infection. We did not detect any competitive inhibition of the microtubule-mediated targeting of parental, incoming HSV-1 capsids to the nucleus. However, a surplus of pUL25, GFPUL25, or UL25GFP or its HCMV or KSHV homologues interfered with the early gene expression of HSV-1, but not with that of vaccinia virus or adenovirus.
Subcellular localization of HSV-1 pUL25.
After synthesis, pUL25 efficiently translocated into the nucleus; associated with newly synthesized mature, nuclear capsids; and remained capsid associated during nuclear egress, assembly, egress, and transport of incoming parental capsids to the nuclei of newly infected cells. Similarly, Conway et al. (13) also reported that pUL25 remains capsid associated during cell entry. We tested several fixation and permeabilization protocols that are particularly suited to maintaining the subcellular organization of the microtubule network. In none of these experiments did we detect any strong targeting of soluble HSV-1 pUL25 to microtubules or NPCs either during infection or after transfection.
Dominant-negative effects of HSV-1 pUL25 during virus cell entry.
Mutants of HSV-1, PrV, or bovine herpesvirus type 1 lacking the entire UL25 gene do not synthesize virions (18, 36, 40, 50, 61). However, not only the absence of a protein of interest can generate informative phenotypes, but also its expression in higher concentrations or at different times than usual. Prominent examples are the dominant-negative effects on dynein-mediated transport after overexpression of dynamitin, a subunit of the dynein cofactor dynactin, or of Rab5, a key regulator of early endosome physiology (9, 20). We, therefore, asked whether a surplus of pUL25 might compete with the capsid-associated pUL25 for host factors, such as dynein, importin β, Nup358/RanBP2, Nup214/CAN, or hCG1, and thus inhibit nuclear targeting of HSV-1 genomes.
After longer expression times and at a higher concentration than reported in this study, pUL25 was toxic (data not shown), suggesting that it can interfere with important host functions, but a transient surplus of pUL25 did not impair early gene expression of vaccinia virus or adenovirus. Thus, pUL25 was not cytotoxic in the amounts analyzed here, but pUL25 and the homologous HCMV pUL77GFP and KSHV-pORF19myc reduced HSV-1 gene expression without inhibiting nuclear targeting of parental HSV-1 capsids. This cell entry phenotype is surprisingly similar to those of HSV-1 ts1249 (pUL25-E233K) and HSV-1 UL25-ΔS578-V580, which also indicates that pUL25 already functions during cell entry (1, 61, 67).
Uncoating and nuclear import of parental HSV-1 genomes.
Surplus pUL25 may have reduced HSV-1 gene expression by impairing HSV-1 genome uncoating or its nuclear import, HSV-1 specific transcription, or nuclear export of HSV-1-specific mRNAs. Our electron microscopy data suggest that HSV-1 capsids had moved to the nucleus and uncoated the genomes in cells expressing GFP or GFPUL25, and to a lesser extent in cells with UL25GFP. However, immunoFISH experiments revealed that there were fewer incoming genomes and/or transcripts in the nucleoplasm but more localized in the cytoplasm and at the nuclear rims. Thus, surplus pUL25 impaired the nuclear import of parental HSV-1 genomes and/or nuclear HSV-1 transcription.
Pasdeloup and collaborators have shown convincingly that the nucleoporins Nup214/CAN and hCG1 can bind to pUL25 and that incoming parental HSV-1 capsids bind overexpressed Nup214/CAN (63). Furthermore, RNA interference (RNAi)-mediated gene silencing of Nup358/RanBP2 or Nup214/CAN reduces HSV-1 gene expression, but not fusion of viral and host membranes (14, 63). Nup358/RanBP2 is required for nuclear protein import mediated by a direct or indirect interaction with members of the karyopherin β family; Nup214/CAN is involved in receptor-mediated import of proteins, CRM1-mediated export of some mRNAs, shuttling of transcription factors, and the nuclear import of adenovirus genomes (29–31, 35, 89). However, our experiments with vaccinia virus and adenovirus indicate that soluble pUL25 did not interfere with all functions of Nup214/CAN, but possibly with some activities specifically required for HSV-1.
HSV-1 capsids require importin β to associate with the NPCs, the inner tegument protein pUL36 may bind to Nup358/RanBP2 that is localized on the cytoplasmic NPC filaments, and capsid-associated pUL25 interacts with Nup214/CAN that is localized on the cytoplasmic NPC surface (14, 62, 63). These interactions may occur either simultaneously or sequentially. The contacts of Nup358/RanBP2 with capsid-associated pUL36, possibly via importin β, may enable an initial docking of incoming HSV-1 capsids to the nuclear pores. Interactions between Nup214/CAN and capsid-associated pUL25 may then contribute to stabilizing the incoming capsids on the NPCs (25, 26, 62, 64, 84). An excess of pUL25 may have prevented an interaction of the incoming parental capsid-associated pUL25 with Nup214/CAN by competitive inhibition. Nevertheless, HSV-1 genome uncoating per se was not impaired, since there were several empty capsids at the NPCs. Possibly, in our experiments, an interaction of the capsid-associated pUL36 with the nuclear pore protein Nup358/RanBP2 was sufficient to target the incoming parental capsids to the NPCs and to initiate viral-genome uncoating.
The DNA genomes of bacteriophages and HSV-1 are released from one penton of the icosahedral capsids, in the case of HSV-1 presumably via the portal pUL6, by an at least initially pressure-driven mechanism (37, 53–55, 74). A pressure-driven ejection mechanisms may push the viral genomes beyond the substantial hydrophobic barrier formed by the FG repeats of many NPC proteins covering the inner channel of the NPCs (17, 42, 65). However, such a pressure-driven ejection mechanism can proceed only until the internal capsid pressure has equilibrated with that of the surroundings (reviewed in references 37 and 74). Theoretical work has suggested that binding of basic proteins, such as transcription factors or histones, to the ejected DNA genomes could constitute a force pulling the genome into the host of the bacteriophage (32) or, in the case of herpesviruses, into the nucleoplasm. Such a scenario is supported by studies showing that naked DNA requires histones and their nuclear import factor transportin for efficient nuclear import in vitro (41). Furthermore, histones, transcription factors, and other nuclear proteins involved in intrinsic antiviral defense, as well as DNA damage repair, associate rapidly with incoming HSV-1 genomes (44, 45, 60).
Hence, the surplus of pUL25 may have competed with incoming capsid-associated pUL25 for efficient binding to Nup214/CAN and hCG1 at the NPCs and thus may have prevented correct positioning of the viral capsids at the nuclear pores. According to our immunoFISH analysis, the HSV-1 genomes still uncoated but did not enter the nucleoplasm efficiently, or at least not in a conformation suitable for transcription. Due to its highly positively charged surface domains and its propensity to bind DNA (7, 59), surplus cytosolic pUL25 may have bound to the ejected HSV-1 genomes and directed them away from the NPCs or may even have competed with nuclear factors, such as histones, for binding to the uncoated genomes.
Our data support the notion that incoming capsid-associated pUL25 fulfills important functions during HSV-1 cell entry. The surplus of pUL25 could have inhibited interactions of incoming capsid-associated pUL25 with host factors that are necessary for efficient viral-genome import into the nucleus. In the presence of surplus pUL25, HSV-1 genome uncoating was still possible, with somewhat reduced efficiency in the case of UL25GFP, but efficient translocation of the HSV-1 genomes into the nucleoplasm or gene expression were inefficient. In summary, surplus pUL25 seems to uncouple the uncoating of HSV-1 genomes from their efficient nuclear import and/or gene expression. Our data suggest that it may be possible to design new pharmacological inhibitors for antiviral therapy that prevent nuclear import and viral-gene expression without impairing the overall functions of the NPCs for the host cell's physiology.
Supplementary Material
ACKNOWLEDGMENTS
We particularly thank Roger Everett (MRC Glasgow) for his advice on in situ hybridization, as well as Alex Schwarz, Anna Buch, Deepika Devadas, Eva Borst, Julia Schipke, and Thalea Koithan (Virology, Hannover Medical School), Amnon Harel (Technion—Israel Institute of Technology, Haifa, Israel), and Wouter Roos and Gijs Wuite (Vrije Universiteit, Amsterdam, Netherlands) for helpful comments on the manuscript. We are very grateful to many colleagues for their generous donations of invaluable viruses, antibodies, and plasmids: E. Cantin (City of Hope National Medical Center, Duarte, CA); G. Cohen and R. Eisenberg (University of Pennsylvania, Philadelphia, PA); A. J. Davison, C. Cunningham, R. Everett, and J. Subak-Sharpe (MRC Virology Unit, Glasgow, United Kingdom); R. Heilbronn (Charité, Berlin, Germany); F. Kreppel (University of Ulm, Ulm, Germany); J. Krijnse-Locker (University of Heidelberg, Heidelberg, Germany); M. Messerle and T. F. Schulz (Hannover Medical School, Hannover, Germany); A. C. Minson and H. Brown (University of Cambridge, Cambridge, United Kingdom); W. Newcomb and J. Brown (University of Virginia, Charlottesville, VA); and D. J. Tenney (Bristol-Myers Squibb, Wallingford, CT). We thank our Laser Microscopy Facility and M. Ballmaier and C. Reimer of our Cell Sorting Core Facility for their support.
Our work was funded by the German Research Council (DFG) (So403/2 and So403/4) and fellowships from the Hannover Biomedical Research School (HBRS-DFG Graduate School of Excellence) to K.R. (GK745) and M.G. (Center of Infection Biology).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 23 February 2011.
REFERENCES
- 1. Addison C., Rixon F. J., Palfreyman J. W., O'Hara M., Preston V. G. 1984. Characterisation of a herpes simplex virus type 1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology 138:246–259 [DOI] [PubMed] [Google Scholar]
- 2. Ali M. A., Forghani B., Cantin E. M. 1996. Characterization of an essential HSV-1 protein encoded by the UL25 gene reported to be involved in virus penetration and capsid assembly. Virology 216:278–283 [DOI] [PubMed] [Google Scholar]
- 3. Baines J. D., Weller S. K. 2005. Cleavage and packaging of herpes simplex 1 viral DNA, p. 135–150 In Catalano C. E. (ed.), Viral genome packaging machines: genetics, structures and mechanism. Landes Biosciences, Austin, TX [Google Scholar]
- 4. Baker M. L., Jiang W., Rixon F. J., Chiu W. 2005. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 79:14967–14970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Batterson W., Furlong D., Roizman B. 1983. Molecular genetics of herpes simplex virus. VIII. further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45:397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Borst E. M., Wagner K., Binz A., Sodeik B., Messerle M. 2008. The essential human cytomegalovirus gene UL52 is required for cleavage-packaging of the viral genome. J. Virol. 82:2065–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bowman B. R., et al. 2006. Structural characterization of the UL25 DNA-packaging protein from herpes simplex virus type 1. J. Virol. 80:2309–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brown J. C., McVoy M. A., Homa F. L. 2002. Packaging DNA into herpesvirus capsids, p. 111–154 In Holzenburg A., Bogner E. (ed.), Structure-function relationships of human pathogenic viruses. Kluwer Academic/Plenum Publishers, New York, NY [Google Scholar]
- 9. Bucci C., et al. 1992. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell 70:715–728 [DOI] [PubMed] [Google Scholar]
- 10. Cockrell S. K., Sanchez M. E., Erazo A., Homa F. L. 2009. Role of the UL25 protein in herpes simplex virus DNA encapsidation. J. Virol. 83:47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cohen G. H., et al. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34:521–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coller K. E., Lee J. I., Ueda A., Smith G. A. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J. Virol. 81:11790–11797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Conway J. F., et al. 2010. Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. J. Mol. Biol. 397:575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Copeland A. M., Newcomb W. W., Brown J. C. 2009. Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment. J. Virol. 83:1660–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cronshaw J. M., Krutchinsky A. N., Zhang W., Chait B. T., Matunis M. J. 2002. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158:915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cunningham C., Davison A. J. 1993. A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology 197:116–124 [DOI] [PubMed] [Google Scholar]
- 17. D'Angelo M. A., Hetzer M. W. 2008. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 18:456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Desloges N., Simard C. 2003. Implication of the product of the bovine herpesvirus type 1 UL25 gene in capsid assembly. J. Gen. Virol. 84:2485–2490 [DOI] [PubMed] [Google Scholar]
- 19. Döhner K., Radtke K., Schmidt S., Sodeik B. 2006. Eclipse phase of herpes simplex virus type 1 infection: efficient dynein-mediated capsid transport without the small capsid protein VP26. J. Virol. 80:8211–8224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Döhner K., et al. 2002. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell 13:2795–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Everett R. D., Murray J. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 79:5078–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Everett R. D., Orr A., Elliott M. 1991. High level expression and purification of herpes simplex virus type 1 immediate early polypeptide Vmw110. Nucleic Acids Res. 19:6155–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gee M. A., Heuser J. E., Vallee R. B. 1997. An extended microtubule-binding structure within the dynein motor domain. Nature 390:636–639 [DOI] [PubMed] [Google Scholar]
- 24. Glass M., Busche A., Wagner K., Messerle M., Borst E. M. 2009. Conditional and reversible disruption of essential herpesvirus proteins. Nat. Methods 6:577–579 [DOI] [PubMed] [Google Scholar]
- 25. Granzow H., Klupp B. G., Mettenleiter T. C. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 79:3200–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Granzow H., et al. 1997. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J. Virol. 71:2072–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guo L., et al. 2008. Interactions of the HSV-1 UL25 capsid protein with cellular microtubule-associated protein. Virologica Sinica 23:211–217 [Google Scholar]
- 28. Homa F. L., Brown J. C. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107–122 [DOI] [PubMed] [Google Scholar]
- 29. Hutten S., Flotho A., Melchior F., Kehlenbach R. H. 2008. The Nup358-RanGAP complex is required for efficient importin alpha/beta-dependent nuclear import. Mol. Biol. Cell 19:2300–2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hutten S., Kehlenbach R. H. 2006. Nup214 is required for CRM1-dependent nuclear protein export in vivo. Mol. Cell. Biol. 26:6772–6785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hutten S., Walde S., Spillner C., Hauber J., Kehlenbach R. H. 2009. The nuclear pore component Nup358 promotes transportin-dependent nuclear import. J. Cell Sci. 122:1100–1110 [DOI] [PubMed] [Google Scholar]
- 32. Inamdar M. M., Gelbart W. M., Phillips R. 2006. Dynamics of DNA ejection from bacteriophage. Biophys. J. 91:411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jovasevic V., Liang L., Roizman B. 2008. Proteolytic cleavage of VP1-2 is required for release of herpes simplex virus 1 DNA into the nucleus. J. Virol. 82:3311–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kaelin K., Dezelee S., Masse M. J., Bras F., Flamand A. 2000. The UL25 protein of pseudorabies virus associates with capsids and localizes to the nucleus and to microtubules. J. Virol. 74:474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kimura T., Hashimoto I., Nagase T., Fujisawa J. 2004. CRM1-dependent, but not ARE-mediated, nuclear export of IFN-alpha1 mRNA. J. Cell Sci. 117:2259–2270 [DOI] [PubMed] [Google Scholar]
- 36. Klupp B. G., Granzow H., Keil G. M., Mettenleiter T. C. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J. Virol. 80:6235–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Knobler C. M., Gelbart W. M. 2009. Physical chemistry of DNA viruses. Annu. Rev. Phys. Chem. 60:367–383 [DOI] [PubMed] [Google Scholar]
- 38. Koslowski K. M., Shaver P. R., Wang X. Y., Tenney D. J., Pederson N. E. 1997. The pseudorabies virus UL28 protein enters the nucleus after coexpression with the herpes simplex virus UL15 protein. J. Virol. 71:9118–9123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kouklis P. D., Merdes A., Papamarcaki T., Georgatos S. D. 1993. Transient arrest of 3T3 cells in mitosis and inhibition of nuclear lamin reassembly around chromatin induced by anti-vimentin antibodies. Eur. J. Cell Biol. 62:224–236 [PubMed] [Google Scholar]
- 40. Kuhn J., et al. 2008. Partial functional complementation of a pseudorabies virus UL25 deletion mutant by herpes simplex virus type 1 pUL25 indicates overlapping functions of alphaherpesvirus pUL25 proteins. J. Virol. 82:5725–5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lachish-Zalait A., et al. 2009. Transportin mediates nuclear entry of DNA in vertebrate systems. Traffic 10:1414–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lim R. Y., Aebi U., Fahrenkrog B. 2008. Towards reconciling structure and function in the nuclear pore complex. Histochem. Cell Biol. 129:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Loret S., Guay G., Lippé R. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82:8605–8618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lu X., Triezenberg S. J. 2010. Chromatin assembly on herpes simplex virus genomes during lytic infection. Biochim. Biophys. Acta 1799:217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lukashchuk V., Everett R. D. 2010. Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J. Virol. 84:4026–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Luxton G. W., et al. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. U. S. A. 102:5832–5837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mabit H., et al. 2002. Intact microtubules support adenovirus and herpes simplex virus infections. J. Virol. 76:9962–9971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marozin S., Prank U., Sodeik B. 2004. Herpes simplex virus type 1 infection of polarized epithelial cells requires microtubules and access to receptors present at cell-cell contact sites. J. Gen. Virol. 85:775–786 [DOI] [PubMed] [Google Scholar]
- 49. Maurer U. E., Sodeik B., Grünewald K. 2008. Native 3D intermediates of membrane fusion in herpes simplex virus 1 entry. Proc. Natl. Acad. Sci. U. S. A. 105:10559–10564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McNab A. R., et al. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mettenleiter T. C., Klupp B. G., Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143:222–234 [DOI] [PubMed] [Google Scholar]
- 52. Nagel C. H., et al. 2008. Nuclear egress and envelopment of herpes simplex virus capsids analyzed with dual-color fluorescence HSV-1(17+). J. Virol. 82:3109–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Newcomb W. W., Booy F. P., Brown J. C. 2007. Uncoating the herpes simplex virus genome. J. Mol. Biol. 370:633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Newcomb W. W., Brown J. C. 1994. Induced extrusion of DNA from the capsid of herpes simplex virus type 1. J. Virol. 68:433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Newcomb W. W., Cockrell S. K., Homa F. L., Brown J. C. 2009. Polarized DNA ejection from the herpesvirus capsid. J. Mol. Biol. 392:885–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Newcomb W. W., Homa F. L., Brown J. C. 2006. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J. Virol. 80:6286–6294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Newcomb W. W., Homa F. L., Brown J. C. 2005. Involvement of the portal at an early step in herpes simplex virus capsid assembly. J. Virol. 79:10540–10546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Newcomb W. W., Thomsen D. R., Homa F. L., Brown J. C. 2003. Assembly of the herpes simplex virus capsid: identification of soluble scaffold-portal complexes and their role in formation of portal-containing capsids. J. Virol. 77:9862–9871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ogasawara M., Suzutani T., Yoshida I., Azuma M. 2001. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J. Virol. 75:1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Oh J., Fraser N. W. 2008. Temporal association of the herpes simplex virus genome with histone proteins during a lytic infection. J. Virol. 82:3530–3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. O'Hara M., et al. 2010. Mutational analysis of the herpes simplex virus type 1 UL25 DNA packaging protein reveals regions that are important after the viral DNA has been packaged. J. Virol. 84:4252–4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ojala P. M., Sodeik B., Ebersold M. W., Kutay U., Helenius A. 2000. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell. Biol. 20:4922–4931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pasdeloup D., Blondel D., Isidro A. L., Rixon F. J. 2009. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J. Virol. 83:6610–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Peng L., Ryazantsev S., Sun R., Zhou Z. H. 2010. Three-dimensional visualization of gammaherpesvirus life cycle in host cells by electron tomography. Structure 18:47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Peters R. 2009. Translocation through the nuclear pore: Kaps pave the way. Bioessays 31:466–477 [DOI] [PubMed] [Google Scholar]
- 66. Phelan A., Dunlop J., Patel A. H., Stow N. D., Clements J. B. 1997. Nuclear sites of herpes simplex virus type 1 DNA replication and transcription colocalize at early times postinfection and are largely distinct from RNA processing factors. J. Virol. 71:1124–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Preston V. G., Murray J., Preston C. M., McDougall I. M., Stow N. D. 2008. The UL25 gene product of herpes simplex virus type 1 is involved in uncoating of the viral genome. J. Virol. 82:6654–6666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Radtke K., et al. 2010. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 6:e1000991 doi10.1371/journal.ppat.1000991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rajcáni J., Andrea V., Ingeborg R. 2004. Peculiarities of herpes simplex virus (HSV) transcription: an overview. Virus Genes 28:293–310 [DOI] [PubMed] [Google Scholar]
- 70. Rao V. B., Feiss M. 2008. The bacteriophage DNA packaging motor. Annu. Rev. Genet. 42:647–681 [DOI] [PubMed] [Google Scholar]
- 71. Reynolds E. S. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Roberts A. P., et al. 2009. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J. Virol. 83:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Roizman B., Knipe D. M., Whitley R. J. 2007. Herpes simplex viruses, p. 2502–2601 In Knipe D. M., Howley P. M. (ed.), Fundamental virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 74. Roos W. H., Ivanovska I. L., Evilevitch A., Wuite G. J. 2007. Viral capsids: mechanical characteristics, genome packaging and delivery mechanisms. Cell. Mol. Life Sci. 64:1484–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Roos W. H., et al. 2009. Scaffold expulsion and genome packaging trigger stabilization of herpes simplex virus capsids. Proc. Natl. Acad. Sci. U. S. A. 106:9673–9678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Salmons T., et al. 1997. Vaccinia virus membrane proteins p8 and p16 are cotranslationally inserted into the rough endoplasmic reticulum and retained in the intermediate compartment. J. Virol. 71:7404–7420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sambrook J., Fritsch E. F., Maniatis T. 1989. Expression of cloned genes in cultured mammalian cells, p. 16.14–16.16 In Sambrook J., Fritsch E. F., Maniatis T. (ed.), Molecular cloning: a laboratory manual, 2nd ed., vol. 3 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 78. Schepis A., Schramm B., de Haan C. A., Krijnse-Locker J. 2006. Vaccinia virus-induced microtubule-dependent cellular rearrangements. Traffic 7:308–323 [DOI] [PubMed] [Google Scholar]
- 79. Schiedner G., Hertel S., Kochanek S. 2000. Efficient transformation of primary human amniocytes by E1 functions of Ad5: generation of new cell lines for adenoviral vector production. Hum. Gene Ther. 11:2105–2116 [DOI] [PubMed] [Google Scholar]
- 80. Scholtes L., Baines J. D. 2009. Effects of major capsid proteins, capsid assembly, and DNA cleavage/packaging on the pUL17/pUL25 complex of herpes simplex virus 1. J. Virol. 83:12725–12737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Schramm B., Krijnse-Locker J. K. 2005. Cytoplasmic organization of Poxvirus DNA replication. Traffic 6:839–846 [DOI] [PubMed] [Google Scholar]
- 82. Simon L. D., Anderson T. F. 1967. The infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope. I. Attachment and penetration. Virology 32:279–297 [DOI] [PubMed] [Google Scholar]
- 83. Slanina H., Weger S., Stow N. D., Kuhrs A., Heilbronn R. 2006. Role of the herpes simplex virus helicase-primase complex during adeno-associated virus DNA replication. J. Virol. 80:5241–5250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sodeik B., Ebersold M. W., Helenius A. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136:1007–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Steven A. C., Heymann J. B., Cheng N., Trus B. L., Conway J. F. 2005. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr. Opin. Struct. Biol. 15:227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Stow N. D. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 75:10755–10765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Thurlow J. K., Murphy M., Stow N. D., Preston V. G. 2006. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J. Virol. 80:2118–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Thurlow J. K., et al. 2005. The herpes simplex virus type 1 DNA packaging protein UL17 is a virion protein that is present in both the capsid and the tegument compartments. J. Virol. 79:150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Trotman L. C., Mosberger N., Fornerod M., Stidwill R. P., Greber U. F. 2001. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell Biol. 3:1092–1100 [DOI] [PubMed] [Google Scholar]
- 90. Trus B. L., et al. 2004. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J. Virol. 78:12668–12671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Trus B. L., Newcomb W. W., Booy F. P., Brown J. C., Steven A. C. 1992. Distinct monoclonal antibodies separately label the hexons or the pentons of herpes simplex virus capsid. Proc. Natl. Acad. Sci. U. S. A. 89:11508–11512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Trus B. L., et al. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol. Cell 26:479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Turcotte S., Letellier J., Lippé R. 2005. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J. Virol. 79:8847–8860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Varnum S. M., et al. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wills E., Scholtes L., Baines J. D. 2006. Herpes simplex virus 1 DNA packaging proteins encoded by UL6, UL15, UL17, UL28, and UL33 are located on the external surface of the viral capsid. J. Virol. 80:10894–10899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wolfstein A., et al. 2006. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic 7:227–237 [DOI] [PubMed] [Google Scholar]
- 97. Zhou F. C., et al. 2002. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 76:6185–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zhu F. X., Li X., Zhou F., Gao S. J., Yuan Y. 2006. Functional characterization of Kaposi's sarcoma-associated herpesvirus ORF45 by bacterial artificial chromosome-based mutagenesis. J. Virol. 80:12187–12196 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.