Abstract
Human cytomegalovirus (HCMV) induces extensive remodeling of the secretory apparatus to form the cytoplasmic virion assembly compartment (cVAC), where virion tegumentation and envelopment take place. We studied the structure of the cVAC by confocal microscopy to assess the three-dimensional distribution of proteins specifically associated with individual secretory organelles. In infected cells, early endosome antigen 1 (EEA1)-positive vesicles are concentrated at the center of the cVAC and, as previously seen, are distinct from structures visualized by markers for the endoplasmic reticulum, Golgi apparatus, and trans-Golgi network (TGN). EEA1-positive vesicles can be strongly associated with markers for recycling endosomes, to a lesser extent with markers associated with components of the endosomal sorting complex required for transport III (ESCRT III) machinery, and then with markers of late endosomes. In comparisons of uninfected and infected cells, we found significant changes in the structural associations and colocalization of organelle markers, as well as in net organelle volumes. These results provide new evidence that the HCMV-induced remodeling of the membrane transport apparatus involves much more than simple relocation and expansion of preexisting structures and are consistent with the hypothesis that the shift in identity of secretory organelles in HCMV-infected cells results in new functional profiles.
INTRODUCTION
Human cytomegalovirus (HCMV; Human herpesvirus 5) is a betaherpesvirus that is a significant cause of disease and disability in immunocompromised patients and in congenitally infected children. The virus has a large genome (∼230 kb) and protracted replication cycle, during which it modulates many cellular processes and immune responses. As for other herpesviruses, HCMV virions consist of the viral genome that is housed in a geometrically well-defined nucleocapsid, a tegument in the space between the nucleocapsid and envelope, and an envelope that is studded with viral glycoproteins and integral membrane proteins. The tegument consists of a collection of viral proteins and RNA that contribute to virion structure and provide functions needed immediately following virion entry (30). Some host proteins have been detected in HCMV virions (8, 55), but their roles in virion assembly and operation are not known. As described by Buchkovich et al. and Moorman et al. (4, 40), HCMV virion assembly involves multiple cellular trafficking pathways and takes place along a highly integrated assembly-egress continuum that extends from nucleocapsid assembly in the nucleus through cytoplasmic tegumentation, final envelopment, and then the regulated release of mature infectious virions at the cell surface.
The HCMV cytoplasmic virion assembly complex (cVAC) is a large discoid structure in which developing virions acquire most of their tegument proteins, are enveloped, and are then transported to the cell surface for release. We previously showed that the underlying structure of the cVAC consists of dramatically rearranged components of the cellular secretory machinery (15, 17). It takes 3 to 4 days of remodeling to form a fully developed cVAC (17), prior to which very little infectious virus is produced. Once the cVAC has formed, its overall architecture appears to be stable for a week or more. The cVAC is located adjacent to the nucleus, which is enlarged in infected cells and is bent into a kidney-shaped form as the result of infection-induced changes in microtubule distribution, dynein activity, and the level and behavior of Sad1p and UNC-84 homology domain (SUN domain) proteins that are responsible for maintaining the architecture of the nuclear membrane; the width of the perinuclear space and the porosity of the nuclear membrane increase in the vicinity of the cVAC (4). The ER chaperone, Bip, is required for virion assembly (6) and interacts with HCMV pp28 and TRS1 in the cVAC (5), and interaction of Bip with HCMV pUL50 is essential for the remodeling of the nuclear lamina that enables nucleocapsid egress from the nucleus (4, 38, 42). HCMV structural and nonstructural proteins are abundant in the cVAC, with individual proteins having unique distributions within the cVAC (15).
Although it was originally thought that viral proteins occlude cellular proteins from the cVAC, work from our and other laboratories has demonstrated that the cVAC consists of components of the secretory apparatus arranged as concentric cylinders. The Golgi apparatus and trans-Golgi network (TGN) form closely interlaced cylindrical rings that surround a cylindrical inner region that consists of vesicles bearing markers of early and recycling endosomes (8, 15, 17, 33), from which nascent virions are transported to the cell surface (54). A network of microtubules radiates outward from a microtubule-organizing center located at the center of the cVAC (27, 49). Thus, the cVAC includes components of the secretory apparatus that have been reconfigured into an unusual arrangement that appears to allow for a generally conventional order of biosynthesis and transport (17). Similar structures have been seen in HCMV-infected fibroblasts and vascular endothelial cells (24).
Several recent studies have provided new information concerning the structure, biogenesis, and function of the cVAC. HCMV pp150 (pUL32) is essential for HCMV replication (18, 60, 61) and is involved in virion maturation and egress (2, 53). pp150 associates with the cellular protein Bicaudal D1, a secretory trafficking regulator that interacts with dynein motors and the Rab6 GTPase. The pp150/Bicaudal D1 interaction is necessary for localization of pp150 to the cVAC and for production of infectious virions. Infection with mutant viruses unable to express pUL71 results in altered cVACs that include large vesicles and marked reduction in production of infectious virions (50, 57). The enlarged vesicles have properties of lysosomes, are rimmed with CD63, and are surrounded by virions that appear to be interrupted during the process of budding into exit vesicles. HCMV UL103 is conserved among the Herpesviridae; ablation of pUL103 expression resulted in the cVAC not forming properly and a reduction in release of enveloped virions and dense bodies (1). Using dominant negative mutants, the endosomal sorting complex required for transport III (ESCRT III) protein, chromatin-modifying protein 1A (CHMP1A), and the ESCRT III-associated AAA ATPase, Vps4A, were found to be required for efficient viral replication; tagged, transfected versions of these proteins localized in or around the cVAC (52). Markers of the late endocytic pathway (cathepsin, CD63, and lysosome-associated membrane protein 1 [LAMP1]) stain vesicles at the cVAC periphery (8, 33), and Rab27a, a regulator of lysosome-related organelle transport, is required for efficient production of infectious virions (19). The SNARE protein syntaxin 3 is present in the cVAC and at the plasma membrane; its depletion results in reduced levels of CD63 and the lysosomal proteins LAMP1 and LAMP2 and a reduction in production of infectious virions (9).
Organelle identity is the net result of molecular composition (lipids, proteins, etc.) and structure, localization within the cell, and biological function(s). Recent work has identified new levels of subcompartmentalization in organelles that are involved in membrane traffic and has shown that proteins and lipids are not uniformly distributed in the various subdomains of organelles of a particular type (reviewed in references 23 and 48). The presence in HCMV virions of proteins from diverse secretory organelles (8, 19) and the numerous observed changes in the localization and arrangement of secretory organelles in HCMV-infected cells suggest the non-mutually exclusive hypotheses that organelle identities are shifted as novel membrane compartments are generated during HCMV infection or that the virion envelope may derive from vesicles involved in TGN/endosomal transport.
Our objective is to understand the process of HCMV virion assembly that takes place in the cytoplasm. Studies of cVAC biogenesis and operation will be facilitated by a better understanding of the composition and three-dimensional (3-D) arrangement of the various substructures that form the cVAC. In previous work, we studied the endoplasmic reticulum (ER)/Golgi apparatus/early endosome axis of the membrane transport apparatus and developed a new model of cVAC structure and operation (17). As part of developing a more complete structural definition of the cVAC, in this work we have compared in uninfected and infected cells the 3-D locations, extents of colocalization, and intracellular volumes of markers for the ER, Golgi apparatus, early and recycling endosomes, multivesicular bodies, and lysosomes, as well as markers associated with the ESCRT III apparatus. Our results confirm and extend prior observations that structural and functional definitions of secretory organelles differ between uninfected and infected cells.
MATERIALS AND METHODS
Virus and cells.
HCMV strain AD169 (American Type Culture Collection, Manassas, VA) was used in these experiments. Virus stocks were prepared in low-passage human foreskin fibroblasts (HFF) in the same medium used for cell propagation, except that 2% serum was used and antibiotics were not. Cells were cultured and propagated in high-glucose Dulbecco's modified Eagle medium (DMEM) with sodium pyruvate plus MEM nonessential amino acids (Invitrogen, Carlsbad, CA), GlutaMAX (Invitrogen), penicillin and streptomycin (50 U/ml each), and 10% fetal bovine serum (HyClone, Logan, UT). Diploid human lung fibroblast cells (MRC-5; ATCC) were used for virus titration. Low-passage human lung fibroblasts (HLF) (a gift from John Stewart, Centers for Disease Control and Prevention, Atlanta, GA) were used for all imaging experiments.
Antibodies.
Primary antibodies used in this study and descriptions of their targets are listed in Table 1. Mouse monoclonal antibodies (MAb) were detected with a secondary antibody labeled with Alexa Fluor 568 (Invitrogen, Carlsbad, CA), which gives red fluorescence. Rabbit polyclonal antibodies were detected with a secondary antibody labeled with Alexa Fluor 488 (Invitrogen, Carlsbad, CA), which gives green fluorescence.
Table 1.
Proteins targeted and antibodies used in this study
| Target proteina | Major/minor localization and function of the target protein | Nature of antibody (source)b | Reference(s)c |
|---|---|---|---|
| Markers of major secretory organelles | |||
| Heat shock 70-kDa protein 5 (HSPA5) (immunoglobulin heavy chain-binding protein [Bip]; 78-kDa glucose-regulated protein [GRP78]; MIF2; FLJ26106) | ER lumen (major); nucleus and cell surface (minor). Molecular chaperone that controls the structural maturation of nascent glycoproteins. Detects hydrophobic patches on improperly folded proteins and initiates the unfolded protein response, thereby serving as an ER stress sensor. | Rabbit polyclonal antibody (Abcam catalog no. ab21685) | 21, 43, 47 |
| Golgin subfamily A member 2 (GOLGA2) (130-kDa cis-Golgi matrix protein I [GM130]; SY11 protein; Golgi autoantigen; Golgin-95; RP11-395P17.5; MGC20672) | cis-Golgi matrix peripheral membrane protein. Functions as a structural element and provides attachment sites for other membranes and proteins of the Golgi apparatus. | MAb, clone 35/GM130 (BD catalog no. 610822) | 3 |
| Mannosidase, alpha, class 2A, member 1 (MAN2A1) (alpha-mannosidase 2 [AMan II]; Golgi alpha-mannosidase II [MANA2]; Golgi integral membrane protein 7 [GOLIM7]; mannosidase, alpha type II [Mann II]; mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase) | cis- and medial-Golgi. Golgi-matrix-interacting enzyme. Mediates interaction of cis- and medial-Golgi compartments. | Rabbit polyclonal antibody (Abcam catalog no. ab12277) | 45, 56 |
| Golgin subfamily A member 4 (Golgin A4) (OTTHUMP00000208897; OTTHUMP00000209805; Golgi autoantigen; Golgin subfamily a, 4; Golgin-240; Golgin-245; protein 72.1; trans-Golgi p230) | TGN GRIP domain protein. Important for biogenesis of distinct populations of vesicles that function in vesicular transport. | MAb, clone 15/p230 trans-Golgi (BD catalog no. 611280) | 20, 59 |
| Early endosome antigen 1 (EEA1) (endosome-associated protein p162; zinc finger FYVE domain-containing protein 2; MST105; MSTP105; ZFYVE2) | Early endosomes. Colocalizes in many cells with Rab5 and phosphatidylinositol 3-phosphate in the early endosomal compartment. Mediates endosome docking and, together with SNARE proteins, initiates membrane fusion. | MAb, clone 14/EEA1 (BD catalog no. 610456); rabbit polyclonal antibody (Abcam catalog no. ab2900) | 13, 28 |
| Markers of late endosomes | |||
| CD63 (melanoma 1 antigen; granulophysin; lysosome-associated membrane glycoprotein 3 [LAMP-3]; melanoma-associated antigen ME491; melanoma-associated antigen MLA1; ocular melanoma-associated antigen [OMA81H]; tetraspanin-30 [tspan-30]) | MVBs and other vesicles. Tetraspanin membrane protein abundant in multivesicular bodies. Cycles between these compartments and the exocytic pathway. | MAb, clone 4X-49.129.5 (SCBT catalog no. 5275) | 32, 46, 58 |
| Lysosome-associated membrane protein 1 (LAMP1) (CD107 antigen-like family member A [CD107a]; lysosome-associated membrane glycoprotein 1; LAMP1A; LGP120) | Lysosomes. Lysosome-associated glycosylated type I membrane protein. | MAb, clone H5G11 (SCBT catalog no. sc-18821) | 10, 35 |
| Markers of recycling endosomes | |||
| Transferrin receptor (TFRC) (TR; p90; TRFR; transferrin receptor protein 1; CD71; TFR; TFR1; OTTHUMP00000208525; T9) | Recycling and sorting endosomes. Recycles between the cell surface and cytoplasmic vesicles, including lysosomes via late endosomes. | MAb, clone b3/25 (SCBT catalog no. sc-65877) | 12, 37 |
| RAB11A, member RAS oncogene family (RAB11A) (RAB11A, member oncogene family; Rab11; MGC1490; YL8) | Recycling endosomes. Located in the cytoplasmic space in the vicinity of several post-Golgi structures, including the TGN and recycling endosomes. Regulates distinct events in the transferrin receptor pathway. Not uniquely associated with any one organelle. | Rabbit polyclonal antibody, clone H-87 (SCBT catalog no. 9020) | 34, 39 |
| Markers for ESCRT III-associated proteins | |||
| Chromatin-modifying protein 1A (CHMP1A) (charged multivesicular body protein 1; charged multivesicular body protein 1a; hVps46-1; procollagen (type III) N-endopeptidase; protease, metallo 1, 33 kD; vacuolar protein sorting-associated protein 46-1; Vps46A; Vps46-1; KIAA0047; PCOLN3; PRSM1) | Required for sorting endosomal vesicles into MVBs and has role in budding of some viruses. Recruits Vps4 and the microtubule-severing enzyme spastin (mammalian DID2B) as part of regulating membrane scission and ESCRT III disassembly. | Rabbit polyclonal antibody, clone FL-196 (SCBT catalog no. 67205) | 25, 26 |
| Vacuolar protein sorting 4 homolog A (vacuolar sorting protein 4 [VPS4]; hVPS4; vacuolar protein sorting factor 4A; vacuolar protein sorting-associated protein 4A; FLJ22197; SKD1 homolog; SKD; SKD1A; SKD2; VPS4-1) | Involved in late endosomal protein transport. Plays role in MVB biogenesis, autophagy, and viral budding. Regulates ESCRT III disassembly. | Rabbit polyclonal antibody, clone H-165 (SCBT catalog no. 32922) | 26, 51 |
Official gene symbols and names. Prior and informal designations are shown in parentheses. Designations used in this paper are shown in bold.
Antibody sources are abbreviated as follows: Abcam, Abcam (Cambridge, MA); BD, BD Biosciences (San Jose, CA); SCBT, Santa Cruz Biotechnologies (Santa Cruz, CA).
References are for papers that describe or summarize functional and localization information for the markers, not necessarily primary citations for the antibody.
Infection and immunofluorescence assays.
Our infection procedure and immunofluorescence assay protocols are similar to those described previously (16). Briefly, 2 × 104 HLF cells were seeded onto 8-well glass chamber slides (LabTek chamber slide; Nunc, Rochester, NY; catalog number 177402) that had been coated with 0.2% gelatin (Sigma, St. Louis, MO; catalog number G9391) in phosphate-buffered saline (PBS; in all instances, PBS lacked Ca2+ and Mg2+) for 1 h at 37°C. The following day, cells were infected at a multiplicity of infection (MOI) of 0.2 in DMEM containing 5% fetal bovine serum. After 120 h, cells were washed with PBS and then fixed with 4% paraformaldehyde in PBS (lacking Ca2+ and Mg2+) at pH 7.4. Autofluorescence was quenched by incubation with 50 mM ammonium chloride for 15 min at room temperature. Cells were permeabilized for 15 min at room temperature using 0.2% Triton X-100 in blocking buffer (10% normal goat serum and 5% glycine in PBS) followed by incubation in blocking buffer for 1 h. All antibody incubations and washes were at room temperature; washes were done with PBS. Primary antibodies diluted with the blocking buffer were incubated on the slides for 1 h, followed by three washes. Slides were then incubated for 1 h with secondary antibodies diluted in blocking buffer and then washed three times. Finally, Vectashield containing DAPI (4′,6-diamidino-2-phenylindole) and antifade (Vector Laboratories Inc., CA) was added to the coverslip, the slide was inverted onto the coverslip, and the sandwich was sealed with clear fingernail polish.
Microscopy.
Z-stack fluorescent images (0.5 μm) were acquired and then processed using a Leica TCS SP5 laser multiphoton confocal microscope (Leica Microsystems Inc., Bannockburn, IL). Volocity software (version 5.4.2; PerkinElmer, Waltham, MA) was used for image manipulation and analysis, including 3-D reconstructions and movies from Z-stack images, stills from the resulting movies, and measurement of colocalization and object volumes and sizes.
RESULTS
Experimental design.
The major purpose of these experiments was to develop a 3-D map of the HCMV cVAC that describes relationships among major membrane transport organelles and the distribution of some of the proteins that play important roles in membrane transport but whose organelle associations can vary depending on the state of the cell. We have identified a set of antibodies that are reliable markers for single major organelles, specifically, the ER, cis- and medial-Golgi, TGN, and early endosomes (Table 1); these antibodies have low levels of background staining and provide strong and consistent staining of a single type of structure, as would be expected from staining a single organelle. In addition, we employed markers for late endocytic and exocytic organelles and for components of the ESCRT III machinery (Table 1). We used the same antibodies to study uninfected cells as a point of reference for the changes induced by HCMV. In previous work, we studied the time course of cVAC development (17). Here, infected cell observations were done at 120 h postinfection (hpi), a time when the cVAC is fully formed and production of infectious virions is well under way. Pairs of antibodies were used to generate 3-D reconstructions that enabled visualization of intracellular structures, as well as quantitation of their volumes and the extent of marker colocalization. The images shown are reproducible and representative; we chose images that represent the range of what was seen. The focus here was on cellular proteins; thus, we did not study the relationship of viral proteins to the cVAC framework. For all markers, videos showing labeled cells rotating in space are provided in the supplemental material. As we previously showed, videos of 3-D reconstructions are much more informative than static images (17), thus the videos are central to our results. Table 1 provides descriptions of the organelles and structures studied and the antibodies we used. Table 2 is a systematic summary of our observations for each marker in uninfected and infected cells.
Table 2.
Comparison of intracellular markers in uninfected and infected HLFs
| Marker (major association) | Intracellular distribution |
Implications | |
|---|---|---|---|
| Uninfected cells | Infected cells (120 hpi) | ||
| Bip (ER) | Present on a large structure(s) appearing to consist of bulbous tubes distributed through much of the cytoplasm. In 3-D images, spaces for other organelles, e.g., the Golgi apparatus, are apparent. | Greatly reduced staining at the center of the cVAC. Little colocalization with a marker for the cis-Golgi (GM130). | Although widely distributed in the cytoplasm, the ER is spatially distinct from other organelles. |
| GM130 (cis-Golgi) | Stains large tubular (∼0.6- to 1.2-μm diam; Fig. 2A) perinuclear structures that extend into the cytoplasm. No visible contact with the nucleus. | Stains large tubular (0.92- to 1.1-μm; Fig. 1C) network that forms a cylinder or cuplike structure around the cVAC. In some cells, a single, well-defined structure is stained. In other cells, the overall staining patterns are similar, but the structures appear to be somewhat fragmented. | The unique and well-defined structure at the cVAC periphery provides an important point of reference for localization of other organelles and structures. |
| Mann II (cis- and medial-Golgi) | Similar in appearance to GM130 staining; sometimes fragmented (Fig. 2A). | Very high, but incomplete colocalization with GM130 (Fig. 4B; image not shown), consistent with it being associated with structures that derive from the cis- and medial-Golgi. Somewhat more granular, fuzzy appearance than that for GM130. | As for GM130. |
| p230 (TGN) | Staining pattern is similar in appearance to GM130 and Mann II, but the p230-positive structures are distinct from structures stained by GM130 and Mann II. | p230-positive structures form a cylindrical ring at the cVAC periphery. These structures are intimately intertwined with similar-appearing but distinct structures labeled by GM130 and Mann II. The structures can be fragmented or continuous (e.g., Fig. 3C, CHMP1A or Vps4 comparisons, respectively). | As for GM130. |
| EEA1 (early endosomes) | Stains nearly spherical ovoid vesicular structures (0.3- to 0.8-μm diam; ∼0.6 μm for the most abundant species) scattered throughout the cytoplasm. | Most EEA1-staining vesicles are located in the cVAC, within the cylinder defined by Golgi and trans-Golgi markers. Some vesicles have rounded structures similar to those in uninfected cells. Other vesicles have similar diameters but are elongated on an axis perpendicular to the growth surface, giving them a cylindrical appearance. Approx 6-fold greater net EEA1-associated vesicle vol than in uninfected cells. | The localization and structure of these perinuclear vesicular structures are unique to the cVAC. |
| TfR (recycling endosomes) | Stains nearly spherical ovoid vesicular structures distributed throughout the cytoplasm. Most vesicles range in diam from 0.3 to 0.6 μm; some are as large as 1.3 μm. High colocalization with EEA1, but the two markers frequently predominantly stain opposite sides of the same structure (or individually stain separate, very closely juxtaposed structures). | In infected cells, TfR-stained vesicles are distributed through much of the cytoplasm and are tightly packed in the cVAC. Vesicle diameters range from 0.2 to 0.7 μm. Very high colocalization with EEA1 in the center of the cVAC. | Tight association between EEA1 and TfR at the cVAC center is consistent with both markers being associated with recycling vesicles that deliver mature virions to the cell surface. |
| Rab11 (recycling endosomes) | Several types of staining: widely scattered small vesicles (<0.3-μm diam), larger areas with an appearance suggestive of tightly packed small vesicles (<0.3-μm diam), scattered orbs similar in size and shape to EEA1-positive vesicles (0.6-μm to 0.9-μm diam). Vesicles strongly stained for Rab11 have little EEA1 staining and vice versa. | In most cases, Rab11 is concentrated at the center of the cVAC. Image resolution is insufficient to distinguish between Rab11 being associated with individual vesicles or it being diffusely distributed in the cytoplasmic space between vesicles. Vesicle diameters range from 0.2 μm to 0.7 μm. Very high levels of colocalization with EEA1 in the center of the cVAC. | Tight association between Rab11 and EEA1 at the cVAC center is consistent with both markers being associated with recycling vesicles that deliver mature virions to the cell surface. |
| CHMP1A (ESCRT III) | Localizes to small (0.3 to 1.1 μm in diam) spherical vesicular structures that concentrate mainly in the small cytoplasmic space above the nucleus; some are scattered throughout the cytoplasm. | Predominantly labels small vesicles (0.3 to 0.9 μm) in and around the cVAC. Much higher levels of colocalization with EEA1 than in uninfected cells. | AC localization and essential role in HCMV replication (52) suggest role in virion maturation. |
| Vps4 (ESCRT III) | Localizes to small vesicles distributed throughout the cytoplasm. Vesicle diameters range from 0.3 to 1.4 μm, with most ranging from 0.3 to 0.4 μm. | Two populations of Vps4-stained objects: small and midsized vesicles at cVAC periphery (0.2- to 1.3-μm diam; many ∼0.7 μm) and diffuse staining at the cVAC center. Infected cells differ in the relative abundances of these populations. There are numerous examples of Vps4-positive “sausages” wrapped in EEA1- or CD63-positive “buns.” | AC localization and essential role in HCMV replication (52) suggest a role in virion maturation. |
| CD63 (MVBs) | Stains nearly spherical ovoid vesicular structures scattered throughout the cytoplasm; distinct from and slightly larger than EEA1-positive vesicles (0.4 to 1.4 μm). | Individual vesicles predominate at the cVAC periphery and range in size from 0.5 to 1.7 μm. Some of the CD63-positive vesicles appear to be part of larger, less sharply bounded structures that are concentrated at the center of the cVAC. More-than-5-fold-greater CD63-associated vesicle vol than that in uninfected cells. Greatly reduced colocalization with EEA1 in infected cells vs that of uninfected cells. | Greatly reduced colocalization between CD63 and EEA1in infected cells vs that in uninfected cells indicates changes in vesicle composition and suggests functional differences. |
| LAMP1 (lysosomes) | Somewhat elongated vesicular structures distributed throughout the cytoplasm. Vesicle diameters range from 0.6 to 1.5 μm. | Staining pattern is markedly different from that of uninfected cells. Diffuse and amorphous or fine-granular staining in the cVAC, with scattered small vesicles (0.5 to 1.3 μm) at the cell periphery and on the opposite side of the nucleus from the cVAC. Greatly reduced levels of colocalization with EEA1 in infected cells vs that in uninfected cells. | Lysosomal function is likely to be very different in infected cells vs that in uninfected cells. |
A clear result from this study is that organelle compositions, and thus details of their function, can differ between infected and uninfected cells. Nonetheless, for convenience we will refer to organelles by their common names.
Relationship between 2-D and 3-D images.
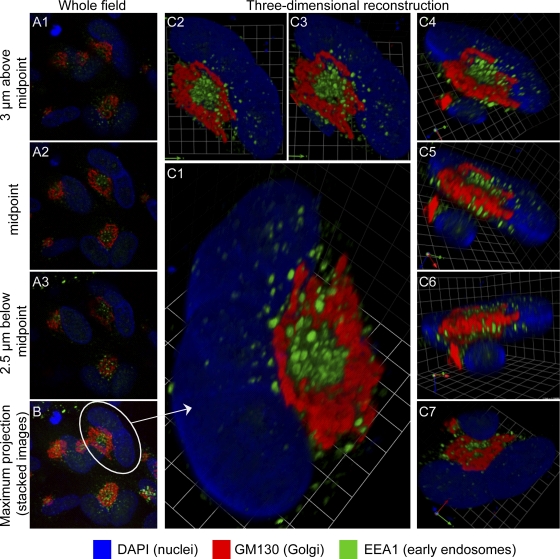
Much of the data provided here is in the form of 3-D images. Because most published images of the HCMV cVAC are from single confocal sections, Fig. 1 illustrates the relationship between such images and 3-D reconstructions based on such images; this demonstrates the enhanced perspective on spatial relationships provided by the 3-D reconstructions. Interpretation of the organelle-specific information from Fig. 1 is presented below.
Fig. 1.
Relationships between images from single confocal planes and reconstructed 3-D images. HCMV(AD169)-infected lung fibroblasts (HLF cells) were stained for the indicated markers at 120 hpi. Serial 0.5-μm confocal sections were obtained with a Leica TCS SP5 laser-scanning confocal microscope. (A1 to A3) Three of the 23 confocal sections that were used to generate the 3-D reconstruction shown in panel C. The location of each section in the Z-series is indicated. (B) Maximum projection image of the entire field containing the cell from which the 3-D reconstruction was made. This shows that many infected cells have related structures. (C to C7) A collection of views of a 3-D reconstruction of a single binucleated infected cell (circled in panel B). A portion of the nucleus of an overlapping adjacent cell is visible in panels C4 through C7. Panel C7 is a view from the bottom. Grid spacing is 3.31 μm. A video showing the reconstruction rotating in space is available as Video S1 in the supplemental material.
ER, Golgi apparatus, TGN, and early endosomes.
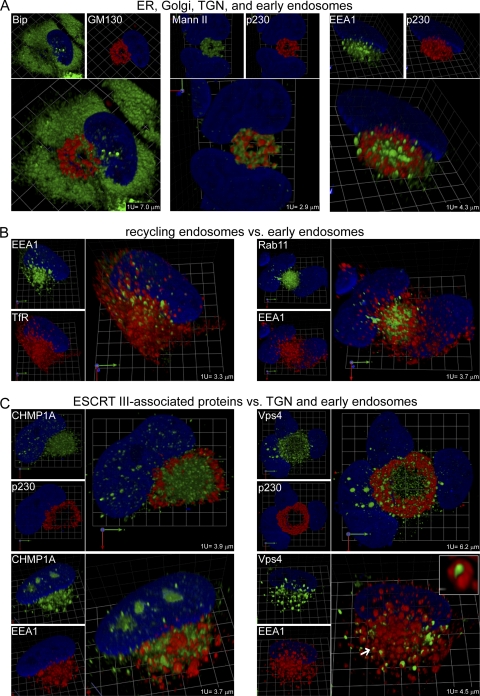
We will begin by considering relationships between major members of the protein secretory and endocytic apparatus. In uninfected cells (Fig. 2A; see also Video S2A in the supplemental material [a higher-resolution version of this video is available at www.med.wayne.edu/micr/gallery/pellett.asp]), antibody against the ER resident protein Bip stained a tightly packed cytoplasmic structure composed of what appear to be interconnected bulbous tubes. The cis-Golgi protein, GM130, has perinuclear localization and forms an interconnected tubular structure that extends well into to the cytoplasm but is structurally distinct from the Bip-defined ER. Bip staining in uninfected cells is similar to that in infected cells (Fig. 3A; see also Video S3A in the supplemental material [a higher-resolution version of this video is available at www.med.wayne.edu/micr/gallery/pellett.asp]), and staining is most prominent in the cytoplasmic space outside the cVAC, as defined in this image by GM130. In Fig. 1 and 3A and Videos S1 and S3A in the supplemental material, the cis-Golgi often appears as a single contiguous tubular structure that is structurally and spatially distinct from the ER. We used a rabbit polyclonal antibody to visualize Bip; this antibody is directed against the C-terminal ER luminal domain of the protein. The distribution of this form of Bip differs from the Bip distribution revealed by other antibodies. For example, a rabbit polyclonal antibody against the 71-to-91 segment (Sigma; ET-21, G9043) reacts with a version of the protein that localizes to the outer periphery of the cVAC (6).
Fig. 2.
Relative localization of markers for major secretory organelles, endocytic organelles, and ESCRT III-associated proteins in uninfected lung fibroblasts (HLF cells). (A) Markers of the ER (Bip), Golgi apparatus (GM130 and Mann II), TGN (p230), and early endosomes (EEA1). (B) Markers of early (EEA1) and recycling (TfR and Rab11) endosomes. Structures whose centers are dually positive for EEA1 and TfR and have TfR-rich domains on their surface are indicated with arrows. (C) Markers of ESCRT III-associated proteins (CHMP1A and Vps4). (D) Markers of late endocytic compartments (CD63 and LAMP1). Rabbit polyclonal antibody staining is shown in green, mouse MAb staining in red, and DAPI staining in blue. Grid spacings are indicated for each image. The corresponding videos, Video S2A and S2BCD, are available in the supplemental material.
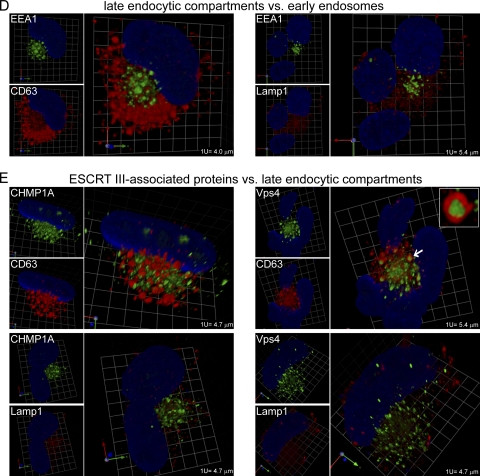
Fig. 3.
Relative localization of markers for major secretory organelles, endocytic organelles, and ESCRT III-associated proteins in HCMV-infected lung fibroblasts (HLF cells). (A) Markers of the ER (Bip), Golgi apparatus (GM130 and Mann II), TGN (p230), and early endosomes (EEA1). (B) Markers of early (EEA1) and recycling (TfR and Rab11) endosomes. (C) Markers of ESCRT III-associated proteins (CHMP1A and Vps4) relative to p230 and EEA1. (D) Markers of late endocytic compartments (CD63 and LAMP1) relative to EEA1. (E) Markers of ESCRT III-associated proteins (CHMP1A and Vps4) versus markers of late endocytic compartments (CD63 and LAMP1). Enlargements of examples of structures described in the text as having the appearance of a sausage wrapped in a bun are provided as insets in the Vps4/EEA1 (C) and Vps4/CD63 (E) images. The objects that were enlarged are indicated with arrows. Rabbit polyclonal antibody staining is shown in green, mouse MAb staining in red, and DAPI staining in blue. Grid spacings are indicated for each image. Corresponding videos, Videos S3A, S3BC, and S3DE, are available in the supplemental material.
Mann II localizes to the cis- and medial-Golgi. In our earlier 2-D work (17), we showed that a mouse monoclonal antibody against GM130 and a rabbit polyclonal antibody against Mann II have highly coincident staining patterns, indicating that they indeed recognize similar structures. Here, we show that Mann II staining and p230 (TGN) staining in uninfected cells look very similar in the side-by-side single-marker panels in Fig. 2A and Video S2A in the supplemental material; this is also true for infected cells (Fig. 3A; see also Video S3A in the supplemental material). However, when the images are merged (Fig. 3A, larger panels; see also the associated videos in the supplemental material), it is obvious that these antibodies stain closely interlaced but distinct tubular structures. In infected cells, the Golgi and TGN markers label ringlike tubular structures that define an outer layer of the cVAC (readily apparent in the supplemental videos). As can be seen in other images, in some instances, the TGN retains its overall shape and distribution but appears to be somewhat fragmented (e.g., see Fig. 3A, EEA1 panel versus p230 panel, and Video S3A in the supplemental material).
In uninfected cells, the early endosomal marker EEA1 stained nearly spherical vesicular structures scattered throughout the cytoplasm in a pattern reminiscent of a starry night (Fig. 2A; see also Video S2A in the supplemental material). As previously shown (17), in infected cells, EEA1-positive vesicles are concentrated at the center of the cVAC and are structurally distinct from those visualized by markers for the ER, Golgi apparatus, and TGN (Fig. 1 and 3A; see also Videos S1 and S3A in the supplemental material).
Early and recycling endosomes.
Early endosomes are protein sorting centers, and recycling endosomes transport cargo between the cytoplasm and plasma membrane. Early endosomes are often marked by phosphatidylinositol 3-phosphate, Rab5, and EEA1, while recycling endosomes are marked by TfR, Rab11, and EEA1. A prior study showed that HCMV is released from actively recycling endosomes (54). EEA1, Rab11, and TfR are associated with similar-appearing structures in infected cells, but these markers have not been studied simultaneously (8, 17, 33).
If examined in isolation, antibodies against TfR and Rab11 have staining patterns in uninfected cells that are highly reminiscent of EEA1 staining (Fig. 2B; see also Video S2BCD in the supplemental material [a higher-resolution version of this video is available at www.med.wayne.edu/micr/gallery/pellett.asp]). When TfR and EEA1 are examined together, it is clear that they mark many of the same vesicles, but some vesicles are marked by TfR and not by EEA1. In some instances, TfR and EEA1 are simultaneously present in the central domain of vesicles, with additional TfR-rich/EEA1-poor domains present on the exterior of these vesicles (arrows). There are more TfR-only objects than EEA1-only objects. With respect to Rab11, few EEA1-positive vesicles have obvious colocalization with Rab11-positive vesicles. Although we did not directly compare them, the large number of vesicles simultaneously stained by EEA1 and TfR and the almost complete absence of vesicles simultaneously stained for EEA1 and Rab11 make it clear that Rab11 and TfR are predominantly associated with different vesicular structures in uninfected fibroblasts.
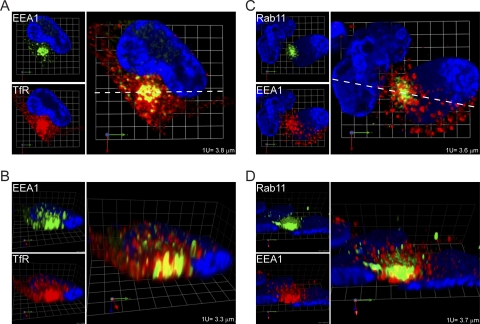
The structural relationships are different in infected cells (Fig. 3B; see also Video S3BC in the supplemental material [a higher-resolution version of this video is available at www.med.wayne.edu/micr/gallery/pellett.asp]). There are more TfR-positive vesicles than EEA1-positive vesicles. While most EEA1-positive vesicles on the cVAC exterior have little colocalization with TfR or Rab11, cross-sectional images reveal very high levels of colocalization at the cVAC center, where most dually stained vesicles are predominantly stained by EEA1 and are surrounded by TfR (Fig. 4A and B). Similarly, Rab11 is present in what appears to be an amorphous mass at the center of the cVAC. In cross-sections (Fig. 4C and D), EEA1 staining is associated with distinct ovoid vesicles, while Rab11 stains the intervesicular space. Figure 4 makes the important point that relationships visible on the exterior of reconstructed structures can be very different from what is happening on the interior. In sum, the strongest associations between EEA1 and TfR or Rab11 are at the center of the cVAC.
Fig. 4.
Regions of high levels of colocalization of early (EEA1) and recycling (TfR and Rab11) endosome markers at the cVAC center. The images are cross-sections through the cVAC of the 3-D reconstructions shown in Fig. 3B, without modification of color intensities. (A and C) Horizontal cross sections (perpendicular to the z-axis) through the 3-D reconstructions (at confocal section 17 of 31 for panel A and 15 of 28 for panel C); (B and D) vertical cross sections (parallel to the z-axis) taken along the dotted white lines in panels A and C. Grid spacings are indicated.
ESCRT III machinery.
CHMP1A regulates membrane scission of intraluminal vesicles and functions as an adaptor molecule for ESCRT III disassembly in the multivesicular body (MVB) pathway. Vps4 is an ESCRT III-associated AAA ATPase that is involved in the membrane constriction that results in formation of the intraluminal vesicles that are hallmarks of MVBs. Both of these molecules have important but undefined roles in the production of infectious HCMV virions (52). In uninfected cells, CHMP1A and Vps4 are present on widely scattered vesicular structures. CHMP1A-positive structures are distinct from EEA1-positive structures, while nearly all vesicles marked by Vps4 are also clearly marked by EEA1. In infected cells, we compared the localization of the ESCRT III-associated markers with p230 (TGN) and EEA1 (Fig. 3C; see also Video S3BC in the supplemental material). CHMP1A and Vps4 are present on structures within and around the cVAC that are distinct from the TGN. CHMP1A is present in the central zone of the cVAC, much of it occupying spaces between EEA1-positive structures, but it is sometimes present on structures that are closely associated with and even surrounded by EEA1-positive structures. As in uninfected cells, nearly all Vps4-labeled structures are closely associated with EEA1-labeled structures. A distinction is that in infected cells, in and around the cVAC, most of the Vps4-positive structures are surrounded by tightly associated EEA1-positive structures. There are numerous examples of Vps4-positive “sausages” wrapped in EEA1-positive “buns” (Fig. 3C, arrow and inset image).
Late endocytic compartment.
The late endocytic compartment consists of late endosomes and lysosomes. Late endosomes are maturational intermediates between early endosomes and lysosomes. Some late endosomes contain numerous small intraluminal vesicles (MBVs), which can be marked by CD63. Lysosomes are marked by LAMP1. In uninfected cells, CD63 and EEA1 stain distinct populations of similar-sized widely distributed vesicles (Fig. 2D; see also Video S2BCD in the supplemental material). In contrast, most EEA1-positive vesicles have coincident LAMP1 staining (Fig. 2D; see also Video S2BCD in the supplemental material). As it does in uninfected cells, in infected cells, CD63 stains populations of vesicles that are distinct from EEA1-stained vesicles. LAMP1 staining is markedly different in infected cells compared to that in uninfected cells, with the infected-cell staining being less intense and localized to smaller, more granular structures that are distinct from EEA1-stained structures. The pattern of LAMP1 staining we observed in uninfected cells is similar to that found by others (8, 57), who also found that LAMP1 antibody stained relatively large structures at the cVAC periphery. The LAMP1 antibody we used stained infected cells in a fine-granular distribution that had no discernible structure or particular relationship with the cVAC.
Because of the role played by ESCRT III machinery in membrane budding events, such as MVB biogenesis and envelopment of some viruses, we studied the relationship between late endocytic compartments and ESCRT III-associated components that are important for HCMV replication (52). As mentioned, in uninfected cells, Vps4 and LAMP1 are both associated with EEA1-positive vesicles, while CHMP1A and CD63 are not present on EEA1-positive structures (Fig. 2C and D; see also Video S2BCD in the supplemental material). In infected cells, CHMP1A and CD63 stain distinct populations of similar-sized vesicles in and around the cVAC (Fig. 3E; see also Video S3DE in the supplemental material [a higher-resolution version of this video is available at www.med.wayne.edu/micr/gallery/pellett.asp]). Likewise, CHMP1A and LAMP1 stain different populations of vesicles in and around the cVAC. As mentioned, LAMP1-positive vesicles are smaller in infected than in uninfected cells. CD63-labeled structures sometimes surround Vps4-positive cylinders, similar to the wrapping of Vps4 cylinders by EEA1-positive structures (Fig. 3E, arrow and inset). The sausage-in-bun structures seen for EEA1/Vps4 and CD63/Vps4 are similar in appearance to complexes that contain syntaxin 3 and glycoprotein H (9). Because CD63 and EEA1 mark distinct populations of vesicles, the CD63/Vps4-positive objects are likely to be different from the similar-appearing EEA1/Vps4 objects. Vps4 and LAMP1 are associated with different objects.
Quantitative analysis of colocalization and organelle volumes.
Thus far, we have described properties of organelle staining that are evident upon visual inspection of the 3-D reconstructions. To put these observations into a quantitative context, we conducted colocalization analyses and measured organelle volumes, comparing infected cells with uninfected cells.
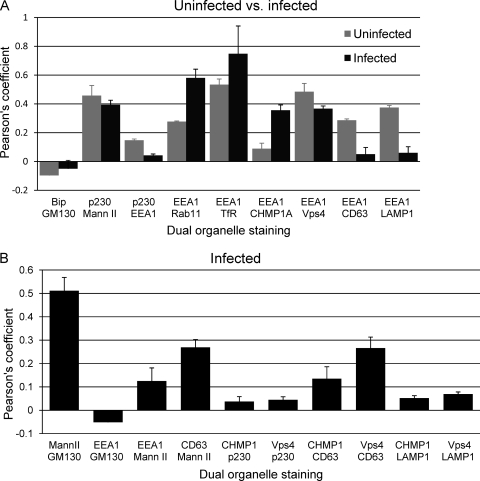
Pearson's threshold colocalization coefficient was measured for pairs of markers (Fig. 5). In uninfected cells, along the ER, Golgi apparatus, TGN, and early endosome axis, Bip (ER) and GM130 (cis-Golgi) have slightly negative colocalization coefficients, Mann II (cis-Golgi) and p230 (TGN) have a relatively high colocalization coefficient, and p230 (TGN) and EEA1 (early endosomes) have low levels of colocalization (Fig. 5A). In comparisons of markers for recycling endosomes with EEA1, Rab11 had moderately high levels of colocalization, while EEA1 and TfR had the highest level of colocalization of any of the pairs of markers tested. EEA1 has a relatively low level of colocalization for one ESCRT III-associated marker (CHMP1A) and a high level of colocalization with the other (Vps4). EEA1 also has a low level of colocalization with markers for late endocytic compartments (CD63 and LAMP1).
Fig. 5.
Colocalization coefficients for pairs of markers. (A) Uninfected versus infected HLF cells. (B) Infected HLF cells. Pearson's threshold colocalization coefficient (as implemented in the Volocity software package) was determined for the indicated pairs of markers. At least 3 cells were evaluated for each pair of markers. Under the analysis conditions used, a voxel (a pixel in three dimensions) was considered positive if both markers were above a specified threshold (36). Under these conditions, voxels that were low positive for one marker and high positive for the other would be considered positive, as would be voxels low or high for both markers. In some instances, images of pairwise comparisons are not shown in the accompanying figures (e.g., Mann II versus GM130 [data not shown]). Mean values are expressed (±standard errors of the means).
Infected cell relationships for some pairs of markers were similar to those for uninfected cells (Bip/GM130, p230/Mann II, and Vps4/EEA1), but the patterns were markedly different for others. Relative to uninfected cells, infected cell colocalizations were greatly reduced for p230 versus that for EEA1, CD63 versus that for EEA1, and LAMP1 versus that for EEA1. Markers for recycling endosomes (Rab11 and TfR) had even higher levels of colocalization with EEA1 than that in uninfected cells. Interestingly, CHMP1A changed from very little colocalization with EEA1 to moderately high levels of colocalization. Having identified the relative localizations in uninfected and infected cells for several major structures involved in membrane transport, we extended the analysis to a broader set of pairwise comparisons in infected cells (Fig. 5B). As we previously showed, the independent Golgi markers, Mann II and GM130, have high levels of colocalization. Both of these markers have low levels of colocalization with EEA1. CD63 and Mann II had moderate levels of colocalization. The ESCRT III-associated markers have little colocalization with p230 (TGN) or LAMP1 (lysosomes). Taken with the results shown in Fig. 5A, the ESCRT III-associated proteins, CHMP1A and Vps4, are associated with independent CD63-positive and EEA1-positive structures. These results provide new evidence that HCMV-induced remodeling of the membrane transport apparatus involves much more than simple relocation or expansion of preexisting structures.
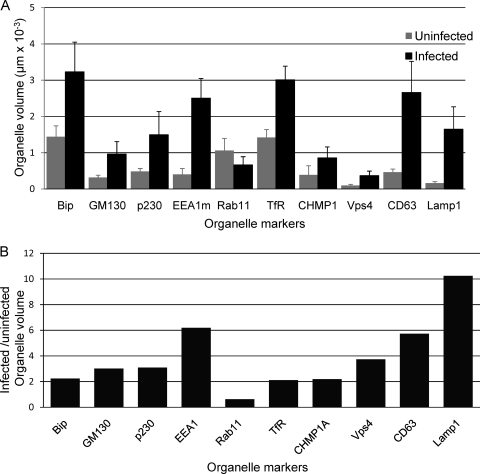
In addition to changes in the distribution of and relationships between markers for various organelles, there are also changes in organelle sizes and volumes in HCMV-infected cells (Fig. 6). Note that the measured volumes are summations of the volumes of the voxels (pixels in three dimensions) that were positive for the marker in question, not the population of “empty” voxels in the vicinity of, or possibly surrounded by, positive voxels. For example, we measured the volume occupied by the tubular structure defined by the Golgi protein GM130 but not the volume of the “empty” space the structure surrounds (Fig. 1). Some of the cells we examined have two or three nuclei, but all have only one cVAC. Because we found an approximately linear relationship between organelle volumes and the number of nuclei in multinucleated cells (data not shown), we expressed organelle volumes on a per-nucleus basis. As might be expected based on the expansion of infected cell volume that occurs as cells become cytomegalic, with the exception of structures marked by Rab11, infected cell organelle volumes were 2- to 10-fold larger than those in uninfected cells.
Fig. 6.
Organelle volumes in uninfected and infected cells. (A) The “Find Objects” algorithm from the Volocity software package was used to identify objects in three-dimensional reconstructions that correspond to objects visible in images such as those shown in Fig. 2 and 3. The net intracellular volume associated with these objects was obtained by summing the volumes of the positive voxels. For EEA1, results were from staining with the mouse MAb. Mean values are expressed (±standard errors of the means). (B) Ratios of organelle volumes in infected cells compared to those in uninfected cells.
DISCUSSION
Herpesvirus virions contain >10,000 individual protein molecules that are the products of ∼40 viral genes, along with some proteins of cellular origin (8, 55). Our objective is to understand the process that ensures production of virions that contain the correct complement (species and number) of these proteins. This will lead to development of new targets for antiviral development, as well as greater understanding of the complex interactions between the virus and its host. In this work, we studied the 3-D distribution in and around the HCMV cVAC of host proteins that have proven to be useful markers for individual organelles or machinery that are important in membrane transport. As a point of reference, we also studied these markers in uninfected cells.
Development of a relational map of the cVAC.
We will begin by summarizing our findings with respect to the different organelle systems we studied. For the ER, Golgi apparatus, TGN, and early endosome axis, the major change for the ER was an increase in its net size, in accordance with the change in cellular volume that is part of cytomegaly. The ER (as defined by staining for Bip) has no colocalization with markers for the Golgi apparatus, and its 3-D distribution is markedly different from that of the other structures studied. The Golgi apparatus and TGN form closely intertwined tubular structures. In many cells they each form single large structures, but in some cells these structures appear to be fragmented while retaining a distribution similar to that of the unitary structures. The cis-Golgi and TGN markers Mann II and p230 stain structures that appear to be distinct from each other upon visual inspection, but when the 3-D data are analyzed quantitatively, they have moderately high colocalization coefficients. This may be explained by a combination of the transfer of Mann II to the TGN during vesicular transport and the possibility of better resolving components of these closely intertwined structures as higher resolution imaging methods become available. Structures stained by antibodies to EEA1 had little colocalization with markers for the Golgi apparatus and TGN.
Sorting events in early endosomes determine whether cargoes, such as proteins or lipids, will be recycled to the plasma membrane, degraded in lysosomes, or delivered to the TGN. We found that EEA1 had moderately high levels of colocalization with markers for recycling endosomes (TfR and Rab11) in uninfected cells, but it was apparent on visual inspection that these markers are present at different ratios on distinct populations of vesicles. Colocalization among these markers increased in infected cells, and many vesicles are clearly simultaneously positive for EEA1 and TfR or Rab11, but some vesicles are much more strongly stained for one marker than for the other. Tooze et al. showed that HCMV virions are released from actively recycling endosomes (54). Consistent with this, soluble transferrin taken up by recycling endosomes from the cell surface colocalizes with the virion glycoprotein gM/gN (33). Cepeda et al. found that TfR colocalizes in the cVAC with the pUL33 G protein-coupled receptor (GPCR); by immunoelectron microscopy, they also found EEA1 and TfR in HCMV virions (8). Further evidence for the importance of recycling endosomes in HCMV maturation and egress is that a dominant negative form of Rab11, a regulator of endocytic recycling, disrupts the cVAC and reduces virus yields by >10-fold (33). We found the highest levels of colocalization between EEA1 and TfR or Rab11 to be at the center of the cVAC, consistent with the possibility that these proteins play active roles in the use of the recycling endosomal pathway for virion egress.
The late endocytic pathway extends from EEA1-positive sorting stations to late endosomes that are marked by CD63 and then to lysosomes (marked by LAMP1). A CD63 antibody and the LAMP1 antibody used here stained structures of very different appearance in infected cells (nearly spherical vesicles of 0.5 to 1.7 μm for CD63 versus amorphous fine-granular staining for LAMP1). In addition, the LAMP1 staining was not confined to the cVAC region but was present at the cell periphery and on the opposite side of the nucleus. We found that intracellular volumes associated with CD63 and LAMP1 both increased substantially in infected cells. EEA1 had moderately high colocalization with CD63 and LAMP1 in uninfected cells, but at 120 hpi, colocalization was nearly absent. Dual staining for CD63 and LAMP1 would be required to learn whether they are staining distinct structures, as opposed to LAMP1 being present on subdomains of larger CD63-positive structures. Our results suggest that lysosome function is likely to be very different in infected cells compared to that in uninfected cells.
ESCRT III machinery is required for membrane scission as part of budding processes that originate in the cytoplasmic space and extend into either the extracellular space or into the lumen of cytoplasmic vesicles. This machinery is involved in the budding of HIV-1, Lassa fever virus, Ebola virus, the paramyxovirus parainfluenza virus 5 (PIV-5), mumps virus, and hepatitis B virus from the cell surface (reviewed in references 11 and 22) and is likely involved in envelopment of herpesviruses, such as human herpesvirus 6 (HHV-6), which egresses by way of MVBs (41), and herpes simplex virus type 1 (HSV-1), which is dependent on Vps4 for its egress (7, 14, 44). For HCMV, dominant negative versions of proteins involved in ESCRT III disassembly, CHMP1A and Vps4, reduce infectious yields, and tagged versions of these proteins localize in the vicinity of the cVAC (52). We found that in infected cells, CHMP1A and Vps4 have low levels of colocalization with markers of the TGN and lysosomes. Consistent with their roles in MVB biogenesis, they had moderately high levels of colocalization in infected cells with the MVB marker, CD63. In uninfected cells, Vps4 but not CHMP1 had high levels of colocalization with EEA1; however, in infected cells, the CHMP1A/EEA1 colocalization increased substantially, to a level comparable to that of Vps4/EEA1. Thus, the two ESCRT III-associated proteins previously shown to be important for a late stage of HCMV replication (52) converge to have similar distributions late in infection, both being associated with vesicles positive for either EEA1 or CD63. This is consistent with the hypothesis that ESCRT III machinery is involved in virion envelopment at early/recycling endosome membranes, via a process similar to generation of intraluminal vesicles in multivesicular bodies.
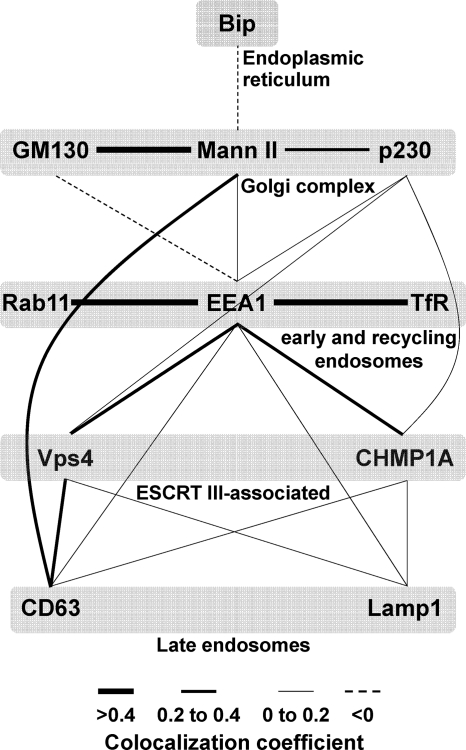
From the set of pairwise colocalization coefficients (Fig. 5), we generated a map of the colocalization network (Fig. 7), which enables simultaneous visualization of multiple relationships and provides information about possible interactions in the complex environment of the cVAC. Several points can be made from this. As might be expected, markers for the cis- and cis/medial-Golgi had high levels of colocalization. Early and recycling endosome markers also had high levels of colocalization. Consistent with the abundance of EEA1-positive vesicles at the cVAC center and our model of cVAC architecture (17), EEA1 had no colocalization with the cis-Golgi marker and low levels of colocalization with the medial-Golgi and TGN markers. EEA1 also colocalized with the ESCRT III-associated proteins and to a lesser extent with markers for late endosomes. The ESCRT III-associated proteins colocalized most strongly with EEA1 but also colocalized with markers for late endosomes and the TGN.
Fig. 7.
Colocalization network map of membrane transport machinery markers in HCMV-infected cells. Pairwise colocalization data from Fig. 5 were used to generate a map of the colocalization network among the various markers studied here. Three categories of line widths were used that are proportional to the extent of the observed Pearson's colocalization coefficient (high = >0.4; medium = 0.2 to 0.4; low = 0 to 0.2); negative colocalization values are represented as dashed lines. All pairwise comparisons from Fig. 5 are represented, but all possible pairwise combinations were not tested.
In summary, each of the organelle markers studied has a 3-D structural identity and colocalization network that markedly differ between uninfected and infected cells. In images obtained as three-dimensional reconstructions from confocal sections, the various tightly packed cVAC-associated secretory organelles retain discrete identities, even though their architectures and compositions are significantly different from those of uninfected cells; these differences can be expected to affect function. Ongoing activities in the cVAC are likely to depend on retention of secretory organelle compartmentalization, as well as on the as-yet-undefined changes in their functions. The relational map described here will be useful for developing hypotheses as to the functions of cVAC-associated viral and cellular proteins.
Organelle identity differs in uninfected and infected cells.
Much of what we report relates to the concept of organelle identity, which derives from a combination of organelle structure, composition, localization, and function. Pathogens can alter organelle identity. For example, some bacteria can cloak themselves within membrane-bound organelles of the secretory systems, a process known as identity theft (29, 31). Our results demonstrate that HCMV induces organelle identity shift rather than identity theft, that is, organelles in and around the cVAC retain much of their essential character, but their localizations, shapes, and sizes markedly differ from those of uninfected cells in ways that are likely to affect their functions. For example, early endosomes are conventionally defined by the presence of Rab5-GTP, EEA1, and PtdIns(3)P. We previously showed that Rab5 and EEA1 are highly colocalized in uninfected cells but not in infected cells (17). In this work, we found that several pairs of markers have lower colocalization coefficients in infected cells (p230/EEA1, EEA1/CD63, EEA1/LAMP1), while CHMP1A and EEA1 are more highly colocalized. The functional differences that result from these changes remain to be determined.
In summary, we provide new data indicating that the membrane transport machinery in the HCMV cVAC has been altered in ways that are likely to provide unique and specialized functions that would be related to, but different from, their operation in uninfected cells. This advances our understanding of the cell biology of membrane transport machinery and further illuminates the magnitude of the cytoplasmic remodeling that is induced by HCMV infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Christina Woodard for her contributions to this work and Mary Olive and Carmel Harkins of the Wayne State University Microscopy and Imaging Resources Laboratory for assistance with the confocal microscopy and analysis.
This work was supported by Wayne State University and NIAID (R21 AI076591-01). The Wayne State University Microscopy and Imaging Resources Laboratory is supported in part by NIH grants P30ES06639, P30CA22453, and U54RR020843, the Burnham Institute, and the Perinatology Research Branch of the National Institute of Child Health and Development.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 6 April 2011.
REFERENCES
- 1. Ahlqvist J., Mocarski E. 2011. Cytomegalovirus UL103 controls virion and dense body egress. J. Virol. 85:5125–5135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. AuCoin D. P., Smith G. B., Meiering C. D., Mocarski E. S. 2006. Betaherpesvirus-conserved cytomegalovirus tegument protein ppUL32 (pp150) controls cytoplasmic events during virion maturation. J. Virol. 80:8199–8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barr F. A., Short B. 2003. Golgins in the structure and dynamics of the Golgi apparatus. Curr. Opin. Cell Biol. 15:405–413 [DOI] [PubMed] [Google Scholar]
- 4. Buchkovich N. J., Maguire T. G., Alwine J. C. 2010. Role of the endoplasmic reticulum chaperone BiP, SUN domain proteins, and dynein in altering nuclear morphology during human cytomegalovirus infection. J. Virol. 84:7005–7017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buchkovich N. J., Maguire T. G., Paton A. W., Paton J. C., Alwine J. C. 2009. The endoplasmic reticulum chaperone BiP/GRP78 is important in the structure and function of the HCMV assembly compartment. J. Virol. 83:11421–11428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buchkovich N. J., et al. 2008. Human cytomegalovirus specifically controls the levels of the endoplasmic reticulum chaperone BiP/GRP78, which is required for virion assembly. J. Virol. 82:31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Calistri A., et al. 2007. Intracellular trafficking and maturation of herpes simplex virus type 1 gB and virus egress require functional biogenesis of multivesicular bodies. J. Virol. 81:11468–11478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cepeda V., Esteban M., Fraile-Ramos A. 2010. Human cytomegalovirus final envelopment on membranes containing both trans-Golgi network and endosomal markers. Cell. Microbiol. 12:386–404 [DOI] [PubMed] [Google Scholar]
- 9. Cepeda V., Fraile-Ramos A. 2011. A role for the SNARE protein syntaxin 3 in human cytomegalovirus morphogenesis. Cell. Microbiol. [Epub ahead of print.] doi:10.1111/j.1462-5822.2011.01583.x [DOI] [PubMed] [Google Scholar]
- 10. Chapuy B., et al. 2008. AP-1 and AP-3 mediate sorting of melanosomal and lysosomal membrane proteins into distinct post-Golgi trafficking pathways. Traffic 9:1157–1172 [DOI] [PubMed] [Google Scholar]
- 11. Chen B. J., Lamb R. A. 2008. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372:221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen J., Wang J., Meyers K. R., Enns C. A. 2009. Transferrin-directed internalization and cycling of transferrin receptor 2. Traffic 10:1488–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Christoforidis S., McBride H. M., Burgoyne R. D., Zerial M. 1999. The Rab5 effector EEA1 is a core component of endosome docking. Nature 397:621–625 [DOI] [PubMed] [Google Scholar]
- 14. Crump C. M., Yates C., Minson T. 2007. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J. Virol. 81:7380–7387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Das S., Pellett P. E. 2007. Members of the HCMV US12 family of predicted heptaspanning membrane proteins have unique intracellular distributions, including association with the cytoplasmic virion assembly complex. Virology 361:263–273 [DOI] [PubMed] [Google Scholar]
- 16. Das S., Skomorovska-Prokvolit Y., Wang F. Z., Pellett P. E. 2006. Infection-dependent nuclear localization of US17, a member of the US12 family of human cytomegalovirus-encoded seven-transmembrane proteins. J. Virol. 80:1191–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Das S., Vasanji A., Pellett P. E. 2007. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J. Virol. 81:11861–11869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dunn W., et al. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223–14228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fraile-Ramos A., Cepeda V., Elstak E., van der Sluijs P. 2010. Rab27a is required for human cytomegalovirus assembly. PLoS One 5:e15318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gleeson P. A., et al. 1996. p230 is associated with vesicles budding from the trans-Golgi network. J. Cell Sci. 109(Pt. 12):2811–2821 [DOI] [PubMed] [Google Scholar]
- 21. Gonzalez-Gronow M., Selim M. A., Papalas J., Pizzo S. V. 2009. GRP78: a multifunctional receptor on the cell surface. Antioxid. Redox Signal. 11:2299–2306 [DOI] [PubMed] [Google Scholar]
- 22. Harrison M. S., Sakaguchi T., Schmitt A. P. 2010. Paramyxovirus assembly and budding: building particles that transmit infections. Int. J. Biochem. Cell Biol. 42:1416–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Holthuis J. C., Levine T. P. 2005. Lipid traffic: floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 6:209–220 [DOI] [PubMed] [Google Scholar]
- 24. Homman-Loudiyi M., Hultenby K., Britt W., Soderberg-Naucler C. 2003. Envelopment of human cytomegalovirus occurs by budding into Golgi-derived vacuole compartments positive for gB, Rab 3, trans-Golgi network 46, and mannosidase II. J. Virol. 77:3191–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Howard T. L., Stauffer D. R., Degnin C. R., Hollenberg S. M. 2001. CHMP1 functions as a member of a newly defined family of vesicle trafficking proteins. J. Cell Sci. 114:2395–2404 [DOI] [PubMed] [Google Scholar]
- 26. Hurley J. H., Hanson P. I. 2010. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat. Rev. Mol. Cell Biol. 11:556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Indran S. V., Ballestas M. E., Britt W. J. 2010. Bicaudal D1-dependent trafficking of human cytomegalovirus tegument protein pp150 in virus-infected cells. J. Virol. 84:3162–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jovic M., Sharma M., Rahajeng J., Caplan S. 2010. The early endosome: a busy sorting station for proteins at the crossroads. Histol. Histopathol. 25:99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kagan J. C., Roy C. R. 2002. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat. Cell Biol. 4:945–954 [DOI] [PubMed] [Google Scholar]
- 30. Kalejta R. F. 2008. Tegument proteins of human cytomegalovirus. Microbiol. Mol. Biol. Rev. 72:249–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Knodler L. A., Steele-Mortimer O. 2003. Taking possession: biogenesis of the Salmonella-containing vacuole. Traffic 4:587–599 [DOI] [PubMed] [Google Scholar]
- 32. Kobayashi T., et al. 2000. The tetraspanin CD63/Lamp3 cycles between endocytic and secretory compartments in human endothelial cells. Mol. Biol. Cell 11:1829–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krzyzaniak M. A., Mach M., Britt W. J. 2009. HCMV-encoded glycoprotein M (UL100) interacts with Rab11 effector protein FIP4. Traffic 10:1439–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lock J. G., Stow J. L. 2005. Rab11 in recycling endosomes regulates the sorting and basolateral transport of E-cadherin. Mol. Biol. Cell 16:1744–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luzio J. P., Pryor P. R., Bright N. A. 2007. Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 8:622–632 [DOI] [PubMed] [Google Scholar]
- 36. Manders E. M. M., Verbeek F. J., Aten J. A. 1993. Measurement of co-localization of objects in dual-color confocal images. J. Microsc. 169:375–382 [DOI] [PubMed] [Google Scholar]
- 37. Maxfield F. R., McGraw T. E. 2004. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5:121–132 [DOI] [PubMed] [Google Scholar]
- 38. Milbradt J., Auerochs S., Sticht H., Marschall M. 2009. Cytomegaloviral proteins that associate with the nuclear lamina: components of a postulated nuclear egress complex. J. Gen. Virol. 90:579–590 [DOI] [PubMed] [Google Scholar]
- 39. Mohrmann K., van der Sluijs P. 1999. Regulation of membrane transport through the endocytic pathway by rabGTPases. Mol. Membr. Biol. 16:81–87 [DOI] [PubMed] [Google Scholar]
- 40. Moorman N. J., Sharon-Friling R., Shenk T., Cristea I. M. 2010. A targeted spatial-temporal proteomics approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation. Mol. Cell. Proteomics 9:851–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mori Y., et al. 2008. Human herpesvirus-6 induces MVB formation, and virus egress occurs by an exosomal release pathway. Traffic 9:1728–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Muranyi W., Haas J., Wagner M., Krohne G., Koszinowski U. H. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854–857 [DOI] [PubMed] [Google Scholar]
- 43. Ni M., Lee A. S. 2007. ER chaperones in mammalian development and human diseases. FEBS Lett. 581:3641–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pawliczek T., Crump C. M. 2009. Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J. Virol. 83:11254–11264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pfeffer S. R. 2001. Constructing a Golgi complex. J. Cell Biol. 155:873–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pols M. S., Klumperman J. 2009. Trafficking and function of the tetraspanin CD63. Exp. Cell Res. 315:1584–1592 [DOI] [PubMed] [Google Scholar]
- 47. Quinones Q. J., de Ridder G. G., Pizzo S. V. 2008. GRP78: a chaperone with diverse roles beyond the endoplasmic reticulum. Histol. Histopathol. 23:1409–1416 [DOI] [PubMed] [Google Scholar]
- 48. Roth M. G. 2004. Phosphoinositides in constitutive membrane traffic. Physiol. Rev. 84:699–730 [DOI] [PubMed] [Google Scholar]
- 49. Sanchez V., Greis K. D., Sztul E., Britt W. J. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schauflinger M., et al. 2011. The tegument protein UL71 of human cytomegalovirus is involved in late envelopment and affects multivesicular bodies. J. Virol. 85:3821–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Scheuring S., et al. 2001. Mammalian cells express two VPS4 proteins, both of which are involved in intracellular protein trafficking. J. Mol. Biol. 312:469–480 [DOI] [PubMed] [Google Scholar]
- 52. Tandon R., AuCoin D. P., Mocarski E. S. 2009. Human cytomegalovirus exploits ESCRT machinery in the process of virion maturation. J. Virol. 83:10797–10807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tandon R., Mocarski E. S. 2008. Control of cytoplasmic maturation events by cytomegalovirus tegument protein pp150. J. Virol. 82:9433–9444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tooze J., Hollinshead M., Reis B., Radsak K., Kern H. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163–178 [PubMed] [Google Scholar]
- 55. Varnum S. M., et al. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Velasco A., et al. 1993. Cell type-dependent variations in the subcellular distribution of alpha-mannosidase I and II. J. Cell Biol. 122:39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Womack A., Shenk T. 2010. Human cytomegalovirus tegument protein pUL71 is required for efficient virion egress. MBio 1:e00282-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Woodman P. G., Futter C. E. 2008. Multivesicular bodies: co-ordinated progression to maturity. Curr. Opin. Cell Biol. 20:408–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yoshino A., et al. 2003. A role for GRIP domain proteins and/or their ligands in structure and function of the trans-Golgi network. J. Cell Sci. 116:4441–4454 [DOI] [PubMed] [Google Scholar]
- 60. Yu D., Silva M. C., Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. U. S. A. 100:12396–12401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zipeto D., Baldanti F., Percivalle E., Gerna G., Milanesi G. 1993. Identification of a human cytomegalovirus mutant in the pp150 matrix phosphoprotein gene with a growth-defective phenotype. J. Gen. Virol. 74:1645–1648 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.