Abstract
HIV-1 circulates within an infected host as a genetically heterogeneous viral population. Viral intrahost diversity is shaped by substitutional evolution and recombination. Although many studies have speculated that recombination could have a significant impact on viral phenotype, this has never been definitively demonstrated. We report here phylogenetic and subsequent phenotypic analyses of envelope genes obtained from HIV-1 populations present in different anatomical compartments. Assessment of env compartmentalization from immunologically discrete tissues was assessed utilizing a single genome amplification approach, minimizing in vitro-generated artifacts. Genetic compartmentalization of variants was frequently observed. In addition, multiple incidences of intercompartment recombination, presumably facilitated by low-level migration of virus or infected cells between different anatomic sites and coinfection of susceptible cells by genetically divergent strains, were identified. These analyses demonstrate that intercompartment recombination is a fundamental evolutionary mechanism that helps to shape HIV-1 env intrahost diversity in natural infection. Analysis of the phenotypic consequences of these recombination events showed that genetic compartmentalization often correlates with phenotypic compartmentalization and that intercompartment recombination results in phenotype modulation. This represents definitive proof that recombination can generate novel combinations of phenotypic traits which differ subtly from those of parental strains, an important phenomenon that may have an impact on antiviral therapy and contribute to HIV-1 persistence in vivo.
INTRODUCTION
Approximately 33 million people globally are infected with HIV-1 group M viruses, with an estimated 2.5 million new infections in 2007 (87). The extreme diversity of HIV-1 is characterized by nine phylogenetically distinct subtypes and a multitude of circulating recombinant forms (CRFs) (6). Such diversity poses a significant obstacle for the development of a successful prophylactic vaccine (21, 82). HIV-1 circulates within an infected host as a genetically heterogeneous viral population. The rapid rate of HIV-1 diversification within an infected individual is driven, in part, by the error-prone nature of HIV-1 reverse transcription and the high replicative rate in vivo that facilitates the incremental accumulation of insertions, deletions (indels), and point mutations in the viral genome (36). The inherent genetic variability of the population leads to humoral (86) and cellular (31) immune escape, resistance to antiretrovirals (9), and altered cytopathogenicity and cell tropism (48–50). Compartmentalization of HIV-1 populations derived from a range of tissues has been reported, including blood versus brain/central nervous system (1, 27, 47, 53, 67), brain versus lymph (4, 42, 68), blood versus female genital tract (51), and blood versus semen (10, 12, 24, 52, 93). In addition, genetic segregation of HIV-1 variants is also apparent between different populations of Langerhans cells (61) and at different sites within the brain (63), spleen (11), and gut (79). Such compartmentalized variation is shaped by a combination of founder effect, restricted cellular/viral trafficking between segregated subpopulations and local environmental selective pressures, including receptor/cellular tropism and tissue-specific immune responses.
A large part of HIV-1's genomic variability is located within the env gene, which encodes the transmembrane glycoprotein gp41 and surface glycoprotein gp120. These glycoproteins exist as trimeric spikes of gp41-gp120 heterodimers on the surface of the viral membrane (34, 92). Progressive evolution of the env gene contributes to escape from host immune responses. However, these processes are restricted by functional constraints associated with receptor and coreceptor binding and membrane fusion. The interaction of gp120 with the primary cellular receptor CD4 results in major conformational changes in the env trimer (34), which facilitates subsequent binding to the chemokine coreceptors CCR5 (8) and/or CXCR4, followed by gp41-mediated membrane fusion.
HIV-1 genetic heterogeneity is also shaped by recombination and mosaicism. HIV-1 is a highly recombinogenic virus, with exchange of genetic material between divergent group M subtypes giving rise to multiple CRFs worldwide (6, 56, 57). Examples of both inter- and intrasubtype recombinants have been reported (2, 6, 35, 58). Molecular epidemiological studies demonstrate rapid dissemination of certain CRFs in specific geographical localities (33, 77), suggesting that in some settings a fitness advantage may be conferred by mosaic genomes (3). However, this has not been definitively demonstrated. Recombination occurs via reverse transcriptase template switching during proviral DNA synthesis. Consequently, this phenomenon can only occur when a target cell is dually infected by genetically discrete viral strains (36, 56, 57). At the intrapatient level, recombination events continually produce mosaic genomes (7, 45). The rate of HIV-1 recombination has been shown to significantly exceed the rate of nucleotide misincorporation by the viral reverse transcriptase per round of genome replication (25, 39). Moreover, rates of HIV-1 recombination have been shown to differ between infected cell types (32). Intrapatient recombination has been proposed to facilitate resistance to targeted antiretroviral therapies (7, 44), as well as alter the tropism of resistant viruses during therapy (69). Exchange of genetic material between viruses isolated from the blood and female genital tract has also been demonstrated (51). However, despite its widespread documentation, no studies have definitively demonstrated that naturally occurring recombination can lead to altered viral phenotype.
To determine the relative contribution of intercompartment recombination to intrahost HIV-1 genetic variability, as well as determine its phenotypic consequences, we characterized HIV-1 env genes, obtained by single genome amplification (SGA), present in different anatomical compartments. Characterization of intrahost diversity by sequence analysis of SGA products excludes polymerase-induced nucleotide misincorporation, amplicon resampling, cloning bias, and the generation of in vitro recombinants via polymerase template switching (62, 70). High-resolution analyses of the sequence data revealed the presence of intercompartment recombinant viruses. Importantly, these recombination events conferred altered and potentially advantageous phenotypes compared to compartmentalized nonrecombinant strains.
MATERIALS AND METHODS
Sample collection and preparation.
Paired blood and semen samples were acquired from four treatment-naive HIV-1+ men attending clinics in Tiko, Cameroon, and Birmingham, United Kingdom. Blood and seminal plasmas were prepared by centrifugation at 1,500 × g for 20 min and stored at −70°C prior to RNA extraction. Seminal fluid mononuclear cells (SFMC) and peripheral blood mononuclear cells (PBMC) were isolated by centrifugation and Ficoll density gradient centrifugation, respectively. Washed PBMC and SFMC pellets were resuspended in 250 μl of saline prior to total DNA extraction (22). Paired samples were collected on the same day for each individual patient and processed within 4 h. Brain and cervical lymph node tissue were obtained at autopsy in Birmingham, United Kingdom, from a single HIV-1+ patient who died of AIDS related complications and stored at −70°C prior to total DNA extraction. Sample collection and subsequent research on all patient tissue utilized in the present study received ethical approval by the West Midlands Local Ethics Committee (United Kingdom) and the National Ethics Committee (Cameroon). All participants gave written informed consent.
Nucleic acid extraction and cDNA synthesis.
Viral RNA was extracted from blood and seminal plasma samples by using a QIAamp viral RNA kit (Qiagen) according to the manufacturer's instructions. Total DNA was extracted from PBMC, SFMC, brain autopsy, and lymph node autopsy samples by using a Stratagene DNA extraction kit according to a previously described, modified version of the manufacturer's protocol (4, 41). A gene-specific primer (20) was used to reverse transcribe cDNA from each RNA sample using a Thermoscript kit (Invitrogen) according to the manufacturer's instructions.
PCR amplification of env from single molecule templates.
Due to high levels of circulating HIV-1 diversity apparent in Cameroon, nested primer pairs shown to be effective at amplifying env from a diverse range of subtypes were used (20) according to previously described conditions (62). Accurate and representative sampling of the env intrahost diversity was achieved via SGA to preclude generation of in vitro artifacts (42, 62, 70). Amplicons were sequenced via BigDye terminator chemistry using an ABI 3100 capillary sequencer. Sequence chromatographs were manually inspected, and amplicons generated from >1 cDNA/provirus molecule were excluded from subsequent analyses. Amplicons that showed evidence for APOBEC3G-induced G-to-A hypermutation (37, 59, 65) or exhibited >1 example of a translation terminating stop codon or a frameshift inducing indel were also omitted. Subtype/CRF designations were determined by using the REGA subtyping tool (13). Sequences generated for the present study have been submitted to GenBank under accession numbers JF706370 to JF706501.
Sequence alignment and phylogenetic reconstruction.
Nucleotide sequences were aligned, with manual editing, according to overlying amino acid sequence using the CLUSTAL W algorithm implemented in MEGA4 software (76). Regions of ambiguous alignment were gap stripped from patient-specific and combined alignments. Pairwise distance matrices were calculated for compartment-specific env alignments via the maximum-composite-likelihood method implemented in MEGA4 software. Values were plotted in Prism GraphPad. A maximum-likelihood (ML) phylogenetic tree was generated for the combined alignment using the PHYML program (23) under the best-fit model of nucleotide substitution calculated by Modeltest (55) and midpoint rooted. Bootstrap values were derived from 1,000 replicate neighbor-joining trees (60), and significant (≥70%) values were assigned to internal ML tree branches.
Recombinant identification.
Patient-specific alignments were subsequently divided into three segments containing equal numbers of variable sites, and 1,000 partition-homogeneity test replicates were conducted using PAUP* v4.0b10 (75). A P value for the congruence of phylogenetic trees derived from different regions of the env gene was subsequently obtained for each patient's viral population (19). ML trees were then generated for each patient's three env region alignments under the best-fit model of nucleotide substitution using PAUP* v4.0b10. Topologies were inspected for significant positional switching of isolates between clades in different regions of env. Amplicons that exhibited shifting between compartment-specific clades were further investigated for mosaicism using a combination of pairwise diversity plots and informative site arrays (56, 72) as implemented in SimPlot version 3.5.1 (35). Breakpoint P values were calculated via 1,000,000 random shuffles of the order of sites of each informative sites array, with the significance test asking how often pseudoreplicate arrays are generated with a breakpoint as high as χ2 values obtained with the real data. Absolute breakpoint coordinates were derived from diversity plot crossover points. Only mosaics composed of identifiable parental strains were characterized.
Cloning of env amplicons for phenotyping.
The phenotypic consequences of intercompartment recombination were subsequently investigated in patients E21 and KM34. Although intercompartment mosaicism was detected in patient KM11, we were unable to clone these env amplicons, despite numerous attempts using a variety of cloning procedures. Therefore, we were unable to perform phenotypic characterization of envelope genes from this patient. Previous phenotypic studies of subtype B primary isolate env genes (86) report successful pseudovirion production using rev1/env cassettes ligated into appropriate expression vectors. Consequently, env amplicons were fused with HXB2 rev1 using an in-house recombinant PCR methodology. Briefly, amplification reactions were set up in 25-μl volumes containing 5 pmol of primer HXB2_R+E_S (5′-CACCCAAAAGCCTTAGGCATCTCC-3′), 5 pmol of primer HXB2_R+E_AS (5′-TCACACTACTTTTTGACCACTTGC-3′), 200 mM deoxynucleoside triphosphates, 0.5 U of Phusion high-fidelity DNA polymerase (Finnzymes), 1× Phusion HF buffer, and 250 pg of HXB2 full-length genome containing plasmid template. The PCR cycling parameters consisted of an initial denaturation step at 98°C for 30 s, followed by 35 cycles of 98°C for 10 s, 66°C for 15 s, and 72°C for 15 s, with a final extension step at 72°C for 1 min. Equimolar amounts of HXB2 rev1 and SGA-derived env amplicons were then used as a template for a recombinant PCR amplification using the primers HXB2_R+E_S and envM (20) according to the reaction setup conditions outlined above. Recombinant PCR was achieved via a two-step cycling strategy. Cycling parameters were an initial denaturation step at 98°C for 30 s, followed by 35 cycles of 98°C for 10 s and 72°C for 45 s, with a final extension step at 72°C for 1 min. The resulting rev/env cassettes were ligated into pcDNA3.1 D-TOPO (Invitrogen), confirmed by sequencing, and transformed in TOP10 or STBL3 cells (Invitrogen).
Cell cultures.
HeLa TZM-BL cells were used to titrate env+ pseudovirions and to evaluate HIV-1 neutralization (54, 85). Macrophage cultures were prepared from elutriated (26) or blood monocytes (48) as described previously. HEK 293T cells (15) were used to prepare env+ pseudovirions by transfection.
Production and titration of env+ pseudovirions.
Both a pNL4.3 construct lacking env and an env+ pcDNA3.1 D-TOPO expression vector were used to produce env+ pseudovirions as described previously (48, 50).
Infectivity assays.
Infectivity assays were performed as previously described (16). Primary macrophages were treated with DEAE dextran (10 μg/ml) prior to infection, before addition of an equal volume of serially diluted env+ pseudovirons and spinoculation (46). HeLa TZM-BL cells were infected without DEAE dextran or spinoculation.
Inhibition and neutralization assays.
Inhibition and neutralization assays for sCD4 (49) and neutralizing monoclonal antibodies b12 (91) and 2G12 (64) were carried out as described previously using HeLa TZM-BL cells as target cells (16, 49). For maraviroc, a CCR5 receptor antagonist (14), cells were treated with 2-fold dilutions in 50 μl for 30 min before adding an equal volume containing 200 FFU of pseudovirus. After 3 h at 37°C, the virus-antibody mixture was removed, growth medium was added, and infected cells were incubated at 37°C for a total of 48 h. To evaluate residual infectivity, the medium was removed, and 100 μl of medium without phenol red was added. The cells were then fixed and solubilized by adding 100 μl of Beta-Glo (Promega, Inc.). Luminescence was then read in a BioTek Clarity luminometer.
RESULTS
Tissue panel and generated env amplicons.
Paired HIV-1+ patient tissue samples included blood and semen from treatment-naive Cameroonian and United Kingdom patients at distinct disease stages, in addition to archival brain and lymph node autopsy samples from a United Kingdom patient who died of AIDS related complications (Table 1). PCR amplification of env genes from single cDNA/proviral template molecules was then performed. A total of 132 env single molecule amplicons were generated, with a range of 20 to 35 amplicons per patient. Multiple sequences from each patient were used to elucidate the infecting subtype/CRF. United Kingdom patients UKBH1 and E21 were infected with subtype B viruses, Cameroonian patients KM11 and KM18 harbored CRF02_AG viruses, while Cameroonian patient KM34 was infected with a group M virus which did not correspond to any described subtype/CRF (Table 1). To ensure patient KM34 was infected with a group M strain, rather than group N or O viruses which also cocirculate in Cameroon, a confirmatory phylogenetic analysis with a panel of HIV-1 group M, N, and O reference strain env genes was conducted. This analysis confirmed KM34 env genes clustered firmly within the group M radiation (data not shown).
Table 1.
Patient and sample information
| Patient | Sampling location | Isolation date (day/mo/yr) | Risk factora | CD4 count (mm3) | Infecting subtype/CRF | Treatment | Sample material | Virus genetic material |
|---|---|---|---|---|---|---|---|---|
| E21 | United Kingdom | 08/04/1991 | MSM | 120 | B | AZT | Lymph node autopsy | DNA |
| Brain autopsy | DNA | |||||||
| UKBH1 | United Kingdom | 18/07/2007 | MSM | 323 | B | None | Blood plasma | RNA |
| Seminal plasma | RNA | |||||||
| Blood PBMC | DNA | |||||||
| Semen SFMC | DNA | |||||||
| KM11 | Cameroon | 17/06/2005 | Hetero | 3 | CRF02 | None | Blood plasma | RNA |
| Seminal plasma | RNA | |||||||
| KM18 | Cameroon | 17/06/2005 | Hetero | 154 | CRF02 | None | Blood plasma | RNA |
| Seminal plasma | RNA | |||||||
| KM34 | Cameroon | 27/03/2006 | Hetero | 251 | Ub | None | Blood plasma | RNA |
| Seminal plasma | RNA |
MSM, men having sex with men; Hetero, heterosexual.
U, the env gene did not correspond to any described group M subtype/CRF.
Length polymorphism and nucleotide sequence divergence.
The variable regions of env exhibit marked length variability and the length of the V1/V2 region has been shown to influence receptor affinity, cellular tropism, and sensitivity to neutralization. Consequently, the median lengths of gp160 proteins encoded by env genes derived from different tissue compartments were compared for each patient's viral population (see Fig. S1A in the supplemental material). Analysis of compartment-specific viral populations from patient E21 showed that the brain-derived population had more compact env coding regions compared to viruses derived from lymph node. In patient KM18 the median length of semen-derived gp160 was greater than blood-derived gp160, while this trend was reversed for patient KM34. The overall length of the env genes for patients UKBH1 and KM11 were not significantly different between anatomic sites.
Patient-specific alignments were then gap stripped, and pairwise nucleotide sequence divergences were calculated for each patient's compartment-specific pool of env variants. The average substitutions per site were calculated for all sequence pairs derived from each tissue compartment and plotted (see Fig. S1B in the supplemental material). Median pairwise distances were generally lower in brain compared to lymph tissue and in semen compared to blood.
Phylogenetic assessment of intrahost compartmentalization.
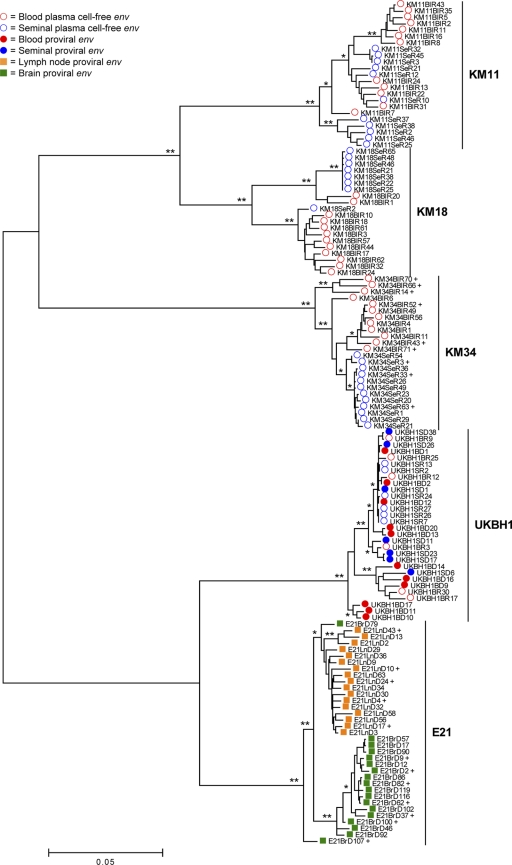
To assess the specific patterns of HIV-1 compartmentalization in each infected individual, phylogenetic analyses were conducted. All env sequences were aligned in a combined data set to enable identification of possible cross-contamination events. The env sequences clustered into five patient-specific monophyletic clades which exhibit various degrees of intrahost compartmentalization (Fig. 1). Patient KM11's sequences exhibited monophyletic blood- and semen-derived HIV-1 populations and a third population containing sequences derived from both compartments. Topologically, the extent of compartmentalization observed in patient's E21 and KM34 was identical, with two distinct sequence populations observed. In both patients, all sequences of one type form a clade within the radiation of sequences of the other type. These distinct sequence populations were largely correlated with tissue compartment. KM18 exhibited almost complete compartmentalization of env variants, although low-level migration of virus and/or infected cells between tissue compartments was also detected. Finally, there was no evidence of compartmentalization of blood- and semen-derived sequences obtained from patient UKBH1.
Fig. 1.
Phylogenetic assessment of env compartmentalization from immunologically discrete tissues. A midpoint-rooted ML phylogenetic reconstruction for the combined HIV-1 env alignment generated using the GTR+I+Γ4 substitution model (74, 88, 89) is shown. Branch lengths are in accordance with the scale bar and are proportional to genetic distance. Significance of clustering of isolates was assessed via bootstrap resampling of the sequence data derived from 1000 replications. *, Values >70%; **, values >99%. Isolates marked with a “+” were selected for subsequent phenotyping.
Intercompartment recombination.
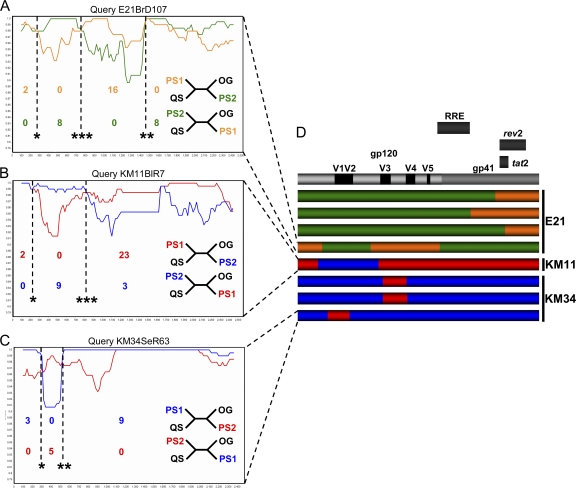
The results of partition incongruence tests (19) showed that different regions of the env gene were topologically incongruent in all patients (P < 0.05), which is indicative of recombination events. Therefore, to assess this further, the trees generated during the incongruence analysis were manually inspected for significant positional shifting of isolates between compartment specific clades in different regions of env: a signature of intercompartment recombination. Utilizing this method, we identified a total of eight putative intercompartment recombinants in 3 patients (E21, n = 4; KM11, n = 1; KM34, n = 3; see Fig. S2 in the supplemental material). There was no compelling evidence for intercompartment recombination in patients UKBH1 and KM18. Assignment of parental sequences was achieved via iterative manual generation of informative site arrays (55, 72), with every possible combination of parental sequences tested against each putative mosaic. The two sequences that in combination gave the highest number of informative sites were designated parental strains of the recombinant virus. The subsequent location of breakpoints and the corresponding P values were determined via a combination of parental strain diversity plot crossover points, and informative site array position which maximized the χ2 value (Fig. 2 and Table 2).
Fig. 2.
Evidence for intercompartment mosaicism. Example similarity plots and informative site arrays were used to characterize intercompartment mosaics. Plots were generated in SimPlot with a sliding window size of 200 bp and a step size of 20 bp and represent a query mosaic compared to two parental sequences. Dotted vertical lines indicate absolute breakpoint positions correlated with diversity plot crossover points. Associated P values (*, P < 0.05; **, P < 0.01; ***, P < 0.001) are derived from informative site arrays (described in Materials and Methods and displayed in Table 2). Relative numbers of informative sites shared by query sequences with parental strains are displayed below regions of differential homology. Four taxon trees consistent with these sites are displayed to the left: QS, query sequence; PS1, parental sequence 1; PS2, parental sequence 2; OG, outgroup. (A) Query mosaic E21BrD107 compared to parental sequences E21BrD90 and E21LnD56. (B) Query mosaic KM11BlR7 compared to parental sequences KM11BlR22 and KM11SeR2. (C) Query mosaic KM34SeR63 compared to parental sequences KM34BlR11 and KM34SeR26. (D) Schematic diagram depicting breakpoint locations in eight characterized mosaics, displayed relative to the positions of functionally defined regions in the HXB2 reference env located above. The patient from which each mosaic was isolated is detailed to the right of each individual recombinant env schematic. Regions of differential homology are color coded.
Table 2.
Putative breakpoints of intercompartment recombinants
| Mosaic sequence | No. of breakpoints | Parental sequences | Regiona | No. of supporting sites | Breakpoint positiona | P |
|---|---|---|---|---|---|---|
| E21BrD46 | 1 | E21BrD82 | 1–1956 | 22 | 2101 | <0.0001 |
| E21LnD10 | 2161–2571 | 6 | ||||
| E21BrD92 | 1 | E21BrD62 | 1–1650 | 22 | 1846 | <0.0001 |
| E21LnD13 | 1931–2571 | 7 | ||||
| E21BrD100 | 1 | E21BrD119 | 1–2127 | 26 | 2210 | <0.0001 |
| E21LnD43 | 2217–2571 | 4 | ||||
| E21BrD107 | 3 | E21BrD90 | 1–253 | 2 | 262 | 0.0442 |
| E21LnD56 | 364–687 | 8 | 775 | <0.0001 | ||
| E21BrD90 | 829–1392 | 16 | 1515 | 0.0056 | ||
| E21LnD56 | 1650–2571 | 8 | ||||
| KM11BlR7 | 2 | KM11BlR22 | 1–93 | 2 | 219 | 0.0365 |
| KM11SeR2 | 394–848 | 9 | 864 | 0.0004 | ||
| KM11BlR22 | 914–2571 | 23 | ||||
| KM34SeR3 | 2 | KM34SeR21 | 1–837 | 17 | 911 | 0.0004 |
| KM34BlR71 | 1017–1080 | 4 | 1164 | 0.0569 | ||
| KM34SeR21 | 1227–2571 | 3 | ||||
| KM34SeR54 | 2 | KM34SeR49 | 1–837 | 14 | 905 | 0.0007 |
| KM34BlR71 | 1017–1080 | 4 | 1173 | 0.0286 | ||
| KM34SeR49 | 1227–2571 | 4 | ||||
| KM34SeR63 | 2 | KM34SeR26 | 1–238 | 3 | 319 | 0.0356 |
| KM34BlR11 | 363–450 | 5 | 551 | 0.0010 | ||
| KM34SeR26 | 557–2571 | 9 |
The coordinates are given relative to the nucleotide position in the HXB2 reference strain env gene.
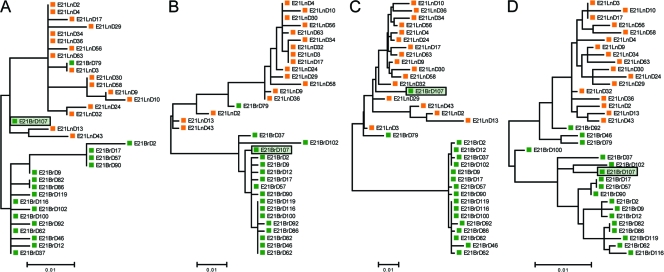
Three variants derived from the brain of patient E21 (E21BrD46, E21BrD92, and E21BrD100) had fragments of the gp41 coding region that were more similar to those present in lymph-derived sequences. A fourth sequence obtained from brain (E21BrD107) was a mosaic virus that possessed fragments containing V1/V2 and gp41 sequences from a brain-derived parental strain and the remaining env coding region representative of lymph-derived variants. Phylogenetic trees corresponding to each of these fragments can be seen in Fig. 3, with the positional shifting of env E21BrD107 between compartment specific clades highlighted. Patient KM11 harbored a recombinant virus present in blood which possessed a V1/V2 loop (plus flanking regions) homologous to sequences observed in the seminal population. Finally, three further recombinants were identified in patient KM34, where semen variants contained either V1/V2 (KM34SeR63) or C3 (KM34SSeR3 and KM34SeR54) sequences corresponding to blood-derived virus (see Fig. 2).
Fig. 3.
Phylogenetic trees corresponding to regions of differential homology in intercompartment mosaic E21BrD107. Patient E21's env alignment was divided into four segments, in agreement with the three breakpoints identified in isolate E21BrD107, and ML phylogenetic trees were constructed for each segment: positions 1 to 262 (A), positions 263 to 774 (B), positions 775 to 1514 (C), and positions 1515 to 2571 (D). Highlighted isolate E21BrD107 exhibits significant positional shifting between compartment specific clades.
Phenotypic characterization.
We next assessed whether there were differences in the phenotypes of viruses present in each tissue compartment and whether the characterized recombination events affected phenotype. Full-length env sequences representative of the tissue compartment specific gp160s, plus the identified intercompartment recombinants from patients E21 and KM34, were cloned into a mammalian expression vector. Resulting gp160 clones were used in fusion and pseudoparticle entry assays to investigate coreceptor usage, macrophage tropism, and sensitivity to a range of entry inhibitors and neutralizing antibodies that block viral entry. A summary of these data can be seen in Table 3. The majority of the clones analyzed used the CCR5 coreceptor. Two clones, one derived from the lymph tissue of E21, which was representative of a minor group of lymph sequences that possessed an unusual V3 loop apical sequence with a triplet amino acid insert (GPG), plus a blood-derived sequence from patient KM34, both utilized CXCR4.
Table 3.
Phenotypic properties of patient-derived gp160s
| gp160a | Sample material | Coreceptor usage | Propertyb |
||||
|---|---|---|---|---|---|---|---|
| Macrophage tropism | Maraviroc sensitivity | sCD4 sensitivity | b12 sensitivity | 2G12 sensitivity | |||
| E21LnD4 | Lymph node | CCR5 | – | + | ++ | +++ | +++ |
| E21LnD10 | Lymph node | CCR5 | – | ++ | ++ | +++ | +++ |
| E21LnD17 | Lymph node | CCR5 | – | + | ++ | +++ | +++ |
| E21LnD24 | Lymph node | CCR5 | – | ++ | +++ | +++ | ++ |
| E21LnD43 | Lymph node | CXCR4 | – | – | + | ++ | ++ |
| E21LnD58 | Lymph node | CCR5 | – | + | + | +++ | +++ |
| E21BrD2 | Brain | CCR5 | ++ | +++ | +++ | – | – |
| E21BrD9 | Brain | CCR5 | – | +++ | +++ | – | – |
| E21BrD37 | Brain | CCR5 | +++ | +++ | +++ | – | – |
| E21BrD82 | Brain | CCR5 | +++ | +++ | +++ | – | – |
| E21BrD62 | Brain | CCR5 | +++ | +++ | +++ | – | – |
| E21BrD100 (R) | Brain | CCR5 | ++ | +++ | +++ | – | – |
| E21BrD107 (R) | Brain | CCR5 | ++ | + | +++ | – | – |
| KM34BlR14 | Blood | CCR5 | – | +++ | +++ | ++ | +++ |
| KM34BlR43 | Blood | CCR5 | + | ++ | +++ | – | – |
| KM34BlR52 | Blood | CCR5 | + | ++ | +++ | – | – |
| KM34BlR66 | Blood | CCR5 | – | ++ | – | – | +++ |
| KM34BlR70 | Blood | CCR5 | – | ++ | – | – | – |
| KM34BlR71 | Blood | CXCR4 | – | – | – | – | – |
| KM34SeR3 (R) | Semen | CCR5 | +++ | ++ | +++ | – | – |
| KM34SeR33 | Semen | CCR5 | +++ | +++ | +++ | – | – |
| KM34SeR63 (R) | Semen | CCR5 | – | +++ | +++ | – | – |
(R), characterized recombinant gp160.
Macrophage tropism is scored as follows: −, <1%; +, 1 to 5%; ++, 5 to 15%; and +++, >15% infectivity in HeLa TZM-BL cells. Maraviroc sensitivity (nM) is scored as follows: +++, IC50 = <0.25; ++, IC50 = 0.25 to 1.0; +, IC50 = >1; and −, CXCR4 user. sCD4, b12, and 2G12 sensitivity (μg/ml) is scored as follows: +++, IC50 = <10; ++, IC50 = 10 to 20; +, IC50 = 20 to 50; and −, >50.
Macrophage tropism.
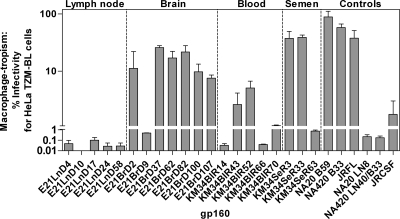
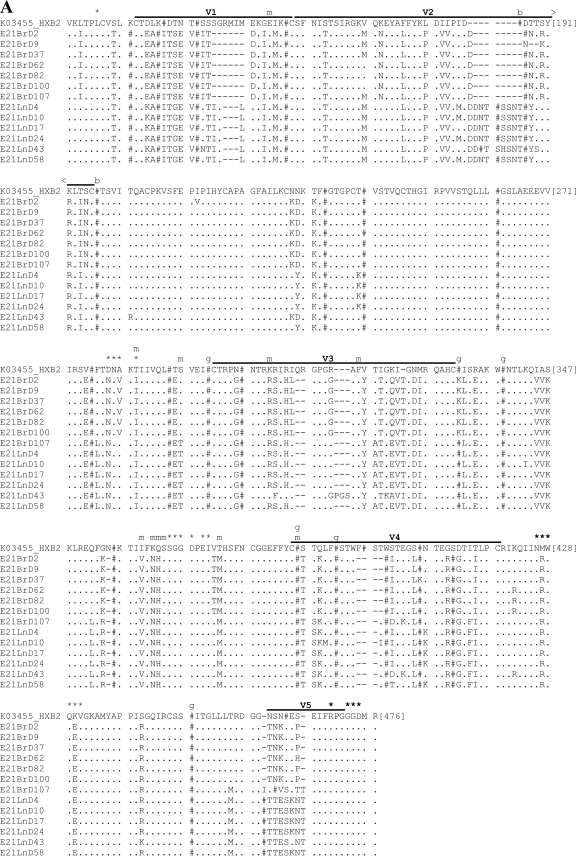
We next investigated how efficiently R5 gp160s could mediate infectivity of primary macrophages. Figure 4 presents macrophage infectivity of pseudovirions as a percentage of infectivity in HeLa TZM-BL cells. A macrophage infectivity of >1% was considered macrophage-tropic. Compartmentalization of macrophage tropism was evident between lymph node/brain- and blood/semen-derived viruses from patients E21 and KM34, respectively. All but one of the E21 brain-derived gp160s conferred higher macrophage infectivity than those derived from lymph node. The exception was E21BrD9, which exhibited substantially reduced macrophage tropism (0.49%) compared to other brain isolates (7.55 to 25.94%). Comparison of its sequence with the other brain-derived sequences showed that the former had an additional two amino acid deletion and loss of the potential N-linked glycosylation (PNG) site at position N186 in the V2 loop, a region which has been previously shown to be important in conferring macrophage-tropism (83). The two recombinant viruses obtained from E21 brain were both macrophage-tropic. R5 macrophage tropism is modulated by polymorphisms in the CD4 contact residues (30, 49) and residues that affect CD4 binding site exposure. These include residues flanking the CD4 binding site (16), an E153G substitution in V1 (43), and the presence of an asparagine at residue 283 (18, 50). Two CD4 contact residues (residues 281 and 455) (30), plus an additional residue (residue 291) thought to modulate macrophage tropism (16), exhibit compartment specific differences between brain- and lymph-derived sequences (Fig. 5A). Recombinant brain-derived gp160 E21BrD107 is highly macrophage-tropic and yet possesses identical residues to those found in the non-macrophage-tropic lymph-derived sequences at these positions. However, large differences in the length and number of PNG sites between lymph node and brain V1/V2 loops were observed. Lymph node V1/V2 loops possessed an average of 70 amino acids and 6 PNG sites, while brain V1/V2s loops were more compact, with an average of 60 amino acids and 5 PNG sites. Together, these data suggest the major determinants of macrophage tropism in patient E21 are located in the V1/V2 loops.
Fig. 4.
Macrophage tropism of HIV-1 gp160s amplified from anatomically discrete tissues. Patient/tissue-specific gp160s were expressed on HIV-1 pseudovirions. env+ pseudovirions were titrated on HeLa TZM-BL cells and on primary macrophages. Macrophage infectivity is expressed as a percentage of HeLa TZM-BL cell infectivity.
Fig. 5.
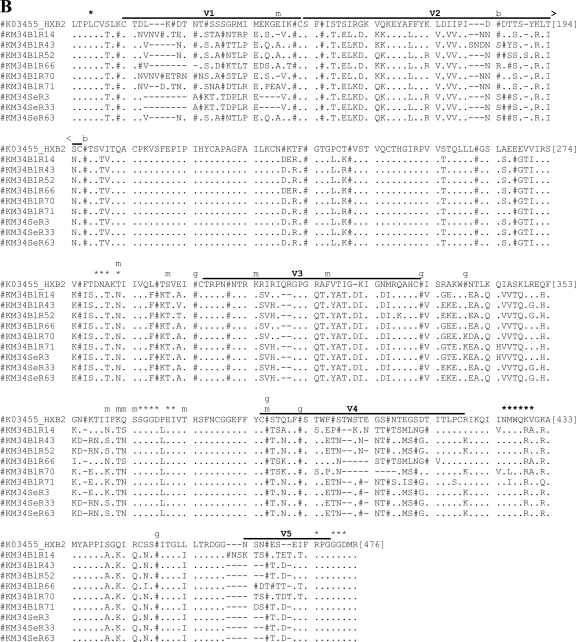
Amino acid sequence alignments for functional gp160s. Functional gp160s from patient E21 (A) and patient KM34 (B) are aligned relative to the HXB2 reference strain, and all residue numbering is relative to homologous positions in HXB2 gp160. Within the alignment, asparagine (N) residues that are likely to be glycosylated have been replaced by “#”. Horizontal lines located above alignment segments indicate the position of the variable regions V1 to V5. Symbols or letters located directly above alignment segments highlight the functionally important residues: *, CD4 contact residue; m, additional residue previously implicated in modulation of macrophage tropism; b, residue previously implicated in modulating sensitivity to b12; and g, residue previously implicated in modulating sensitivity to 2G12.
Blood-derived gp160s from patient KM34 were generally non-macrophage-tropic or conferred low levels of macrophage tropism, while semen-derived gp160s were either highly macrophage-tropic or non-macrophage-tropic (Fig. 4). Despite differences in macrophage tropism, residue N283, CD4 contact residues, and other residues implicated in modulating macrophage tropism did not exhibit compartment specific differences, although some variability was observed. Inspection of the encoded gp160 proteins revealed the observed range of tropism in patient KM34 is likely to be modulated by differences in the V1/V2 region, where substantial differences in the length and number of PNG sites between blood- and semen-derived V1/V2 loops were observed (Fig. 5B). Blood V1/V2 loops possess an average of 66 amino acids with 6 PNG sites, while semen V1/V2 loops are more compact, with an average of 57 amino acids and 5 PNG sites (see Fig. 5B). Recombinant semen envelope KM34SeR63 had a blood-like V1 loop and exhibited a 50-fold reduction in macrophage tropism (0.76%) compared to semen-derived KM34SeR3 (recombinant, blood-like C3 region, 37.40%) and KM34SeR33 (nonrecombinant, 39.11%). Indeed, nonrecombinant seminal isolateKM34SeR33 is identical to recombinant KM34SeR63 at all gp120 residues outside the V1 loop (see Fig. 5B). These data indicate a blood-like V1 loop in a seminal background renders gp160 KM34SeR63 non-macrophage-tropic and demonstrate cellular tropism modulation due to intercompartment env recombination.
Maraviroc sensitivity.
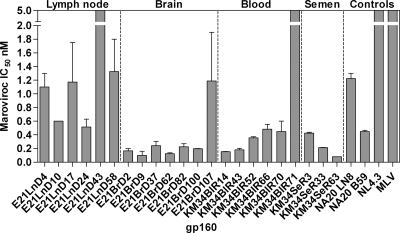
The CCR5 antagonist maraviroc demonstrates potent antiviral activity against R5-tropic primary isolates derived from diverse clades (14). As expected, the X4 gp160s KM34BIR71 and E21lnD43 were highly resistant to this entry inhibitor (Table 3 and Fig. 6). In contrast all of the R5 gp160s tested exhibited sensitivity to maraviroc (50% inhibitory concentration [IC50] range, 0.08 to 1.33 nM), regardless of tissue origin. In patient E21, compartmentalization of sensitivity to maraviroc was observed. Nonrecombinant brain-derived gp160s were highly sensitive (mean IC50 = 0.18 nM), while lymph node R5 isolates exhibited ∼5-fold less sensitivity (mean IC50 = 1.06 nM). Sensitivity to maraviroc can be modulated by sequence differences in the V3 loop, which is the principle determinant for CCR5 coreceptor binding. Amino acid residues that differed between brain- and lymph-derived sequences were located at positions L309I, G315R, T319A, and Q322E (Fig. 5A). Intercompartment mosaic E21BrD107 contains a lymph node-like V3 loop (see Fig. 2) and possesses lymph node-like sensitivity to maraviroc (IC50 = 1.19 nM, Table 3 and Fig. 6). Together, these data indicate compartment specific differences in sensitivity to maraviroc in patient E21 and demonstrate maraviroc sensitivity switching due to intercompartment env mosaicism. In patient KM34, there was limited variability in the V3 loop sequences of blood- or semen-derived isolates (Fig. 5B), and all of the R5 gp160s derived from this patient had similar maraviroc sensitivity.
Fig. 6.
Sensitivity of HIV-1 gp160s to maraviroc. Patient/tissue-specific env+ pseudovirions were tested for sensitivity to entry inhibition by maraviroc.
Soluble CD4 sensitivity.
Previously, we demonstrated that R5 macrophage tropism is significantly correlated with increased sensitivity to sCD4, which is compatible with increased exposure of the CD4 binding site and/or substitutions which confer tighter CD4 binding (49). The macrophage-tropic gp160s derived from brain and semen samples of patients E21 and KM34 were highly sensitive to entry inhibition by sCD4, whereas the non-macrophage-tropic gp160s derived from corresponding lymph node and blood were demonstrably less sensitive to sCD4 entry inhibition (Table 3). Intercompartment recombination had minimal effect on the sensitivity to sCD4 inhibition.
Sensitivity to broadly neutralizing antibodies IgG1-b12 (b12) and 2G12.
The monoclonal antibody b12 binds to a conformational conserved epitope on gp120 overlapping a discrete subset of CD4 contact residues (91). Previous data has shown that subtype B macrophage-tropic brain-derived gp160s are more sensitive to b12 than non-macrophage-tropic lymph tissue-derived gp160s (17, 49). In complete contrast to these findings, the non-macrophage-tropic lymph-derived gp160s from patient E21 were highly sensitive to b12 neutralization, while brain gp160s exhibited b12 resistance (Table 3). Regulation of b12 sensitivity, albeit in CRF01_AE viruses, has been linked to two N-linked glycan sites at amino acid positions 186 and 197 in the V2 and C2 regions (78) (Fig. 5A). Mosaic envelope E21BrD107 is similar to lymph-derived gp120s across nearly all regions except the V1V2 region (Fig. 2) and yet maintains a brain-like b12-resistant phenotype. Together, these data suggest the determinants contributing to b12 resistance/sensitivity in patient E21 gp160s are located in one or more of the V1/V2/C2 regions. The gp160s from patient KM34 (non-subtype B) were largely resistant to b12 inhibition, irrespective of their macrophage tropism or source.
The monoclonal antibody 2G12 targets a carbohydrate epitope, primarily in subtype B viruses, composed of high-mannose/hybrid glycans in the C2, C3, C4, and V4 domains (64). Lymph node-derived gp160s from patient E21 were susceptible to 2G12 neutralization, while brain-derived gp160s were resistant (Table 3). PNG sites at N295, N332, N339, N386, N392, and N448 are reportedly important for 2G12 binding (38, 49, 64, 66). The PNG site at position 332 is absent from E21 brain-derived gp160s but present in lymph node-derived gp160s. However, the brain-derived mosaicE21BrD107 has a PNG site at position N332 and yet a 2G12-resistant phenotype, suggesting that 2G12 resistance exhibited by E21 brain-derived gp160s is associated with determinants other than those previously described. Indeed, lymph node gp160s contain an extra PNG site in the V2 loop between N186 and N197 (see Fig. 5A), which may confer susceptibility to 2G12. Mosaic envelope E21BrD107 was similar to brain isolates across V1/V2 and therefore lacked this extra glycan and had a 2G12-resistant phenotype. Together, these data indicate compartmentalization of sensitivity to 2G12 in patient E21 and suggest that contributory determinants of 2G12 resistance are located in the V2 loop. While the majority of gp160s from KM34 were 2G12 resistant, two blood-derived viruses (KM34BlR14 and KM34BlR66) were highly sensitive to entry inhibition by this antibody (Table 3). Both of these gp160s possess an additional PNG site in the V5 region compared to other KM34 isolates. These data suggest sensitivity to 2G12 in patient KM34 is correlated to an additional PNG site in the V5 region.
DISCUSSION
Previous studies have demonstrated HIV-1 intrahost genetic compartmentalization between numerous tissues (1, 4, 10, 12, 24, 27, 42, 47, 51–53, 63, 67, 79, 93). HIV-1 recombination at the intersubtype (6), intrasubtype (58), and intrahost level (7, 45) has also been described. However, studies assessing HIV-1 intrahost compartmentalization have largely failed to investigate the contribution of recombination to the evolution of genetically discrete strains within an individual patient. Moreover, the impact of intrapatient genetic mosaicism on viral phenotype, particularly within the envelope gene, has previously been entirely overlooked. We hypothesized that intercompartment env mosaic viruses would arise in vivo. Also, given the divergent nature of compartmentalized sequences, we reasoned that any putative intercompartment mosaics would be readily detectable. In the present study we have shown that intercompartment recombination is frequent in the evolution of HIV-1 env in infected patients. In addition, we report that some of these recombination events confer new and potentially advantageous combinations of phenotypic traits upon the recombinant progeny compared to nonrecombinant parental strains.
Many previous studies assessing intrahost compartmentalization have utilized bulk amplification, cloning and sequencing strategies or have utilized viruses selected for growth in vitro. These methods are subject to numerous biases and in vitro artifacts which directly affect datasets generated and the subsequent interpretation of patterns of sequence evolution (62, 73, 80, 81). In addition, compartmentalization studies often utilize relatively small fragments of env (V1-V3 or C2-V5), reducing the chances of detecting putative recombination events in highly similar sequences. To minimize any experimentally induced biases and maximize the phylogenetic signal apparent in our data set, as well as facilitating phenotypic analyses, we utilized full-length env sequences amplified from single molecule templates derived directly from paired patient tissue samples. This methodology has been demonstrated to minimize polymerase induced misincorporations, reducing artificial inflation of intrahost viral diversity. If introduced in the initial rounds of amplification, the signatures of polymerase induced errors are easily identifiable since they appear as dual peaks in the sequence chromatographs. If introduced in the later rounds of amplification, misincorporations remain undetected since they represent only a minor component of generated amplicons (62). Critically, viral sequences generated in this manner are not subject to in vitro recombination events, which occur via polymerase template switching during bulk amplification from multiple starting templates (42, 62, 70).
Numerous phylogenetic and statistical tests can be used to assess degrees of HIV-1 intrahost compartmentalization. However, genetic mosaicism has been shown to adversely affect the results of these tests (90). Furthermore, identifying recombinant sequences in datasets derived from highly related variants is challenging. Automated procedures for the detection of recombination implemented in GARD (28, 29) identified numerous significant breakpoints in all datasets, as defined by discordant topologies, but lacked the facility to identify parental strains requiring subsequent manual investigation of the sequence data. Intercompartment mosaics breakpoints and parental strains could be correctly detected in via tests used in RDP2 (40). However, disparate results were obtained using the various tests used in this program, which required some prior knowledge of the likely recombinants and parental strains for correct interpretation of the output data. Consequently, we utilized an iterative approach involving manual inspection for topological incongruence in trees derived from different regions of env. Pairwise diversity plots and informative site arrays then enabled high-resolution identification of breakpoints and parental sequences. A number of breakpoints within env were identified across the intercompartment recombinant forms we characterized. Although specific breakpoint hot spots were not obvious, breakpoint locations generally occurred in the more conserved regions of env or were located at gp120 conserved/variable region boundaries. Breakpoint locations were generally in agreement with intersubtype breakpoint hot spots identified by in vitro modeling and in vivo observations (71). This is despite the fact that predicted constraints, such as maintenance of protein fold, are likely to be less severe at the intrahost level compared to the intersubtype level: viral variants circulating within a host infected with a single strain possess limited diversity while different group M subtypes can differ by up to 38% in envelope amino acid identity (82). All of the recombinant gp160s tested were shown to be functional in pseudotype assays, indicating that the characterized recombination events resulted in no deleterious disruption of gp160 tertiary or quaternary conformation and stability. In addition, it is likely that genomic constraints may also influence patterns of HIV-1 env recombination. Secondary RNA structures apparent in the env gene region (84) may favor/inhibit certain recombination events in naturally circulating virus. Variable loop regions (V1 to V5) in gp120 are reported to contain largely unstructured genomic RNA but are bordered by evolutionary conserved RNA structures in the conserved regions (84). Our identified breakpoints were generally observed in conserved regions or at conserved/variable region boundaries. Mosaic genomes that result in disruption to gp160 protein conformation or evolutionary conserved genomic RNA elements will be deleterious and removed from populations via purifying selection.
Viruses circulating in the peripheral blood or present in lymph nodes evolve to occlude critical regions, such as receptor binding sites, from host neutralizing antibodies. The blood-brain barrier and blood-testes barrier restrict the passage of neutralizing antibodies, making the brain and testes immunoprivileged tissues (5). Similarly, the type and frequency of HIV-1 permissive cells differs between sites, and the relative availability of CD4 and coreceptors on these target cells is likely to influence their permissiveness for different strains (49, 50). Consequently, viruses replicating in the brain or semen are likely to evolve in response to their unique environment, acquiring tropism and receptor affinity appropriate to each niche. Our analyses show that compartmentalization can be associated with altered macrophage tropism and sensitivity to entry inhibitors and monoclonal neutralizing antibodies. Crucially, our data has shown that these phenotypes can be altered by intercompartment recombination events. For example, recombinant brain isolate E21BrD107, which possesses a lymph node-like C2-C5 region, maintained the macrophage tropism of nonrecombinant brain-derived gp160s but acquired a reduced sensitivity to maraviroc through acquisition of V3 loop represented in lymph node-derived gp160. In addition, in patient KM34, blood-derived viruses were non-macrophage-tropic or poorly macrophage-tropic, whereas semen-derived gp160s were highly macrophage-tropic. However, recombinant semen envelope KM34SeR63, which possessed a region of gp120 encompassing the V1 loop present in blood-derived gp160s, was non-macrophage-tropic.
At the population level, the wide global dissemination of multiple intra- and intersubtype CRFs indicates recombinant genomes may possess a fitness advantage compared to pure subtypes in certain environmental settings. At the intrahost level, intercompartment recombination represents an additional molecular mechanism for generating viable and potentially fitter viral variants. Our study reveals the genetic signatures of intercompartment mosaicism are readily detectable in viruses isolated from different tissues, indicating this process occurs frequently in infected individuals. Indeed, although the present study was on a relatively small scale, with five patient's viral populations investigated, intercompartment mosaicism was detected in three individuals, with recombinant env genes comprising between 2 to 11% of generated amplicons. Our study was limited to viruses amplified from two tissue compartments per patient derived from a single time point, and characterization of recombinant isolates was restricted to those where both parental strains were represented in the population. Consequently, as HIV-1 is reported to be present in multiple tissue types, it is likely that intercompartment recombination occurs at rates in excess of those reported herein. Investigation of the functional consequences of the shuffling of phenotypic traits suggests this process may facilitate viral immune evasion and development of resistance to entry inhibitors. Viral progeny generated in this manner possessing advantageous combinations of traits will be rapidly swept to fixation within an infected host.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gerry Gillean for indispensable help with sample acquisition.
This study was supported by NIH grant R01HD049273-02.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 6 April 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Abbate I., et al. 2005. Cell membrane proteins and quasispecies compartmentalization of CSF and plasma HIV-1 from aids patients with neurological disorders. Infect. Genet. Evol. 5:247–253 [DOI] [PubMed] [Google Scholar]
- 2. Abecasis A. B., et al. 2007. Recombination confounds the early evolutionary history of human immunodeficiency virus type 1: subtype G is a circulating recombinant form. J. Virol. 81:8543–8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aulicino P. C., Holmes E. C., Rocco C., Mangano A., Sen L. 2007. Extremely rapid spread of human immunodeficiency virus type 1 BF recombinants in Argentina. J. Virol. 81:427–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ball J. K., Holmes E. C., Whitwell H., Desselberger U. 1994. Genomic variation of human immunodeficiency virus type 1 (HIV-1): molecular analyses of HIV-1 in sequential blood samples and various organs obtained at autopsy. J. Gen. Virol. 75(Pt. 4):67–79 [DOI] [PubMed] [Google Scholar]
- 5. Bart J., et al. 2002. An oncological view on the blood-testis barrier. Lancet Oncol. 3:357–363 [DOI] [PubMed] [Google Scholar]
- 6. Buonaguro L., Tornesello M. L., Buonaguro F. M. 2007. Human immunodeficiency virus type 1 subtype distribution in the worldwide epidemic: pathogenetic and therapeutic implications. J. Virol. 81:10209–10219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Charpentier C., Nora T., Tenaillon O., Clavel F., Hance A. J. 2006. Extensive recombination among human immunodeficiency virus type 1 quasispecies makes an important contribution to viral diversity in individual patients. J. Virol. 80:2472–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choe H., et al. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135–1148 [DOI] [PubMed] [Google Scholar]
- 9. Clavel F., Hance A. J. 2004. HIV drug resistance. N. Engl. J. Med. 350:1023–1035 [DOI] [PubMed] [Google Scholar]
- 10. Curran R., Ball J. K. 2002. Concordance between semen-derived HIV-1 proviral DNA and viral RNA hypervariable region 3 (V3) envelope sequences in cases where semen populations are distinct from those present in blood. J. Med. Virol. 67:9–19 [DOI] [PubMed] [Google Scholar]
- 11. Delassus S., Cheynier R., Wain-Hobson S. 1992. Nonhomogeneous distribution of human immunodeficiency virus type 1 proviruses in the spleen. J. Virol. 66:5642–5645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Delwart E. L., et al. 1998. Human immunodeficiency virus type 1 populations in blood and semen. J. Virol. 72:617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Oliveira T., et al. 2005. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics 21:3797–3800 [DOI] [PubMed] [Google Scholar]
- 14. Dorr P., et al. 2005. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob. Agents Chemother. 49:4721–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DuBridge R. B., et al. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duenas-Decamp M. J., Peters P. J., Burton D., Clapham P. R. 2009. Determinants flanking the CD4 binding loop modulate macrophage tropism of human immunodeficiency virus type 1 R5 envelopes. J. Virol. 83:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dunfee R. L., Thomas E. R., Gabuzda D. 2009. Enhanced macrophage tropism of HIV in brain and lymphoid tissues is associated with sensitivity to the broadly neutralizing CD4 binding site antibody b12. Retrovirology 6:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dunfee R. L., et al. 2006. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc. Natl. Acad. Sci. U. S. A. 103:15160–15165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Farris J. S., Kallersjo M., Kluge A. G., Bult C. 1994. Testing significance of incongruence. Cladistics 10:315–319 [Google Scholar]
- 20. Gao F., et al. 1996. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. WHO and NIAID Networks for HIV Isolation and Characterization. J. Virol. 70:1651–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gaschen B., et al. 2002. Diversity considerations in HIV-1 vaccine selection. Science 296:2354–2360 [DOI] [PubMed] [Google Scholar]
- 22. Ghosn J., et al. 2004. Evidence of genotypic resistance diversity of archived and circulating viral strains in blood and semen of pretreated HIV-infected men. AIDS 18:447–457 [DOI] [PubMed] [Google Scholar]
- 23. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 24. Gupta P., et al. 2000. Human immunodeficiency virus type 1 shedding pattern in semen correlates with the compartmentalization of viral quasi-species between blood and semen. J. Infect. Dis. 182:79–87 [DOI] [PubMed] [Google Scholar]
- 25. Jetzt A. E., et al. 2000. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 74:1234–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kalter D. C., et al. 1991. Enhanced HIV replication in macrophage colony-stimulating factor-treated monocytes. J. Immunol. 146:298–306 [PubMed] [Google Scholar]
- 27. Korber B. T., et al. 1994. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J. Virol. 68:7467–7481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kosakovsky Pond S. L., Posada D., Gravenor M. B., Woelk C. H., Frost S. D. 2006. Automated phylogenetic detection of recombination using a genetic algorithm. Mol. Biol. Evol. 23:1891–1901 [DOI] [PubMed] [Google Scholar]
- 29. Kosakovsky Pond S. L., Posada D., Gravenor M. B., Woelk C. H., Frost S. D. 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 22:3096–3098 [DOI] [PubMed] [Google Scholar]
- 30. Kwong P. D., et al. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leslie A. J., et al. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282–289 [DOI] [PubMed] [Google Scholar]
- 32. Levy D. N., Aldrovandi G. M., Kutsch O., Shaw G. M. 2004. Dynamics of HIV-1 recombination in its natural target cells. Proc. Natl. Acad. Sci. U. S. A. 101:4204–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liao H., et al. 2009. Phylodynamic analysis of the dissemination of HIV-1 CRF01_AE in Vietnam. Virology 391:51–56 [DOI] [PubMed] [Google Scholar]
- 34. Liu J., Bartesaghi A., Borgnia M. J., Sapiro G., Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lole K. S., et al. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Malim M. H., Emerman M. 2001. HIV-1 sequence variation: drift, shift, and attenuation. Cell 104:469–472 [DOI] [PubMed] [Google Scholar]
- 37. Mangeat B., et al. 2003. Broad antiretroviral defense by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103 [DOI] [PubMed] [Google Scholar]
- 38. Manrique A., et al. 2007. In vivo and in vitro escape from neutralizing antibodies 2G12, 2F5, and 4E10. J. Virol. 81:8793–8808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mansky L. M., Temin H. M. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087–5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martin D. P., Williamson C., Posada D. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260–262 [DOI] [PubMed] [Google Scholar]
- 41. McClure P., Curran R., Boneham S., Ball J. K. 2000. A polymerase chain reaction method for the amplification of full-length envelope genes of HIV-1 from DNA samples containing single molecules of HIV-1 provirus. J. Virol. Methods 88:73–80 [DOI] [PubMed] [Google Scholar]
- 42. McCrossan M., et al. 2006. An immune control model for viral replication in the CNS during presymptomatic HIV infection. Brain 129:503–516 [DOI] [PubMed] [Google Scholar]
- 43. Musich T., et al. 2011. A conserved determinant in the V1 loop of HIV-1 modulates the V3 loop to prime low CD4 use and macrophage infection. J. Virol. 85:2397–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nora T., et al. 2007. Contribution of recombination to the evolution of human immunodeficiency viruses expressing resistance to antiretroviral treatment. J. Virol. 81:7620–7628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Novitsky V., et al. 2009. Evolution of proviral gp120 over the first year of HIV-1 subtype C infection. Virology 383:47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. O'Doherty U., Swiggard W. J., Malim M. H. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074–10080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ohagen A., et al. 2003. Genetic and functional analysis of full-length human immunodeficiency virus type 1 env genes derived from brain and blood of patients with AIDS. J. Virol. 77:12336–12345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peters P. J., et al. 2004. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusigenicity for macrophages. J. Virol. 78:6915–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Peters P. J., et al. 2008. Variation in HIV-1 R5 macrophage-tropism correlates with sensitivity to reagents that block envelope: CD4 interactions but not with sensitivity to other entry inhibitors. Retrovirology 5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Peters P. J., et al. 2006. Non-macrophage-tropic human immunodeficiency virus type 1 R5 envelopes predominate in blood, lymph nodes, and semen: implications for transmission and pathogenesis. J. Virol. 80:6324–6332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Philpott S., et al. 2005. Human immunodeficiency virus type 1 genomic RNA sequences in the female genital tract and blood: compartmentalization and intrapatient recombination. J. Virol. 79:353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pillai S. K., et al. 2005. Semen-specific genetic characteristics of human immunodeficiency virus type 1 env. J. Virol. 79:1734–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pillai S. K., et al. 2006. Genetic attributes of cerebrospinal fluid-derived HIV-1 env. Brain 129:1872–1883 [DOI] [PubMed] [Google Scholar]
- 54. Platt E. J., Wehrly K., Kuhmann S. E., Chesebro B., Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophage-tropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Posada D., Crandall K. A. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818 [DOI] [PubMed] [Google Scholar]
- 56. Robertson D. L., Hahn B. H., Sharp P. M. 1995. Recombination in AIDS viruses. J. Mol. Evol. 40:249–259 [DOI] [PubMed] [Google Scholar]
- 57. Robertson D. L., Sharp P. M., McCutchan F. E., Hahn B. H. 1995. Recombination in HIV-1. Nature 374:124–126 [DOI] [PubMed] [Google Scholar]
- 58. Rousseau C. M., et al. 2007. Extensive intrasubtype recombination in South African human immunodeficiency virus type 1 subtype C infections. J. Virol. 81:4492–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sadler H. A., Stenglein M. D., Harris R. S., Mansky L. M. 2010. APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. J. Virol. 84:7396–7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 61. Sala M., et al. 1994. Spatial discontinuities in human immunodeficiency virus type 1 quasispecies derived from epidermal Langerhans cells of a patient with AIDS and evidence for double infection. J. Virol. 68:5280–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Salazar-Gonzalez J. F., et al. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 82:3952–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Salemi M., et al. 2005. Phylodynamic analysis of human immunodeficiency virus type 1 in distinct brain compartments provides a model for the neuropathogenesis of AIDS. J. Virol. 79:11343–11352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sanders R. W., et al. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sato K., et al. 2010. Remarkable lethal G-to-A mutations in vif-proficient HIV-1 provirus by individual APOBEC3 proteins in humanized mice. J. Virol. 84:9546–9556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Scanlan C. N., Offer J., Zitzmann N., Dwek R. A. 2007. Exploiting the defensive sugars of HIV-1 for drug and vaccine design. Nature 446:1038–1045 [DOI] [PubMed] [Google Scholar]
- 67. Schnell G., Spudich S., Harrington P., Price R. W., Swanstrom R. 2009. Compartmentalized human immunodeficiency virus type 1 originates from long-lived cells in some subjects with HIV-1-associated dementia. PLoS Pathog. 5:e1000395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sheehy N., Desselberger U., Whitwell H., Ball J. K. 1996. Concurrent evolution of regions of the envelope and polymerase genes of human immunodeficiency virus type 1 during zidovudine (AZT) therapy. J. Gen. Virol. 77(Pt. 5):1071–1081 [DOI] [PubMed] [Google Scholar]
- 69. Shi B., et al. 2010. Evolution and recombination of genes encoding HIV-1 drug resistance and tropism during antiretroviral therapy. Virology 404:5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Simmonds P., Balfe P., Ludlam C. A., Bishop J. O., Brown A. J. 1990. Analysis of sequence diversity in hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J. Virol. 64:5840–5850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Simon-Loriere E., et al. 2009. Molecular mechanisms of recombination restriction in the envelope gene of the human immunodeficiency virus. PLoS Pathog. 5:e1000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Smith J. M. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34:126–129 [DOI] [PubMed] [Google Scholar]
- 73. Spira A. I., Ho D. D. 1995. Effect of different donor cells on human immunodeficiency virus type 1 replication and selection in vitro. J. Virol. 69:422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sullivan J., Swofford D. L., Naylor G. J. P. 1999. The effect of taxon sampling on estimating rate heterogeneity parameters of maximum-likelihood models. Mol. Biol. Evol. 16:1347–1356 [Google Scholar]
- 75. Swofford D. L. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA [Google Scholar]
- 76. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software, version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 77. Tee K. K., et al. 2009. Estimating the date of origin of an HIV-1 circulating recombinant form. Virology 387:229–234 [DOI] [PubMed] [Google Scholar]
- 78. Utachee P., et al. 2010. Two N-linked glycosylation sites in the V2 and C2 regions of human immunodeficiency virus type 1 CRF01_AE envelope glycoprotein gp120 regulate viral neutralization susceptibility to the human monoclonal antibody specific for the CD4 binding domain. J. Virol. 84:4311–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. van Marle G., et al. 2007. Compartmentalization of the gut viral reservoir in HIV-1-infected patients. Retrovirology 4:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. van Opijnen T., de Ronde A., Boerlijst M. C., Berkhout B. 2007. Adaptation of HIV-1 depends on the host-cell environment. PLoS One 2:e271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. von Briesen H., et al. 1999. Selection of HIV-1 genotypes by cultivation in different primary cells. AIDS 13:307–315 [DOI] [PubMed] [Google Scholar]
- 82. Walker B. D., Burton D. R. 2008. Toward an AIDS vaccine. Science 320:760–764 [DOI] [PubMed] [Google Scholar]
- 83. Walter B. L., et al. 2005. Role of low CD4 levels in the influence of human immunodeficiency virus type 1 envelope V1 and V2 regions on entry and spread in macrophages. J. Virol. 79:4828–4837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Watts J. M., et al. 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460:711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wei X., et al. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wei X., et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 87. WHO/UNAIDS 2007. AIDS epidemic update. WHO/UNAIDS, Geneva, Switzerland: http://data.unaids.org/pub/epislides/2007/2007_epiupdate_en.pdf [Google Scholar]
- 88. Yang Z. 1994. Estimating the pattern of nucleotide substitution. J. Mol. Evol. 39:105–111 [DOI] [PubMed] [Google Scholar]
- 89. Yang Z. 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J. Mol. Evol. 39:306–314 [DOI] [PubMed] [Google Scholar]
- 90. Zarate S., Pond S. L., Shapshak P., Frost S. D. 2007. Comparative study of methods for detecting sequence compartmentalization in human immunodeficiency virus type 1. J. Virol. 81:6643–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhou T., et al. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhu P., et al. 2006. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441:847–852 [DOI] [PubMed] [Google Scholar]
- 93. Zhu T., et al. 1996. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J. Virol. 70:3098–3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.