Abstract
HIV-1 viruses and virus-like particles (VLPs) bear nonnative “junk” forms of envelope (Env) glycoprotein that may undermine the development of antibody responses against functional gp120/gp41 trimers, thereby blunting the ability of particles to elicit neutralizing antibodies. Here, we sought to better understand the nature of junk Env with a view to devising strategies for its removal. Initial studies revealed that native trimers were surprisingly stable in the face of harsh conditions, suggesting that junk Env is unlikely to arise by trimer dissociation or gp120 shedding. Furthermore, the limited gp120 shedding that occurs immediately after synthesis of primary HIV-1 isolate Envs is not caused by aberrant cleavage at the tandem gp120/gp41 cleavage sites, which were found to cleave in a codependent manner. A major VLP contaminant was found to consist of an early, monomeric form of gp160 that is glycosylated in the endoplasmic reticulum (gp160ER) and then bypasses protein maturation and traffics directly into particles. gp160ER was found to bind two copies of monoclonal antibody (MAb) 2G12, consistent with its exclusively high-mannose glycan profile. These findings prompted us to evaluate enzyme digests as a way to remove aberrant Env. Remarkably, sequential glycosidase-protease digests led to a complete or near-complete removal of junk Env from many viral strains, leaving trimers and viral infectivity largely intact. “Trimer VLPs” may be useful neutralizing antibody immunogens.
INTRODUCTION
The encouraging results of a recent phase IIb trial suggest that an HIV-1 vaccine may be possible (63). Optimal efficacy may require a component that induces broadly neutralizing antibodies (bNAbs) that have the rare ability to bind to the native Env spikes on particle surfaces, thus interfering with receptor engagement and virus infection (28, 35, 59).
Env spikes consist of trimers of gp120/gp41 heterodimers, in which gp120 is the surface subunit and gp41 is the transmembrane-anchoring subunit. These derive from a gp160 precursor that is glycosylated cotranslationally in the endoplasmic reticulum (ER), where it is also thought to oligomerize (22). In the Golgi compartment, cleavage at the gp120/gp41 junction occurs via the action of furin. The resulting spikes are compact and highly glycosylated, features that allow the virus to evade neutralization (23, 50).
The glycans that decorate HIV-1 Env are unusual in that a fraction of them fail to fully mature. In normal circumstances, glycan synthesis (summarized in Fig. 1B of reference 4) begins in the endoplasmic reticulum, where high-mannose (HM) precursors are transferred cotranslationally to the free amide of the asparagine of a glycan signal sequence, or sequon (30). Terminal glucose and mannose moieties are then trimmed to form a Man5GlcNAc2 intermediate (where Man is mannose and GlcNac is N-acetylglucosamine). The transfer of another GlcNAc moiety to this intermediate results in a hybrid glycan. Further mannose trimming, the addition of more GlcNAc moieties, and additional modifications result in mature complex glycans (39).
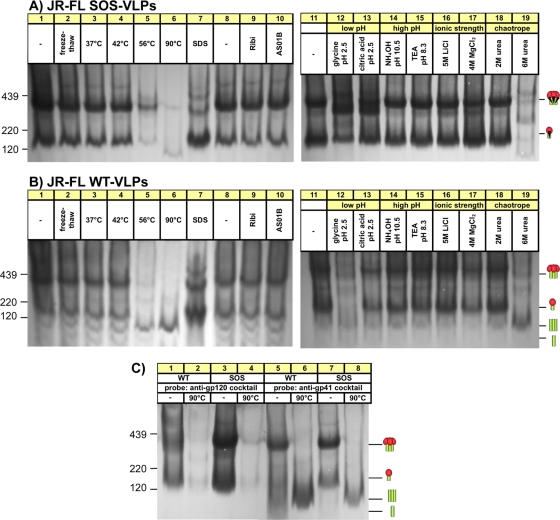
Fig. 1.
Effects of harsh conditions on native JR-FL trimer stability. SOS-VLPs and WT-VLPs were subjected to five freeze-thaw cycles, a 10-min incubation at various temperatures, or exposure to 0.5% SDS, adjuvants, or various harsh buffers, as indicated. VLPs were then washed (SDS treatment excepted), resuspended in PBS, and then analyzed by BN-PAGE and Western blotting, probing with the standard MAb cocktail. Trimeric and monomeric forms of gp120/gp41 and gp41 stumps are indicated by cartoons in which gp120s are depicted as red blobs (one or three for the monomer or trimer, respectively) and gp41s are shown as green sticks. Molecular mass markers for ferritin (220 kDa and 439 kDa) and gp120 are indicated. (C) SOS-VLPs and WT-VLPs were subjected to a 10-min 90°C incubation, as indicated. Blots were then probed separately with anti-gp120 or anti-gp41 cocktails. TEA, triethanolamine.
In the exceptional case of HIV-1 Env, a patch of glycans at the immunologically “silent face” is so dense that steric constraints limit mannosidase trimming, such that the glycans fail to mature (19, 84). This high-mannose patch is recognized by the equally unusual neutralizing monoclonal antibody (MAb) 2G12 (12, 64, 66, 75). The close interactions between the component subunits of mature trimeric spikes appear to further limit glycan maturation compared to monomeric gp120, leading to an even greater proportion of high-mannose glycans (24, 31, 53). Indeed, the glycans present on trimers are so densely packed that most glycosidases cannot excise them, in contrast to the glycosidase-sensitive glycans of monomeric gp120 (47, 51, 64).
The receptor binding sites of the neutralizing face of gp120 are protected by a relatively sparse “forest” of complex glycans (68). This partial coverage probably reflects a trade-off between a requirement to protect underlying functional domains from NAb attack while providing sufficient flexibility for the refolding events associated with receptor binding and fusion (29).
To date, all Env-based vaccine candidates have failed to induce NAbs of significant breadth and potency. Instead, they largely induce antibodies directed to determinants that are occluded on authentic spikes (78). This may be because the immunogens insufficiently resemble native Env trimers and therefore lack the stringency to selectively elicit exquisite neutralizing specificities. Fully authentic Env trimers might therefore fare better as immunogens since any antibodies they induce in a vaccine setting should, in theory, neutralize. Attempts to produce authentic trimers in a soluble form have been hampered by their instability (13). This problem can be solved by various stabilizing mutations, usually in gp41. However, these manipulations tend to affect conformation such that soluble trimers no longer fully resemble native spikes.
Presentation of trimers in a membrane context may naturally enhance their stability without a need for stabilizing mutations. This idea has been attempted in the form of particle and cell-based vaccines (14, 17, 34, 40, 61, 77). However, success in eliciting NAbs by these strategies has, to date, been limited, perhaps because the Env is not biochemically homogeneous. In addition to native trimers, particles also exhibit various nonfunctional forms of Env, including uncleaved gp160 and gp41 stumps (55). Hypothetically, this “junk” could interfere with the development of NAb responses. In support of this idea, junk Env is antigenically promiscuous, as demonstrated by the efficient capture of virus particles by monoclonal Abs that neither bind trimers nor neutralize (43, 55, 57, 60). The superior immunogenicity of junk Env is also suggested by the observation that virus-immune complexes form shortly after infection, long before the development of NAbs (74). Vaccine interference is, in fact, a common phenomenon and may be exacerbated when the components are antigenically related (42, 76, 79). However, there is presently no information on how genetically identical but conformationally distinct proteins might interfere in a vaccine setting (73, 76).
The true potential of native trimers as vaccine immunogens may be revealed only if we can eliminate antigenic interference (55). A better understanding of junk Env may assist in developing new strategies for its removal. One form of junk Env, namely, gp41 stumps, is generated when gp120 dissociates from particles, presumably due to the instability of noncovalently associated gp120-gp41 complexes (54). This phenomenon is commonly referred to as gp120 shedding. Previously, we showed that gp120 shedding could be eliminated by the introduction of an intermolecular gp120-gp41 disulfide bridge, termed the SOS mutant (7). Virus-like particles (VLPs) carrying this mutation (SOS-VLPs) were found to be infectious and neutralization resistant, exactly like wild-type (WT) particles (WT-VLPs), suggesting that authentic conformation is preserved (1, 5, 6, 18). However, SOS-VLPs fared little better than WT-VLPs as immunogens (17), suggesting the possibility of still other forms of Env junk.
An analysis of SOS-VLPs by blue native PAGE (BN-PAGE) revealed distinct Env trimers and monomers (55). Two possibilities might account for the monomer. First, it may be a by-product of trimer dissociation in the lateral plane. Alternatively, it could be an immature form of Env that somehow contaminates particles. The latter possibility is supported by findings that Env synthesis is fraught with inefficiencies. Thus, while some gp120/gp41 trimers reach the cell surface with a half-life (t1/2) of ∼2 h after translation, another population of gp160 remains in the endoplasmic reticulum much longer (2, 20, 23, 41, 80), perhaps due to inefficient signal peptide removal (41, 46) or poor gp120/gp41 processing (2, 8, 20, 23, 41, 50, 80, 81). Potentially, this could lead to a breakdown of normal protein maturation and the incorporation of immature Env into particles. Although early studies suggested that all gp160 on particle surfaces is fully processed into gp120/gp41 (20, 71), others suggest that there are no checkpoints to exclude uncleaved gp160 (20, 55, 62, 81). A recent study reported that the Env glycans of HIV-1 virions are comprised almost entirely (>98%) of the high-mannose variety (21). An immature form of junk Env might explain this unexpected bias of high-mannose glycans. Here, we investigated the nature of particulate Env in depth and sought to develop new methods to selectively remove the junk Env with a view to manufacturing “trimer VLPs.”
MATERIALS AND METHODS
MAbs and monomeric gp120.
A panel of MAbs included the following: b12, directed to epitopes overlapping the CD4 binding site (CD4bs) of gp120 (11, 82); 2G12, directed to a unique glycan-dependent epitope of gp120 (64, 66); E51, directed to a CD4-inducible (CD4i) epitope of gp120 (83); LA21, F2A3, Co11, and 39F reactive with clade B gp120 V3 loop sequences (18, 58, 69); 5.8C, 1.4E, 3074, and 3869 reactive with non-clade B V3 loop sequences (36); 2F5 and 4E10, directed to the gp41 membrane-proximal ectodomain region (MPER) (56); and 7B2 and 2.2B, directed to the gp41 cluster I and II epitopes, respectively. Recombinant monomeric JR-FL gp120 produced in CHO cells was a gift from Progenics Pharmaceuticals (Tarrytown, NY).
Virus preparations and infectivity assays.
VLPs were produced by the cotransfection of 293T cells or N-acetylglucosamine transferase I-deficient (GnTI−) 293S cells (4, 24) with an Env-expressing plasmid and the Env-deficient HIV-1 genomic backbone plasmids pNL-LucR-E− (5, 44, 45). Plasmids for expressing Env were all made in the pCAGGS vector and included those expressing SOS, uncleaved (UNC) SOS, WT, and UNC WT mutants of JR-FL gp160 with a truncation of the cytoplasmic tail (gp160ΔCT). Mutants were made by QuikChange mutagenesis (Stratagene). Most of the other Env plasmids were derived from clade B and C reference panels (16, 44, 45). A KNH1144 Env plasmid has been described previously (37). VLPs were purified as described previously (55). Concentrated and inactivated HIV-1 isolates BaL, MN, and ADA, produced in SupT1-CCR5 CL.30 cells and purified over sucrose gradients, were provided by Julian Bess and Jeff Lifson (National Cancer Institute [NCI]). Single-round infection assays were performed as described previously (55).
SDS-PAGE and Western blotting.
Env was resolved by reducing SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Western blots were probed with an anti-gp120 cocktail, consisting of MAbs CO11, F2A3, LA21, and 39F or a gp41 cocktail, consisting of MAbs 2F5 and 4E10. Each MAb was used at 1 μg/ml. Clade C Envs were probed with a cocktail of MAbs 5.8C, 1.4E, 3074, and 3869 at 1 μg/ml each. An alkaline phosphatase-labeled anti-human Fc conjugate was used to detect MAb binding (Jackson), and the signal was developed using SigmaFast BCIP/NBT (5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium) substrate (Sigma). Band densities were measured using UN-SCAN-IT software (Silk Scientific) (16).
Native PAGE and Western blotting.
Blue native PAGE (BN-PAGE) was performed as described previously (16–18, 55). Briefly, VLPs were treated in various ways, including freeze-thaw cycling, 10-min incubations at various temperatures, or the use of various buffers, MAbs, 0.5% SDS, or adjuvants Ribi, Ras3C, or AS01B. VLPs were then washed, as necessary, and then gently solubilized in 0.12% Triton X-100 in 1 mM EDTA–1.5 M aminocaproic acid with a protease inhibitor cocktail containing 4-(2-aminoethyl)benzenesulfonyl fluoride, E-64, bestatin, leupeptin, aprotinin, and sodium EDTA (P-2714; Sigma). An equal volume of 2× sample buffer (100 mM morpholinepropanesulfonic acid [MOPS], 100 mM Tris-HCl, pH 7.7, 40% glycerol, and 0.1% Coomassie blue) was added. Samples were then loaded onto a 4 to 12% Bis-Tris NuPAGE gel (Invitrogen) and separated at 4°C for 3 h at 100 V. Ferritin (Amersham) was used as a size standard. The gel was then blotted onto polyvinylidene difluoride, destained, immersed in blocking buffer (4% nonfat milk in phosphate-buffered saline [PBS]), and probed with an anti-gp120 cocktail (MAbs 2G12, b12, E51, and 39F at 1 μg/ml each), an anti-gp41 cocktail (MAbs 2F5, 4E10, 7B2, and 2.2B at 1 μg/ml each), or a standard MAb cocktail consisting of both the anti-gp120 and anti-gp41 cocktails. Non-clade B Envs were probed with a cocktail of anti-gp120 and -gp41 MAbs as above, substituting clade B V3 MAbs for non-clade B V3 MAbs (clade C MAb cocktail). Alternatively, a clade C plasma pool was used consisting of BB75, BB81, and BB86 (6) at 1:5,000 each. Blots were then probed by an anti-human Fc alkaline phosphatase conjugate (Jackson) and developed using SigmaFast BCIP/NBT substrate (Sigma).
Enzyme digests.
In glycosidase-protease digest reactions, recombinant gp120 monomer (1 μg) or VLPs (typically, 5 μl of VLPs concentrated 1,000-fold from transfection supernatants [1,000×]) were incubated at 37°C with glycosidases: 500 U of endoglycosidase H (endo H; New England BioLabs, Ipswitch, MA) or a deglycosylation mix consisting of 250 U of peptide N-glycosidase F (PNGase F), 25 U of neuraminidase, 4 U of β(1–4)-galactosidase, 20,000 U of endo-α-N-acetylgalactosaminidase, 1.6 U of β-N-acetylglucosaminidase (New England BioLabs, Ipswitch, MA), and/or 1.8 mU of fucosidase (Prozyme). Glycosidase digests were followed by a 1-h digest with 1 μg of trypsin, chymotrypsin, subtilisin, and/or proteinase K (Sigma) at 37°C. In some cases, particles were denatured and reduced by boiling for 5 min in SDS and beta-mercaptoethanol (45) prior to enzyme treatment. In other instances, digests were inhibited using a protease inhibitor cocktail (P-2714; Sigma).
RESULTS
Particulate Env trimers are remarkably stable.
Here, we sought to characterize particulate Env junk, with a view to developing approaches for its removal. We first looked at trimer stability to address the possibility that junk Env derives from trimer dissociation (55). The JR-FL Env clone was selected as a prototype here because of its unusually efficient gp160 processing. We along with others previously resolved gp120 shedding (54, 55) by introducing an intermolecular disulfide bridge between gp120 and gp41, termed the SOS mutant (7). SOS-VLPs are infectious and exhibit a similar neutralization resistance profile as the parent wild-type (WT) VLPs, suggesting that native conformation is preserved (1, 5, 18). Notably, SOS-VLPs do not use the isoleucine-to-proline (IP) mutation of SOSIP (65), which is a helix-breaking gp41 mutation used to stabilize soluble SOS gp140 trimers. We expressed Envs with a truncated gp41 tail, termed gp160ΔCT. This affects neither VLP infectivity nor its neutralization sensitivity (6, 21). We reasoned that the higher expression levels and increased gp120/gp41 processing efficiency of gp160ΔCT might improve our chances of characterizing junk Env.
To assess trimer stability, JR-FL VLPs were subjected to a variety of conditions (Fig. 1A and B). SOS trimers resisted freeze-thaw treatments, temperatures up to 42°C, adjuvants Ribi or AS01B, high-pH buffers, 5 M lithium chloride, and 2 M urea (Fig. 1A). However, high temperatures and 6 M urea caused a global loss in staining (Fig. 1A, lanes 5 and 19). SDS caused trimers to dissociate into monomers. Some SDS-resistant oligomers might be due to the aggregation or the presence of intermolecular disulfide bonds (Fig. 1A, lane 7). Magnesium chloride induced some trimer dissociation. Low-pH buffers induced changes that may reflect partial denaturation or aggregation.
WT trimers were similarly stable (Fig. 1B), with the exception that they were more sensitive to glycine at pH 2.5. Most treatments did not promote gp120 shedding. However, a 56°C incubation caused a build-up of trimeric gp41 stumps. This effect was accentuated by a 90°C incubation or exposure to 6 M urea (Fig. 1B, lanes 6 and 19). Exposure of SOS-VLPs to these conditions had similar effects (Fig. 1A, lanes 6 and 19). This was unexpected, considering the disulfide bond that links gp120 and gp41. We therefore investigated the identity of the treatment-induced band by probing duplicate blots separately with anti-gp41 and anti-gp120 MAb cocktails (Fig. 1C). This confirmed that the 90°C stress-induced band consists of gp41 stumps. Thus, extreme conditions can break the gp120-gp41 SOS covalent bond even in the absence of a reducing agent.
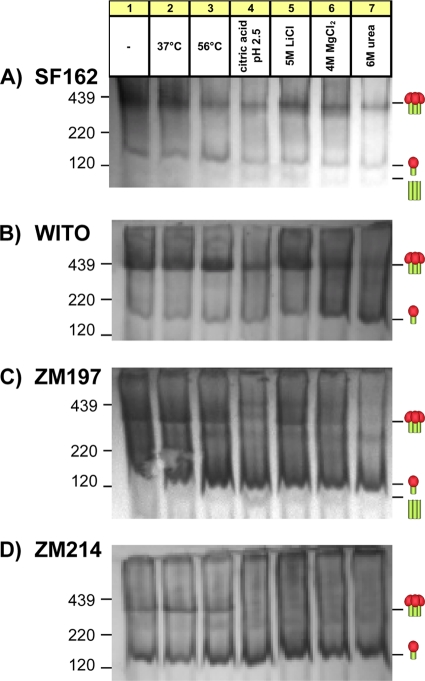
We next examined the stability of Env trimers from clade B isolates SF162 and WITO and clade C isolates ZM197 and ZM214. Each trimer exhibited a distinct resistance profile (Fig. 2). All four Envs survived incubation at 37°C. In surprising contrast to JR-FL (Fig. 1), WITO and ZM214 trimers survived a 56°C incubation, and SF162 and ZM197 trimers partially survived (Fig. 2A and C). Like JR-FL, SF162 and WITO trimers partially survived citric acid (Fig. 2A and B), but clade C trimers dissociated (Fig. 2C and D). SF162, WITO, and ZM197 trimers largely survived lithium chloride, but ZM214 trimers did not. As above, magnesium chloride was a somewhat harsher ionic-strength buffer and at least partially denatured all four trimers. Surprisingly, SF162 and WITO trimers partially survived an exposure to 6 M urea, while clade C trimers were completely sensitive. Unlike JR-FL, urea did not induce an obvious build-up of gp41 stumps, due to gp120 shedding, although citric acid treatment mildly increased the amount of gp41 stumps of ZM197. The faint staining of gp41 stumps in all four isolates (Fig. 2) suggests that the gp120 shedding is even lower than that for JR-FL (Fig. 1). Taken together, these data contest the widely held notion that HIV-1 Env trimers are labile.
Fig. 2.
Stability of various native Env trimers of various HIV-1 isolates. VLPs bearing Envs from various isolates as indicated were subjected to a stability analysis under selected conditions and were then probed by Western BN-PAGE blotting, as described in the legend of Fig. 1. SF162 and WITO were probed with the standard MAb cocktail, ZM197 was probed with the clade C MAb cocktail, and ZM214 was probed with the clade C plasma pool. Cartoons are as defined in the legend of Fig. 1.
gp120 shedding is not a progressive phenomenon.
The common perception that native trimers are unstable is partly based on early observations of gp120 shedding from HIV-1 isolates cultured in T cell lines (54). However, for tier 2 viruses, gp120 shedding appears rather limited (Fig. 1 and 2) (15). A report by Earl et al. showed that during Env synthesis, gp120 shedding occurs immediately after Env reaches the membrane surface but does not progress further (23), consistent with our observations that harsh conditions do not typically promote gp120 shedding (Fig. 1 and 2).
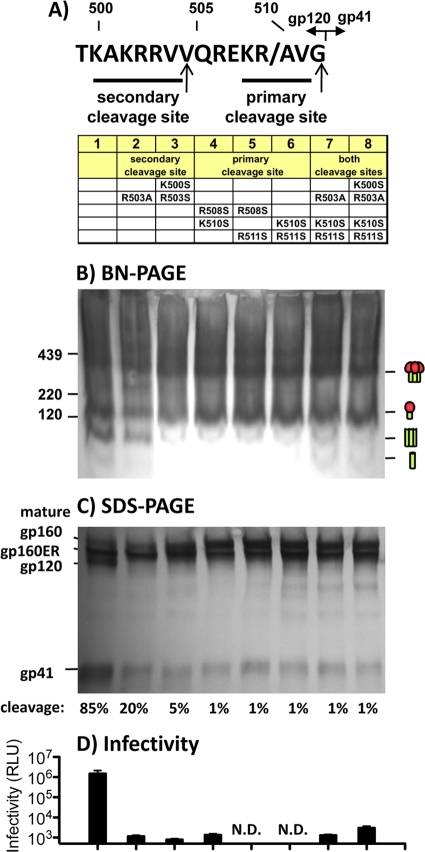
gp120/gp41 processing could potentially occur at two sites (indicated by arrows in Fig. 3A). Cleavage at the primary site is thought to generate functional virus, while the role of the second site is unclear (10, 25, 38, 50). Thus, it is not known if functional trimers are cleaved at both sites or only at the primary site. Irregular cleavage may underlie gp120 shedding. For example, if cleavage at both sites is necessary to generate functional spikes, then cleavage at only one site might yield a product with a propensity to shed gp120.
Fig. 3.
Analysis of gp120/gp41 cleavage site mutants. (A) The primary sequence of the JR-FL gp120-gp41 cleavage site. Residues are numbered according to the HXB2 sequence. Arrows indicate putative cleavage sites at residues 504 and 511. (B and C) BN-PAGE and SDS-PAGE with Western blot analysis of WT VLP cleavage site mutants. Blots were probed with both anti-gp120 and gp41 cocktails. Separate blots were performed with either cocktail alone to facilitate band identification (Fig. 4 and data not shown). Cartoons indicate gp120/gp41 trimers and monomers and gp41 stumps. (D) Infectivity of cleavage site mutants. ND, not done. Cartoons are as defined in the legend of Fig. 1.
To better understand gp160 processing and possibly prevent gp120 shedding, we investigated various cleavage site mutants. BN-PAGE analysis of WT-VLPs reveals two forms of gp41 stump (Fig. 1B) (55). The larger of these could arise from gp120/gp41 trimer dissociation, while the smaller stumps could arise from gp120/gp41 monomer dissociation (55). Unexpectedly, all cleavage mutants led to a dramatic decrease in gp41 trimers in BN-PAGE, regardless of whether they affected the primary or secondary site. Of these, the R503A mutant had the weakest effect and the K500S R503A K510S R511S mutant had the strongest effect (Fig. 3B, lanes 2 and 8). The effect of mutants on the gp41 monomer was unclear, due to its very faint staining (Fig. 3B, lane 1). Uncleaved gp160ΔCT (referred to here as gp160 for simplicity) resolved as a monomer and at least two oligomeric species (Fig. 3B, lanes 2 to 8).
By SDS-PAGE and Western blotting, all mutants exhibited diminished quantities of gp120 and gp41 compared to the control (Fig. 3C). Furthermore, two forms of gp160 were observed (Fig. 3C). All mutants except for the R503A mutant enhanced the proportion of the upper gp160 band, namely, mature gp160, while the smaller band, termed gp160ER, was unaffected. gp160ER is discussed in more detail below. The percent Env cleavage was measured as a factor of gp41 band density divided by the sum of gp41 and mature gp160 densities (Fig. 3C).
Interestingly, all the mutants involving the primary cleavage site resulted in a slightly larger gp41 species (Fig. 3C). Thus, in the experiment shown in Fig. 3C, the gp41 band in lanes 4 to 8 migrated more slowly than that in lanes 1 to 3. This could be because in lanes 4 to 8, although cleavage at the primary site is essentially blocked, cleavage at the secondary site can still occur even though in some cases (lanes 7 and 8) this second site is also mutated. Cleavage at only the secondary site would be expected to lead to a modestly larger gp41 species, due to the additional 7 amino acids (504 to 511) that adds about 5% mass to the ∼20-kDa gp41ΔCT (Fig. 3A, arrows marking the two cleavage sites).
In infection assays, none of the mutants were appreciably functional (Fig. 3D). Thus, cleavage at both sites is necessary for the formation of functional spikes. In fact, their processing is inextricably linked: eliminating cleavage at one site dramatically diminishes cleavage at the other. Thus, it appears unlikely that aberrant cleavage causes gp120 shedding.
A high-mannose form of gp160 contaminates VLPs.
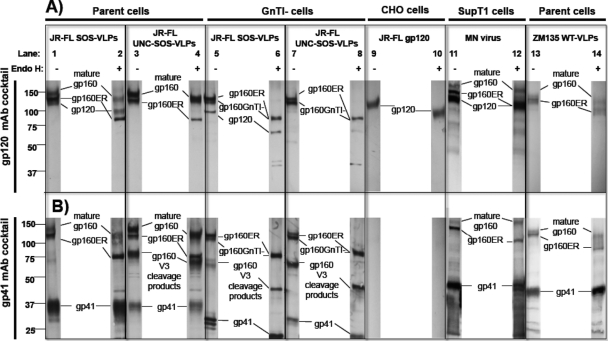
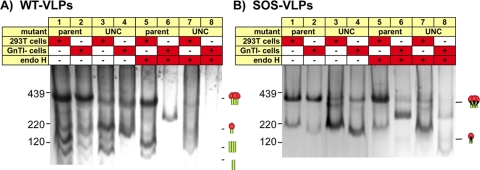
The above findings suggest that junk Env is not a product of trimer dissociation but, rather, is a distinct specie(s), perhaps an aberrant product of Env synthesis (23, 25). To investigate this possibility, we examined VLP Env by SDS-PAGE and Western blotting (Fig. 4). To help identify each band, duplicate blots were probed separately with anti-gp120 and anti-gp41 MAb cocktails (Fig. 4A and B). We also assessed the effect of endoglycosidase H (endo H), which selectively digests immature high-mannose glycans (39). VLPs were produced in 293T (parent) cells or cells defective for N-acetylglucosamine transferase I (GnTI−), an enzyme involved in converting trimmed Man5GlcNAc2 into hybrid glycans (24). Expression in these cells leads to the replacement of complex glycans with Man5GlcNAc2.
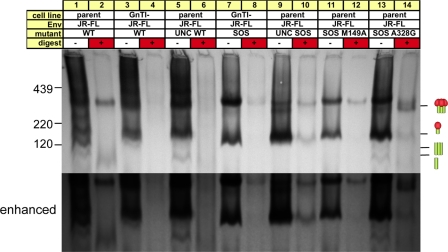
Fig. 4.
Reducing SDS-PAGE-Western blot analysis of particulate Env and monomeric gp120. SOS-VLPs and UNC SOS-VLPs (R510S R511S mutant) produced in parent 293T cells (lanes 1 to 4) or GnTI− cells (lanes 5 to 8), monomeric gp120 (lanes 9 and 10), inactivated MN virus grown in SupT1 cells (lanes 11 and 12), and ZM135 WT-VLPs (lanes 13 and 14) were compared in reducing SDS-PAGE and Western blotting. VLPs were digested with endo H after denaturation, as indicated. Duplicate blots in panels A and B were probed with anti-gp120 or anti-gp41 MAb cocktails, respectively. ZM135 VLPs were probed with modified gp120 cocktail containing V3 MAbs that recognize clade C Env.
Parent SOS-VLPs exhibited gp160, gp120, and gp41 bands of generally greater mass than their GnTI− equivalents (Fig. 4A and B, compare lanes 1 and 5). This reflects the addition of complex glycans that add ∼1.5 kDa per glycan compared to the exclusively high-mannose glycans that decorate their GnTI− counterparts (4, 24). The ∼15-kDa difference in parent and GnTI− gp120 molecular masses (Fig. 4A, lanes 1 and 5) therefore suggests ∼10 complex glycans on parent gp120. Parent Env bands were generally more diffuse than their GnTI− equivalents (Fig. 4, compare lanes 1 to 4 and 5 to 8), in line with the more variable nature of complex glycans (19, 26, 32, 53). Endo H dramatically increased the mobility of GnTI− Env bands, consistent with the removal of high-mannose glycans (Fig. 4, lanes 5 to 8). In contrast, parent Env and recombinant gp120 were more resistant due to the presence of endo H-resistant complex glycans (Fig. 4, lanes 1 to 4, 9, and 10).
Unexpectedly, an identical form of gp160 decorated exclusively with immature high-mannose glycans was present in all VLPs, regardless of the mutant or cell line in which the virus was expressed. We term this species gp160ER, distinguishing it from the cell line-specific forms of gp160 (mature gp160 and gp160 GnTI−), where ER stands for endoplasmic reticulum and refers to the untrimmed high-mannose glycans added after protein translation in this compartment. Like gp160 GnTI−, gp160ER is highly sensitive to endo H. In fact, the gp160 GnTI− and gp160ER bands coalesced after endo H treatment (Fig. 4, lanes 5 and 6). gp160 GnTI− is somewhat smaller, presumably due to a degree of mannosidase trimming that does not occur for gp160ER (4, 24).
In Fig. 4A, lane 1, gp160ER staining is relatively prominent compared to the gp120 band that likely derives from native trimers. This suggests that gp160ER constitutes a surprisingly dominant fraction of total VLP Env (55). In contrast, mature gp160 stained modestly (Fig. 4, lanes 1 and 3), consistent with the efficient cleavage observed in Fig. 3C (lane 1). Mature gp160 is somewhat larger than gp160ER, due to the presence of complex glycans, which also render it relatively resistant to endo H. The larger gp41 of parent 293T cells was also endo H resistant (Fig. 4B, compare lanes 1 to 4 to lanes 5 to 8). The estimated ∼6-kDa size difference suggests that the glycans that occupy the four putative sequons in native gp41 are likely to be normally complex in nature, as suggested previously (26, 32). Staining of UNC SOS-VLPs with an anti-gp41 cocktail revealed an additional band that was not observed with anti-gp120 MAb staining (using V3 MAbs) (compare Fig. 4A and B, lanes 3, 4, 7, and 8). This species was also faintly stained in lanes loaded with SOS-VLPs produced in GnTI− cells but not in SOS-VLPs produced from parent cells (Fig. 4B, lanes 1, 2, 5, and 6). The size of this band and its lack of staining with anti-V3 MAbs (Fig. 4A) suggest that this is a digestive product of protease cleavage at the V3 loop (49). The dominance of this species in UNC VLPs may stem from increased exposure of the V3 loop that renders it more sensitive to proteases.
Similar methods were used to examine other VLPs and live HIV-1 preparations. WT VLPs exhibited gp160ER, similar to SOS-VLPs (not shown). Live inactivated MN and ADA viruses and clade C ZM214 and ZM135 WT VLPs also exhibit more than one gp160 species, one of which was endo H sensitive, suggesting that it is gp160ER (Fig. 4, lane 11 to 14; also data not shown). The gp41 of these viruses was larger than that of SOS-VLPs (Fig. 4B, compare lanes 1 and 2 to lanes 11 to 14), consistent with full-length gp41, in contrast to the gp160ΔCT of our JR-FL VLPs. Taken together, these data reveal that gp160ER consistently contaminates HIV-1 particles, regardless of the producer cells, purification protocol, or strain. Moreover, gp160ER provides a new explanation for the aberrant monomer we observed in BN-PAGE (Fig. 1).
Characterization of VLP Env by BN-PAGE.
BN-PAGE analysis of VLPs reveals various forms of junk Env (55), including gp160 monomers and gp41 stumps (Fig. 1). We sought to better understand these contaminants by comparing different versions of JR-FL-based VLPs, namely, WT and SOS Envs presented in either a cleavage-competent (parent) or UNC format and expressed in either 293T or GnTI− cells (Fig. 5). Cleavage-competent Envs formed trimers, while UNC Env was largely monomeric (Fig. 5A and B, compare lanes 1 and 2 to lanes 3 and 4). Thus, native trimer assembly appears to require gp120/gp41 cleavage. The small amount of trimer could reflect a small degree of gp160 cleavage, consistent with traces of gp41 stumps (Fig. 3 and 5A, compare lanes 1 and 3).
Fig. 5.
Effects of endoglycosidase H on VLP Env. WT VLPs and SOS-VLPs and cleavage-defective mutants thereof produced in either parent 293T or GnTI− cells were incubated with endo H overnight and then analyzed by BN-PAGE and Western blotting. The UNC SOS-VLP mutant was K510S R511S and the UNC WT VLP mutant was K500S R503A K510S R511S. Cartoons are as defined in the legend of Fig. 1, with the addition of dark triangles that represent the SOS band.
GnTI− cells expressed lower quantities of VLPs, consistent with earlier studies (Fig. 5, lanes 2 and 4) (4). gp41 stumps were particularly faint in GnTI− cell-produced WT-VLPs (Fig. 5A, compare lanes 1 and 2) and were undetectable in GnTI− UNC WT VLPs, in stark contrast with the parent UNC WT-VLPs (Fig. 5A, compare lanes 3 and 4). This suggests the intriguing possibility that gp120 shedding is promoted by glycan maturation.
Previous studies have shown that endo H can remove high-mannose (HM) glycans present on the gp120 outer domain. However, silent domain glycans are largely endo H resistant due to the steric restrictions imposed by their high density—the same steric restrictions that also limit their maturation (4). Mature native Env trimers expressed in parent cells exhibit complex glycans on the outer domain and are therefore largely resistant to endo H. Conversely GnTI− trimers bear high-mannose glycans on the outer domain and are therefore endo H sensitive. To simplify further description, from hereon we refer to gp120 bearing complex outer domain glycans and a high-mannose silent domain as having a C/HM glycan profile, and those with high mannose on both domains are termed HM/HM.
Effects of endo H digestion.
Endo H digestion could potentially have two effects on gp120: to eliminate 2G12 detection in BN-PAGE–Western blotting and to reduce Env mass. Thus, an affected band might migrate faster, with a reduced signal. Endo H digests diminished the staining of the parent WT monomer, consistent with it being comprised at least partly of gp160ER (HM/HM) (Fig. 5A, compare lanes 1 and 5). The gp41 monomer, but not the gp41 trimer, was also partially depleted. All forms of GnTI− Env (HM/HM) were endo H sensitive (Fig. 5A, compare lanes 2 and 4 with lanes 6 and 8). Thus, GnTI− WT-VLP trimers underwent a dramatic drop in size (Fig. 5A, compare lanes 2 and 6) (4), while parent trimers were only marginally affected, in keeping with a C/HM glycan profile (Fig. 5A, compare lanes 1 and 5) (4). GnTI− UNC WT-VLP Env was undetectable after endo H digest (Fig. 5A, compare lanes 4 and 8). The resistance of 293T cell-derived UNC WT-VLPs (Fig. 5A, compare lanes 7 and 8) may be due to the presence of endo H-resistant mature gp160 (as observed in Fig. 3C).
Like their WT equivalents, parent SOS trimers completely resisted endo H (Fig. 5B, compare lanes 1 and 5). However, unlike the WT monomer, the SOS monomer was enzyme resistant, as was the UNC SOS monomer (Fig. 5B, compare lanes 1 and 3 to 5 and 7). This suggests that the SOS bridge shields glycans from endo H, a point we return to in more detail below.
As expected, GnTI− SOS Env was relatively sensitive to endo H, due to its HM/HM glycan profile (Fig. 5B, compare lanes 5 and 6). Thus, endo H completely digested GnTI− SOS monomers and induced a drop in size of the GnTI− SOS trimer similar to that observed with the WT (Fig. 5B, lane 6).
Together, the above findings improve our understanding of particulate Env (55). Functional Env from parent cells consists of endo H-resistant gp120/gp41 with a C/HM glycan profile. Uncleaved gp160 is a monomer, largely consisting of endo H-sensitive gp160ER. Trimeric gp41 stumps are endo H resistant, probably because they derive from native trimer shedding. Conversely, monomeric gp41 stumps are endo H sensitive, probably because they largely derive from cleavage of gp160ER. Finally, the SOS bond protects glycans from endo H.
SOS gp160ER monomer exhibits two binding sites for MAb 2G12.
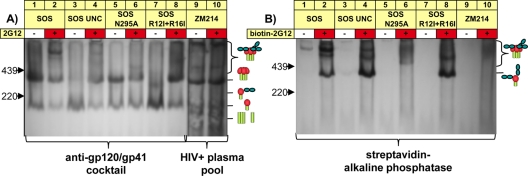
Previously, it was observed that gp120 produced under conditions that prevent complex glycan maturation can bind two molecules of MAb 2G12 (67). This may be because an HM/HM glycan profile creates a second high-mannose glycan cluster in the outer domain that is recognized by 2G12. Perhaps related to this finding, our earlier work suggested that 2G12 induces a supershift of gp120/gp41 monomers by BN-PAGE (Fig. 6A, lane 2) (16), consistent with the idea that the monomer is largely an HM/HM species, i.e., gp160ER, and binds two copies of 2G12. A degree of uncertainty about this interpretation, however, stems from the nearly identical sizes of the unliganded trimer and the 2G12 supershifted monomer. We therefore investigated this point further using two BN-PAGE shift assay formats. In the standard format, we incubated MAb with VLPs, washed away unbound MAb, and probed blots with our standard MAb cocktail. In a second format, VLPs were mixed with biotinylated 2G12, and blots were probed with a streptavidin conjugate.
Fig. 6.
The SOS-VLP Env monomer in BN-PAGE exhibits two 2G12 binding sites. SOS-VLPs, mutants thereof, and ZM214 WT-VLPs produced in parent 293T cells were incubated with or without 2G12 or biotinylated 2G12, as indicated, and then resolved by BN-PAGE and Western blotting. Mutant SOS-VLPs included UNC (K510S R511S), the N295A mutant, and the R12I R16I mutant. Blots were detected with the full anti-gp120 and anti-gp41 cocktail and a clade C HIV+ plasma cocktail (for ZM214) (A) or with streptavidin-alkaline phosphatase (B). Cartoons are as defined in the legend of Fig. 1, with the addition of dark green blobs that represent bound 2G12.
As before, 2G12 induced a supershift in the SOS and UNC SOS monomer, adding an estimated ∼300 kDa in mass and rendering it similar in size to the trimer (Fig. 6A, lanes 1 to 4), indicating the binding of two 2G12 molecules. A band of identical size was observed using biotinylated 2G12 in the second assay format (Fig. 6B, lanes 2 and 4), suggesting that the monomer consists largely of gp160ER, whose HM/HM glycan profile allows it to bind two copies of 2G12.
In further experiments, we examined the dependency of the second 2G12 binding site on the primary site. Two scenarios were considered. First, we ablated the primary 2G12 site with an N295A mutation in JR-FL Env. We also examined a clade C virus that naturally lacks the glycan at residue N295 important for 2G12 binding (9, 33). 2G12 was unable to shift the monomer in either case (Fig. 6A and B, lanes 6 and 10), suggesting that the second 2G12 binding site depends on an intact primary 2G12 binding site. Possibly, the absence of the N295 glycan in the primary site leads to a loss in glycan density, affecting the compactness of the secondary site and therefore 2G12 binding.
Mutation of the Env signal peptide increases gp160 monomer contamination.
The above results suggest that the monomer observed in BN-PAGE consists primarily of gp160ER. This could be related to previous observations that intracellular gp160 trafficking is abnormally inefficient, perhaps due to slow removal of the signal peptide (46, 48). To investigate this possibility, we mutated the gp160 signal peptide to render its removal even more inefficient. If this leads to a further accumulation of monomer on VLPs, it would provide further evidence of a role of inefficient protein trafficking as a basis for junk Env.
Positively charged residues at the N terminus of the signal peptide are crucial for protein trafficking (46, 48). A double knockout mutant (R12I R16I) of the Env signal peptide led to the expression of SOS-VLPs bearing only monomer, similar to UNC SOS (Fig. 6A, compare lanes 3 and 7). Like UNC SOS, this monomer bound to two copies of 2G12 (Fig. 6A and B, compare lanes 4 and 8). This suggests that the naturally slow process of gp160 signal peptide cleavage promotes VLP contamination with junk Env.
Effects of enzyme digests on VLPs as visualized by SDS-PAGE and Western blotting.
The above insights suggested that it might be possible to remove junk Env by enzyme digestion, a possibility supported by our observation that UNC Env is susceptible to V3 cleavage (Fig. 4B, lanes 3, 4, 7, and 8). Protease digests might be more effective if the glycan shell is first partly dismantled by glycosidases. Like monomeric gp120, junk Env is largely endo H sensitive while native Env trimers are largely resistant to glycosidases, except those that target glycan termini such as neuraminidase and mannosidases (51, 64) (Fig. 5).
We initially used SDS-PAGE and Western blotting to investigate enzyme digests using UNC VLPs as substrates essentially consisting of all junk Env (Fig. 7). Denaturing VLPs before digests allowed us to assess the maximum digestive effect. Proteinase K completely digested all denatured WT Env (Fig. 7, lane 3). As shown in Fig. 4, endo H induced a precipitous drop in the size of WT gp160ER but had a milder effect on mature gp160 and no effect on the faint gp41 band (Fig. 7, lane 2). Under native conditions, endo H also digested UNC WT gp160ER efficiently (Fig. 7, compare lanes 2 and 4), suggesting few constraints to digestion. In contrast, SOS gp160ER was incompletely sensitive to endo H (data not shown), consistent with inefficient digestion of the SOS monomer (Fig. 5B, lanes 5 and 7).
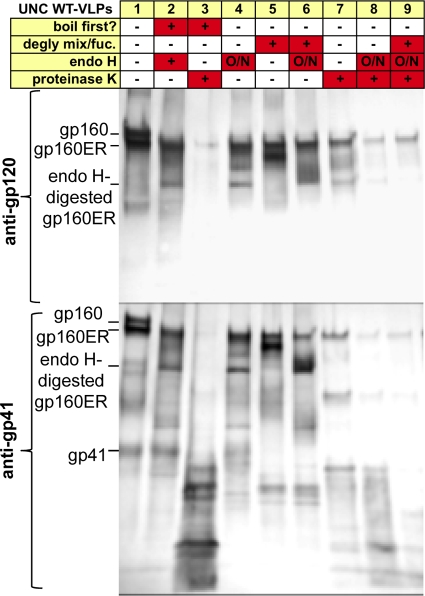
Fig. 7.
SDS-PAGE analysis of glycosidase-protease digests of UNC VLPs. UNC WT-VLPs produced in 293T cells were enzyme digested, as indicated, and then analyzed by SDS-PAGE and Western blotting. Blots were probed separately with anti-gp120 or anti-gp41 cocktails. Digests with endo H were incubated overnight (O/N), those with deglycosylation mix/fucosidase were incubated for 2 h, and proteinase K digests were incubated for 1 h. In some cases, VLPs were boiled for 5 min before digests. Washes were performed between each digest reaction.
We next investigated the effects of a deglycosidase mix containing PNGase F, neuraminidase, β(1–4)-galactosidase, endo-α-N-acetylgalactosaminidase, β-N-acetylglucosaminidase, and fucosidase. As expected, this digested mature gp160 and gp41, consistent with the removal of complex glycans (Fig. 7, lane 5). When the deglycosylation mix and endo H were used together, all gp160 species were digested. This effect can be seen more clearly in the lower blot probed with the anti-gp41 cocktail (Fig. 7, compare lane 6 to lanes 4 and 5).
We next looked at the effects of proteases. Proteinase K alone dramatically depleted both gp160 species and gp41 (Fig. 7, lane 7). This effect was enhanced by endo H priming (Fig. 7, lane 8). Shorter endo H priming incubations were less effective (data not shown). Additional priming by the deglycosylation mix led to improved digestion of gp41 (Fig. 7, compare lanes 7 to 9). Taken together, this suggests that proteinase K clears junk forms of Env, and glycosidase priming augments this effect.
Effects of digests on VLPs visualized by BN-PAGE–Western blotting.
We next investigated the effects of enzyme digests by BN-PAGE. As shown in Fig. 5A, endo H depleted the staining of the WT gp160 monomer, while the trimer and gp41 stumps were unaffected (Fig. 8, compare lanes 1 and 2). Chymotrypsin had less effect on the monomer but caused gp41 trimer stumps to smear (Fig. 8, lane 3). An endo H digest followed by a chymotrypsin digest led to a further gp41 smearing and depletion of the gp160 monomer while the trimer was unaffected (Fig. 8, lane 4). Substitution of chymotrypsin with stronger proteases led to even more gp41 smearing and depletion of gp160 monomer, but native trimers remained intact (Fig. 8, lane 5 and 6). The inclusion of fucosidase had little further effect, save for a slight depletion of the native trimer staining in some lanes (Fig. 8, compare lanes 7 to 9 to lanes 4 to 6). Priming with the deglycosidase mix or PNGase F/neuraminidase depleted all contaminant bands to nearly undetectable levels. However, this came at a price: the native trimer band became rather diffuse (Fig. 8, compare lanes 10 to 15 to lanes 7 to 9), perhaps due to PNGase F-mediated removal of complex glycans from the trimer and/or neuraminidase-mediated removal of sialic acids from complex glycans.
Fig. 8.
BN-PAGE analysis of glycosidase-protease digests of WT and SOS-VLPs. WT-VLPs were treated with various combinations of glycosidases and proteases, as indicated, and then resolved by BN-PAGE and Western blotting. Degly. mix/fuc, deglycosylation mix/fucosidase; NA, neuraminidase; prot. K, proteinase K. Cartoons are as defined in the legend of Fig. 1.
The effects of key digests were next evaluated on SOS-VLPs (Fig. 9). Overall, digests were less effective, consistent with the relative endo H resistance observed above (Fig. 5). Nevertheless, some combinations of endo H with strong proteases were quite effective (Fig. 9, compare lane 1 to lanes 5 to 7). In contrast to WT trimers, endo H digests caused a slight increase in SOS trimer mobility, perhaps due to removal of high-mannose glycans from the silent domain. This did not increase susceptibility to proteases (Fig. 9, lanes 4 to 7). As for WT-VLP digests, deglycosylation mix and PNGase/neuraminidase efficiently depleted junk Env but caused trimer smearing (Fig. 9, lanes 6 and 7).
Fig. 9.
BN-PAGE analysis of glycosidase-protease digests of SOS-VLPs. SOS-VLPs were treated with various combinations of glycosidases and proteases, as indicated, and then resolved by BN-PAGE and Western blotting. Cartoons are as defined in the legend of Fig. 1.
Protease inhibitors protect gp41 stumps from protease digestion.
In Fig. 8 and 9, the controls in lane 1 were neither exposed to enzymes nor incubated for an extended period at 37°C. It is possible that long incubations directly affect Env, perhaps due to the presence of cellular enzymes that copurify with VLPs. Therefore, we performed a control experiment in which an overnight incubation at 37°C was compared to VLPs freshly thawed from −70°C. Furthermore, to confirm the specificity of digests, we tested whether protease inhibitors can protect junk Env. In this experiment, we chose WT VLPs as a substrate and used the modestly effective digest conditions of endo H/chymotrypsin (Fig. 8, lane 4).
A control incubation at 37°C improved the clarity of VLP Env (Fig. 10, lanes 1 and 2). The gp160 monomer was slightly depleted, as were monomeric gp41 stumps. However, the native trimer and trimeric gp41 stumps were largely unaffected. This suggests that cellular proteases copurified with VLPs affect some forms of junk Env. The inclusion of endo H in this overnight digest did not have much additional effect on junk Env (Fig. 10, compare lanes 2 and 3). Thus, the depletion of gp160 monomer observed by endo H digests (Fig. 5, 8, and 9) has more to do with the long incubation itself than the effects of the enzyme. In this particular blot, a marginal increase in trimer mobility was observed with endo H treatment, apparently due to partial removal of high-mannose glycans. An overnight control digest followed by a 1-h chymotrypsin digest increased the mobility of gp41 stumps but had little additional effect on the monomer (Fig. 10, lane 4). The inclusion of endo H in the overnight digest accentuated the effect on gp41 (Fig. 10, lane 5). As shown in Fig. 8, a powerful protease cocktail removed junk Env more effectively (Fig. 10, lane 10). From here on, we refer to this as our standard digest protocol.
Fig. 10.
Reversal of digests using protease inhibitors. WT-VLPs were digested with endo H and/or chymotrypsin in the presence or absence of a protease inhibitor cocktail, as indicated. The reactions used for lanes 2 to 10 all included an overnight incubation at 37°C. In some cases, endo H was added during these incubations. In others, the incubation was essentially a mock treatment with no added endo H. When protease inhibitors were used, they were present for the entire digestion course. Cartoons are as defined in the legend of Fig. 1.
Protease inhibitors (Fig. 10, lanes 6 to 9) protected trimeric gp41 stumps from chymotrypsin (best observed by comparing the matched conditions in lanes 5 and 9). In contrast, the gp160 monomer and monomeric gp41 stumps were not preserved. Thus, gp41 trimers were specifically digested by added proteases while other junk Env was partly removed by traces of cellular enzymes. The latter digestive effects appear to be difficult to inhibit, perhaps because of the inherent sensitivity of the substrate.
Stability of Env trimers after digests.
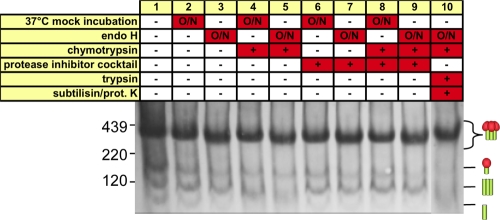
To investigate the effects of digests on trimer stability, we digested WT VLPs using our standard protocol (Fig. 10, lane 10) and subjected them to some of the harsh conditions used in the experiment shown in Fig. 1. WT Env trimers exhibited a resilient profile similar to their undigested counterparts (compare Fig. 11 to 1B). Thus, digestions do not affect trimer stability.
Fig. 11.
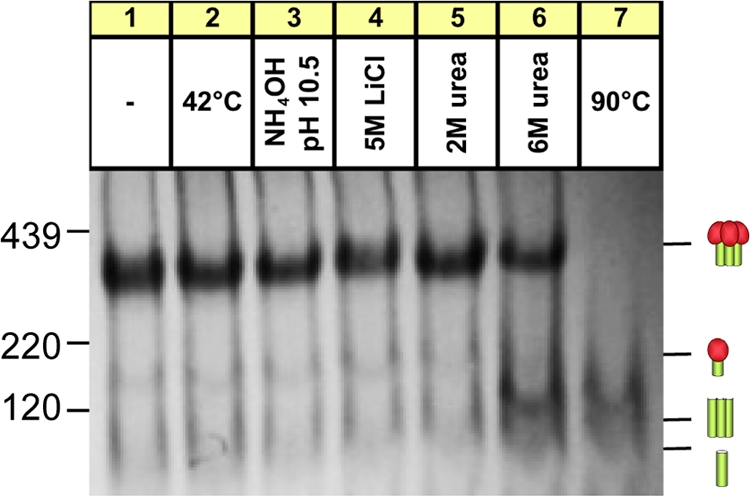
Stability of Env trimers after VLP digests. WT-VLPs were digested with endo H, chymotrypsin, trypsin, subtilisin, and proteinase K (compare Fig. 8, lane 6), and trimer stability was then determined under the harsh conditions indicated. Samples were then analyzed by BN-PAGE and Western blotting and probed with anti-gp120/gp41 MAb cocktails. Cartoons are as defined in the legend of Fig. 1.
Enzyme-mediated removal of junk Env is broadly applicable.
We further investigated the effect of digests on different VLPs using our standard protocol (Fig. 10, lane 10). Digests of WT VLPs substantially depleted junk Env, while the trimer was retained (Fig. 12, lanes 1 and 2). Probing duplicate blots separately with anti-gp120 cocktail or -gp41 cocktail revealed that the remaining junk Env smear consisted entirely of gp41 (data not shown). Digests of GnTI− WT-VLPs and UNC WT-VLPs completely obliterated all forms of Env (Fig. 12, lanes 3 to 6). However, faint traces of trimer remained after digestion of GnTI− SOS-VLPs and parent UNC SOS-VLPs (Fig. 12, lanes 7 to 10). In the latter case, the trimer may have formed from the small amount of cleavage that occurs in the UNC mutant. Digests completely removed the junk from M149A mutant SOS-VLPs (Fig. 12, lane 12), but the trimer survived. Adjusting the contrast of this blot (lower panel) confirms that little, if any, junk Env remains. In contrast, A328G SOS-VLP trimers became a smear after digestion, and junk Env was still detected.
Fig. 12.
Glycosidase-protease digests remove nonfunctional Env from JR-FL-based VLP variants. VLPs produced in parent or GnTI− cells bearing various JR-FL Envs, and mutants were digested with endo H, chymotrypsin, trypsin, subtilisin, and proteinase K and then resolved by BN-PAGE-Western blotting. The UNC SOS mutant used was K510S R511S. The UNC WT mutant used was K500S R503A K510S R511S. The blot was probed with the standard MAb cocktail. Cartoons are as defined in the legend of Fig. 1.
In further experiments, we digested a panel of seven clade B Envs using the standard protocol. As for JR-FL WT-VLPs, the gp160 monomer was removed completely in each case, leaving a gp41 smear (Fig. 13). In all cases, native trimer clearly survived. Clade A and C trimers (KNH144, ZM109, ZM135, ZM197, and ZM214) also survived digests (Fig. 13, lanes 15 and 16; also data not shown). In contrast, recombinant gp120 monomer was fully sensitive to digests (Fig. 13, lanes 19 and 20).
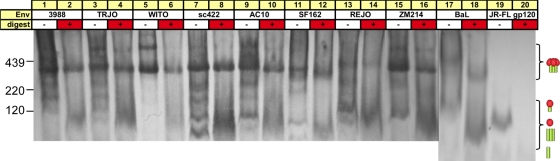
Fig. 13.
Enzymes remove junk Env from various Env VLPs and inactivated virus substrates. VLPs produced in parent cells and bearing clade B or C Envs, inactivated BaL viruses, and recombinant JR-FL gp120 were digested with endo H, chymotrypsin, trypsin, subtilisin, and proteinase K and then resolved by BN-PAGE and Western blotting. Blots were probed with anti-gp120/gp41 MAb cocktails, except ZM214, in which clade B V3 MAbs were substituted for nonclade B V3 MAbs 5.8C, 1.4E, 3074, and 3869. Cartoons are as defined in the legend of Fig. 1.
In several cases, trimers became smaller after digests. This effect was particularly marked for the trimers of inactivated viruses ADA, MN, and BaL grown in T cell lines and, to a lesser extent, AC10 and SF162 (Fig. 13 and data not shown). The most likely explanation is the partial removal of high-mannose glycans by endo H, an effect that may be more pronounced for neutralization-sensitive isolates that may also be more enzyme accessible. In many cases, trimer staining was also weaker, perhaps because the partial removal of high-mannose glycans affects 2G12 detection.
Effect of enzyme digests on viral infectivity.
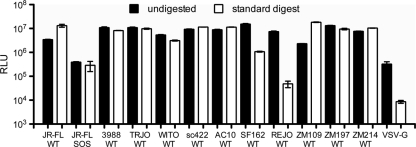
Finally, we assessed the effect of standard digests on viral infectivity. Controls were incubated at 37°C for a matched time period without enzymes. VLPs were then washed and assessed for their ability to infect CF2 CD4 CCR5 cells. The infectivity of the HIV Env VLPs was preserved in the face of digests. WT VLP infection increased modestly (2- to 3-fold) relative to controls, while SOS-VLP infectivity decreased slightly (within 2-fold of controls) (Fig. 14). The infectivities of all but one of the other Env VLPs were preserved after digests. An exception was REJO, whose trimer stained weakly and appeared to be quite sensitive to enzyme digests (Fig. 13). Vesicular stomatitis virus G protein (VSV-G) Env VLP infectivity was also reduced to background levels. Overall, these results are consistent with the preservation of native HIV-1 Env trimer function in the face of digests.
Fig. 14.
Preservation of viral infectivity in the face of enzyme digests. Various VLPs were subjected to mock or standard enzyme treatments, washed with PBS, and then assessed for their ability to infect CF2 CD4 CCR5 cells. RLU, relative light units.
DISCUSSION
The work presented here sheds new light on the nature of junk Env, the mechanisms that underlie its incorporation on HIV-1 particle surfaces, and a method for its removal. The observation that native trimers are stable challenges popular perceptions (Fig. 1 and 2). Previously, junk Env was thought to be a by-product of trimer dissociation, an assumption based partly on early findings that HIV-1 particles shed gp120 (54). However, gp120 shedding of primary field isolates is, in fact, minimal (15). Furthermore, harsh treatments do not cause gp41 stumps to accumulate, consistent with their failure to promote gp120 shedding (Fig. 1 and 2). Our results are more in line with the idea that gp120 shedding occurs during Env synthesis and manifests at the moment Env reaches particle surfaces and does not progress further (15, 23).
We hypothesized that aberrant gp120/gp41 processing at one of the two putative gp120/gp41 cleavage sites might play a role in gp120 shedding but did not find any evidence to support this idea (Fig. 3). Unexpectedly, both sites were important for proper processing (Fig. 3) (10), as has been observed for other viral coat proteins containing multiple cleavage sites (72).
The observation that GnTI− VLPs exhibit few gp41 stumps suggests the intriguing possibility that glycans influence gp120 shedding (Fig. 5A, lane 2). Complex glycans are somewhat heterogeneous in nature, as evidenced by the observation that the Env bands of VLPs expressed in parent 293T cells are more diffuse than those expressed in GnTI− cells (Fig. 4). The increased gp120 shedding from parent VLPs might therefore be due to the maturation of certain complex glycans that decrease gp120-gp41 stability.
To gain further insights into junk Env, we analyzed VLP Env by SDS-PAGE and Western blotting. Unexpectedly, this revealed gp160ER as a major form of junk Env (Fig. 4). The observation that two copies of MAb 2G12 bound to gp160ER (Fig. 6) suggests that the high-mannose glycans of its outer domain are essentially untrimmed, consistent with an early form of Env that is not subject to the normal protein maturation.
There appear to be three possible explanations for the presence of gp160ER on VLPs: (i) gp160ER could populate vesicles that contaminate particles, (ii) gp160ER could be released from dead cells and then coat virions via the hydrophobic gp41 subunit (43), or (iii) gp160ER could bypass maturation and leak directly into particles from the ER. The first possibility is effectively ruled out by observations that infectious virus can be captured by MAbs that are unable to bind to native spikes (55). Regarding the second possibility, we have found that SOS Env in the supernatants of cells transfected with a plasmid (pCAGGS SOS gp160ΔCT) coats particle surfaces very inefficiently, making this an unlikely scenario (data not shown).
The third hypothesis is supported by previous studies that suggested a side pathway in Env synthesis (23, 25). Although some Env reaches the membrane surface within 2 h of initial expression, vast quantities of gp160 accumulate in the ER (23, 41, 46) that may cause ER stress (52), a phenomenon that leads to a breakdown in normal protein processing and apoptosis. This could cause leakage of gp160ER into budding particles, as supported by our observation that slowing down protein processing by mutating the signal peptide enriches high-mannose gp160 monomer on VLPs (Fig. 6). These surprising downstream effects of inefficient processing are not unprecedented: the Ebola virus glycoprotein similarly fails to traffic effectively from the endoplasmic reticulum and may contribute to the cytopathic effects of Ebola infection (3).
In contrast to particulate Env, soluble forms of Env, e.g., gp120 monomer, are not contaminated with HM/HM species similar to gp160ER (24). Various factors may contribute to this difference. The lack of a transmembrane domain could accelerate protein trafficking, as might the widespread use of hybrid signal peptides. Furthermore, structural proteins of the virus may exacerbate the ER stress caused by Env (52), favoring gp160ER contamination.
Recently, ∼98% of particulate Env was found to be of the high-mannose variety (21), contrasting sharply with the ∼30% found on the recombinant gp120 monomer. It was suggested that native trimers largely bear high-mannose glycans, apparently with only one or two complex glycans at most per spike (21). However, evidence developed here and elsewhere challenges this interpretation and instead shows that native trimers expressed in normal parent cells exhibit a C/HM profile, as follows: (i) Env trimers expressed in 293T cells, like recombinant gp120, are partially resistant to endo H, contrasting with the sensitivity of those expressed in GnTI− cells (Fig. 5) (4); (ii) endo H induces a rapid loss in GnTI− virus infectivity but not parent 293T virus infectivity (4); (iii) parent virus is relatively resistant to sCD4 compared to GnTI− virus, consistent with the protective effects of larger complex glycans lining the receptor binding site (4); (iv) neuraminidase enhances parent virus infectivity, consistent with the presence of sialic acids of complex glycans (4, 51); (v) gp41 stumps bear exclusively complex glycans (Fig. 4) (26, 32); and (vi) gp120 released in virus cultures bears a substantial fraction of complex glycans (21, 31). Both of the latter complex glycan-rich products are likely to stem gp120 shedding of native trimers.
Taken together, the above observations overwhelmingly support the view that gp160ER rather than native trimers accounts for the prevalence of high-mannose glycans in the aforementioned study (21). Assuming that the C/HM profile of native trimers resembles that of gp120 (30% high-mannose glycans), the observed bias of 98% high-mannose glycans on particulate Env suggests that gp160ER is in significant excess over native spikes, as supported by the intense staining of gp160ER in SDS-PAGE and Western blotting (Fig. 4).
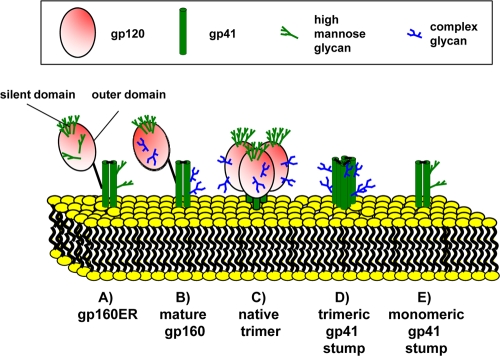
Collectively, our studies provide a clearer picture of particulate Env, key forms of which are depicted in Fig. 15. The most dominant contaminant, gp160ER, is largely or completely monomeric and bears high-mannose glycans on both the outer and silent domains of gp120 (Fig. 15A). Mature gp160 is generated when the glycans of the gp120 outer domain and gp41 mature (Fig. 15B). Cleavage leads to oligomerization, resulting in native trimers (Fig. 15C). A small proportion of these trimers shed gp120 apparently during synthesis, leaving behind trimeric gp41 stumps (Fig. 15D). Some gp120 shedding may also occur before trimerization, resulting in monomeric gp41 stumps (Fig. 15E).
Fig. 15.
Schematic depiction of particulate Env.
The promiscuous antigenic properties of junk Env and its prevalence (especially in the case of gp160ER) may interfere with the development of antitrimer responses. If so, “trimer VLPs” bearing only native trimers may be useful vaccine immunogens. Our improved understanding of junk Env led us to develop enzyme digestion strategies for its removal (J. M. Binley, U.S. patent application 61/360,067). We reasoned that compact trimers might resist enzymes in the same way they resist the binding of nonneutralizing MAbs while the antigenically promiscuous junk Env might be relatively sensitive (16, 18, 55). Indeed, previous studies indicated that trimers resist most glycosidases while soluble forms of Env tend to be sensitive (24, 51, 64). The prior observation that the low-pH form of influenza virus HA1 is trypsin sensitive while its precursor is resistant (70) provided a precedent for the idea that different forms of the same viral coat protein might be differentially sensitive to proteases.
In practice, enzyme digests effectively removed junk Env, leaving native trimers behind. Viral infectivity was largely unaffected, suggesting that digests do not adversely affect trimers (Fig. 14). We were thus able to make pure trimer VLPs in two cases, namely, SOS GnTI− and the M149A mutant (Fig. 12). These successes may simply relate to relatively small amounts of starting monomer that facilitated cleaner digests (Fig. 12). Therefore, similarly “clean” digests of other VLPs may be possible simply by reducing the VLP input. Other improvements may require a better understanding of why some junk Env is enzyme resistant. Incomplete digestion of gp41 stumps was a consistent problem for WT-VLP substrates (Fig. 13). This might be resolved in several ways: (i) PNGase F priming to remove the complex glycans on gp41 stumps, allowing for improved protease digests (Fig. 8); (ii) removal of gp41 glycosylation sites by mutation; (iii) VLP expression in cell lines with modified glycosylation machinery to help prevent the appearance of gp41 stumps (Fig. 5); and (iv) use of the SOS mutation to prevent gp120 shedding. Ultimately, the resistant gp41 smear may not be a problem for immunogenicity studies as the gp160 monomer is probably the main culprit in deflecting B cell responses from the trimer. Thus, upon gp160 removal, NAb responses targeting the gp120 component of trimers may be free to develop without immune interference.
Aside from acting as an immune decoy, junk Env may have other biological consequences. For example, it could account for the antiviral effects of nonneutralizing MAbs that mediate anti-HIV-1 effector functions or neutralize virus in macrophage cultures (27). It could also increase local anti-Env Ab concentrations at the virus surface, affecting neutralization, although preliminary data suggest little effect on VLP neutralization sensitivity (T. Tong, E. T, Crooks, and J. M. Binley, unpublished data).
Previously, VLP immunizations elicited high titers of nonneutralizing anti-gp120 antibodies that were probably due in large part to interference by gp160ER (17). Thus, it will be of interest to evaluate trimer VLPs as immunogens in which this interference is eliminated. Since trimer binding correlates with neutralization (16, 28, 55), any anti-Env responses generated should—at least in theory—be neutralizing.
ACKNOWLEDGMENTS
This work was supported by grants AI58763 and AI84714 (NIH/NIAID/DAIDS), the Bill and Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery (CAVD-VIMC grant 38619), and the Torrey Pines Institute's AIDS and Infectious Disease Science Center.
We thank Rogier Sanders, David Montefiori, John Mascola, Michael Zwick, and Chil-Yong Kang for advice; Jeff Lifson, Julian Bess, and Larry Arthur for sharing inactivated virus preparations; Dennis Burton, James Robinson, Susan Zolla-Pazner, and the NIH AIDS Research and Reference Reagent Repository for providing MAbs; and Glaxo for providing AS01B.
Footnotes
Published ahead of print on 6 April 2011.
REFERENCES
- 1. Abrahamyan L. G., Markosyan R. M., Moore J. P., Cohen F. S., Melikyan G. B. 2003. Human immunodeficiency virus type 1 Env with an intersubunit disulfide bond engages coreceptors but requires bond reduction after engagement to induce fusion. J. Virol. 77:5829–5836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berman P. W., Nunes W. M., Haffar O. K. 1988. Expression of membrane-associated and secreted variants of gp160 of human immunodeficiency virus type 1 in vitro and in continuous cell lines. J. Virol. 62:3135–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bhattacharyya S., Hope T. J. 2011. Full-length Ebola glycoprotein accumulates in the endoplasmic reticulum. Virol. J. 8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Binley J., et al. 2010. Role of complex carbohydrates in human immunodeficiency virus type 1 infection and resistance to antibody neutralization. J. Virol. 84:5637–5655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Binley J. M., et al. 2003. Redox-triggered infection by disulfide-shackled human immunodeficiency virus type 1 pseudovirions. J. Virol. 77:5678–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Binley J. M., et al. 2008. Profiling the specificity of neutralizing antibodies in a large panel of HIV-1 plasmas from subtype B and C chronic infections. J. Virol. 82:11651–11668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Binley J. M., et al. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Binley J. M., et al. 2002. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 76:2606–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Binley J. M., et al. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232–13252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bosch V., Pawlita M. 1990. Mutational analysis of the human immunodeficiency virus type 1 env gene product proteolytic cleavage site. J. Virol. 64:2337–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Burton D. R., et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027 [DOI] [PubMed] [Google Scholar]
- 12. Calarese D. A., et al. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065–2071 [DOI] [PubMed] [Google Scholar]
- 13. Center R. J., et al. 2002. Oligomeric structure of the human immunodeficiency virus type 1 envelope protein on the virion surface. J. Virol. 76:7863–7867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen X., et al. 2005. Pseudovirion particle production by live poxvirus human immunodeficiency virus vaccine vector enhances humoral and cellular immune responses. J. Virol. 79:5537–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chertova E., et al. 2002. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 76:5315–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Crooks E. T., et al. 2008. Relationship of HIV-1 and SIV envelope glycoprotein trimer occupation and neutralization. Virology 377:364–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Crooks E. T., et al. 2007. A comparative immunogenicity study of HIV-1 virus-like particles bearing various forms of envelope proteins, particles bearing no envelope and soluble monomeric gp120. Virology 366:245–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Crooks E. T., et al. 2005. Characterizing anti-HIV monoclonal antibodies and immune sera by defining the mechanism of neutralization. Hum. Antibodies 14:101–113 [PMC free article] [PubMed] [Google Scholar]
- 19. Cutalo J. M., Deterding L. J., Tomer K. B. 2004. Characterization of glycopeptides from HIV-I(SF2) gp120 by liquid chromatography mass spectrometry. J. Am. Soc. Mass Spectrom. 15:1545–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dewar R. L., Vasudevachari M. B., Natarajan V., Salzman N. P. 1989. Biosynthesis and processing of human immunodeficiency virus type 1 envelope glycoproteins: effects of monensin on glycosylation and transport. J. Virol. 63:2452–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Doores K. J., et al. 2010. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc. Natl. Acad. Sci. U. S. A. 107:13800–13805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Earl P. L., Doms R. W., Moss B. 1990. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 87:648–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Earl P. L., Moss B., Doms R. W. 1991. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J. Virol. 65:2047–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eggink D., et al. 2010. Lack of complex N-glycans on HIV-1 envelope glycoproteins preserves protein conformation and entry function. Virology 401:236–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fenouillet E., Gluckman J. C. 1992. Immunological analysis of human immunodeficiency virus type 1 envelope glycoprotein proteolytic cleavage. Virology 187:825–828 [DOI] [PubMed] [Google Scholar]
- 26. Fenouillet E., Miquelis R., Drillien R. 1996. Biological properties of recombinant HIV envelope synthesized in CHO glycosylation-mutant cell lines. Virology 218:224–231 [DOI] [PubMed] [Google Scholar]
- 27. Forthal D. N., Moog C. 2009. Fc receptor-mediated antiviral antibodies. Curr. Opin. HIV AIDS 4:388–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fouts T. R., Binley J. M., Trkola A., Robinson J. E., Moore J. P. 1997. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J. Virol. 71:2779–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Frost S. D., et al. 2005. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc. Natl. Acad. Sci. U. S. A. 102:18514–18519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gagneux P., Varki A. 1999. Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology 9:747–755 [DOI] [PubMed] [Google Scholar]
- 31. Geyer H., Holschbach C., Hunsmann G., Schneider J. 1988. Carbohydrates of human immunodeficiency virus. Structures of oligosaccharides linked to the envelope glycoprotein 120. J. Biol. Chem. 263:11760–11767 [PubMed] [Google Scholar]
- 32. Go E. P., et al. 2008. Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopes' accessibility. J. Proteome Res. 7:1660–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gray E. S., Moore P. L., Pantophlet R. A., Morris L. 2007. N-linked glycan modifications in gp120 of human immunodeficiency virus type 1 subtype C render partial sensitivity to 2G12 antibody neutralization. J. Virol. 81:10769–10776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hammonds J., et al. 2005. Induction of neutralizing antibodies against human immunodeficiency virus type 1 primary isolates by Gag-Env pseudovirion immunization. J. Virol. 79:14804–14814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haynes B. F., Montefiori D. C. 2006. Aiming to induce broadly reactive neutralizing antibody responses with HIV-1 vaccine candidates. Expert Rev. Vaccines 5:347–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hioe C. E., et al. 2010. Anti-V3 monoclonal antibodies display broad neutralizing activities against multiple HIV-1 subtypes. PLoS One 5:e10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kang Y. K., et al. 2009. Structural and immunogenicity studies of a cleaved, stabilized envelope trimer derived from subtype A HIV-1. Vaccine 27:5120–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kieny M. P., et al. 1988. Improved antigenicity of the HIV env protein by cleavage site removal. Protein Eng. 2:219–225 [DOI] [PubMed] [Google Scholar]
- 39. Kornfeld R., Kornfeld S. 1985. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54:631–664 [DOI] [PubMed] [Google Scholar]
- 40. LaCasse R. A., et al. 1999. Fusion-competent vaccines: broad neutralization of primary isolates of HIV. Science 283:357–362 [DOI] [PubMed] [Google Scholar]
- 41. Land A., Zonneveld D., Braakman I. 2003. Folding of HIV-1 envelope glycoprotein involves extensive isomerization of disulfide bonds and conformation-dependent leader peptide cleavage. FASEB J. 17:1058–1067 [DOI] [PubMed] [Google Scholar]
- 42. Larke N., et al. 2007. Combined single-clade candidate HIV-1 vaccines induce T cell responses limited by multiple forms of in vivo immune interference. Eur. J. Immunol. 37:566–577 [DOI] [PubMed] [Google Scholar]
- 43. Leaman D. P., Kinkead H., Zwick M. B. 2010. In-solution virus capture assay helps deconstruct heterogeneous antibody recognition of human immunodeficiency virus type 1. J. Virol. 84:3382–3395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li M., et al. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li M., et al. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J. Virol. 80:11776–11790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li Y., et al. 1996. Effects of inefficient cleavage of the signal sequence of HIV-1 gp 120 on its association with calnexin, folding, and intracellular transport. Proc. Natl. Acad. Sci. U. S. A. 93:9606–9611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li Y., Luo L., Rasool N., Kang C. Y. 1993. Glycosylation is necessary for the correct folding of human immunodeficiency virus gp120 in CD4 binding. J. Virol. 67:584–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li Y., Luo L., Thomas D. Y., Kang C. Y. 2000. The HIV-1 Env protein signal sequence retards its cleavage and down-regulates the glycoprotein folding. Virology 272:417–428 [DOI] [PubMed] [Google Scholar]
- 49. Ling H., Xiao P., Usami O., Hattori T. 2004. Thrombin activates envelope glycoproteins of HIV type 1 and enhances fusion. Microbes Infect. 6:414–420 [DOI] [PubMed] [Google Scholar]
- 50. McCune J. M., et al. 1988. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell 53:55–67 [DOI] [PubMed] [Google Scholar]
- 51. Means R. E., Desrosiers R. C. 2000. Resistance of native, oligomeric envelope on simian immunodeficiency virus to digestion by glycosidases. J. Virol. 74:11181–11190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Michalski C. J., Li Y., Kang C. Y. 2010. Induction of cytopathic effects and apoptosis in Spodoptera frugiperda cells by the HIV-1 Env glycoprotein signal peptide. Virus Genes 41:341–350 [DOI] [PubMed] [Google Scholar]
- 53. Mizuochi T., et al. 1990. Diversity of oligosaccharide structures on the envelope glycoprotein gp120 of human immunodeficiency virus 1 from the lymphoblastoid cell line H9. Presence of complex-type oligosaccharides with bisecting N-acetylglucosamine residues. J. Biol. Chem. 265:8519–8524 [PubMed] [Google Scholar]
- 54. Moore J., McKeating J., Weiss R., Sattentau Q. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250:1139–1142 [DOI] [PubMed] [Google Scholar]
- 55. Moore P. L., et al. 2006. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 80:2515–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nelson J. D., et al. 2007. An affinity-enhanced neutralizing antibody against the membrane-proximal external region of human immunodeficiency virus type 1 gp41 recognizes an epitope between those of 2F5 and 4E10. J. Virol. 81:4033–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nyambi P. N., et al. 1998. Mapping of epitopes exposed on intact human immunodeficiency virus type 1 (HIV-1) virions: a new strategy for studying the immunologic relatedness of HIV-1. J. Virol. 72:9384–9391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pantophlet R., Aguilar-Sino R. O., Wrin T., Cavacini L. A., Burton D. R. 2007. Analysis of the neutralization breadth of the anti-V3 antibody F425-B4e8 and re-assessment of its epitope fine specificity by scanning mutagenesis. Virology 364:441–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Phogat S., Wyatt R. T., Karlsson Hedestam G. B. 2007. Inhibition of HIV-1 entry by antibodies: potential viral and cellular targets. J. Intern. Med. 262:26–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Poignard P., et al. 2003. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J. Virol. 77:353–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Poon B., et al. 2005. Induction of humoral immune responses following vaccination with envelope-containing, formaldehyde-treated, thermally inactivated human immunodeficiency virus type 1. J. Virol. 79:4927–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Provine N. M., Puryear W. B., Wu X., Overbaugh J., Haigwood N. L. 2009. The infectious molecular clone and pseudotyped virus models of human immunodeficiency virus type 1 exhibit significant differences in virion composition with only moderate differences in infectivity and inhibition sensitivity. J. Virol. 83:9002–9007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rerks-Ngarm S., et al. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209–2220 [DOI] [PubMed] [Google Scholar]
- 64. Sanders R. W., et al. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sanders R. W., et al. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 76:8875–8889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Scanlan C. N., et al. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306–7321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Scanlan C. N., et al. 2007. Inhibition of mammalian glycan biosynthesis produces non-self antigens for a broadly neutralising, HIV-1 specific antibody. J. Mol. Biol. 372:16–22 [DOI] [PubMed] [Google Scholar]
- 68. Schief W. R., Ban Y. E., Stamatatos L. 2009. Challenges for structure-based HIV vaccine design. Curr. Opin. HIV AIDS 4:431–440 [DOI] [PubMed] [Google Scholar]
- 69. Schulke N., et al. 2002. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J. Virol. 76:7760–7776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Skehel J. J., et al. 1982. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 79:968–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Stein B. S., Engleman E. G. 1990. Intracellular processing of the gp160 HIV-1 envelope precursor. Endoproteolytic cleavage occurs in a cis or medial compartment of the Golgi complex. J. Biol. Chem. 265:2640–2649 [PubMed] [Google Scholar]
- 72. Thomas G. 2002. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 3:753–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tobin G. J., et al. 2008. Deceptive imprinting and immune refocusing in vaccine design. Vaccine 26:6189–6199 [DOI] [PubMed] [Google Scholar]
- 74. Tomaras G. D., et al. 2008. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 82:12449–12463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Trkola A., et al. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vidor E. 2007. The nature and consequences of intra- and inter-vaccine interference. J. Comp. Pathol. 137(Suppl. 1):S62–S66 [DOI] [PubMed] [Google Scholar]
- 77. Vzorov A. N., Lea-Fox D., Compans R. W. 1999. Immunogenicity of full-length and truncated SIV envelope proteins. Viral Immunol. 12:205–215 [DOI] [PubMed] [Google Scholar]
- 78. Walker L. M., Burton D. R. 2010. Rational antibody-based HIV-1 vaccine design: current approaches and future directions. Curr. Opin. Immunol. 22:358–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang X., et al. 2010. The nature and combination of subunits used in epitope-based Schistosoma japonicum vaccine formulations affect their efficacy. Parasit. Vectors 3:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Willey R. L., Bonifacino J. S., Potts B. J., Martin M. A., Klausner R. D. 1988. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc. Natl. Acad. Sci. U. S. A. 85:9580–9584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Willey R. L., Klimkait T., Frucht D. M., Bonifacino J. S., Martin M. A. 1991. Mutations within the human immunodeficiency virus type 1 gp160 envelope glycoprotein alter its intracellular transport and processing. Virology 184:319–329 [DOI] [PubMed] [Google Scholar]
- 82. Wu X., et al. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Xiang S. H., Doka N., Choudhary R. K., Sodroski J., Robinson J. E. 2002. Characterization of CD4-induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res. Hum. Retroviruses 18:1207–1217 [DOI] [PubMed] [Google Scholar]
- 84. Zhu X., Borchers C., Bienstock R. J., Tomer K. B. 2000. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry 39:11194–11204 [DOI] [PubMed] [Google Scholar]