Abstract
Domestic cats endure infections by all three subfamilies of the retroviridae: lentiviruses (feline immunodeficiency virus [FIV]), gammaretroviruses (feline leukemia virus [FeLV]), and spumaretroviruses (feline foamy virus [FFV]). Thus, cats present an insight into the evolution of the host-retrovirus relationship and the development of intrinsic/innate immune mechanisms. Tetherin (BST-2) is an interferon-inducible transmembrane protein that inhibits the release of enveloped viruses from infected cells. Here, we characterize the feline homologue of tetherin and assess its effects on the replication of FIV. Tetherin was expressed in many feline cell lines, and expression was induced by interferons, including alpha interferon (IFN-α), IFN-ω, and IFN-γ. Like human tetherin, feline tetherin displayed potent inhibition of FIV and HIV-1 particle release; however, this activity resisted antagonism by either HIV-1 Vpu or the FIV Env and “OrfA” proteins. Further, as overexpression of complete FIV genomes in trans could not overcome feline tetherin, these data suggest that FIV lacks a functional tetherin antagonist. However, when expressed stably in feline cell lines, tetherin did not abrogate the replication of FIV; indeed, syncytium formation was significantly enhanced in tetherin-expressing cells infected with cell culture-adapted (CD134-independent) strains of FIV (FIV Fca-F14 and FIV Pco-CoLV). Thus, while tetherin may prevent the release of nascent viral particles, cell-to-cell spread remains efficient in the presence of abundant viral receptors and tetherin upregulation may enhance syncytium formation. Accordingly, tetherin expression in vivo may promote the selective expansion of viral variants capable of more efficient cell-to-cell spread.
INTRODUCTION
Innate resistance to retroviral infection and replication is induced by interferons (IFNs). IFN-inducible factors restrict-ing viral replication include the cytidine deaminaseAPOBEC3G (40, 60) and the E3 ubiquitin ligase TRIM5 (1), both of which target replication primarily during the process of viral entry. A third IFN-inducible activity, tetherin (BST-2/CD317/HM1.24), acts to restrict viral release (13, 35, 36, 41, 62). The importance of these factors in controlling viral replication is underlined by the requirement for lentiviral genomes to encode trans-acting countermeasures; lentiviral Vif proteins (33, 54, 55) and spumaviral Bet proteins (28, 42, 51) counteract APOBECs whereas HIV-1 Vpu, HIV-2 Nef, and HIV-2 and simian immunodeficiency virus (SIV) Envs may counteract tetherins (15, 18, 26, 35, 36, 62, 65).
Tetherin is a type II single-pass transmembrane protein. It is anchored to the cell membrane by both N-terminal transmembrane domain and C-terminal glycophosphatidylinositol (GPI) anchors that are linked by an extracellular coiled-coil domain that promotes dimerization of adjacent tetherin molecules. Accordingly, tetherin in both the cell membrane and the envelope of the budding virus can prevent virion release either by direct cross-linking or by the formation of dimers between adjacent coiled-coil domains (41). The primary role for tetherin remains unclear; however, it is likely that, by trapping enveloped viruses at the cell surface, tetherin prevents the further dissemination of nascent virions. However, given the constitutive high-level expression of tetherin on plasmacytoid dendritic cells (pDC [type I IFN-producing cells]) (5), tetherin may play a more fundamental role in the initiation and perpetuation of a virus-specific immune response (58).
The domestic cat lineage has faced multiple invasions by viruses from the family Retroviridae. In addition to an exogenous gammaretrovirus (feline leukemia virus [FeLV]), a lentivirus (feline immunodeficiency virus [FIV]), and a spumavirus (feline foamy virus [FFV]), cats also harbor the endogenous RD114 gamma retrovirus (47, 48) and full-length endogenous FeLVs (50). While lentiviruses have spread throughout the Felidae, from lions in Africa to pumas in North America and Pallas cats in Mongolia (61), the gamma retroviruses are restricted solely to domestic cats (3, 4, 47, 48), although occasional cross-species transmission events have been recorded in Florida panthers (37) and Iberian lynxes (30). The limited distribution of the gamma retroviruses among felids suggests that they entered the domestic cat population after the divergence of the Felis lineage from the other felids circa 6.2 million years ago (19). The presence of three exogenous members and one endogenous member of the Retroviridae in domestic cats offers an intriguing insight into the retrovirus-host interaction. As cats express a truncated TRIM5 lacking a capsid-binding B30.2/SPRY domain (29), their ability to suppress retroviral replication may be impaired. If tetherin is to have a major role in the control of retroviral replication in any species, the cat would seem a likely example.
In this study we describe the identification and characterization of a feline tetherin (feTHN) homologue and examine its activity against the FIV feline lentivirus. As with the activities of tetherin homologues in other species, feline tetherin prevented the release of FIV in transient assays. However, in assays of viral replication, feline tetherin displayed little suppression of viral growth. Indeed, syncytium formation mediated by strains of FIV capable of interacting directly with CXCR4 (CD134-independent strains) was enhanced in the presence of tetherin. As CD134-independent viruses emerge in vivo in FIV-infected animals, we speculate that an unexpected consequence of the induction of tetherin expression may be enhanced cell-cell spread in CD134-negative cells.
MATERIALS AND METHODS
Identification and cloning of feline tetherin.
The genomic sequence for the prospective feline homologue of tetherin was identified by comparing the sequences of human (Homo sapiens) tetherin (GenBank accession no. NM_004335) and dog (Canis familiaris) tetherin (GenBank accession no. XM_847295 and XM_860510) to those of the 2X domestic cat (Felis catus) genome by the use of megaBLAST (algorithm of Zhang et al. [66]). A candidate feline tetherin gene was identified on Felis catus c430601298.contig 1 (GenBank accession no. ACBE01053987). Oligonucleotide primers were synthesized corresponding to the predicted start (forward primer 5′-ATGGCACCTGCTTTTTTACCAC-3′) and stop (reverse primer 5′-TCAGGCCAGCAGAGCAACGAA-3′) codons of feline tetherin. Mya-1 cells were lysed (QIAshredder; Qiagen Ltd., Crawley, United Kingdom), and total RNA was prepared by guanidine-isothiocyanate lysis using an RNeasy Minikit (Qiagen Ltd., Crawley, United Kingdom) and reverse transcribed using a Transcriptor High Fidelity cDNA synthesis kit and oligo(dT)12 primer (Roche Applied Science, Burgess Hill, United Kingdom). The feline tetherin (feTHN) cDNA was amplified using Phusion DNA polymerase [Finnzymes; New England BioLabs (UK) Ltd., Hitchin, United Kingdom] and its nucleic acid sequence determined using BigDye Terminator v1.1 cycle sequencing (Applied Biosystems, Life Technologies, Paisley, United Kingdom) followed by analysis performed using an Applied Biosystems 3730xl genetic analyzer and chromatogram analysis performed using the Chromas version 1.45 software package (Technelysium Pty. Ltd., Tewantin, Australia). The tetherin cDNA was then reamplified to incorporate SalI and NotI restrictions sites and inserted into VR1012 eukaryotic expression vector (Vical Inc., San Diego, CA), generating feTHN-VR1012.
Quantification of tetherin by real-time PCR.
Cells were seeded in 25-cm2 culture flasks to achieve 80% confluence 24 h postseeding. Cells were then treated with 1,000 IU/ml recombinant feline omega IFN (IFN-ω) (Virbac Limited, Bury St Edmonds, United Kingdom) or feline IFN-α and IFN-γ (R&D Systems Europe Ltd., Abingdon, United Kingdom) for 24 h, harvested, and lysed, and total RNA was prepared as described above. Total RNA (1 μg) was used to synthesize first-strand cDNA (Transcriptor First Strand cDNA synthesis kit; Roche Applied Science). A 1-μl volume (1/20) of the first-strand cDNA synthesis product was used as a template for real-time PCR and combined with either the feline tetherin primer and probe set consisting of primer FcTHN-Fwd (5′-GAGAAGGCCCAGAGCCAGGAG-3′), primer FcTHN-Rev (5′-GCAACGAAGGCCAGGAGCAG-3′), and probe 5′-FAM (6-carboxyfluorescein)-TGCAGAACGCTTCGGTGGAGGTGGAAAGACTGAGAAA-TAMRA (6-carboxytetramethylrhodamine)-3′ or with the feline rRNA set consisting of primer 343-Fwd (5′-CCATTCGAACGTCTGCCCTA-3′), primer 409-Fwd (5′-TCACCCGTGGTCACCATG-3′), and probe 5′-FAM-CGATGGTAGTCGCCGTGCCTA-TAMRA-3′. Probes, primers, and templates were combined in TaqMan Universal Master Mix (Applied Biosystems, a division of Life Technologies, Paisley, United Kingdom) in MicroAmp Optical 96-well reaction plates (Applied Biosystems, Paisley, United Kingdom) and analyzed on an Applied Biosystems 7500 real-time PCR system. ΔCT values (where “CT” represents “threshold cycle”) were calculated by subtracting the mean CT for the abundant rRNA from the CT for the feline tetherin mRNA. For analyses of tetherin mRNA expression in transiently transfected 293T cells, RNA was pretreated with DNase prior to cDNA synthesis and additional control reactions were included in which the cDNA synthesis step prior to real-time PCR was omitted.
Cells.
Human 293T cells, and feline cell lines AH927 and FEA, and CRFK (clone CRFK) fibroblasts were maintained in Dulbecco's modification of Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), 2 mM glutamine, and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin) at 37°C and 5% CO2. The feline MYA-1 T-cell line (32) was maintained in RPMI 1640, supplemented with 10% FBS, 2 mM glutamine, antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin), 10 mM HEPES, 1 mM sodium pyruvate, 0.05 mM 2-mercaptoethanol, and conditioned medium from a murine cell line (L2.3), and transfected with a human IL-2 expression construct (equivalent to 100 U/ml of recombinant human interleukin-2 (IL-2). Stable ectopic expression of CD134 has been described previously (56), and cells expressing CD134 from the pDON-AI construct were maintained in Geneticin/G418 (Invitrogen, Paisley, United Kingdom). All media and supplements were obtained from Invitrogen Ltd. (Paisley, United Kingdom).
Assay for tetherin activity.
293T cells were seeded in 12-well plates at a density of 2 × 105 cells per well and allowed to adhere overnight. Cells were then transfected using either SuperFect (Qiagen Ltd., Crawley, United Kingdom) or Fugene 6 (Roche Applied Science, Mannheim, Germany) according to the manufacturer's recommendations. HIV-1 pseudotypes were prepared using HIV-1-derived 8.91 packaging plasmid (Gag-Pol, Tat, and Rev) or 8.2 packaging plasmid (Gag-Pol, Tat, Rev, Vpr, Vpu, Vif, and Nef), CSGW (green fluorescent protein [GFP]-encoding HIV-1 genome), and MDG vesicular stomatitis virus G glycoprotein (VSV-G)-encoding vector (2, 34, 67). FIV pseudotypes were prepared using FIV-based vectors FP93 (Gag-Pol) and pGiNSin (GFP). FP93 (52) was derived from the 34TF10 molecular clone (59) of FIV Petaluma and thus shares 99.6% nucleic acid sequence identity over this region with the F14 molecular clone (39) of FIV Petaluma from which the FIV-Fca F14 virus stock was prepared. The viral expression vectors were cotransfected with 1 μg of feTHN-VR1012 and, in the case of HIV-1, 1 μg of either HIV-1 Vpu-cDNA3 or empty vector (pcDNA3). In experiments where the level of feline tetherin was varied, feTHN-VR1012 was serially diluted from 600 ng to 50 ng and VR1012 was used to balance the amount of DNA. In the case of HIV-1, the amount of HIV-Vpu was kept constant at 600 ng per well. In competition experiments, constructs bearing FIV OrfA or entire FIV genomes were cotransfected with the tetherin construct during pseudotype production (0.25 μg OrfA and 0.5 μg feTHN). OrfA expression constructs consisted of VR1012-OrfA (prepared by amplifying OrfA from FIV-GL8 and cloning into VR1012) or codon-optimized FIV OrfA, 1S-5RL (44), or CMVG8MΔpol4, a derivative of pCT5b (45) in which a PacI-NdeI fragment was exchanged with the equivalent fragment from the GL8MYA molecular clone. The resulting clone, CMV-G8M, was then modified by deleting a fragment of pol by PacI/SanDI digestion followed by treatment with the Klenow fragment of DNA polymerase I prior to religation. CMVG8MΔpol4 thus produces all FIV-encoded proteins under the control of a cytomegalovirus (CMV) promoter but is not replication competent.
Stable expression of feline tetherin.
For stable expression, FcTHN was inserted into pDON-AI-2 Neo expression vector (Takara Bio Europe, S.A.S. Saint-Germain-en-Laye, France) with NotI and BamHI. CRFK fibroblasts stably expressing feTHN were generated by retroviral vector transduction. Tetherin-containing Moloney murine leukemia virus (MLV) pseudotypes were produced by seeding 1 × 106 293T cells into 100-mm-diameter cell culture dishes and cotransfecting them with pCMVi (MLV Gag-Pol), pMDG (VSV-G), and feTHN-pDON-AI-2 Neo or empty vector (pDON-AI-2 Neo). These pseudotypes were then used to infect CRFK cells. At 72 h postinfection, stably transduced cells were selected in G418 (800 μg/ml). For confocal microscopy experiments, tetherin was labeled with an internal hemagglutinin (HA) tag by amplification of two fragments by the use of primer pair FcTHN-HA-FWD (5′-TACCCATACGACGTCCCAGACTACGTCGTCGCGTCTGCCA-3′) and FcTHN-REV-BamHI (5′-ACAGGATCCTCAGGCCAGCAGAGCAACGAAG-3′) and primer pair FcTHN-FWD-NotI (5′-ATCGGCGGCCGCATGGCACCTGCTTTTTACCAC-3′) and FcTHN-HA-REV (5′-GACGTAGTCTGGGACGTCGTATGGGTATTCCTTTTTCTTGC-3′), with feline tetherin cDNA used as the template. The products were then purified, combined, and used as a template for a second round of amplification using primers FcTHN-FWD-NotI and FcTHN-REV-BamHI. The product was then cloned into pDON-AI-2 Neo and transduced into CRFK cells as described above.
Virion release assays.
A total of 1 × 106 293T cells were seeded in 100-mm-diameter cell culture dishes, and virion release was assayed by transient transfection with 7.5 μg of HIV-1-GFP or FIV-GFP expression vectors (as described above) and 2.5 μg of FcTHN-VR1012. At 72 h posttransfection, the released viral pseudotypes were harvested. The cells were washed with phosphate-buffered saline (PBS) and pelleted for immunoblot analysis. Virion-containing supernatants were clarified by centrifugation and filtered using pores 0.2 μm in size. A 1-ml volume of supernatant was used to infect 1 × 105 293T cells per well in 12-well plates in triplicate experiments. At 72 h postinfection, infected cells were washed with PBS and harvested for flow cytometric analysis. The remainder of the filtered virion-containing supernatant (9 ml) was layered on a 20% sucrose cushion and centrifuged at 20,000 × g for 2 h. The virion yield was determined by immunoblotting.
In order to confirm FIV OrfA expression, additional experiments were performed in which VSV-G-pseudotyped, yellow fluorescent protein (YFP)-encoding HIV-1 was prepared as previously described (15). Briefly, 1 × 106 293T cells per well were seeded in six-well plates and transfected using 6 μl Fugene-6 (Roche) with p8.91 Gag-Pol expression vector (3 μg), pMDG (VSV-G) (300 ng), and HIV-1 vector encoding YFP (450 ng). Tetherin constructs (100 ng) were cotransfected along with either 500 ng of HIV-1 NL-43 Vpu or 1S5RL OrfA or empty vector (pCDNA3.1; Invitrogen). After 48 h, the supernatant was harvested and filtered and 293T cell titers were determined as described previously (15).
Immunoblotting.
Cells were lysed in buffer comprising Tris-HCl (pH 7.5), 150mM NaCl, and CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} zwitterionic detergent (1% [wt/vol]) and supplemented with protease inhibitor cocktail (Complete protease inhibitor; Roche). Cell lysates and corresponding pelleted virions were separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on 12% SDS-PAGE gels. Separated proteins were then transferred onto nitrocellulose membranes, blocked with 5% dried milk powder in PBS containing 0.1% (vol/vol) Tween for 1 h at room temperature, and incubated with the appropriate primary antibody for 1 h. HIV-1 p24 was detected using 183-H12-5C anti-HIV-1 CA (NIH AIDS Research and Reference Reagent Program), and FIV p24 was detected using vpg50 antibody. Both antibodies were used as hybridoma supernatants at a dilution of 1:100 in PBS–0.1% Tween. Primary antibodies were detected with biotinylated anti-mouse IgG secondary antibody (Vector Laboratories, Peterborough, United Kingdom) at a dilution of 1:1,000 in PBS–0.1% Tween for 1 h at room temperature and subsequent chromogenic development using a Vectastain ABC system (Vector Laboratories Ltd., Peterborough, United Kingdom) and 5-bromo-4-chloro-3-indolyl phosphate–nitroblue tetrazolium (BCIP/NBT; Vector Laboratories Ltd., Peterborough, United Kingdom) as a substrate.
To confirm FIV OrfA expression, immunoblotting was performed as described previously (15). HIV-1 CA was detected using 183-H12-5C monoclonal antibody (MAb), and membranes were then stripped and reprobed for β-actin and HA (antibody 3F10; Roche Applied Sciences, Burgess Hill, United Kingdom). Cell extracts were made by lysing cells in radioimmunoprecipitation assay (RIPA) buffer (15). Cleared lysate was then added to Laemmli (24) buffer. Supernatants containing virions were pelleted and resuspended in Laemmli buffer, and all samples were then boiled before separation by SDS-PAGE and immunoblotting. Bound antibodies were detected using sheep anti-mouse horseradish peroxidase (HRP) conjugate followed by Amersham ECL Plus (both from GE Healthcare Life Sciences, Little Chalfont, United Kingdom).
Virus replication assays.
Viral growth was assayed by nonisotopic lentiviral reverse transcriptase (RT) assays using a LentiRT kit (Cavidi AB, Uppsala, Sweden) per the manufacturer's instructions.
Confocal microscopy.
CRFK cells stably expressing HA-tagged feline tetherin were seeded onto 13-mm-diameter glass coverslips in 24-well plates and incubated overnight. The cells were then infected with FIV or F14 for 3 days. Cells were next fixed with 4% paraformaldehyde–PBS (pH 7.4) for 15 min and then neutralized with 0.1 M glycine–PBS for 5 min. Where appropriate, cells were then permeabilized with 0.2% Triton X-100–PBS for 20 min. Cells were next blocked by incubation with 2% fetal bovine serum (FBS)–PBS for 20 min before incubation with a mixture of primary antibodies comprising VP71.2 mouse anti-FIV Env and rabbit anti-HA (Sigma-Aldrich, Poole, Dorset, United Kingdom) for 1 h at room temperature in 1% FBS–PBS. Cells were then washed three times with PBS, including incubation for 5 min with each wash. Secondary antibodies [an Alexa-Fluor 594-conjugated F(ab′)2 fragment of goat anti-rabbit IgG from Invitrogen/Molecular Probes, Paisley, United Kingdom, and a fluorescein isothiocyanate (FITC)-conjugated F(ab′)2 fragment of goat anti-mouse IgG from Abd Serotec, Oxford, United Kingdom] were then added, and the mixtures were incubated for 45 min in 1% FBS–PBS at room temperature. Cells were washed three times with PBS (including incubation for 5 min with each wash), removed from the 24-well plates, and mounted onto glass slides in mounting medium containing DAPI (4′,6-diamidino-2-phenylindole) (Vectashield; Vector Laboratories, Peterborough, United Kingdom). Slides were then analyzed on a Leica TCS SP2 confocal microscope.
Electron microscopy.
Monolayer cultures of FIV-infected CRFK cells were fixed in situ for 2 h at room temperature with 4% paraformaldehyde–0.1 M sodium phosphate buffer (pH 7) and stored at 4°C in 0.2% paraformaldehyde. Prior to processing for electron microscopy, the cells were exposed to 2.5% glutaraldehyde overnight at 4°C. The cells were then scraped and pelleted by centrifugation followed by 1% osmium tetroxide fixation. Fixed cells were then resuspended and pelleted through 1% SeaPlaque agarose (Lonza, Slough, United Kingdom). The cell pellets were dehydrated through a graded alcohol series and embedded in Epon 812 resin. Sections approximately 120 nm in thickness were cut with a UC6 ultramicrotome (Leica Microsystems, Germany) and stained with saturated uranyl acetate (in 50% ethanol) and Reynolds lead citrate (49). Sections were observed and photographed with JEOL 1200 EX and JEOL JEM-2200FS transmission electron microscopes, and images were recorded on a Gatan Ultrascan camera.
Nucleotide sequence accession number.
The sequence of the feline tetherin homologue has been deposited in GenBank (accession no. HM461970).
RESULTS
Identification of a feline tetherin homologue.
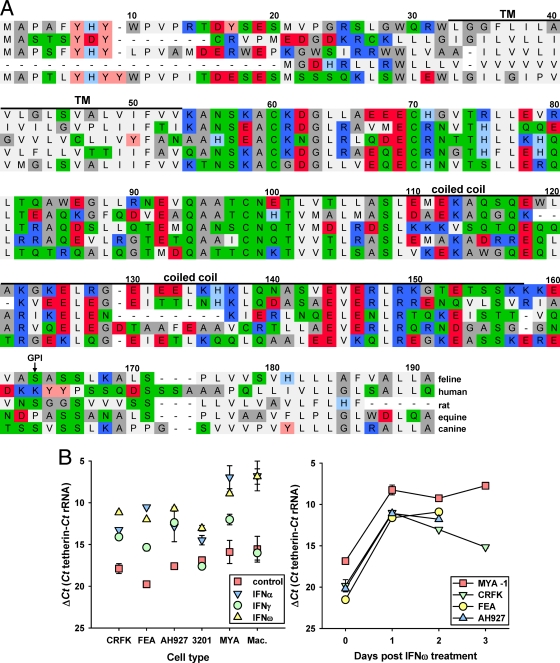
Screening the 1.8X feline genome identified a genomic sequence with significant homology to tetherin/BST-2. Oligonucleotide primers based on this sequence amplified a 561-bp product from IFN-ω-stimulated feline IL-2-dependent T cell (MYA-1) cDNA with 77% nucleic acid and 60% amino acid identity to canine BST-2, transcript variant 2 (XM860510), a sequence determined from the canine (boxer) genome by automated computational analysis (Gnomon; NCBI). Translation predicted an amino acid sequence of 184 amino acids with an amino-terminal transmembrane domain, an extracellular loop with three conserved cysteines (C59, C69, and C97), and a coiled-coil domain (Fig. 1A). The big-π predictor (10) revealed a potential site for a glycophosphatidylinositol anchor at serine 161 (Fig. 1A) followed by a carboxy-terminal hydrophobic domain. The conservation of amino acid sequence and structural features was consistent with the product being the feline homologue of tetherin. Modeling the three-dimensional structure of the feline tetherin homologue on the basis of the published crystal structure of the extracellular domain of human tetherin indicated conservation of the structure (data not shown).
Fig. 1.
Identification of a feline homologue of tetherin. (A) Alignment of the predicted amino acid sequences of feline, human, murine (rat), equine, and canine tetherins. Black bars denote transmembrane (TM) and coiled-coil regions, and an arrow marks a predicted site for glycosylphosphatidylinositol (GPI) anchor attachment. Residues are color coded as follows: light gray, hydrophobic FPMVLI; dark gray, amphiphilic WAG; green, hydrophilic neutral NQTSC; light blue, slightly basic H; blue, basic K and R; pink, slightly acidic Y; red, acidic DE. (B) Quantification of feline tetherin RNA by real-time PCR and effect of interferon treatment. IL-2-dependent CD4+ T cells, macrophages, and the FEA, AH927, CRFK, and 3201 cell lines were cultured overnight with or without feline IFN-α, IFN-γ, and IFN-ω at 103 U/ml prior to RNA extraction, cDNA preparation, and tetherin cDNA quantification. Results are expressed as mean copy numberss ± SE per cell as determined in three independent experiments (n = 3).
In the mouse, tetherin is expressed constitutively with B cells and plasmacytoid dendritic cells; however, expression may be upregulated with many cell types following exposure to both type I (5, 21, 31, 35, 38) and type II (5) interferons. We treated four feline cell lines, AH927 (46) (fibroblast cells), CRFK (8) (kidney epithelioid cells), FEA (17) (fetal embryo fibroblast-like cells), and 3201 (57) (thymic lymphosarcoma cells), and two primary cell cultures (IL-2-dependent CD4+ T cells [MYA-1 cells] and monocyte-derived macrophages) with type I (α and ω) and type II (γ) interferon and examined tetherin mRNA expression levels by quantitative RT-PCR (qRT-PCR) (Fig. 1). The cell lines showed various levels of basal tetherin mRNA expression, with the lowest levels on FEA (ΔCT = 21.26 ± 0.39, representing the means ± standard errors [SE] of the results of three independent analyses, with three replicate experiments per analysis). CRFK, AH927, and 3201 expressed broadly similar levels of tetherin at ΔCT = 19.83 ± 0.80, ΔCT = 17.48 ± 0.55, and ΔCT = 18.75 ± 0.54, respectively. The highest level of basal expression, CT = 15.93 ± 0.32, was seen with the IL-2-dependent CD4+ T MYA-1 cell line. Treatment of feline cells with the type I interferons IFN-α and IFN-ω (a type I interferon widely used in the treatment of viral infections in domestic cats) increased tetherin expression for all of the cell types examined. A similar increase in tetherin expression was also noted with the FEA, AH927, and CRFK cells treated with IFN-γ. A more modest induction of tetherin expression was observed with 3201 cells following treatment with IFN-α and IFN-ω, while IFN-γ did not have a significant effect on 3201 tetherin expression. IL-2-dependent CD4+ T cells and monocyte-derived macrophages responded to IFN-α and IFN-ω treatment by increasing expression of tetherin, whereas IFN-γ had a modest effect on CD4+ T cells and no effect on macrophages. Therefore, as with previous observations with human tetherin, feline tetherin expression may be induced by both type I and II interferons and is expressed on the known in vivo cellular targets for FIV, activated T cells and macrophages.
Feline tetherin inhibits the release of lentiviruses.
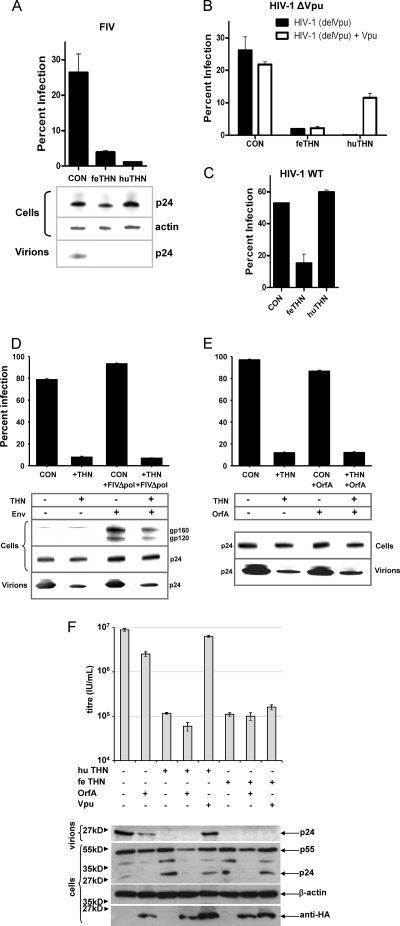
293T cells were transfected with expression vectors bearing feline or human tetherin in conjunction with constructs to produce FIV(VSV) GFP pseudotypes. Pseudotypes were plated onto uninfected 293T cells and the viral titers assessed by flow cytometry as previously described (15) (Fig. 2A). Both feline and human tetherin reduced the titer of the FIV(VSV) pseudotypes significantly, suggesting inhibition of particle release from the transfected cells. In agreement with this notion, viral p24 released into the culture supernatant was reduced by tetherin expression to levels below the limit of detection. Similar activity of feline tetherin against HIV-1 lacking a functional Vpu [HVI-1 (delVpu)] was detected; however, unlike human tetherin, antiviral activity was not counteracted by the addition of HIV-1 Vpu in trans (Fig. 2B). Similarly, feline tetherin prevented the release of wild-type HIV-1 encoding an intact vpu open reading frame, whereas human tetherin had no effect (Fig. 2C). In the absence of an open reading frame with significant homology to vpu in FIV, we asked whether the FIV env or orfA gene products were able to counteract the activity of feline tetherin, in analogy to the activities ascribed to the SIV Nef and Env proteins (15, 18, 26, 35, 36, 62, 65). Coexpression of a replication-defective molecular clone of FIV(CMVG8MΔpol4) in which expression of the entire FIV molecular clone is enhanced by a cytomegalovirus (CMV) promoter, thus increasing expression of all FIV proteins, had no effect on the ability of feline tetherin to counteract FIV release from transfected cells (Fig. 2D). Similarly, cotransfection of a codon-optimized FIV OrfA expression vector (43) (Fig. 2E and F), or of a GL8 orfA cloned in VR1012 eukaryotic expression vector (not shown), had no effect on the activity of feline tetherin against the release of FIV from transfected cells.
Fig. 2.
Effect of feline tetherin (THN) on the release of FIV and HIV. (A) FIV(VSV-G) pseudotypes bearing a GFP marker gene were prepared by transfection of 293T cells in the presence of human or feline tetherin, and viral release was quantified by titration on 293T cells. Virus production was also assessed by immunoblotting cell lysates and pelleted virions for the capsid protein p24. CON, control. (B) HIV(VSV) pseudotypes derived from a Vpu-deleted clone of HIV-1 [HIV-1 (delVpu)] were prepared in the presence of either feline or human tetherin, and HIV-1 Vpu was provided in trans. Virus release was assessed by infection of 293T cells. (C) HIV pseudotypes were also prepared from a construct with an intact Vpu (HIV-1 WT) in the presence of either feline or human tetherin, and infectivity was assessed by infection of 293T cells. (D) Ability of FIV-encoded proteins to counteract feline tetherin. FIV(VSV-G)/GFP pseudotypes were prepared in the presence or absence of both feline tetherin and a replication-defective molecular clone of FIV, CMVG8MΔpol4 (+FIVΔpol). Expression of FIV Env from CMVG8MΔpol4 was confirmed by immunoblotting with MAb VPG71.2. (E) FIV(VSV-G)/GFP pseudotypes were prepared in the presence or absence of both feline tetherin and a codon-optimized FIV OrfA, and infectivity was assayed on 293T cells. (F) HIV(VSV-G)/YFP pseudotypes were prepared in the presence or absence of both feline tetherin and human tetherin and either a codon-optimized FIV OrfA or HIV-1 Vpu, and infectivity was assayed using 293T cells.
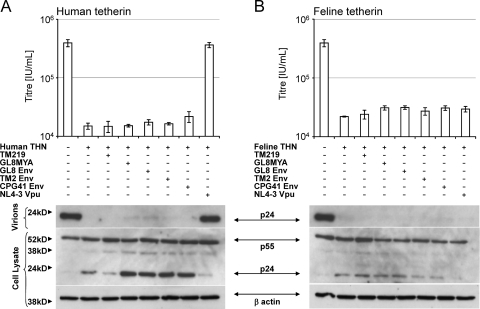
Further confirmation of the absence of tetherin-counteracting activity on the feline genome was provided by cotransfection of FIV Env expression vectors expressing the GL8, TM2, or CPG41 Env or the GL8 and TM219 molecular clones, none of which had a discernible effect on the ability of human (Fig. 3A) or feline (Fig. 3B) tetherin to inhibit HIV-1 particle release. In contrast, under identical conditions, cotransfection of HIV-1 Vpu counteracted human but not feline tetherin activity. Thus, under standard experimental conditions that demonstrate Vpu or Env antagonism of primate tetherins (15) we could find no evidence for tetherin-counteracting activity in the FIV genome.
Fig. 3.
Absence of a specific tetherin antagonist in the genome of FIV. HIV(VSV) pseudotypes derived from a Vpu-deleted clone of HIV-1 were prepared in the presence of the FIV TM219 and GL8MYA molecular clones (subtypes B and A), Env GL8, TM2, and CPG41 VR1012 expression vectors (subtypes A, B, and C), or HIV-1 (NL-43) Vpu expression vector in the presence of human tetherin (A) or feline tetherin (B). Titers of viral supernatants were determined for 293T cells and infectious titers plotted (means ± SD [n = 3]). Viral protein production in the transfected cells and virion release into the culture supernatant were monitored by immunoblotting.
Effect of feline tetherin on retroviral replication.
Given the potency of feline tetherin for preventing the release of FIV in a single-cycle assay, and the apparent absence of tetherin-counteracting activity in the FIV genome, we investigated whether feline tetherin inhibited FIV replication in a spreading infection. CRFK cells were stably transduced with a retroviral vector expressing feline tetherin. Real-time PCR analysis indicated that the drug-selected cells expressed abundant tetherin mRNA (ΔCT = 8.95 ± 0.25) compared with the vector-only selected control cells (ΔCT = 16.68 ± 0.34). Thus, the stably transduced cells achieved levels of tetherin expression that exceeded those attained following IFN-ω treatment of control CRFK cells (ΔCT = 11.17 ± 0.17) (Fig. 1). In comparison, when 293T cells were transfected transiently with either VR1012 or VR1012-feTHN (see Fig. 2 and 3), ΔCT increased from 23.43 ± 0.43 (VR1012) to 5.97 ± 0.39 (VR1012-feTHN), representing an order of magnitude higher than the level achieved in stably transduced CRFK (considering that, following transient transfection of 293T cells, only a proportion of the cells would take up DNA) and perhaps indicating why the inhibitory effect of tetherin on virion release from 293T is so marked.
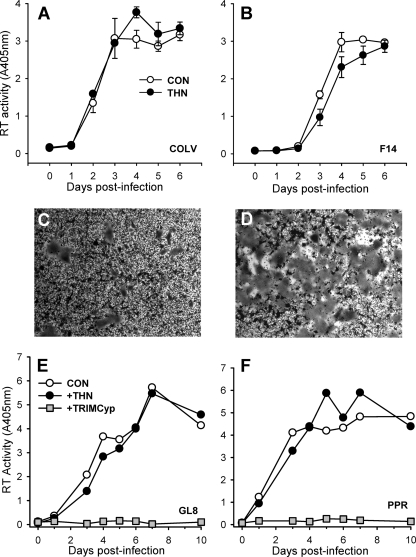
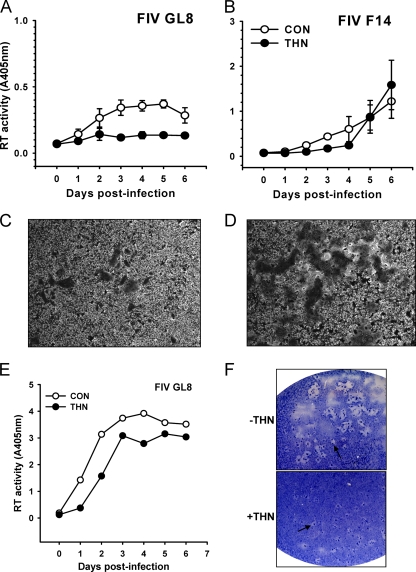
Cells stably expressing feline tetherin were infected with CRFK-tropic strains of FIV-Pco (COLV; Fig. 4A) or FIV-Fca (F14; Fig. 4B), and virus spread was monitored by an RT assay of the culture supernatant. In contrast to the marked inhibitory effect of tetherin on lentiviral pseudotype release, ectopic expression of tetherin did not inhibit virus production from FIV-infected CRFK cells. As the FIV lentiviral vectors used to generate FIV pseudotypes (Fig. 2) and the F14 strain of FIV share an origin (FIV Petaluma), the data indicate that single-cycle release assays and assays of productive infection yield conflicting results. Furthermore, syncytium formation following FIV F14 infection was enhanced significantly in the tetherin-expressing cells (Fig. 4D) compared with control cells (Fig. 4C). This observation suggests that functional levels of tetherin are expressed and is consistent with the presence of trapped virions at the cell surface promoting cell-cell spread, a phenomenon proposed recently for HIV-1 (20).
Fig. 4.
Effect of stable expression of feline tetherin on FIV replication. CRFK cells were stably transduced with a retroviral vector bearing feline tetherin. (A and B) Cells were infected with the cell culture-adapted FIV-Pco COLV (A) and FIV-Fca F14 (B) strains, and virus replication was monitored by RT assay of the supernatant (means ± SE [n = 3]). (C and D) Representative fields displaying enhanced syncytium formation in tetherin-expressing cells following FIV-Fca F14 infection (D) compared with control cells (C). (E and F) Infection of CRFK cells stably expressing CD134 and either feline tetherin or synthetic feline TRIMCyp with primary isolates FIV-Fca GL8 (E) and FIV-Fca PPR (F). There was no inhibition of viral replication by tetherin in comparison with potent inhibition by TRIMCyp (mean [n = 2]).
We next considered the effect of feline tetherin on the replication of primary strains of FIV. CRFK cells stably transduced with an expression vector bearing the CD134 primary viral receptor (56) were transduced with retroviral vectors bearing either feline tetherin or a synthetic feline TRIMCyp (9) (as a positive control for viral restriction). CD134-expressing CRFK cells were permissive of the replication of both the GL8 (Fig. 4E) and PPR (Fig. 4F) strains of FIV, and tetherin expression had no inhibitory effect. In contrast, feline TRIMCyp completely prevented viral replication. These data suggest that stable ectopic expression of feline tetherin does not restrict the replication of primary or cell culture-adapted strains of FIV.
Formally, it is possible that low levels of surface expression of tetherin in the stably modified cell lines are insufficient to block viral release and that this explains their inability to block spreading FIV infection. To control for this possibility, we took advantage of the fact that the GL8 primary FIV isolate cannot replicate in CRFK cells unless they express the CD134 coreceptor. Transfection of CRFK cells with GL8 therefore does not lead to a spreading infection but allows us to measure the amount of GL8 released into the supernatant by assaying for reverse transcriptase. CRFK cells transfected with GL8 released virus into the supernatant, whereas CRFK cells expressing tetherin did not (Fig. 5A), which is consistent with tetherin preventing virus release. As reported above, spreading infection of CD134-independent FIV F14 was not inhibited by tetherin expression in these cells, and once again, enhanced syncytium formation was noted in the tetherin-expressing cells (Fig. 5D) in comparison to control cells (Fig. 5C). We next expressed the CD134 coreceptor in the same CRFK cells in order to render them permissive for GL8 infection. CD134-expressing CRFK cells were permissive for GL8 replication whether or not they expressed tetherin (Fig. 5E). Tetherin expression in these cells reduced replication by about 2-fold, leading to a delay of approximately 1 day in achieving peak virus loads, but tetherin was unable to block replication in a manner analogous to that seen with TRIMCyp. This effect was confirmed by analysis of the degree of syncytium formation at the end of the experiment (day 6); while the absolute numbers of syncytia in the GL8-infected cells were similar, the syncytia in the tetherin-expressing cells were smaller (Fig. 5F), which is consistent with a partial retardation of viral growth.
Fig. 5.
Effect of tetherin expression on virus release and syncytium formation. (A and B) CRFK cells expressing feline tetherin were transfected with molecular clones of FIV GL8 (A) or FIV F14 (B). Virus release was monitored by RT assays (means ± SE [n = 3]). (C and D) Syncytium formation in FIV F14-transfected cells in the absence (C) or presence (D) of tetherin (representative fields observed microscopically in methylene blue/basic fuchsin-stained monolayers). (E and F) Measurement of FIV GL8 growth (E) and syncytium formation (F) in the absence or presence of tetherin. Virus release was monitored by RT assays of supernatant (means [n = 2]), while syncytium formation was observed macroscopically following staining (as described above). Representative syncytia are indicated (arrows).
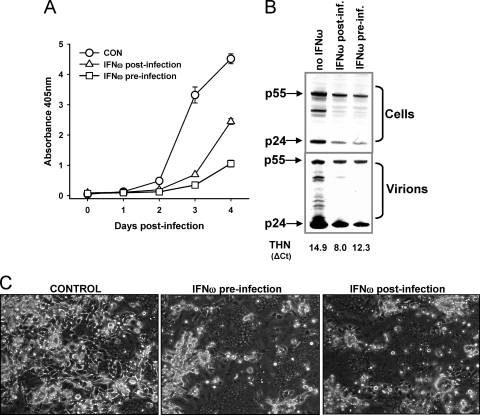
Thus, we observed two contrasting outcomes following infection with GL8 and F14; GL8 infection was partially retarded, with a reduction in syncytium formation, whereas F14 infection was unaffected and syncytium formation was enhanced. Enhanced syncytium formation following increased tetherin expression would be detrimental to the host and counterintuitive with respect to the idea of a role for tetherin in restricting viral growth. We therefore asked whether such a scenario would occur following upregulation of endogenously expressed tetherin by interferon. CRFK cells were infected with the F14 (CD134-independent) strain of FIV. As expected, treatment of the cells with interferon-ω either 24 h prior to infection or 24 h postinfection suppressed virus production (Fig. 6A). Immunoblotting confirmed both the reduced release of virus into the culture supernatant and reduced viral protein production within the cells (Fig. 6B), which is consistent with the pleiotropic antiviral activities of type-I interferons suppressing viral growth. As would be expected, addition of IFN-ω 24 h postinfection suppressed viral replication to a lesser extent than preincubation with IFN-ω. As real-time PCR analysis of tetherin transcripts confirmed that tetherin expression had been induced (Fig. 6B) following interferon stimulation, we asked whether, in the face of the antiviral activities of IFN-ω, we could discern evidence for the enhancement of syncytium formation that we had observed in the cells stably expressing tetherin. When the cells were examined microscopically (Fig. 6C), a marked enhancement of syncytium formation was noted in the CRFK cells infected with FIV F14 and treated with IFN-ω (Fig. 6C), irrespective of whether the interferon was added before or after viral infection. These data suggest that the results observed in cells stably expressing tetherin are recapitulated in cells in which tetherin expression is induced by IFN-ω.
Fig. 6.
Effect of interferon-ω on FIV production and syncytium formation. CRFK cells were infected with FIV-F14, and virus production was monitored by RT assays. Cells were left untreated (CON), pretreated with IFN-ω 24 h prior to infection, or treated with IFN-ω 24 h postinfection. (B) Virus protein levels were monitored in lysates of the infected cells and in culture supernatants at day 4 postinfection by immunoblotting. Induction of tetherin expression was confirmed by real-time PCR (THN CT value). (C) Representative images of syncytium formation at day 3 postinfection (phase-contrast microscopy). Dark areas packed with nuclei represent syncytia.
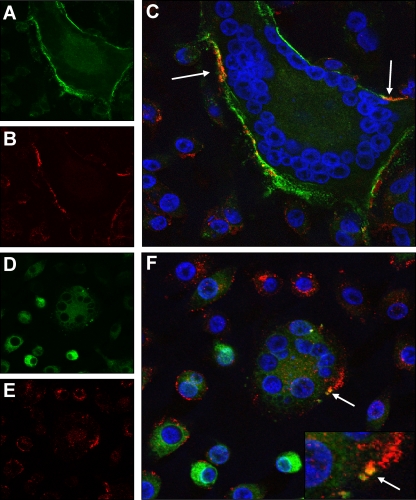
In order to examine the cellular localization of FIV and tetherin in infected cells, CRFK cells were stably transduced with a retroviral vector expressing feline tetherin tagged in the extracellular domain with an internal HA tag. Confocal analysis of Env expression on intact (nonpermeabilized) cells demonstrated enrichment of Env staining along the entire perimeter of syncytia (Fig. 7A). While tetherin expression was detected with the majority of single cells where expression appeared punctuate (Fig. 7B), expression on the perimeter of syncytia followed a pattern similar to that seen with Env, and in several regions, marked overlap of Env and tetherin was noted (Fig. 7C). In permeabilized cells, Env expression was diffuse throughout the cytoplasm of the cells (Fig. 7D) whereas tetherin expression appeared largely punctuate (Fig. 7E). It was notable that, in some Env-expressing cells, tetherin appeared to be concentrated at the periphery of the cell (Fig. 7F, lower left), whereas in cells lacking Env, tetherin appeared to be perinuclear (Fig. 7E, top center). Occasionally, syncytia contained regions of colocalization of tetherin and Env, possibly indicating the presence of intracellular bodies rich in both Env and tetherin.
Fig. 7.
Distribution of FIV in infected cells. CRFK cells stably expressing feline tetherin incorporating an internal HA tag were fixed and stained for expression of FIV Env (FITC [green]) (A and D) or tetherin (Alexa Fluor 594 [red]) (B and E). As shown in merged images (C and F), nuclei were also visualized with DAPI (blue). Cells were stained either in intact form (A to C) or following detergent permeabilization (D to F). Images are representative of at least five separate fields; arrows indicate regions where Env and tetherin expression coincided.
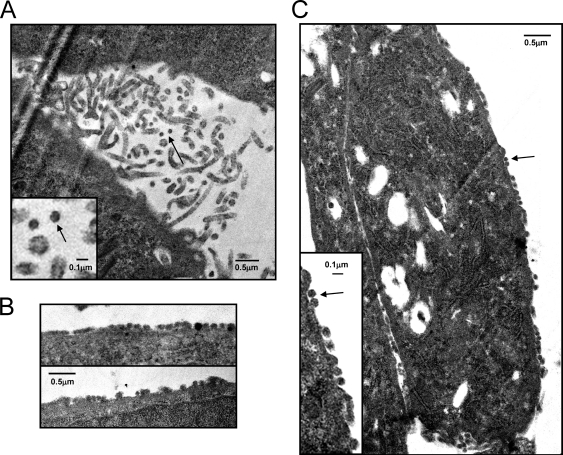
Finally, we examined FIV-infected tetherin-expressing CRFK cells for evidence of the retention of virus particles on the cell surface. FIV-infected CRFK cells produced abundant microvilli in the presence or absence of tetherin (Fig. 8A) with a morphology similar to that described previously for HeLa cells (12). While 0.1-μm-diameter particles were noted in the regions rich in microvilli, the resolution of the CRFK-derived images made it difficult to distinguish conclusively electron-dense virus-like particles from a cross-section through the tip of a microvillus (Fig. 8A); previous reports have described mouse mammary tumor virus budding from the tip of microvilli in CRFK cells (25). However, aggregates of 0.1-μm-diameter particles aligned on the surface of cells were unique to the FIV-infected tetherin-expressing cells (Fig. 8C and D), a feature we were unable to identify in the FIV-infected cells not expressing tetherin despite screening multiple fields. The uniformity and alignment of these particles on the cell surface were consistent with the presence of trapped virions and may represent evidence indicating the same regions identified by confocal microscopy in which tetherin and Env were coexpressed at a high level.
Fig. 8.
Electron microscopy of CRFK cells infected with FIV. (A) Microvillus-rich regions in FIV-infected control cells with occasional ∼0.1-μm-diameter particles (inset, two particles [arrow] adjacent to a cross-section of a microvillus for comparison). (B and C) Aggregates of ∼0.1-μm-diameter particles aligned on the surface of tetherin-expressing cells infected with FIV. Arrows indicate ∼0.1-μm-diameter particles associated with the cell surface.
DISCUSSION
Primate lentiviruses have developed several strategies for the evasion of host restriction factors: Vif counteracts APOBECs, mutations in CA prevent TRIM5s binding to the incoming capsid, and HIV-1 Vpu, HIV-2 Env, and SIV Nef and Env counteract tetherin. The diverse mechanisms employed by primate lentiviruses to evade the antiviral activities of tetherin suggest a prominent role for tetherin in controlling lentiviral replication. Pandemic HIV-1 M strain genes encode Vpu proteins with potent antitetherin activity, a property they share with SIVgsn, SIVmus, and SIVmon strains (53). In contrast, Vpus from nonpandemic O or N strains is a poor tetherin antagonist, while SIVcpz, the nearest ancestor of HIV-1, uses Nef to counteract tetherin (53), a feature it shares with SIVsmm, SIVmac, SIVsyk, and SIVagm (18, 53, 65, 65). A five-amino-acid deletion in the N-terminal cytoplasmic tail of human tetherin has rendered it refractory to antagonism by SIVcpz Nefs, and there is evidence for strong positive selective pressure in favor of escape from Nef binding (27). These data provide compelling evidence for an ongoing struggle between host and pathogen and have led to speculation that it is the ability to evade tetherin that renders HIV-1 M strains able to achieve pandemic spread in the human population (16, 53).
Like primates, felids coexist with a lentivirus, FIV. Thus, they provide a useful comparator for the likely role of tetherin in the control of lentiviral infections. In this study, we demonstrated that a feline homologue of tetherin inhibits the release of FIV from infected cells in vitro as potently as human tetherin. However, both primary and cell culture-adapted strains of FIV overcame restriction by tetherin in a spreading infection assay with relative ease, even in the absence of identifiable antitetherin activity. Indeed, tetherin expression promoted enhanced syncytium formation in adherent cells upon infection with cell-culture-adapted FIV, as would be expected with virion trapping, hence increasing levels of envelope protein expression at the cell surface. In comparison, synthetic feline TRIMCyp (9) blocked FIV replication completely when expressed under identical conditions. What is the significance of the enhanced syncytium formation seen with cell culture-adapted strains of virus? Cell culture-adapted strains of FIV are similar to, and are frequently derived from, CD134-independent strains of FIV that arise in chronic infection in vivo. They may be considered to be analogous to the X4 strains of HIV that emerge in the later stages of HIV infection. Initially, these viruses were thought to be the consequence of cell culture adaptation, a selection for adaptation for growth in CD134-negative cell lines such as CRFK. Indeed, Petaluma F14 and 34TF10, the prototypic molecular clones of FIV, bear mutations in their V3 loops consistent with CRFK adaptation and CD134-independent infection. We have investigated the primary isolate of FIV Petaluma that had been maintained solely in peripheral blood mononuclear cells and have identified CD134-independent variants among the viral quasispecies (data not shown). Combined with our own observations in which CD134-independent strains of FIV arose in vivo following infection with the CD134-dependent GL8 strain of FIV (23), it would appear that there is selective pressure in vivo for the generation of CD134-independent strains of virus. We have shown previously that the humoral immune response can exert a selective pressure that is capable of modulating the virus-receptor interaction (64). The activities of feline tetherin that we have observed in vitro may exhert an additional selective pressure on receptor use in vivo. While interferon production would upregulate tetherin expression and thus prevent the release of nascent virions, CD134-independent viruses may counteract the effects of tetherin by spreading cell to cell and in so doing may overcome its antiviral effects. Why should F14 syncytium formation be enhanced when GL8 syncytium formation is modestly reduced? It is likely that the switch to CD134-independence facilitates a more efficient cell-cell fusion mediated by use of the abundant coreceptor CXCR4 alone.
Tetherin clearly inhibits virus release; however, there may be other roles for tetherin that do not involve the inhibition of viral growth. Tetherin may downmodulate IFN production following an interaction with the ITL7 receptor on plasmacytoid dendritic cells (pDCs), also known as type-I interferon-producing cells (IPCs) (6), and viral modulation of the regulation of IFN production may also modulate the innate response to infection (reviewed in reference 11). We propose that an additional function of tetherin might be related to the cell type on which it is expressed in abundance in vivo, i.e., pDCs. Tetherin is known to target virions for endocytosis and degradation, and this might lead to antigen processing for subsequent presentation to T cells. While they were initially thought to have a primary role in immunomodulation resulting from the secretion of large amounts of type I interferon, subsequent studies have indicated that pDCs are capable of presenting antigen to both CD4 and CD8+ T cells and may have a potential role in antigen capture, processing, and presentation to T cells at sites of infection and in lymph nodes (reviewed in reference 63). Expression of tetherin on nonprofessional antigen-presenting cells would also trap virions at the cell surface, marking the cells for destruction by antibody-dependent cellular cytotoxicity. Evasion of either of these outcomes would be vastly beneficial to a virus, whether lentivirus, herpesvirus, or filovirus, all of which have evolved means of counteracting the activity of tetherin.
Do our findings contradict those of previous studies conducted with other retroviruses? Many of the studies considering tetherin function performed to date have used single-cycle assays focusing on virus release following transfection of nonsusceptible cells such as HEK-293 or infection of nonsusceptible cells such as HeLa with HIV(VSV-G)-pseudotypes, but few have examined viral replication in susceptible cells, and those that have done so yielded conflicting results. Tetherin was found to inhibit the cell-to-cell transmission of ΔVpu HIV and to impair the transmission of wild-type HIV from donor HeLa, 293T, or CD4+ T cells to recipient CD4+ T cells or Jurkat cells (7). In contrast, a second study found that ΔVpu HIV disseminated more efficiently by cell-to-cell contact when tetherin was expressed and that depletion of tetherin using small interfering RNA (siRNA) impaired cell-to-cell transmission (20). The observations we describe here concerning feline tetherin and the felid lentiviruses are consistent with the findings of latter study in that, in assays in which we assessed the growth of replication-competent viruses, we found that feline tetherin has a relatively minor effect on virus growth. When we quantified tetherin gene expression in 293T cells transiently transfected with a tetherin expression vector, we found that expression exceeded by an order of magnitude that achieved in either stably transfected cells or interferon-treated cells, suggesting supraphysiological levels of gene expression. Thus, we observed potent inhibition of virus release in 293T cells, whereas there was a weak effect on virus growth but an enhancement of syncytium formation in stably transfected CRFK cells. The latter observation is particularly striking in that, prior to the discovery of tetherin, previous studies reported an increase in the size of syncytia in HeLa-CD4 cells transfected with Vpu-deleted HIV (22), while enhanced cell-to-cell spread and the formation of giant syncytia in Jurkat cells infected with a mutant virus bearing a frameshift in vpu were noted (14). These data are consistent with the increase in syncytium size we observed in tetherin-expressing cells following infection with a CD134-independent virus. Further studies are required to distinguish the role of tetherin in preventing virus release, enhancing antigen presentation, and preventing virus spread both from cell to cell and within a population of permissive individuals. The study of tetherin promises to continue to reveal important insights into cellular and viral biology.
ACKNOWLEDGMENTS
This work was funded by a program grant from The Wellcome Trust to B.J.W. and M.J.H., Wellcome Trust Senior Fellowship no. 090940 to G.J.T., and grants from the Medical Research Council and the National Institute of Health UCL/UCLH Comprehensive Biomedical Research Centre.
We are grateful to our colleagues in the MRC—University of Glasgow Centre for Virus Research and VDS for their guidance regarding electron microscopy (David Bhella, Fraser Rixon, and Lisa Moir) and confocal microscopy (Claudio Murgia). We also thank Martin Löchelt (Heidelberg) for many helpful discussions, John Elder (Scripps Institute) for an anti-OrfA polyclonal serum, and Mauro Pistello (Universita di Pisa) for the codon-optimized OrfA expression vector.
Footnotes
Published ahead of print on 13 April 2011.
REFERENCES
- 1. Asaoka K., et al. 2005. A retrovirus restriction factor TRIM5alpha is transcriptionally regulated by interferons. Biochem. Biophys. Res. Commun. 338:1950–1956 [DOI] [PubMed] [Google Scholar]
- 2. Bainbridge J. W., et al. 2001. In vivo gene transfer to the mouse eye using an HIV-based lentiviral vector; efficient long-term transduction of corneal endothelium and retinal pigment epithelium. Gene Ther. 8:1665–1668 [DOI] [PubMed] [Google Scholar]
- 3. Benveniste R. E., Todaro G. J. 1974. Evolution of C-type viral genes: inheritance of exogenously acquired viral genes. Nature 252:456–459 [DOI] [PubMed] [Google Scholar]
- 4. Benveniste R. E., Todaro G. J. 1975. Segregation of RD-114 AND FeL-V-related sequences in crosses between domestic cat and leopard cat. Nature 257:506–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blasius A. L., et al. 2006. Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J. Immunol. 177:3260–3265 [DOI] [PubMed] [Google Scholar]
- 6. Cao W., et al. 2009. Regulation of TLR7/9 responses in plasmacytoid dendritic cells by BST2 and ILT7 receptor interaction. J. Exp. Med. 206:1603–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Casartelli N., et al. 2010. Tetherin restricts productive HIV-1 cell-to-cell transmission. PLoS Pathog. 6:e1000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crandell R. A., Fabricant C. G., Nelson-Rees W. A. 1973. Development, characterization, and viral susceptibility of a feline (Felis catus) renal cell line (CRFK). In Vitro 9:176–185 [DOI] [PubMed] [Google Scholar]
- 9. Dietrich I., et al. 2010. Potent lentiviral restriction by a synthetic feline TRIM5 cyclophilin A fusion. J. Virol. 84:8980–8985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eisenhaber B., Bork P., Yuan Y., Loffler G., Eisenhaber F. 2000. Automated annotation of GPI anchor sites: case study C. elegans. Trends Biochem. Sci. 25:340–341 [DOI] [PubMed] [Google Scholar]
- 11. Evans D. T., Serra-Moreno R., Singh R. K., Guatelli J. C. 2010. BST-2/tetherin: a new component of the innate immune response to enveloped viruses. Trends Microbiol. 18:388–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fisher H. W., Cooper T. W. 1967. Electron microscope studies of the microvilli of HeLa cells. J. Cell Biol. 34:569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goffinet C., Schmidt S., Kern C., Oberbremer L., Keppler O. T. 2010. Endogenous CD317/tetherin limits replication of HIV-1 and MLV in rodent cells and is resistant to antagonists from primate viruses. J. Virol. 84:11374–11384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gummuluru S., Kinsey C. M., Emerman M. 2000. An in vitro rapid-turnover assay for human immunodeficiency virus type 1 replication selects for cell-to-cell spread of virus. J. Virol. 74:10882–10891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gupta R. K., et al. 2009. Simian immunodeficiency virus envelope glycoprotein counteracts tetherin/BST-2/CD317 by intracellular sequestration. Proc. Natl. Acad. Sci. U. S. A. 106:20889–20894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gupta R. K., Towers G. J. 2009. A tail of Tetherin: how pandemic HIV-1 conquered the world. Cell Host Microbe 6:393–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jarrett O., Laird H. M., Hay D. 1973. Determinants of the host range of feline leukaemia viruses. J. Gen. Virol. 20:169–175 [DOI] [PubMed] [Google Scholar]
- 18. Jia B., et al. 2009. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 5:e1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnson W. E., et al. 2006. The late Miocene radiation of modern Felidae: a genetic assessment. Science 311:73–77 [DOI] [PubMed] [Google Scholar]
- 20. Jolly C., Booth N. J., Neil S. J. 2010. Cell-cell spread of human immunodeficiency virus type 1 overcomes tetherin/BST-2-mediated restriction in T cells. J. Virol. 84:12185–12199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kawai S., et al. 2008. Interferon-alpha enhances CD317 expression and the antitumor activity of anti-CD317 monoclonal antibody in renal cell carcinoma xenograft models. Cancer Sci. 99:2461–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klimkait T., Strebel K., Hoggan M. D., Martin M. A., Orenstein J. M. 1990. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J. Virol. 64:621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kraase M., et al. 2010. Feline immunodeficiency virus env gene evolution in experimentally infected cats. Vet. Immunol. Immunopathol. 134:96–106 [DOI] [PubMed] [Google Scholar]
- 24. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 25. Lasfargues E. Y., Lasfargues J. C., Dion A. S., Greene A. E., Moore D. H. 1976. Experimental infection of a cat kidney cell line with the mouse mammary tumor virus. Cancer Res. 36:67–72 [PubMed] [Google Scholar]
- 26. Le Tortorec A., Neil S. J. 2009. Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J. Virol. 83:11966–11978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lim E. S., Malik H. S., Emerman M. 2010. Ancient adaptive evolution of tetherin shaped the functions of Vpu and Nef in human immunodeficiency virus and primate lentiviruses. J. Virol. 84:7124–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Löchelt M., et al. 2005. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. U. S. A. 102:7982–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McEwan W. A., et al. 2009. Truncation of TRIM5 in Feliformia explains the absence of retroviral restriction in cells of the domestic cat. J. Virol. 83:8270–8275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meli M. L., et al. 2009. Feline leukemia virus and other pathogens as important threats to the survival of the critically endangered Iberian lynx (Lynx pardinus). PLoS One 4:e4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miyagi E., Andrew A. J., Kao S., Strebel K. 2009. Vpu enhances HIV-1 virus release in the absence of Bst-2 cell surface down-modulation and intracellular depletion. Proc. Natl. Acad. Sci. U. S. A. 106:2868–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miyazawa T., et al. 1989. Establishment of a feline T-lymphoblastoid cell line highly sensitive for replication of feline immunodeficiency virus. Arch. Virol. 108:131–135 [DOI] [PubMed] [Google Scholar]
- 33. Münk C., et al. 2008. Functions, structure, and read-through alternative splicing of feline APOBEC3 genes. Genome Biol. 9:R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Naldini L., et al. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267 [DOI] [PubMed] [Google Scholar]
- 35. Neil S. J., Sandrin V., Sundquist W. I., Bieniasz P. D. 2007. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe 2:193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Neil S. J., Zang T., Bieniasz P. D. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430 [DOI] [PubMed] [Google Scholar]
- 37. Nolen R. S. 2004. Feline leukemia virus threatens endangered panthers. J. Am. Vet. Med. Assoc. 224:1721–1722 [PubMed] [Google Scholar]
- 38. Ohtomo T., et al. 1999. Molecular cloning and characterization of a surface antigen preferentially overexpressed on multiple myeloma cells. Biochem. Biophys. Res. Commun. 258:583–591 [DOI] [PubMed] [Google Scholar]
- 39. Olmsted R. A., et al. 1989. Molecular cloning of feline immunodeficiency virus. Proc. Natl. Acad. Sci. U. S. A. 86:2448–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Peng G., Lei K. J., Jin W., Greenwell-Wild T., Wahl S. M. 2006. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J. Exp. Med. 203:41–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Perez-Caballero D., et al. 2009. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139:499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perkovic M., et al. 2009. Species-specific inhibition of APOBEC3C by the prototype foamy virus protein bet. J. Biol. Chem. 284:5819–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pistello M., et al. 2006. AIDS vaccination studies with an ex vivo feline immunodeficiency virus model: analysis of the accessory ORF-A protein and DNA as protective immunogens. J. Virol. 80:8856–8868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pistello M., et al. 2005. Evaluation of feline immunodeficiency virus ORF-A mutants as candidate attenuated vaccine. Virology 332:676–690 [DOI] [PubMed] [Google Scholar]
- 45. Poeschla E. M., Wong-Staal F., Looney D. J. 1998. Efficient transduction of nondividing human cells by feline immunodeficiency virus lentiviral vectors. Nat. Med. 4:354–357 [DOI] [PubMed] [Google Scholar]
- 46. Rasheed S., Gardner M. B. 1980. Characterisation of cat cell cultures for expression of retrovirus, FOCMA and endogenous sarc genes, p. 393–400 In Essex M., McClelland A. J. (ed.), Feline leukemia virus. Proceedings of the 3rd International Feline Leukaemia Virus Meeting Elsevier-North Holland, Amsterdam, Netherlands [Google Scholar]
- 47. Reeves R. H., Nash W. G., O'Brien S. J. 1985. Genetic mapping of endogenous RD-114 retroviral sequences of domestic cats. J. Virol. 56:303–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reeves R. H., O'Brien S. J. 1984. Molecular genetic characterization of the RD-114 gene family of endogenous feline retroviral sequences. J. Virol. 52:164–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reynolds E. S. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Roca A. L., Pecon-Slattery J., O'Brien S. J. 2004. Genomically intact endogenous feline leukemia viruses of recent origin. J. Virol. 78:4370–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Russell R. A., et al. 2005. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J. Virol. 79:8724–8731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saenz D. T., Poeschla E. M. 2004. FIV: from lentivirus to lentivector. J. Gene Med. 6(Suppl. 1):S95–S104 [DOI] [PubMed] [Google Scholar]
- 53. Sauter D., et al. 2009. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6:409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650 [DOI] [PubMed] [Google Scholar]
- 55. Sheehy A. M., Gaddis N. C., Malim M. H. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404–1407 [DOI] [PubMed] [Google Scholar]
- 56. Shimojima M., et al. 2004. Use of CD134 as a primary receptor by the feline immunodeficiency virus. Science 303:1192–1195 [DOI] [PubMed] [Google Scholar]
- 57. Snyder H. W., Hardy W. D., Zuckerman E. E., Fleisner E. 1978. Characterisation of a tumour-specific antigen on the surface of feline lymphosarcoma cells. Nature 275:656. [DOI] [PubMed] [Google Scholar]
- 58. Swiecki M., Colonna M. 2010. Unraveling the functions of plasmacytoid dendritic cells during viral infections, autoimmunity, and tolerance. Immunol. Rev. 234:142–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Talbott R. L., et al. 1989. Nucleotide sequence and genomic organization of feline immunodeficiency virus. Proc. Natl. Acad. Sci. U. S. A. 86:5743–5747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tanaka Y., et al. 2006. Anti-viral protein APOBEC3G is induced by interferon-alpha stimulation in human hepatocytes. Biochem. Biophys. Res. Commun. 341:314–319 [DOI] [PubMed] [Google Scholar]
- 61. Troyer J. L., et al. 2005. Seroprevalence and genomic divergence of circulating strains of feline immunodeficiency virus among Felidae and Hyaenidae species. J. Virol. 79:8282–8294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Van Damme N., et al. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Villadangos J. A., Young L. 2008. Antigen-presentation properties of plasmacytoid dendritic cells. Immunity 29:352–361 [DOI] [PubMed] [Google Scholar]
- 64. Willett B. J., et al. 2010. Modulation of the virus-receptor interaction by mutations in the V5 loop of feline immunodeficiency virus (FIV) following in vivo escape from neutralising antibody. Retrovirology 7:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang F., et al. 2009. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6:54–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang Z., Schwartz S., Wagner L., Miller W. 2000. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7:203–214 [DOI] [PubMed] [Google Scholar]
- 67. Zufferey R., Nagy D., Mandel R. J., Naldini L., Trono D. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15:871–875 [DOI] [PubMed] [Google Scholar]