Abstract
Deletion of the small hydrophobic (SH) protein of certain paramyxoviruses has been found to result in attenuation, suggesting that the SH protein is a virulence factor. To investigate the role of the mumps virus (MuV) SH protein in virulence, multiple stop codons were introduced into the open reading frame (ORF) of a MuV molecular clone (r88-1961SHstop), preserving genome structure but precluding production of the SH protein. No differences in neurovirulence were seen between the wild-type and the SHstop viruses. In contrast, upon deletion of the SH gene, significant neuroattenuation was observed. These data indicate that the MuV SH protein is not a neurovirulence factor and highlight the importance of distinguishing gene deletion effects from protein-specific effects.
TEXT
Mumps virus (MuV), a rubulavirus of the Paramyxoviridae family, contains tandemly linked transcription units encoding the nucleo- (N), V/phospho-/I (V/P/I), matrix (M), fusion (F), small hydrophobic (SH), hemagglutinin-neuraminidase (HN), and large (L) protein genes (4, 6). The MuV SH protein is a 57-amino-acid type III integral membrane protein. Certain MuV strains contain a point mutation in the F gene polyadenylation signal that results in an F-SH bicistronic mRNA of which only the upstream cistron (F) is translated (12, 22, 23), demonstrating that the SH protein is not essential for virus replication, a conclusion supported by studies of related paramyxoviruses (2, 3, 8).
The amino acid sequence of the MuV SH protein is the most hypervariable in the genome, with differences between some strains exceeding 20% (23), further indicating its nonessential role in the life cycle of the virus. Nonessential proteins typically serve a luxury function, and in the case of the MuV SH protein, this appears to be the blocking of apoptosis in certain cell types (9, 13, 26). Multiple reports have also suggested that the paramyxovirus SH proteins may play an ancillary role in virus-mediated cell fusion (10, 21), aid virus transport through the secretory pathway (19), or act as a viroporin, a group of small, highly hydrophobic proteins that can oligomerize and form ion channels at the cell membrane to modulate membrane permeability (18). In vivo studies of SH gene deletion mutants of related viruses, such as human respiratory syncytial virus (hRSV) and avian metapneumovirus, have been interpreted to indicate a role of the protein in replication, virulence, and immunogenicity (3, 11, 14, 24, 25). However, in these studies, because the SH mutants were produced by deletion of the gene, it was not possible to discern if the observed altered phenotypes are related to the absence of the SH protein itself or if they are a consequence of altering the genome structure (e.g., a decrease in genome length, an altered untranslated region, a change in transcription gradient of downstream genes, or other yet-to-be-identified effects).
To investigate the role of the MuV SH protein in virus neurovirulence, we generated two modified versions of a molecular clone of the highly neurovirulent wild-type MuV clinical isolate 88-1961WT, one wherein the SH gene was deleted (r88-1961ΔSH) and one wherein three stop codons were introduced in the SH open reading frame (ORF) (r88-1961SHstop) to abrogate SH protein expression without altering the structure of the genome.
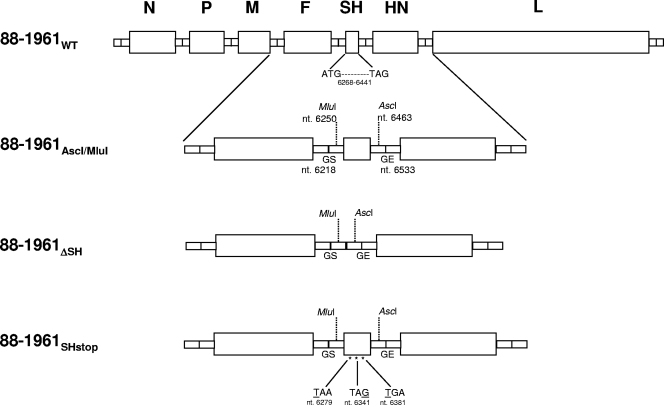
The plasmid p88-1961WT (15), which encodes the 15,384-nucleotide positive-strand (antigenomic) RNA sequence of 88-1961WT, was modified by site-directed mutagenesis (SDM) to introduce the MluI and AscI restriction sites in the intergenic regions flanking the SH gene, resulting in plasmid p88-1961AscI/MluI (Fig. 1). The restriction sites in p88-1961AscI/MluI were subsequently used to introduce a linker, comprising only the 5′ and 3′ SH intergenic sequences, into the 88-1961 genome in place of the ORF, resulting in plasmid p88-1961ΔSH (Fig. 1). Deletion of the SH ORF (174 nucleotides) maintained the genome length at an even multiple of six. The detailed cloning strategy for this construct and for those described below can be obtained from the authors upon request. Plasmid p88-1961ΔSH was used to produce three independently rescued virus stocks of r88-1961ΔSH from BHK BSR-T7/5 cells (15), which were amplified once on Vero cells. The genetic identities of the cDNA-derived r88-1961ΔSH viruses were confirmed by sequencing a 1,366-bp region encompassing the deleted SH ORF. The restriction sites in p88-1961AscI/MluI were also used to introduce a modified SH ORF containing three translational stop codons at positions 13, 73, and 115 of the SH ORF (genome positions 6279, 6341, and 6381) into the 88-1961 genome, resulting in plasmid p88-1961SHstop (Fig. 1), which was used to produce two independently rescued virus stocks of r88-1961SHstop from BHK BSR-T7/5 cells, amplified once on Vero cells. The genetic identities of the cDNA-derived r88-1961SHstop viruses were confirmed by sequencing a 1,540-bp region encompassing the 173-bp SH ORF containing the stop codons and all sequences in the plasmid that had been produced by PCR. No mutations were identified other than those specifically introduced by SDM.
Fig. 1.
Construction of 88-1961 variant plasmids to abrogate SH protein expression. Schematic diagram showing stepwise construction of p88-1961AscI/MluI, p88-1961ΔSH, and p88-1961SHstop from p88-1961WT. Nucleotide numbers are in genome sense.
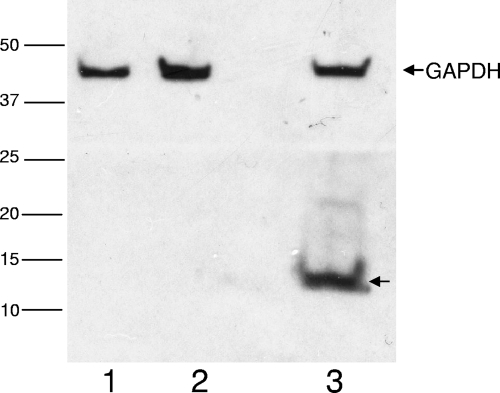
Our ability to confirm the effectiveness of the three stop codons was hindered by the lack of an antibody against the MuV SH protein. Therefore, the SHWT and SHSTOP sequences were isolated from p88-1961WT and p88-1961SHstop, respectively, and subcloned into the pCAGGS vector (kindly provided by J. C. de la Torre, La Jolla, CA) (17) in-frame with a hemagglutinin (HA) tag at the carboxyl terminus. Following transfection of cells, lysates were evaluated by Western blotting. Expression of HA-tagged SH protein was detected in cells transfected with pSHWT-HA (Fig. 2, lane 3) but not in cells transfected with pSHstop-HA, (Fig. 2, lane 1), indicating, albeit indirectly, that the SH protein is not translated in the recombinant virus containing the three stop codons. Of note, insertion of the HA tag into the full-length recombinant viruses themselves was not performed, given the demonstration by others of HA tag interference with SH protein function (26).
Fig. 2.
Western blot analysis of SHWT and SHSTOP expression. SH-HA expression (anti-HA antibody [1/1,000]; Roche, Indianapolis, IN) was observed in cells transfected with pSHWT-HA (lane 3) but not in cells transfected with pSHstop-HA (lane 1). Vero extracts isolated from cells transfected with the pCAGGS vector served as the assay control (lane 2). Expression of a cellular marker, glyceraldehyde-3-phosphate dehydrogenase (GAPDH; anti-GAPDH antibody [1/5,000]; Abcam, Cambridge, MA), was detectable in all samples.
To analyze the in vitro growth characteristics and to demonstrate the replication competence of r88-1961SHstop and r88-1961ΔSH relative to those of r88-1961WT, we determined cumulative virus production during the first 7 days postinfection (p.i.) for each virus on Vero cells as described previously (15) (Fig. 3). A one-way analysis of variance (ANOVA) did not reveal statistical differences between the replication kinetics of r88-1961WT and r88-1961SHstop (P = 0.95); however, differences between the replication kinetics of r88-1961ΔSH and 88-1961WT were significant (P = 0.01). A post hoc Tukey's multiple-comparison test found differences between 88-1961WT and r88-1961ΔSH R1 and between 88-1961WT and r88-1961ΔSH R2 but not between 88-1961WT and r88-1961ΔSH R3. However, the three rescues of r88-1961ΔSH replicated similarly to each other (P = 0.09; one-way ANOVA), and importantly, all were replication competent. Relative to that for r88-1961WT, the onset of cytopathic effects (abnormal cell morphology, syncytial formation, and cell lysis) was more rapid in cells infected with r88-1961ΔSH but not in cells infected with r88-1961SHstop (data not shown). The faster destruction of the cell monolayer by r88-1961ΔSH may account for the lower replication kinetics relative to that of r88-1961WT. However, unpublished observations from our laboratory suggest that there is no correlation between the extent of virus fusogenicity or other in vitro cytopathic effects and virus virulence in our rat model.
Fig. 3.
Comparison of the in vitro growth kinetics of the 88-1961 variant virus strains. Confluent monolayers of Vero cells were infected with r88-1961WT, r88-1961SHstop (rescues 1 and 2), and r88-1961ΔSH (rescues 1, 2, and 3) at a multiplicity of infection (MOI) of 0.05. Cumulative virus production over a period of 7 days was determined. The data represent mean values ± standard errors of the means (SEMs) from two independent experiments. Note that the error bars, while present, are not resolvable on the figure at the scale presented. The limit of detection of the assay is 40 PFU/ml.
Having confirmed that the modified 88-1961 viruses were replication competent, we compared the neurovirulence potentials of 88-1961ΔSH, r88-1961SHstop, and r88-1961WT in vivo in a rat neurovirulence assay (20). No significant differences were apparent in the mean neurovirulence score of r88-1961WT (17.6 ± 0.9) and that of r88-1961SHstop (17.2 ± 0.7; P = 0.72; unpaired t test) (Fig. 4). Maintenance of the stop codons in the recombinant viruses from each rescue of r88-1961SHstop following replication in rat brain was confirmed by extraction of total RNA from brain homogenates at day 3 p.i., the time of maximal virus production (16), followed by reverse transcription (RT)-PCR and sequencing (data not shown). In contrast, the mean neurovirulence score of 88-1961 ΔSH (9.6 ± 0.6) was significantly lower than that of r88-1961WT (17.6 ± 0.9; P = < 0.0001; unpaired t test). These data indicate the possibility that the attenuation observed by others may not be due to the absence of the SH protein per se but rather a consequence of disturbing the genome structure as a result of removing the SH transcription unit. The most likely cause of this would be an altered transcription/translation gradient, but this has proved difficult to assess. He et al. (8) and Wilson et al. (26) examined the effect of the removal of the SH gene from parainfluenza virus 5 (PIV-5) and were unable to reproducibly detect differences in levels of protein expression between the two viruses. Fuentes et al. attempted a similar analysis with an SH deletion mutant of hRSV and found variable results ranging from no effect to slight increases in expression levels of proteins both upstream and downstream of the deleted transcription unit (7). Thus, it is not clear from the literature if protein levels are affected by removal of the SH genes and, if they are, if such differences are detectable by Western blotting. While one could examine mRNA levels of the genes of interest, this has proven not to be predictive of protein levels (3). Whether altered levels of mRNA transcripts can affect virus phenotype without affecting protein levels is not known but seems unlikely. In the case of MuV, analyzing differences in gene-specific mRNA levels between two viruses is particularly complicated, given the abundance of polycistronic message species resulting in a laddering of transcripts that are not easily quantifiable (1, 5). Despite our inability to pinpoint the mechanism behind the observation that gene deletion alters virus phenotype, it is clear from our studies that the MuV SH protein per se is not a virulence factor in our animal model, although we cannot rule out that the MuV SH protein might be a virulence factor in a natural setting (i.e., respiratory infection in humans).
Fig. 4.
Severity of hydrocephalus in rats inoculated with variant MuV strains. Newborn rats were inoculated intracerebrally (i.c.) with the indicated viruses and euthanized 30 days later. Brains were removed, and hydrocephalus scores were calculated. Error bars represent the SEM. An asterisk (*) denotes cumulative historic data from testing of the parental 88-1961WT and Jeryl Lynn vaccine strains, respectively. Previously published data for the highly neuroattenuated Jeryl Lynn strain (15) are included to emphasize the ability of the assay to determine MuV neuroattenuation.
Acknowledgments
Salary support for C. Wolbert, K. Werner, L. Ngo, and C. Zhang was provided by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration.
We thank K. Conzelmann and U. Buchholz for providing the BHK BSR-T7/5 cells.
The findings and conclusions in this article have not been formally disseminated by the Food and Drug Administration and should not be construed to represent any agency determination or policy.
Footnotes
Published ahead of print on 6 April 2011.
REFERENCES
- 1. Afzal M. A., Elliott G. D., Rima B. K., Orvell C. 1990. Virus and host cell-dependent variation in transcription of the mumps virus genome. J. Gen. Virol. 71:615–619 [DOI] [PubMed] [Google Scholar]
- 2. Biacchesi S., et al. 2005. Infection of nonhuman primates with recombinant human metapneumovirus lacking the SH, G, or M2-2 protein categorizes each as a nonessential accessory protein and identifies vaccine candidates. J. Virol. 79:12608–12613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bukreyev A., Whitehead S. S., Murphy B. R., Collins P. L. 1997. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J. Virol. 71:8973–8982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Elango N., Varsanyi T. M., Kovamees J., Norrby E. 1988. Molecular cloning and characterization of six genes, determination of gene order and intergenic sequences and leader sequence of mumps virus. J. Gen. Virol. 69:2893–2900 [DOI] [PubMed] [Google Scholar]
- 5. Elliott G. D., Afzal M. A., Martin S. J., Rima B. K. 1989. Nucleotide sequence of the matrix, fusion and putative SH protein genes of mumps virus and their deduced amino acid sequences. Virus Res. 12:61–75 [DOI] [PubMed] [Google Scholar]
- 6. Elliott G. D., et al. 1990. Strain-variable editing during transcription of the P gene of mumps virus may lead to the generation of non-structural proteins NS1 (V) and NS2. J. Gen. Virol. 71:1555–1560 [DOI] [PubMed] [Google Scholar]
- 7. Fuentes S., Tran K. C., Luthra P., Teng M. N., He B. 2007. Function of the respiratory syncytial virus small hydrophobic protein. J. Virol. 81:8361–8366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. He B., Leser G. P., Paterson R. G., Lamb R. A. 1998. The paramyxovirus SV5 small hydrophobic (SH) protein is not essential for virus growth in tissue culture cells. Virology 250:30–40 [DOI] [PubMed] [Google Scholar]
- 9. He B., Lin G. Y., Durbin J. E., Durbin R. K., Lamb R. A. 2001. The SH integral membrane protein of the paramyxovirus simian virus 5 is required to block apoptosis in MDBK cells. J. Virol. 75:4068–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heminway B. R., et al. 1994. Analysis of respiratory syncytial virus F, G, and SH proteins in cell fusion. Virology 200:801–805 [DOI] [PubMed] [Google Scholar]
- 11. Jin H., et al. 2000. Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2-2 genes are attenuated in vitro and in vivo. Virology 273:210–218 [DOI] [PubMed] [Google Scholar]
- 12. Künkel U., Schreier E., Siegl G., Schultze D. 1994. Molecular characterization of mumps virus strains circulating during an epidemic in eastern Switzerland 1992/93. Arch. Virol. 136:433–438 [DOI] [PubMed] [Google Scholar]
- 13. Li Z., et al. 2011. Function of the small hydrophobic protein of J paramyxovirus. J. Virol. 85:32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ling R., et al. 2008. Deletion of the SH gene from avian metapneumovirus has a greater impact on virus production and immunogenicity in turkeys than deletion of the G gene or M2-2 open reading frame. J. Gen. Virol. 89:525–533 [DOI] [PubMed] [Google Scholar]
- 15. Malik T. H., et al. 2007. A single nucleotide change in the mumps virus F gene affects virus fusogenicity in vitro and virulence in vivo. J. Neurovirol. 13:513–521 [DOI] [PubMed] [Google Scholar]
- 16. Malik T. H., Wolbert C., Nerret L., Sauder C., Rubin S. 2009. Single amino acid changes in the mumps virus haemagglutinin-neuraminidase and polymerase proteins are associated with neuroattenuation. J. Gen. Virol. 90:1741–1747 [DOI] [PubMed] [Google Scholar]
- 17. Niwa H., Yamamura K., Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199 [DOI] [PubMed] [Google Scholar]
- 18. Perez M., Garcia-Barreno B., Melero J. A., Carrasco L., Guinea R. 1997. Membrane permeability changes induced in Escherichia coli by the SH protein of human respiratory syncytial virus. Virology 235:342–351 [DOI] [PubMed] [Google Scholar]
- 19. Rixon H. W., et al. 2004. The small hydrophobic (SH) protein accumulates within lipid-raft structures of the Golgi complex during respiratory syncytial virus infection. J. Gen. Virol. 85:1153–1165 [DOI] [PubMed] [Google Scholar]
- 20. Rubin S. A., et al. 2005. The rat-based neurovirulence safety test for the assessment of mumps virus neurovirulence in humans: an international collaborative study. J. Infect. Dis. 191:1123–1128 [DOI] [PubMed] [Google Scholar]
- 21. Saikia P., Shaila M. S. 2008. Identification of functional domains of phosphoproteins of two morbilliviruses using chimeric proteins. Virus Genes 37:1–8 [DOI] [PubMed] [Google Scholar]
- 22. Takeuchi K., Tanabayashi K., Hishiyama M., Yamada A. 1996. The mumps virus SH protein is a membrane protein and not essential for virus growth. Virology 225:156–162 [DOI] [PubMed] [Google Scholar]
- 23. Takeuchi K., Tanabayashi K., Hishiyama M., Yamada A., Sugiura A. 1991. Variations of nucleotide sequences and transcription of the SH gene among mumps virus strains. Virology 181:364–366 [DOI] [PubMed] [Google Scholar]
- 24. Tripp R. A., et al. 1999. Respiratory syncytial virus G and/or SH protein alters Th1 cytokines, natural killer cells, and neutrophils responding to pulmonary infection in BALB/c mice. J. Virol. 73:7099–7107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Whitehead S. S., et al. 1999. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J. Virol. 73:3438–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wilson R. L., et al. 2006. Function of small hydrophobic proteins of paramyxovirus. J. Virol. 80:1700–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]