Abstract
Human cytomegalovirus (HCMV) selectively relocalizes many DNA repair proteins, thereby avoiding a potentially detrimental damage response. In the present study, we evaluated interactions between HCMV and the homology-directed repair (HDR) pathway. In permissive human foreskin fibroblasts, a fluorescence-based double-stranded break repair assay was used to determine that HCMV stimulated HDR. Repair of both stably integrated and extrachromosomal reporter substrates was observed to increase. HDR was also stimulated through individual expression of the viral immediate-early protein IE1-72, mimicking full virus infection. These experiments further demonstrate HCMV's role in modulating critical cellular processes during a permissive infection.
TEXT
Replication of the genome of human cytomegalovirus (HCMV) is thought to proceed in a biphasic manner (24). Initially, single-copy origin-specific replication of the circularized genome takes place. Later, replication switches to a rolling-circle mechanism, where large concatemers are formed (37, 38). Homologous recombination (HR) occurs along the concatameric DNA, as evidenced by inversions of genome segments in successive monomeric units. HR also occurs between free cleaved monomers and the concatameric structure, producing branched intermediate structures observed at late times postinfection (p.i.) (9, 35, 36, 40). It has been suggested that these recombinogenic structures trigger a host cell DNA damage response (DDR) during herpesvirus replication (17, 25, 28, 44).
Previous studies have shown that the host cell's ability to mount and complete various types of DDR can be subverted by many viruses (6, 11, 20, 39, 49). In HCMV-infected human foreskin fibroblasts (HFFs), DDR was activated at the time of viral deposition and during late-phase replication (17, 28). However, incomplete DDR occurred and checkpoint activation and apoptosis were subverted due to selective relocalization of host cell DDR proteins into viral replication centers (28). No specific repair processes were examined in depth in these studies.
Our previous studies have shown that HCMV can inflict site-specific chromosomal damage in the form of double-strand breaks (DSBs) (13, 30). DSBs can be repaired without error by homology-directed repair (HDR) or by error-prone mechanisms such as nonhomologous end-joining or single-strand annealing (34, 42). In this study, we investigated the effects of HCMV infection on HDR in HFFs.
Introduction of a recombination substrate to assess HDR.
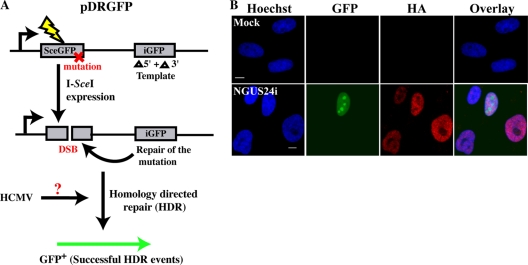
Our studies utilized the reporter substrate pDRGFP (a gift from Maria Jasin [31]), which is composed of two green fluorescent protein (GFP) cassettes: (i) a nonfunctional mutant GFP, SceGFP, engineered to contain an 18-bp restriction site for the rare endonuclease I-SceI, and (ii) an internal GFP (iGFP), which contains a 5′- and 3′-truncated (internal) fragment capable of correcting the mutation in the SceGFP cassette (Fig. 1A). Introduction of I-SceI into cells containing pDRGFP induces a DSB in the restriction site contained in SceGFP. Reconstitution of functional GFP (only possible via recombination with the iGFP donor fragment) indicates successful HDR of I-SceI-induced DSBs. In this study, these events were quantified microscopically for GFP positivity. Several researchers have used this substrate to study HDR successfully (10, 12, 18, 21, 48).
Fig. 1.
Efficient expression of I-SceI from an Ad-based system in HFFs with an integrated pDRGFP substrate. (A) Schematic diagram of reporter plasmid pDRGFP, depicting I-SceI-induced DSB-initiated HDR to produce functional GFP. (B) Representative IF staining of mock- or NGUS24i-infected clone 4. Cells were seeded onto glass coverslips and mock or Ad infected. Coverslips were harvested and processed at 72 hpi as described previously (27). GFP+ cells were scored as HDR competent. Cells were also stained with HA-specific Ab (mouse monoclonal antibody [MAb; IgG2b] 12CA5; Abcam) to detect HA-I-SceI. In all IF experiments, isotype-specific secondary Abs conjugated with either tetramethyl rhodamine isothiocyanate (TRITC; Jackson ImmunoResearch Laboratories) or Alexa Fluor 488 (Molecular Probes) were used to detect mouse primary Abs. Nuclei were counterstained in all experiments with Hoechst dye. Scale bar, 5 μm for all panels in all figures.
Primary HFFs (propagated as described previously [33]) were electroporated with pDRGFP in a Bio-Rad Gene Pulser II using the following conditions: 300 V/cm, 75 Ω, 2,500 μF, 2-mm gap cuvettes, and 1 × 106 cells suspended in 400 μl of tissue culture medium containing 100 mM HEPES (pH 7.4) and 10 μg pDRGFP. Two individual stable clones were generated through continued growth in puromycin (1 μg/ml). The results obtained using both clones were consistent, indicating, as was previously demonstrated (3, 14), that random integration of pDRGFP among the clones (and potential variable copy numbers) did not affect the frequency of HDR. We therefore report only representative studies carried out in HFFpDRGFP stable clone 4 (hereafter referred to as clone 4).
Clone 4 was synchronized in G0 (as described in reference 7), and depending upon the experiment, cells were replated at 5 × 105 or 1 × 106 cells/60-mm dish containing glass coverslips. After allowing 2 h for attachment, we infected cells with one of two recombinant adenoviruses (Ad): dL70-3, used as a control, or NGUS24i, which encodes the I-SceI enzyme (gifts from Frank Graham and Philip Ng, McMaster University [1, 5]). Cells were mock infected or infected with dL70-3 or NGUS24i at various multiplicities of infection (MOIs; 10, 25, 50, 100, and 200) for 30 min in 0.2 ml of medium (or in phosphate-buffered saline [PBS] containing 0.01% CaCl2 and 0.01% MgCl2 for transient pDRGFP introduction). An additional 1 ml of medium was added, and cells were analyzed for efficiency of I-SceI production at different times p.i. The I-SceI protein possesses a hemagglutinin (HA) tag, and its expression can be detected by immunofluorescence (IF) using an anti-HA antibody (Ab). One hundred percent of cells infected with NGUS24i were HA positive (HA+), and no HA staining was observed in mock-infected (Fig. 1B) or dL70-3-infected (data not shown) cells.
Clone 4 was examined for spontaneous HDR events (GFP-positive [GFP+] cells); none were found in mock-infected (Fig. 1B) or dL70-3-infected (data not shown) cells. However, GFP+ cells appeared following infection with NGUS24i, indicating that I-SceI-induced DSBs had been successfully repaired by HDR. In our hands, NGUS24i infection effects were greatest using an MOI of 200 and a 96-h incubation. The Ad-based enzyme delivery system and pDRGFP construct constituted an elegant means of studying HDR in our cells.
HDR was stimulated in HCMV-infected HFFs.
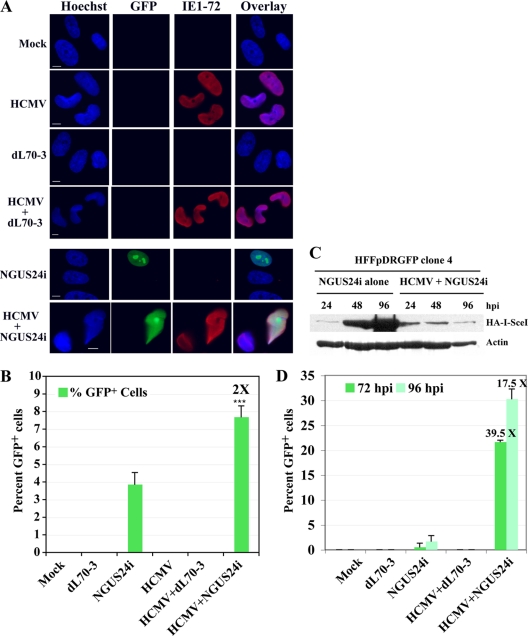
Our earlier studies (28) led us to suspect that HDR would be downregulated in HCMV-infected HFFs. To test this hypothesis, we G0 synchronized clone 4, replated it, and then infected it with HCMV (Towne strain; ATCC VR 977) at an MOI of 5 for 4 h. These cells were subsequently superinfected with dL70-3 or NGUS24i as described above and harvested 96 h after Ad infection. Cells were stained for the immediate-early (IE) protein IE1-72 to monitor HCMV infection. As expected, all cells were IE1-72 positive (IE1-72+) at the time of harvest (∼100 h p.i. [hpi]) (Fig. 2A). Cells were also scored for GFP positivity (Fig. 2A and B). NGUS24i-infected cells displayed an average of 3.8% GFP+ cells, while no GFP+ cells were observed in either dL70-3-infected or mock-infected cells (Fig. 2B). In HCMV-infected cells subsequently superinfected with NGUS24i, an average of 8.0% GFP+ cells was detected, an ∼2-fold increase. Although modest, this increase was reproducible and statistically significant, indicating that HCMV infection enhanced HDR.
Fig. 2.
HCMV infection enhanced HDR in HFFs in the presence of both integrated and transiently expressed pDRGFP. (A and B) Clone 4 was seeded onto glass coverslips and mock infected, infected with HCMV or Ad alone, or dually infected with HCMV and Ad in succession. Coverslips were harvested 96 h after Ad superinfection. GFP+ cells were scored as HDR competent. Clone 4 was also stained by IF for the presence of IE1-72 to monitor HCMV infection, using a mouse MAb (IgG2a; a gift from Bill Britt, University of Alabama, Birmingham). (A) Representative IF staining for GFP+ and IE1-72+ cells. (B) Experimental results represent the averages of three experiments. In each experiment, 200 to 500 cells per coverslip were counted and scored. Values at the tops of the bars indicate fold increases in percentages of GFP+ cells in all figures. Error bars represent ± one standard deviation in all figures. The asterisks (***) indicate statistical significance at the level of P < 0.05, as determined by the unpaired Student t test. (C) Representative protein profile for steady-state levels of HA-tagged I-SceI in clone 4 infected with NGUS24i alone or dually infected with HCMV and NGUS24i. Cells were synchronized in G0 prior to seeding and infection. Total cell lysates derived from equivalent numbers of cells were used for each lane (7, 28). Actin was used as a loading control and was detected using an anti-pan actin mouse MAb (IgG1; NeoMarkers). Secondary for both Abs was horseradish peroxidase (HRP)-linked sheep anti-mouse Ab (GE Healthcare, United Kingdom). (D) Percentages of GFP+ cells in HFFs transiently expressing pDRGFP, as described in the text and above. Data represent the averages of two experiments.
To ensure that this stimulation was not due to increased production of I-SceI, we examined steady-state levels of I-SceI in the presence and absence of HCMV. I-SceI expression levels were determined by immunoblot analysis via its HA tag. Clone 4 was G0 synchronized, mock or HCMV infected, and then superinfected with Ad NGUS24i for 24, 48, or 96 h. Lysates derived from an equivalent number of cells were loaded in each lane. Although at 24 hpi dual infections with HCMV and NGUS24i produced steady-state levels of HA-I-SceI slightly above the basal level of NGUS24i alone, at 48 and 96 hpi HA-I-SceI steady-state levels were dramatically higher during infections with NGUS24i alone (Fig. 2C). This finding indicated that the increase in HDR found during HCMV infection was not correlated with increased expression of I-SceI.
In clone 4, increases in HDR were consistent but modest. Would unchromatinized DNA display similar results? To answer this question, we transiently expressed pDRGFP by electroporation into HFFs. In a parallel experiment using a pEGFP reporter plasmid, electroporation produced approximately 50% GFP+ cells. Twenty-four to forty h postelectroporation, HFFs transiently expressing pDRGFP were mock or HCMV infected at an MOI of 5 for 2 to 8 h and then superinfected with dL70-3 or NGUS24i. Cells were then harvested at 72 and 96 h after Ad infection and analyzed (Fig. 2D).
Following infection with NGUS24i alone, an average of 0.55% GFP+ (72 h) and 1.7% GFP+ (96 h) cells were detected. No GFP+ cells were observed in control infections. These percentages and those found in clone 4 constituted baseline regeneration of functional GFP similar to results previously described (10). In HCMV-infected HFFs superinfected with NGUS24i, averages of 21.7% GFP+ (72 h) and 30.3% GFP+ (96 h) cells were detected, an ∼39-fold or ∼17-fold increase, respectively, over those detected in cells infected with NGUS24i alone. Thus, HCMV infection greatly enhanced HDR of a readily accessible unchromatinized pDRGFP reporter construct.
Steady-state level changes occurred in HDR proteins in HCMV-infected HFFs.
The above increases in HDR prompted investigation of any correlated changes in steady-state levels of HDR proteins. Our earlier studies found that steady-state levels of Rad51, a central regulator of HDR, were dramatically increased in HCMV-infected HFFs at late times p.i. (28). In this study, we analyzed two additional HDR proteins, Rad51C and FANCG, in HCMV-infected HFFs.
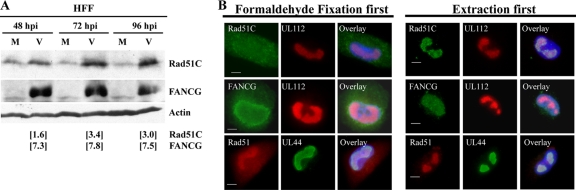
Rad51C contributes to the protection of genome integrity by transducing DNA damage signals. Rad51C plays a direct role in Holliday junction resolution during HDR (26) and may also be involved in trafficking recombination proteins from the cytoplasm into the nucleus (14). Also, recent investigation of the HDR pathway has linked it to Fanconi anemia (FA) (43), and cell lines deficient in Fanconi anemia complementation group D2 protein (FANCD2) or FANCG displayed defective HR (46, 47). We observed that steady-state levels of Rad51C rose only slightly above mock-infected HFF levels at 48 hpi; however, significant increases were observed at 72 and 96 hpi (Fig. 3A). FANCG levels increased dramatically between 48 and 96 hpi. A shift to a slower-migrating form of FANCG was observed at all times p.i. (Fig. 3A). This slower-migrating form may indicate a phosphorylation event of FANCG, as has been reported elsewhere (15, 45). Fold increases above normalized mock-infected values are shown below the blots (Fig. 3A).
Fig. 3.
Following HCMV infection, steady-state levels of HDR proteins were increased in HFFs and were available for cellular DNA repair. (A) G0-synchronized HFFs were mock or HCMV infected at an MOI of 5. Cells were harvested at the indicated times p.i. Total cellular lysates derived from equivalent numbers of mock-infected and virus-infected cells were separated by SDS-PAGE, transferred to Protran, and probed with anti-Rad51C MAb (IgG1) or anti-FANCG MAb (IgG2a) (Abnova) followed by HRP-linked sheep anti-mouse secondary Ab. Representative protein profiles are shown. Actin was used as a loading control. Quantitation of this immunoblot was performed using Bio-Rad Quantity One software, and numbers underneath each lane represent the fold change relative to the value for the normalized mock-infected cells at a given time point. All lanes were normalized to their actin control. (B) Rad51 (mouse MAb 14B4; Abcam Ab213), Rad51C, and FANCG were localized during HCMV infection of HFFs at 48 hpi using either standard formaldehyde fixation (27) or extraction-first (28) conditions to reveal more tightly associated protein localization. Viral replication centers were localized using mouse MAbs to either UL44 (IgG1; Rumbaugh-Goodwin Institute for Cancer Research, Inc.) or UL112 (a gift from Misaki Shirakata, Japan).
Rad51, Rad51C, and FANCG were localized by IF using standard formaldehyde fixation procedures (27). None of these proteins were exclusively localized/sequestered into viral replication centers in the nucleus (Fig. 3B), and all, to various degrees, were also found in the cytoplasm, as previously described (4, 16). This localization was unsurprising given our observation that these proteins were all available, at least to some degree, for repair of cellular substrates such as the integrated pDRGFP cassette. FANCG remained broadly nuclear when coverslips were extracted first (as described in reference 28), while Rad51 and Rad51C were both found to be more tightly associated with the viral replication centers. This could indicate more selective repair of the viral genome under certain circumstances.
The virus-specific functions of these proteins in stimulating HDR in HCMV-infected cells are unknown. However, we speculate that during its replication in HFFs, the virus may exploit the cellular HDR machinery to enhance virus genome replication efficiency and fidelity. Several other viruses recruit Rad51 to replication compartments (6, 23, 44). Additionally, in both simian virus 40 (SV40) and Epstein-Barr virus (EBV), the knockdown of Rad51 causes a reduction in viral genome synthesis, suggesting that HDR is necessary for efficient replication.
Further support for the involvement of both viral and cellular proteins in efficient viral replication comes from the Weller lab. They have utilized a strand exchange assay to study recombination events in the context of in vitro herpes simplex virus 1 (HSV-1) infection. They have shown that HSV-1 UL12 and ICP8 jointly mediate strand exchange (32). In a more recent study, Weller and colleagues found direct interaction between UL12 and the Mre11, Rad50, and Nbs1 complex, important cellular factors in the ataxia telangiectasia mutated protein (ATM)-dependent HR pathway (2).
Transient expression of IE1-72 was sufficient to enhance HDR in HFFs.
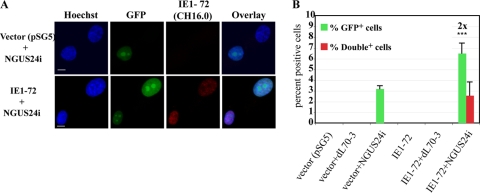
In HCMV-infected cells, rapid and robust expression of viral IE genes is a hallmark for productive infection. Several studies have suggested that IE1-72 plays a pivotal role in the activation of early gene expression (8, 19). The IE proteins also directly modulate components of the cell cycle machinery. In fact, solely overexpressing IE1-72 has, in some circumstances, induced cells to enter S phase (8). Such cells, primed for replication, could be expected to exhibit an enhanced capacity for HDR. These findings prompted us to determine whether IE proteins (IE1-72, in particular) were involved in increased HDR. Clone 4 was therefore nucleofected with 3 μg of vector (pSG5) or pSG5IE1-72 (5 × 105 cells; Amaxa Biosystems program A-033) (22). Twenty-four h after nucleofection, the cells were either mock or Ad infected and were then scored for GFP positivity at 72 hpi. Cells were also stained and scored for IE1-72 (Fig. 4A).
Fig. 4.
Transient expression of IE1-72 alone was enough to enhance HDR in HFFs. Cells were nucleofected with either pSG5 or pSG5IE1-72 and seeded onto glass coverslips. After 24 h, cells were mock or Ad infected. Coverslips were harvested 72 h later. Cells were stained by IF for the presence of IE1-72 using an anti-IE1 and anti-IE2 MAb Ch16.0 (IgG1; Rumbaugh-Goodwin Institute for Cancer Research, Inc.). (A) Representative IF staining for IE1-72+ and GFP+ cells. (B) Percentages of GFP+ and double-positive (GFP+ plus IE1-72+) cells. Bars represent the averages of four experiments. The differences in the percentages of GFP+ cells were considered statistically significant (*** = P < 0.05).
An average of 3.2% GFP+ cells were detected in cells nucleofected with vector alone and then infected with NGUS24i (Fig. 4B). No GFP+ cells were observed in any control experiments. In cells nucleofected with the IE1-72 construct and then infected with NGUS24i, an average of 6.5% GFP+ cells were scored, a 2-fold increase. This increase was statistically significant and equivalent to the increase found in HCMV-infected clone 4. More importantly, the 3.3% increase (from 3.2 to 6.5%) in GFP+ cells observed in the IE1-72-nucleofected/NGUS24i-infected population was almost entirely accounted for by the subpopulation of cells that were double positive for GFP and IE1-72 (∼2.6%) (Fig. 4B). Thus, transient expression of IE1-72 alone was capable of recapitulating the HCMV infection results.
Other reports in the literature complement our finding that a single viral protein can modulate HDR (29, 41). Importantly, this study has identified a novel function for HCMV IE1-72, namely, that sole expression of it can upregulate HDR. Our future studies will focus on specific interactions between IE1-72 and the HDR machinery. We will attempt to elucidate the specific mechanism of this enhancement through the identification of IE1-72 binding partners via coimmunoprecipitation in both HCMV-infected and IE1-72-expressing cells. We hope that studying interactions between HCMV and the cellular DDR machinery will elucidate the mechanisms by which HCMV utilizes and subverts this machinery for its own gain and likely to the detriment of the host.
Acknowledgments
This work was supported by NIH grants RO1-AI51463 and P20 RR015587 (COBRE program) to E.A.F.
We thank John O'Dowd for critical reading of the manuscript.
Footnotes
Published ahead of print on 13 April 2011.
REFERENCES
- 1. Anglana M., Bacchetti S. 1999. Construction of a recombinant adenovirus for efficient delivery of the I-SceI yeast endonuclease to human cells and its application in the in vivo cleavage of chromosomes to expose new potential telomeres. Nucleic Acids Res. 27:4276–4281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balasubramanian N., Bai P., Buchek G., Korza G., Weller S. K. 2010. Physical interaction between the herpes simplex virus type 1 exonuclease, UL12, and the DNA double-strand break-sensing MRN complex. J. Virol. 84:12504–12514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bechter O. E., Zou Y., Shay J. W., Wright W. E. 2003. Homologous recombination in human telomerase-positive and ALT cells occurs with the same frequency. EMBO Rep. 4:1138–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bennett B. T., Knight K. L. 2005. Cellular localization of human Rad51C and regulation of ubiquitin-mediated proteolysis of Rad51. J. Cell. Biochem. 96:1095–1109 [DOI] [PubMed] [Google Scholar]
- 5. Bett A. J., Haddara W., Prevec L., Graham F. L. 1994. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc. Natl. Acad. Sci. U. S. A. 91:8802–8806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boichuk S., Hu L., Hein J., Gjoerup O. V. 2010. Multiple DNA damage signaling and repair pathways deregulated by simian virus 40 large T antigen. J. Virol. 84:8007–8020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Casavant N. C., et al. 2006. Potential role for p53 in the permissive life cycle of human cytomegalovirus. J. Virol. 80:8390–8401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castillo J. P., Kowalik T. F. 2002. Human cytomegalovirus immediate early proteins and cell growth control. Gene 290:19–34 [DOI] [PubMed] [Google Scholar]
- 9. Courcelle C. T., Courcelle J., Prichard M. N., Mocarski E. S. 2001. Requirement for uracil-DNA glycosylase during the transition to late-phase cytomegalovirus DNA replication. J. Virol. 75:7592–7601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cuozzo C., et al. 2007. DNA damage, homology-directed repair, and DNA methylation. PLoS Genet. 3:e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dahl J., You J., Benjamin T. L. 2005. Induction and utilization of an ATM signaling pathway by polyomavirus. J. Virol. 79:13007–13017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dronkert M. L., et al. 2000. Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol. Cell. Biol. 20:3147–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fortunato E. A., Dell'Aquila M. L., Spector D. H. 2000. Specific chromosome 1 breaks induced by human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 97:853–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. French C. A., Tambini C. E., Thacker J. 2003. Identification of functional domains in the RAD51L2 (RAD51C) protein and its requirement for gene conversion. J. Biol. Chem. 278:45445–45450 [DOI] [PubMed] [Google Scholar]
- 15. Futaki M., Watanabe S., Kajigaya S., Liu J. M. 2001. Fanconi anemia protein, FANCG, is a phosphoprotein and is upregulated with FANCA after TNF-alpha treatment. Biochem. Biophys. Res. Commun. 281:347–351 [DOI] [PubMed] [Google Scholar]
- 16. Garcia-Higuera I., Kuang Y., Naf D., Wasik J., D'Andrea A. D. 1999. Fanconi anemia proteins FANCA, FANCC, and FANCG/XRCC9 interact in a functional nuclear complex. Mol. Cell. Biol. 19:4866–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gaspar M., Shenk T. 2006. Human cytomegalovirus inhibits a DNA damage response by mislocalizing checkpoint proteins. Proc. Natl. Acad. Sci. U. S. A. 103:2821–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Golding S. E., et al. 2004. Double strand break repair by homologous recombination is regulated by cell cycle-independent signaling via ATM in human glioma cells. J. Biol. Chem. 279:15402–15410 [DOI] [PubMed] [Google Scholar]
- 19. Greaves R. F., Mocarski E. S. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J. Virol. 72:366–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hein J., et al. 2009. Simian virus 40 large T antigen disrupts genome integrity and activates a DNA damage response via Bub1 binding. J. Virol. 83:117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kikuchi K., et al. 2005. Fen-1 facilitates homologous recombination by removing divergent sequences at DNA break ends. Mol. Cell. Biol. 25:6948–6955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klucher K. M., Sommer M., Kadonaga J. T., Spector D. H. 1993. In vivo and in vitro analysis of transcriptional activation mediated by the human cytomegalovirus major immediate-early proteins. Mol. Cell. Biol. 13:1238–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kudoh A., et al. 2009. Homologous recombinational repair factors are recruited and loaded onto the viral DNA genome in Epstein-Barr virus replication compartments. J. Virol. 83:6641–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lehman I. R., Boehmer P. E. 1999. Replication of herpes simplex virus DNA. J. Biol. Chem. 274:28059–28062 [DOI] [PubMed] [Google Scholar]
- 25. Lilley C. E., Carson C. T., Muotri A. R., Gage F. H., Weitzman M. D. 2005. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. U. S. A. 102:5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu Y., Masson J. Y., Shah R., O'Regan P., West S. C. 2004. RAD51C is required for Holliday junction processing in mammalian cells. Science 303:243–246 [DOI] [PubMed] [Google Scholar]
- 27. Luo M. H., Fortunato E. A. 2007. Long-term infection and shedding of human cytomegalovirus in T98G glioblastoma cells. J. Virol. 81:10424–10436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Luo M. H., Rosenke K., Czornak K., Fortunato E. A. 2007. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection. J. Virol. 81:1934–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nakai-Murakami C., et al. 2007. HIV-1 Vpr induces ATM-dependent cellular signal with enhanced homologous recombination. Oncogene 26:477–486 [DOI] [PubMed] [Google Scholar]
- 30. Nystad M., Fagerheim T., Brox V., Fortunato E. A., Nilssen O. 2008. Human cytomegalovirus (HCMV) and hearing impairment: infection of fibroblast cells with HCMV induces chromosome breaks at 1q23.3, between loci DFNA7 and DFNA49—both involved in dominantly inherited, sensorineural, hearing impairment. Mutat. Res. 637:56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pierce A. J., Johnson R. D., Thompson L. H., Jasin M. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13:2633–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reuven N. B., Staire A. E., Myers R. S., Weller S. K. 2003. The herpes simplex virus type 1 alkaline nuclease and single-stranded DNA binding protein mediate strand exchange in vitro. J. Virol. 77:7425–7433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rosenke K., Samuel M. A., McDowell E. T., Toerne M. A., Fortunato E. A. 2006. An intact sequence-specific DNA-binding domain is required for human cytomegalovirus-mediated sequestration of p53 and may promote in vivo binding to the viral genome during infection. Virology 348:19–34 [DOI] [PubMed] [Google Scholar]
- 34. Rothkamm K., Kruger I., Thompson L. H., Lobrich M. 2003. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 23:5706–5715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Severini A., Scraba D. G., Tyrrell D. L. 1996. Branched structures in the intracellular DNA of herpes simplex virus type 1. J. Virol. 70:3169–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Severini A., Sevenhuysen C., Garbutt M., Tipples G. A. 2003. Structure of replicating intermediates of human herpesvirus type 6. Virology 314:443–450 [DOI] [PubMed] [Google Scholar]
- 37. Skaliter R., Lehman I. R. 1994. Rolling circle DNA replication in vitro by a complex of herpes simplex virus type 1-encoded enzymes. Proc. Natl. Acad. Sci. U. S. A. 91:10665–10669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Skaliter R., Makhov A. M., Griffith J. D., Lehman I. R. 1996. Rolling circle DNA replication by extracts of herpes simplex virus type 1-infected human cells. J. Virol. 70:1132–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stracker T. H., Carson C. T., Weitzman M. D. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348–352 [DOI] [PubMed] [Google Scholar]
- 40. Strang B. L., Stow N. D. 2005. Circularization of the herpes simplex virus type 1 genome upon lytic infection. J. Virol. 79:12487–12494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Trojanek J., et al. 2006. T-antigen of the human polyomavirus JC attenuates faithful DNA repair by forcing nuclear interaction between IRS-1 and Rad51. J. Cell. Physiol. 206:35–46 [DOI] [PubMed] [Google Scholar]
- 42. Valerie K., Povirk L. F. 2003. Regulation and mechanisms of mammalian double-strand break repair. Oncogene 22:5792–5812 [DOI] [PubMed] [Google Scholar]
- 43. Wang W. 2007. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 8:735–748 [DOI] [PubMed] [Google Scholar]
- 44. Wilkinson D. E., Weller S. K. 2004. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J. Virol. 78:4783–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wilson J. B., et al. 2008. FANCG promotes formation of a newly identified protein complex containing BRCA2, FANCD2 and XRCC3. Oncogene 27:3641–3652 [DOI] [PubMed] [Google Scholar]
- 46. Yamamoto K., et al. 2005. Fanconi anemia protein FANCD2 promotes immunoglobulin gene conversion and DNA repair through a mechanism related to homologous recombination. Mol. Cell. Biol. 25:34–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yamamoto K., et al. 2003. Fanconi anemia FANCG protein in mitigating radiation- and enzyme-induced DNA double-strand breaks by homologous recombination in vertebrate cells. Mol. Cell. Biol. 23:5421–5430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yatagai F., Suzuki M., Ishioka N., Ohmori H., Honma M. 2008. Repair of I-SceI induced DSB at a specific site of chromosome in human cells: influence of low-dose, low-dose-rate gamma-rays. Radiat. Environ. Biophys. 47:439–444 [DOI] [PubMed] [Google Scholar]
- 49. Zhao X., et al. 2008. Ataxia telangiectasia-mutated damage-signaling kinase- and proteasome-dependent destruction of Mre11-Rad50-Nbs1 subunits in simian virus 40-infected primate cells. J. Virol. 82:5316–5328 [DOI] [PMC free article] [PubMed] [Google Scholar]