Abstract
Expression of a retroviral protein, Gag, in mammalian cells is sufficient for assembly of immature virus-like particles (VLPs). VLP assembly is mediated largely by interactions between the capsid (CA) domains of Gag molecules but is facilitated by binding of the nucleocapsid (NC) domain to nucleic acid. We have investigated the role of SP1, a spacer between CA and NC in HIV-1 Gag, in VLP assembly. Mutational analysis showed that even subtle changes in the first 4 residues of SP1 destroy the ability of Gag to assemble correctly, frequently leading to formation of tubes or other misassembled structures rather than proper VLPs. We also studied the conformation of the CA-SP1 junction region in solution, using both molecular dynamics simulations and circular dichroism. Consonant with nuclear magnetic resonance (NMR) studies from other laboratories, we found that SP1 is nearly unstructured in aqueous solution but undergoes a concerted change to an α-helical conformation when the polarity of the environment is reduced by addition of dimethyl sulfoxide (DMSO), trifluoroethanol, or ethanol. Remarkably, such a coil-to-helix transition is also recapitulated in an aqueous medium at high peptide concentrations. The exquisite sensitivity of SP1 to mutational changes and its ability to undergo a concentration-dependent structural transition raise the possibility that SP1 could act as a molecular switch to prime HIV-1 Gag for VLP assembly. We suggest that changes in the local environment of SP1 when Gag oligomerizes on nucleic acid might trigger this switch.
INTRODUCTION
Retrovirus particles are roughly spherical, somewhat pleomorphic objects with diameters of approximately 120 nm. Expression of a single virus-coded protein, Gag, in mammalian cells is sufficient for efficient particle assembly (61). HIV assembly occurs at the plasma membrane, where accumulation of Gag is first observed as electron-dense patches under the membrane. The initial product of assembly, or immature particle, contains several thousand Gag molecules arranged radially under the surface of the sphere, with their N-terminal matrix (MA) domains in contact with the lipid bilayer surrounding the particle and their C termini facing the interior of the particle (11, 27, 67, 69, 71). While the mechanistic details of the assembly process are still unknown, assembly is facilitated by interaction with RNA or short oligonucleotides (13–15, 31, 32, 47, 48, 51, 72); lack of this interaction can be compensated for by adding an additional oligomerization domain to the protein (2, 19, 37, 73). Thus, protein oligomerization, either upon binding nucleic acid or mediated by additional protein interaction domains, appears to be an early and crucial step in virus assembly.
Following virus assembly and release from the virus-producing cell, the viral protease normally cleaves Gag into a series of cleavage products to form the mature virion. In HIV-1, these cleavage products are, from the N to C terminus, MA, capsid (CA), SP1, nucleocapsid (NC), SP2, and p6 (35). One important function of the MA domain in particle assembly is targeting of Gag to the plasma membrane. The CA domain appears to play a principal role in protein-protein association during assembly of the immature particle. The NC domain is involved in interactions of Gag with RNA, while p6 contains the “late domain” (26), which mediates interactions of Gag with cellular factors leading to release of the assembled particle from the cell (61).
Several studies have shown that particles closely resembling authentic immature virions (except for the absence of a lipid membrane) can be assembled from purified, recombinant Gag in a fully defined system in vitro (13–15, 31, 32, 47, 72). Purified recombinant Gag protein, which is normally soluble, undergoes rapid and spontaneous assembly into virus-like particles (VLPs) upon binding nucleic acid and, in some cases, an additional small molecule, inositol pentakisphosphate (IP5). The structural similarity of in vitro assembled particles and authentic immature particles suggests that VLP structure is a direct consequence of the size and shape of the Gag molecule and the precise ways that it interacts with other Gag molecules upon binding to nucleic acid. The mechanism by which nucleic acid binding triggers VLP assembly is unclear; gaining an insight to this mechanism is crucial to our understanding of virus assembly.
The three-dimensional structure of HIV-1 Gag has not been determined. However, nuclear magnetic resonance (NMR) and crystallographic structures have been reported for free MA (36, 49), the two domains within CA (28–30, 68), and NC in complexes with small RNA stem-loops (6, 23). The structure of a fragment containing both MA and the N-terminal domain of CA has also been determined by NMR (62). One major obstacle to structural studies of HIV-1 Gag is the fact that it contains several flexible regions; solution properties of Gag are fully consistent with this idea (21).
The present report is concerned with the function of the SP1 domain in HIV-1 assembly (see Fig. 1A). SP1 is only 14 residues long; its sequence is highly conserved among HIV-1 isolates. Deletions or alterations within this 14-residue stretch of Gag can be extremely deleterious for correct particle assembly (1, 34, 40, 44, 66). Protein structure prediction programs suggest that the last 7 residues of the CA domain and the first 7 residues of SP1 might form an α-helix (1). In fact, NMR data show that the SP1 domain exhibits only a slight propensity for helix formation (52), but this tendency is dramatically enhanced in 30% trifluoroethanol (TFE), a helix-promoting solvent (50).
Fig. 1.
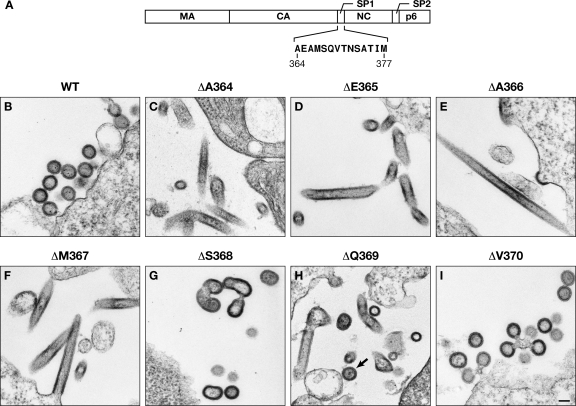
(A) Schematic representation of HIV-1 Gag, showing location and sequence of SP1. (B) Thin-section EM images of 293T cells following transfection with the indicated Gag expression plasmids. In panel H, the arrow indicates a proper VLP. Scale bar, 100 nm.
We now present the results of a detailed mutational study of the role of the N-terminal portion of SP1 in HIV-1 immature particle assembly. In addition, we have investigated the structure of SP1 using both molecular dynamics (MD) simulations and circular dichroism (CD) analysis. These studies further demonstrate the extreme dependence of virus assembly on the SP1 sequence and also reveal the exquisite sensitivity of SP1 conformation to environmental conditions, including molecular crowding. Based on these studies, we propose that local concentration changes upon binding of Gag to nucleic acid can lead to a change in SP1 conformation; in turn, this structural change might be a molecular switch which triggers virus assembly.
MATERIALS AND METHODS
Plasmids.
Except where otherwise noted, all experiments were performed with derivatives of pCMV55M1-10, the plasmid directing the Rev-independent expression in mammalian cells of HXB2 Gag (56). This plasmid was a kind gift of Barbara Felber (National Cancer Institute [NCI]). For infectivity assays, pNL4-3 lacking vpr and containing the firefly luciferase gene in place of env was used together with a vesicular stomatitis virus G protein (VSV-G) expression plasmid. The former plasmid was a kind gift of Vineet KewalRamani and Alok Mulky (NCI). We also used the Gag-Z plasmid, in which NC, SP2, and p6 of pCMV55M1-10 have been replaced by a leucine zipper motif (19) and a plasmid directing bacterial expression of a Gag protein lacking residues 16 to 99 and all of p6 (Δ16–99 Gag) (13, 32). All deletions and point mutations were generated using the QuikChange technique as directed by the manufacturer (Stratagene).
Cells and viruses.
All experiments involving mammalian cells were conducted in 293T cells. Twenty-four-hour harvests of culture fluid were collected 48 and 72 h after transfection. The fluids were first filtered through 0.45-μm-pore-size filters, and virus-like particles (VLPs) were then collected by centrifugation at 25,000 rpm in an SW28 rotor (Beckman) for 1 h through a cushion of 20% (wt/wt) sucrose. Transfections, infectivity assays, immunoblotting, and electron microscopy (EM) of thin sections of transfected cells were as described in Crist et al. (19); reverse transcriptase (RT) activity was assayed using a Quant-T-RT kit (GE Healthcare) according to the manufacturer's instructions. At least 20 fields were examined in each EM analysis. The cells used in the infectivity assays expressed the ecotropic murine leukemia virus receptor MCAT although this is unnecessary for infection by the virions bearing the VSV-G protein used here.
In vitro assembly.
All in vitro studies were with derivatives of Δ16–99 Δp6 Gag protein. Recombinant protein was purified for in vitro assembly as described previously (20, 22). Assembly experiments were performed by adding various amounts of Saccharomyces cerevisiae tRNA (Ambion) to 1 mg/ml of purified protein in assembly buffer (0.1 M NaCl–0.02 M Tris [pH 8.0]–5 mM β-mercaptoethanol). To quantitate assembly, the mixture was incubated for 1 h on ice and then centrifuged for 30 min at 4°C at ∼20,000 × g. Pellets were dissolved in 0.02 M Tris (pH 8.0)–6 M guanidine hydrochloride. Absorbance of these solutions was measured at 260 and 280 nm, and amounts of pelleted protein were estimated using the technique of Warburg and Christian (65). Both supernatants and pellets from this centrifugation were also analyzed by SDS-PAGE and Coomassie brilliant blue staining to ensure that equal amounts of total protein (supernatant plus pellet) were present in all samples. The pelleted material was also characterized by negative-stain electron microscopy using 2% uranyl acetate.
Molecular dynamics simulations.
Conformations of wild-type (WT) and mutant (E365P) peptides were analyzed by a combination of molecular mechanics-based modeling techniques including exhaustive molecular dynamics (MD) and replica exchange MD trajectories. We obtained a highly redundant sampling of peptide conformation space for peptides in solvent mixtures of variable dimethyl sulfoxide (DMSO)-water composition. The techniques are described in detail in material posted at http://home.ncifcrf.gov/hivdrp/Rein_publications/JV2011DattaSupplemental.pdf.
Circular dichroism.
The peptide spanning P356 to T373 (peptide P356-T373; PGHKARVLAEAMSQVTNT) was obtained from New England Peptide LLC (Gardner, MA) and further purified by reverse-phase high-performance liquid chromatography (HPLC) on a C18 column with a linear gradient of water and acetonitrile containing 0.1% trifluoroacetic acid. After confirmation of homogeneity by matrix-assisted laser desorption ionization (MALDI) mass spectroscopy, the purified peptide was lyophilized to a powder. The lyophilized peptide was rehydrated overnight in deionized water at 4°C to make a stock solution at 20 mg/ml (10.4 mM). Peptide concentration was determined by UV absorbance spectroscopy at 205 nm (57) and confirmed by amino acid analysis. Circular dichroism spectra of samples at 25°C (±0.1°C) were acquired using an AVIV 202 CD spectrometer (Aviv Instruments). The peptide at 20 mg/ml (10.4 mM) in water was diluted to 0.052 mM or 0.52 mM in various water-TFE or water-ethanol (EtOH) mixtures containing 10 mM sodium borate, pH 8.0. For concentration dependence studies, the peptide was diluted into 0.5 mM sodium borate, pH 8.0. CD spectrum acquisition was initiated within 1 min of peptide dilution. Quartz cells (Hellma) of 1-cm, 1-mm, and 0.1-mm path lengths were used to acquire CD spectra in the region of 180 to 260 nm or 190 to 260 nm (wavelength step, 0.5 nm; averaging time, 1.000 s; settling time, 0.330 s). The secondary structural composition was estimated from the spectra using Pepfit (55), which uses a peptide-specific basis set. A Microsoft Excel-ported version of Pepfit (7), a kind gift from Michael A. Amon, was used for convenience. The results from Pepfit were corroborated using CONTINLL (54) and CDSSTR (17) with the basis set 7 (60), which includes 48 proteins (five denatured), all accessed at the Dichroweb site (http://dichroweb.cryst.bbk.ac.uk) (46).
RESULTS
Deletions of single residues in SP1.
Fig. 1A shows the sequence of SP1 and its position within the HIV-1 Gag protein. Several prior studies (1, 34, 40, 45, 66) indicate that residues in SP1 are critical for proper VLP assembly. To identify these residues as specifically as possible, we individually deleted each of the N-terminal 7 residues of SP1. The assembly properties of these seven deletion mutants were tested by expressing them in 293T cells and analyzing Gag-specific structures in the cells by transmission electron microscopy. No normal VLPs were formed by Gag proteins lacking any of the first 4 residues (Fig. 1C to F). Rather, these proteins predominantly assembled into tubular structures. While the structures formed by Gag lacking the fifth residue (ΔS368) were not normal, some of them did exhibit some features, such as radius of curvature, in common with normal VLPs (Fig. 1G). Strikingly, Gag lacking residue 6 of SP1 assembled into a mixture of apparently normal VLPs and abnormal structures such as tubes (Fig. 1H, ΔQ369), while Gag lacking residue 7 formed fully normal-looking VLPs (Fig. 1I). Thus, each of the first 4 residues of SP1 is essential for VLP assembly; residues 5 and 6 are required for fully normal assembly, but Gag proteins lacking either of these residues do form structures with recognizable similarity to normal VLPs; and residue 7 is unnecessary for normal assembly.
Mutations in SP1.
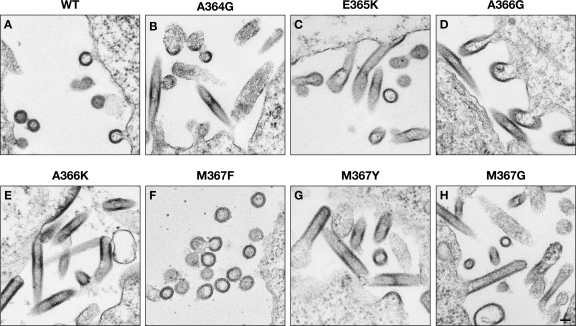
In order to further define the sequences in SP1 required for proper particle assembly, we also tested a series of point mutants at its N-terminal 4 residues. The mutations were chosen to provide insight into the range of variation consistent with proper assembly. Electron micrographs of selected mutants are shown in Fig. 2, and the morphological and other properties of the entire set are summarized in Table 1. Electron micrographs of all of the remaining mutants are presented in Fig. S1 posted at http://home.ncifcrf.gov/hivdrp/Rein_publications/JV2011DattaSupplemental.pdf. Replacements of A364, the first residue of SP1, included A364E, A364K, A364P, A364G, A364F, and A364W mutants. Briefly, we found that the morphology of A364W particles was indistinguishable from that of wild-type particles, that A364F produced a mixture of normal-looking VLPs and a variety of aberrant structures, and that the remaining A364 mutants all produced predominantly tubular structures. An example is shown in Fig. 2B.
Fig. 2.
Thin-section EM images of 293T cells following transfection with Gag expression plasmids as indicated. Scale bar, 100 nm.
Table 1.
Properties of mutantsa
| WT amino acid | Mutant amino acid | Characteristic(s)b | Reference or source |
|---|---|---|---|
| A364 | V | Infectious | 3, 44 |
| E | Tubes, Gag on uncurved PM, some proper VLPs (?) | This report | |
| K | Tubes, Gag on uncurved PM, knobs on PM | This report | |
| W | Proper VLPs | This report | |
| P | Tubes, misshapen blobs | This report | |
| F | Tubes, misshapen blobs, Gag on uncurved PM, proper VLPs | This report | |
| G | Tubes | This report | |
| E365 | Q | Infectious | 1 |
| A | Some infectivity | 1, 44 | |
| D | Proper VLPs | This report | |
| G | Reduced release | 1 | |
| P | Reduced release | 1 | |
| K | Tubes, proper VLPs | This report | |
| F | Proper VLPs, blobs, Gag on uncurved PM | This report | |
| W | VLPs, blobs | This report | |
| A366 | V | Poor assembly | 3, 44 |
| T | Little release | 3 | |
| E | Tubes, Gag on uncurved PM | This report | |
| G | Blobs | This report | |
| K | Tubes | This report | |
| P | Tubes, Gag on uncurved PM, very heterogeneous | This report | |
| F | Proper VLPs, Gag on uncurved PM, blobs | This report | |
| W | Proper VLPs, some infectivity | This report | |
| M367 | A | Tubes, balloons, Gag on uncurved PM | This report |
| G | Tubes, blobs, Gag on uncurved PM | This report | |
| E | Tubes, blobs | This report | |
| K | Tubes, Gag on uncurved PM | This report | |
| F | Proper VLPs, some blobs | This report | |
| P | Tubes, blobs, balloons | This report | |
| Y | Tubes, blobs, balloons | This report | |
| L | Proper VLPs | This report | |
| W | Some infectivity | This report |
Expression plasmids for mutant and wild-type Gag were transfected into 293T cells and then processed for thin-section electron microscopy.
PM, plasma membrane; blobs, released particles lacking the regular shape and radius of curvature of WT (proper) VLPs; knobs, buds on the plasma membrane lacking the regular radius of curvature of WT buds; balloons, densely stained membrane (putatively coated with Gag) not in contact with cytoplasm. The table also incorporates descriptions of mutant phenotypes from the literature.
Several mutants of the second SP1 residue, E365, have already been described by other laboratories (1, 44). We found that replacement of E365 with aspartate resulted in normal assembly. However, E365K (Fig. 2C), E365W, and E365F mutants were all unable to assemble correctly.
We also replaced A366 with a variety of other amino acids, including glutamate, glycine, lysine, proline, phenylalanine, and tryptophan. We found that VLPs formed by A366W were indistinguishable from WT VLPs. A366F assembled into some normal-looking VLPs (not shown) and some misassembled structures. A366G (Fig. 2D), A366K (Fig. 2E), A366E, and A366P proteins all formed tubular structures; A366E and A366P also formed a pleomorphic mixture of other structures.
Finally, M367 was replaced by alanine, glycine, glutamate, lysine, tryptophan, phenylalanine, tyrosine, leucine, and proline. We found that VLPs assembled from M367L and M367W were not detectably different in morphology from WT VLPs. M367F (Fig. 2F) produced VLPs closely resembling WT VLPs but also irregular budding structures. M367Y (Fig. 2G) assembled into tubes, irregularly shaped VLPs, and “balloons,” in which Gag-coated plasma membrane seemed to form a large sheet disconnected from the cell. M367A, M367E, M367K, M367G (Fig. 2H), and M367P mutants all formed tubular structures; in addition, balloons were observed with M367A and M367P, as well as M367Y, while M367A, M367G, M367E, M367K, M367P, and M367Y proteins all formed irregularly shaped VLPs and/or failed to induce curvature when coating the plasma membrane.
Since the immature particles produced by A366W and M367W were morphologically similar to WT VLPs, it was of interest to determine whether virions containing these alterations in SP1 could be infectious. We therefore generated these mutations in an HIV-1-derived luciferase vector. Virus was produced by transient transfection of 293T cells and assayed for luciferase-inducing activity on naive 293 cells. RT activity was also assayed so that the specific infectivities of the mutant virus preparations could be compared with the specific infectivity of WT virus. The results of these assays (Table 2) showed that both mutations caused some reduction in the amount of virus produced, as measured by the RT assay, and an additional decrease in the specific infectivity of these virions. The effects of A366W on both parameters were significantly more severe than those of M367W.
Table 2.
Infectivity of SP1 mutants
| Virus | Activity profilea |
||||
|---|---|---|---|---|---|
| Luc | Relative Luc | RT | Relative RT | Relative Luc/RT | |
| WT | 338,697 | 1 | 22,390 | 1 | 1 |
| A366W | 3,443 | 0.01 | 4,547 | 0.2 | 0.05 |
| M367W | 31,350 | 0.09 | 8,864 | 0.4 | 0.23 |
Virus was collected following transfection into 293T cells of a luciferase-containing vector with WT or mutant SP1. The deletion in Env in the vector was complemented by cotransfection with VSV-G. The released particles were assayed for RT activity and for their ability to induce luciferase activity in susceptible cells. Relative activity was determined with respect to the level of the WT virus. Luc, luciferase.
In vivo assembly properties of ΔΑ364.
In an effort to better understand the role of SP1 in virus assembly, we characterized the assembly properties of one of the mutants, i.e., ΔA364 (Fig. 1C), in more detail.
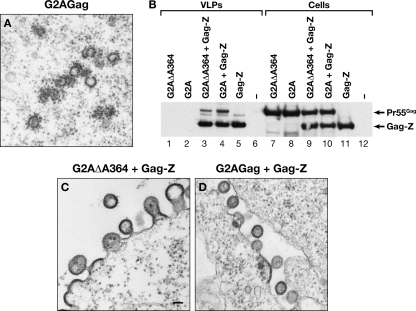
VLP assembly normally occurs at the plasma membrane. Wild-type HIV-1 Gag is targeted to the plasma membrane, in part because its N-terminal glycine residue is modified with a myristate moiety. However, assembly is not absolutely dependent upon membrane association: under our transient transfection conditions, G2A Gag, which is not myristylated because the N-terminal glycine is replaced by alanine, assembles into roughly spherical structures within the cytoplasm (Fig. 3A). In contrast, no such structures were seen in cells expressing G2A ΔA364 Gag; indeed, electron micrographs of cells containing this protein could not be distinguished from those of cells transfected with empty plasmid, despite extensive examination (data not shown). Although the cells contained no visible virus-specific structures, they contained easily detectable levels of the protein, as shown below (Fig. 3B, lane 7). Thus, assembly of ΔA364 Gag appears significantly more dependent on membrane association than that of WT Gag.
Fig. 3.
Assembly properties of G2A ΔA364 Gag. (A) Thin-section EM image of cells expressing G2A Gag. In contrast, EM images of cells expressing G2A ΔA364 Gag could not be distinguished from those of mock-transfected cells (data not shown). (B) Inefficient coassembly of G2A ΔA364 Gag with Gag-Z. Cells were transfected with the indicated Gag expression plasmids, and VLPs and cell lysates were then analyzed by immunoblotting with anti-p24 antiserum. (C) Thin-section EM image of cells expressing both Gag-Z and G2A ΔA364 Gag. (D) Thin-section EM image of cells expressing both Gag-Z and G2A Gag. Scale bar, 100 nm.
We also tested the ability of ΔA364 Gag to coassemble with a protein resembling WT Gag. In the latter protein, all domains distal to SP1, i.e., NC, SP2, and p6, had been replaced by a leucine zipper motif. These Gag-Z chimeras assemble efficiently in mammalian cells, forming particles nearly identical in morphology to authentic immature particles (19). We have previously shown (19) that Gag-Z can “rescue” G2A Gag into VLPs; the two proteins can be distinguished in immunoblots by their different molecular weights. We therefore coexpressed Gag-Z with G2A ΔA364 Gag and tested the VLPs for the presence of the latter protein by immunoblotting. As shown in Fig. 3B, the G2A ΔA364 Gag was detectable in the VLPs (lane 3) but at a somewhat lower level than G2A Gag in the control (lane 4).
The cells coexpressing Gag-Z and G2A ΔA364 Gag were also examined by electron microscopy. Some normal-looking VLPs were seen, but there were also many fields in which Gag was budding from the plasma membrane, but without a consistent radius of curvature. An example is shown in Fig. 3C. In contrast, the cells coexpressing G2A Gag and Gag-Z, like those with Gag-Z alone, were producing normal-looking VLPs (Fig. 3D). Taken together, these results imply that Gag molecules lacking residue A364 can interfere with correct VLP assembly by Gag-Z.
Assembly properties of a ΔE365 leucine zipper chimera.
Free HIV-1 Gag protein monomers have a relatively weak tendency to dimerize, with a Kd (dissociation constant) of ∼10−5 M (22). In contrast, Gag-Z proteins form dimers with significantly higher affinity (19). It seemed possible that enhancing the affinity of Gag monomers for each other would correct the assembly defect caused by mutations in SP1. To test this possibility, we placed an SP1 mutation, i.e., ΔE365, into Gag-Z. When this construct was expressed in 293T cells, it was found to misassemble into highly aberrant structures, with an overall appearance similar to those formed by ΔE365 itself (data not shown). Thus, the defective assembly of the SP1 mutant appears to be independent of the dimeric affinity of Gag.
In vitro assembly properties of ΔA364 and ΔV370.
Purified recombinant Gag protein lacking most of the MA domain and all of p6 (Δ16–99 Gag) assembles into VLPs when mixed with nucleic acid (32). One way of monitoring this assembly is by centrifugation: the VLPs can readily be pelleted while the free protein remains in the supernatant. We tested whether deletion of residue A364 would interfere with in vitro assembly in this system. As a control, we also analyzed ΔV370 Gag, which forms normal-looking VLPs in mammalian cells (Fig. 1I). When increasing amounts of tRNA were added to ΔA364 Gag, the protein tended to assemble into pelletable structures. However, at each tRNA concentration, the amount of ΔA364 Gag pelleted was lower than that of the ΔV370 Gag; the titration pattern of the latter was indistinguishable from that of the WT control (Fig. 4A).
Fig. 4.
In vitro assembly properties of Δ16–99 ΔA364 Gag and Δ16–99 ΔV370 Gag. (A) Δ16–99 ΔA364 Gag (ΔA364) does not assemble efficiently upon addition of nucleic acid. Increasing amounts of tRNA were added to Δ16–99 ΔA364 Gag or Δ16–99 ΔV370 Gag (ΔV370), and the reactions were fractionated into supernatant and pellet. Protein in the pellets was quantitated by absorbance measurements as described in Materials and Methods. (B) Negative-stain EM image of pellet from Δ16–99 ΔA364 Gag plus tRNA. (C) Negative-stain EM image of pellet from Δ16–99 ΔV370 Gag plus tRNA. Scale bar, 100 nm.
VLPs formed in vitro by Δ16-99 Gag closely resemble authentic immature VLPs (except for the absence of a lipid membrane around the particle) (11, 32). Therefore, we also analyzed the products of in vitro assembly by electron microscopy. We found that pelleted material obtained by mixing the ΔA364 protein with nucleic acid was completely amorphous, with no regular structures discernible (Fig. 4B). In contrast, the ΔV370 protein formed spherical VLPs (Fig. 4C) which were indistinguishable from WT control VLPs (not shown). Thus, although ΔA364 can form some pelletable structures upon addition of nucleic acid, these structures are aggregates, not correctly assembled VLPs.
SP1 conformation: molecular dynamics simulations.
It is clear from the results presented above that the ability of Gag to assemble correctly is critically dependent on the sequence of its SP1 domain. Unfortunately, the three-dimensional structure of SP1 is not yet clear. Its sequence is compatible with α-helix formation (1). However, NMR studies indicate that SP1 has only a slight propensity for helix formation in aqueous buffer (52) although a helix is formed in a medium containing 30% trifluoroethanol (TFE) (50), a helix-promoting solvent.
The nature of the transition between these two conformations is of considerable interest. As one approach to this problem, we have analyzed SP1 structure by molecular dynamics (MD) simulations in solvents over a range of dielectric constants. Since the physical characteristics of dimethyl sulfoxide (DMSO)-water mixtures are particularly well characterized, we performed simulations on the sequence between P356 and Q386 over a range of DMSO concentrations (70). We found (Fig. 5A) that the average helicity (as assessed by the dihedral angles of the peptide bonds [63]) of the stretch from residue P356 to Q386 increased monotonically between 0% and 80% DMSO, with a particularly steep rise between 50 and 60% DMSO.
Fig. 5.
(A) Result of MD simulation of the helical content of residues P356 to Q386 of Gag in a series of water-DMSO mixtures. Points on the graph represent the mean helical contents; 50% of the events are contained within the error bars. (B) Helical content of individual residues between A364 and M378 in water (open symbols) or 80% DMSO (closed symbols). (C) Helical wheel depiction of residues P356 to A378. Polar residues are black, and nonpolar residues are gray.
We also assessed the helical propensity of each position in the WT sequence between residues P356 and Q386. Interestingly, we found (Fig. 5B) that the entire stretch between A364 and M378 (essentially, the SP1 region of Gag) had a relatively uniform, high helical content in 80% DMSO and a very low helical content in water.
It is interesting that if the stretch of Gag between L363 and M378 were to form an α-helix, the helix would have a strikingly amphipathic character, with a hydrophobic face and a polar face, as shown in Fig. 5C. Since it has been suggested (11, 69) that SP1 forms a six-helix bundle in immature HIV-1 VLPs, we searched for indications that the helical structures arising in our MD simulations could form dimers or hexamers. We performed simulations in which two or six molecules of the stretch A360 to Q379 and A360 to Q386 were incubated together in silico in 100% DMSO. However, despite the amphipathic character of the predicted helices (Fig. 5C), no stable oligomeric structures were observed in these simulations (data not shown); thus, we have not yet been able to model the putative hexamer, or dimers that could associate into a hexamer, using these approaches.
SP1 conformation: circular dichroism.
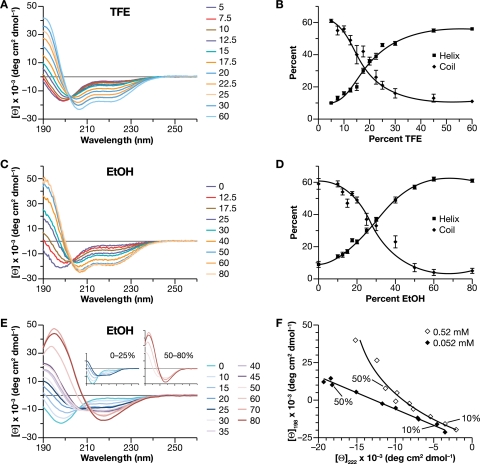
Finally, we used circular dichroism (CD) spectroscopy to analyze the structure of a peptide, P356-T373, consisting of the last 8 residues of CA and the first 10 residues of SP1. (The prediction of a helical conformation applies to residues G357 to V370, spanning the CA-SP1 junction [1]). As absorbance by DMSO makes this solvent impractical for CD measurements, we varied the medium using TFE. The peptide was first studied at a low concentration (0.052 mM [0.1 mg/ml]) over a range of TFE concentrations. The spectra are shown in Fig. 6A. In the absence of organic solvent, the peptide has a spectrum dominated by a single negative peak with a minimum at 198 nm, characteristic of unstructured coils. With increasing TFE concentrations, the strength of the negative peak decreases and the minimum red-shifts to 206 nm, with a concomitant increase in ellipticity between 210 and 230 nm suggestive of increased structure in the peptide. It is notable that the molar residual ellipticities at different TFE concentrations all cross at the isodichroic point of ∼202 nm, suggestive of a two-state system in which the peptide switches between two conformations as a function of TFE concentration. The spectra were deconvoluted using Pepfit, and the percent helix and percent coil from this analysis are shown in Fig. 6B. The error bars depict the standard error between the calculated estimates for the five best fits. It can be seen that the peptide converts from largely coil to largely helix as the TFE concentration increases. Maximal change is observed in the range of 15% to 25% TFE.
Fig. 6.
Solvent-dependent change in secondary structure of the P356-T373 peptide, determined by CD spectroscopy as described in Materials and Methods. (A) CD spectra of peptide at 0.052 mM as a function of TFE concentration. (B) Helix and coil contents from panel A, estimated using Pepfit. (C) CD spectra of peptide at 0.052 mM as a function of EtOH concentration. (D) Helix and coil contents from panel C. (E) CD spectra of peptide at 0.52 mM as a function of EtOH concentration. Insets show magnified spectra from 0 to 25% EtOH and 50 to 80% EtOH. (F) Ellipticities at 198 nm versus ellipticities at 222 nm for peptide at 0.052 mM (from panel C) and 0.52 mM (from panel E) over a range of EtOH concentrations. Values from 10% and 50% EtOH are indicated. deg, degree.
While TFE and other halogenated alcohols are widely used to promote α-helix formation, the mechanisms underlying this effect are still not fully understood (12). Indeed, in certain cases TFE induces helical structure in proteins which are predominantly β-structured (58). Therefore, we also altered the dielectric constant with ethanol (EtOH) rather than TFE. We found that the CD spectrum of the peptide responds to addition of EtOH (Fig. 6C) in a manner very similar to the TFE response, with an isodichroic point of 202 nm. Analysis of these spectra using Pepfit gave the results shown in Fig. 6D: like TFE, ethanol appears to induce a transition in the peptide from predominantly coil conformation to predominantly helix conformation, approximately linearly between 10% and 50% ethanol. To eliminate any algorithm-related anomaly in our analysis, the data were also analyzed by CONTIN and CDSSTR; these results, which are shown in Fig. S2A and B posted at http://home.ncifcrf.gov/hivdrp/Rein_publications/JV2011DattaSupplemental.pdf, show that the algorithms also indicate a shift from coil to helix with increasing EtOH concentrations. Similar results were also obtained with isopropanol (data not shown).
Concentration-dependence of the CD spectra.
Gag molecules are at an extremely high local concentration in an assembling VLP. It was therefore of interest to determine whether the concentration of the free peptide might affect its conformation. We repeated the EtOH titrations with the peptide at 0.52 mM, i.e., a concentration 10 times higher than that used in the experiments shown in Fig. 6C and D. The results are shown in Fig. 6E. Strikingly, we found that the spectra are quite different at this peptide concentration. Two distinct phases are observed in the titration. At low EtOH concentrations (Fig. 6E, left inset), there is a decrease in ellipticity at 198 nm and an increase between 210 and 230 nm. This early part of the titration shows a clear isodichroic point (206 nm). The spectrum at 25% EtOH already shows minima at 208 nm and 222 nm, characteristic of α-helices (18). Increasing the EtOH concentration above 50%, however, yields spectra with a single minimum at 218 nm (Fig. 6E, right inset), characteristic of β-sheet-rich structures (18). This concentration-dependent shift in the response to EtOH is highlighted in Fig. 6F, in which the ellipticity at 198 nm (from Fig. 6C and E) is plotted against the ellipticity at 222 nm for the two peptide concentrations. It can be seen that at 0.052 mM peptide, there is a linear relationship between the two values, suggestive of a simple two-state transition; in contrast, at 0.52 mM peptide, the plot is curved, reflecting further changes beyond a simple coil-helix transition.
We also examined the CD spectra of the peptide at several concentrations in an aqueous environment. As shown in Fig. 7, when the peptide is diluted to 2.6 mM instead of 0.052 mM, there is a shift in the wavelength of minimum ellipticity from ∼198 nm to ∼204 nm and appearance of a dichroic signal between 210 and 230 nm, suggestive of structuring of the peptide. However, the CD spectrum acquired immediately after diluting the peptide to a higher concentration of 5 mM displays a classic α-helical CD spectrum, with minimum ellipticities at both ∼211 and ∼223 nm. The ratio of ellipticity at 222 nm and 208 nm of ∼1 in this case suggests associating helices (42, 74). Acquisition of the spectrum at such a high concentration also shows some absorbance flattening, suggestive of interpeptide interactions (64). Over time, the CD spectrum displays a gradual, time-dependent change in the spectrum, suggesting a gradual loss of helical content at this concentration (data not shown). Experiments further characterizing these changes are now under way.
Fig. 7.
Concentration-dependent changes in secondary structure of the P356-T373 peptide. Representative CD spectra of the peptide diluted to 0.052 mM, 2.6 mM, and 5.2 mM from a 10.4 mM stock solution.
DISCUSSION
The SP1 region of HIV-1 Gag is crucial for VLP assembly (40) and is proposed to form a six-helix bundle in assembled virions (69). This report documents two fundamental properties of the SP1 region of HIV-1 Gag protein. First, immature particle assembly by HIV-1 Gag is extraordinarily sensitive to changes in the first ∼6 residues of SP1. Even minute changes in this region, especially in the first 4 residues, completely destroy the ability of the protein to assemble correctly in mammalian cells; rather, these mutant proteins misassemble into tubes and, frequently, a variety of other structures. Our survey (summarized in Table 1), along with prior results (1, 3, 44), indicates that proper assembly probably requires a hydrophobic residue at position 364 (the first position in SP1); that E365 can be replaced with aspartate, glutamine, or alanine but not by glycine, proline, lysine, phenylalanine, or tryptophan; that A366 can be replaced by tryptophan (A366V and A366T are replication competent only in the presence of the drug PA-457 [4]) but not phenylalanine or a series of more disparate amino acids; and that either leucine or tryptophan can replace methionine at position 367, but phenylalanine here produces a mixture of proper VLPs and irregular buds while all other substitutions tested at this position were incompatible with proper assembly. Both A366W and M367W retain some infectivity (Table 2). Single-residue deletions of the first 4 residues of SP1, which would affect the amphipathic nature of the helix, destroy correct assembly. Moreover, helix-breakers such as glycine and proline were not tolerated in any of the positions tested. The sensitivity of assembly to subtle changes in the SP1 sequence is exemplified by the difference between M367F, which can produce some normal-looking VLPs, and M367Y, which apparently cannot: evidently, addition of a hydroxyl group to the phenylalanine renders the protein completely incapable of correct assembly (Table 1; see also Fig. S1 posted at http://home.ncifcrf.gov/hivdrp/Rein_publications/JV2011DattaSupplemental.pdf). Single-residue deletions in SP1 have previously been tested for their effects on virus replication capacity; the results correspond in general to our observations on virus morphology (43).
Several observations suggested that many alterations of the SP1 sequence not only prevent proper assembly but also reduce the strength of the Gag-Gag interactions underlying assembly. Thus, we also found that under our experimental conditions, assembly in vivo of ΔA364 Gag into pleomorphic structures is apparently completely dependent upon N-terminal myristylation and membrane association while, in contrast, unmyristylated Gag with a WT SP1 sequence can assemble into nearly spherical structures in the cytoplasm (Fig. 3). Similarly, recombinant ΔA364 Δ16–99 Gag protein forms large pelletable structures upon addition of nucleic acid, but these structures are amorphous aggregates, not VLPs (Fig. 4B); interestingly, they are also formed less efficiently than those assembled from control Δ16–99 Gag protein (Fig. 4A). ΔV370 Δ16–99 Gag resembles Δ16–99 Gag in both of these respects (Fig. 4A and C). ΔA364 Gag is also capable of somewhat inefficient coassembly with WT Gag when the two proteins are coexpressed in vivo (Fig. 3). The misassembly of a similar mutant, ΔE365, is not corrected by replacement of NC, SP2, and p6 with a leucine zipper motif (data not shown). Taken together, these observations highlight the fact that the correct interactions between Gag proteins leading to VLP formation depend upon the correct sequence of the N-terminal region of SP1.
The second basic result presented here is the sensitivity of SP1 structure to environmental conditions. This was evident both from MD simulations of the SP1 sequence over a range of DMSO concentrations and from CD spectroscopy on an 18-residue peptide spanning the CA-SP1 junction in a series of TFE or EtOH concentrations or as a function of peptide concentration in an aqueous environment. The MD simulations suggest that the sequence would undergo a concerted coil-helix transition as a function of environmental dielectric conditions. Interestingly, the residues in SP1 (A364 to M378) all have equally high propensities for helix formation. The ability to adopt an α-helical conformation in TFE is consistent with the prior work of Morellet et al. (50). However, our experiments underscore the intrinsic helix-forming properties of the sequence, since they were performed with a short peptide under dilute conditions, and extend the observation to EtOH, which in general does not promote α-helix formation as potently as TFE. If SP1 were to adopt a helical structure, it would be amphipathic (Fig. 5C) and have a strong helical hydrophobic moment (25). Such a sequence would be especially sensitive to environmental conditions, and in aqueous environments the hydrophobic face of the helix would promote self-association of the peptide along with helix formation. The ratio of the ellipticity at 222 nm and 208 nm for the peptide at 0.052 mM in TFE (60%) and ethanol (80%) is <0.86; this is indicative of the formation of isolated helices (42, 74). The conversion to an α-helical conformation in solutions containing EtOH was enhanced at elevated peptide concentrations (Fig. 6E and F), where a helical structure is formed at intermediate EtOH concentrations and then a β-sheet-like structure is formed at high EtOH. The ratio of ellipticity at 222 nm and 208 nm at 40% EtOH is ∼1, as observed with associating helices and coiled coils (42, 74). Indeed, when the peptide was diluted to 10 mg/ml (i.e., ∼5 mM) in a purely aqueous buffer, it underwent a remarkable structural transition, displaying a classic α-helical CD profile (Fig. 7); this suggests that in the absence of external influences, self-association and helical formation go hand in hand.
The structure of immature HIV-1 particles has recently been analyzed in detail by electron cryotomography (11, 69). These studies found that the Gag molecules are arranged in a hexameric lattice in the particles, with the CA domains in the lattice forming a ring around a central hole. A pillar of density is present just “beneath” (i.e., toward the interior of the particle) this hole; it was suggested that this density is a six-helix bundle of SP1 domains. To our knowledge, the only six-helix bundle described to date is in cucumber mosaic virus particles, with leucine residues on the inner “face” of each helix (59). Our data do not address the lattice structure directly, but the remarkable dependence of HIV-1 assembly upon the precise sequence of the N-terminal 5 to 6 residues of SP1 (Fig. 1 to 4 and Table 1) strongly implies that the side chains in this region of the Gag protein are in intimate contact with each other in assembled particles. Further, the change in conformation seen at high concentrations of the isolated SP1 peptide (Fig. 7) is a strong indication of peptide-peptide interactions.
Why might SP1 be so important for proper assembly? It should be noted that HIV-1 assembly generally requires the presence of nucleic acid (14); this function can be fulfilled in vitro by oligonucleotides (14), which are presumably only long enough to bind a few Gag molecules. Further, the nucleic acid requirement can be eliminated if the NC domain (the principal nucleic acid-binding domain) is replaced with a leucine zipper motif (2, 19, 37, 73). These observations imply that the function of the nucleic acid in assembly is to induce oligomerization of Gag molecules at or near their C termini. The interfaces leading to assembly are probably all within the CA and SP1 domains (5, 10, 19, 41). Thus, exposure of these interfaces apparently depends upon transmission of a signal, triggered by oligomerization, from the C-terminal region of Gag toward the CA domain. Similarly, in the alpharetrovirus Rous sarcoma virus (RSV), the C-terminal domain of CA in Gag protein appears to adopt an assembly-competent conformation upon interaction of Gag with short oligonucleotides (47, 48). It seems possible that the SP1 domain in HIV, which is clearly capable of assuming alternative conformations (Fig. 5 to 7), acts as a switch whose structure changes when the C-terminal region of Gag oligomerizes. The abrupt shift in CD spectrum in aqueous buffer when the peptide was tested at 5.2 mM, rather than lower concentrations (Fig. 7), is completely consistent with this idea. It is intriguing that somewhat analogous behavior has also been described in the p12 protein of Mason-Pfizer monkey virus (MPMV), a betaretrovirus (39).
One model consistent with the data is that association of Gag molecules near their C termini induces a conformational change in SP1, as shown in Fig. 7; in turn, this change could be “propagated” into the C-terminal subdomain of the CA domain. The latter region of the protein contains crucial interfaces responsible for immature particle structure, and a recent MD study suggests that it plays a preeminent role in stabilizing the Gag lattice in the immature particle (8, 9).
Finally, it is of interest to consider the possible generality of these phenomena. A recent comparison of the region corresponding to SP1 in immature lattices of HIV, RSV, and MPMV reveals somewhat different geometries (24). However, in all retroviruses tested, including avian leukosis virus (37, 38), bovine immunodeficiency virus (33), and murine leukemia virus (MLV) (16, 53), as well as HIV-1, changes at or near the C terminus of the CA domain are extremely detrimental for correct particle assembly. In MLV, which does not contain a spacer between CA and NC, the C-terminal end of CA consists of an extraordinary run of charged residues, termed the “charged assembly helix” or “electric wire” motif (16). If this motif were to adopt an α-helical conformation, the positively and negatively charged residues would be positioned as bands on the helix. Remarkably, deletions which would remove integral numbers of turns from this helix are frequently compatible with correct assembly and infectivity while other deletions drastically disrupt assembly (16). (We also tested an analogous set of deletions in HIV-1 SP1, but none of them assembled correctly [data not shown]). Thus, although the nature of the residues at this position in MLV is quite different from the residues in HIV-1 and although there is only a single maturation cleavage between CA and NC in this virus, these results suggest that in MLV, as well as in HIV-1, the ability to form an amphipathic helix connecting the “body” of the CA domain with the NC domain is critical for assembly. Further studies are necessary to investigate whether the molecular switch-like behavior observed with SP1 is a broadly applicable mechanism in retroviral assembly.
ACKNOWLEDGMENTS
We thank Barbara Felber, Vineet KewalRamani, and Alok Mulky for reagents and Hirsh Nanda and Adam Zlotnick for helpful discussions. We thank Raymond Sowder for purifying the commercially procured peptide. The assistance of Sergey Tarasov and Marzena Dyba of the Biophysics Resource in the Structural Biophysics Laboratory, CCR, in acquiring CD spectra is gratefully acknowledged.
This work was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN26120080001E.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print on 16 February 2011.
REFERENCES
- 1. Accola M. A., Hoglund S., Gottlinger H. G. 1998. A putative α-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J. Virol. 72:2072–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Accola M. A., Strack B., Gottlinger H. G. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adamson C. S., et al. 2006. In vitro resistance to the human immunodeficiency virus type 1 maturation inhibitor PA-457 (Bevirimat). J. Virol. 80:10957–10971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adamson C. S., Waki K., Ablan S. D., Salzwedel K., Freed E. O. 2009. Impact of human immunodeficiency virus type 1 resistance to protease inhibitors on evolution of resistance to the maturation inhibitor bevirimat (PA-457). J. Virol. 83:4884–4894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ako-Adjei D., Johnson M. C., Vogt V. M. 2005. The retroviral capsid domain dictates virion size, morphology, and coassembly of Gag into virus-like particles. J. Virol. 79:13463–13472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Amarasinghe G. K., et al. 2000. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 301:491–511 [DOI] [PubMed] [Google Scholar]
- 7. Amon M. A., et al. 2008. Kinetic and conformational properties of a novel T-cell antigen receptor transmembrane peptide in model membranes. J. Pept. Sci. 14:714–724 [DOI] [PubMed] [Google Scholar]
- 8. Ayton G. S., Voth G. A. 2010. Multiscale computer simulation of the immature HIV-1 virion. Biophys. J. 99:2757–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Borsetti A., Ohagen A., Gottlinger H. G. 1998. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J. Virol. 72:9313–9317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Briggs J. A., Johnson M. C., Simon M. N., Fuller S. D., Vogt V. M. 2006. Cryo-electron microscopy reveals conserved and divergent features of Gag packing in immature particles of Rous sarcoma virus and human immunodeficiency virus. J. Mol. Biol. 355:157–168 [DOI] [PubMed] [Google Scholar]
- 11. Briggs J. A., et al. 2009. Structure and assembly of immature HIV. Proc. Natl. Acad. Sci. U. S. A. 106:11090–11095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Buck M. 1998. Trifluoroethanol and colleagues: cosolvents come of age. Recent studies with peptides and proteins. Q. Rev. Biophys. 31:297–355 [DOI] [PubMed] [Google Scholar]
- 13. Campbell S., et al. 2001. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc. Natl. Acad. Sci. U. S. A. 98:10875–10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Campbell S., Rein A. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Campbell S., Vogt V. M. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487–6497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cheslock S. R., et al. 2003. Charged assembly helix motif in murine leukemia virus capsid: an important region for virus assembly and particle size determination. J. Virol. 77:7058–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Compton L. A., Johnson W. C., Jr 1986. Analysis of protein circular dichroism spectra for secondary structure using a simple matrix multiplication. Anal. Biochem. 155:155–167 [DOI] [PubMed] [Google Scholar]
- 18. Creighton T. E. 1993. Proteins, 2nd ed. W. H. Freeman and Co., New York, NY [Google Scholar]
- 19. Crist R. M., et al. 2009. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: implications for retrovirus assembly. J. Virol. 83:2216–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Datta S. A., Rein A. 2009. Preparation of recombinant HIV-1 gag protein and assembly of virus-like particles in vitro. Methods Mol. Biol. 485:197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Datta S. A. K., et al. 2007. Conformation of the HIV-1 Gag protein in solution. J. Mol. Biol. 365:812–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Datta S. A. K., et al. 2007. Interactions between HIV-1 Gag molecules in solution: an inositol phosphate-mediated switch. J. Mol. Biol. 365:799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. De Guzman R. N., et al. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279:384–388 [DOI] [PubMed] [Google Scholar]
- 24. de Marco A., et al. 2010. Conserved and variable features of Gag structure and arrangement in immature retrovirus particles. J. Virol. 84:11729–11736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eisenberg D., Weiss R. M., Terwilliger T. C. 1982. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature 299:371–374 [DOI] [PubMed] [Google Scholar]
- 26. Freed E. O. 2002. Viral late domains. J. Virol. 76:4679–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fuller S. D., Wilk T., Gowen B. E., Krausslich H. G., Vogt V. M. 1997. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr. Biol. 7:729–738 [DOI] [PubMed] [Google Scholar]
- 28. Gamble T. R., et al. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285–1294 [DOI] [PubMed] [Google Scholar]
- 29. Gamble T. R., et al. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849–853 [DOI] [PubMed] [Google Scholar]
- 30. Gitti R. K., et al. 1996. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273:231–235 [DOI] [PubMed] [Google Scholar]
- 31. Gross I., Hohenberg H., Krausslich H. G. 1997. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur. J. Biochem. 249:592–600 [DOI] [PubMed] [Google Scholar]
- 32. Gross I., et al. 2000. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 19:103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guo X., Hu J., Whitney J. B., Russell R. S., Liang C. 2004. Important role for the CA-NC spacer region in the assembly of bovine immunodeficiency virus Gag protein. J. Virol. 78:551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guo X., Roldan A., Hu J., Wainberg M. A., Liang C. 2005. Mutation of the SP1 sequence impairs both multimerization and membrane-binding activities of human immunodeficiency virus type 1 Gag. J. Virol. 79:1803–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Henderson L. E., Bowers M. A., Sowder R. C., 2nd, et al. 1992. Gag protein of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J. Virol. 66:1856–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hill C. P., Worthylake D., Bancroft D. P., Christensen A. M., Sundquist W. I. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. U. S. A. 93:3099–3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johnson M. C., Scobie H. M., Ma Y. M., Vogt V. M. 2002. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 76:11177–11185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Keller P. W., Johnson M. C., Vogt V. M. 2008. Mutations in the spacer peptide and adjoining sequences in Rous sarcoma virus Gag lead to tubular budding. J. Virol. 82:6788–6797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Knejzlik Z., et al. 2009. Conformational changes of the N-terminal part of Mason-Pfizer monkey virus p12 protein during multimerization. Virology 393:168–176 [DOI] [PubMed] [Google Scholar]
- 40. Krausslich H. G., Facke M., Heuser A. M., Konvalinka J., Zentgraf H. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Krishna N. K., Campbell S., Vogt V. M., Wills J. W. 1998. Genetic determinants of Rous sarcoma virus particle size. J. Virol. 72:564–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lau S. Y., Taneja A. K., Hodges R. S. 1984. Synthesis of a model protein of defined secondary and quaternary structure. Effect of chain length on the stabilization and formation of two-stranded alpha-helical coiled-coils. J. Biol. Chem. 259:13253–13261 [PubMed] [Google Scholar]
- 43. Li F., et al. 2006. Determinants of activity of the HIV-1 maturation inhibitor PA-457. Virology 356:217–224 [DOI] [PubMed] [Google Scholar]
- 44. Liang C., et al. 2002. Characterization of a putative alpha-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 Gag. J. Virol. 76:11729–11737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liang C., Hu J., Whitney J. B., Kleiman L., Wainberg M. A. 2003. A structurally disordered region at the C terminus of capsid plays essential roles in multimerization and membrane binding of the Gag protein of human immunodeficiency virus type 1. J. Virol. 77:1772–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lobley A., Whitmore L., Wallace B. A. 2002. DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics 18:211–212 [DOI] [PubMed] [Google Scholar]
- 47. Ma Y. M., Vogt V. M. 2004. Nucleic acid binding-induced Gag dimerization in the assembly of Rous sarcoma virus particles in vitro. J. Virol. 78:52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ma Y. M., Vogt V. M. 2002. Rous sarcoma virus Gag protein-oligonucleotide interaction suggests a critical role for protein dimer formation in assembly. J. Virol. 76:5452–5462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Massiah M. A., et al. 1994. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J. Mol. Biol. 244:198–223 [DOI] [PubMed] [Google Scholar]
- 50. Morellet N., Druillennec S., Lenoir C., Bouaziz S., Roques B. P. 2005. Helical structure determined by NMR of the HIV-1 (345–392)Gag sequence, surrounding p2: implications for particle assembly and RNA packaging. Protein Sci. 14:375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Muriaux D., Mirro J., Harvin D., Rein A. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. U. S. A. 98:5246–5251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Newman J. L., Butcher E. W., Patel D. T., Mikhaylenko Y., Summers M. F. 2004. Flexibility in the P2 domain of the HIV-1 Gag polyprotein. Protein Sci. 13:2101–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Oshima M., et al. 2004. Effects of blocking individual maturation cleavages in murine leukemia virus Gag. J. Virol. 78:1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Provencher S. W., Glockner J. 1981. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20:33–37 [DOI] [PubMed] [Google Scholar]
- 55. Reed J., Reed T. A. 1997. A set of constructed type spectra for the practical estimation of peptide secondary structure from circular dichroism. Anal. Biochem. 254:36–40 [DOI] [PubMed] [Google Scholar]
- 56. Schneider R., Campbell M., Nasioulas G., Felber B. K., Pavlakis G. N. 1997. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol. 71:4892–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Scopes R. K. 1974. Measurement of protein by spectrophotometry at 205 nm. Anal. Biochem. 59:277–282 [DOI] [PubMed] [Google Scholar]
- 58. Shiraki K., Nishikawa K., Goto Y. 1995. Trifluoroethanol-induced stabilization of the alpha-helical structure of beta-lactoglobulin: implication for non-hierarchical protein folding. J. Mol. Biol. 245:180–194 [DOI] [PubMed] [Google Scholar]
- 59. Smith T. J., Chase E., Schmidt T., Perry K. L. 2000. The structure of cucumber mosaic virus and comparison to cowpea chlorotic mottle virus. J. Virol. 74:7578–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sreerama N., Woody R. W. 2000. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287:252–260 [DOI] [PubMed] [Google Scholar]
- 61. Swanstrom R., Wills J. W. 1997. Synthesis, assembly, and processing of viral proteins, p. 263–334 In Coffin J. M., Hughes S. H., Varmus H. E. (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 62. Tang C., Ndassa Y., Summers M. F. 2002. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 9:537–543 [DOI] [PubMed] [Google Scholar]
- 63. Vila J. A., Ripoll D. R., Scheraga H. A. 2000. Physical reasons for the unusual alpha-helix stabilization afforded by charged or neutral polar residues in alanine-rich peptides. Proc. Natl. Acad. Sci. U. S. A. 97:13075–13079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wallace B. A., Mao D. 1984. Circular dichroism analyses of membrane proteins: an examination of differential light scattering and absorption flattening effects in large membrane vesicles and membrane sheets. Anal. Biochem. 142:317–328 [DOI] [PubMed] [Google Scholar]
- 65. Warburg O., Christian W. 1942. Isolierung und Kristallisation des Garungsferments Enolase. Biochem. Z. 310:384–421 [Google Scholar]
- 66. Wiegers K., et al. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 72:2846–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wilk T., et al. 2001. Organization of immature human immunodeficiency virus type 1. J. Virol. 75:759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Worthylake D. K., Wang H., Yoo S., Sundquist W. I., Hill C. P. 1999. Structures of the HIV-1 capsid protein dimerization domain at 2.6 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 55:85–92 [DOI] [PubMed] [Google Scholar]
- 69. Wright E. R., et al. 2007. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 26:2218–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yang L. J., Yang X. Q., Huang K. M., Jia G. Z., Shang H. 2009. Dielectric properties of binary solvent mixtures of dimethyl sulfoxide with water. Int. J. Mol. Sci. 10:1261–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yeager M., Wilson-Kubalek E. M., Weiner S. G., Brown P. O., Rein A. 1998. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc. Natl. Acad. Sci. U. S. A. 95:7299–7304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yu F., et al. 2001. Characterization of Rous sarcoma virus Gag particles assembled in vitro. J. Virol. 75:2753–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhang Y., Qian H., Love Z., Barklis E. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 72:1782–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhou N. E., Kay C. M., Hodges R. S. 1992. Synthetic model proteins. Positional effects of interchain hydrophobic interactions on stability of two-stranded alpha-helical coiled-coils. J. Biol. Chem. 267:2664–2670 [PubMed] [Google Scholar]