Abstract
Alphaviruses are small, spherical, enveloped, positive-sense, single-stranded, RNA viruses responsible for considerable human and animal disease. Using microinjection of preassembled cores as a tool, a system has been established to study the assembly and budding process of Sindbis virus, the type member of the alphaviruses. We demonstrate the release of infectious virus-like particles from cells expressing Sindbis virus envelope glycoproteins following microinjection of Sindbis virus nucleocapsids purified from the cytoplasm of infected cells. Furthermore, it is shown that nucleocapsids assembled in vitro mimic those isolated in the cytoplasm of infected cells with respect to their ability to be incorporated into enveloped virions following microinjection. This system allows for the study of the alphavirus budding process independent of an authentic infection and provides a platform to study viral and host requirements for budding.
INTRODUCTION
Alphaviruses are positive-sense RNA viruses belonging to the family Togaviridae and are responsible for considerable disease worldwide. The alphaviruses have been studied in great detail for the past 30 years, primarily using Sindbis virus (SINV) and Semliki Forest virus (SFV), but many details about the virus life cycle are yet to be elucidated (7). Structurally, the alphavirus virion is an enveloped particle with T=4 icosahedral symmetry (2). The viral envelope is derived from the host plasma membrane and contains 240 copies each of the E2 and E1 envelope proteins and substoichiometric amounts of the 6K protein. E2 and E1 are arranged as 80 trimers of heterodimers and are referred to as spikes (10). The viral membrane surrounds the nucleocapsid (NC), which is composed of 240 copies of capsid protein associated with a single copy of the viral genomic RNA. The NC also has icosahedral T=4 symmetry (2, 10).
In the infected cell, cytoplasmic assembly of genomic RNA and capsid protein produces the NCs that associate with viral envelope proteins at the plasma membrane, resulting in release of virions. NC assembly requires interactions between capsid proteins and the genomic RNA as well as interactions between individual capsid proteins (6, 13). NC assembly occurs rapidly after capsid protein synthesis (19), and yet only a fraction of synthesized capsid protein leaves the cell in the form of virion particles (16).
Little is known about the exact details and requirements of the budding process. At the plasma membrane, capsid protein molecules in the NC interact with the cytoplasmic domain of the E2 (cdE2) envelope protein, providing the free energy for membrane envelopment and budding of virions from the cell (22, 26). Mutations in capsid protein or cdE2 that perturb the NC-cdE2 interactions lead to a failure of NCs to bud (12, 22). The involvement of host factors in the budding process has been relatively unexplored. Precise details regarding the transit of the NCs to the site of budding are still not known, and in fact, the formation of distinct NCs in the cytoplasm is not required for virus release (4). In short, the cellular pathway undertaken by alphavirus NCs from their site of synthesis to their site of budding is poorly understood, and a system to probe NC trafficking would provide a wealth of knowledge.
In vitro assembly of Sindbis virus NCs is possible by utilizing purified capsid protein and single-stranded nucleic acid. Structurally, in vitro-assembled NCs, or core-like particles (CLPs), resemble NCs assembled in the cytoplasm of an infected cell (9). However, the ability of CLPs to functionally imitate authentic NCs during the egress and entry processes of alphavirus infections has not been explored.
Here, we describe the establishment of a system to study the budding process of alphaviruses in the absence of a native infection. Baby hamster kidney (BHK) cells in culture expressing the structural envelope proteins of SINV were microinjected with NCs either derived from the cytoplasm of an infected cell or assembled in vitro. We demonstrate that both species of NCs are capable of being incorporated into infectious virus-like particles (VLPs) after microinjection and thus demonstrate that the “membrane-enwrapping” model of alphavirus budding is biologically feasible (5). We also demonstrate for the first time that in vitro-assembled Sindbis virus-derived CLPs can act as functional surrogates for cytoplasmic NCs, yielding infectious virions. Our results are similar to those recently described for a Ross River virus-based system using in vitro-assembled cores and alternative transfection techniques (1). The data described raise several interesting and novel questions regarding the alphavirus budding process. The system characterized herein can be exploited to elucidate the viral and host factors required for alphavirus budding, and the methodology can be used to probe putative antivirals aimed at disrupting the budding process.
MATERIALS AND METHODS
Cell culture.
BHK-15 cells obtained from the American Type Culture Collection were maintained in minimal essential medium (MEM) (Life Technologies) supplemented with 10% fetal bovine serum (FBS), except for enzyme-linked immunosorbent assay (ELISA) studies in which the FBS was omitted.
NC purification.
Cytoplasmic NCs were purified by infecting 5- by 150-mm plates of confluent BHK cells with SINV derived from the Toto64 cDNA clone at a multiplicity of infection (MOI) of 20. Infection was allowed to proceed for 8 h, at which time media were removed and cells were washed 3 times with phosphate-buffered saline (PBS) and then scraped off the cell culture dish using 10 ml PBS. Cells were pelleted for 10 min at 750 × g and resuspended in 2.25 ml of hypotonic buffer (10 mM NaCl, 10 mM Tris, pH 7.4, 20 mM EDTA) to swell for 20 min on ice. Five hundred microliters of 20% Triton X-100 was added. Lysed cell debris and nuclei were pelleted at 2,000 × g for 10 min. The solution was then loaded onto a continuous 0 to 30% iodixanol gradient in TNE + Triton X-100 (50 mM Tris, pH 7.4, 100 mM NaCl, 1 mM EDTA, 0.1% Triton X-100) and centrifuged at 32,000 rpm in a Beckman SW-41 rotor for 2.5 h. The NC band was concentrated, and buffer was exchanged with 50 μl of TNE with Amicon 100-kDa centrifugal filter units. To gauge purified NC concentrations, bovine serum albumin (BSA) standards of known protein concentrations were electrophoresed in parallel with NC preparations on a 10% continuous SDS-PAGE gel, stained with Coomassie blue R-250, and scanned with a Li-Cor Odyssey infrared imager. In vitro-assembled NCs were prepared as described earlier (23), using an arbitrary single-stranded DNA (ssDNA) 109-mer (5′ GATCCGAAAGCGCGCCGTGAGTGCCTGACGCCATACGCCCTGGCCCCAAACGCCGTAATCCCAGCGGCCGCTGCCGCCGCGTGCTGCGTTAGGTCGGCCAATGCTTAGC 3′).
Rate zonal centrifugation.
Postmicroinjection medium (1 ml) was applied to a continuous 10-ml 0 to 30% iodixanol gradient in TNE (50 mM Tris, pH 7.4, 100 mM NaCl, 1 mM EDTA) and centrifuged at 32,000 rpm in a Beckman SW-41 rotor for 2 h. At the end of the run, the gradient was fractionated such that the top 6 ml was removed in 1-ml fractions and the bottom 5 ml was removed in 500-μl fractions. The amount of infectious material in each fraction was titrated on BHK cells by plaque assay.
Transient transfection and expression of SINV envelope proteins.
The SINV sequence encoding the envelope proteins (E3, red fluorescent protein [RFP], E2, 6K, and E1 [E2 contained an N-terminal RFP fusion]) was cloned into the pcDNA4/TO/myc/his/A mammalian expression vector (Invitrogen) (pcDNA4/SINV/ENV). BHK cells were seeded into 35-mm culture dishes at ∼70% confluence and then transiently transfected with 4 μg of plasmid with Lipofectamine 2000 (Invitrogen) 24 h prior to microinjection with NCs or RNA.
Microinjection.
Dextran-Cascade Blue conjugates (3 kDa; Invitrogen) were diluted to 2 mg/ml in NC or RNA samples to be microinjected. Samples were loaded into a glass capillary attached to an Eppendorf Femtojet microinjection unit. Injections were carried out using a PatchMan NP 2 micromanipulator (Eppendorf). BHK cells plated in 35-mm culture dishes that demonstrated RFP-derived fluorescence were microinjected with 100-hPa injection pressure while compensation pressure was set to 40 hPa. Each cell was estimated to receive 0.1 to 1 pl of sample per injection. Both viral RNA (vRNA) and NCs were diluted to a concentration of 8,000 molecules/pl before injection. About 30 cells were microinjected from each plate.
RT-PCR.
Five microliters of cytoplasmic core preparation was subjected to reverse transcriptase PCR (RT-PCR) to check for exogenous RNA. The cMaster RTplusPCR system (Eppendorf) one-step RT-PCR method was used according to the manufacturer's recommendations. Nucleotides 7466 to 8620 of the SINV genome were amplified using the following primers: 5′-AGTGGCGGTGACCACCCGG (positive) and 5′-GCTTTTGCTGCTGCCAGACGATCCGCACCGCAATATGGC (negative).
PCR.
Detection of the ssDNA 109-mer in cell lysates was accomplished using routine PCR amplification with 109-mer-specific primers. Briefly, cells were lysed with TRIzol (Invitrogen), and total DNA was purified from the organic fraction, according to the manufacturer's instructions. The resultant DNA pellet was resuspended in 100 μl Milli-Q water, and 32.5 μl of this DNA was subjected to PCR using Platinum Pfx DNA polymerase and 109-mer-specific sense and antisense primers in a 50-μl reaction mixture.
Antibody neutralization.
Samples were diluted to 100 PFU/ml in PBS containing 1% FBS plus 10 mM CaCl2 and 10 mM MgCl2. A 1:10 dilution of neutralizing E2 antibody (monoclonal antibody [MAb] 202) or neat anticapsid 82CY polyclonal antibody (PAb) was added to 250 μl of diluted virus at volumes ranging from 5 μl to 0.1 μl, and the number of infectious units was quantified by plaque assay.
Plaque assay.
Tenfold serial dilutions of culture media or virus were made in PBS containing 1% FBS plus 10 mM CaCl2 and 10 mM MgCl2. BHK cells at ∼90% confluence were inoculated with 250 μl of the dilutions for 1 h at room temperature and then overlaid with MEM containing 5% FBS and 1% agarose. Cells were incubated at 37°C for 48 h and stained with neutral red.
ELISA.
Two milliliters of postmicroinjection media were concentrated to 100 μl using Amicon 100-kDa centrifugal filters and applied to a 96-well Polysorp ELISA plate (Sigma) overnight at 4°C. The samples were blocked with 5% BSA in PBS for 2 h followed by three washes with PBS-T (PBS + 0.1% Tween 20). A 1:500 dilution of a monoclonal anti-E2 antibody (202) or a 1:1,000 dilution of a polyclonal anti-RFP antibody (DS Red; Clontech) was applied to the sample for 2 h, followed by three washes with PBS-T. A 1:1,000 dilution of a horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody (E2) or a 1:1,500 dilution of HRP-conjugated goat anti-rabbit (RFP) secondary antibody was applied to the sample for 1 h, followed by three PBS-T washes. The tetramethylbenzidine (TMB) peroxidase detection system (KPL) was used to develop the ELISA according to the manufacturer's instructions. The optical density (OD) was determined at 450 nm.
Fluorescence microscopy.
Cells to be microinjected were grown on glass coverslips and were microinjected with NCs in which the N terminus of the capsid protein was fused to the enhanced yellow fluorescent protein (eYFP) (clone constructed as described in reference 11). Cells were fixed at various time points with 3.7% formaldehyde in PBS and examined using confocal microscopy (MRC Bio-Rad 1024).
Flow cytometry.
For each experiment, 1 million BHK cells in 100 μl PBS-5% calf serum were stained with 1:100 dilutions of a monoclonal anti-E2 primary antibody (SV127) and a goat anti-mouse fluorescein isothiocyanate (FITC) secondary antibody either 24 h post-transfection of pcDNA4/SINV/ENV or 8 h post-infection with Sindbis virus. Flow analysis was performed on a Beckman Coulter FC500 flow cytometer.
RESULTS
Detection of infectious units in the culture media after microinjection of cytoplasm-derived NCs.
Microinjection provides a method for delivering molecules directly into a cell. We examined whether SINV NCs could be microinjected into BHK cells and, if so, whether these NCs could be enveloped by SINV glycoproteins expressed by the microinjected cell. This provides a unique approach to dissect the transport and assembly pathway of SINV as well as the final step of virion budding.
Given the large diameter of the SINV NCs (∼400 Å), we first ascertained the feasibility of delivering SINV NCs into a mammalian cell by microinjection. BHK cells expressing the SINV envelope proteins (as described below) were microinjected with eYFP-tagged SINV NCs assembled in vitro and then fixed with formaldehyde at multiple intervals postmicroinjection. The presence of NCs within the cell was established using confocal microscopy (see Fig. S1 in the supplemental material). Given the ability to deliver SINV NCs into BHK cells, we next attempted to recover infectious units from the media of cells expressing the SINV envelope proteins after they had been microinjected with SINV NCs purified from the cytoplasm of infected cells.
BHK cells were transiently transfected with a plasmid encoding the SINV envelope proteins E3, an N-terminal RFP:E2 fusion, 6K, and E1. Cryo-electron microscopy (cryo-EM) reconstruction of the N-terminal RFP:E2 fusion shows that the RFP moiety lies laterally adjacent to each E2 molecule in a heterotrimer spike, thus allowing for maintenance of E2-receptor interaction (J. Jose and R. J. Kuhn, unpublished data). The E2 coding region of the RFP:E2 fusion was present either as wild type (wt) or as a double mutant, Y400A/L402A. The double mutant has been previously shown to be defective in budding because of an altered E2-NC interaction (12, 31). In the context of SINV containing the RFP:E2 fusion, the infectivity and release kinetics mimic those of the wild-type virus (Jose and Kuhn, unpublished). Accordingly, we sought to express in BHK cells the SINV envelope proteins containing this RFP:E2 fusion for two reasons: (i) the presence of the RFP fluorescence during microinjection allowed selective microinjection into cells expressing significant amounts of SINV envelope proteins and (ii) infectious units released after microinjection of NCs or RNA (at equal concentrations) would be expected to contain the RFP moiety.
Transfection of cells with the SINV envelope protein-expressing construct was performed 24 h prior to microinjection. Flow cytometry analysis of cell surface envelope protein levels indicated that at 24 h posttransfection E2 levels were equivalent to those of cells infected with virus for 8 h (see Table S1 in the supplemental material). Thirty RFP-expressing cells were selected from each plate, and wild-type (wt) SINV RNA or cytoplasmic NCs purified from cells infected with wt SINV were microinjected into them at concentrations of 8,000 NC particles/pl. NCs purified from the cytoplasm were relatively free of proteins other than the capsid protein (see Fig. S2), and when added directly to naïve cells, these purified NCs were incapable of themselves initiating an infection. In addition, prior to microinjection, cytoplasmic NC preparations were checked for the presence of exogenous copurified viral RNA using a temperature-dependent RT-PCR. Briefly, cytoplasmic NCs were subjected to parallel RT-PCRs in which the reverse transcriptase reaction was performed at either 38°C or 45°C. Above 42°C, cytoplasmic NCs begin to irreversibly denature (data not shown), exposing the encapsidated RNA to the reverse transcriptase enzyme. In contrast, at 38°C, RNA present within NCs remains unavailable to the RT enzyme. Thus, the reaction at 38°C acts as a detection for exogenous RNA, and the reaction at 45°C acts as an amplification control. Using this assay, no free RNA was detected in the NC samples used for microinjection.
Immediately prior to microinjection, some aliquots of cytoplasmic NCs were incubated at 55°C for 3 min in order to completely denature the NC structure, releasing the encapsidated RNA prior to microinjection. Because the encapsidated viral RNA is fully infectious, and the NCs were irreversibly denatured, the release of infectious units after microinjection of these aliquots could arise only from the translation of the released viral RNA and the consequent initiation of an infection within the cell.
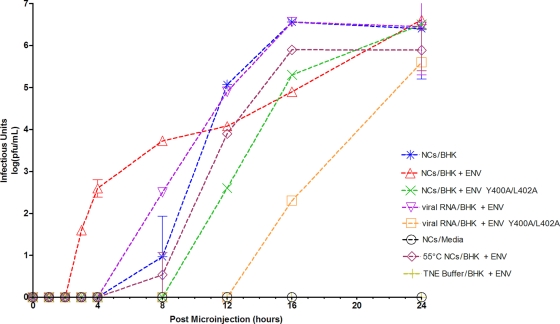
At multiple time points after microinjection, the culture medium was removed and replaced with fresh culture medium, as done in a one-step growth curve. The isolated culture media were subjected to a plaque assay to detect the presence of infectious units in the media, and the results are shown in Fig. 1. For all sample types that were microinjected, infectious units were never found in media removed immediately after microinjection (0-h time point). When cytoplasmic NCs were microinjected into cells expressing the viral envelope proteins, infectious units were detected in the media by 3 h after microinjection (Fig. 1). Conversely, when cytoplasmic NCs were microinjected into naïve BHK cells, infectious units were not detected until 8 h after microinjection, likely due to the disassembly of the microinjected cores and the translation and replication of the released RNA genome. Similarly, microinjection of viral RNA or of 55°C heat-denatured NCs, at a concentration of 8,000 molecules/pl or 8,000 NCs/pl, respectively, did not show the presence of infectious units in the culture medium until the 8-h time point. When medium from microinjected cells was overlaid onto naïve BHK cells for 24 h prior to plaque assay, in order to amplify the presence of released virions, infectious units were detected as early as 1 h after microinjection of cytoplasmic NCs into cells expressing wild-type envelope proteins; however, infectious units were not detected until >4 h after microinjection of viral RNA (see Fig. S3 in the supplemental material).
Fig. 1.
Detection of infectious unit release after microinjection of cells with nucleocapsids. Nucleocapsids (NCs) or viral RNAs were microinjected into the cell type as indicated. Cells expressed wild-type envelope proteins containing an RFP:E2 fusion (BHK + ENV), the envelope proteins containing the E2 Y400A/L402A mutations in addition to the RFP:E2 fusion (BHK + ENV Y400A/L402A), or no viral envelope proteins (BHK). In some cases, NCs were heat shocked at 55°C for 3 min immediately before microinjection. At the time points indicated on the x axis, media were removed from cells and replaced with fresh media. Media were assayed by plaque assay for the presence of infectious units, and the titer [log(PFU/ml)] was plotted as a function of time postmicroinjection. Microinjection of NC buffer (TNE) into BHK cells expressing the envelope proteins represented a mock sample, and NCs microinjected directly into the media provided an additional negative control.
Microinjection of either cytoplasmic NCs or viral RNA into BHK cells expressing the viral envelope proteins containing the E2 Y400A/L402A substitutions showed a marked delay in release of infectious units, with respect to that of the wild-type E2, with detection of infectious units first occurring (Fig. 1) at the 12-h time point for NCs and at 16 h for viral RNAs. SINV E2 residues Y400 and L402 are known to be critical residues involved in maintaining the structural integrity between the inner nucleocapsid and the outer envelope glycoproteins, through a 1:1 interaction between each molecule of capsid protein and E2 within the virion (10). E2 Y400 and L402 are also suggested to be important for the budding process itself (12). Microinjecting TNE buffer into cells, or microinjecting NCs into the culture medium rather than into cells, did not yield infectious units over the duration of the experiment (Fig. 1).
Infectivity of units in the culture medium can be neutralized using Sindbis virus-specific antibodies.
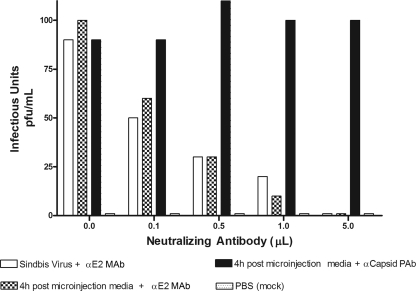
In order to gain insight into the properties of the infectious units detected after microinjection, we probed for protein content using antibodies directed against viral structural proteins. Medium isolated after microinjection of cytoplasmic NCs into envelope protein-expressing cells 4 h postmicroinjection was diluted to a concentration of 100 PFU/ml. This sample, along with purified SINV similarly diluted to 100 PFU/ml, was incubated with increasing volumes of either a neutralizing anti-SINV E2 monoclonal antibody or a nonneutralizing anti-SINV capsid antibody. As shown in Fig. 2, both the infectivity of the postmicroinjection medium and that of the purified virus decreased in similar fashions as the amount of neutralizing antibody increased. Addition of the nonneutralizing anti-SINV capsid antibody, which is directed against a capsid protein epitope not accessible in the enveloped virion, had no effect on infectivity. Thus, the released infectious units detected after microinjection of cytoplasmic NCs contain and require the presence of the E2 glycoprotein for their infectivity.
Fig. 2.
Neutralization of infectious units released after microinjection of nucleocapsids. The ability of infectious units in the postmicroinjection media to initiate an infection can be reduced by incubation with a known SINV-neutralizing antibody. One hundred PFU of infectious particles (from postmicroinjection media) or purified virus stock was incubated with different volumes of neutralizing antibody anti-E2 MAb, and the resultant infectivity was analyzed by plaque assay. Incubation with nonneutralizing anticapsid antibody, anticapsid PAb, was used as a negative control, and the addition of PBS to cells, rather than infectious media, represents a mock sample.
Heterologous infectious units are detected at early time points after microinjection.
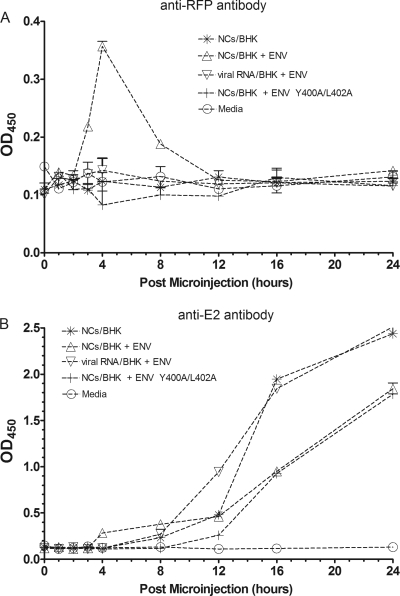
Virions released from cells after microinjection would be expected to encapsidate a wild-type genome because the microinjected NCs were derived from cells infected with wild-type virus. However, if the envelope proteins expressed in the microinjected cells were used for the budding of these microinjected NCs, then the virions should contain RFP:E2 proteins in their viral envelope. An ELISA was established to detect the presence of such particles containing the wild-type genome but also RFP:E2 molecules. Postmicroinjection medium was adsorbed to 96-well ELISA plates. Tenfold serial dilutions of purified wild-type SINV and of SINV containing the RFP:E2 fusion, both of predetermined titers, were used to generate standard curves. ELISA plates were probed with either an anti-RFP or an anti-E2 antibody, and signal was generated with HRP-conjugated secondary antibodies directed against the species of the primary antibody.
When samples were probed with the anti-RFP antibody, significant signal was found for the 3-h, 4-h, and 8-h time points, corresponding to the samples isolated after microinjection of cytoplasmic NCs into SINV envelope protein-expressing cells (Fig. 3A). Of interest is the decreased signal for the 8-h time point, relative to the 3-h and 4-h time points, given that plaque assay results demonstrate more infectious particles in the medium at 8 h than at other times. All other samples generated signals comparable to that of the mock control. However, when probed with an anti-E2 antibody (Fig. 3B), all samples yielded a signal that, compared to the standard curve, mimicked the plaque assay results shown in Fig. 1. Of particular interest is that the 3- and 4-h-postmicroinjection time points for the samples isolated after microinjection of cytoplasmic NCs into SINV envelope protein-expressing cells generated an above-background signal, relative to all other sample types, which mimics the results in Fig. 1. Sequencing of RNA isolated from postmicroinjection culture medium revealed the presence of the wild-type E2 gene in all cases.
Fig. 3.
ELISA detects release of RFP-containing VLPs after microinjection. Media isolated at various time points after microinjection of cytoplasm-derived NCs into cells expressing the viral envelope proteins were concentrated and applied to 96-well ELISA plates. Plates were probed with either an anti-RFP (DS Red) primary antibody (A) or an anti-E2 primary antibody (B) followed by an HRP-conjugated secondary antibody. The signal was measured optically at 450 nm. Medium isolated from cells expressing the envelope proteins, but not microinjected with NCs, was used as a negative control.
Released infectious units sediment similarly to Sindbis virions.
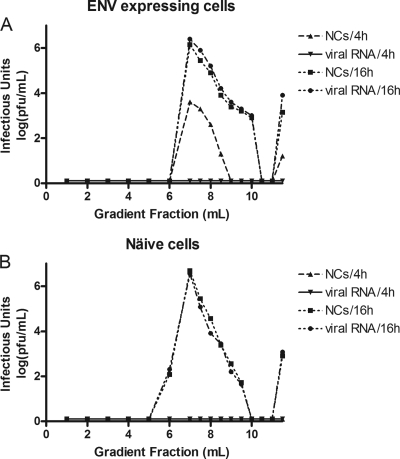
To further examine the properties of the infectious units detected in medium after microinjection, we applied aliquots of postmicroinjection medium to 0 to 30% continuous iodixanol gradients in the ultracentrifuge and subjected the resultant fractions taken from the gradients to infectious unit titration using plaque assay. Analysis was performed on medium taken at 4 h or 16 h after microinjection of either viral RNA or NCs (at concentrations identical to those in previous experiments) into either SINV envelope protein-expressing BHK cells or naïve BHK cells. Similar to a direct plaque assay of postmicroinjection medium (Fig. 1), no infectivity in any fraction was identified from postmicroinjection medium taken 4 h after microinjection of viral RNA into SINV envelope protein-expressing BHK cells. However, analysis of postmicroinjection medium taken 4 h after microinjection of NCs demonstrated infectious units in fractions 7 through 9, and primarily in fraction 7. The titer of infectious units seen was on the order of 103 PFU/ml in this fraction. Analysis of medium taken 16 h after microinjection of either viral RNA or NCs into SINV envelope protein-expressing cells showed a similar peak of infectivity in fraction 7, with a decrease in titer in subsequent fractions (Fig. 4A). In a similar experiment, in which molecules were microinjected into naïve BHK cells, an absence of infectivity was noted in medium taken 4 h postmicroinjection for both viral RNA and NCs, but the 16-h medium contained infectious units after microinjection of either viral RNA or NCs (Fig. 4B).
Fig. 4.
Gradient analysis of infectious units. Postmicroinjection media (1 ml) were applied to a continuous 10-ml 0 to 30% iodixanol gradient and centrifuged at 32,000 rpm in a Beckman SW-41 rotor for 2 h. At the end of the run, the gradient was fractionated such that the top 6 ml was removed in 1-ml fractions and the bottom 5 ml was removed in 500-μl fractions. The amount of infectious material in each fraction was assayed on BHK cells by plaque assay titration. Nucleocapsids (NCs) or viral RNAs were microinjected into BHK cells expressing the Sindbis virus envelope proteins (A) or naïve BHK cells (B). Postmicroinjection media were isolated at either 4 h or 16 h and applied to the gradient.
Microinjection of in vitro-assembled CLPs generates infectious units.
Assembly of NCs in vitro has been established using capsid protein isolated either from the cytoplasm of infected cells (28) or from Escherichia coli-expressed and purified capsid protein (23). Both single-stranded nucleic acids and other anionic polymers can be encapsidated. Structurally, in vitro-assembled NCs, referred to as core-like particles (CLPs), mimic their authentic counterparts (9). It is not known, however, if in vitro-assembled CLPs can functionally substitute for NCs assembled in virus-infected cells. Using E. coli-expressed and purified capsid protein, we assembled CLPs in vitro with a random ssDNA 109-mer and microinjected these particles into cells transiently expressing the viral envelope proteins (as described above). Importantly, the CLPs described here do not contain an infectious nucleic acid. Thus, if particles from the postmicroinjection medium are capable of entering overlaid naïve cells, they must have resulted from the direct budding of CLPs.
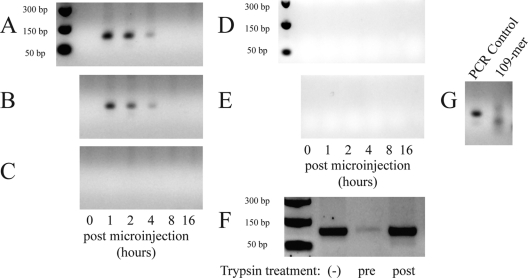
At multiple time points postmicroinjection, the culture media were harvested and replaced with fresh media. Naïve BHK cells were incubated with 250 μl of each sample of culture medium along with antibody to either SINV capsid protein or SINV E2 protein at 20°C (for antibody adsorption) or 4°C (for trypsin treatment), to allow maximum adsorption of any infectious units in the medium, and then shifted to 37°C for 20 min to allow internalization of any bound infectious units. Media were removed, and cells were washed with PBS and then lysed with TRIzol (Invitrogen). Total cellular DNA was purified from the lysates per the manufacturer's protocol. Purified DNA was subjected to a standard PCR amplification using primers specific to the 109-mer used to assemble NCs in vitro. As shown in Fig. 5A, infectious units were detected in the 1-h-, 2-h-, and, to a lesser extent, the 4-h-postmicroinjection media, but not in the 0-h, 8-h, or 16-h samples. As demonstrated for infectious units released after microinjection of cytoplasmic NCs, a neutralizing SINV antibody directed to the E2 glycoprotein was able to inhibit the entry into naïve cells by infectious units released after microinjection of in vitro-assembled CLPs, whereas an anticapsid antibody was not (Fig. 5B and C).
Fig. 5.
Detection of infectious unit release after microinjection of cells with in vitro-assembled CLPs. Media were harvested at various time points after BHK cells expressing the Sindbis virus envelope proteins were microinjected with in vitro-assembled CLPs containing 109-mer ssDNA oligonucleotides. Two hundred fifty microliters of medium was adsorbed to naïve BHK cells for 1 h at room temperature with or without addition of antibody. Naïve cells were then shifted to 37°C for 20 min to allow endocytosis of any bound virions. Cells were then lysed with TRIzol reagent, and total DNA was purified. PCR was used to screen for the presence of the ssDNA 109-mer (encapsidated within released virions) inside the naïve cells. PCRs were electrophoresed on a 2% Tris-acetate-EDTA agarose gel. (A) Media were incubated with PBS prior to adsorption on naïve cells. (B) Media were incubated with a nonneutralizing anticapsid antibody prior to adsorption on naïve cells. (C) Media were incubated with a neutralizing anti-E2 antibody prior to adsorption on naïve cells. (D) CLPs were microinjected into naïve cells (not expressing Sindbis virus envelope proteins). (E) The 109-mer was microinjected into cells expressing Sindbis virus envelope proteins. (F) Postmicroinjection media from panel A were adsorbed onto naïve BHK cells for 30 min at 4°C, and then, prior to cell lysis, cells were either shifted to 37°C for 20 min (−), treated with 4°C trypsin for 3 min before being shifted to 37°C for 20 min (pre), or shifted to 37°C for 20 min and then treated with 4°C trypsin for 3 min (post). (G) Controls: a PCR amplification control using the 109-mer molecule as a template (left) and electrophoresis of the actual 109-mer oligonucleotide itself (right). Note that panels A to E and G contain cropped portions of a single gel.
Furthermore, if the 109-mer molecule alone was microinjected into SINV envelope-expressing cells, or if microinjection of in vitro-assembled CLPs assembled with the 109-mer into naïve (not expressing SINV envelope proteins) cells was performed, infectious units were not detected in the postmicroinjection media (Fig. 5D and E). Sequencing the PCR product confirmed that it was derived from the 109-mer template, and attempts to amplify the same product from a naïve cell lysate were negative.
If cells were briefly (3 min) treated with trypsin after adsorption of postmicroinjection media but before return to 37°C to allow endocytosis of adsorbed VLPs, the number of infectious units entering naïve cells was reduced, whereas trypsin treatment of cells after the shift to 37°C had no effect on 109-mer VLP internalization relative to that of a control (Fig. 5F). Thus, taken together, microinjection of in vitro-assembled CLPs yields release of E2-containing particles capable of infecting naïve cells.
DISCUSSION
This report demonstrates that microinjection of Sindbis virus NCs into cells transiently expressing the structural envelope proteins can produce infectious virus-like particles (VLPs). Furthermore, the results outlined here demonstrate, for the first time, that in vitro-assembled Sindbis virus CLPs can act as functional surrogates for cytoplasmic NCs during the assembly and budding processes. This system allows for the study of the alphavirus assembly and budding processes in the absence of an authentic viral infection and permits facile manipulation of the viral components.
When NCs and viral RNA were microinjected in parallel and at identical concentrations, both plaque assays and ELISA showed the release of infectious units much earlier for NC microinjection than for viral RNA microinjection. Introducing the NCs directly into the culture medium did not yield infectious units, confirming that naked NCs are not infectious. Thus, the presence of infectious units shortly after microinjection requires introduction of intact NCs into cells expressing the Sindbis virus envelope proteins.
The experiments described here provide insight into several unanswered questions related to alphavirus budding.
First, our results indicate that cytoplasmic NCs preformed within the cell are not dead-end products. Two models for alphavirus budding have been proposed (5). In the first, a “membrane-enwrapping” model, capsid protein and viral genomic RNA self-assemble into distinct NCs that subsequently interact with the cytoplasmic domains of the viral envelope proteins, specifically E2, and bud from the cell. In the second model, capsid protein and viral genomic RNA are loosely associated at the plasma membrane, and formation of the icosahedral NCs, as well as budding, is mediated by lateral interactions of the envelope proteins residing in the plasma membrane. This second model is supported by evidence that preformation of NCs within the cytoplasm is not a requirement for release of infectious virus. More explicitly, Forsell et al. suppressed the ability of SFV capsid protein to associate into cytoplasmic NCs by deleting the sequence responsible for CP-CP interaction, yet virus was released at nearly wild-type levels (4). Other groups reached similar conclusions, confounding the previously held dogma that NCs found within a virion are solely derived from cytoplasmic NCs (8, 18). Our results indicate that budding of preassembled cytoplasmic NCs, as well as of in vitro-assembled CLPs, is possible. However, our results do not exclude the possibility that budding is associated with nucleocapsid formation at the plasma membrane.
A second interesting observation of the current work is that only a small proportion of microinjected NCs appear to be incorporated into VLPs. Based on the number of cells microinjected and the concentration of microinjected NCs, we estimate that 104 to 105 NCs were microinjected per plate, yet the number of VLPs detected by plaque assay at early time points was about 103, or ≤10% of the number of NCs microinjected. It is known that only a small amount of synthesized capsid protein is incorporated into released virions (16), and yet the bulk of capsid protein is rapidly incorporated into NCs (19). Perhaps the majority of NCs formed within the cytoplasm are dead-end products, assembled into a conformation not amenable to budding. The small percentage of NCs released seen in these experiments may be due to a number of different factors. The titer of infectious units may be underestimated in that some released VLPs are likely to bind to and enter naïve cells and are not collected when the medium is removed at intervals postmicroinjection. Other factors, such as disassembly of a large fraction of NCs after microinjection, heterogeneity of the pool of cytoplasmic NCs isolated and subsequently microinjected, inefficiency of the budding process involving intact NCs, or the requirement that budding of NCs involve early interaction with newly synthesized E2, can also be the cause of lower numbers of detected budded particles after microinjection.
With regard to the latter point, it is possible that NCs associate with the viral envelope proteins at some point prior to arrival of the envelope proteins at the plasma membrane. This association is perhaps mediated by the interaction of the cytoplasmic domain of E2 with individual capsid protein molecules within the NCs. Garoff et al. have postulated a scenario in which capsid protein and RNA associate near replication complexes, with the envelope proteins transiting to the plasma membrane only after visiting these sites, thereby providing an initial interaction point for NCs and the envelope proteins (5). Indeed, a review of the ELISA (Fig. 3) data may support this model. Microinjection of viral RNA into cells expressing the RFP:E2 variant of SINV envelope proteins yielded release of virions containing only wild-type E2, although cell surface envelope protein levels at the time of microinjection are similar to that of a cell infected for 8 h with virus (see Fig. S1 in the supplemental material) and, additionally, the RFP:E2 variant remains at the cell surface throughout the duration of the experiment (data not shown). This suggests that the NCs created during the infection initiated by the microinjected viral RNA fail to use the envelope proteins expressed by the mammalian expression plasmid, and thus, NC assembly and subsequent interaction with the cytoplasmic domain of E2 may be temporally and spatially linked.
A third result, of interest to work on both alphavirus assembly and disassembly, is that microinjected NCs appear to be unstable within the cell. Specifically, microinjection of cytoplasmic NCs into naïve BHK cells initiates a productive infection after a brief lag period (Fig. 1). Although the structural conformation of an encapsidated NC likely changes after envelopment (5), it is curious that NCs within the cytoplasm of an infected cell appear stable, whereas the same NCs disassemble rapidly after being introduced into the cytoplasm of a naïve cell during infection (27). In this light, it is surprising to see the presence of infectivity in cell media after microinjection of NCs into naïve cells. As described above, NCs were assayed for the presence of exogenous viral RNA prior to microinjection; thus, the source of the infection seen after microinjection of NCs into naïve cells should be the viral RNA encapsidated within the NCs. At 8 h, the titer of infectious units in cell media is about 100 times lower for NCs microinjected into naïve cells than for viral RNA (vRNA) that has been microinjected (Fig. 1). However, by 12 h after microinjection, the titers are nearly identical; thus, between 8 and 12 h after microinjection of NCs into naïve cells, a large pool of viral RNA apparently has become available for translation, or perhaps even directly for encapsidation. As the time frame of SINV infection in cell culture is on the order of several hours (21), it is not known if the half-life of an NC during a normal infection—in which the NC is delivered to the cytoplasm via the endosomal pathway, rather than directly via microinjection—is of the same order. Nor is it known if an authentic alphavirus infection prevents some cellular factor from disassembling newly formed NCs during infection, whereas such a factor is available for NC disassembly in the context of our system, in which the neither the capsid protein nor the viral nonstructural proteins are produced.
Wengler et al. proposed that ribosomes can bind to the capsid proteins and facilitate disassembly of incoming NCs in the cytoplasm of a newly infected cell, whereas the ribosomes of an infected cell are saturated with capsid protein and thus unable to participate in the disassembly of newly assembled cytoplasmic NCs (29, 30). It has also been shown that NCs entering the cytoplasm of cells harboring SFV replicons did not disassemble (17), suggesting that the alphavirus nonstructural proteins inhibit some cellular process capable of NC disassembly. Thus, alphavirus NCs may be inherently unstable in the naïve cell. We cannot rule out, however, the possibility that the microinjection or NC purification processes may cause mechanical disruption of some NCs, allowing release of the encapsidated RNA to the cell and thus allowing an infection to arise in naïve cells.
ELISA results demonstrated that VLPs released at early time points after microinjection of cytoplasmic NCs into cells expressing wild-type envelope proteins originated from budding and not the generation of virus from translation of the encapsidated RNA. Specifically, probing adsorbed virions with an anti-RFP antibody showed the presence of RFP within the infectious VLPs at the 3-h-, 4-h-, and 8-h-postmicroinjection time points, whereas no RFP:E2s are detected in virions at later times. Ekstrom et al. (3) have developed a system to study alphavirus budding in which cells producing two different species of SFV E2 were examined. Analysis of released particles showed that virions can indeed incorporate two different species of E2 (3). However, in this case, expression of the envelope proteins was driven by the viral replicase whereas, in the system described here, one set of envelope proteins is transcribed from a plasmid, thus being provided in trans. At any rate, it is clear that the infectious units detected after microinjection of NCs are composed of two populations: the first due to budding of microinjected NCs, yielding VLPs, and the second due to initiation of a novel infection either from VLPs entering naïve cells or from disassembly of microinjected NCs and translation of the viral genome.
Interestingly, although fluorescence microscopy demonstrated that RFP:E2 is present in the cell plasma membrane throughout the course of the experiment (data not shown), RFP:E2 is not detected in released virions after microinjection of viral RNA into cells expressing the RFP:E2-tagged SINV envelope proteins. This may indicate that interaction of NCs and envelope proteins occurs early after and in close proximity to translation of the structural protein polypeptide, before the envelope proteins reach the plasma membrane. Indeed, recently published data on the structure of alphavirus cytopathic vacuole type II (CPV-II) vacuoles, which are believed to transport the envelope proteins to the site of budding, show that these vacuoles may be decorated with NCs (20).
Microinjection of NCs or viral RNA into cells expressing the envelope proteins containing the E2 Y400A/L402A double mutant shows both a marked delay in release and decreased titer of virus after microinjection. Within a virion, Y400 and L402 of E2 interact 1:1 with a hydrophobic pocket present on the capsid protein within the NC (10). When the E2 Y400A/L402A double mutant is expressed from full-length viral RNA, NCs are found throughout the cell rather than associated with cellular membranes, and release of virus is inhibited, almost certainly because the E2-capsid interaction is disturbed (12). Thus, it was expected that formation of infectious units following microinjection of NCs into cells expressing the E2 Y400A/L402A double mutant would be delayed relative to that into cells expressing the wild-type envelope proteins, likely mimicking the release profile of NCs microinjected into naïve cells. However, it was surprising that there was a significantly longer delay in release, and decreased titer, of virus after microinjection into the E2 Y400A/L402A-expressing cells. These results may indicate that, even after translation of wild-type envelope proteins from RNA from microinjected NCs or viral RNA, the defective E2 Y400A/L402A may still be capable of interfering with virus release, perhaps because through their intact lateral interactions they form patches of close-packed regions that make more difficult the wrapping of the capsid by wt E2 proteins. Virions released from cells expressing the E2 Y400A/L402A double mutant were not examined for their composition, and it is unknown if any molecules of E2 Y400A/L402A are incorporated into released virions. Based on a comparison between the plaque assay (Fig. 1) and ELISA results (Fig. 3), the specific infectivity of the particles that are released from the E2 Y400A/L402A-expressing cells does not appear to be decreased.
A system was recently described in which an alphavirus replicon expressed the vesicular stomatitis virus G protein (VSV-G) and initiated a self-propagating infection (14, 15). However, it was found that the infectious material was likely of vesicular origin, containing alphavirus replicases enveloped in VSV-G-containing membranes. Given some of the parallels of our approach with this previously described system (overexpression of viral envelope proteins and introduction of infectious materials into cells), we examined the sedimentation properties of the VLPs generated in our approach. The sedimentation property of VLPs released from cells microinjected with cytoplasm-derived nucleocapsids was examined, and it was found that VLPs migrated in a continuous gradient in a fashion very similar to that of authentic Sindbis virions derived from cells microinjected with full-length viral RNA (Fig. 4). Namely, the bulk of infectivity for VLPs was derived from particles that migrated down into the fraction, like virions.
CLPs have proven a useful tool for studying the synthesis and assembly process of alphavirus NCs (24, 25). Authentic NCs and CLPs have been shown to be structurally similar; however, the capacity with which CLPs can take part in the virion assembly and disassembly processes has not been explored. To address this question and to simultaneously utilize an encapsidated nucleic acid incapable of initiating an infection, we tested the ability of CLPs to bud from cells. The data presented in Fig. 5 show that CLPs can be incorporated into virus-like particles that are competent for egress from the cell. Indirectly, the assay also demonstrates that these virus-like particles (VLPs) can bind to and enter naïve cells. Thus, it appears that CLPs mimic the cytoplasmic NCs not only in structure but also in function. Of interest is the apparent “acceleration” in release of VLPs containing CLPs relative to that of VLPs containing cytoplasm-derived NCs. Specifically, the bulk of VLPs containing CLPs appear in the culture media within 2 h, whereas the majority of VLPs released after microinjection of cells with cytoplasmic NCs occur after 2 h (Fig. 1 and 4). The nature of this differential VLP release kinetics is not known but could potentially be explained if the pool of purified cytoplasmic NCs used for microinjection contains many NCs assembled in a conformation not amenable to budding and thus they remain in the cell.
The data presented here describe a system to study the viral budding process independent of a viral infection. The ability to differentially label the components that assemble into a virion provides a powerful set of tools to dissect the process of assembly in vivo. This system provides a new platform to study viral and host requirements for budding, and the data described here provide additional insight into the alphavirus budding process. Of particular interest is that (i) microinjected, fully intact NCs derived from the cytoplasm of infected cells are capable of budding from otherwise-naïve, glycoprotein-expressing cells; (ii) in vitro-assembled CLPs appear to be able to act as functional surrogates for their cytoplasmic assembled counterparts, in terms of their budding competence; and (iii) microinjected, fully intact NCs result in the initiation of infection in naïve cells. Although not investigated, this system should also prove amenable to study the process of NC disassembly.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rushika Perera for useful discussions and Justin Meyers for assistance with flow cytometry experiments. We acknowledge the use of the Flow Cytometry and Cell Separation Facility of the Bindley Bioscience Center.
We also acknowledge support from the NIH through NIGMS award GM56279 (to R.J.K.), from the NSF through award CHE07-14411 (to C.M.K. and W.M.G.), from NIH Biophysics Training Grant 5T32GM008296-21 to J.E.S., from NIH Virology Training Grant 5T32AI060567-07 to O.A., and from NIH/NCRR grant RR025761 to the Flow Cytometry and Cell Separation Facility.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 6 April 2011.
REFERENCES
- 1. Cheng F., Mukhopadhyay S. 18 February 2011. Generating enveloped virus-like particles with in vitro assembled cores. Virology doi:10.1016/j.virol.2011.02.001 [DOI] [PubMed] [Google Scholar]
- 2. Cheng R. H., et al. 1995. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 80:621–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ekstrom M., Liljestrom P., Garoff H. 1994. Membrane protein lateral interactions control Semliki Forest virus budding. EMBO J. 13:1058–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Forsell K., Griffiths G., Garoff H. 1996. Preformed cytoplasmic nucleocapsids are not necessary for alphavirus budding. EMBO J. 15:6495–6505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garoff H., Sjoberg M., Cheng R. H. 2004. Budding of alphaviruses. Virus Res. 106:103–116 [DOI] [PubMed] [Google Scholar]
- 6. Geigenmuller-Gnirke U., Nitschko H., Schlesinger S. 1993. Deletion analysis of the capsid protein of Sindbis virus: identification of the RNA binding region. J. Virol. 67:1620–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jose J., Snyder J. E., Kuhn R. J. 2009. A structural and functional perspective of alphavirus replication and assembly. Future Microbiol. 4:837–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee S., et al. 1996. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure 4:531–541 [DOI] [PubMed] [Google Scholar]
- 9. Mukhopadhyay S., Chipman P. R., Hong E. M., Kuhn R. J., Rossmann M. G. 2002. In vitro-assembled alphavirus core-like particles maintain a structure similar to that of nucleocapsid cores in mature virus. J. Virol. 76:11128–11132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mukhopadhyay S., et al. 2006. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure 14:63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Orvedahl A., et al. 2010. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe 7:115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Owen K. E., Kuhn R. J. 1997. Alphavirus budding is dependent on the interaction between the nucleocapsid and hydrophobic amino acids on the cytoplasmic domain of the E2 envelope glycoprotein. Virology 230:187–196 [DOI] [PubMed] [Google Scholar]
- 13. Perera R., Owen K. E., Tellinghuisen T. L., Gorbalenya A. E., Kuhn R. J. 2001. Alphavirus nucleocapsid protein contains a putative coiled coil alpha-helix important for core assembly. J. Virol. 75:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rolls M. M., Webster P., Balba N. H., Rose J. K. 1994. Novel infectious particles generated by expression of the vesicular stomatitis virus glycoprotein from a self-replicating RNA. Cell 79:497–506 [DOI] [PubMed] [Google Scholar]
- 15. Rose N. F., Publicover J., Chattopadhyay A., Rose J. K. 2008. Hybrid alphavirus-rhabdovirus propagating replicon particles are versatile and potent vaccine vectors. Proc. Natl. Acad. Sci. U. S. A. 105:5839–5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scheele C. M., Pfefferkorn E. R. 1969. Kinetics of incorporation of structural proteins into Sindbis virions. J. Virol. 3:369–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Singh I. R., Suomalainen M., Varadarajan S., Garoff H., Helenius A. 1997. Multiple mechanisms for the inhibition of entry and uncoating of superinfecting Semliki Forest virus. Virology 231:59–71 [DOI] [PubMed] [Google Scholar]
- 18. Skoging-Nyberg U., Liljestrom P. 2001. M-X-I motif of Semliki Forest virus capsid protein affects nucleocapsid assembly. J. Virol. 75:4625–4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Soderlund H. 1973. Kinetics of formation of the Semliki Forest virus nucleocapsid. Intervirology 1:354–361 [DOI] [PubMed] [Google Scholar]
- 20. Soonsawad P., et al. 2010. Structural evidence of glycoprotein assembly in cellular membrane compartments prior to alphavirus budding. J. Virol. 84:11145–11151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Strauss J. H., Strauss E. G. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Suomalainen M., Liljestrom P., Garoff H. 1992. Spike protein-nucleocapsid interactions drive the budding of alphaviruses. J. Virol. 66:4737–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tellinghuisen T. L., Hamburger A. E., Fisher B. R., Ostendorp R., Kuhn R. J. 1999. In vitro assembly of alphavirus cores by using nucleocapsid protein expressed in Escherichia coli. J. Virol. 73:5309–5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tellinghuisen T. L., Kuhn R. J. 2000. Nucleic acid-dependent cross-linking of the nucleocapsid protein of Sindbis virus. J. Virol. 74:4302–4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tellinghuisen T. L., Perera R., Kuhn R. J. 2001. In vitro assembly of Sindbis virus core-like particles from cross-linked dimers of truncated and mutant capsid proteins. J. Virol. 75:2810–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tzlil S., Deserno M., Gelbart W. M., Ben-Shaul A. 2004. A statistical-thermodynamic model of viral budding. Biophys. J. 86:2037–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wengler G. 2009. The regulation of disassembly of alphavirus cores. Arch. Virol. 154:381–390 [DOI] [PubMed] [Google Scholar]
- 28. Wengler G., Boege U., Bischoff H., Wahn K. 1982. The core protein of the alphavirus Sindbis virus assembles into core-like nucleoproteins with the viral genome RNA and with other single-stranded nucleic acids in vitro. Virology 118:401–410 [DOI] [PubMed] [Google Scholar]
- 29. Wengler G., Gros C. 1996. Analyses of the role of structural changes in the regulation of uncoating and assembly of alphavirus cores. Virology 222:123–132 [DOI] [PubMed] [Google Scholar]
- 30. Wengler G., Wurkner D. 1992. Identification of a sequence element in the alphavirus core protein which mediates interaction of cores with ribosomes and the disassembly of cores. Virology 191:880–888 [DOI] [PubMed] [Google Scholar]
- 31. Zhao H., Lindqvist B., Garoff H., von Bonsdorff C. H., Liljestrom P. 1994. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 13:4204–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.