Abstract
Adenovirus has a linear, double-stranded DNA genome that is perceived by the cellular Mre11-Rad50-Nbs1 (MRN) DNA repair complex as a double-strand break. If unabated, MRN elicits a double-strand break repair response that blocks viral DNA replication and ligates the viral genomes into concatemers. There are two sets of early viral proteins that inhibit the MRN complex. The E1B-55K/E4-ORF6 complex recruits an E3 ubiquitin ligase and targets MRN proteins for proteasome-dependent degradation. The E4-ORF3 protein inhibits MRN through sequestration. The mechanism that prevents MRN recognition of the viral genome prior to the expression of these early proteins was previously unknown. Here we show a temporal correlation between the loss of viral core protein VII from the adenovirus genome and a gain of checkpoint signaling due to the double-strand break repair response. While checkpoint signaling corresponds to the recognition of the viral genome, core protein VII binding to and checkpoint signaling at viral genomes are largely mutually exclusive. Transcription is known to release protein VII from the genome, and the inhibition of transcription shows a decrease in checkpoint signaling. Finally, we show that the nuclease activity of Mre11 is dispensable for the inhibition of viral DNA replication during a DNA damage response. These results support a model involving the protection of the incoming viral genome from checkpoint signaling by core protein VII and suggest that the induction of an MRN-dependent DNA damage response may inhibit adenovirus replication by physically masking the origins of DNA replication rather than altering their integrity.
INTRODUCTION
Adenovirus (Ad) has a linear, double-stranded DNA genome with inverted terminal repeats at each end that contain the origins of viral DNA replication. The cellular Mre11-Rad50-Nbs1 (MRN) complex can recognize the termini of the linear Ad genome as double-strand breaks (DSBs) and elicit a DNA damage response (reviewed in reference 42). The repair program leads to activation of a checkpoint signaling cascade and the ligation of the Ad genomes into concatemers (6, 7, 18, 33, 41). There are several reasons this response may inhibit viral DNA replication. First, Ad genome concatenation would bury the viral origins of DNA replication within the multimer, inhibiting efficient replication (11). Second, deletion of viral DNA sequences at the concatemeric junctions resulted in the loss of functional origins of replication (18). Finally, multimeric Ad genomes would be too large to package into the viral capsid (27).
There are three Ad proteins that function to inactivate the MRN complex and thereby inhibit the DNA damage response. The E4-ORF3 protein induces the rearrangement of promyelocytic leukemia (PML) nuclear bodies into track structures in the nucleus and sequesters the MRN complex in these tracks by 6 h postinfection (hpi) (12, 33), a time prior to the onset of viral DNA replication. The viral proteins E1B-55K and E4-ORF6 work in concert to recruit a CUL5-containing E3 ubiquitin ligase complex that targets specific cellular proteins for degradation, such as p53, Mre11, Rad50, Nbs1, DNA ligase IV, and integrin α3 (2, 10, 14, 28). Either mechanism of inhibition of the MRN complex is sufficient to allow efficient viral DNA replication. However, the deletion of E4-ORF3 and either E4-ORF6 or E1B-55K results in activation of an MRN-mediated DNA damage response and a significant inhibition of viral DNA replication (4, 13, 16, 31, 33). This replication block is alleviated in cells that lack Mre11 or Nbs1 (ataxia-telangiectasia-like disorder [ATLD] or Nijmegen breakage syndrome [NBS] cells, respectively) (13, 20, 25). In cells infected with E4-ORF3/E4-ORF6 or E4-ORF3/E1B-55K double mutant viruses, checkpoint signaling was indicated by the induction of phospho-ATM (pATM) nuclear foci (18). These foci resemble ionizing radiation-induced foci (IRIF) observed following the induction of DNA damage (24) and were not observed in cells infected with wild-type (WT) Ad5 or E4-ORF3, E4-ORF6, or E1B-55K single mutant viruses that retain the ability to inhibit the MRN complex (18).
IRIF are generally induced by recognition of a DSB by the MRN complex, followed by recruitment of the protein kinase ATM (38). Intermolecular autophosphorylation dissociates dimers of ATM into active monomers, and phosphorylation of downstream effectors occurs within minutes of the introduction of the DSB (3). The histone variant H2AX becomes phosphorylated and can be found in the γH2AX form up to megabases away from the DNA lesion (5, 29). The mediator of DNA damage checkpoint 1 (Mdc1) binds to γH2AX and serves as a protein bridge that can bind to other proteins involved in checkpoint signaling and DNA repair (35). These proteins are necessary for sustained foci of the MRN complex; however, the initial recognition of the DSB by the MRN complex appears to be independent of other proteins and is thought to occur in response to even one DSB (8, 22, 32).
We previously found that IRIF-like foci did not form during an Ad E4-ORF3/E4-ORF6 or E4-ORF3/E1B-55K double mutant virus infection until around 10 hpi (18), well after the peak of early gene expression and around the onset of viral DNA replication with wild-type Ad5. Since a single DSB can induce checkpoint signaling, the incoming Ad genomes would be anticipated to induce a DNA damage response early after infection, yet this did not occur. The Ad terminal protein (TP), covalently attached at each genomic 5′ terminus, may provide protection for the viral genome and prevent MRN recognition, yet this is clearly not sufficient to block a DNA damage response in the context of viral E4 and E1B mutants. Mre11 is known to have both 3′-to-5′ exonucleolytic activity and single-stranded DNA endonucleolytic activity and is able to cleave Spo11 from meiosis-specific DSBs in Saccharomyces cerevisiae (26). This led to speculation that Mre11 may have a similar function to cleave TP from the viral genome, allowing the MRN complex to recognize the termini as DSBs (34, 42). However, we found that TP remains attached to the viral genome in the context of a DNA damage response, suggesting that TP is unable to prevent activation of MRN (18).
Several viral processes occur around 10 hpi that could be associated with the initiation of checkpoint signaling seen at this time. DNA replication begins around this time during wild-type Ad5 infection, and newly replicated DNA could trigger MRN activation. Also, early gene transcription triggers the release of viral core protein VII from the genome around this time (9). Viral protein VII is a basic protein that forms a chromatin-like structure on the viral DNA and is found to be associated with the viral genome within the virion and during the early phase of infection (9, 30, 36, 39). We hypothesized that viral protein VII could protect the incoming viral genome from recognition by the MRN complex until other early viral proteins have effectively inhibited the complex. Here, we show that incoming Ad genomes do not trigger checkpoint signaling during the early phase of virus infection. We demonstrate a temporal and functional correlation between the loss of core protein VII from the Ad genome due to viral transcription and the activation of checkpoint signaling. We analyzed whether the nuclease activity of Mre11 is required to inhibit Ad DNA replication and found that nuclease-deficient Mre11 (1) was able to inhibit DNA replication of an E4-ORF3/E4-ORF6 double mutant virus. These results suggest a new model whereby the induction of an MRN-dependent DNA damage response may inhibit Ad replication by physically masking the origins of DNA replication rather than altering their integrity.
MATERIALS AND METHODS
Cells, viruses, and infections.
A549 cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% calf serum at 37°C in 5% CO2. Immortalized ATLD1 cells (7), kindly provided by Matthew Weitzman (Salk Institute), and human diploid fibroblasts (HDF) were grown in DMEM supplemented with 10% fetal bovine serum at 37°C in 5% CO2. E1/E4 double mutant viruses were propagated on 911-E4 cells (15) grown in DMEM containing 10% FetalClone III serum (HyClone Laboratories) at 37°C in 5% CO2. Virus particles were purified on CsCl equilibrium gradients. The viruses used include dl355/inORF3 (E4-ORF6 and E4-ORF3 double mutant) (16), dl312 (E1A mutant) (17), dl366/ΔE1-CMV (E4/E1 deletion mutant; deletion of E1 nucleotides 450 to 3330, with a cytomegalovirus [CMV] promoter/enhancer in place of the E1 region) (12), and dl366/ΔE1-Mre11-WT and dl366/ΔE1-Mre11-3 viruses, same as dl366/ΔE1-CMV but with FLAG-tagged Mre11-WT and Mre11-3 (1) coding sequences, respectively, downstream of the CMV promoter/enhancer. Infections were performed at a multiplicity of infection of 200 (A549 cells) or 1,000 (ATLD1 cells) virus particles per cell for 1 h, followed by replacement with medium and incubation at 37°C in 5% CO2.
Immunofluorescence.
A549 or ATLD1 cells were seeded on glass coverslips and infected as described above. In some cases, the cells were treated with cytosine arabinoside (AraC) (25 mg/ml) or α-amanitin (20 mg/ml) immediately following infection. At appropriate time points, the cells were washed with phosphate-buffered saline (PBS). A549 cells were fixed with −20°C methanol for 5 min and washed with PBS. ATLD1 cells were fixed with 4% paraformaldehyde for 15 min at room temperature, permeabilized with PBS containing 0.25% Triton X-100 for 10 min at room temperature, and washed with PBS. The cells were blocked with PBS containing 10% goat serum and then incubated with primary antibodies diluted in blocking buffer. The antibodies used were anti-pVII rabbit polyclonal antibody (generously provided by Dan Engel, University of Virginia) at 1:1,000, anti-phospho-ATM mouse monoclonal antibody (Cell Signaling Technology) at 1:1,000, anti-phospho-ATM-S1981 rabbit polyclonal antibody (Rockland Immunochemicals) at 1:300, anti-γH2AX (Upstate Biotechnology) at 1:300, anti-DBP monoclonal antibody B6 (generously provided by Arnold Levine, Princeton University) at 1:100 or anti-rabbit polyclonal antibody (generously provided by Peter van der Vleit, University of Utrecht) at 1:2,500, and anti-Mre11 (GeneTex GTX70212) at 1:1,000. The cells were washed and incubated with Alexa Fluor 350 goat anti-mouse or -rabbit IgG (Molecular Probes), fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse or -rabbit IgG (Zymed), and tetramethylrhodamine isothiocyanate (TRITC)-conjugated goat anti-mouse or -rabbit IgG (Zymed). The cells were washed a final time and mounted on slides with Immu-Mount (Thermo Shandon). The microscope used was a Zeiss Axiovert 200M digital deconvolution microscope fitted with a Chroma filter set and an ApoTome, and images were captured with a Peltier-cooled charge-coupled-device AxioCam HRm camera and analyzed with AxioVision 4.5 software.
For the Click-iT 5-ethynyl-2′-deoxyuridine (EdU) assay (Invitrogen), dl355/inORF3 virus was grown by infecting cells and replacing the medium at 18 hpi with medium containing a final concentration of 10 mM EdU. Virions were purified and used to infect cells on coverslips. Following fixation as described above, the cells were incubated with the recommended Click-iT reaction cocktail with Alexa Fluor 488 azide for 30 min as described by the manufacturer. Then, extensive washing with PBS was performed, followed by incubation in blocking buffer. Antibody staining was performed as described above.
Immunoblots and quantitative PCR.
Whole-cell extracts were prepared by resuspending cell pellets in SDS lysis buffer (1.2% SDS, 150 mM Tris [pH 6.8], 30% glycerol) and boiling for 10 min. The samples were centrifuged at 16,100 × g for 30 min, supernatants were collected, and the total protein concentration was determined using the Pierce bicinchoninic acid (BCA) method. Standardized amounts of protein were subjected to 12.5% SDS-PAGE and transferred to nitrocellulose. Mre11 was detected using an anti-Mre11 mouse monoclonal antibody (GeneTex), and E1A was detected using an anti-E1A rabbit polyclonal antibody (Santa Cruz Biotechnology) and enhanced chemiluminescence (Millipore) according to the manufacturer's instructions. Quantitative PCR was performed as previously described (18).
RESULTS
pATM foci formed in cells infected with mutant viruses do not colocalize with core protein VII.
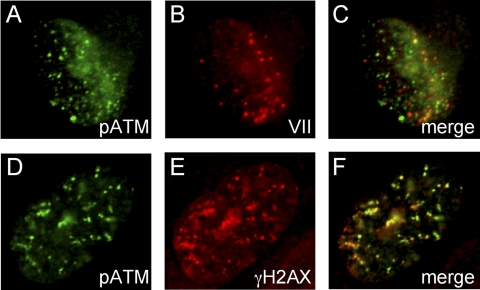
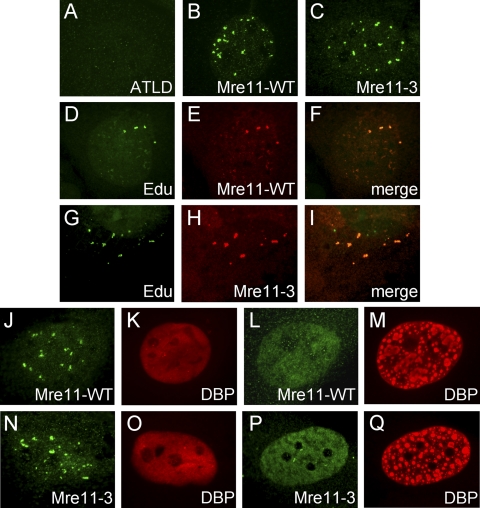
We previously found that infection of A549 cells with an E4-ORF3/E4-ORF6 mutant virus induced the formation of nuclear pATM foci without viral DNA replication (18). These results suggested that the input viral genomes were sufficient to trigger a DNA damage response. To test this idea, A549 cells infected with dl355/inORF3 virus were used for immunofluorescence with antibodies against pATM and viral core protein VII. Core protein VII binds to the viral genome during the early phase of infection and may be used as an indicator of Ad genome localization (9). Unexpectedly, we found that these two proteins did not colocalize with the majority of pATM or protein VII foci that were detected (Fig. 1A to C). However, the pATM foci did colocalize with γH2AX, as expected (Fig. 1D to F), suggesting that the pATM foci represent normal IRIF-like foci. These data show that pATM and protein VII do not overlap spatially and suggest that pATM and protein VII bind the Ad genome in a largely mutually exclusive manner.
Fig. 1.
Mutually exclusive binding interactions of core protein VII and pATM to Ad genomes. A549 cells were infected for 10 h with dl355/inORF3 mutant virus, fixed, and immunostained for pATM (FITC) (A) and protein VII (TRITC) (B). A merge of panels A and B is shown in panel C. dl355/inORF3 virus-infected cells were also stained for pATM (FITC) (D) and γH2AX (TRITC) (E). A merge of panels D and E is shown in panel F.
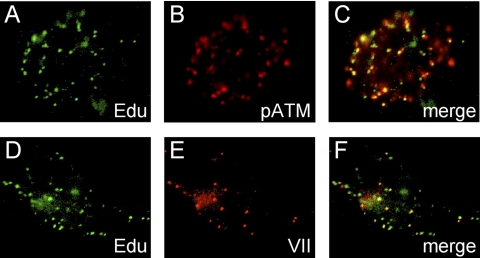
Protein VII foci are representative only of viral genomes that still are bound by this protein at the particular time point of infection. To visualize the Ad genomes by an alternative means, the Click-iT EdU imaging assay was used. In short, dl355/inORF3 virus was prepared where the genomes contained EdU molecules incorporated during replication. A549 cells were infected with this mutant EdU-containing virus, and at 10 hpi, the cells were fixed and a Click-iT reaction was used to create a covalent bond between an Alexa Fluor-conjugated azide and the alkyne-containing EdU molecule. Cells were then immunostained for either pATM (Fig. 2A to C) or core protein VII (Fig. 2D to F). In each case, some but not all of the EdU-labeled Ad genomes colocalized with pATM or protein VII foci. The dots detected with the Click-iT approach appear to represent individual viral genomes since the numbers varied in direct relation to the multiplicities of infection. At 10 hpi, nearly all of the input viral genomes were nuclear. There were a few pATM foci that did not appear to colocalize with an EdU-labeled viral genome, which may be due to a low level of viral DNA replication that has generated daughter genomes lacking EdU or due to the low level of pATM foci that were occasionally seen in uninfected cells. Thus, pATM foci were forming largely at sites of viral genome localization. These data support the conclusion that while both core protein VII and pATM colocalize with dl355/inORF3 mutant viral genomes at 10 hpi, they do not colocalize with each other and most likely represent mutually exclusive interactions with viral DNA.
Fig. 2.
pATM foci form on the Ad genome. A549 cells were infected for 10 h with dl355/inORF3 mutant virus containing genome incorporation of EdU and fixed, and a Click-iT reaction was performed to label EdU-incorporated viral genomes (A, D). Subsequently, the cells were immunostained for pATM (TRITC) (B). A merge of panels A and B is shown in panel C. dl355/inORF3 virus-infected cells were also immunostained for protein VII (TRITC) (E). A merge of panels D and E is shown in panel F.
Temporal correlation between the loss of protein VII and the gain of pATM foci.
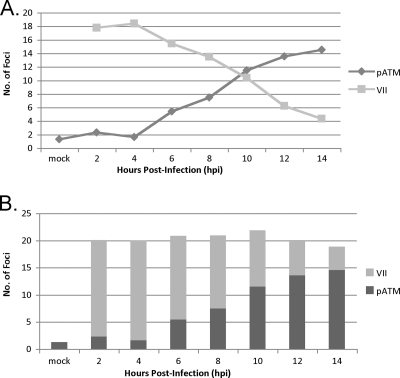
Core protein VII is released from viral genomes that are undergoing transcription (9). As the infection progresses, the number of VII foci decreases until all of the protein VII foci disappear; there is weak and diffuse nucleoplasmic protein VII staining by 14 hpi (9). We performed a time course experiment by using the dl355/inORF3 double mutant virus and staining for pATM and protein VII from 2 to 14 hpi of A549 cells. The number of foci containing each protein was determined in the same 50 cells in triplicate experiments, and the averages were plotted in two graphs using the same data (Fig. 3). As the infection progressed, the number of protein VII foci decreased, as expected, and the number of pATM foci increased at an inverse rate, with the highest number of foci being 14 and 18 for pATM and pVII, respectively (the multiplicity of infection was 200 particles/cell, corresponding to 10 to 20 infectious viruses/cell) (Fig. 3A). When these data are viewed in a stacked bar graph, it is worth noting that the total number of foci remained nearly the same (Fig. 3B), indicating a conversion of protein VII-containing viral genomes into pATM-containing viral genomes, supporting the conclusion that protein VII and pATM bind the Ad genome in mutually exclusive manners and that pATM foci form on viral genomes as protein VII is released.
Fig. 3.
An inverse correlation is observed between a decrease in protein VII foci and an increase in pATM foci. (A) A549 cells were mock infected or infected with dl355/inORF3 virus over a time course of 2 to 14 h. Cells were fixed and immunostained for protein VII and pATM. The number of foci for each protein was counted in the same 50 cells for each experiment in triplicate, and the average numbers are plotted. (B) The same data from panel A are presented as a stacked bar graph, with the total number of foci detected per time point plotted in comparison to protein VII and pATM foci.
Inhibition of transcription inhibits the formation of pATM foci on Ad genomes.
The increase in pATM foci was initiated around 6 hpi, suggesting that it was an event occurring at this time that triggered the recognition of the viral genomes by the MRN complex. Early region transcription is active at this time, and several lines of evidence were provided to conclude that transcription releases core protein VII from the viral genome (9). Viral DNA replication with wild-type Ad5 begins between 8 and 10 hpi.
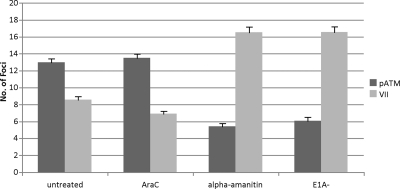
We performed immunofluorescence experiments to determine whether transcription or DNA replication was involved in the increase in the formation of pATM foci in dl355/inORF3 virus-infected cells. A549 cells were infected for 10 h with dl355/inORF3 virus; cells were left untreated or were treated immediately after infection with AraC to inhibit viral DNA replication or with α-amanitin to inhibit viral transcription. Alternatively, cells were infected with dl312 (E1A-deleted virus), a mutant that is defective in the activation of early gene expression. The number of pATM and protein VII foci were counted as described for Fig. 3 under each condition (Fig. 4). In the untreated dl355/inORF3 virus-infected cells, the average number of pATM foci was 13 and the number of protein VII foci was 8 at 10 hpi. Similar results were observed after treatment with AraC, indicating that viral DNA replication is not required for the transition between protein VII and pATM foci. Interestingly, treatment of dl355/inORF3 virus-infected cells with α-amanitin, or infection with dl312, resulted in a reversal of these results, with an increase in the number of protein VII foci (about 16 under both conditions) and a decrease in the number of pATM foci (5 to 6 under both conditions). Control experiments were performed to verify that the drugs used had the intended consequences. AraC treatment of cells reduced viral DNA replication more than 300-fold, as measured using quantitative PCR. α-Amanitin significantly reduced E1A gene expression, as measured by Western blot analysis (Fig. 5A). These results show that the inhibition of transcription, but not the inhibition of viral DNA replication, reduced recognition of the viral genome by pATM and, presumably, the prerequisite recognition of viral DNA by the MRN complex. Since transcription causes the release of core protein VII from the Ad genome, these results suggest that the presence of protein VII on viral DNA protects the viral genome from recognition by MRN and/or the recruitment of pATM.
Fig. 4.
Inhibition of transcription during Ad infection inhibits the formation of pATM foci. A549 cells were infected with dl355/inORF3 or dl312 virus for 10 h. Cells infected with dl355/inORF3 virus were left untreated, or AraC or α-amanitin was added to the medium immediately after infection. Cells were fixed and immunostained for protein VII and pATM. The number of foci for each protein was counted in 50 cells each in triplicate experiments, and the average numbers are plotted. The error bars represent 95% confidence intervals, and the values for cells treated with α-amanitin or infected with dl312 virus are statistically significant (P < 0.001).
Fig. 5.
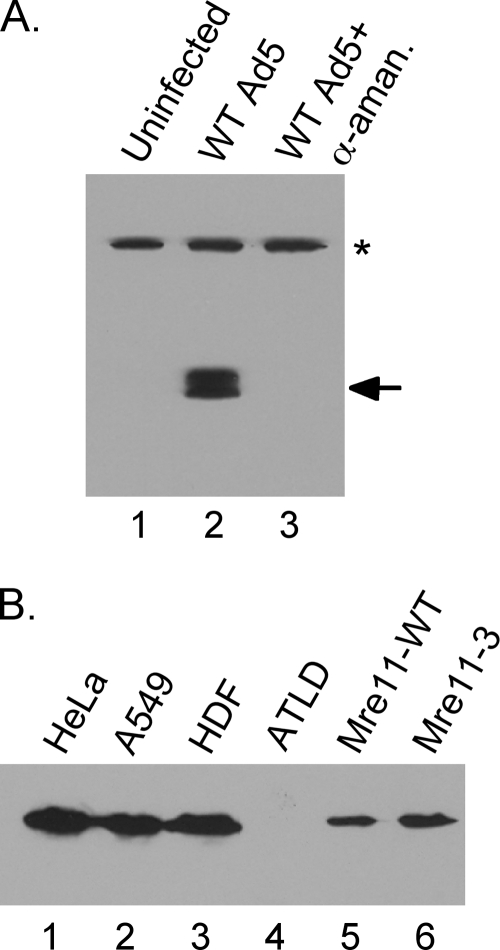
Expression of E1A and Mre11. (A) Whole-cell extracts were prepared from uninfected A549 cells (lane 1) or A549 cells infected with wild-type Ad5 in the absence (lane 2) or presence (lane 3) of α-amanitin in the culture medium. Equal amounts of protein extracts were separated by SDS-PAGE, and Western blot analyses were performed using an antibody that recognizes E1A (arrow). The band labeled with an asterisk represents a cross-reactive cellular protein that serves as a loading control. (B) Whole-cell extracts were prepared from uninfected HeLa (lane 1) and A549 (lane 2) cells and human diploid fibroblasts (HDF) (lane 3) and ATLD1 cells (lane 4) as well as from ATLD1 cells infected with adenoviruses that express Mre11-WT (lane 5) or Mre11-3 (lane 6). Equal amounts of protein extracts were separated by SDS-PAGE, and Western blot analyses were performed using an antibody that recognizes Mre11.
The nuclease activity of Mre11 is not necessary to inhibit Ad DNA replication.
The results of earlier studies showed that TP was covalently attached to the termini of dl355/inORF3 viral genomes at a time (10 hpi) when viral DNA replication was significantly inhibited by the cellular DNA damage response (18). These results suggested that the endonucleolytic digestion of the viral genome by Mre11 was not involved in the inhibition of dl355/inORF3 virus replication, but the assay did not assess whether the exonuclease activity of Mre11 was involved in this process. As a 3′-to-5′ exonuclease, Mre11 could degrade the template strand required for viral DNA replication (21). To determine whether the nuclease activity of Mre11 is required to inhibit viral DNA replication during a DNA damage response, the nuclease-defective Mre11 mutant, Mre11-3 (1), was used. Mre11-3 retains the ability to bind DNA, Rad50, and Nbs1 as well as trigger the formation of IRIF, but the mutation disrupts Mre11 nuclease activity. Mutant viruses were constructed using dl366 (E4 deleted) as a backbone with either the wild type or the Mre11-3 mutant expressed from the CMV promoter in place of the E1 genes. These recombinant viruses lack E1A, E1B, and E4 and thus are unable to inhibit the MRN complex. Mre11-WT and Mre11-3 were expressed using these viral vectors in Mre11-deficient ATLD1 cells, which have a hypomorphic mutation in Mre11 (7), at levels that were slightly reduced compared to those of HeLa and A549 cell lines and human diploid fibroblasts (Fig. 5B).
These viruses were used to infect ATLD1 cells on coverslips. The multiplicity of infection used, 1,000 particles/cell, was sufficient to infect ∼50% of the cells. The cells were fixed at 48 hpi and immunostained for Mre11 (Fig. 6); this time point was used since Western blot analysis showed that the accumulation of Mre11 increased during this period (data not shown). No Mre11 signal was observed in uninfected cells (Fig. 6A). Mre11 foci were evident in cells infected with viruses expressing both Mre11-WT and Mre11-3 (Fig. 6B and C). Mre11 foci that correspond to sites of Ad DNA localization were previously described (25). In order to determine whether these Mre11 foci colocalized with Ad genomes, the EdU-labeled dl355/inORF3 virus was employed. ATLD1 cells were infected with the Mre11-expressing viruses and at 48 hpi were reinfected with EdU-labeled dl355/inORF3 virus. At 6 hpi, the cells were fixed and immunostained for Mre11, and the Click-iT reaction was employed to label viral genomes. Mre11 foci were found to costain with dl355/inORF3 viral genomes (Fig. 6D to I), indicating recognition of viral genomes by the MRN complex, as would be expected based on the results described above.
Fig. 6.
Mre11 nuclease activity is not required to inhibit virus replication during a DNA damage response. ATLD1 cells were mock infected (A) or infected with recombinant adenoviruses that express Mre11-WT (B) or Mre11-3 (C). At 48 hpi, cells were fixed and immunostained for Mre11. ATLD1 cells were infected with recombinant adenoviruses that express Mre11-WT (D to F) or Mre11-3 (G to I). At 48 hpi, cells were reinfected with dl355/inORF3 virus containing genome incorporation of EdU. (D, G) Cells were fixed at 6 hpi with dl355/inORF3 virus, and a Click-iT reaction was performed to label EdU-incorporated viral genomes. (E, H) Subsequently, the cells were immunostained for Mre11. A merge of panels D and E and panels G and H are shown in panels F and I, respectively. ATLD1 cells were infected with recombinant adenoviruses that express Mre11-WT (J to M) or Mre11-3 (N to Q). At 48 hpi, cells were reinfected with dl355/inORF3 virus. Cells were fixed at 48 hpi with dl355/inORF3 virus and immunostained for Mre11 (J, L, N, and P) and Ad DBP (K, M, O, and Q).
The Ad DNA binding protein (DBP) is an early viral protein that is diffusely localized throughout the nucleus prior to viral DNA replication. Upon DNA replication, large replication centers form that colocalize with viral DNA and represent active sites of viral DNA replication. It was previously shown that DBP remains diffusely localized in cells that have a block of viral DNA replication (37, 40). Thus, viral DNA replication may be assessed in single-cell assays by the DBP immunofluorescence pattern. We utilized this assay to assess replication of dl355/inORF3 virus in ATLD1 cells in which Mre11-WT or Mre11-3 was expressed. Single-cell assays were used because high multiplicities of infection are required to coinfect all ATLD1 cells in a population with two viruses (data not shown); this was not suitable since E4 mutant viruses exhibit multiplicity leakiness (16).
ATLD1 cells on coverslips were infected with the E1/E4-deleted viruses expressing either Mre11-WT or Mre11-3. At 48 hpi, the cells were reinfected with dl355/inORF3 virus. We previously demonstrated that the growth of this double mutant virus was rescued in ATLD1 cells in comparison to that in primary IMR90 cells that express a functional MRN complex (13). At 48 hpi, the cells were fixed and immunostained for Mre11 and Ad DBP. These time points were used to allow for Mre11 expression and to allow dl355/inORF3 virus sufficient time to establish viral DNA replication centers, if evident. Fifty to 75 cells per coverslip in triplicate experiments were blindly scored for the formation of Mre11 foci and then scored for the status of Ad DBP localization. Approximately 25% of the cells in the population were infected with both viruses. A striking result became apparent in most of the coinfected cells. Most coinfected cells that exhibited Mre11 foci displayed diffuse DBP (90.6% for Mre11-WT and 91.1% for Mre11-3), whereas a much smaller percentage of the cells with Mre11 foci exhibited Ad replication centers (9.4% for Mre11-WT and 8.9% for Mre11-3). Similar results were found with Mre11-WT and Mre11-3 (Fig. 6J, K, N, and O). In contrast, most cells that displayed active Ad replication centers showed low or no Mre11 expression and lacked Mre11 foci (examples shown in Fig. 6L, M, P, and Q). These cells could represent either coinfected cells with weak Mre11 expression or cells singly infected with dl355/inORF3 virus. We conclude from these results that the nuclease-defective Mre11 mutant is capable of inhibiting the replication of a mutant virus that is unable to block MRN activity. Mre11-WT and Mre11-3 proteins did associate with the expression vector genomes in single infections but were not scored in our assays since we counted only DBP-positive cells and the E1-deleted expression vectors do not express DBP due to the lack of E1A.
DISCUSSION
Adenovirus has evolved redundant mechanisms to inhibit the MRN complex and the cellular DNA damage response because this response is highly detrimental to viral DNA replication (42). Yet both of these mechanisms are functions of early proteins, E4-ORF3, E4-ORF6, and E1B-55K, whose expression does not occur until several hours after infection. The MRN complex is able to recognize DSBs rapidly in the context of cellular DNA damage (23), while it takes hours for the Ad early genes to be expressed to levels sufficient to effectively inhibit the MRN complex (approximately 5 to 6 h in the case of the E4-ORF3 protein and around the onset of viral DNA replication with the E1B-55K/E4-ORF6 complex). Ad TP may help to protect the Ad genome from recognition by the MRN complex. There are several characteristics of TP that make this a reasonable suggestion. One is that TP forms a covalent linkage at the 5′ termini of the viral genomes, which would be the region of DNA that would elicit a checkpoint signaling response such that TP may mask the genomic termini. TP is also linked to the Ad genome within the virion, during translocation into the nucleus, and throughout the early phase of infection. It had been hypothesized that eventually, in a mutant virus infection in which MRN is not inhibited, Mre11 nuclease activity would cleave the termini in a fashion similar to the way it cleaves Spo11 from meiosis-specific DSBs in S. cerevisiae (34, 42) and cleave TP off the DNA along with the terminal nucleotides. If TP were the protective factor, this would leave the viral genome vulnerable to checkpoint signaling due to MRN recognition. The nucleolytic activity of Mre11 could degrade the termini of the viral genome, thereby destroying the origins of replication and preventing DNA replication complex formation. Supporting this idea is the fact that large deletions are found at the junctions of the Ad genome concatemers that form following nonhomologous end joining of the viral genomes (18).
The fact that TP is still linked to the genome of E1B and E4 mutant viruses at a time when viral DNA replication was inhibited (18) raised questions about these ideas. Interestingly, when the pATM foci that form during an E4-ORF3/E4-ORF6 mutant virus infection were visualized in relation to viral genomes detected by association with core protein VII, an unexpected result was obtained—pATM and protein VII foci did not colocalize in the majority of cases. In fact, our results suggest that the associations of protein VII and pATM with the Ad genome are largely mutually exclusive. When dl355/inORF3 virus genomes were detected using anti-VII antibody, most of the genomes that were detected did not colocalize with pATM foci (Fig. 1), whereas when the same genomes were detected using a reagent to visualize viral DNA directly, a clear colocalization of Ad DNA with pATM foci was evident (Fig. 2). It also stands to reason that γH2AX foci are forming at viral genomes since they colocalize with pATM during the mutant virus infection (Fig. 1). The binding of histones to the Ad genome and the status of viral chromatin during the early phase of infection have been controversial topics. Recently, it was reported that viral core proteins help to recruit histones to the Ad genome (19). Our results do not address this conclusion but are not inconsistent with this idea. Chen et al. discovered that core protein VII remains bound to the Ad genome during the early phase of infection and is subsequently released due to transcription (9). When a time course experiment was performed in which the number of protein VII and pATM foci were quantified, we found an inverse correlation in which pATM foci increased as the protein VII foci decreased. The total number of foci per cell was constant, indicating that pATM binds to viral genomes as protein VII is released (Fig. 3). Together, these results suggest mutually exclusive binding interactions of core protein VII and pATM to viral genomes and, by extension, the accumulation of other DNA repair proteins, such as MRN. Finally, we determined that the inhibition of transcription, but not DNA replication, inhibited pATM foci formation as well as the release of protein VII from dl355/inORF3 virus genomes (Fig. 4). This further suggests that the release of core protein VII is connected to checkpoint signaling since both are initiated by transcription. We cannot rule out the possibility that transcription itself somehow alerts the MRN complex to the viral genome or that transcription of an early gene plays a role in eliciting a checkpoint signaling response, but we consider these possibilities less likely to explain these results. Our results support the model in which protein VII can protect the Ad genome from recognition by the MRN complex, but once released, MRN can recognize the viral genome and elicit DNA damage and checkpoint signaling responses. We envision that protein VII masks the Ad genomic termini to prevent MRN recognition.
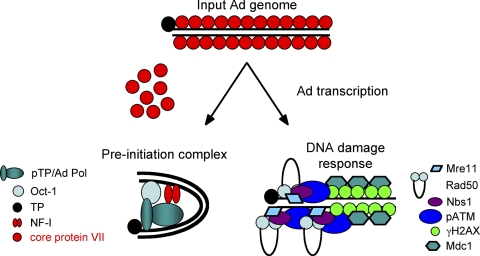
An attractive model that is supported by these results is that there is a competition between Ad replication proteins and components of the DNA damage response, such as MRN, for binding to Ad terminal sequences (Fig. 7). With wild-type Ad infection, MRN signaling is inhibited by E1B and E4 protein mechanisms such that as core protein VII is released from viral DNA, Ad replication proteins are synthesized and can recognize the viral origins of DNA replication at the genomic termini to form the preinitiation complex. We speculate that in contrast, when cells are infected with mutant viruses that are not able to inhibit MRN, such as dl355/inORF3 virus, the DNA damage response results in the recruitment of DNA repair and checkpoint signaling proteins to viral genomic termini, excluding Ad DNA replication proteins from binding. A major prediction of this model is that the nuclease activity of Mre11 would not be required for a DNA damage response to inhibit Ad mutant replication. In fact, exactly this result was obtained when wild-type Mre11 or a nuclease-deficient Mre11 mutant was used to reconstitute Mre11 activity in ATLD1 cells. Here we found that the Mre11 nuclease-deficient mutant was as effective as wild-type Mre11 in inhibiting the replication of an E4-ORF3/E4-ORF6 mutant virus (Fig. 6; Table 1). These data confirm that the DNA damage response does not inhibit Ad mutant replication by altering the structure of the viral origins of DNA replication but rather support the idea that the DNA damage response inhibits Ad mutant replication by physically masking the viral origins of replication from recognition by Ad replication proteins to block formation of the preinitiation complex (Fig. 7). It has been shown that Mre11 and Nbs1 are necessary to inhibit Ad E4 mutant virus DNA replication, while ATM and ATR are not (20). This could be due to the fact that Mre11 is necessary for viral DNA degradation while Nbs1 is necessary for proper localization of the MRN complex or that the entire MRN complex is required to occupy the termini of the viral genomes. Our results support the latter possibility. Finally, while the nuclease activity of Mre11 is not required to inhibit Ad E4 mutant virus DNA replication, it is clear that eventually the nuclease activity of Mre11, or another cellular component, does degrade viral terminal DNA sequences since small and large deletions were evident when junctions present in Ad concatemers were sequenced (18). We believe, however, that this is a late event in the Ad replication cycle since concatemers are formed in E4 mutant virus-infected cells well after the peak of viral DNA replication (41).
Fig. 7.
Competition model for Ad replication. The viral genome enters the nucleus bound to core protein VII, which masks the viral genomic termini from recognition by MRN. When early gene expression takes place, core protein VII is released from the genome, exposing the genomic termini. If E1B and E4 proteins inhibit MRN activity, then the replication preinitiation complex can form and replication ensues. If a DNA damage response takes place, then MRN and associated DNA repair components bind to the Ad genomic termini and physically mask the origins of DNA replication. Pol, polymerase; NF-I, nuclear factor I.
Overall, these studies have clarified different aspects of the Ad replication cycle that had formerly been left to speculation. We show that TP is not the viral protein that prevents MRN recognition during the initial phase of infection. Our data suggest that the viral protein that serves this function is instead viral core protein VII. We also show that the inhibition of viral DNA replication is not due to the degradation of the origins of replication of the viral genomes. The nuclease activity of Mre11 was shown to be dispensable for inhibition of viral DNA replication, but the MRN complex is necessary. This leads to a competition model that suggests that the presence of the MRN complex, perhaps in conjunction with other DNA repair proteins, at the termini is sufficient to prevent DNA replication complex formation and thus inhibit virus DNA replication if unabated. We propose that the DNA damage response results in the masking of the Ad origins of DNA replication such that viral DNA replication proteins are unable to gain access. This idea is consistent with the observation that while E4 mutant viruses show a dramatic decrease in viral DNA replication, they do replicate to a limited extent but with delayed kinetics compared to those of the wild type (4, 16, 18). If the origins of DNA replication with E4 mutants were nucleolytically digested during the DNA damage response, then we would anticipate that they would be completely unable to replicate. Instead, we suggest that a small fraction of the E4 mutant virus genomes are relieved from the DNA damage response and replicate accordingly.
ACKNOWLEDGMENTS
We thank Arnold Levine and Peter van der Vleit for antibodies against DBP, Dan Engel for antibody against protein VII, and Matthew Weitzman for ATLD1 cells. We acknowledge the excellent technical assistance of Ilana Shoshani. We thank members of our laboratory for informed discussions.
This work was supported by NIH grant CA122677. K.K. was supported by NIH training grant AI007539.
Footnotes
Published ahead of print on 23 February 2011.
REFERENCES
- 1. Arthur L. M., et al. 2004. Structural and functional analysis of Mre11-3. Nucleic Acids Res. 32:1886–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baker A., Rohleder K. J., Hanakahi L. A., Ketner G. 2007. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 81:7034–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bakkenist C. J., Kastan M. B. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421:499–506 [DOI] [PubMed] [Google Scholar]
- 4. Bridge E., Ketner G. 1989. Redundant control of adenovirus late gene expression by early region 4. J. Virol. 63:631–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burma S., Chen B. P., Murphy M., Kurimasa A., Chen D. J. 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276:42462–42467 [DOI] [PubMed] [Google Scholar]
- 6. Carson C. T., et al. 2009. Mislocalization of the MRN complex prevents ATR signaling during adenovirus infection. EMBO J. 28:652–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carson C. T., et al. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 22:6610–6620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Celeste A., et al. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5:675–679 [DOI] [PubMed] [Google Scholar]
- 9. Chen J., Morral N., Engel D. A. 2007. Transcription releases protein VII from adenovirus chromatin. Virology 369:411–422 [DOI] [PubMed] [Google Scholar]
- 10. Dallaire F., Blanchette P., Groitl P., Dobner T., Branton P. E. 2009. Identification of integrin alpha3 as a new substrate of the adenovirus E4orf6/E1B 55-kilodalton E3 ubiquitin ligase complex. J. Virol. 83:5329–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Jong R. N., van der Vleit P. C., Brenkman A. B. 2003. Adenovirus DNA replication: protein priming, jumping back and the role of the DNA binding protein DBP. Curr. Top. Microbiol. Immunol. 236:1–12 [DOI] [PubMed] [Google Scholar]
- 12. Evans J. D., Hearing P. 2003. Distinct roles of the adenovirus E4 ORF3 protein in viral DNA replication and inhibition of genome concatenation. J. Virol. 77:5295–5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Evans J. D., Hearing P. 2005. Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication. J. Virol. 79:6207–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harada J. N., Shevchenko A., Shevchenko A., Pallas D. C., Berk A. J. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76:9194–9206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He T. C., et al. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. U. S. A. 95:2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang M. M., Hearing P. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 63:2605–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jones N., Shenk T. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17:683–689 [DOI] [PubMed] [Google Scholar]
- 18. Karen K. A., Hoey P. J., Young C. S., Hearing P. 2009. Temporal regulation of the Mre11-Rad50-Nbs1 complex during adenovirus infection. J. Virol. 83:4565–4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Komatsu T., Haruki H., Nagata K. 2011. Cellular and viral chromatin proteins are positive factors in the regulation of adenovirus gene expression. Nucleic Acids Res. 39:889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lakdawala S. S., et al. 2008. Differential requirements of the C terminus of Nbs1 in suppressing adenovirus DNA replication and promoting concatemer formation. J. Virol. 82:8362–8372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lamarche B. J., Orazio N. I., Weitzman M. D. 2010. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 584:3682–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lukas C., et al. 2004. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 23:2674–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lukas J., Lukas C., Bartek J. 2004. Mammalian cell cycle checkpoints: signalling pathways and their organization in space and time. DNA Repair 3:997–1007 [DOI] [PubMed] [Google Scholar]
- 24. Maser R. S., Monsen K. J., Nelms B. E., Petrini J. H. 1997. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol. 17:6087–6096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mathew S. S., Bridge E. 2007. The cellular Mre11 protein interferes with adenovirus E4 mutant DNA replication. Virology 365:346–355 [DOI] [PubMed] [Google Scholar]
- 26. Moreau S., Ferguson J. R., Symington L. S. 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19:556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ostapchuk P., Hearing P. 2005. Control of adenovirus packaging. J. Cell. Biochem. 96:25–35 [DOI] [PubMed] [Google Scholar]
- 28. Querido E., et al. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rogakou E. P., Boon C., Redon C., Bonner W. M. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sergeant A., Tigges M. A., Raskas H. J. 1979. Nucleosome-like structural subunits of intranuclear parental adenovirus type 2 DNA. J. Virol. 29:888–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shepard R. N., Ornelles D. A. 2003. E4orf3 is necessary for enhanced S-phase replication of cell cycle-restricted subgroup C adenoviruses. J. Virol. 77:8593–8595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shroff R., et al. 2004. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 14:1703–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stracker T. H., Carson C. T., Weitzman M. D. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348–352 [DOI] [PubMed] [Google Scholar]
- 34. Stracker T. H., et al. 2005. Serotype-specific reorganization of the Mre11 complex by adenoviral E4orf3 proteins. J. Virol. 79:6664–6673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stucki M., Jackson S. P. 2006. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair 5:534–543 [DOI] [PubMed] [Google Scholar]
- 36. Tate V. E., Philipson L. 1979. Parental adenovirus DNA accumulates in nucleosome-like structures in infected cells. Nucleic Acids Res. 6:2769–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ullman A. J., Reich N. C., Hearing P. 2007. Adenovirus E4 ORF3 protein inhibits the interferon-mediated antiviral response. J. Virol. 81:4744–4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Uziel T., et al. 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 22:5612–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vayda M. E., Rogers A. E., Flint S. J. 1983. The structure of nucleoprotein cores released from adenovirions. Nucleic Acids Res. 11:441–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Voelkerding K., Klessig D. F. 1986. Identification of two nuclear subclasses of the adenovirus type 5-encoded DNA-binding protein. J. Virol. 60:353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weiden M. D., Ginsberg H. S. 1994. Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proc. Natl. Acad. Sci. U. S. A. 91:153–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weitzman M. D., Ornelles D. A. 2005. Inactivating intracellular antiviral responses during adenovirus infection. Oncogene 24:7686–7696 [DOI] [PubMed] [Google Scholar]