Abstract
CD200 is a cell surface glycoprotein that binds an inhibitory receptor (CD200R) on myeloid cells. CD200 orthologues are present in many species of virus, and we show that the rat cytomegalovirus CD200 orthologue (e127) is expressed at the cell surface on infected cells. It binds the host CD200R with the same affinity as that of the host protein, and thus this protein acts as a close mimic of the host protein and has the potential to downregulate immune responses to the virus.
TEXT
The Herpesviridae and Poxviridae have large genomes with more than 100 open reading frames (ORFs). A number of these have been acquired from their hosts and are involved in subverting the immune system (6, 13). These include CD200-like sequences that have been identified in many viruses, including gammaherpesviruses (human herpesvirus 8 [HHV-8]), betaherpesviruses (HHV-6, HHV-7, and the English isolate of rat cytomegalovirus [RCMV-E]), poxviruses (Yaba-like disease virus and myxoma virus), and duck adenovirus (see references 5 and 16). CD200 is a membrane protein containing two IgSF (immunoglobulin superfamily) domains expressed by many host cell types that interacts with a receptor, CD200R, that is highly expressed on myeloid cells and can mediate anti-inflammatory signals (1, 2, 7, 8, 10, 11, 15, 17).
The CD200 orthologues in HHV-6, HHV-7, and HHV-8 (14) have been shown to bind CD200R, and in one case the HHV-8 K14 protein has been shown to bind with the same kinetics as those of the host protein (5). The use of recombinant viral CD200 orthologues has indicated that these can give downregulatory signals to myeloid cells, including macrophages and basophils in vitro in HHV-8 (5, 14) and in rhesus rhadinovirus (9). The poxvirus orthologues show less similarity to the host than these herpesvirus orthologues, and they have only one IgSF domain. However, there are functional data for myxoma virus consistent with an interaction with the CD200R (4, 18). In this report, we identify CD200R as the receptor for the RCMV-E CD200 orthologue e127 (16). The latter protein binds the inhibitory CD200 receptor with the same affinity and kinetics as those of the host CD200 and is expressed during viral infection.
The e127 ORF predicts a transmembrane protein with 56% amino acid sequence identity with rat CD200 (16). In order to characterize its receptor on host cells, a recombinant protein was constructed with the two IgSF domains of RCMV e127 coupled to CD4 domains 3 and 4 as an antigenic tag and a peptide sequence at the C terminus that allows biotinylation in vitro (e127CD4) (3). A comparable CD200CD4 protein had been used to identify the receptor for host CD200 by coupling the protein onto fluorescent beads (17). The e127CD4 beads bound rat peritoneal cells in a manner identical to that of host CD200CD4 beads, as shown by flow cytometry (Fig. 1). The receptor was confirmed as the CD200R, as binding of both reagents was blocked if the peritoneal cells were preincubated with monoclonal antibody (MAb) specific for CD200R (OX102) but not with an isotype-matched control (Fig. 1). This indicates that the e127 ORF codes for a protein that binds the same receptor as that of host CD200, confirming that it is a structural orthologue of CD200 in addition to showing sequence similarity.
Fig. 1.
RCMV e127 binds CD200R on rat peritoneal macrophages. Flow cytometry shows that RCMV e127CD4 (A)- or rat CD200CD4 (B)-coated fluorescent beads bound rat peritoneal macrophages (green line) compared to rat CD4-coated control beads (purple). The specificity for the CD200R was shown by the complete blocking of bead binding with rat CD200R MAb (OX102) (pink dashed lines are indistinguishable from control beads [purple]) but not with isotype-matched control MAb (blue dotted lines).
A direct interaction between purified e127 and CD200R proteins was shown by surface plasmon resonance, and the affinity was determined by injecting a series of different concentrations of purified monomeric e127CD4 (Fig. 2, left) or rat CD200CD4 injected over immobilized rat CD200RCD4 (Fig. 2, right) and a negative control (CD4) in separate flow cells on a streptavidin-coated chip in a BIAcore 2000 apparatus (BIAcore AB, St. Albans, United Kingdom) as described previously (17). The binding is represented by the difference in response units (RU) observed in the rat CD200R and control flow cells (Fig. 2A) once equilibrium has been reached. The equilibrium binding affinities (KD) were measured at 37°C and calculated by both nonlinear curve fitting of the Langmuir binding (Fig. 2B) and Scatchard transformations of the binding data (Fig. 2B, insets), giving similar values. The rat CD200 and the e127 bound to rat CD200R with identical equilibrium binding affinities (KD of 1.5 μM at 37°C). Clearly, the kinetic parameters of this interaction are important and are mimicked in this virus and the orthologue in HHV-8 (5). There are data for binding to CD200R of other CD200 viral orthologues in rhadinovirus, HHV-6, and HHV-7 (5, 14).
Fig. 2.
The viral RCMV e127 protein interacts with rat CD200R with equilibrium affinities and kinetics similar to those of the rat CD200 protein. The left panels show data for the RCMV e127, and the right panels show data for rat CD200. (A) The indicated concentrations (μM) of viral ligand RCMV e127CD4 (left) or host ligand rat CD200CD4 (right) were injected at 15 μl/min, at 37°C, through flow cells with 1,526 response units (RU) of CD200RCD4bio (CD200R) or 3,095 RU of CD4-biotin (control) immobilized. The amount of rat CD200CD4 or RCMV e127CD4 that bound at each concentration was calculated from the response at equilibrium in the CD200RCD4 and control flow cell, and these are plotted as binding curves in panel B. The inset shows SDS-PAGE analysis of the two chimeric proteins RCMV e127CD4 (left) and rat CD200CD4 (right) used in these studies. (B) The curved line in the main plots is a nonlinear curve fit of the data and corresponds to an affinity of 1.5 μM for both, the host and the viral ligand. Scatchard transformations to the same binding data are shown in the insets, and the linear fit corresponds to a KD of 1.5 μM for both ligands.
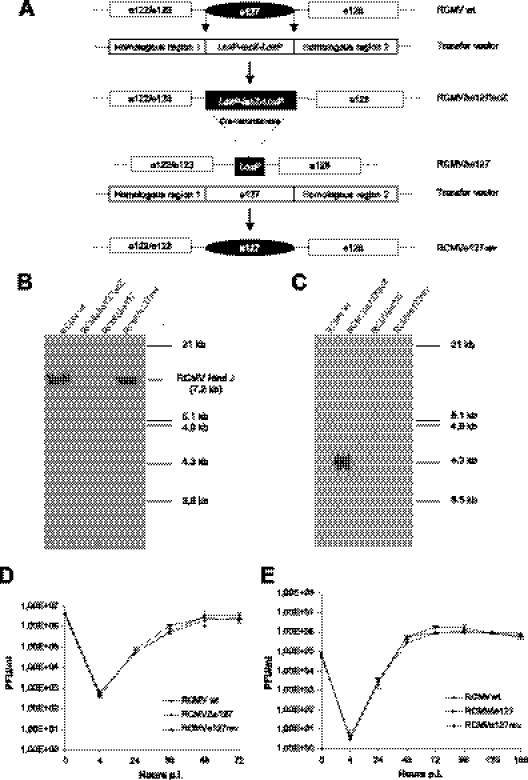
The molecular data point to a role for the e127 protein mimicking the host CD200 in giving a downregulatory signal to myeloid cells. To investigate if e127 is involved in downregulation of macrophage activity in vitro or in vivo, we constructed a mutant virus lacking the e127 ORF by inserting a lacZ gene flanked by LoxP sites into the e127 ORF in the RCMV wild type (RCMV wt) by homologous recombination in rat embryo fibroblast (REF) cells (Fig. 3A). Disruption of the e127 ORF was verified by Southern blot analysis (Fig. 3B, C) and sequencing. To repair the RCMVΔe127lacZ virus, virion DNA was cotransfected with the RCMV HindIII J fragment containing the e127 ORF (16), and the revertant virus (RCMVe127rev) was isolated. The RCMVΔe127lacZ-negative virus was generated by passage of the original mutant virus in REF cells expressing Cre recombinase as described previously (12), and the RCMVΔe127 virus was isolated with one LoxP site remaining (Fig. 3 A). The replication abilities of the viruses were compared in single- and multistep growth experiments by infecting cells with a low multiplicity of infection (MOI) of 0.1 and a high MOI of 5, respectively. No significant differences of viral replication in cell culture could be detected at these two multiplicities of infection (Fig. 3D, E). This is consistent with a role for e127 subverting the immune response rather than affecting virus replication.
Fig. 3.
Construction and characterization of recombinant viruses. (A) Construction of recombinant viruses following homologous recombination in REF cells. In the initially obtained mutant virus, e127 was replaced by lacZ flanked by LoxP sites. The resultant RCMVΔe127lacZ virion DNA was cotransfected with Cre recombinase into REF cells to generate the RCMVΔe127 virus. All viruses were double-plaque purified. Recombinant viruses were analyzed by HindIII digest and Southern blot analysis (using a probe directed against e127 [B] or lacZ [C]). Virus growth capacities of RCMV wt, RCMVΔe127, and RCMVe127rev viruses were analyzed by infecting REF cells with each virus at either a high MOI of 5 (D) or a low MOI of 0.1 (E).
If the e127 protein functions as a CD200 orthologue, then it should be expressed at the cell surface during infection. This was tested using the OX2 MAb that recognizes rat CD200 and cross-reacted on the e127 protein. Infection of REF cells that lack CD200 expression confirmed that e127 was expressed at the protein level during RCMV wt or RCMVe127rev infection but not with the virus in which e127 was deleted (Fig. 4A to D). In contrast to RCMV wt infection in REF cells, e127 could not be detected on the surface after infection of the rat macrophage cell line NR8383 (Fig. 4E), and virus spread was inefficient among macrophages (Fig. 4F).
Fig. 4.
OX2 (CD200) MAb recognizes RCMV e127 on infected cells but not the rat macrophage cell line NR8383. REF cells were mock infected (A) or infected with the RCMV wt (B), RCMVΔe127 (C), or RCMVe127rev (D) virus. (E) e127 is not expressed on infected NR8383 cells. REF cells and macrophages were infected at MOIs of 5 and 10, respectively. Shaded histogram, no staining; dotted line, avidin-fluorescein isothiocyanate (FITC); solid line, OX2-biotin followed by avidin-FITC staining (F). Growth analysis of RCMV wt, RCMVΔe127, and RCMVe127rev viruses using the rat macrophage cell line NR8383. Following macrophage infection, supernatants were harvested at the times indicated and titers were determined on REF cells.
The possibility that e127 caused downregulation of inflammatory responses was tested by measuring inducible nitric oxide synthase (iNOS) and tumor necrosis factor alpha (TNF-α) production from activated macrophages by cell-cell contact with cells expressing e127. To test this, an assay was devised to culture the macrophage cell line NR8383 in the presence of REF cells expressing viral e127 and then monitor macrophage activity by TNF-α and iNOS production. REF cells were infected with the RCMV wt, RCMVΔe127, and RCMVe127rev viruses. After 24 h, NR8383 was added to the infected REF cells and then activated with a combination of 10 ng/ml recombinant rat IFN-γ and 20 ng/ml lipopolysaccharide (LPS). Supernatants were harvested at different time points and quantitatively assayed for TNF-α by enzyme-linked immunosorbent assay (ELISA; eBioscience) and iNOS production by a Griess assay kit (Promega; see Fig. S1A and B in the supplemental material). In other approaches, NR8383 was preactivated with 10 ng/ml gamma interferon (IFN-γ) and 20 ng/ml LPS for 24 h prior to adding them to the infected REFs (see Fig. S1C and D in the supplemental material), or NR8383 was mixed and incubated with the infected REFs for 24 h and then activated with IFN-γ and LPS (see Fig. S1E and F in the supplemental material). However, even under these different stimulation approaches and times, no differences between RCMV wt and the e127 mutant virus could be detected. To further investigate if the expression of CD200R on the macrophages was altered by activation of the cells, we analyzed CD200R expression by flow cytometry; however, this did not reveal any differences (see Fig. S2 in the supplemental material).
In addition, we infected different cohorts of rats with this set of viruses to investigate if the presence of e127 correlates with lower levels of myeloid cell activity detected by production of iNOS and other markers in the spleen. Spleens of rats infected with either wild-type or mutant virus showed slight differences in iNOS and major histocompatibility complex (MHC) class II expression, but the variation between animals prevented a definitive conclusion as to whether inflammation was affected by the e127 protein. Statistically significant changes were not observed, and it is likely that e127 serves a subtle effect not evident in these assays. The role of CD200 as a negative regulator of myeloid cells together with the fact that CD200 orthologues are expressed by a wide range of viruses indicates that capture of host CD200 provides an evolutionary advantage to the virus and indicates a role in their defense against the immune system.
Supplementary Material
Acknowledgments
We are grateful to Debbie Hatherley for help in the analysis of CD200R-like proteins and Jakob Ettinger and Sigrun Schmähling for help with macrophage experiments.
The work was supported by the Medical Research Council (grant G9826026) and the Georg und Agnes Blumenthal Stiftung.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 6 April 2011.
REFERENCES
- 1. Akkaya M., Barclay A. N. 2010. Heterogeneity in the CD200R paired receptor family. Immunogenetics 62:15–22 [DOI] [PubMed] [Google Scholar]
- 2. Boudakov I., et al. 2007. Mice lacking CD200R1 show absence of suppression of lipopolysaccharide-induced tumor necrosis factor-alpha and mixed leukocyte culture responses by CD200. Transplantation 84:251–257 [DOI] [PubMed] [Google Scholar]
- 3. Brown M. H., et al. 1998. 2B4, the natural killer and T cell immunoglobulin superfamily surface protein, is a ligand for CD48. J. Exp. Med. 188:2083–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cameron C. M., Barrett J. W., Liu L., Lucas A. R., McFadden G. 2005. Myxoma virus M141R expresses a viral CD200 (vOX-2) that is responsible for down-regulation of macrophage and T-cell activation in vivo. J. Virol. 79:6052–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Foster-Cuevas M., Wright G. J., Puklavec M. J., Brown M. H., Barclay A. N. 2004. Human herpesvirus 8 K14 protein mimics CD200 in down-regulating macrophage activation through CD200 receptor. J. Virol. 78:7667–7676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Griffin B. D., Verweij M. C., Wiertz E. J. 2010. Herpesviruses and immunity: the art of evasion. Vet. Microbiol. 143:89–100 [DOI] [PubMed] [Google Scholar]
- 7. Hoek R. M., et al. 2000. Down-regulation of the macrophage lineage through interaction with OX2 (CD200). Science 290:1768–1771 [DOI] [PubMed] [Google Scholar]
- 8. Jenmalm M. C., Cherwinski H., Bowman E. P., Phillips J. H., Sedgwick J. D. 2006. Regulation of myeloid cell function through the CD200 receptor. J. Immunol. 176:191–199 [DOI] [PubMed] [Google Scholar]
- 9. Langlais C. L., Jones J. M., Estep R. D., Wong S. W. 2006. Rhesus rhadinovirus R15 encodes a functional homologue of human CD200. J. Virol. 80:3098–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mihrshahi R., Brown M. H. 2010. Downstream of tyrosine kinase 1 and 2 play opposing roles in CD200 receptor signaling. J. Immunol. 185:7216–7222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mukhopadhyay S., et al. 2010. Immune inhibitory ligand CD200 induction by TLRs and NLRs limits macrophage activation to protect the host from meningococcal septicemia. Cell Host Microbe 8:236–247 [DOI] [PubMed] [Google Scholar]
- 12. Sandford G. R., Brock L. E., Voigt S., Forester C. M., Burns W. H. 2001. Rat cytomegalovirus major immediate-early enhancer switching results in altered growth characteristics. J. Virol. 75:5076–5083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seet B. T., et al. 2003. Poxviruses and immune evasion. Annu. Rev. Immunol. 21:377–423 [DOI] [PubMed] [Google Scholar]
- 14. Shiratori I., et al. 2005. Down-regulation of basophil function by human CD200 and human herpesvirus-8 CD200. J. Immunol. 175:4441–4449 [DOI] [PubMed] [Google Scholar]
- 15. Simelyte E., Alzabin S., Boudakov I., Williams R. 2010. CD200R1 regulates the severity of arthritis but has minimal impact on the adaptive immune response. Clin. Exp. Immunol. 162:163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Voigt S., Sandford G. R., Hayward G. S., Burns W. H. 2005. The English strain of rat cytomegalovirus (CMV) contains a novel captured CD200 (vOX2) gene and a spliced CC chemokine upstream from the major immediate-early region: further evidence for a separate evolutionary lineage from that of rat CMV Maastricht. J. Gen. Virol. 86:263–274 [DOI] [PubMed] [Google Scholar]
- 17. Wright G. J., et al. 2000. Lymphoid/neuronal cell surface OX2 glycoprotein recognizes a novel receptor on macrophages implicated in the control of their function. Immunity 13:233–242 [DOI] [PubMed] [Google Scholar]
- 18. Zhang L., et al. 2009. Inhibition of macrophage activation by the myxoma virus M141 protein (vCD200). J. Virol. 83:9602–9607 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.