Abstract
The fourth component of human complement (C4) plays an important role in innate immune function. C4 activity has been observed to be significantly lower in patients with chronic hepatitis C virus (HCV) infections, although the mechanism remains unknown. In this study, we have examined the mechanisms of C4 regulation by HCV. Liver biopsy specimens from patients with chronic HCV infections displayed significantly lower C4 mRNA levels than liver tissue samples from patients with unrelated liver disease. Further, C4 mRNA levels of the two isoforms (C4A and C4B) were significantly reduced in hepatocytes transfected with RNA from HCV genotype 1a or 2a. Subsequently, a significant C4 regulatory role of HCV core or NS5A upon C4 promoter activity was observed. HCV core or NS5A transgenic mice displayed a reduction in C4 mRNA. Gamma interferon (IFN-γ)-induced C4 promoter activation was also impaired in the presence of HCV proteins. We further demonstrated that HCV core reduced the expression of upstream stimulating factor 1 (USF-1), a transcription factor important for basal C4 expression. On the other hand, the expression of interferon regulatory factor 1 (IRF-1), which is important for IFN-γ-induced C4 expression, was inhibited by hepatocytes expressing HCV NS5A. These results underscore the roles of HCV proteins in innate immune regulation in establishing a chronic infection.
INTRODUCTION
The complement system is a set of biochemical pathways that helps to clear pathogens from an organism as part of the innate and acquired immunity programs. Activation of the complement system triggers a wide range of cellular responses ranging from apoptosis to opsonization (27). The complement system plays a critical role in the pathogenesis of a variety of chronic human diseases. Viruses have been shown to attenuate complement activation. Recently, the nonstructural protein (NS1) from flaviviruses (dengue fever virus, West Nile virus, and yellow fever virus) has been reported to attenuate complement activation by interacting with several complement components (1, 8). Results from another study suggest that the complement system is also involved in the pathogenesis of a variety of liver disorders, including liver injury and repair, fibrosis, viral hepatitis, alcoholic liver disease, and liver ischemia/reperfusion injury (24). The fourth component of the complement system, C4, plays a vital role in mounting a proper immune response against infection (35). Chronic hepatitis C virus (HCV) infection is a leading cause of progressive liver disease. The mechanisms responsible for HCV persistence are not well understood, although the interactions between HCV and host cells appear to play an important role. Dumestre-Perard et al. (11) reported in a study of a large cohort that in patients with chronic HCV infections, the blood level of specific complement components is depleted, and C4 activity is significantly lower. A decrease in specific C4 activity was reported among relapsers compared with sustained responders, before and during therapy, suggesting its role in viral clearance. These results pointed to a potential predictive function of C4 specific activity to monitor the response to therapy. No significant correlations were found between C4 specific activity values and virology, enzymology, and liver histological parameters. Thus, the mechanism for C4 regulation by HCV remains unknown.
C4 is a polymorphic serum protein consisting of two isoforms, C4A and C4B. These isoforms are encoded by two separate genes that are located 10 kb apart within the central portion of the major histocompatibility complex (MHC) on chromosome 6p (6). The promoter regions of the C4 gene are conserved in C4A and C4B (32). As with other components of the complement system, C4 is expressed primarily in the liver and in macrophages, and its expression is induced in response to acute inflammation or tissue injury (13). Indeed, high plasma C4 concentration is primarily due to liver synthesis, while macrophages deliver complement locally. Because of these specific features, the C4 gene is an excellent model for understanding the transcriptional regulation of the complement genes. Unlike most other acute-phase reactants, gamma interferon (IFN-γ) is the only known inducer of C4 expression in the liver and serves to increase mRNA and protein expression by 50% (21). In HepG2 cells, induction by IFN-γ occurs predominantly through stabilization of C4 mRNA.
In this study, we have examined the mechanism for regulation of C4 complement synthesis in hepatocytes by HCV. Our results suggest that HCV core and NS5A proteins transcriptionally downregulate C4 expression by modulating the expression of upstream stimulating factor 1 (USF-1) and interferon regulatory factor 1 (IRF-1), respectively. Thus, HCV infection appears to play a significant role in innate immune modulation, favoring establishment of chronic infection and liver disease progression.
MATERIALS AND METHODS
Patient materials.
Paired serum samples and liver biopsy specimens from patients with chronic HCV infection (n = 12) were used in this study; their clinical data are listed in Table 1. Clinical specimens were collected after approval of the research protocol by the Saint Louis University Institutional Review Board. All subjects gave written informed consent. Patients were seropositive for anti-HCV and HCV RNA. Viral load was assessed and recorded within 3 months of biopsy as determined by commercial assays and expressed in international units per milliliter (IU/ml). HCV genotypes were similarly determined by widely available clinical assays. Liver biopsy specimens were read by an experienced liver pathologist, and the severity of hepatitis was graded and staged according to a system described by Scheuer (29). Other causes of chronic liver disease were ruled out. Control liver biopsy specimens were obtained from HCV-negative individuals undergoing hepatic resection for colorectal cancer metastasis. Sera from healthy volunteers were used as controls.
Table 1.
Clinical and virological parameters of 12 HCV-infected patients examined in this study
| Liver biopsy specimena | Sexb | Fibrosis stage | Grade of activity | HCV viral load (IU/ml) | Rheumatoid factorc | Total C4 level (ng/ml) (mean ± SD) |
|---|---|---|---|---|---|---|
| 158 | M | 1 | 6,687,000 | − | 696.6 ± 15.2 | |
| 164 | M | 2 | Mild activity grade 2 | 1,095,730 | − | 1,083.3 ± 60.2 |
| 165 | F | 3 | Mild activity grade 2 | 1,001,400 | − | 1,366.6 ± 35.1 |
| 167 | F | 0 | 928,400 | − | 1,230.0 ± 73.2 | |
| 170 | M | 0 | 69,000,000 | − | 1,206.6 ± 15.2 | |
| 171 | F | 3 | Mild activity grade 2 | 2,690,000 | − | 1,290.0 ± 55.6 |
| 173 | F | 2 | Periportal fibrosis, mild activity grade 2 | 432,700 | + (240) | 1,133.3 ± 83.2 |
| 176 | M | 1 | Stage 1 activity | 1,872,412 | − | 959.6 ± 15.5 |
| 180 | M | 1 | Stage 2 activity | 3,624,000 | − | 1,004.0 ± 89.1 |
| 181 | F | 3 | Stage 2 activity | 3,929,434 | − | 829.0 ± 69.5 |
| 183 | F | 1 | Mild activity grade 2 | 160,700 | + (480) | 619.3 ± 11.0 |
| 186 | M | 2 | Stage 2 activity | 5690,000 | − | 1,088.6 ± 2.3 |
All 12 patients were infected with hepatitis C virus (HCV) genotype 1a.
The sex of the patient is indicated as follows: M, male; F, female.
The presence (+) or absence (−) of rheumatoid factor in blood and the amount found (in international units per milliliter).
Cells and transfections.
Huh7 cells were transfected with plasmid DNA from a mammalian expression vector (pcDNA3) containing HCV-specific genomic region, HCV full-length (FL) genome, or a clone with a mutation in the NS5B region (pCI-neo-HCV FL) under the control of a cytomegalovirus promoter using Lipofectamine 2000 (Life Technologies, Inc.). Stable cell colonies were selected using neomycin and pooled for subsequent studies to avoid artifactual results from clonal variation (4). Parental Huh7 cells transfected with empty vector were used in parallel as a control. Stable transfectants of Huh7 cells were maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal calf serum and a lower dose of the selection antibiotic (400 μg of neomycin/ml). Immortalized human hepatocytes (IHH) were generated and maintained in SABM medium (Lonza, MD) supplemented with 5% heat-inactivated fetal calf serum as previously described (25). HCV genotype 1a (clone H77) was grown in IHH as previously described (15).
BB7 cells, a derivative of Huh7 cells, supporting replication of subgenomic HCV genotype 1b replicon (5) was kindly provided by Charles Rice (Rockefeller University, NY). BB7 cells were grown in medium containing 800 μg/ml of G418 (Geneticin; Gibco/Invitrogen, Carlsbad, CA). Human fibrosarcoma epithelium-like HT1080 cells and human U3A cells were kindly provided by George Stark (Cleveland Clinic Foundation Research Institute, OH). The human cell line U3A is a derivative of the HT1080 cell line, which lacks endogenous Stat-1 (19, 22). These cells were maintained in DMEM supplemented with 10% fetal calf serum.
Construction of C4 promoter luciferase reporter gene and luciferase assay.
A XhoI/HindIII fragment of the C4 promoter containing nucleotides −1148 to +44 (34) was amplified using a forward primer (5′-CTTCTCGAGAACAACCCGGGAGCA-3′) and a reverse primer (5′-CAGAGAAGCTTCATGGCTGGA-3′). Underlined sequences indicate restriction enzyme sites. The amplified gene was cloned into a pGL3-basic vector (Promega Corp., WI). A deletion construct (pGL3-310) was generated by incorporating an XhoI site in the forward primer (5′-CCTGTACTCGAGGGCCATGTT-3′) and a HindIII site in the reverse primer (5′-CAGAGAAGCTTCATGGCTGGA-3′). Huh7 cells were grown to ∼70 to 85% confluence in a 24-well plate overnight and transfected with a C4 promoter-luciferase reporter plasmid (500 ng) with or without the plasmid construct encoding the HCV FL gene, the specific genomic region, or 50 nM specific small interfering RNA (siRNA). The cells were incubated at 37°C for 36 h, treated with IFN-γ (1,000 U/ml) for 12 h or not treated with IFN-γ, and lysed with reporter lysis buffer (Promega). The relative luciferase activity values are from at least three independent experiments.
Real-time PCR.
A quantitative real-time PCR analysis was performed for total C4, C4A, and C4B mRNAs using specific TaqMan primers and probes (Applied Biosystems, Foster City, CA). RNA was isolated from Huh7 cells, IHH, or liver biopsy specimens using Trizol (Invitrogen). cDNA synthesis was done using random hexamers and the Superscript III first-strand synthesis kit (Invitrogen). Total C4 mRNA was evaluated using specific oligonucleotide primers (Applied Biosystems assay identification [ID] Hs00416393-g1 and Mm00550309-m1;) normalizing with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Hs99999905-m1 and Mm99999915-g1) or 18S RNA (Hs03928992-g1).
Two separate reactions were used for quantifying the C4A and C4B genes. Both reaction mixtures contained 6 μM forward (5′ GCA GGA GAC ATC TAA CTG GCT TCT 3′) and 6 μM reverse (5′ CCG CAC CTG CAT GCT CCT 3′) primers, 1× TaqMan universal PCR master mix (No AmpErase UNG) and genomic DNA template. Each reaction mixture contained the C4A-specific TaqMan probe (5′ FAM-ACC CCT GTC CAG TGT TAG-MGB 3′ where FAM stands for 6-carboxyfluorescein and MGB stands for minor groove binding) and the FAM-labeled C4B specific TaqMan probe (5′ FAM-ACC TCT CTC CAG TGA TAC-MGB 3′) separately. The boldface letters in the probe sequences display differences in the two probes corresponding to the nucleic acid variations that distinguish the C4A and C4B genes. The FAM-labeled 1× GAPDH gene expression assay was used as an endogenous control in a total volume of 25 μl. All reactions were performed in triplicate with an ABI Prism 7700 analyzer. Thermocycling was initiated by incubating the reaction mixture at 95°C for 10 min to denature cDNA and to activate AmpliTaq Gold DNA polymerase, followed by 40 cycles, with 1 cycle consisting of two steps, 95°C for 15 s and 60°C for 1 min. The fluorescence intensity was measured during the 60°C step. Samples were analyzed at least 3 times.
The levels of upstream stimulating factor 1 (USF-1) and interferon regulatory factor 1 (IRF-1) mRNA in liver biopsy specimens were analyzed similarly using specific TaqMan gene expression assay (Applied Biosystems assay ID Hs00273038-m1 and Hs00971960-m1).
C4 ELISA.
HCV-infected patient sera or hepatocyte cultures were used for total C4 estimation using a commercially available kit (Abnova, Taiwan). Culture fluid from HCV-infected hepatocytes was replaced with fresh culture medium after 2 days of infection, and the cultures were incubated for another 2 days prior to collection for enzyme-linked immunosorbent assay (ELISA).
Luciferase assay.
Huh7 or Stat-1 knockout cells (U3A) were transfected with the pGL3-C4 promoter construct (100 ng/well) alone or together with HCV structural protein (core), nonstructural proteins (NS2, NS3, or NS5A), or full-genome length construct (500 ng/well) in a 24-well plate. The cells were lysed with reporter lysis buffer 48 h after transfection, and luciferase activity was measured. Whenever necessary, IRF-1 or USF-1 siRNA (Santa Cruz Biotechnology) was transfected into Huh7 cells to knock down IRF-1 or USF-1 prior to transfection of the luciferase construct.
Western blot analysis.
Proteins in cell lysates were resolved by SDS-PAGE, transferred onto a nitrocellulose membrane, and blotted with the appropriate primary antibody. Positive signals were detected using a peroxidase-conjugated secondary antibody. Protein bands were visualized by using an enhanced chemiluminescence Super Signal West detection kit (Thermo Chemical Company, IL). Cellular actin was detected in a similar manner to compare the protein load in each lane.
Fluorescence and confocal microscopy.
The cells were fixed with formaldehyde (4%) and incubated with monoclonal antibody to HCV core (Thermo Fischer), NS5A (kindly provided by Chen Liu, University of Florida, Gainesville), and/or polyclonal antibody to USF-1 or IRF-1 (Santa Cruz Biotechnology). The cells were stained with a secondary antibody conjugated to a fluorochrome (Molecular Probes, Carlsbad, CA). The cells were stained with DAPI (4′,6′-diamidino-2-phenylindole) for nuclear staining (Molecular Probes) and mounted for confocal microscopy (Olympus FV1000). Whenever necessary, the images were merged digitally to monitor localization of two different proteins.
Statistical analysis.
Results were expressed as the mean ± standard deviation (SD), and statistical analyses were performed using two-tailed unpaired Student t test or one-way analysis of variance (ANOVA) in GraphPad Prism 5 (GraphPad, La Jolla, CA). A P value of <0.05 was considered statistically significant.
RESULTS
HCV infection inhibits the expression of C4 complement component.
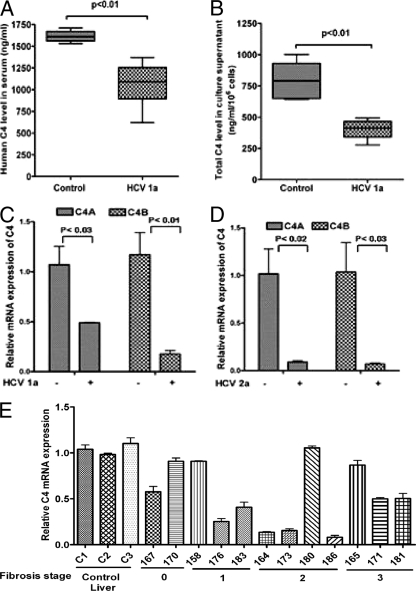
The complement component C4 contributes to the clearance of several viral pathogens through its functions as an opsonin and by virtue of its central role in the classical and lectin complement activation pathways (27). C4 is one of the major targets for viruses to evolve strategies to overcome the immune response of the host and consequently establish persistent infection. The liver is the main source of C4, and constitutive expression of C4 is enhanced during the acute-phase response. In order to investigate the role of HCV infection on C4 expression, we first evaluated the total level of C4 protein in patient sera by ELISA. Five sera from healthy individuals were included as a control. HCV-infected patient sera exhibited a significant reduction of total C4 level (1,055.0 ± 24.0 ng/ml) compared to control sera (1,610.0 ± 6.1 ng/ml) (P < 0.001) (Table 1 and Fig. 1A). A significant reduction in C4 protein level in HCV genotype 1a (clone H77)-infected hepatocyte culture supernatant was also observed (Fig. 1B). C4 exists in two isoforms, C4A and C4B, and we measured mRNA expression for both isoforms. The levels of C4A and C4B mRNA expression were reduced in cells transfected with HCV genotype 1a (clone H77) RNA in immortalized human hepatocytes (IHH) and genotype 2a (clone JFH1) RNA in Huh7.5 cells (Fig. 1C and D).
Fig. 1.
HCV infection represses C4 expression. (A) There was a significant reduction in total C4 expression displayed in sera from patients infected with HCV genotype 1a (HCV 1a) (n = 12) compared with sera from healthy controls (n = 6). (B) Comparison of total C4 protein level in immortalized human hepatocytes (IHH) and IHH infected with HCV genotype 1a. (C and D) IHH were electroporated with HCV genotype 1a (C) or Huh7.5 cells were electroporated with HCV genotype 2a (D), and the cells were allowed to grow for 6 days. Cellular RNA was extracted, and the levels of mRNA expression of two isoforms of C4 (C4A and C4B) were measured by real-time PCR using specific TaqMan primers and probes. The values are means plus standard deviation (error bars). (E) C4 mRNA expression was measured in HCV genotype 1a-infected paired liver tissue samples (marked by 3-digit numbers) by real-time PCR. Results were normalized to endogenous 18S RNA and compared with non-HCV-infected control liver tissues (C1, C2, and C3). C4 mRNA expression in control liver specimen C1 was arbitrarily set at 1, and the comparative results are shown from individual samples (P < 0.0001 for the values for the group of experimental samples compared to the values for the controls using one-way ANOVA). The values for samples 158, 165, 170, and 180 are not statistically significantly different (P > 0.05) from the values for the controls by Dunnett's multiple-comparison test. The stage of fibrosis of the patient liver is shown at the bottom.
The level of C4 mRNA in paired HCV-infected liver biopsy specimens was evaluated. To do this, RNA was extracted from liver biopsy samples of 12 HCV genotype 1a-infected patients, and C4 mRNA expression was measured by real-time PCR. Eight out of 12 RNA samples from infected liver specimens exhibited a significant reduction in C4 mRNA expression compared with three samples from patients with unrelated liver disease (Fig. 1E). Further, changes in the C4 level did not correlate with the liver fibrosis stage or whether rheumatoid factor was present in blood. Together, these results suggested that HCV infection inhibits C4 complement component expression at the transcriptional level within the hepatocytes, which is also reflected in the total C4 protein level in patient sera. Next, we determined whether the observed effects are due to cross talk between cellular genes and HCV proteins.
HCV core and NS5A proteins attenuate C4 promoter activity and reduce C4 expression in vivo.
Since we observed regulation of C4 at the mRNA level, we assumed that a HCV protein(s) regulates C4 complement component at the transcriptional level. In order to identify the viral proteins that are involved in C4 regulation, we constructed a C4 promoter (nucleotides −1148 to +44) with a luciferase reporter gene (33) and studied the effect of HCV protein expression in a permissive cell culture system. Plasmid containing HCV core, NS2, NS3/NS4A, or NS5A DNA or the full-length (FL) genome was separately transfected along with the C4 promoter construct in Huh7 cells. The cells were lysed 48 h after transfection, and promoter activation was measured as a function of luciferase activity. Our results suggested that transfection of the FL genome reduced basal C4 promoter activity compared to mock-transfected control cells. Further, core or NS5A expression significantly reduced C4 promoter activity. However, transfection with the plasmid containing NS2 and NS3/NS4A DNA had no significant effect on C4 promoter activity (Fig. 2A). Further study revealed that downregulation of C4 promoter activation occurred in a dose-dependent manner in cells transfected with HCV core or NS5A DNA (Fig. 2B). To delineate the regulatory element in the C4 promoter sequence, we constructed a C4 deletion mutant (nucleotides −310 to +44). The luciferase activity of the C4 deletion mutant was similar to that of cells containing the full-length C4 promoter (Fig. 2C). Thus, the results agree with the previous report (33) suggesting that the region from nucleotides −310 to +44 contains essential elements responsible for C4 promoter activation. IFN-γ is the only known inducer of C4 expression in the liver. Next, we examined the effect of viral protein expression on IFN-γ-induced C4 promoter activity. Our results suggested that IFN-γ-induced C4 promoter activity is significantly attenuated in both HCV core- and NS5A-expressing cells (Fig. 2D).
Fig. 2.
HCV core and NS5A significantly inhibit C4 promoter activity. (A) HCV polyprotein expression from full-length (FL) genome construct, core, or NS5A significantly suppressed C4 promoter activity in a luciferase reporter assay. However, nonstructural protein NS2 or NS3 did not affect C4 promoter activity. The results obtained were statistically significantly different (P < 0.0001) from the values for the vector control by the unpaired two-tailed t test. (B) Core and NS5A proteins downregulated C4 promoter activity in a dose-dependent manner (0.2 μg and 0.5 μg [the amount is indicated by the height of the black triangle below the bars]). The results obtained were statistically significantly different (P < 0.0001) from the value for the vector control by unpaired two-tailed t test. (C) Core and NS5A proteins exhibited similar levels of suppression of luciferase activity in cells transfected with a plasmid with the wild-type (WT) C4 promoter and in cells transfected with a plasmid in which the C4 promoter had been deleted (pGL3-C4-310). (D) HCV FL, core, or NS5A protein also suppressed IFN-γ-induced C4 promoter activation.
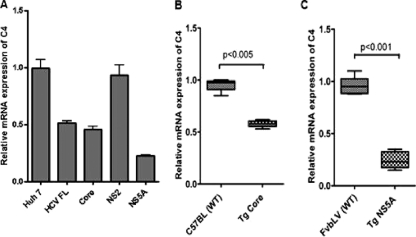
Hepatocytes expressing HCV FL, core, and NS5A displayed reduced levels of C4 mRNA expression (Fig. 3A). On the other hand, in a similar experiment in which NS2 was used as a specific control, the C4 mRNA expression level did not change compared to the vector control. We further verified C4 regulation at the transcriptional level in an in vivo system using core or NS5A (18, 31) transgenic mouse liver (Fig. 3B and C). Our results suggested that C4 mRNA expression was significantly decreased in mouse liver expressing core or NS5A proteins. Thus, HCV core and NS5A proteins played an important role in C4 downregulation in transgenic mouse liver and in human hepatocytes.
Fig. 3.
HCV protein expression downregulates C4 mRNA expression in human hepatocytes and in transgenic mouse liver. (A) Total C4 mRNA level was suppressed in Huh7 cells transfected with HCV FL, core, or NS5A DNA (stable transfectants), while cells transfected with NS2 and used as a negative control did not display any significant effect on C4 mRNA level. GAPDH was used as an endogenous control. C4 mRNA expression in parental hepatocytes (Huh7) was arbitrarily set at 1; the other results shown are from stable transfectants and the values are shown relative to C4 mRNA expression in Huh7 cells (P < 0.0001 for the values for the group of experimental samples using one-way ANOVA). (B and C) C4 mRNA level was measured by real-time PCR and revealed downregulation in transgenic mouse liver expressing HCV core (Tg Core) or NS5A protein (Tg NS5A). Liver RNA from nontransgenic littermates with the same genetic background (C57BL or FvbLV) was used as a control for comparison. Results show the average value of C4 mRNA expression from 6 individual transgenic mice. GAPDH was used an endogenous control.
HCV core protein suppresses USF-1 expression in hepatocytes.
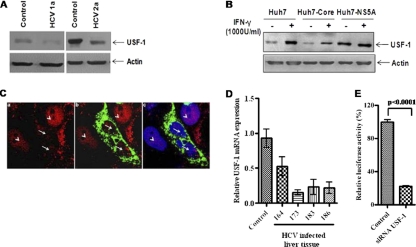
The basic helix-loop-helix leucine zipper transcription factor USF-1 binds to characteristic E-box elements via base-specific DNA contacts with its basic region (9). The C4 promoter contains an E-box element that is important for basal C4 promoter activation. Several different DNA-binding assays indicated that the C4 E-box motif is recognized by USF-1 (13). Binding of USF-1 to the conserved E-box motif CACGTG (E-C4) element correlates with its ability to regulate the C4 complement promoter. To understand the role of USF-1 in HCV protein-expressing cells, we studied the basal expression level of USF-1 in HCV-infected hepatocytes. Western blot analysis suggested that USF-1 expression was suppressed in hepatocytes infected with HCV genotype 1a or 2a (Fig. 4A). Basal USF-1 expression was also low in Huh7 cells transfected with plasmid containing HCV core DNA compared to cells transfected with plasmid containing NS5A DNA (Fig. 4B). Treatment of Huh7 cells with IFN-γ increased USF-1 expression. However, core-expressing Huh7 cells displayed a modest increase in USF-1 upon IFN-γ treatment. USF-1 expression in parental Huh7 cells and NS5A-expressing cells was not altered upon IFN-γ treatment. Actin levels were presented in paired lanes, implying similar protein loads. Thus, a reduced level of USF-1 in HCV core-expressing cells may account for C4 suppression.
Fig. 4.
USF-1 expression status and C4 promoter activation. (A) Western blot analysis to determine the level of expression of USF-1 in hepatocytes infected with HCV genotype 1a or 2a. (B) Western blot analysis to determine the level of expression of USF-1 in IFN-γ-treated (+) or non-IFN-γ-treated (−) mock-transfected Huh7 cells or Huh7 cells transfected with HCV core or NS5A. Cellular actin was used as an internal control to compare the protein load in each lane. (C) Localization of USF-1 in core-expressing cells by immunofluorescence. Huh7 cells were transiently transfected with HCV core plasmid. The cells were stained after 72 h of transfection with antibody to USF-1 (red) tagged with anti-rabbit Alexa Fluor 594 (a), along with antibody to core (green) tagged with anti-mouse Alexa Fluor 488 (b); merged panels were stained with DAPI (c). Fluorescence, shown in panels a and b, displayed a reduced nuclear expression of USF-1 in HCV core-expressing hepatocytes (white arrows). Cells with no HCV core protein displayed a strong nuclear localization of USF-1 (white arrowheads). (D) USF-1 mRNA expression from selected liver biopsy specimens from HCV-infected patients in comparison to liver biopsy specimens from controls by real-time PCR (P < 0.01 for the values for experimental samples compared to controls using one-way ANOVA). (E) Knockdown of USF-1 by siRNA attenuates IFN-γ-induced C4 promoter activation in Huh7 cells.
On the basis of these results, we next performed an immunofluorescence study to understand cellular localization of USF-1 transcription factor in the presence and absence of IFN-γ. Transient transfection of cells with plasmid containing HCV core DNA did not alter nuclear localization of USF-1, although the level of nuclear USF-1 appeared to decrease from that in untransfected parental Huh7 cells (Fig. 4C). IFN-γ treatment of core-transfected or parental Huh7 cells did not display altered nuclear localization of USF-1 (data not shown). Together, these results suggested that HCV core protein modulates USF-1 expression. We further determined the level of USF-1 mRNA by real-time PCR from selected liver biopsy specimens from HCV-infected patients showing the highest level of C4 repression (Fig. 1E). In all samples tested, a significant decrease in USF-1 mRNA level was observed, suggesting a link between C4 regulation and USF-1 expression (Fig. 4D). The binding of USF-1 to the E box was suggested to correlate with its ability to regulate the C4 complement promoter. This was further verified by using USF-1 siRNA, which knocked down C4 promoter activity (Fig. 4E). Taken together, our results suggest that HCV core protein modulates C4 promoter activity by suppressing USF-1 expression.
HCV NS5A modulates IRF-1 protein expression and inhibits C4 promoter activation.
The C4 promoter region contains interferon response elements. Stat-1 is an important transcription factor in the IFN-γ signaling pathway, although its precise role in C4 expression remains unknown. We observed an inhibition of phospho-Stat-1 in IFN-γ-treated hepatocytes expressing NS5A (Fig. 5A). Our data are in agreement with repression of the C4 promoter by NS5A. To further understand the role of Stat-1 in C4 regulation, we transfected a C4 promoter containing a luciferase gene construct into a human fibroblast (U3A) cell line lacking Stat-1 and measured C4 promoter activation in the presence and absence of IFN-γ (Fig. 5B). The results suggested that IFN-γ treatment did not induce C4 promoter activation in Stat-1 knockout cells as observed in the parental control (HT1080) cell line. However, basal C4 promoter activation was observed in Stat-1 knockout cells, which suggests that Stat-1 plays a role in IFN-γ-induced C4 regulation. The fact that Stat-1 plays a role in regulation of the C4 complement promoter was also verified by using Stat-1 siRNA, which knocked down C4 promoter activity (Fig. 5C).
Fig. 5.
HCV NS5A modulates Stat-1 and suppresses C4 promoter activation. (A) Stat-1 activation was examined by transient transfection of HCV NS5A into Huh7 cells. The inhibitory effect of HCV NS5A on IFN-γ-induced Stat-1 phosphorylation is shown. pStat-1, phosphorylated Stat-1. (B) C4 promoter activity was examined in Stat-1 knockout (KO) (U3A) cells. IFN-γ treatment did not induce C4 promoter activation compared to HT1080 cells. (C) Knockdown of Stat-1 by siRNA attenuates IFN-γ-induced C4 mRNA expression in Huh7 cells.
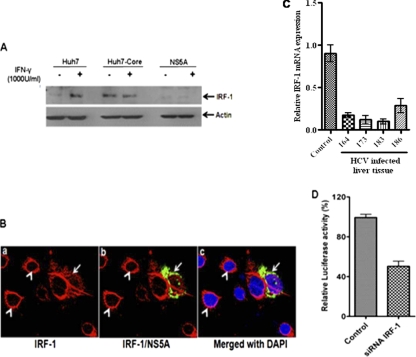
IRF-1 is one of the downstream targets for Stat-1. Therefore, we investigated the effects of HCV proteins on IRF-1 expression. Our Western blot data suggested that IRF-1 expression was almost undetectable in IFN-γ-treated or untreated Huh7 cells stably transfected with plasmid containing HCV NS5A DNA compared to HCV core-expressing cells (Fig. 6A). Similar observations were noted using Huh7 cells bearing the subgenomic replicon. Further, we determined the level of IRF-1 mRNA by real-time PCR from selected liver biopsy specimens of HCV-infected patients showing the highest level of C4 repression. In all samples tested, a significant decrease in the level of IRF-1 mRNA was observed, suggesting a link between C4 regulation and IRF-1 expression (Fig. 6C). Introduction of NS5A by transient transfection diminished the intensity of perinuclear IRF-1 compared to parental Huh7 cells (Fig. 6B). Unlike transcription factor USF-1, treatment of cells with IFN-γ translocated most of the IRF-1 into the nucleus (data not shown). The results from this study suggested that IRF-1 is differentially regulated by HCV core and NS5A proteins.
Fig. 6.
IRF-1 expression and C4 promoter activation. (A) Huh7 cells stably transfected with HCV core or NS5A were analyzed for expression of IRF-1 by Western blotting. (B) IRF-1 expression in NS5A-expressing cells by immunofluorescence. Huh7 cells were transiently transfected with HCV NS5A plasmid. (a and b) The cells were stained after 72 h of transfection with antibody to IRF-1 (red) tagged with anti-rabbit Alexa Fluor 594 (a) or antibody to NS5A (green) tagged with anti-mouse Alexa Fluor 488 merged with IRF-1 (b). (c) A reduced expression level of IRF-1 that partially colocalized with NS5A in DAPI-stained cells (shown by arrow) was detected. The absence of HCV NS5A protein was displayed as a clear ring like the perinuclear appearance of IRF-1 (white arrowheads). (C) IRF-1 mRNA expression from selected liver biopsy specimens from HCV-infected patients and comparison to liver biopsy specimens from controls by real-time PCR (P < 0.001 for the values for experimental samples compared to the control values by one-way ANOVA). (D) Knockdown of IRF-1 attenuates IFN-γ-induced C4 promoter activation in Huh7 cells. The results obtained were statistically significantly different (P < 0.0003) from the values for the control by unpaired two-tailed t test.
Since the phosphorylation of Stat-1 and expression of IRF-1 are inhibited by HCV infection and by the presence of NS5A protein (17, 26), we examined the effect of IFN-γ on C4 promoter activation in Huh7 cells in which IRF-1 had been knocked down. To do this, siRNA directed against IRF-1 was transfected into Huh7 cells. Transfection of the C4 promoter into cells where IRF-1 had been knocked down exhibited ∼50% suppression in IFN-γ-induced C4 promoter activation compared to control Huh7 cells (Fig. 6D). Together, these results suggested that HCV NS5A suppresses IRF-1 expression by modulating Stat-1 phosphorylation and that it inhibits C4 expression.
DISCUSSION
The complement system has been shown to contribute to host protection from virus infection. C4 plays a vital role in mounting an effective immune response. In this study, we have shown a significant reduction of C4 mRNA expression in liver tissue samples and sera from patients with chronic HCV infections. C4 reduction did not appear to be related to the stage of liver disease or viral load in patients. A reduced level of C4 mRNA expression was also observed in vitro in cells expressing HCV protein. Our results suggested that expression of HCV core and NS5A protein significantly reduced basal and IFN-γ-induced C4 promoter activity. Further in-depth analysis suggested that HCV core and NS5A proteins repress C4 expression by modulating upstream stimulatory factor 1 (USF-1) and interferon regulatory factor 1 (IRF-1) expression, respectively.
The transcriptional response of the host to HCV infection can vary considerably (30). Suppression of other complement components, C1 proteins, C6, and C9, was also observed in our HCV patient liver biopsy specimens (data not shown). Interestingly, C6 and C9 genes were found to be downregulated in a study examining tissue samples from patients with cirrhosis that found a cancer-associated molecular signature that could be used to separate patients with chronic liver disease into a high-risk group and a low-risk group for hepatocellular carcinoma (16). However, when tested, the levels of many other complement components primarily regulated by hepatocytes in the liver, such as C5, C8, factor B, MASP-1, or MBL-2, were not reduced in liver biopsy specimens. Further studies should clarify whether and to what extent complement regulation is linked to liver dysfunction. We have previously shown that transcription of HCV core and NS5A proteins plays a regulatory role on a number of cellular genes (2), and our current observations could also be due to the effect of HCV proteins on complement-related genes via a common or distinct transcriptional regulatory control. Future in-depth study on other complement-related genes should provide novel information on their regulation by HCV proteins.
Activation of complement gene expression plays a major role in the host response to pathogens (24). Immune defects associated with a deficiency in the classical complement pathway lie at the level of B cell priming and failure to form germinal centers (12). IgG1 restriction, low titer, and a delayed antibody appearance are observed during persistent HCV infection (7). HCV assembles intracellularly and is most likely released from the endoplasmic reticulum of cells by a secretory mechanism. As a result, it is unlikely that HCV would be cleared by antibody-dependent cellular cytotoxicity, although an efficient neutralization of extracellular virus is likely to occur in the presence of serum complement. We observed previously that the addition of serum complement leads to a significant enhancement of pseudotype neutralization by antibodies (20). In patients with chronic HCV infections, the level of specific complement components is depleted, and C4 activity is significantly lower in blood (10, 11). A recent report suggested that other flaviviruses use their nonstructural protein, NS1, to attenuate complement activation by directly interacting with C4, thus favoring viral persistence (1).
The induction of complement synthesis occurs primarily in the liver and in macrophages and is mediated, at least in part, by increased transcription of the complement genes. The C4 promoter region contains an interferon gene regulatory element (IRE) sequence, and transcription factors that respond to IFN-γ are involved in promoter activation. Earlier studies on the C4 promoter identified a conserved E-box motif CACGTG (E-C4) at positions −75 to −70, and point mutations within this motif reduced transcription by up to 90% compared to transcription in cells with the full-length promoter (33, 34), indicating that it plays a major role in maintaining C4 basal activity. The E-C4 region is also recognized by USF-1 and may play a major role in C4 regulation. The basic helix-loop-helix leucine zipper transcription factor USF-1 binds to the characteristic E-box elements via base-specific DNA contacts with its basic region (9). Binding of USF-1 to the E-C4 element correlates with its ability to regulate the C4 complement promoter (13). Our results suggested that USF-1 expression is suppressed in Huh7 cells expressing HCV core protein, as well as in liver biopsy samples from patients with HCV infections. USF-1 participates in this immune response phase by regulating C4 complement component gene expression (13). Therefore, a reduction of USF-1 expression by HCV core protein might be involved in C4 suppression.
Stat-1 phosphorylation is suppressed in hepatocytes from patients with HCV infections (26) and in cells expressing HCV NS5A (17). IFN-γ-induced C4 activation requires functional Stat-1. Interferon regulatory factor (IRF) expression is upregulated by Stat-1, which in turn binds to IRE and drives C4 activation. Our results suggested that knockdown of interferon regulatory factor 1 (IRF-1) attenuates IFN-γ-induced C4 promoter activation, which suggests that it plays a role in C4 regulation. NS5A appeared to negatively regulate IRF-1 expression, which might be in part responsible for a decrease in downstream gene expression (14). NS5A alone has been shown to be sufficient to block both the activation of IRF-1 and the induction of IRF-1-dependent cellular promoter activity by double-stranded RNA (dsRNA) (23). Our results suggested that IRF-1 expression was significantly reduced in NS5A-expressing cells but remains unaffected in core-expressing cells, indicating a more pronounced effect of NS5A on C4 regulation. This observation is in agreement with our previous reports that HCV core protein does not activate IRF-1 or class II transactivator (CIITA) promoter activity (3, 28). We have also observed that the basal IRF-1 expression at the mRNA level was not altered in the presence or absence of IFN-γ compared to control hepatocytes. This observation corroborates with the Stat-1 status. However, we observed a lower level of IRF-1 and its partial colocalization with HCV NS5A, not facilitated by proteosomal degradation (unpublished observation). Together, our study suggests that HCV infection suppresses basal C4 expression by modulating USF-1 and IRF-1 expression (Fig. 7). USF-1 and IRF-1 are common transcription factors and may participate in the downregulation of other complement promoter regulatory activities. Further studies should offer additional intricate understanding of complement regulation by HCV for establishment and maintenance of chronic infection.
Fig. 7.
Schematic diagram depicting the possible mechanism for HCV core- or NS5A-induced suppression of C4 complement component. HCV core inhibits USF-1 expression, thus inhibiting a basal reduction of C4 expression. On the other hand, IFN-γ-induced C4 activation is attenuated by HCV NS5A via a reduction of IRF-1 expression. P, phosphate.
ACKNOWLEDGMENTS
We thank Charles Rice, Young Hahn, and Chen Liu for providing research materials; Bob Belshe and Mike Diamond for helpful suggestions, Patricia Osmack for technical assistance, Sandip Bose for helping with data analysis and presentation, and Lin Cowick for preparation of the manuscript.
This work was supported by research grant U54-AI057160 from the NIAID to the Midwest Regional Center of Excellence (MRCE) for Biodefense and Emerging Infectious Diseases Research and by research grant DK80812 from the NIDDK (R.R.) and grant DK081817 from the NIDDK (R.B.R.).
Footnotes
Published ahead of print on 23 February 2011.
REFERENCES
- 1. Avirutnan P., et al. 2010. Antagonism of the complement component C4 by flavivirus nonstructural protein NS1. J. Exp. Med. 207:793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Banerjee A., Ray R. B., Ray R. 2010. Oncogenic potential of hepatitis C virus proteins. Viruses 2:2108–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Basu A., Meyer K., Ray R. B., Ray R. 2001. Hepatitis C virus core protein modulates the interferon-induced transacting factors of Jak/Stat signaling pathway but does not affect the activation of downstream IRF-1 or 561 gene. Virology 288:379–390 [DOI] [PubMed] [Google Scholar]
- 4. Basu A., Meyer K., Ray R. B., Ray R. 2002. Hepatitis C virus core protein is necessary for the maintenance of immortalized human hepatocytes. Virology 298:53–62 [DOI] [PubMed] [Google Scholar]
- 5. Blight K. J., Kolykhalov A. A., Rice C. M. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972–1974 [DOI] [PubMed] [Google Scholar]
- 6. Carroll M. C., Campbell R. D., Bentley D. R., Porter R. R. 1984. A molecular map of the human major histocompatibility class III region linking complement genes C4, C2 and factor B. Nature 307:237–241 [DOI] [PubMed] [Google Scholar]
- 7. Chen M., et al. 1999. Limited humoral immunity in hepatitis C virus infection. Gastroenterology 116:135–143 [DOI] [PubMed] [Google Scholar]
- 8. Chung K. M., et al. 2006. West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory protein factor H. Proc. Natl. Acad. Sci. U. S. A. 103:19111–19116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Corre S., Galibert M. D. 2005. Upstream stimulating factors: highly versatile stress-responsive transcription factors. Pigment Cell Res. 18:337–348 [DOI] [PubMed] [Google Scholar]
- 10. Daoud M. S., et al. 1996. Chronic hepatitis C, cryoglobulinemia, and cutaneous necrotizing vasculitis. Clinical, pathologic, and immunopathologic study of twelve patients. J. Am. Acad. Dermatol. 34:219–223 [DOI] [PubMed] [Google Scholar]
- 11. Dumestre-Perard C., et al. 2002. Complement C4 monitoring in the follow-up of chronic hepatitis C treatment. Clin. Exp. Immunol. 127:131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fischer M. B., et al. 1996. Regulation of the B cell response to T-dependent antigens by classical pathway complement. J. Immunol. 157:549–556 [PubMed] [Google Scholar]
- 13. Galibert M. D., Boucontet L., Goding C. R., Meo T. 1997. Recognition of the E-C4 element from the C4 complement gene promoter by the upstream stimulatory factor-1 transcription factor. J. Immunol. 159:6176–6183 [PubMed] [Google Scholar]
- 14. Jung C. R., Choi S., Im D. S. 2007. The NS5A protein of hepatitis C virus represses gene expression of hRPB10alpha, a common subunit of host RNA polymerases, through interferon regulatory factor-1 binding site. Virus Res. 129:155–165 [DOI] [PubMed] [Google Scholar]
- 15. Kanda T., et al. 2006. Generation of infectious hepatitis C virus in immortalized human hepatocytes. J. Virol. 80:4633–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim J. W., et al. 2004. Cancer-associated molecular signature in the tissue samples of patients with cirrhosis. Hepatology 39:518–527 [DOI] [PubMed] [Google Scholar]
- 17. Lan K., et al. 2007. HCV NS5A inhibits interferon-α signaling through suppression of STAT1 phosphorylation in hepatocyte-derived cell lines. J. Hepatol. 46:759–767 [DOI] [PubMed] [Google Scholar]
- 18. Majumder M., et al. 2003. Expression of hepatitis C virus non-structural 5A protein in the liver of transgenic mice. FEBS Lett. 555:528–532 [DOI] [PubMed] [Google Scholar]
- 19. McKendry R., et al. 1991. High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc. Natl. Acad. Sci. U. S. A. 88:11455–11459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meyer K., et al. 2002. Complement-mediated enhancement of antibody function for neutralization of pseudotype virus containing hepatitis C virus E2 chimeric glycoprotein. J. Virol. 76:2150–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mitchell T. J., et al. 1996. IFN-gamma up-regulates expression of the complement components C3 and C4 by stabilization of mRNA. J. Immunol. 156:4429–4434 [PubMed] [Google Scholar]
- 22. Müller M., et al. 1993. Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-alpha and -gamma signal transduction pathways. EMBO J. 12:4221–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pflugheber J., et al. 2002. Regulation of PKR and IRF-1 during hepatitis C virus RNA replication. Proc. Natl. Acad. Sci. U. S. A. 99:4650–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qin X., Gao B. 2006. The complement system in liver diseases. Cell. Mol. Immunol. 3:333–340 [PubMed] [Google Scholar]
- 25. Ray R. B., Meyer K., Ray R. 2007. Hepatitis C virus core protein promotes immortalization of primary human hepatocytes. Virology 271:193–204 [DOI] [PubMed] [Google Scholar]
- 26. Raychoudhuri A., et al. 2010. Hepatitis C virus infection impairs IRF-7 translocation and interferon-alpha synthesis in immortalized human hepatocytes. J. Virol. 84:10991–10998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ricklin D., Hajishengallis G., Yang K., Lambris J. D. 2010. Complement: a key system for immune surveillance and homeostasis. Nat. Immunol. 11:785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saito K., et al. 2008. Hepatitis C virus inhibits cell surface expression of HLA-DR, prevents dendritic cell maturation, and induces interleukin-10 production. J. Virol. 82:3320–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scheuer P. J. 1991. Classification of chronic viral hepatitis: a need for reassessment. J. Hepatol. 13:372–374 [DOI] [PubMed] [Google Scholar]
- 30. Smith M. W., et al. 2006. Gene expression patterns that correlate with hepatitis C and early progression to fibrosis in liver transplant recipients. Gastroenterology 130:179–187 [DOI] [PubMed] [Google Scholar]
- 31. Soguero C., et al. 2002. Hepatitis C virus core protein leads to immune suppression and liver damage in a transgenic murine model. J. Virol. 76:9345–9354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ulgiati D., Abraham L. J. 1996. Extensive conservation of upstream C4 promoter sequences: a comparison between C4A and C4B. Tissue Antigens 48:600–603 [DOI] [PubMed] [Google Scholar]
- 33. Ulgiati D., Subrata L. S., Abraham L. J. 2000. The role of Sp family members, basic Kruppel-like factor, and E box factors in the basal and IFN-gamma regulated expression of the human complement C4 promoter. J. Immunol. 164:300–307 [DOI] [PubMed] [Google Scholar]
- 34. Vaishnaw A. K., Mitchell T. J., Rose S. J., Walport M. J., Morley B. J. 1998. Regulation of transcription of the TATA-less human complement component C4 gene. J. Immunol. 160:4353–4360 [PubMed] [Google Scholar]
- 35. Yang Y., et al. 2003. Diversity in intrinsic strengths of the human complement system: serum C4 protein concentrations correlate with C4 gene size and polygenic variations, hemolytic activities, and body mass index. J. Immunol. 171:2734–2745 [DOI] [PubMed] [Google Scholar]