Abstract
African swine fever virus (ASFV) is a highly infectious disease of domestic pigs, with virulent isolates causing a rapidly fatal hemorrhagic fever. In contrast, the porcine species endogenous to Africa tolerate infection. The ability of the virus to persist in one host while killing another genetically related host implies that disease severity may be, in part, modulated by host genetic variation. To complement transcription profiling approaches to identify the underlying genetic variation in the host response to ASFV, we have taken a candidate gene approach based on known signaling pathways that interact with the virus-encoded immunomodulatory protein A238L. We report the sequencing of these genes from different pig species and the identification and initial in vitro characterization of polymorphic variation in RELA (p65; v-rel reticuloendotheliosis viral oncogene homolog A), the major component of the NF-κB transcription factor. Warthog RELA and domestic pig RELA differ at three amino acids. Transient cell transfection assays indicate that this variation is reflected in reduced NF-κB activity in vitro for warthog RELA but not for domestic pig RELA. Induction assays indicate that warthog RELA and domestic pig RELA are elevated essentially to the same extent. Finally, mutational studies indicate that the S531P site conveys the majority of the functional variation between warthog RELA and domestic pig RELA. We propose that the variation in RELA identified between the warthog and domestic pig has the potential to underlie the difference between tolerance and rapid death upon ASFV infection.
INTRODUCTION
African swine fever (ASF) virus (ASFV) is a pathogen of the Suidae (domestic and wild pig species), which may be transmitted directly or via an arthropod vector in the form of Ornithodorus ticks (35). ASFV is highly infectious, with virulent isolates causing an acute, rapidly fatal hemorrhagic fever in domestic pigs (Sus scrofa) (10, 34). This is thought, in part, to be the result of a proinflammatory cytokine storm driven by infected macrophages (9, 15, 16, 42–44, 56). Initiation of a systemic inflammatory response results in severe hematological and vascular perturbations, ultimately leading to cardiovascular collapse in a manner not dissimilar to septic shock (3, 17, 18, 20, 21, 40, 52, 53). In addition to hemorrhage, severe widespread apoptosis of infected macrophages and uninfected lymphocytes is a prominent feature of the disease; this is also likely related to markedly elevated proinflammatory cytokine levels (19, 33, 37, 43). In comparison to the severe disease which occurs in domestic pigs, in its natural hosts, warthogs (Phacochoerus sp.) and bushpigs (Potamochoerus sp.), ASF is subclinical and persistent (2, 32, 50, 51).
ASFV is notifiable to the World Organization for Animal Health (OIE), placing it in the highest category of infectious animal pathogens. It exhibits remarkable potential for transboundary spread, and outbreaks in domestic pig populations have a serious socioeconomic impact worldwide. Furthermore, ASF is considered to be the major limiting factor to pig production in Africa (34). ASFV is a large, double-stranded DNA virus and the only member of the Asfarviridae family (12), suggesting that it may carry novel genes that are not carried by other virus families. Furthermore, the ability of the virus to persist in one host while killing another genetically related host alludes to the possibility that disease severity may, in part, be modulated by host genetic variation.
Several candidate ASFV-encoded immune modulatory factors have been identified, including homologues of CD2 (8-DR/CD2v) (5, 6, 41), IAP (A224L) (31, 39), Bcl-2 (A179L; 5-HL) (1, 7, 8, 30), and IκBα (A238L; 5-EL) (36, 49). Of these, A238L shares 40% sequence homology and 20% identify with domestic pig IκBα (NFKBIA) and substitutes for NFKBIA by binding to the RELA (p65; v-rel reticuloendotheliosis viral oncogene homolog A) subunit of NF-κB. Thus, A238L reduces the ability of NF-κB to be activated (36, 49). In addition to inhibiting host NF-κB, A238L also suppresses calcineurin phosphatase activation of NFAT signaling by the following two mechanisms: direct binding to calcineurin phosphatase 3, β isoform (PPP3CB), and binding to the immunophilin carrier cyclophilin A (PPIA) in a manner similar to that of the immunosuppressive drug cyclosporine A (28, 29).
Various groups have initiated transcription profiling of host genes implicated in ASFV infection (15, 16, 42–44, 56). These studies identified numerous upregulated host genes, but to date, all are limited to analysis in domestic pig cells. In this study, we take a complementary approach to this question by the testing of variation in targeted candidate genes. Clearly A238L represents a novel and versatile immunoregulatory mechanism by which ASFV can inhibit both the NF-κB and NFAT signaling pathways (11, 28, 29, 36, 49). We therefore consider the three A238L target proteins, RELA, PPP3CB, and PPIA, and the two proteins it mimics, NFKBIA and NFATC1, as candidates for the genetic variation between pig species which may contribute to species-specific responses to ASFV infection. We now report the sequencing of these genes from different pig species and identification and initial in vitro characterization of polymorphic variation in one of them.
MATERIALS AND METHODS
mRNA isolation, cDNA synthesis, and DNA sequencing.
Whole blood (5 ml) was collected into EDTA from a domestic pig (Sus scrofa, commercial pig; United Kingdom), common warthog (Phacochoerus africanus; Rotterdam Zoo, Holland), and babirusa (Babyrousa babyrussa; Marwell Zoo, United Kingdom) and transferred immediately into DNA/RNA stabilization reagent for blood/bone marrow (Roche Diagnostics). This was processed using an mRNA isolation kit for white blood cells (Roche Diagnostics). Initially, cDNA libraries were synthesized using the SMART RACE (rapid amplification of cDNA ends) cDNA amplification kit (Clontech) to enable partial sequencing and design of species-specific primers (Table 1) for synthesis of individual cDNAs using proofreading PCR using a 1-μl sample in a 25-μl PCR mixture consisting of 20 pmol of each primer in 2 mM MgCl2 and 2 mM deoxynucleoside triphosphates (dNTPs) with 0.7 U High-Fidelity DNA polymerase (Roche Diagnostics). The PCR cycling conditions used were optimized for each gene (data not shown). PCR products were resolved on a 1% agarose gel, excised with a scalpel blade, and extracted using a QIAquick gel extraction kit (Qiagen). These were cloned into pGEM-T Easy (Promega) and sequenced in both directions. To achieve the full coding sequence for NFKBIA, 5′-end fragments were amplified directly from genomic DNA.
Table 1.
Gene-specific PCR primers
| Gene/domain | Primer sequence (5′–3′) |
|
|---|---|---|
| Sense | Antisense | |
| PPIA (cDNA) | ATC TNT CAG TGC TGC TCA GC | CAG AAG GAA TGG TCT GAT GG |
| NFKBIA (cDNA) | AAG GAG CGG CTA CTG GAC G | CAT GGT CTT TTA GAC ACT TTC CA |
| NFKBIA (5′-end DNA) | CTC ATC GCA GGG AGT TTC TC | TCC TCG TCC TTC ATG GAG TC |
| NFATC1 (cDNA) | ATC TCA GCT GTT GGG TCA GC | AGT GAG GGT GAG TGG TCC AG |
| PPP3CB (cDNA) | CCC AAC ACA TCG TTT GAC AT | ATG TGA GAG TCC CTG GGA AG |
| RELA (cDNA) | GAC CTC TTC CCC CTC ATC TT | CCC CTT AGG AGC TGA TCT GA |
| RELA transactivation domains (DNA) | GGA AGG GAC ACTGAC AGA GG | TCA GAA GGG CTG AGA AGT CC |
Genomic DNA isolation and sequencing.
Samples of skeletal muscle were collected into 20% dimethyl sulfoxide (DMSO)-saturated salt (NaCl) solution and stored at −70°C. DNA was extracted from 0.2 g muscle using the BACC2 extraction kit for blood and cell cultures (Nucleon Biosciences). For PCR, 50 ng genomic DNA was used as template in a 25-μl PCR mixture consisting of 20 pmol of each primer in 2 mM MgCl2 and 2 mM dNTPs, with 0.7 U High-Fidelity DNA polymerase (Roche Diagnostics).
Plasmid construction.
Restriction sites were introduced into RELA products from the above-described sequencing study by nested PCR; this enabled insertion of domestic pig and warthog RELA genotypes into the multiple cloning site of the pFLAG-CMV-4 vector (Sigma-Aldrich). PCR products were diluted to 1:500 in sterile water, and 1 μl was used as a template in a 25-μl PCR mixture consisting of 20 pmol of each primer (forward HindIII [5′-CCA AGC TTG ACC TCT TCC CCC TCA TCT T-3′] and reverse NotI [5′-GCG CGG CCG CTT AGG AGC TGA TCT GA-3′]) in 2 mM MgCl2 and 2 mM dNTPs, with 0.7 U High-Fidelity DNA polymerase (Roche Diagnostics). Restriction sites are underlined. Each ∼1.6-kbp PCR product was resolved on a 1% agarose gel, excised with a scalpel blade, and extracted using the QIAquick gel extraction kit (Qiagen). Following HindIII and NotI restriction digestion of the vector and RELA PCR products, the products were ligated into the open plasmid. These constructs allowed constitutive expression of warthog RELA and domestic pig RELA driven by the cytomegalovirus (CMV) promoter. In addition, an N-terminal eight-amino-acid FLAG sequence is incorporated into the protein, which is recognized by an anti-FLAG monoclonal antibody. Site-directed mutagenesis was preformed using a QuikChange II site-directed mutagenesis kit (Stratagene).
Plasmid transient transfection was performed using Lipofectamine2000 (Invitrogen) in ∼1 × 105 cells/well of a 12-well plate in culture medium at 37°C and 5% CO2. COS-7 cells were cultured in Glasgow minimal essential medium (GMEM; Sigma), containing 10% fetal bovine serum (Invitrogen), 1% l-glutamine (Invitrogen), 1% sodium pyruvate (Invitrogen), 1% nonessential amino acids (Invitrogen), and 0.2% β-mercaptoethanol (Invitrogen). Mouse embryonic fibroblasts (MEFs) and RELA−/− MEFs were cultured in Dulbecco's modified Eagle medium (DMEM; Sigma), containing 10% fetal bovine serum (Invitrogen). Luciferase activity was determined in triplicate using the dual reporter assay system (Promega) and analyzed with Excel (Microsoft), with total protein measured with the bicinchoninic acid (BCA) protein assay kit (Pierce). The data are presented as the means ± standard deviations; statistical significance was evaluated using the unpaired Student t test, with a difference between groups being considered statistically significant if the P value of the comparison was <0.05.
Western blotting.
For Western blot analysis, denatured protein (10 μg) was run in Tris-glycine-SDS running buffer (National Diagnostics) on precast NuPage 12% Tris-glycine gels (Invitrogen) before transfer to a nitrocellulose membrane (National Diagnostics). Proteins were visualized using anti-FLAG M2-peroxidase (horseradish peroxidase [HRP]) (Sigma)-conjugated primary antibody and mouse β-actin primary antibody with goat anti-mouse IgG HRP (Sigma)-conjugated secondary antibody and detected with Immobilon Western (Millipore) chemiluminescent HRP substrate.
RESULTS
Limited sequence variation in candidate porcine genes.
cDNAs were produced for PPIA, NFKBIA, NFATC1 (regulatory domain), PPP3CB, and RELA. These were obtained from the following three pig species: the domestic pig (Sus scrofa), common warthog (Phacochoerus africanus), and babirusa (Babyrousa babyrussa). The domestic pig and warthog represent ASFV-susceptible and ASFV-tolerant species, respectively. The “outspecies” is the babirusa, which is considered to be the most ancient extant species of pig; its range is restricted to the island of Sulawesi in the Indonesian archipelago. It has no common ancestor with the domestic pig more recently than approximately 10 to 19 million years ago (38). It is not known how this pig species would respond regarding ASFV infection. We generated sequence data (deposited in the EMBL database) (Table 2) and aligned all sequences to those of the human homologues which we used as reference sequences.
Table 2.
Sequences deposited into the EMBL database
Limited sequence variation was observed in this study (data not shown), with the following genes displaying complete homology at the translated protein level between the domestic pig and warthog: PPIA, NFATC1 regulatory domain, and NFKBIA. In the warthog, PPP3CB contained two insertions with respect to the domestic pig. However, these were located outwith the known functional domains. Furthermore, these correlate with splice variants described in human PPP3CB (22, 27). Only in the RELA subunit of NF-κB were potentially significant coding differences between the domestic pig and warthog identified (Fig. 1).
Fig. 1.
RELA primary protein sequence minus the first two amino acids of the porcine sequences. The Rel homology domain is underlined, the transactivation 2 domain is dashed underlined, and the transactivation 1 domain is double underlined. The nuclear localization signal (KRKR) is dotted underlined. The T448A, S485P, and S531P variations between the domestic pig and warthog are highlighted. SS, Sus scrofa; PA, Phacochoerus africanus; BB, Babyrousa babyrussa; HS, Homo sapiens (M62399).
Sequence of the porcine RELA gene.
The RELA open reading frame (ORF) is 1,662 nucleotides in the domestic pig and warthog and is slightly smaller at 1,556 nucleotides in the babirusa. These encode proteins which are 554 and 552 amino acids in length, respectively. Human RELA ORF and protein are the same lengths as the babirusa sequences. Due to insufficient primer binding sites in the 5′ untranslated region (UTR), we sequenced all but the first 6 nucleotides for all three pig species (Fig. 1).
Although the babirusa and human sequences are both 6 nucleotides shorter than the other pig sequences, the nucleotide deletions occur at different locations. The babirusa sequence has a single 6-nucleotide deletion, in relation to those of the other pig species, located between the Rel homology domain and the transactivation 2 domain. Human RELA has two 3-nucleotide deletions, one 20 nucleotides upstream from the babirusa deletion and the other in the transactivation 2 domain.
Of the 13 nucleotide differences between the domestic pig and warthog, only three are nonsynonymous and result in codon changes, T448A, S485P, and S531P. The threonine at 448 in the domestic pig occurs as an alanine in the warthog and babirusa and is absent in human RELA. The serine at 485 in the domestic pig sequence is a proline in the other porcine sequences and in human RELA. The serine at 531 in the domestic pig is also found in babirusa and human but is a proline in warthog RELA. All three amino acid differences occur outside the Rel homology domain, with amino acids 448 and 485 located within transactivation domain 2 and amino acid 531 within transactivation domain 1. To confirm these sequence differences, a 268-nucleotide region was amplified directly from genomic DNA and sequenced for an additional 5 domestic pigs and 11 warthogs (domestic pig sequences, EMBL accession numbers FN424224 to FN424228; warthog sequences, EMBL accession numbers FN424229 to FN424239). All domestic pig sequences were identical to each other over this region, and similarly, all warthog sequences were identical to each other over this region.
Predicted structural changes as a consequence of RELA sequence variation.
Given that even single amino acid changes can have significant effects on protein structure, we performed in silico analyses on the identified changes in RELA in an attempt to determine the structural consequences of the amino acid substitutions. At the three sites, the greater hydrophobicity was conferred by the threonine at 448 in the domestic pig sequence and the prolines at 385 and 531 in the warthog sequence. No difference in structural disorder was evident (Phyre analysis) (25), and the only site predicted to be highly likely (0.994) to undergo phosphorylation was the domestic pig serine at 531 (485S = 0.072; 448T = 0.0142) (NetPhos2.0) (4). However, the Phospho.ELM database infers that the threonine at 448 could also be a target for phosphorylation (23).
Basal activity of porcine RELA variants.
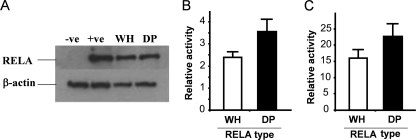
To determine whether the identified sequence variation between domestic pig RELA and warthog RELA affects NF-κB activity, we established a cell transfection assay. Cells were transiently transfected in duplicate using a triple plasmid cotransfection strategy involving the following: (i) 1 μg expression vector for FLAG-tagged domestic pig or warthog RELA or an empty vector control (pFLAG-CMV4; Sigma Aldrich); (ii) 1 μg NF-κB reporter plasmid comprising 4 copies of the NF-κB consensus binding sequence, driving expression of firefly luciferase (pNFκB-Luc; BD Biosciences, Clontech); and (iii) 1 μg transfection control vector expressing Renilla luciferase driven off the herpes simplex virus (HSV) thymidine kinase (TK) promoter (pRL-TK; Promega). Cells were harvested at 24 h posttransfection, and a dual luciferase assay (firefly luciferase activity relative to Renilla luciferase activity) was performed on quantified protein extracts. Luciferase activity values for each RELA type were averaged, and fold differences were calculated against background values for empty vector transfections. This strategy resulted in the delivery of an equivalent amount of either domestic pig or warthog RELA, as determined by Western blotting (Fig. 2A). Initially, we established the assay in COS-7 cells, which have a functional endogenous RELA gene. Repeat transfection experiments (n = 5) demonstrated that domestic pig RELA is 50% more active than warthog RELA (Fig. 2B). A similar (44%) differential activity was also evident in RELA−/− mouse embryonic fibroblasts (MEFs) that lacked endogenous RELA activity (derived from RELA−/− murine embryos) using the same transfection regime (n = 3) (Fig. 2C).
Fig. 2.
Effect of polymorphic RELA on basal NF-κB activity. (A) Western blot of COS-7 cells at 24 h after transfection with 1 μg FLAG-tagged warthog (WH) or domestic pig (DP) RELA or empty FLAG vector (−ve) and established RELA-FLAG expressing COS-7 cells (+ve). (B) Fold difference between NF-κB-luciferase activity in COS-7 cells after 24 h of transient transfection with 1 μg warthog (WH) or 1 μg domestic pig (DP) RELA. Activity is presented as the fold difference between RELA-induced NF-κB—luciferase activity normalized to cotransfected TK-Renilla luciferase activity relative to that of the empty vector. Error bars = standard deviations from the means. P < 0.03 between WH and DP. (C) Fold difference between NF-κB-luciferase activities in RELA−/− MEFs after 24 h of transient transfection with warthog (WH) and domestic pig (DP) RELA genes. Activity is presented as the fold difference between RELA-induced NF-κB-luciferase activity normalized to cotransfected TK-Renilla luciferase activity relative to that of the empty vector normalized to cotransfected TK-Renilla luciferase activity. Error bars = standard deviations from the means. P < 0.05 between WH and DP.
We attempted to produce stably transfected RELA−/− MEFs for the domestic pig and warthog RELA genes. Although we could generate numerous colonies for warthog RELA, we were reproducibly unsuccessful in generating those for domestic pig RELA. It is possible that the higher activity level of domestic pig RELA may not be compatible with survival in these cells; a similar scenario may have occurred during a study investigating phosphorylation at the equivalent site (S529) in human RELA (54).
Effect of RELA sequence variation on induced NF-κB activity.
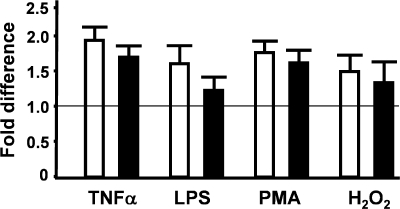
To determine if the ability to induce NF-κB activity was altered by the different pig RELA proteins, we stimulated transiently transfected RELA−/− MEFs with known inducers of NF-κB signaling (9, 14, 15, 16, 42–44, 56). Induction with tumor necrosis factor alpha (TNF-α) nearly doubled (90% increase) warthog RELA activity, whereas domestic pig RELA showed a 70% increase in activity (Fig. 3). We tested a further three known NF-κB-inducing agents, lipopolysaccharide, phorbol-12-myristate-13-acetate, and hydrogen peroxide. For all three additional agents, the fold induction observed for warthog RELA was always greater than that of domestic pig (Fig. 3). FLAG immunostaining of COS-7 cells transfected with either domestic pig or warthog RELA and treated with phorbol-12-myristate-13-acetate did not identify any gross differences in cytoplasm-nuclear transit time (RELA of both species was in the nucleus within 1 min and back in the cytoplasm after 20 min) (data not shown).
Fig. 3.
Induction of RELA allelic variants. RELA−/− MEFs were transiently cotransfected with 1 μg warthog (clear box) or 1 μg domestic pig (filled box) RELA. Cells were treated with TNF-α (30 ng/ml), lipopolysaccharide (LPS; 10 μg/ml), phorbol-12-myristate 13-acetate (PMA; 20 mM), and hydrogen peroxide (10 μM) immediately after and for the duration of the transfection. Cells were harvested at 24 h. RELA-induced NF-κB–luciferase (1 μg) activity was normalized to cotransfected TK-Renilla luciferase (1 μg) activity and presented as the fold induction above nondrug-treated cells (relative value of 1 depicted by the line).
Identification of functional mutations in porcine RELA.
In our sequence data, we identified three amino acid differences between domestic pig RELA and warthog pig RELA. To determine if the in vitro difference in domestic pig and warthog RELA activities was a cumulative effect of these three differences, or due to one or other individual mutations, we generated versions of RELA carrying the following single-base changes: T448A, S485P, and S531P. Transfection of these RELA variants into RELA−/− MEFs demonstrated that the majority of the reduced basal activity observed for warthog RELA compared to that observed for domestic pig RELA was attributed to the S531P mutation (Fig. 4).
Fig. 4.
Comparison of individual allelic RELA variation on NF-κB activity. Comparison of NF-κB–luciferase activity in RELA−/− MEFs after 24 h transient transfection with 1 μg warthog (WH) or 1 μg domestic pig (DP) RELA or 1 μg of RELA variants encoding the individual amino acid substitutions T448A, S485P, and S531P. Activity is presented as the fold difference between RELA-induced NF-κB–luciferase activity normalized to cotransfected TK-Renilla luciferase activity relative to that of the empty vector normalized to cotransfected TK-Renilla luciferase activity. Error bars = standard deviations from the means, *, P < 0.08 versus DP.
DISCUSSION
In this study, we have taken a candidate approach, identifying genes which may affect the severity of the host response to ASFV infection in the highly susceptible domestic pig and ASFV-tolerant warthog. The ASFV immunomodulatory factor A238L is known to interact with components of both NF-κB and NFAT host signaling pathways (11, 28, 29, 36, 49). We have sequenced five key factors in these pathways, including three A238L-targeted proteins, RELA, PPP3CB, and PPIA, and the two proteins it mimics, NFKBIA and NFATC1 (regulatory domain). Modest sequence differences have been identified at the cDNA (mRNA) level between the domestic pig and warthog; however, the majority of these are synonymous (silent) and do not alter the resulting amino acid sequences. We also sequenced the same genes from the ancient babirusa and observed a small number of amino acid differences between this species and the other two species of pig. These differences likely reflect the long evolutionary distance that exists between these species (38).
Despite the high degree of conservation observed between warthog and domestic pig PPP3CB-, PPIA-, NFKBIA-, and NFATC1-translated protein sequences, significant variation was detected in RELA. RELA is the predominant member of the heterodimeric transcription factor NF-κB (47). Moreover, the sequence variation includes a phosphorylation site in transactivation domain 1 (position 531), which is highly conserved across mammals and has been demonstrated to modulate the activity of human RELA (equivalent to S529) (54, 55). The function of approximately one-third of all eukaryote proteins is controlled by phosphorylation; thus, the observed S531P sequence variation represents an intriguing candidate regulator for the reduced pathology observed in African pigs infected with ASFV. We demonstrate that the genetic variation in the RELA sequence between the domestic pig and warthog is reflected in NF-κB activity in vitro, with warthog RELA displaying significantly reduced basal and induced NF-κB activity. We discuss three (not mutually exclusive) scenarios of how the genetic variation we have identified between the domestic pig and warthog may underlie the dramatic phenotypic difference in how these two pig species respond to ASFV.
First, as suggested by our in vitro assays, warthog RELA is inherently less active than the domestic pig RELA, exhibiting lower basal and induced levels. In a study of macrophage transcription profiles following ASFV infection in vitro, several factors within the NF-κB signaling pathway displayed elevated expression (i.e., NFKB1, NFKBIA); however, RELA expression was not observed to be altered (56). The impact of ASFV infection on the warthog RELA expression level is not known, since this study used only domestic pig macrophages. Therefore, although we have not determined whether the porcine RELA variants are differentially phosphorylated, it would appear that variation in domestic pig RELA activity may not be determined by altered expression levels. Alternatively, the warthog will have to function with reduced basal NF-κB activity, which would presumably indicate an adapted NF-κB-dependent transcriptome between the two species.
In the second scenario, phosphorylation of domestic pig RELA at S531 activates a set(s) of genes which are not activated by warthog RELA, as it lacks this phosphorylation site. Much is known about activation of the NF-κB pathway, with data coming primarily from studies of human RELA (47). NF-κB activity is regulated by two different mechanisms. The classical canonical pathway involves inhibitors (e.g., NFKBIA) that sequester this transcription factor in the cytoplasm until they are proteolytically degraded by the ubiquitin pathway (24). The alternative pathway revolves around posttranscriptional modifications, predominantly phosphorylation of RELA (46). At least 8 inducible phosphorylation sites have been identified in human RELA, which enable transcription of subsets of NF-κB-dependent genes (26, 45). One such site is S529, which is equivalent to domestic pig S531, suggesting that this site may perform a similar role. Indeed, differential expression of subsets of immune and inflammatory proteins as a result of S531 phosphorylation could play a role in determining ASFV pathogenesis in the domestic pig. Furthermore, phosphorylation not only controls specific transcription profiles but also can underlie the developmental timing in gene activation (13). It is tempting to speculate that if a similar mechanism is applied through the domestic pig S531 site, then it could also play a role in the gross physical differences that characterize the various pig species.
In the third scenario, we consider whether the S531P variation in RELA results in different outcomes of interaction with A238L during ASFV infection. Phosphorylation of human S529 (equivalent to domestic pig S531) is inhibited by the interaction of RELA with NFKBIA; only upon activation and degradation of NFKBIA can S529 phosphorylation occur (55). During ASFV infection, NFKBIA is degraded and replaced by A238L, which mimics NFKBIA but is not susceptible to proteolytic degradation (36, 49). As a result, in domestic pig cells infected with ASFV, S531 may not be exposed for phosphorylation. In comparison, warthogs lack this phosphorylation site; therefore, the ability of A238L to block phosphorylation at this site is irrelevant. The role of NFKBIA is to prevent nuclear translocation of NF-κB (24). We did not observe any gross temporal differences in nuclear transportation rates upon stimulation between cells expressing domestic pig RELA and warthog RELA; likewise, phosphorylation of human S529 also does not affect nuclear translocation (54). Furthermore, studies using recombinant ASFV lacking A238L indicate that neither nuclear import nor export of RELA is affected by this immune modulator (48). This suggests that the immunomodulatory functions of A238L are not the result of preventing nuclear translocation in a manner similar to that used by NFKBIA. Instead, A238L may inhibit NF-κB-mediated transcription by other mechanisms, for example, by preventing phosphorylation of RELA in domestic pigs, as discussed above. Whether such differences in how domestic pig RELA and warthog RELA interact with A238L exist will require further investigation. Likewise, elucidating how different mechanisms governing RELA activity and NF-κB-mediated transcription have evolved in these species, and determining the full extent of their functional implications on the immune system and wider transcriptome, will require additional study.
ASFV is highly infectious, with virulent isolates causing an acute, rapidly fatal hemorrhagic fever in domestic pigs (10, 34). In contrast, the porcine species endogenous to Africa tolerate the virus. Outbreaks have significant economic repercussions in addition to welfare concerns. In Africa, ASFV limits the use of the genetically improved breeding pig stock that is pervasive in the Eurasian landscape. Furthermore, ASF poses a constant threat to Europe and Asia, as documented by the list of outbreaks that have occurred over the last 30 to 40 years. As our world climate changes and the international movement of pork products continues to rise, this risk may well increase. No effective vaccine has been developed, so many look to genetic strategies to mitigate the geographical limits of pig breeding imposed by ASFV. We have demonstrated that a polymorphic RELA variant found in warthogs has the potential to underlie the difference between tolerance and rapid death upon infection with ASFV.
ACKNOWLEDGMENTS
This work was supported by the BBSRC (United Kingdom) and a Genus/PIC Postgraduate Studentship. C.J.P. was also funded by a Postgraduate Centenary Fellowship (University of Edinburgh). L.G. was funded through a Faraday-BBSRC CASE studentship.
RELA−/− MEFs were kindly donated by Ron Hay (University of Dundee). The pcDNA-A238L expression vector was kindly donated by Linda Dixon (Institute for Animal Health, Pirbright). We are grateful to Susan Rhind, Liz Glass, Sarah Howie, and Paul Hopwood (University of Edinburgh) for their support of this work.
Footnotes
Published ahead of print on 30 March 2011.
REFERENCES
- 1. Afonso C. L., Neilan J. G., Kutish G. F., Rock D. L. 1996. An African swine fever virus Bc1-2 homolog, 5-HL, suppresses apoptotic cell death. J. Virol. 70:4858–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson E. C., Hutchings G. H., Mukarati N., Wilkinson P. J. 1998. African swine fever infection of the bushpig (Potamochoerus porcus) and its significance to the epidemiology of the disease. Vet. Microbiol. 62:1–15 [DOI] [PubMed] [Google Scholar]
- 3. Anderson E. C., Williams S. M., Fisher-Hoch S. P., Wilkinson P. J. 1987. Arachidonic acid metabolites in the pathophysiology of thrombocytopenia and haemorrhage in acute African swine fever. Res. Vet. Sci. 42:387–394 [PubMed] [Google Scholar]
- 4. Blom N., Gammeltoft S., Brunak S. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294:1351–1362 [DOI] [PubMed] [Google Scholar]
- 5. Borca M. V., et al. 1998. Deletion of a CD2-like gene, 8-DR, from African swine fever virus affects viral infection in domestic swine. J. Virol. 72:2881–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Borca M. V., et al. 1994. An African swine fever virus gene with similarity to the T-lymphocyte surface antigen CD2 mediates hemadsorption. Virology 199:463–468 [DOI] [PubMed] [Google Scholar]
- 7. Brun A., Rivas C., Esteban M., Escribano J. M., Alonso C. 1996. African swine fever virus gene A179L, a viral homologue of bcl-2, protects cells from programmed cell death. Virology 225:227–230 [DOI] [PubMed] [Google Scholar]
- 8. Brun A., Rodriguez F., Escribano J. M., Alonso C. 1998. Functionality and cell anchorage dependence of the African swine fever virus gene A179L, a viral bcl-2 homolog, in insect cells. J. Virol. 72:10227–10233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carrasco L., et al. 2002. African swine fever: expression of interleukin-1 alpha and tumour necrosis factor-alpha by pulmonary intravascular macrophages. J. Comp. Pathol. 126:194–201 [DOI] [PubMed] [Google Scholar]
- 10. Colgrove G. S., Haelterman E. O., Coggins L. 1969. Pathogenesis of African swine fever in young pigs. Am. J. Vet. Res. 30:1343–1359 [PubMed] [Google Scholar]
- 11. Dixon L. K., et al. 2004. African swine fever virus proteins involved in evading host defence systems. Vet. Immunol. Immunopathol. 100:117–134 [DOI] [PubMed] [Google Scholar]
- 12. Dixon L. K., et al. 2005. Family Asfarviridae, p. 135–143 In Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, London, England [Google Scholar]
- 13. Gavalda N., Gutierrez H., Davies A. M. 2009. Developmental switch in NF-kappaB signalling required for neurite growth. Development 136:3405–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ghosh S., May M. J., Kopp E. B. 1998. NF-κB and Rel proteins: evolutionary conserved mediators of immune responses. Annu. Rev. Immunol. 16:225–260 [DOI] [PubMed] [Google Scholar]
- 15. Gil S., et al. 2003. Expression at mRNA level of cytokines and A238L gene in porcine blood-derived macrophages infected in vitro with African swine fever virus (ASFV) isolates of different virulence. Arch. Virol. 148:2077–2097 [DOI] [PubMed] [Google Scholar]
- 16. Gomez del Moral M., et al. 1999. African swine fever virus infection induces tumor necrosis factor alpha production: implications in pathogenesis. J. Virol. 73:2173–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gomez-Villamandos J. C., et al. 1997. African swine fever virus infection of bone marrow: lesions and pathogenesis. Vet. Pathol. 34:97–107 [DOI] [PubMed] [Google Scholar]
- 18. Gomez-Villamandos J. C., et al. 1998. Thrombocytopenia associated with apoptotic megakaryocytes in a viral haemorrhagic syndrome induced by a moderately virulent strain of African swine fever virus. J. Comp. Pathol. 118:1–13 [DOI] [PubMed] [Google Scholar]
- 19. Gomez-Villamandos J. C., et al. 1995. Experimental African swine fever: apoptosis of lymphocytes and virus replication in other cells. J. Gen. Virol. 76(Pt. 9):2399–2405 [DOI] [PubMed] [Google Scholar]
- 20. Gomez-Villamandos J. C., et al. 1995. A pathological study of the perisinusoidal unit of the liver in acute African swine fever. Res. Vet. Sci. 59:146–151 [DOI] [PubMed] [Google Scholar]
- 21. Gomez-Villamandos J. C., et al. 1995. Pathological changes in the renal interstitial capillaries of pigs inoculated with two different strains of African swine fever virus. J. Comp. Pathol. 112:283–298 [DOI] [PubMed] [Google Scholar]
- 22. Guerini D., Klee C. B. 1989. Cloning of human calcineurin A: evidence for two isozymes and identification of a polyproline structural domain. Proc. Natl. Acad. Sci. U. S. A. 86:9183–9187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hammet A., et al. 2003. FHA domains as phospho-threonine binding modules in cell signaling. IUBMB Life 55:23–27 [DOI] [PubMed] [Google Scholar]
- 24. Karin M. 2006. Nuclear factor-kappaB in cancer development and progression. Nature 441:431–436 [DOI] [PubMed] [Google Scholar]
- 25. Kelley L. A., Sternberg M. J. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 26. Law M., Corsino P., Parker N. T., Law B. K. 2010. Identification of a small molecule inhibitor of serine 276 phosphorylation of the p65 subunit of NF-kappaB using in silico molecular docking. Cancer Lett. 291:217–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McPartlin A. E., Barker H. M., Cohen P. T. 1991. Identification of a third alternatively spliced cDNA encoding the catalytic subunit of protein phosphatase 2B beta. Biochim. Biophys. Acta 1088(2):308–310 [DOI] [PubMed] [Google Scholar]
- 28. Miskin J. E., Abrams C. C., Dixon L. K. 2000. African swine fever virus protein A238L interacts with the cellular phosphatase calcineurin via a binding domain similar to that of NFAT. J. Virol. 74:9412–9420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miskin J. E., Abrams C. C., Goatley L. C., Dixon L. K. 1998. A viral mechanism for inhibition of the cellular phosphatase calcineurin. Science 281:562–565 [DOI] [PubMed] [Google Scholar]
- 30. Neilan J. G., et al. 1993. An African swine fever virus gene with similarity to the proto-oncogene bcl-2 and the Epstein-Barr virus gene BHRF1. J. Virol. 67:4391–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nogal M. L., et al. 2001. African swine fever virus IAP homologue inhibits caspase activation and promotes cell survival in mammalian cells. J. Virol. 75:2535–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oura C. A., Powell P. P., Anderson E., Parkhouse R. M. 1998. The pathogenesis of African swine fever in the resistant bushpig. J. Gen. Virol. 79:1439–1443 [DOI] [PubMed] [Google Scholar]
- 33. Oura C. A., Powell P. P., Parkhouse R. M. 1998. African swine fever: a disease characterized by apoptosis. J. Gen. Virol. 79:1427–1438 [DOI] [PubMed] [Google Scholar]
- 34. Penrith M.-L., Thomson G. R., Bastos A. D. S. 2004. African swine fever, p. 1088–1119 In Coetzer J. A. W., Tustin R. C. (ed.), Infectious diseases of livestock, 2nd ed., vol. 2 Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 35. Plowright W., Parker J., Pierce M. A. 1969. The epizootiology of African swine fever in Africa. Vet. Rec. 85:668–674 [PubMed] [Google Scholar]
- 36. Powell P. P., Dixon L. K., Parkhouse R. M. 1996. An IkappaB homolog encoded by African swine fever virus provides a novel mechanism for downregulation of proinflammatory cytokine responses in host macrophages. J. Virol. 70:8527–8533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ramiro-Ibanez F., Ortega A., Brun A., Escribano J. M., Alonso C. 1996. Apoptosis: a mechanism of cell killing and lymphoid organ impairment during acute African swine fever virus infection. J. Gen. Virol. 77(Pt. 9):2209–2219 [DOI] [PubMed] [Google Scholar]
- 38. Randi E., Lucchini V., Hoong Diong C. 1996. Evolutionary genetics of the Suiformes as reconstructed using mtDNA sequencing. J. Mamm. Evol. 3:163–194 [Google Scholar]
- 39. Rodriguez C. I., et al. 2002. African swine fever virus IAP-like protein induces the activation of nuclear factor kappa B. J. Virol. 76:3936–3942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rodriguez F., Fernandez A., Martin de las Mulas J. P., Sierra M. A., Jover A. 1996. African swine fever: morphopathology of a viral haemorrhagic disease. Vet. Rec. 139:249–254 [DOI] [PubMed] [Google Scholar]
- 41. Rodriguez J. M., Yanez R. J., Almazan F., Vinuela E., Rodriguez J. F. 1993. African swine fever virus encodes a CD2 homolog responsible for the adhesion of erythrocytes to infected cells. J. Virol. 67:5312–5320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Salguero F. J., et al. 2002. Changes in macrophages in spleen and lymph nodes during acute African swine fever: expression of cytokines. Vet. Immunol. Immunopathol. 90:11–22 [DOI] [PubMed] [Google Scholar]
- 43. Salguero F. J., Sanchez-Cordon P. J., Nunez A., Fernandez de Marco M., Gomez-Villamandos J. C. 2005. Proinflammatory cytokines induce lymphocyte apoptosis in acute African swine fever infection. J. Comp. Pathol. 132:289–302 [DOI] [PubMed] [Google Scholar]
- 44. Salguero F. J., et al. 2004. Apoptosis of thymocytes in experimental African swine fever virus infection. Histol. Histopathol. 19:77–84 [DOI] [PubMed] [Google Scholar]
- 45. Sasaki C. Y., Barberi T. J., Ghosh P., Longo D. L. 2005. Phosphorylation of RelA/p65 on serine 536 defines an I{kappa}B{alpha}-independent NF-{kappa}B pathway. J. Biol. Chem. 280:34538–34547 [DOI] [PubMed] [Google Scholar]
- 46. Schmitz M. L., Bacher S., Kracht M. 2001. IκB-independent control of NF-κB activity by modulatory phosphorylations. Trends Biochem. Sci. 26(3):186–190 [DOI] [PubMed] [Google Scholar]
- 47. Schmitz M. L., Mattioli I., Buss H., Kracht M. 2004. NF-kappaB: a multifaceted transcription factor regulated at several levels. Chembiochem 5:1348–1358 [DOI] [PubMed] [Google Scholar]
- 48. Silk R. N., Bowick G. C., Abrams C. C., Dixon L. K. 2007. African swine fever virus A238L inhibitor of NF-kappaB and of calcineurin phosphatase is imported actively into the nucleus and exported by a CRM1-mediated pathway. J. Gen. Virol. 88:411–419 [DOI] [PubMed] [Google Scholar]
- 49. Tait S. W. G., Reid E. B., Greaves D. R., Wileman T. E., Powell P. P. 2000. Mechanism of inactivation of NF-κB by a viral homologue of IκBα. J. Biol. Chem. 275:34656–34664 [DOI] [PubMed] [Google Scholar]
- 50. Thomson G. R. 1985. The epidemiology of African swine fever: the role of free-living hosts in Africa. Onderstepoort J. Vet. Res. 52:201–209 [PubMed] [Google Scholar]
- 51. Thomson G. R., Gainaru M. D., Van Dellen A. F. 1980. Experimental infection of warthogs (Phacochoerus aethiopicus) with African swine fever virus. Onderstepoort J. Vet. Res. 47:19–22 [PubMed] [Google Scholar]
- 52. Villeda C. J., Williams S. M., Wilkinson P. J., Vinuela E. 1993. Consumption coagulopathy associated with shock in acute African swine fever. Arch. Virol. 133:467–475 [DOI] [PubMed] [Google Scholar]
- 53. Villeda C. J., Williams S. M., Wilkinson P. J., Vinuela E. 1993. Haemostatic abnormalities in African swine fever a comparison of two virus strains of different virulence (Dominican Republic '78 and Malta '78). Arch. Virol. 130:71–83 [DOI] [PubMed] [Google Scholar]
- 54. Wang D., Baldwin A. S., Jr 1998. Activation of nuclear factor-κB-dependent transcription by tumor necrosis factor-α is mediated through phosphorylation of RelA/p65 on serine 529. J. Biol. Chem. 273:29411–29416 [DOI] [PubMed] [Google Scholar]
- 55. Wang D., Westerheide S. D., Hanson J. L., Baldwin A. S., Jr 2000. Tumor necrosis factor α-induced phosphorylation of RelA/p65 on Ser529 is controlled by casein kinase II. J. Biol. Chem. 275:32592–32597 [DOI] [PubMed] [Google Scholar]
- 56. Zhang F., et al. 2006. Macrophage transcriptional responses following in vitro infection with a highly virulent African swine fever virus isolate. J. Virol. 80:10514–10521 [DOI] [PMC free article] [PubMed] [Google Scholar]