Abstract
The presentation of viral peptide epitopes to host cytotoxic T lymphocytes (CTLs) is crucial for adaptive cellular immunity to clear the virus infection, especially for some chronic viral infections. Indeed, hosts have developed effective strategies to achieve this goal. The ideal scenario would be that the peptide epitopes stimulate a broad spectrum of CTL responses with diversified T-cell receptor (TCR) usage (the TCR repertoire). It is believed that a diversified TCR repertoire requires a “featured” peptide to be presented by the host major histocompatibility complex (MHC). A featured peptide can be processed and presented in a number of ways. Here, using the X-ray diffraction method, the crystal structures of an antigenic peptide derived from rinderpest virus presented by bovine MHC class I N*01801 (BoLA-A11) have been solved, and two distinct conformations of the presented peptide are clearly displayed. A detailed analysis of the structure and comparative sequences revealed that the polymorphic amino acid isoleucine 73 (Ile73) is extremely flexible, allowing the MHC groove to adopt different conformations to accommodate the rinderpest virus peptide. This makes the peptide more featured by exposing different amino acids for T-cell recognition. The crystal structures also demonstrated that the N*01801 molecule has an unusually large A pocket, resulting in the special conformation of the P1 residue at the N terminus of the peptide. We propose that this strategy of host peptide presentation might be beneficial for creating a diversified TCR repertoire, which is important for a more-effective CTL response.
INTRODUCTION
Viral diseases and other infections caused by intracellular pathogens of cattle result in large global economic losses every year. In the past several decades, rinderpest virus (RPV) (23, 40), foot-and-mouth disease virus (FMDV) (7), and bovine tuberculosis infection (62) have brought disastrous consequences, including social panic and even a threat to human health. Nearly 6 decades ago, most countries were committed to developing more-effective vaccines (46), and remarkable achievements were made in controlling threatening bovine viral infectious diseases and other zoonoses (23, 46). In the 1960s, British virologist Walter Plowright developed a live attenuated vaccine against rinderpest virus, which was widely used in rinderpest eradication efforts (40). However, it still remains extremely challenging for humans to completely eradicate a virus. The questions of how the bovine attenuated vaccine induces immune responses and how the viral peptides are presented and recognized by host immune molecules are still largely unanswered. Therefore, studies on bovine immunity against the rinderpest virus may provide clues for our battle against other viral infections.
Generally speaking, effective vaccines have common characteristics: they are rich in T-cell and B-cell epitopes and can induce the immune system to generate protective immune responses (3, 36, 54). During this process, major histocompatibility complex (MHC) molecules play a pivotal role in the host. MHC class I (MHC I) molecules form a heterotrimer complex composed of the heavy chain of MHC, the antigenic peptide, and β2-microglobulin (β2m). Generally, MHC I loads endogenous peptides, including the virus-derived peptides. Briefly, at the initial stage of virus infection, antigenic proteins are processed in a proteasome-dependent or -independent manner. The resultant short peptides are then transported to the endoplasmic reticulum and are loaded onto the peptide-binding grooves of MHC I molecules. Consequently, the peptide-loading MHC I complexes are translocated to the cell surface and are recognized by cytotoxic T lymphocytes (CTLs) with specific T-cell receptors (TCRs) (49, 63). This immune recognition induces an MHC-restricted CD8+ T-cell response that is characterized by the killing and elimination of the infected cells by effector T cells. The majority of virus-derived peptides are processed and presented through this pathway and consequently trigger the CTL-specific immune responses. These virus-derived peptides presented by MHC I molecules are known as antigenic CTL-specific epitopes.
Based on structural studies, the antigenicity of a peptide is partially dependent on the characteristics of the peptide related to its presentation by MHC molecules (31, 33, 34, 53, 55). Generally, peptides are divided into featured, featureless, and bulged peptides according to their presentation properties (25, 57). Featured peptides are those with solvent-exposed, prominent side chains or harmonious bulged conformations, which always correspond to a diverse repertoire of TCRs (32, 37, 38, 58). In contrast, featureless peptides have fewer or no solvent-exposed residues with prominent side chains. Bulged peptides are usually long peptides (>12 amino acids) that contain an extreme bend in the middle of the peptide chain binding to the MHC I molecule. The structural landscapes of featureless and bulged peptides result in an immune T-cell repertoire of limited diversity (58–60). Therefore, investigation of the structural characteristics of a peptide presented by MHC I molecules can improve the understanding of the antigenicity of that peptide and aid in the rational development and modification of peptide-based vaccines (33).
Bovine MHC I, which is called BoLA-I, has a critical role in presenting virus-derived antigen peptides and dominating bovine antiviral-specific CTL immune responses. In the 1970s, BoLA-I protein was first identified and genotyped by use of a specific serum antibody. BoLA-I was first cloned in 1988 (20), and since then, BoLA-I related research has entered the gene era. Thus far, hundreds of BoLA-I cDNA sequences have been registered at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/), and 58 complete BoLA-A sequences have been uploaded to the Immuno Polymorphism Database (IPD) (http://www.ebi.ac.uk/ipd/) (5). BoLA is located on the 23rd bovine chromosome (22), including at least six expressed gene loci (29). Further research has demonstrated that there are one to three gene loci that can be transcribed and expressed in one BoLA haplotype (16, 17). The fact that a breed of BoLA haplotypes expresses only one BoLA-A gene locus (6, 15) provides an important experimental model for controlling bovine viral diseases in cattle with a single MHC I gene locus.
Due to polymorphisms in BoLA-I, there are more than 50 serotypes of one BoLA-I allele locus (4, 13). The BoLA-A11 serotype specifically indicates that the BoLA-A11/pBoLA-19 (D18.3) gene product N*01801 is expressed (18, 28, 47, 48). Hegde and colleagues obtained the N*01801 peptide binding motif by monoclonal antibody purification of MHC molecules and subsequent acid elution of peptides (28). The majority of the peptides that occupied the binding groove of N*01801 were nonamers. There was clear evidence that position 2 of the peptides was occupied by Pro, and the C-terminal amino acid anchor was Ile/Val. Among the alleles whose motifs have been reported, the peptide motif of N*01801 is similar to those of H-2Ld, HLA-B7, HLA-B*3501, HLA-B*5101, HLA-B*5102, HLA-B*5103, HLA-B*7801, and HLA-Cw*0401 (21, 45). Sequence comparison of the bovine MHC I allele N*01801 to these MHC I molecules demonstrates that key residues share similar characteristics (and are conserved in some cases), which are involved in accommodating the similar anchor residues in P2 and P9. This was the first report of a BoLA allele-specific peptide motif (ASPM; 28). To identify bovine-restricted CTL epitopes derived from pathogens, according to N*01801 ASPMs, dozens of potential bovine CTL epitopes have been synthesized, and their antigenicity was preliminarily analyzed by CTL assays (27). In 2004, one of the CTL epitopes with a typical N*01801 ASPM (peptide IPA) was identified in rinderpest virus hemagglutinin (H) protein amino acids 408 to 416 (IPAYGVLTI) (51). Recently, Guzman et al. confirmed that there is an MHC-restricted CD8+ T-cell response after FMDV infection (26). However, the mechanism of bovine antiviral-specific T-cell immunity remains unclear, partially due to the lack of clues as to the binding and presentation of the antigenic peptides by BoLA-I.
To describe the landscape of peptide presentation by bovine MHC I N*01801 and to understand the immune basis of the eradication of rinderpest virus, we determined the crystal structures of N*01801 complexed to the rinderpest virus-derived peptide IPA. Analysis of the structures we determined suggests that compared to the structures of other MHC I molecules, including that of bovine N*01301, which was determined recently by Macdonald and colleagues (34), the structure of bovine MHC I N*01801 is distinctive. The structure of bovine N*01801 illuminates a novel presentation strategy for featured epitopes among mammalian MHCs and may lead to improved understanding of the structural basis of CTL-based immune responses (9).
MATERIALS AND METHODS
Peptide synthesis.
The peptides used in this study (Table 1), including peptide IPA (IPAYGVLTI), derived from rinderpest virus attachment glycoproteins (51), were synthesized and purified by reverse-phase high-performance liquid chromatography (HPLC) (SciLight Biotechnology). The peptide purity was determined to be ∼90% by analytical HPLC and mass spectrometry. These peptides were stored in lyophilized aliquots at −80°C after synthesis and were dissolved in dimethyl sulfoxide (DMSO) before use.
Table 1.
Peptides used in this study
| Name | Derived protein | Position | Sequencea | Reference |
|---|---|---|---|---|
| IPAb | Rinderpest virus H protein | 408–416 | IPAYGVLTI | 51 |
| APA | BHV-Ic BICP0 protein | 482–490 | APAPISTMI | 27 |
| NPM | BHV-I glycoprotein B precursor | 780–788 | NPMKALYPI | 27 |
| TPG | BHV-I glycoprotein C precursor | 18–26 | TPGATTPV | 27 |
Underlined boldface residues are the typical primary anchors of the peptides presented by N*01801.
Peptide IPA was used in the structural determination of the bovine MHC class I N*01801.
BHV-I, bovine herpesvirus 1.
Preparation of the bovine MHC I N*01801-β2m-IPA complexes.
Reverse transcription-PCR (RT-PCR) was used to amplify the full-length cDNA of N*01801 (A11) and bovine β2m from bovine kidney cells. Details of the primers used can be found in Table S1 in the supplemental material. The extracellular region of N*01801 (amino acids 2 to 275) was amplified from the cDNA according to its nucleotide sequence (GenBank accession no. BC151402) using primers 1 and 2. Bovine β2m (amino acids 1 to 98) was cloned from bovine kidney cell cDNA according to its nucleotide sequence (GenBank accession no. BC118352) with primers 3 and 4. Mouse β2m (amino acids 1 to 99) was cloned from BALB/c mouse spleen cDNA according to its nucleotide sequence (GenBank accession no. M84364.1) with primers 5 and 6. The amplified products were ligated into a pET21a vector (Novagen) and were transformed into Escherichia coli strain BL21(DE3). The recombinant proteins were expressed as inclusion bodies and were then purified as described previously (8).
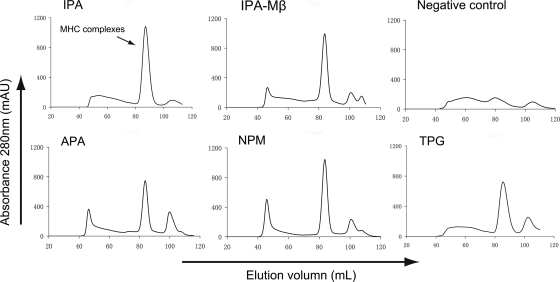
The N*01801-β2m-peptide complexes were prepared essentially with refolding assays as described by Zhou et al. in 2004 (64). Briefly, the N*01801 heavy chain and bovine or murine β2m inclusion bodies were separately dissolved in a solution of 10 mM Tris-HCl (pH 8.0) and 8 M urea. The N*01801 heavy chain, β2m, and the peptide in a 1:1:3 molar ratio were refolded by gradual dilution. After 48 h of incubation at 4°C, the soluble portion of the complexes was concentrated and then purified by size exclusion chromatography on a Superdex 200 16/60 column. The comparison of the absorbance peaks of the refolded complexes using different peptides is presented in Fig. 1. If the complex was prepared for crystallization, it was further purified by Resource-Q anion-exchange chromatography (GE Healthcare).
Fig. 1.
Binding of peptides to bovine MHC I N*01801 indicated via in vitro refolding. Peptides with the ability to bind to N*01801 can help the N*01801 heavy chain and β2m refold in vitro. After correct refolding, we found that high absorbance peaks of the MHCs with the expected molecular mass of 45 kDa eluted at the estimated volume of 16 ml on a Superdex 200 column (GE Healthcare). Complexes were formed in refolding with peptides IPA, APA, NPM, and TPG (which have the typical N*01801-restricted motif). IPA also aided in the renaturation of the N*01801 heavy chain with the murine β2m (IPA-Mβ). No binding peaks appeared when there was no peptide in the refolding system.
Crystallization and data collection.
The BoLA complexes were ultimately concentrated to 12 mg/ml in crystallization buffer (4.0 mM sodium formate), mixed with reservoir buffer at a 1:1 ratio, and crystallized by the hanging-drop vapor diffusion technique at 291 K. A Crystal Screen kit (Hampton Research, Riverside, CA) was used to screen for optimal crystal growth conditions. After several days, crystals of N*01801 complexed with the IPA peptide and bovine β2m (N*01801) were obtained with Crystal Screen solution 33 (4.0 mM sodium formate). Diffraction data were collected to a resolution of 2.7 Å using an in-house X-ray source (Rigaku MicroMax007 desktop rotating anode X-ray generator with a Cu target operated at 40 kV and 30 mA) and an R-AXIS IV++ imaging-plate detector at a wavelength of 1.5418 Å. Similarly, the crystals of N*01801 complexed with the IPA peptide and murine β2m (N*01801-Mβ) were grown in 0.2 M ammonium sulfate, 0.1 M Tris (pH 8.5), and 25% (wt/vol) polyethylene glycol 3350 (PEG 3350) at a concentration of 12 mg/ml, and the resolution of the diffraction data was 1.9 Å. The crystals were first soaked in reservoir solution containing 25% glycerol as a cryoprotectant and were then flash-cooled in a stream of gaseous nitrogen at 100 K (42). The collected intensities were indexed, integrated, corrected for absorption, scaled, and merged using the HKL2000 package (41).
Structure determination and refinement.
The structures of the BoLA complexes were solved by molecular replacement using the MOLREP program with HLA-B*5101 (Protein Data Bank [PDB] code 1E27) as a search model. Extensive model building was performed by hand with COOT (19), and restrained refinement was performed using REFMAC5. Additional rounds of refinement were performed using the phenix refine program implemented in the PHENIX package (2) with isotropic atomic displacement parameter (ADP) refinement and bulk solvent modeling. The stereochemical quality of the final model was assessed with the PROCHECK program (30).
Protein structure accession numbers.
The coordinates and structure factors of N*01801 and N01801-Mβ have been deposited in the Protein Data Bank with accession numbers 3PWV and 3PWU, respectively.
RESULTS
Typical peptides bound to bovine MHC I molecule N*01801 revealed by in vitro refolding.
A series of bovine MHC I allele N*01801-restricted CD8+ T-cell epitopes derived from different pathogens have recently been screened and characterized (14, 27, 51); from these epitopes, the peptide motif of this MHC I molecule was defined. However, no rapid and efficient methods to evaluate the ability of a peptide with a given motif to bind to bovine MHC I molecules in vitro have been developed yet. As indicated in previous studies (31–33, 55), the refolding experiments can semiquantitatively reflect the binding capability of peptides for MHCs. Four previously identified N*01801-restricted CD8+ T-cell epitopes (Table 1) were synthesized and used in refolding assays to evaluate their abilities to bind to N*01801 molecules and to define the candidates used for determination of the N*01801 structure. These four N*01801-restricted peptides helped the heavy chain of N*01801 and bovine β2m refold in vitro, as evidenced by the absorbance peaks of the complexes (Fig. 1). All four peptides contain a proline at position P2, while the Pc (C terminus of the peptide) positions were occupied by either isoleucine or valine. This peptide motif was partially demonstrated previously (27).
Overall structure of bovine MHC I N*01801-peptide complexes.
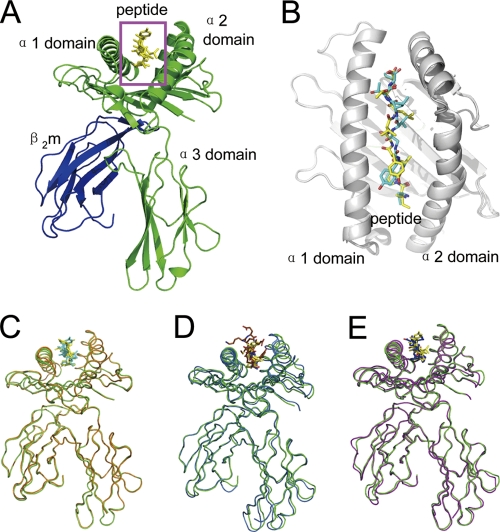
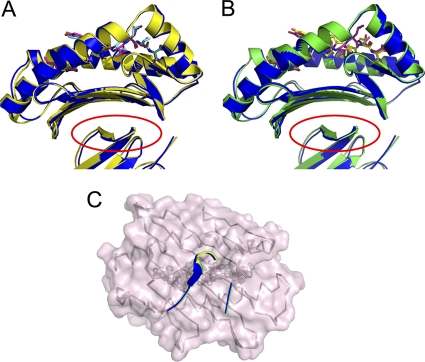
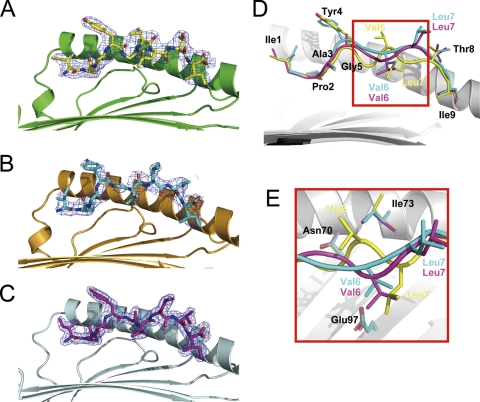
The bovine MHC I N*01801 complexed with peptide IPA was crystallized in the P212121 orthorhombic space group with a resolution of 2.7 Å (Table 2). The overall structure of N*01801 consists of the characteristic α1 and α2 domains of a heavy chain underpinned by α3 domains and β2m (Fig. 2A). The peptide lies along the peptide binding groove formed by the α1 and α2 domains (Fig. 2B). Within one asymmetric unit, there are two N*01801 molecules (referred to below as molecule 1 and molecule 2) that have similar overall conformations, with a root mean square difference (RMSD) of 0.300 Å (0.267 and 0.137 Å for the heavy chain and β2m, respectively) (Fig. 2C). The overall structure of N*01801 is similar to those of other class I MHC molecules. Structural alignment of molecule 1 of N*01801 with the structures of the bovine MHC I molecule N*01301 and human MHC HLA-B*5101 yielded RMSD values of 0.563 and 0.533 Å, respectively (Fig. 2D and E). Among all human MHC I molecules, HLA-B*5101 is sequentially the most similar to N*01801, displaying 80.29% identity (Fig. 3). Notably, this is at the same level as the identity between the intraspecies alleles of cattle: N*01801 and N*01301 (82.9%).
Table 2.
X-ray diffraction data processing and refinement statistics
| Statistic | Value for: |
|
|---|---|---|
| N*01801 | N*01801-Mβ | |
| Data processing | ||
| Space group | P41212 | P212121 |
| Cell parameters | ||
| a (Å) | 83.9 | 47.9 |
| b (Å) | 83.9 | 70.0 |
| c (Å) | 153.0 | 120.8 |
| α (°) | 90.0 | 90.0 |
| β (°) | 90.0 | 90.0 |
| γ (°) | 90.0 | 90.0 |
| Resolution range (Å) | 50.0–2.7 (2.8–2.7)a | 50.0–1.9 (1.97–1.9) |
| Total reflections | 201,829 | 228,256 |
| Unique reflections | 28,833 | 15,065 |
| Completeness (%) | 99.4 (98.9) | 99.9 (100.0) |
| Rmerge (%)b | 14.2 (54.8) | 6.6(53.5) |
| I/σ | 16.2 (4.3) | 29.2 (3.1) |
| Refinement | ||
| Rwork (%)c | 18.8 | 19.2 |
| Rfree (%) | 23.2 | 23.0 |
| RMSD | ||
| Bond lengths (Å) | 0.006 | 0.005 |
| Bond angles (°) | 0.892 | 0.909 |
| Average B factor | 44.6 | 28.7 |
| Ramachandran plot quality | ||
| Most favored (%) | 88.9 | 90.2 |
| Disallowed (%) | 0 | 0 |
Values in parentheses are given for the highest-resolution shell.
Rmerge = ∑hkl∑i | Ii − 〈I〉 | ∑hkl∑iIi, where Ii is the observed intensity and 〈I〉 is the average intensity of multiple observations of symmetry-related reflections.
R = ∑hkl ‖ Fobs | − k | Fcal | | ∑hkl | Fobs |, where Rfree is calculated for a randomly chosen 5% of reflections and Rwork is calculated for the remaining 95% of reflections used for structure refinement.
Fig. 2.
Overview of the structure of bovine MHC I N*01801. (A) Overview of the structure of the bovine MHC I N*01801 represented by molecule 1 in the asymmetric unit. (B) The peptides (yellow for the peptide of molecule 1 and cyan for the peptide of molecule 2) are presented in the peptide-binding clefts. (C) Superposition of the two molecules with one asymmetric unit of N*01801 (green for molecule 1 and orange for molecule 2). (D) Structural superposition of the bovine MHC I N*01801 molecule 1 (green) with N*01301 (blue) (PDB code 2XFX). (E) Structural alignments of the MHC heavy chain and β2m show the similar overview conformations of N*01801 molecule 1 (green) and HLA-B*5101 (purple) (PDB code 1E27). This figure and subsequent figures showing molecule models were generated using PyMol (www.pymol.org).
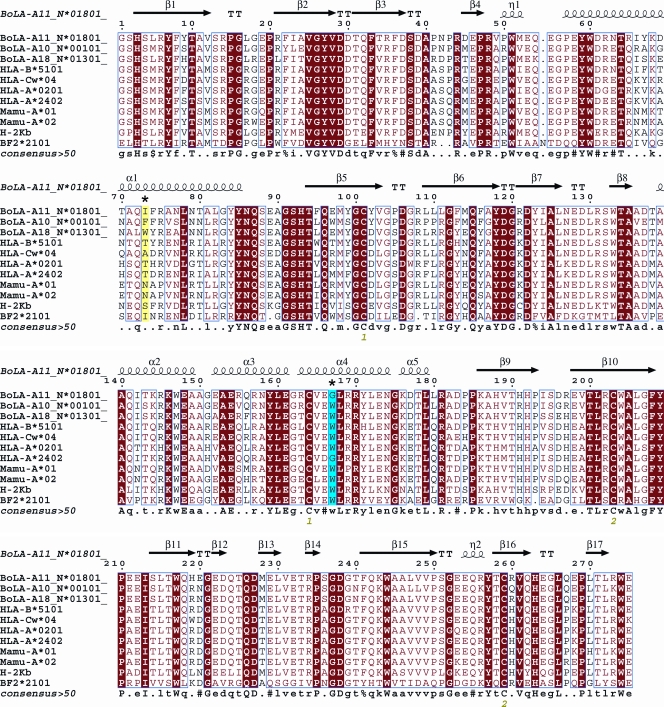
Fig. 3.
Structure-based sequence alignment of N*01801 and eight different types of MHC I molecules. Cylinders indicate α-helices, and black arrows indicate β-strands. Residues highlighted in red are completely conserved, and boxed residues are highly (>80%) conserved. Residues that play a critical role in peptide presentation are asterisked (residue 73, highlighted in yellow, and residue 167, highlighted in cyan). The sequence alignment was generated with Clustal X (56) and ESPript (24).
Due to the high cost of cattle for experimental use, it is necessary to investigate the possibility of utilizing mice as a model (e.g., MHC heavy-chain-transgenic mice) with which to study bovine-specific cellular immunity. To elucidate whether the peptide-presenting features of a bovine MHC I molecule can be influenced by binding to murine β2m, we solved the structure of the N*01801 heavy chain complexed with murine β2m and peptide IPA (referred to as N*01801-Mβ below to distinguish it from the above-mentioned bovine β2m complex) at a resolution of 1.9 Å. There is only one molecule in an asymmetric unit. The α1 and α2 domains, which form the peptide binding groove of N*01801-Mβ, are quite similar to the corresponding portions of N*01801, with RMSDs of 0.337 and 0.345 Å to molecule 1 and molecule 2, respectively (Fig. 4). This may indicate that the structure of the peptide loaded in the groove formed by the α1 and α2 domains of N*01801 was not affected by the substitution of the β2m subunit. In the consideration of a higher resolution of N*01801-Mβ (with murine β2m), we use this structure together with the structure of N*01801 (with bovine β2m) to analyze peptide presentation by bovine MHC I.
Fig. 4.
Similar binding of murine β2m and bovine β2m to the α1α2 domains of the N*01801 heavy chain. (A and B) N*01801-Mβ (light blue), which was formed by the N*01801 heavy chain and murine β2m, is superposed on molecule 1 (green) (A) and molecule 2 (brown) (B) of the N*01801 complex, which was renatured with the N*01801 heavy chain and bovine β2m, respectively. The red circles indicate the similar conformations of residues in the interface of the murine and bovine β2m during binding to the α1α2 domains of the bovine MHC I N*01801 heavy chain. (C) Similar conformations of key residues of β2m in the binding to the α1α2 domains of N*01801. Residues 31 to 34 and 52 to 60 from murine β2m are presented as blue strands and loops, and the corresponding residues 31 to 34 and 51 to 59 from bovine β2m are shown as green (molecule 1) and yellow (molecule 2) strands and loops. The β-strands within the α1α2 domains of N*01801 (gray ribbons) remain in the same conformation, which does not affect the binding of the peptides (gray spheres). A transparent surface of the α1α2 domains of the N*01801 heavy chain in the structure of N*01801-Mβ is shown in light purple.
Two distinct conformations of peptide IPA presented by bovine MHC I N*01801.
In the BoLA binding grooves of molecule 1 and molecule 2, the electron densities around the IPA peptides are well defined and clearly reveal two distinct conformations of the peptide. For simplicity, the peptides presented by molecule 1 and molecule 2 are referred to as IPA-M1 and IPA-M2 below; they are shown in Fig. 5A and B, respectively. IPA-M1 and IPA-M2 have an RMSD of 1.284 Å. However, the conformation of IPA-M2 is quite similar to that of the IPA peptide in the N*01801-Mβ structure, with an RMSD of 0.474. Furthermore, the interactions, including the hydrogen bonds and the nonpolar contacts, of IPA-M2 in the structure of N*01801 are also similar to those of the IPA peptide in the N*01801-Mβ structure. The IPA peptide presented by N*01801-Mβ is referred to as IPA-Mβ and is shown in Fig. 5C. The B factors of the peptides in molecule 1 and N*01801-Mβ are 39.1 and 25.2, respectively, indicating the structural accuracy of these two distinct conformations. The structure of N*01801-Mβ has a higher resolution (1.9 Å) than that of N*01801. Thus, we describe only the differences between the peptide conformations of molecule 1 and N*01801-Mβ in detail here.
Fig. 5.
Two distinct conformations of peptides presented by N*01801. (A to C) Electron density at the 1σ contour level clearly shows the peptide conformations of the bovine MHC-specific epitope within molecule 1 (yellow) (A), molecule 2 (cyan) (B), and N*01801-Mβ (purple) (C). (D) Peptide alignment according to the superposition of the α1α2 domains of molecule 1, molecule 2, and Mβ represents two different conformations of IPA. One is the “double-M” conformation of peptide IPA-M1 in molecule 1 (yellow), with Gly5 and Leu7 located deep in the groove and the side chain of Val6 protruding out of the groove. The other is that of molecule 2 (IPA-M2) (cyan) and Mβ (IPA-Mβ) (purple), which have similar M-shaped conformations, with Val6 as a secondary anchor residue in the middle of the peptide and the side chain of Leu7 solvent exposed. (E) This phenomenon is associated mainly with the conformational change of Ile73 in the α1 domain of N*01801. In molecule 1, Ile73 (yellow) points upward, creating space for the middle of the peptide (yellow) of molecule 1 to bulge out of the groove. In contrast, Ile73 of molecule 2 (cyan) or Mβ (purple) protrudes toward the groove and suppresses the middle of the peptide of molecule 2 (cyan) or Mβ (purple) down toward the groove.
The residues at both ends of the peptides (Ile1, Pro2, and Ala3 at the N terminus and Thr8 and Ile9 at the C terminus) remain the same. In contrast, the major differences between the two peptides occur in the central region (Tyr4, Gly5, Val6, and Leu7) of the peptides (Fig. 5D). Within the groove of molecule 1, the main chain of IPA-M1 turns sharply into the floor of the groove at the P5 (Gly5) position and then rises at P6 (Val6). In position P7, the Cα of Leu7 again falls into the binding groove, while Thr8 in position P8 lifts upward. The tortuous main chain of the peptide presents a “double-M” conformation, which we believe to be unique among the MHC I structures solved thus far. Gly5 and Leu7 of IPA-M1 are located deep in the groove of molecule 1 as two secondary anchor residues. Conversely, the side chain of Val6 is solvent exposed and may function in TCR docking. However, IPA-M1 and IPA-Mβ display a totally different conformation in this region. The main chain of the peptide falls only once in the middle, with the side chain of Val6 protruding into the groove of the α1α2 “bed” as a secondary anchor in the M-shaped peptide. The M-shaped conformation of IPA-Mβ is more common among the structures of mammalian MHC I molecules (12, 35).
An unusually flexible Ile73 in the α1-helix.
Superposition of all of the residues making up the peptide binding groove of molecule 1 and N*01801-Mβ shows that only Ile73 in the α1-helix of the heavy chain differs (Fig. 5E). In molecule 1, the side chain of Ile73 points up into the solvent and leaves enough space for the raised conformation of the P6 valine in IPA-M1. Inversely, the side chain of Ile73 in N*01801-Mβ protrudes toward the α2-helix and occupies the space above the middle region of IPA-Mβ. In this sense, the distinct conformations of Ile73 may contribute to the two different presentation conformations of the IPA peptide. Indeed, residues at position 73 that locate in the flanking side of the peptide binding groove are a key factor for the peptide conformation in other MHC I molecules (11, 34). For different MHC molecules, residues have diverse polymorphism in position 73 (Fig. 3). Therefore, the residues in this position have different impacts on peptide presentation. In a recently solved structure of the bovine MHC I N*01301 molecule, peptide Tp1214–224 displays an unusual bulged C-terminal conformation, which is affected by Trp73 beneath the peptide (34). Another factor may also influence the flexibility of the IPA peptide. Both of the bovine MHCs possess E97 in the bottom of the peptide binding groove. While the secondary anchor residue of K5 of Tp1214–224 forms a stable salt bridge with E97 (34), IPA does not interact with E97 due to the hydrophobicity of the middle residues of the peptide (Gly5, Val6, and Leu7). Thus, the lack of a salt bridge may partially contribute to the flexibility of IPA.
Different exposed areas and buried residues formed by the two IPA conformations.
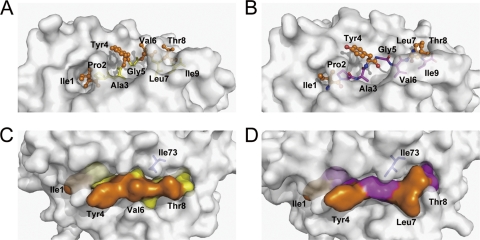
The two distinct conformations of IPA in the two molecules of the asymmetric unit of the bovine MHC I N*01801 structures may contribute to the exposure of different surfaces of the peptides to the solvent (Fig. 6). With the “double-M” conformation, IPA-M1 protrudes from the side chains of all of the even-numbered residues (Tyr4, Val6, and Thr8) of the C-terminal portion of the peptide (P3 to P9), which conspicuously presents their side chains for potential TCR docking (Fig. 6A). The side chain of Ile73 protrudes into the solvent, leaving enough space for the exposure of the middle region of IPA-M1 (Fig. 6C). Calculation of the accessible areas shows that the exposed surface of the three residues occupies most (87.7%) of the total exposed area of IPA-M1. The odd-numbered residues in this portion of the peptide (Ala3, Gly5, Leu7, and Ile9) bury themselves deeply into the groove. In the structure of N*01801-Mβ, P4 Tyr, P7 Leu, and P8 Thr are the most prominent residues and are likely important for TCR interaction (Fig. 6B). The exposed surface of these three residues (Tyr4, Leu7, and Thr8) accounts for 81.0% of the total exposed surface of IPA-Mβ (Fig. 6D). In addition, the main chains of P5 Gly and P6 Val also partially contribute to the exposed area of IPA-Mβ. However, the side chain of Ile73 extends over the middle region of IPA-Mβ, which may affect the direct docking of the TCR to the main chain of P5 Gly and P6 Val. These distinct characteristics of the exposed area of the peptide presented by N*01801 may be involved in recognition by a greater range of the TCR repertoire.
Fig. 6.
Exposed area of peptide IPA presented by bovine MHC I N*01801. (A) Peptide IPA-M1 presentation in the groove of N*01801 molecule 1. The residues exposed to the solvent are highlighted by an orange ball-and-stick representation (Ile1, Tyr4, Val6, and Thr8). The other residues, shown as yellow sticks, are located deep in the groove and act as anchors. (B) Molecule 2 (IPA-M2) and N*01801-Mβ (IPA-Mβ) present the IPA peptide (N*01801-Mβ shown here) in a different way than molecule 1. Leu7 rather than Val6 (as in molecule 1) protrudes out of the groove. Here, Val6 acts as a secondary anchor residue. (C) The exposed area of IPA-M1 in molecule 1 is largely covered by residues Tyr4, Val6, and Thr8. Residue Ile73 (blue) of the N*01801 heavy chain points toward the solvent, leaving space in the middle of the peptide exposed. (D) For molecule 2 and N*01801-Mβ (N*01801-Mβ shown here), the IPA peptide exposes Tyr4, Leu7, and Thr8 for potential TCR docking. Residue Ile73 (blue) hangs over the main chain of the peptide and pushes Val6 into the groove.
Peptide presentation characteristics of bovine N*01801 compared to those of other MHC I molecules.
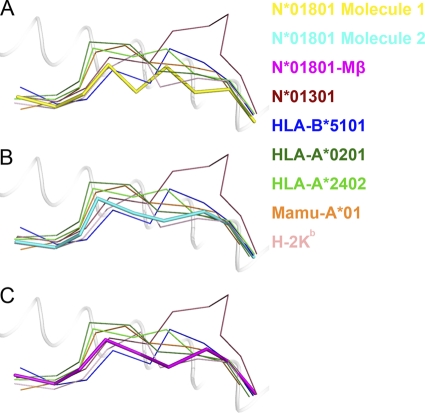
Superposition of the peptide presented by bovine MHC I N*01801 with N*01301 and MHC I molecules from other species revealed key characteristics of N*01801-restricted peptide presentation. Figure 7A shows the Cα backbone of IPA-M1 with other MHC I-presented nonameric peptides (excluding the 13-mer peptide presented by N*01301). Compared to the nonamer presented by HLA-B*5101, the main chain of the N-terminal portion (P1 to P4) of the peptide IPA has a similar conformation. In P5 of the nonamer, valine acts as a secondary anchor residue with its side chain inserted deep into the groove of HLA-B*5101. In N*01801, P5 is a glycine and thus lacks a side chain. However, the highly buried area of Gly5 indicates that this residue also acts as a secondary anchor for the peptide N*01801. Together with Leu7, whose side chain protrudes into the E pocket of the groove, we can conclude that peptide IPA contains two secondary anchor residues including Gly5 in the C-terminal portion of the IPA-M1 peptide (P4 to Pc). This “double-M” conformation is seldom observed among the nonameric peptides presented by MHC I molecules. The IPA-M2 (Fig. 7B) and IPA-Mβ (Fig. 7C) peptides have similar “M-shaped” conformations, which are common among peptides presented by other MHC molecules. The peptides within other MHC grooves possess either the M-shaped conformation with one secondary anchor residue (HLA-B*5101, HLA-A*0201, Mamu-A*01, and H-2Kb) or a bulged conformation without any secondary anchor in the P5-to-Pc region (for HLA-A*2402 and N*01301).
Fig. 7.
Peptide presentation characteristics of bovine MHC I N*01801 compared to those of other MHC molecules. Shown are IPA conformations (thick sticks) presented by N*01801 molecule 1 (yellow) (A) and molecule 2 (cyan) (B) and by N*01801-Mβ (purple) (C). All of the other peptides, which are depicted as thin sticks, are nonameric peptides presented by other mammalian MHC alleles, except for the 11-mer peptide with bovine N*01301 restriction. The structures used here are bovine N*01301 (PDB code 2XFX), HLA-B*5101 (1E27), HLA-A*0201 (3I6G), HLA-A*2402 (3I6L), Mamu-A*01 (1ZLN), and H-2Kb (2VAB).
Another characteristic resulting from the “double-M” conformation of peptide IPA-M1 is that the main chain of the peptide is deeply located in the groove of N*01801. The Cα backbone of the entire chain of the peptide lies deeper in the peptide binding groove than that in other MHC I molecules. Scrutinizing the available MHC I structures, we found that the main chain of the nonameric peptide presented by H-2Kb is also buried deep in the groove. However, as shown in Fig. 7A, the Cα atoms of the most exposed residues (Tyr4 and Val6) of IPA-M1 are still below the Cα of P5 of the nonamer presented by H-2Kb. In the other conformation of peptide IPA (IPA-M1 and IPA-Mβ), the entire peptide is also located deep in the groove (Fig. 7B and C). Moreover, the highest Cα atom in the backbone of the peptide is still lower than the Cα atom of P5 of the nonamer presented by H-2Kb. The buried main chain and exposed side chains of the middle residues of IPA may dictate the specific TCR recognition of the peptide presented by N*01801.
Several factors may contribute to the low positioning of IPA in both of the conformations. First, the shorter residue Ile73 may allow the portion near the C terminus of the peptide to fall into the binding groove. In contrast, Trp73 of BoLA-A18, which forms a bulged ridge in the groove, pushes the peptide up into the solvent (34). Second, the large E pocket of N*01801 may allow the residues in the middle of the peptide to be inserted deeply into the groove (Val6 for IPA-M1; Leu7 for IPA-M2 and IPA-Mβ).
Pockets in the N*01801 peptide binding groove.
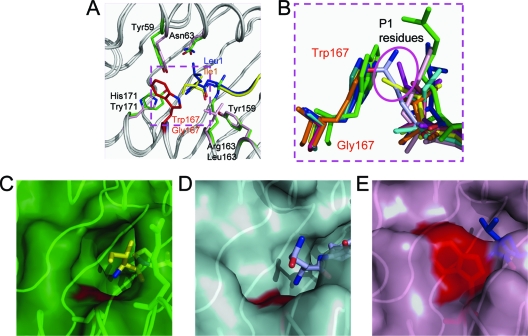
Residues Tyr59, Asn63, Tyr159, Arg163, Gly167, and Tyr171 form the A pocket of the peptide binding groove of N*01801 (Fig. 8A). The P1 Ile of the peptide is located in the A pocket and points its side chain out of the groove. Considering MHC I molecules from different species (Fig. 3), Gly167 of N*01801 is rare among all of the MHC I molecules (i.e., it is found only in certain alleles, such as HLA-A*2402). In the N*01801 structure, the A pocket has the small residue Gly in position 167, making the space in the A pocket larger (Fig. 8B and C), which is also seen in the structure of HLA-A*2402 (Fig. 8D) (32). The side chain of Ile1 of the IPA-M1 peptide in molecule 1 points toward the N terminus (or toward the right side if observed from the α2 domain of the MHC). The different conformation of the side chain of Ile1 in molecule 2 compared to molecule 1 reveals the flexibility of Ile1 in the N*01801 structure (Fig. 5A, B, and D), which reflects the large space of the A pocket. The most common residue at position 167 in other MHC alleles (in human, monkey, mouse, chicken, and bovine BoLA-A18) is Trp (Fig. 3), which forms the right side of the A pocket (observed from the α2 domain of the MHC). Trp167 of these MHC I alleles reduces the size of the A pocket and acts as a wall to prevent the side chain of the P1 residue of the peptide from pointing toward the N terminus (Fig. 8B and E). Further, this characteristic of the N*01801 A pocket may influence the recognition of the P1 residue by TCRs (61).
Fig. 8.
Unexpectedly large A pocket of bovine MHC I N*01801. (A) Residue comparison of the A pockets of N*01801 (green) and HLA-B*5101 (light purple). When different residues are in the same position, the lower residue is from N*01801 and the upper residue is from HLA-B*5101. The most important residue influencing the space of the A pocket is highlighted in red (Gly167 for N*01801 and Trp167 for HLA-B*5101). The P1 residue (Ile1) (yellow) of N*01801 is closer to the N terminus of the peptide than the P1 residue (Leu1) (blue) of HLA-B*5101. (B) Superposition of the residues in position 167 of different MHC I alleles and their peptides. Trp167 acts like a wall preventing P1 residues from pointing toward the N termini of the peptides. All of the side chains of the P1 residues protrude upward. Residue 167 is glycine in N*01801 and the human MHC allele HLA-A*2402. The peptides of N*01801 molecule 1 (yellow), N*01801-Mβ (purple), and HLA-A*2402 (light blue) point toward the N termini (toward the right side observed from the α2 domain of the MHC) of the peptides (highlighted in a purple circle). (C) The A pocket of N*01801 molecule 1 in green surface representation. Gly167 (red surface) leaves a large space for the P1 residue of the peptide (yellow). Arg163 hangs over the main chain of the peptide. (D) The A pocket of HLA-A*2402 (peptide in deep purple) in cyan surface representation. The amino acid in position 167 is also a glycine (red surface), as in N*01801. (E) The A pocket of HLA-B*5101 is shown in purple. The surfaces of Trp167, shown in red, block the N terminus of the peptide groove, and thus, the peptide (blue) points upward. The structures used here are bovine N*01301 (PDB code 2XFX), HLA-B*5101 (1E27), HLA-A*0201 (3I6G), HLA-A*2402 (3I6L), Mamu-A*01 (1ZLN), H-2Kb (2VAB), and BF2*2101 (2BEW).
The structure of N*01801 (Fig. 6) reveals Pro2 as a primary anchor residue fitted into a small B pocket surrounded by Ile66, Tyr67, and Tyr8. These residues are similar to the corresponding residues (Ile66, Phe67, and Tyr8) of HLA-B*5101 (Fig. 3), which also accommodates Pro (or other small residues, such as Gly or Ala) as the P2 anchor. Structural comparison of these residues demonstrated very similar conformations, with an RMSD of 0.099 Å. This may lead to the common preference of N*01801 and HLA-B*5101 for Pro as the P2 anchor. In the F pocket of N*01801, the peptide Ile9 is surrounded by hydrophobic amino acids: Ala81, Tyr84, Phe95, Phe116, and Try147 (Fig. 6; see also Fig. S1 in the supplemental material). However, the space in the F pocket is larger than the side chain of Ile9, which indicates that the F pocket may be able to accommodate larger residues. Aside from small aliphatic residues (e.g., Val, Ile, and Leu), in previous studies on the peptide motif of N*01801, aromatic residues (e.g., Tyr) were also defined as the C-terminal residues of N*01801-restricted peptides. The structure of N*01801 complexed with IPA clearly confirmed the peptide motifs of N*01801 that were defined by pool sequencing of eluted peptides in previous reports (14, 27, 51).
DISCUSSION
To determine the structural characteristics of peptide binding and presentation by bovine MHC I N*01801 (BoLA-A11), we crystallized N*01801 in complex with a CTL epitope from rinderpest virus. Additionally, we also determined the structure of N*01801 complexed with the same peptide but with murine β2m instead of bovine β2m. Interspecies β2m replacement of MHC complexes has been widely exploited in structural studies of MHC molecules. The β2m does not directly interact with the peptide and has no observable effect on peptide binding and presentation (1, 10, 43, 50). Here, the peptide binding grooves formed by the N*01801 α1α2 domains were similar in the two structures determined (Fig. 4). The interspecies structure may also veritably reflect the peptide binding of N*01801 (at a higher resolution) in addition to the structure of N*01801 complexed with bovine β2m.
As revealed by peptide elution from an N*01801-transfected cell line, nonameric peptides account for most of the naturally separated peptides (28). Proline is found almost exclusively at the P2 position of these peptides, and as shown in the N*01801 structure, the relatively small, bowl-shaped, hydrophobic B pocket perfectly accommodates a proline at this position. The B pocket of N*01801 is formed by residues similar to those in the B pocket of HLA-B*5101, which may explain the same preference for proline as the prevalent anchor residue in the P2 position. In the C-terminal position of the IPA peptide, isoleucine and valine act as the primary anchor residues, inserting into the hydrophobic F pocket of N*01801, as indicated in the structure of N*01801 and the refolding assays. The space within the F pocket appears larger than the side chain of isoleucine, which may indicate that larger hydrophobic and even aromatic side chains can be accommodated. This is coincident with the peptide motif studies of N*01801, which indicated that Ile/Val and a small proportion of Leu/Tyr occupied the C termini of the naturally processed peptides of N*01801 (28).
Studies of CD8+ T-cell epitopes indicate that different peptides elicit diverse T-cell responses, corresponding to distinct T-cell repertoires. Recent studies have also demonstrated that even minor modification of an epitope can lead to a profound effect on the antigenicity of the peptide (37, 58). Peptides with a relatively rigid conformation may result in limited types of TCR docking, leading to a specific T-cell repertoire with limited diversity. Recently, this has been thoroughly investigated by Macdonald and colleagues in structural and function studies of bovine MHC I N*01301 (34). Hence, a peptide with a flexible structure presented in the binding groove of an MHC molecule may lead to a diverse TCR profile. In a number of reported cases, a broader T-cell repertoire for the pathogen-specific epitope may facilitate a more effective host immune response against the invading pathogen and may prevent the emergence of immune escape mutants (39, 44, 52). The IPA peptide in our structure unexpectedly appears to be presented by N*01801 in two distinct conformations. The completely different exposed areas in the middle of the peptide may indicate different manners of TCR docking. Further study should focus on whether the two distinct conformations of the IPA peptide correspond to different T-cell repertoires. The contribution of the uncommon peptide presentation strategy of N*01801 to the CTL-specific responses of N*01801+ cattle to pathogens also needs further exploration.
Generally, a featured peptide contains exposed residues protruding out of the landscape of the MHC complexed to peptide, and this can be implemented via two different strategies. First, epitopes with characteristic long side chain residues, which are solvent exposed, may act as featured peptides to elicit diverse T-cell repertoires (58). Second, moderately bulged peptides help the short side chains rise to a suitable level for TCR docking (32). In the structure of bovine MHC I N*01801 complexed with IPA, N*01801 presents the same peptide in two distinct conformations, which is associated with the flexibility of Ile73 in the peptide binding groove. Different exposed residues and secondary anchor residues result from these conformational changes. This adjustable strategy of host antigen presentation may represent a novel type of featured peptide and may expand our understanding of the crucial role of T-cell epitopes in antipathogen immunity.
BoLA-A11 is one of the most common global haplotypes in dairy cattle. Studies of T-cell-specific responses against pathogens have identified a large number of CD8+ T-cell epitopes with the BoLA-A11 restriction. Here, we demonstrated the distinctive characteristics of peptide presentation by BoLA-A11 through the structural determination of bovine MHC I N*01801 (one of the most common alleles of BoLA-A11 haplotypes) complexed with a rinderpest virus-derived IPA peptide. Our study may lead to further definition of featured T-cell epitopes in a structural manner and, moreover, may pave the way for rational CTL-based design of vaccines for cattle or other species.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Ministry of Science and Technology of China (Transgenic special grant 2009ZX08009-150B; Project 973 grant 2007CB815805) and a Special Grant for Protein Science of the National 973 Project (grant 2010CB911902). G.F.G. is a leading principal investigator of the National Natural Science Foundation of China (NSFC) Innovative Research Group (grant 81021003). The funders had no role in study the design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Joel Haywood, Yi Shi, and Hao Cheng of the Institute of Microbiology, Chinese Academy of Sciences, for their excellent suggestions on this study.
The authors declare no financial or commercial conflict of interest.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 30 March 2011.
REFERENCES
- 1. Achour A., et al. 2006. Structural basis of the differential stability and receptor specificity of H-2Db in complex with murine versus human β2-microglobulin. J. Mol. Biol. 356:382–396 [DOI] [PubMed] [Google Scholar]
- 2. Adams P. D., et al. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948–1954 [DOI] [PubMed] [Google Scholar]
- 3. Ahmed N., Gottschalk S. 2009. How to design effective vaccines: lessons from an old success story. Expert Rev. Vaccines 8:543–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amorena B., Stone W. H. 1978. Serologically defined (SD) locus in cattle. Science 201:159–160 [DOI] [PubMed] [Google Scholar]
- 5. Babiuk S., et al. 2007. BoLA class I allele diversity and polymorphism in a herd of cattle. Immunogenetics 59:167–176 [DOI] [PubMed] [Google Scholar]
- 6. Bensaid A., et al. 1991. Identification of expressed bovine class I MHC genes at two loci and demonstration of physical linkage. Immunogenetics 33:247–254 [DOI] [PubMed] [Google Scholar]
- 7. Birtley J. R., et al. 2005. Crystal structure of foot-and-mouth disease virus 3C protease. New insights into catalytic mechanism and cleavage specificity. J. Biol. Chem. 280:11520–11527 [DOI] [PubMed] [Google Scholar]
- 8. Chen W., et al. 2010. Crystal structure of a bony fish β2-microglobulin: insights into the evolutionary origin of immunoglobulin superfamily constant molecules. J. Biol. Chem. 285:22505–22512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen Y., Shi Y., Cheng H., An Y. Q., Gao G. F. 2009. Structural immunology and crystallography help immunologists see the immune system in action: how T and NK cells touch their ligands. IUBMB Life 61:579–590 [DOI] [PubMed] [Google Scholar]
- 10. Chu F., et al. 2007. First glimpse of the peptide presentation by rhesus macaque MHC class I: crystal structures of Mamu-A*01 complexed with two immunogenic SIV epitopes and insights into CTL escape. J. Immunol. 178:944–952 [DOI] [PubMed] [Google Scholar]
- 11. Ciatto C., et al. 2001. Zooming in on the hydrophobic ridge of H-2Db: implications for the conformational variability of bound peptides. J. Mol. Biol. 312:1059–1071 [DOI] [PubMed] [Google Scholar]
- 12. Cole D. K., et al. 2006. Crystal structure of HLA-A*2402 complexed with a telomerase peptide. Eur. J. Immunol. 36:170–179 [DOI] [PubMed] [Google Scholar]
- 13. Davies C. J., et al. 1994. Polymorphism of bovine MHC class I genes. Joint report of the Fifth International Bovine Lymphocyte Antigen (BoLA) Workshop, Interlaken, Switzerland, 1 August 1992. Eur. J. Immunogenet. 21:239–258 [DOI] [PubMed] [Google Scholar]
- 14. De Groot A. S., et al. 2003. T cell epitope identification for bovine vaccines: an epitope mapping method for BoLA A-11. Int. J. Parasitol. 33:641–653 [DOI] [PubMed] [Google Scholar]
- 15. Ellis S. 2004. The cattle major histocompatibility complex: is it unique? Vet. Immunol. Immunopathol. 102:1–8 [DOI] [PubMed] [Google Scholar]
- 16. Ellis S. A., Ballingall K. T. 1999. Cattle MHC: evolution in action? Immunol. Rev. 167:159–168 [DOI] [PubMed] [Google Scholar]
- 17. Ellis S. A., Staines K. A., Morrison W. I. 1996. cDNA sequence of cattle MHC class I genes transcribed in serologically defined haplotypes A18 and A31. Immunogenetics 43:156–159 [DOI] [PubMed] [Google Scholar]
- 18. Ellis S. A., Staines K. A., Stear M. J., Hensen E. J., Morrison W. I. 1998. DNA typing for BoLA class I using sequence-specific primers (PCR-SSP). Eur. J. Immunogenet. 25:365–370 [DOI] [PubMed] [Google Scholar]
- 19. Emsley P., Lohkamp B., Scott W. G., Cowtan K. 2004. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ennis P. D., Jackson A. P., Parham P. 1988. Molecular cloning of bovine class I MHC cDNA. J. Immunol. 141:642–651 [PubMed] [Google Scholar]
- 21. Falk K., et al. 1995. Peptide motifs of HLA-B51, -B52 and -B78 molecules, and implications for Behcet's disease. Int. Immunol. 7:223–228 [DOI] [PubMed] [Google Scholar]
- 22. Fries R., Hediger R., Stranzinger G. 1986. Tentative chromosomal localization of the bovine major histocompatibility complex by in situ hybridization. Anim. Genet. 17:287–294 [DOI] [PubMed] [Google Scholar]
- 23. Gilbert N. 2009. Cattle disease faces total wipeout. Nature 462:709. [DOI] [PubMed] [Google Scholar]
- 24. Gouet P., Robert X., Courcelle E. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31:3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gras S., Kjer-Nielsen L., Burrows S. R., McCluskey J., Rossjohn J. 2008. T-cell receptor bias and immunity. Curr. Opin. Immunol. 20:119–125 [DOI] [PubMed] [Google Scholar]
- 26. Guzman E., Taylor G., Charleston B., Skinner M. A., Ellis S. A. 2008. An MHC-restricted CD8+ T-cell response is induced in cattle by foot-and-mouth disease virus (FMDV) infection and also following vaccination with inactivated FMDV. J. Gen. Virol. 89:667–675 [DOI] [PubMed] [Google Scholar]
- 27. Hegde N. R., Deshpande M. S., Godson D. L., Babiuk L. A., Srikumaran S. 1999. Bovine lymphocyte antigen-A11-specific peptide motif as a means to identify cytotoxic T-lymphocyte epitopes of bovine herpesvirus 1. Viral Immunol. 12:149–161 [DOI] [PubMed] [Google Scholar]
- 28. Hegde N. R., et al. 1995. Peptide motif of the cattle MHC class I antigen BoLA-A11. Immunogenetics 42:302–303 [DOI] [PubMed] [Google Scholar]
- 29. Holmes E. C., Roberts A. F., Staines K. A., Ellis S. A. 2003. Evolution of major histocompatibility complex class I genes in Cetartiodactyls. Immunogenetics 55:193–202 [DOI] [PubMed] [Google Scholar]
- 30. Laskowski R. A., Moss D. S., Thornton J. M. 1993. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231:1049–1067 [DOI] [PubMed] [Google Scholar]
- 31. Liu J., et al. 2010. The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes. J. Infect. Dis. 202:1171–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu J., et al. 2010. Novel immunodominant peptide presentation strategy: a featured HLA-A*2402-restricted cytotoxic T-lymphocyte epitope stabilized by intrachain hydrogen bonds from severe acute respiratory syndrome coronavirus nucleocapsid protein. J. Virol. 84:11849–11857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu J., Zhang S., Tan S., Zhen B., Gao G. F. 2011. Revival of the identification of CTL epitopes for immunological diagnosis, therapy and vaccine development. Exp. Biol. Med. doi:10.1258/ebm.2010.010278 [DOI] [PubMed] [Google Scholar]
- 34. Macdonald I. K., et al. 2010. MHC class I bound to an immunodominant Theileria parva epitope demonstrates unconventional presentation to T cell receptors. PLoS Pathog. 6:e1001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maenaka K., et al. 2000. Nonstandard peptide binding revealed by crystal structures of HLA-B*5101 complexed with HIV immunodominant epitopes. J. Immunol. 165:3260–3267 [DOI] [PubMed] [Google Scholar]
- 36. Markel H. 2005. The search for effective HIV vaccines. N. Engl. J. Med. 353:753–757 [DOI] [PubMed] [Google Scholar]
- 37. Maryanski J. L., et al. 1997. The diversity of antigen-specific TCR repertoires reflects the relative complexity of epitopes recognized. Hum. Immunol. 54:117–128 [DOI] [PubMed] [Google Scholar]
- 38. Meijers R., et al. 2005. Crystal structures of murine MHC Class I H-2Db and Kb molecules in complex with CTL epitopes from influenza A virus: implications for TCR repertoire selection and immunodominance. J. Mol. Biol. 345:1099–1110 [DOI] [PubMed] [Google Scholar]
- 39. Messaoudi I., Guevara Patino J. A., Dyall R., LeMaoult J., Nikolich-Zugich J. 2002. Direct link between MHC polymorphism, T cell avidity, and diversity in immune defense. Science 298:1797–1800 [DOI] [PubMed] [Google Scholar]
- 40. Normile D. 2008. Rinderpest. Driven to extinction. Science 319:1606–1609 [DOI] [PubMed] [Google Scholar]
- 41. Otwinowski Z., Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 42. Parkin S., Hope H. 1998. Macromolecular cryocrystallography: cooling, mounting, storage and transportation of crystals. J. Appl. Crystallogr. 31:945–953 [Google Scholar]
- 43. Pedersen L. O., et al. 1995. The interaction of β2-microglobulin (β2m) with mouse class I major histocompatibility antigens and its ability to support peptide binding. A comparison of human and mouse β2m. Eur. J. Immunol. 25:1609–1616 [DOI] [PubMed] [Google Scholar]
- 44. Price D. A., et al. 2004. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity 21:793–803 [DOI] [PubMed] [Google Scholar]
- 45. Rammensee H. G., Friede T., Stevanoviic S. 1995. MHC ligands and peptide motifs: first listing. Immunogenetics 41:178–228 [DOI] [PubMed] [Google Scholar]
- 46. Rodriguez L. L., Grubman M. J. 2009. Foot and mouth disease virus vaccines. Vaccine 27(Suppl. 4):D90–D94 [DOI] [PubMed] [Google Scholar]
- 47. Russell G. C., Oliver R. A., Sawhney S. M. 1996. Cloning, transfection, and DNA sequence of a second gene from the BoLA-A11 haplotype. Immunogenetics 44:315–318 [DOI] [PubMed] [Google Scholar]
- 48. Sawhney S. M., et al. 1995. Transfection, expression, and DNA sequence of a gene encoding a BoLA-A11 antigen. Immunogenetics 41:246–250 [DOI] [PubMed] [Google Scholar]
- 49. Shastri N., Schwab S., Serwold T. 2002. Producing nature's gene-chips: the generation of peptides for display by MHC class I molecules. Annu. Rev. Immunol. 20:463–493 [DOI] [PubMed] [Google Scholar]
- 50. Shields M. J., Moffat L. E., Ribaudo R. K. 1998. Functional comparison of bovine, murine, and human β2-microglobulin: interactions with murine MHC I molecules. Mol. Immunol. 35:919–928 [DOI] [PubMed] [Google Scholar]
- 51. Sinnathamby G., Seth S., Nayak R., Shaila M. S. 2004. Cytotoxic T cell epitope in cattle from the attachment glycoproteins of rinderpest and peste des petits ruminants viruses. Viral Immunol. 17:401–410 [DOI] [PubMed] [Google Scholar]
- 52. Slifka M. K., Whitton J. L. 2001. Functional avidity maturation of CD8+ T cells without selection of higher affinity TCR. Nat. Immunol. 2:711–717 [DOI] [PubMed] [Google Scholar]
- 53. Sliz P., et al. 2001. Crystal structures of two closely related but antigenically distinct HLA-A2/melanocyte-melanoma tumor-antigen peptide complexes. J. Immunol. 167:3276–3284 [DOI] [PubMed] [Google Scholar]
- 54. Subbarao K., Murphy B. R., Fauci A. S. 2006. Development of effective vaccines against pandemic influenza. Immunity 24:5–9 [DOI] [PubMed] [Google Scholar]
- 55. Sun Y., et al. 2010. Identification and structural definition of H5-specific CTL epitopes restricted by HLA-A*0201 derived from the H5N1 subtype of influenza A viruses. J. Gen. Virol. 91:919–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Turner S. J., Doherty P. C., McCluskey J., Rossjohn J. 2006. Structural determinants of T-cell receptor bias in immunity. Nat. Rev. Immunol. 6:883–894 [DOI] [PubMed] [Google Scholar]
- 58. Turner S. J., et al. 2005. Lack of prominent peptide-major histocompatibility complex features limits repertoire diversity in virus-specific CD8+ T cell populations. Nat. Immunol. 6:382–389 [DOI] [PubMed] [Google Scholar]
- 59. Tynan F. E., et al. 2005. High resolution structures of highly bulged viral epitopes bound to major histocompatibility complex class I. Implications for T-cell receptor engagement and T-cell immunodominance. J. Biol. Chem. 280:23900–23909 [DOI] [PubMed] [Google Scholar]
- 60. Tynan F. E., et al. 2005. T cell receptor recognition of a ‘super-bulged’ major histocompatibility complex class I-bound peptide. Nat. Immunol. 6:1114–1122 [DOI] [PubMed] [Google Scholar]
- 61. Tynan F. E., et al. 2007. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat. Immunol. 8:268–276 [DOI] [PubMed] [Google Scholar]
- 62. Vordermeier H. M., et al. 2009. Adjuvants induce distinct immunological phenotypes in a bovine tuberculosis vaccine model. Clin. Vaccine Immunol. 16:1443–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yewdell J. W., Haeryfar S. M. 2005. Understanding presentation of viral antigens to CD8+ T cells in vivo: the key to rational vaccine design. Annu. Rev. Immunol. 23:651–682 [DOI] [PubMed] [Google Scholar]
- 64. Zhou M., et al. 2004. Complex assembly, crystallization and preliminary X-ray crystallographic studies of MHC H-2Kd complexed with an HBV-core nonapeptide. Acta Crystallogr. D Biol. Crystallogr. 60:1473–1475 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.