Abstract
Anterograde transport of herpes simplex virus (HSV) from neuronal cell bodies into, and down, axons is a fundamentally important process for spread to other hosts. Different techniques for imaging HSV in axons have produced two models for how virus particles are transported in axons. In the Separate model, viral nucleocapsids devoid of the viral envelope and membrane glycoproteins are transported in axons. In the Married model, enveloped HSV particles (with the viral glycoproteins) encased within membrane vesicles are transported in the anterograde direction. Earlier studies of HSV-infected human neurons involving electron microscopy (EM) and immunofluorescence staining of glycoproteins and capsids supported the Separate model. However, more-recent live-cell imaging of rat, chicken, and mouse neurons produced evidence supporting the Married model. In a recent EM study, a mixture of Married (75%) and Separate (25%) HSV particles was observed. Here, we studied an HSV recombinant expressing a fluorescent form of the viral glycoprotein gB and a fluorescent capsid protein (VP26), observing that human SK-N-SH neurons contained both Separate (the majority) and Married particles. Live-cell imaging of rat superior cervical ganglion (SCG) neuronal axons in a chamber system (which oriented the axons) also produced evidence of Separate and Married particles. Together, our results suggest that one can observe anterograde transport of both HSV capsids and enveloped virus particles depending on which neurons are cultured and how the neurons are imaged.
INTRODUCTION
Herpes simplex virus (HSV) and other alphaherpesviruses establish latency in the sensory nervous system. Periodic reactivation leads to the production of infectious virus in sensory ganglia, followed by virus transport in neuronal axons to epithelial tissues. Repetitive infection of the cornea causes scarring, which represents the major infectious cause of blindness. Anterograde transport (from neuronal cell bodies to axon termini) is a fundamentally important property of alphaherpesviruses, essential for long-term survival in the form of spread to other hosts. Early studies of HSV infection in human fetal neurons involving electron microscopy (EM), immuno-EM, and immunofluorescence analyses led to the conclusion that capsids are transported in the anterograde direction separately from vesicles containing viral glycoproteins (9, 17). More-recent studies from the same laboratory showed that there was virus envelopment (assembly of capsids with glycoproteins) at relatively numerous varicosities and at growth cones in cultured human neurons (18). Studies in our laboratory also supported what we termed the “Separate” model for HSV anterograde transport, i.e., transport of unenveloped capsids separately from viral glycoproteins (20–22). In this model, envelopment occurs at axon termini. In human neuroblastoma (SK-N-SH) cells differentiated to produce neurites, HSV glycoproteins stained with a panel of different antibodies (against gB, gD, gE, or gI) were observed as puncta that were separate from capsids stained with different antibodies specific to capsid proteins (20–22).
Early studies of the anterograde transport of the porcine alphaherpesvirus pseudorabies virus (PRV) involving fixed, antibody-stained rat superior cervical ganglion (SCG) or chicken dorsal route ganglion (DRG) neurons supported Separate transport of capsids and glycoproteins (19, 26). However, subsequent studies involving EM of rat neurons (4) and live-cell analyses of a “two-color” recombinant PRV expressing both a fluorescent small capsid protein (VP26-monomeric red fluorescent protein [mRFP]) and glycoprotein gD fused to green fluorescent protein (gD-GFP) in chick neurons (2) concluded that enveloped PRV particles are transported in the anterograde direction in neurons. In this so-called “Married” model, envelopment occurs in neuron cell bodies, and enveloped particles within vesicles are transported in axons. This model was also supported by EM studies showing numerous PRV enveloped capsids in axons (13). Thus, it has been suggested that PRV and HSV might differ in the mechanisms by which capsids are transported in neuronal axons (6, 13, 20, 21), with PRV using the Married mechanism and HSV using the Separate mechanism. Given the fundamentally important nature of alphaherpesvirus transport in neurons, the notion that PRV and HSV differ in this process was surprising.
More-recent EM studies of HSV-infected neurons have produced other results. In one study of HSV-infected rat SCG neurons, 25% of capsids were Separate and 75% Married (15). A second study reported only Married HSV particles in rat neuronal axons (10). We note that one of the earlier EM studies involving HSV-infected human neurons described a fraction of the total particles that were Married (2 of a total of 15 particles in axons distant from varicosities) (18).
There are difficulties with the interpretation of both the antibody-staining and the EM experiments described above. First, fixed neurons do not produce information on whether capsids or glycoproteins are being actively transported in the anterograde direction versus the retrograde direction or are stalled. This is especially problematic if capsids that are imaged are assumed to be undergoing anterograde transport but instead have entered the neuron following “secondary infection.” Secondary infection will be defined here as a virus, derived from another neuron or a nonneuronal cell, that reaches extracellular spaces and then enters the neuron under study. On entry into the neuron under study, there is deenvelopment, producing a capsid (free of glycoproteins) that moves in the retrograde direction in axons, toward the nucleus (1). Second, most EM studies have reported the presence of relatively few HSV particles in axons, compared with EM studies of PRV (13, 15, 17, 18), making interpretation of the results difficult.
Investigations into HSV anterograde transport have also involved live-cell analyses of HSV recombinants expressing either the capsid protein VP26 fused to GFP or gD fused to yellow fluorescent protein (YFP), designated “one-color” viruses here (21, 22). These studies involved primarily SK-N-SH neuroblastoma cells, although we note that the central conclusions were also confirmed in rat trigeminal ganglion (TG) neurons. With these one-color recombinants expressing VP26-GFP (22) or VP26-mCherry capsids (A. Snyder, unpublished data), it was possible to image more capsids than the EM studies did and to measure the kinetics of transport. However, it was necessary to fix and stain neurons to assess the colocalization of capsids with glycoproteins, leading to criticisms that those puncta analyzed for colocalization might not be moving. There are several arguments against the notion that the capsids characterized in these studies were produced by secondary infection (and thus were moving retrogradely). First, we used low multiplicities of infection (MOIs) (1 to 2 PFU/neuron) (20–22), in contrast to other studies involving PRV and HSV, in which high MOIs (100 to 1,000 PFU/neuron) were apparently used (1, 12, 19). Second, there was marked accumulation of HSV capsids at axon hillocks from 12 to 17 h after infection, followed by a burst of capsid transport into and down axons with both SK-N-SH cells and rat TG neurons (22), as had been previously reported with human DRG neurons (14). The kinetics of this capsid accumulation followed by transport into axons was not consistent with secondary infection. Third, studies of HSV VP26-GFP-labeled capsids clearly showed anterograde transport of a significant fraction of capsids (22).
Recently, Antinone et al. (3) constructed a “two-color” HSV recombinant expressing both a fluorescent capsid protein (VP26-mRFP) and a fluorescent glycoprotein (GFP-gB) that allowed imaging of capsids and a glycoprotein. Infection of chicken and rat neurons and mouse neuroblastoma cells produced evidence that 64 to 70% of capsids were in the Married form while undergoing anterograde transport (3). However, no GFP-gB signal was detected in 42% of extracellular virus particles produced by their HSV recombinant (particles that should have an envelope). They concluded that all or most anterograde transport of HSV involves enveloped (Married) particles. Here, we constructed a similar HSV recombinant expressing mRFP-gB and VP26-Venus (a variant form of the enhanced yellow fluorescent protein known as VenusA260K). Imaging of human SK-N-SH neurons and rat neurons in isolator chambers produced evidence of both Married and Separate anterograde transport.
MATERIALS AND METHODS
Cells and viruses.
Vero cells were propagated in Dulbecco's minimum essential medium (DMEM) (Invitrogen) with 7 to 8% fetal bovine serum (FBS) (ISC BioExpress, Kaysville, UT). HSV recombinants were propagated, and their titers determined, on Vero cells.
Construction of an HSV recombinant expressing mRFP-gB and VP26-Venus.
To construct a protein in which HSV-1 gB sequences were fused to monomeric red fluorescent protein, approaches similar to those used to construct a gB fusion with enhanced cyan fluorescent protein (ECFP) were used (23). A plasmid designated pBS-mRFP1-UL27, containing gB sequences fused to mRFP1 coding sequences (a generous gift from R. Y. Tsien), was generated by an in-frame insertion of the mRFP1 coding sequences into a NotI site of the UL27 gene in pBS-UL27. This placed the mRFP at the N terminus of gB between the signal sequence and the N terminus of mature gB. A recombinant HSV, YK613, that expressed the gB fusion protein was derived by transfecting rabbit skin cells with intact YK304 viral DNA (wild-type HSV-1 strain DNA derived from a bacterial artificial chromosome [BAC]) and plasmid pBS-mRFP1-UL27 as described previously (23, 24). A second recombinant, YK614, which expresses mRFP-gB and VP26-Venus, was produced by coinfecting Vero cells with YK613 and another HSV recombinant, YK60, expressing VP26-VenusA206K, which was described previously (23). YK614 was selected by screening for viruses that expressed both mRFP and Venus fluorescence as described previously (23). The presence of mRFP-gB and VP26-Venus sequences in the HSV recombinant YK614 was confirmed by PCR amplification and sequencing.
Extracellular HSV particles.
Vero cells were infected with YK614 (using 5 PFU/cell). After 22 h, cell culture supernatants were harvested and cell debris removed by low-speed centrifugation at 1,000 × g for 15 min, followed by 10,000 × g for 5 min. These virus particles were adsorbed onto glass coverslips that had been coated with poly-d-lysine (30 μg/ml) and laminin (2 μg/ml) for 16 h at 37°C.
Neuron cell cultures.
SK-N-SH neuroblastoma cells (American Type Culture Collection) were propagated as described previously (20–22). A separate line of these cells was derived in our laboratory by repeated, selective removal of a fraction of cells from dishes using brief incubation with 100 mM Na citrate buffer, pH 7.3. Rat superior cervical ganglion (SCG) neurons were prepared from rat (embryonic day 18 [E18]) embryos as described previously (12). These neurons (10 to 40,000 cells) were plated in microfluidic chambers (23 mm diameter; Xona Microfluidics, Temecula, CA), consisting of small plastic chambers connected by 10-μm-wide microgrooves or channels (12, 16). These chambers were mounted in poly-d-lysine- and laminin-coated glass-bottom 35-mm-diameter dishes (WillCo). Neurons were plated in the somal chamber; they extended axons into the channels and subsequently into the adjacent axonal chamber. The axons in channels can be imaged from below. These cells were incubated in growth medium for 7 to 14 days of culture and were then infected with HSV using 3 to 12 PFU/neuron (based on the number of cells originally plated) by introducing the virus into the somal chamber.
Imaging of virus particles adhering to glass coverslips and within SK-N-SH neurites.
YK614 extracellular particles adsorbed onto glass coverslips were fixed with 4% paraformaldehyde and were permeabilized with 0.2% Triton X-100 as described previously (22). The particles were then stained with various antibodies, followed by secondary fluorescent antibodies as described previously (20–22). SK-N-SH neurons were plated onto Lab-Tek Permanox chamber slides (Nalge Nunc) coated with poly-d-lysine (30 μg/ml; Sigma) and laminin (2 μg/ml; Sigma) or rat tail collagen (5 μg/ml; Roche) and were differentiated using retinoic acid as described previously (20). After 10 to 14 days, approximately 60 to 70% of the cells exhibited neurites, and the cells were infected with YK614 (1 to 3 PFU/cell) for 19 to 23 h; then longer neurites that were not in association with other cells were imaged. Images were acquired on a high-resolution wide-field DeltaVision Core system (Applied Precision) utilizing an Olympus IX71 inverted microscope with a proprietary XYZ stage enclosed in a controlled environment chamber, transmitted-light differential interference contrast (DIC), and a solid-state module for fluorescence. A Nikon CoolSNAP ES2 HQ camera was used to acquire images as optical axis images in a 512-by-512 BIN of 2 format with a 60× (numerical aperture, 1.42) Plan Apo N objective in 2 colors, Venus and mRFP. The images were deconvolved using softWorX Explorer software with the appropriate optical transfer function (OTF) using an iterative algorithm of 10 iterations. Histograms were optimized for the most positive image and were applied to all the other images from that experiment before the images were saved as 24-bit merged tagged-image format files (TIFF).
Live-cell imaging of rat SCG axons in microfluidic chambers.
Rat neurons in microfluidic chambers were infected with YK614 using 3 to 12 PFU/cell for 19 to 23 h and were then imaged while at 37°C under 6% CO2 in an enclosed chamber using an inverted objective and the deconvolution system described above. Axons were observed for 1 to 2.5 min using frames of 0.5 s. Again, images were deconvolved using softWorX Explorer.
Radiolabeling, immunoprecipitation, and gel electrophoresis of viral proteins.
Vero cells were infected with various HSV strains for 5 to 6 h. Then the cells were washed with DMEM lacking methionine and cysteine and were incubated in this medium containing [35S]methionine-cysteine (50 μCi/ml) for 2 to 3 h. Cell extracts were made using Nonidet P-40 (NP-40)–deoxycholate (DOC) buffer, comprising 50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate containing 2 mg/ml bovine serum albumin (BSA), and 0.5% phenylmethylsulfonyl fluoride (PMSF). Radiolabeled cell extracts were centrifuged at 40,000 rpm in a Beckman benchtop ultracentrifuge for 40 min to remove insoluble proteins and were then mixed with the gB-specific monoclonal antibody (MAb) 15βB2 or GFP-specific rabbit antibodies. Then protein A-Sepharose was added for an additional 90 min as described previously (27). The protein A-Sepharose was washed 3 times with NP-40–DOC buffer; proteins were eluted by boiling in buffer containing 2% sodium dodecyl sulfate and 2% β-mercaptoethanol; and the mixture was then subjected to electrophoresis using polyacrylamide gels as described previously (27).
RESULTS
Construction and characterization of an HSV recombinant expressing mRFP-gB and VP26-Venus.
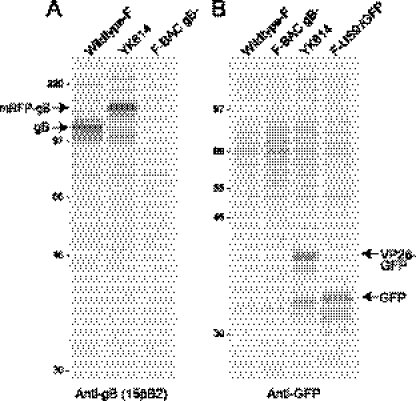
To address the mode of transport of HSV further, we constructed an HSV expressing a gB protein fused to monomeric red fluorescent protein (designated mRFP-gB) and VenusA260K fused to the C terminus of the small capsid protein VP26 (designated VP26-Venus). The construction of the gB fusion protein was similar to that of gB fused to monomeric enhanced cyan fluorescent protein (ECFP), which was described previously (23). This mRFP-gB construct was recombined into wild-type HSV-1 strain F DNA, producing recombinant virus YK613, as described in Materials and Methods. A recombinant expressing both mRFP-gB and VP26-Venus (23), designated YK614, was constructed by coinfecting Vero cells with YK613 and another HSV recombinant, YK601, which expresses VP26-VenusA206K (23), and screening for viruses that expressed both mRFP and Venus fluorescence as described previously (23). The presence of mRFP-gB and VP26-Venus sequences was confirmed by PCR amplification and sequencing. In addition, VP26 and gB were immunoprecipitated from YK614-infected Vero cells that had been radiolabeled with [35S]methionine-cysteine. YK614 expressed a gB glycoprotein that was substantially larger than the gB glycoprotein expressed by wild type HSV-1 and had the size expected given the fusion of gB with mRFP (Fig. 1A). YK614-infected cells also expressed a protein that was immunoprecipitated with GFP-specific antibodies (which cross-react with the Venus protein) and had the size expected for a VP26-Venus fusion protein (Fig. 1B).
Fig. 1.
Immunoprecipitation of mRFP-gB and VP26-Venus from YK614-infected cells. Vero cells were infected with either wild-type HSV-1 strain F, YK614 (carrying the mRFP-gB and VP26-Venus sequences), F-BAC gB−, which does not express gB (7), or F-US9−/GFP, which expresses GFP (21, 22). After 6 h, the cells were radiolabeled with [35S]methionine-cysteine for 150 min; then the cells were lysed in NP-40–DOC buffer containing 2 mg/ml BSA and 1 mM PMSF. Extracts were centrifuged at high speed to remove insoluble debris and were then mixed with anti-gB monoclonal antibody (MAb) 15βB2 (A) or with rabbit anti-GFP antibodies (B). Then protein A-Sepharose was added, and gB or GFP was immunoprecipitated as described previously (27). The positions of gB, mRFP-gB, GFP, and VP26-GFP, as well as those of molecular mass markers of 220, 97, 66, 46, and 30 kDa, are indicated.
The replication of YK614 (mRFP-gB, VP26-Venus), YK613 (mRFP-gB), and YK304 (wild-type HSV-1 derived from a BAC) was compared using rat SCG neurons. The neurons were infected using 5 PFU/cell, and the combined cells and cell culture supernatants were harvested after 2, 15, 10, and 26 h. Summarizing these data, YK614 produced ≈10-fold-less infectious virus than YK613 and wild-type HSV after 26 h (not shown). Previously, we found that replication of an HSV expressing VP26-GFP produced ≈10-fold-less infectious virus in neurons than wild type HSV-1, and many fewer capsids were transported into axons (22). Thus, the fusion of GFP or Venus to VP26 had substantial effects on virus replication and capsid transport, while the fusion of mRFP to gB had less of an effect on virus replication.
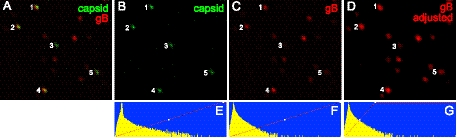
There can be variability in the incorporation of certain glycoproteins fused to fluorescent proteins into the virion envelope. As many as 15% of PRV extracellular particles did not contain detectable gD-GFP (2), and 42% of HSV extracellular particles contained no detectable gB-GFP (3). The vast majority of extracellular PRV and HSV particles are enveloped, and thus, it has been concluded that there can be reduced incorporation of glycoprotein fusion proteins into the virion envelope. We also noted some proteolytic cleavage of gD-YFP in previous studies, although 95% of those puncta that stained with anti-gD polyclonal antibodies also exhibited gD-YFP fluorescence (20). In the present studies, there was no evidence of proteolysis of mRFP-gB (Fig. 1A). We prepared YK614 extracellular particles from Vero cells, harvesting cell culture supernatants at 22 h, removing cell debris at low speed (1,000 × g for 15 min, followed by 10,000 × g for 5 min), and then allowing these HSV particles to adhere to glass coverslips coated with poly-d-lysine and laminin. The particles were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and imaged using an Olympus inverted wide-field microscope with a solid-state module for fluorescence, and images were deconvolved. As in previous studies, there were relatively numerous mRFP-gB puncta that did not exhibit VP26-Venus fluorescence, consistent with L particles or membrane blebs (Fig. 2). However, most of the VP26-Venus-labeled capsids (green) exhibited mRFP-gB fluorescence. As before, there was variability in the amounts of mRFP-gB fluorescence associated with different capsids. In Fig. 2, the particles numbered 1, 2, and 4 displayed relatively strong mRFP-gB fluorescence (Fig. 2C). Particle number 5 showed lower mRFP-gB fluorescence (Fig. 2C) and appeared as an entirely green particle in Fig. 2A. Particle number 3 had no detectable mRFP-gB fluorescence in Fig. 2C. However, it was possible to enhance the mRFP-gB signal by increasing the minimum fluorescent intensity detected and thus to detect signals from particles 3 and 5 (Fig. 2D). With enhancement of the mRFP-gB signal, we could detect gB associated with 80% of VP26-Venus capsids (Table 1). The differences between these data and previous studies involving a similar HSV recombinant expressing VP26-mRFP and GFP-gB in which 42% of capsids contained no detectable gB (1) may reflect our use of solid-state illumination in deconvolution microscopy, which may surpass standard wide-field microscopy (1), although detection systems also influence the capacity to detect low levels of fluorescence.
Fig. 2.
Characterization of YK614 extracellular particles. Vero cells were infected for 22 h with YK614; then cell culture supernatants were harvested, and cell debris was removed by low-speed centrifugation at 1,000 × g for 15 min, followed by 10,000 × g for 5 min. These supernatants were incubated with polylysine- and laminin-coated glass coverslips for 16 h in a humidified incubator at 37°C. Virus particles that stuck to coverslips were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and then imaged by deconvolution microscopy as described previously (22). (A) mRFP-gB fluorescence superimposed on VP26-Venus fluorescence. (B and C) VP26-Venus and mRFP-gB fluorescence, respectively. (D) mRFP-gB fluorescence enhanced by raising the minimum intensity that was assigned maximum fluorescence, so that the red puncta numbered 3 and 5 were more intense than those in panel C. (E to G) Histograms of the number of pixels on the y axis versus the relative pixel intensity on the x axis for panels B to D. Panel G differs from panel F in that any pixel to the right of the diagonal red line has been assigned maximum intensity. VP26-Venus puncta 1 to 5 are labeled in all the images.
Table 1.
Colocalization of fluorescence in YK614 virus particles from cell culture supernatants of Vero cellsa
| Fluorescence | Colocalizationb |
|---|---|
| mRFP-gB | 221/276 (80) |
| R#68 (rabbit polyclonal anti-gB) | 152/210 (72) |
| DL6 (mouse anti-gD MAb) | 165/211 (78) |
| 3B6 and H1.4 (anti-VP5 MAb) | 222/263 (84) |
| NC-1 (rabbit polyclonal anti-VP5) | 197/225 (88) |
| NC-7 (rabbit polyclonal anti-VP26) | 178/198 (90) |
YK614 (expressing both mRFP-gB and VP26-Venus) particles from cell culture supernatants were allowed to adhere to polylysine- and laminin-coated glass coverslips; then they were fixed with 4% PFA and permeabilized with 0.2% Triton X-100 before being imaged directly (VP26-Venus versus mRFP-gB) or stained with various antibodies.
Expressed as the number of capsids detected by VP26-Venus fluorescence that exhibited mRFP-gB fluorescence or stained with the anti-gB, anti-gD, anti-VP5, or anti-VP26 antibodies/total number of capsids detected by VP26-Venus fluorescence (percentage).
Previously we characterized the transport of wild-type HSV capsids using two anti-VP5 (large capsid protein) MAbs: 3B6 and H1.4 (21, 22). Antinone et al. (3) used a mixture of these two antibodies under similar conditions and were unable to recognize approximately half of the extracellular capsids released from Vero cells. They suggested that our studies might have missed Married particles in axons, because these MAbs were less able to access capsid epitopes in enveloped particles. Here, we addressed this concern by staining YK614 extracellular particles with various anticapsid and antiglycoprotein antibodies. Capsids adsorbed onto polylysine- and laminin-coated glass coverslips, fixed with 4% PFA, and permeabilized with 0.2% Triton X-100 were stained with antibodies as described previously (22). Under these conditions, MAbs 3B6 and H1.4 recognized 84% of VP26-Venus-labeled capsids (Table 1). Rabbit NC-1 sera, specific for VP5 (5), and NC-7 sera, specific for VP26 (5), recognized 88% and 90% of capsids, respectively. Rabbit polyclonal antibodies specific for gB recognized 72% of these particles, and a mouse MAb specific to gD recognized 78% of the particles. While the two VP5-specific MAbs produced lighter staining than the polyclonal antibodies, there was clear staining with these VP5 MAbs, so that masking of epitopes in enveloped particles, in our experiments, was not likely. More likely, the differences between our results and those of Antinone et al. relate to the instability of the anti-VP5 MAbs (these antibodies appear to be prone to denaturation during freeze-thaw cycles) or to our use of more-sensitive means to detect fluorescence.
Analyses of anterograde transport of capsids and glycoproteins in SK-N-SH neuroblastoma cells.
Many of our previous studies of HSV anterograde transport involved SK-N-SH neuroblastoma cells differentiated with retinoic acid to produce neurites (20–22). We note that our SK-N-SH cultures are different from those of the American Type Culture Collection because we optimized for cells that produce more-extensive and longer neurites and that transport HSV capsids well. This involved repeated selection of a fraction of cells from dishes by incubation with 100 mM Na citrate buffer, pH 7.3. Following differentiation, 60 to 80% of these SK-N-SH cells produced neurites. It is theoretically possible that nonneuronal cells present in these cultures after differentiation could produce HSV progeny causing secondary infection of neuronal cells, so that there is retrograde transport in neurites. To reduce the level of secondary infection, SK-N-SH cells were plated at less dense concentrations in the presence of neutralizing antibodies and were infected at low MOIs (1 to 2 PFU/cell). In addition, HSV capsids accumulated in axon hillocks until about 16 to 19 h and were subsequently moved into axons, and imaging of VP26-GFP capsids was consistent with substantial anterograde spread. These properties were not consistent with secondary infection.
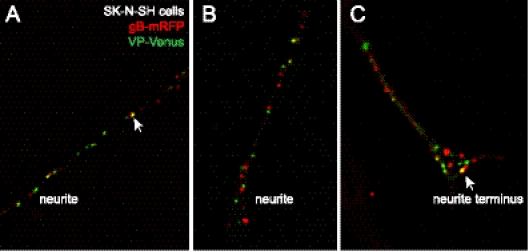
SK-N-SH neurons infected with YK614 for 19 to 23 h were fixed with 4% PFA, and neurites were examined by deconvolution microscopy (20–22). The majority of puncta corresponding to capsids (VP26-Venus) (green) were not colocalized with detectable mRFP-gB (red) in SK-N-SH neurites (Fig. 3A and B). However, there was clear colocalization of VP26-Venus and mRFP-gB fluorescence (involving concentric fluorescence) in some cases (arrow in Fig. 3A). This imaging involved levels of fluorescence enhancement similar to those used for Fig. 2D, so it was unlikely that we were unable to detect mRFP-gB, if present, in more than 20% of the capsids. Enumerating more than 150 VP26-Venus puncta, we observed more than 80% that did not contain detectable mRFP-gB fluorescence. Differentiated SK-N-SH neurons produced flaring at neurite termini, similar to growth cones, and there was more often colocalization of capsids and gB there (arrow in Fig. 3C). Presumably this colocalization corresponds to secondary envelopment.
Fig. 3.
Imaging of capsids and gB in SK-N-SH neurons infected with YK614. SK-N-SH neuroblastoma cells were differentiated to produce neurites as described previously (22). The cells were infected with YK614 using 1 to 2 PFU/cell; then, after 19 to 23 h, the cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and imaged by deconvolution microscopy (22). Red fluorescence (mRFP-gB) and green fluorescence (VP26-Venus) are superimposed. (A and B) Neurites distal from cell bodies. (C) Neurite with a flared axon terminus similar to a neuronal growth cone. The arrows in panels A and C point to puncta with colocalization of mRFP-gB and VP26-Venus fluorescence.
Live-cell analyses of capsids transported in YK614-infected SK-N-SH neurites proved to be difficult for a number of reasons. First, there were relatively low numbers of capsids in neurites, as was noted by Antinone et al. with a similar HSV recombinant (3). Second, a relatively high fraction of the YK614 capsids did not move or moved relatively short distances during the period of observation (1 to 3 min). There were examples of both Married and Separate capsid transport in these live-cell analyses. However, these problems in live-cell analyses and the possibility of secondary infection led us to move these studies into rat neurons growing in microfluidic chambers.
Analyses of anterograde transport in rat neurons using microfluidic chambers.
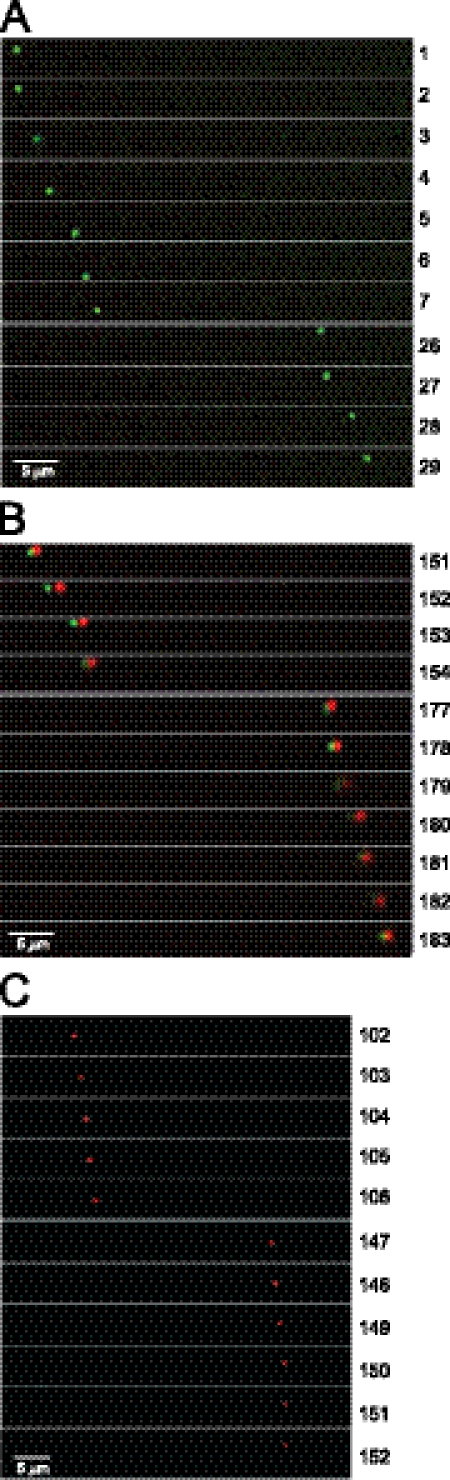
Liu et al. described microfluidic chambers that were used to characterize anterograde spread of PRV (12). These are small plastic chambers connected by 10-μm-wide microgrooves or channels (16). Neurons plated in the somal chamber extend axons into the channels and subsequently into the adjacent axonal chamber. Hydrostatic pressure between the somal and axonal chambers prevents movement of fluids or virus between chambers, isolating the axons and reducing secondary infection. We prepared rat SCG neurons as described previously (12) and plated 10 to 40,000 cells in somal chambers mounted in poly-d-lysine- and laminin-coated glass-bottom 35-mm-diameter dishes. After 7 to 14 days of culture, the cells were infected with YK614 using 3 to 12 PFU/neuron (based on the number of cells originally plated) by introducing virus into the somal chamber. After 19 to 23 h, neuronal axons within the channels were imaged from below using live-cell deconvolution microscopy. Puncta corresponding to mRFP-gB (red) and capsids (green) were followed over the course of 1 to 2.5 min. We observed three types of particles that moved in the anterograde direction. First, there were particles that exhibited VP26-Venus (green) fluorescence with no detectable mRFP-gB (red) fluorescence (Fig. 4A). Second, there were Married capsids, i.e., VP26-Venus capsids and mRFP-gB nearly superimposed on one another (Fig. 4B). The small displacement of green and red signals relates to the need to switch filters to detect different fluorescent wavelengths. Third, there were vesicles containing mRFP-gB and no obvious VP26-Venus capsids (Fig. 4C), consistent with previous observations of glycoprotein vesicles (1, 2, 20, 22). In Fig. 4 we installed gaps (e.g., between frames 7 and 26 in Fig. 4A) in order to describe more distance and time in the transport monitored. In some instances, puncta were lost from the plane of focus for several frames (each frame is ≈0.5 s), because axons were not entirely in the same horizontal plane and puncta moved out of the focal plane. However, every punctum that was enumerated could be accurately followed. The capsid in Fig. 4A was transported ≈35 μm in 14.5 s, averaging 2.4 μm/s, which is similar to the speed reported for PRV (19).
Fig. 4.
Imaging of capsids and gB in axons of SCG rat neurons present in the connecting channels between microfluidic chambers. Rat SCG neurons were plated in the somal chambers of microfluidic devices (20,000 neurons per chamber) for 14 days. The cells were infected with YK614 using 3 PFU/cell and were then imaged by live-cell deconvolution microscopy after 23 h. (A) VP26-Venus-labeled capsid (Separate particle) moving in the anterograde direction, from left to right. (B) VP26-Venus labeled capsid with colocalized mRFP-gB fluorescence (Married particle) moving in the anterograde direction. The small displacement of green and red signals relates to the need to switch filters in order to detect different fluorescent wavelengths. (C) Puncta containing mRFP-gB moving in the anterograde direction. Gaps were introduced, e.g., in panel A between frames 7 and 26, in order to show longer distances of travel. Frames of 0.5 s are numbered along the right side. Bars, 5 μm.
Three representative movies of the live-cell analyses are presented as supplemental material. Movie S1 shows a green punctum, i.e., a Separate capsid, that begins one-third of the distance from the left at time zero and moves to the right (from the axonal toward the somal chamber), largely disappearing from focus twice and then reappearing. This capsid moved ≈35 μm in approximately 17 s, corresponding to 2 μm/s. Following these first 17 s of the movie, there was an absence of puncta for approximately a minute. This illustrates the relatively low levels of capsid transport in some axons, especially when fewer axons are present in the channels. Beginning at 1 min, 18 s, a Married capsid, with intense mRFP-gB fluorescence, moved from the extreme left side of the frame toward the middle over the course of 27 s. Again, there was a small lag in detecting the green fluorescence relative to the red fluorescence, related to the need to change filters. Movie S2 in the supplemental material shows a Married particle (indicated by an open arrow) with stronger VP26-Venus fluorescence and less-intense mRFP-gB fluorescence that begins near the middle of the frame at time zero, moves over or under a red punctum, and continues to the right, moving slowly until 22 s and then moving more rapidly to the right, with a total of 28 μm of movement from 0 to 1.0 min. To the left of this Married capsid there was a green punctum (indicated by a filled arrow), i.e., a Separate capsid, that was initially stalled, then moved about 8 μm over 10 to 15 s, stalled again, after 1.0 min moved out of focus (1.0 to 1.1 min), and then moved ≈10 μm from 1.1 to 1.2 min before going out of focus. There were also numerous vesicles containing gB (red) that moved, while others were stalled. These images illustrated common observations of stalling, followed by shorter periods of transport. Movie S3 in the supplemental material shows a channel with more neurons (plated at 40,000 neurons per somal chamber) and numerous capsids and glycoprotein vesicles. The majority (70 to 80%) of capsids that moved either from left to right (in the anterograde direction) or from right to left (in the retrograde direction) were Separate. There was again substantial stalling, followed by transport. For example, there was a bright green punctum (indicated by the open arrow) approximately 13 to 14 μm from the left that was largely stalled from 0 to 4 s; then the capsid moved ≈10 μm from 4 to 15 s, slowed, stalled until 20 s, and then went out of focus. From 0 to 15 s there was a Married particle (arrowhead) that began just left of center and then moved more slowly ≈20 to 25 μm to the right. This movie also represented another important observation that likely explains the substantial right-to-left movement of capsids and glycoproteins. At time zero, there was a bright green punctum (indicated by the filled arrow) in the middle of the frame. This capsid was stalled for the first 5 s; then it jiggled and moved slowly, initially to the right, then downwards in an arch from 6 to 15 s, changing direction, and ultimately moving left (toward the somal chamber) and out of the frame from 15 to 42 s, with some periods of stalling. We observed other examples of red puncta that initially moved from right to left altering course and moving from left to right in axons. Thus, it appeared that these axons initially grew in the direction of the axonal chamber but then turned back, in an arc or semicircle, toward the somal chamber. These observations suggest that some of the puncta that moved right to left (toward the somal chamber) were actually moving in the anterograde direction in turned axons. Note that these turns were relatively rare even when neurons were plated more densely (40,000 cells/well) and were not observed at all when neurons were less dense.
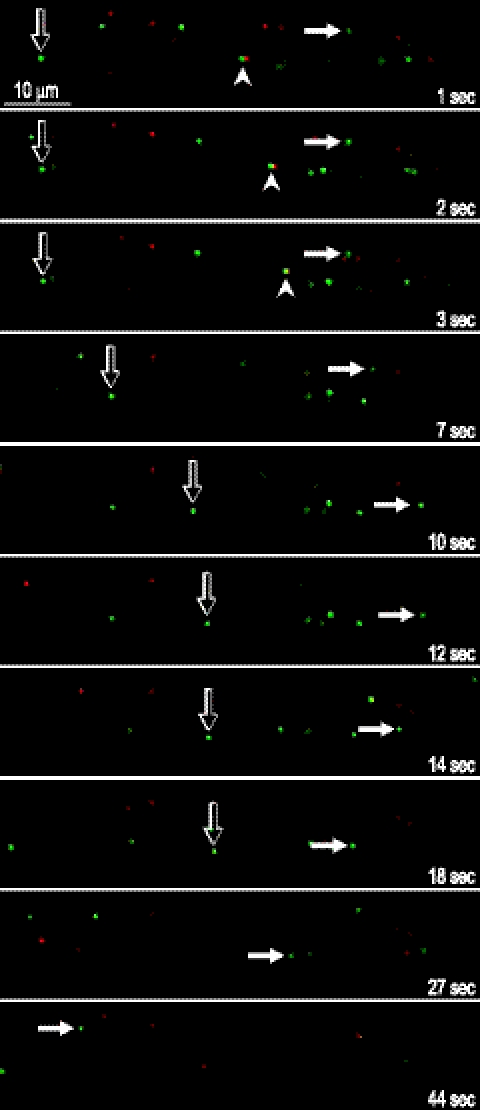
Figure 5 shows representative still images derived from movie S3 in the supplemental material. The open arrow points to a Separate capsid that moved in the anterograde direction (right) slowly at first and then more rapidly between 4 to 14 s, was stalled until 18 s, and then moved right and out of focus at 23 s (not shown). The filled arrowhead indicates a Married particle (green and red fluorescence) that moved in the anterograde direction (right) over ≈10 μm until 5 s (not shown) and disappeared from the focal plane at 6 s (not shown). The filled arrow documents a capsid that was stalled at first (1 to 3 s) and then moved right (3 to 6 s), then in an arc from 6 to 15 s (compare 7 to 14 s), and then rapidly leftward (toward the stromal chamber) over 40 to 45 μm from 14 to 44 s.
Fig. 5.
Still images derived from live-cell analyses (shown in movie S3 in the supplemental material). Images of several rat neuronal axons within a channel connecting the somal and axonal chambers are shown. The number of seconds following the initiation of live-cell imaging is given. Open arrows follow a Separate capsid (green) that moved slowly in the anterograde direction (rightward) from 1 to 4 s and then rapidly to the right over ≈10 μm from 4 to 14 s, after which it was largely stalled from 14 to 18 s and then moved right again and out of focus at 23 s (not shown). Filled arrowheads point to a Married particle (green and red fluorescence) at 1 s that moved to the right over ≈10 μm until 5 s (not shown) and disappeared at 6 s (not shown). Filled arrows point to a bright Separate capsid that was stalled from 1 to 3 s, moved slowly to the right from 3 to 6 s (compare 3 to 7 s), then moved downwards in an arc from 6 to 15 s (compare 7 to 12 s), and changed directions, moving rapidly left (toward the stromal chamber) over ≈40 to 45 μm from 14 to 44 s (14, 18, 27, and 44 s shown), followed by a period of stalling.
Table 2 summarizes the transport of capsids in four experiments involving different concentrations of neurons (15 to 40,00 neurons in the somal chamber), different culture periods (7 to 14 days after neuron plating), and different MOIs (3 to 12 PFU/cell). In experiment 1, 70% of capsids that moved in the anterograde direction (entirely left to right during the whole observation period) were Separate. Note that we enhanced the gB signal in the images as much as possible and erred on the conservative side: if there was there was any detectable mRFP-gB fluorescence associated with a capsid, the particle was labeled Married. The neurons in experiment 1 were relatively more concentrated, and we observed two axons that turned back toward somal chambers but classified all right-to-left movement as retrograde in Table 2, although, as discussed above, several of these capsids were likely moving in the anterograde direction. In experiment 1, there were also substantial numbers of capsids (25% of the total capsids) that were static for the entire period of observation (1.5 to 2.0 min), and these capsids were also largely (93%) Separate. Given their location, these particles likely moved before and after the relatively short observation period. Experiment 2 involved less-dense neurons with many fewer axons in the channels, and most (89%) of the anterograde capsids were Separate. In experiment 2 and experiment 3, there were fewer neurons and fewer retrograde capsids, again consistent with less turning of axons when neurons were less dense. In experiment 4, there were moderate bundles, and over half of the capsids that moved in the anterograde direction were Married. Similarly, about half the static capsids were Married. Totaling all the capsids that moved in the anterograde direction in these rat neurons, 67% of the capsids were Separate.
Table 2.
Summary of the anterograde and retrograde transport of HSV capsids in SCG rat neurons in microfluidic chambers
| Expt no. | Expt descriptiona | No. of capsidsb |
|||||
|---|---|---|---|---|---|---|---|
| Anterogradec |
Retrograde |
Static |
|||||
| Married | Separate | Married | Separate | Married | Separate | ||
| 1 | 40,000 neurons, large bundles of axons, 7 days in culture before infection using 3 PFU/cell, imaging begun at 20 h p.i. | 20 | 46 | 3 | 55 | 4 | 38 |
| 2 | 15,000 neurons, fewer axons in channels, 14 days in culture before infection using 3 PFU/cell, imaging begun at 19 h p.i. | 4 | 31 | 0 | 0 | 3 | 1 |
| 3 | 20,000 neurons, fewer axons in channels, 14 days in culture before infection using 12 PFU/cell, imaging begun at 22 h p.i. | 35 | 61 | 1 | 6 | 0 | 6 |
| 4 | 20,000 neurons, moderate bundling of axons in channels, 14 days in culture before infection using 3 PFU/cell, imaging begun at 23 h p.i. | 16 | 14 | 1 | 2 | 41 | 37 |
YK614 was used for infection in all experiments. p.i., postinfection.
Capsids detected by VP26-Venus fluorescence that exhibited mRFP-gB fluorescence (Married) or that showed no detectable mRFP-gB fluorescence (Separate) were enumerated. Anterograde, capsids that moved exclusively in the anterograde direction; retrograde, capsids that moved in the retrograde direction during the majority of their transport; static, capsids that remained static for all of the observation period.
A total of 75 capsids (33%) moving in the anterograde direction were Married, and 152 (67%) were separate, in the four experiments taken together.
DISCUSSION
Our studies with the YK614 HSV recombinant expressing mRFP-gB and VP26-Venus provided evidence for both Married and Separate anterograde transport. In SK-N-SH cells, 80% of the capsids observed in fixed neurites showed no detectable mRFP-gB. Given that we could not detect mRFP-gB fluorescence in ≈20% of HSV extracellular particles, it was likely that the majority of the capsids in SK-N-SH neurites were transported as Separate particles. Live-cell imaging of SK-N-SH neurites encountered two problems: (i) lower levels of capsid transport during the relatively short periods of observation and (ii) difficulties in interpreting the data based on the possibility of secondary infection. In SK-N-SH neurons, there were often numerous capsids that were stalled. For reasons described in the introduction and the second section of Results, secondary infection (resulting in retrograde transport) is unlikely to account for these stalled capsids. These capsids managed to reach distal regions of SK-N-SH axons by some mechanism, and the capsids were clearly inside neuronal axons. Thus, the more likely possibility is that these capsids and also glycoproteins moved in the anterograde direction but were not observed in motion during the short (1- to 3-min) observation periods. Substantial stalling was also observed in rat neurons. However, we also believe that transport of HSV particles in SK-N-SH axons is less efficient, i.e., there are more capsids stalled at any given time, than that in embryonic neurons, which likely have active transport systems. That said, SK-N-SH neurons do transport HSV capsids and glycoproteins into axons well, i.e., these structures get there, but live-cell analyses more readily capture transport in embryonic neurons.
Given these considerations, we performed live-cell analyses using rat SCG axons in microfluidic chambers. In these neurons, 67% of HSV capsids that were moving in the anterograde direction were Separate. This figure must be considered in terms of the 20% of extracellular enveloped virus particles that do not contain detectable mRFP-gB. However, again, we concluded that a large fraction, or even a majority, of YK614 capsids that moved in the anterograde direction in both SK-N-SH and rat SCG neurons were transported as Separate particles. We also compared YK614 to the HSV recombinant GS2843, described by Antinone et al. (3), which expresses GFP-gB and VP26-mRFP. Rat SCG neurons growing in microfluidic chambers infected with GS2843 displayed both Separate and Married particles, although the ratios of these two types of particles were not accurately quantified (not shown).
Notwithstanding observations of Separate transport, the present studies clearly showed Married particles, observations that we had not made previously. In previous studies, the fraction of Married particles observed in wild-type HSV-infected SK-N-SH neurons and stained with antibodies was low (5 to 10%) (20, 22). In contrast, the studies reported here with the two-color HSV recombinant in fixed SK-N-SH neurites (that were not stained with antibodies) produced evidence for as many as 20% Married particles. In rat SCG axons there were even more Married particles (33%). These differences might, in part, reflect the fact that YK614 expresses mutant forms of both gB and VP26 with large protein sequences fused to viral proteins, whereas the previous studies were done using wild-type HSV or one-color recombinants. Both YK614 and an HSV expressing VP26-GFP (22) produced 10-fold-lower quantities of infectious HSV. More importantly, we reported that HSV recombinants expressing VP26-GFP or gD-YFP displayed reduced numbers of capsids that were transported into axons and that this transport of capsids was significantly delayed (20, 22). Antinone et al. (3) also observed relatively few HSV capsids in axons with their GFP-gB–VP26-mRFP recombinant in chicken, rat, and mouse neurons, as we did with YK614. Thus, the addition of bulky fluorescent proteins to VP26 reduces capsid transport, and this might also be the case with gB, although decreased titers were not observed with YK613. Moreover, it is conceivable that addition of fluorescent proteins to both capsids and glycoproteins could alter the ratio of Married to Separate particles. At the very least, these two-color recombinants should be considered mutants. Another result might also explain some of the differences in Married versus Separate particles described here and elsewhere. In the present studies, rat neurons plated at higher densities produced axons that were more often bundled or arranged in fascides, and there tended to be a higher proportion of Married particles in these bundles (Table 2). When neurons were plated at lower densities, there were fewer bundles, and as many as 90% of the capsids moving in the anterograde direction were Separate. We never or infrequently imaged bundles of axons in previous studies of SK-N-SH or rat neurons (20, 22).
The differences we observed in the ratios of Separate to Married particles when comparing rat SCG neurons with human SK-N-SH neurons might also be pertinent. HSV is obviously a human virus and may more prominently transport capsids in human neurons. EM and immunofluorescence studies by Cunningham and colleagues involving human fetal dorsal route neurons infected with a clinical HSV strain have consistently produced evidence of Separate HSV capsids in axons, although 2 of a total of 15 particles in axons distant from varicosities were Married on one study (9, 14, 18). Recent EM studies involving three HSV lab strains in rat SCG neurons observed both Married (75%) and Separate (25%) particles. Antinone et al. (3), in their studies of an HSV strain expressing VP26-mRFP and GFP-gB, observed that 64 to 70% of capsids moving in the anterograde direction were Married in rat DRG, chicken, and mouse neurons. They could not detect gB-GFP in as many as 42% of extracellular HSV particles. One possibility is that there were lower levels of GFP-gB incorporated into extracellular virions, compared with the incorporation of mRFP-gB in YK614 (20% of particles without mRFP-gB). Alternatively, given our observations involving the anti-VP5 MAb, it is also possible that our detection of mRFP-gB and other fluorophores was more sensitive. Perhaps there was a small fraction of Separate particles that was obscured by the imaging conditions in the studies of Antinone et al. It may be that we missed a fraction of Married particles in SK-N-SH neuroblastoma cells (20, 22), although those studies involved wild-type or one-color HSV, which may be different. Perhaps HSV interacts differently with components of the human anterograde transport machinery than with those in rodent or chicken neurons. There are cases of HSV proteins that do not function well in rodent cells. For example, the HSV ICP47 protein blocks the human transporter for antigen presentation (TAP) but not mouse or rat TAP (11, 25).
In summary, there is growing evidence that HSV can transport capsids in the anterograde direction by either of two mechanisms. Kinesin motors can apparently transport both capsids coated with tegument proteins and vesicles containing enveloped capsids. This would not be too surprising, given the broad diversity of kinesins and the large numbers of structures they transport (8). If true, this opens up the fascinating possibility that there are multiple viral proteins that cannibalize the transport systems in neurons.
Supplementary Material
ACKNOWLEDGMENTS
We are especially indebted to Aurelie Snyder at the Advance Light Microscopy Core at the Jungers Center, OHSU, for her skill in performing deconvolution and live-cell imaging. We are grateful to Tiffani Howard (HowardInk.com), who was invaluable in graphic design, modifying live-cell images, and who did all the rat surgery. We are also indebted to Lynn Enquist for advice and for helping us gain experience with rat surgery. We thank Roselyn Eisenberg and Gary Cohen for their generous gifts of antibodies.
This work was supported by National Institutes of Health grant EY018755 (to D.C.J.), a Grant for Scientific Research in Priority Areas (to Y.K.), and a contract research fund for the Japan Initiative for Global Research Network on Infectious Diseases from the Ministry of Education, Culture, Science, Sports and Technology (MEXT) of Japan (to Y.K.).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 30 March 2011.
REFERENCES
- 1. Antinone S. E., Smith G. A. 2010. Retrograde axon transport of herpes simplex virus and pseudorabies virus: a live-cell comparative analysis. J. Virol. 84:1504–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Antinone S. E., Smith G. A. 2006. Two modes of herpesvirus trafficking in neurons: membrane acquisition directs motion. J. Virol. 80:11235–11240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Antinone S. E., Zaichick S. V., Smith G. A. 2010. Resolving the assembly state of herpes simplex virus during axon transport by live-cell imaging. J. Virol. 84:13019–13030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ch'ng T. H., Enquist L. W. 2005. Neuron-to-cell spread of pseudorabies virus in a compartmented neuronal culture system. J. Virol. 79:10875–10889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cohen G. H., et al. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34:521–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Curanovic D., Enquist L. 2009. Directional transneuronal spread of alpha-herpesvirus infection. Future Virol. 4:591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Farnsworth A., et al. 2007. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc. Natl. Acad. Sci. U. S. A. 104:10187–10192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hirokawa N., Noda Y., Tanaka Y., Niwa S. 2009. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10:682–696 [DOI] [PubMed] [Google Scholar]
- 9. Holland D. J., Miranda-Saksena M., Boadle R. A., Armati P., Cunningham A. L. 1999. Anterograde transport of herpes simplex virus proteins in axons of peripheral human fetal neurons: an immunoelectron microscopy study. J. Virol. 73:8503–8511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang J., Lazear H. M., Friedman H. M. 2011. Completely assembled virus particles detected by transmission electron microscopy in proximal and mid-axons of neurons infected with herpes simplex virus type 1, herpes simplex virus type 2 and pseudorabies virus. Virology 409:12–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jugovic P., Hill A. M., Tomazin R., Ploegh H., Johnson D. C. 1998. Inhibition of major histocompatibility complex class I antigen presentation in pig and primate cells by herpes simplex virus type 1 and 2 ICP47. J. Virol. 72:5076–5084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu W. W., Goodhouse J., Jeon N. L., Enquist L. W. 2008. A microfluidic chamber for analysis of neuron-to-cell spread and axonal transport of an alpha-herpesvirus. PLoS One 3:e2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maresch C., et al. 2010. Ultrastructural analysis of virion formation and anterograde intraaxonal transport of the alphaherpesvirus pseudorabies virus in primary neurons. J. Virol. 84:5528–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miranda-Saksena M., Armati P., Boadle R. A., Holland D. J., Cunningham A. L. 2000. Anterograde transport of herpes simplex virus type 1 in cultured, dissociated human and rat dorsal root ganglion neurons. J. Virol. 74:1827–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Negatsch A., et al. 2010. Ultrastructural analysis of virion formation and intraaxonal transport of herpes simplex virus type 1 in primary rat neurons. J. Virol. 84:13031–13035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park J. W., Vahidi B., Taylor A. M., Rhee S. W., Jeon N. L. 2006. Microfluidic culture platform for neuroscience research. Nat. Protoc. 1:2128–2136 [DOI] [PubMed] [Google Scholar]
- 17. Penfold M. E., Armati P., Cunningham A. L. 1994. Axonal transport of herpes simplex virions to epidermal cells: evidence for a specialized mode of virus transport and assembly. Proc. Natl. Acad. Sci. U. S. A. 91:6529–6533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saksena M. M., et al. 2006. Herpes simplex virus type 1 accumulation, envelopment, and exit in growth cones and varicosities in mid-distal regions of axons. J. Virol. 80:3592–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith G. A., Gross S. P., Enquist L. W. 2001. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc. Natl. Acad. Sci. U. S. A. 98:3466–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Snyder A., Bruun B., Browne H. M., Johnson D. C. 2007. A herpes simplex virus gD-YFP fusion glycoprotein is transported separately from viral capsids in neuronal axons. J. Virol. 81:8337–8340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Snyder A., Polcicova K., Johnson D. C. 2008. Herpes simplex virus gE/gI and US9 proteins promote transport of both capsids and virion glycoproteins in neuronal axons. J. Virol. 82:10613–10624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Snyder A., Wisner T. W., Johnson D. C. 2006. Herpes simplex virus capsids are transported in neuronal axons without an envelope containing the viral glycoproteins. J. Virol. 80:11165–11177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sugimoto K., et al. 2008. Simultaneous tracking of capsid, tegument, and envelope protein localization in living cells infected with triply fluorescent herpes simplex virus 1. J. Virol. 82:5198–5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tanaka M., Kagawa H., Yamanashi Y., Sata T., Kawaguchi Y. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 77:1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tomazin R., et al. 1996. Stable binding of the herpes simplex virus ICP47 protein to the peptide binding site of TAP. EMBO J. 15:3256–3266 [PMC free article] [PubMed] [Google Scholar]
- 26. Tomishima M. J., Enquist L. W. 2001. A conserved alpha-herpesvirus protein necessary for axonal localization of viral membrane proteins. J. Cell Biol. 154:741–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wisner T., Brunetti C., Dingwell K., Johnson D. C. 2000. The extracellular domain of herpes simplex virus gE is sufficient for accumulation at cell junctions but not for cell-to-cell spread. J. Virol. 74:2278–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.