Abstract
In budding yeast and humans, cohesion establishment during S phase requires the acetyltransferase Eco1/Esco1-2, which acetylates the cohesin subunit Smc3 on two conserved lysine residues. Whether Smc3 is the sole Eco1/Esco1-2 effector and how Smc3 acetylation promotes cohesion are unknown. In fission yeast (Schizosaccharomyces pombe), as in humans, cohesin binding to G1 chromosomes is dynamic and the unloading reaction is stimulated by Wpl1 (human ortholog, Wapl). During S phase, a subpopulation of cohesin becomes stably bound to chromatin in an Eso1 (fission yeast Eco1/Esco1-2)-dependent manner. Cohesin stabilization occurs unevenly along chromosomes. Cohesin remains largely labile at the rDNA repeats but binds mostly in the stable mode to pericentromere regions. This pattern is largely unchanged in eso1Δ wpl1Δ cells, and cohesion is unaffected, indicating that the main Eso1 role is counteracting Wpl1. A mutant of Psm3 (fission yeast Smc3) that mimics its acetylated state renders cohesin less sensitive to Wpl1-dependent unloading and partially bypasses the Eso1 requirement but cannot generate the stable mode of cohesin binding in the absence of Eso1. Conversely, nonacetylatable Psm3 reduces the stable cohesin fraction and affects cohesion in a Wpl1-dependent manner, but cells are viable. We propose that Psm3 acetylation contributes to Eso1 counteracting of Wpl1 to secure stable cohesin interaction with postreplicative chromosomes but that it is not the sole molecular event by which this occurs.
INTRODUCTION
Following DNA replication in S phase, sister DNA molecules are linked together by cohesin. Thereafter, cohesion between sister chromatids persists throughout the G2 phase and until mitosis, where it allows chromosome biorientation on the mitotic spindle (16). Defects in this process have been linked to aneuploidy and tumor progression. Cohesin is a multisubunit protein complex made of a dimer of long, flexible Smc subunits, which form a ring-shaped structure stabilized by the binding of a kleisin subunit (Scc1/Rad21 in the mitotic cycle; Rec8 in meiosis) (2, 25). Kleisin cleavage by separase destroys the ring and allows chromatid separation at anaphase (28, 43, 58). The ring shape of the complex suggested that cohesin ensures cohesion by topological trapping of sister DNA molecules, and strong experimental evidence supports this model (24), although other modes of cohesin-DNA interaction might coexist (29, 40).
One key aspect of the cohesion cycle is how cohesion is made during S phase. Cohesin is first deposited on unreplicated chromosomes in a reaction requiring ATP hydrolysis by the Smc heads and the cohesin-loading complex Scc2/Scc4 (4, 5, 15, 61). In an unperturbed cell cycle, cohesion is made exclusively during S phase, and except in the event of a DNA double-strand break (DSB), cohesin loading after DNA replication does not result in functional cohesion (26, 34, 51, 60). Numerous studies have shown that mutations in nonessential factors associated with the replication fork machinery affect sister chromatid cohesion, leading to the notion that cohesion is created in a reaction coupled with replication fork progression (3, 20, 27, 32, 35, 36, 44). One crucial factor associated with the replisome is the acetyltransferase Eco1/Ctf7 (30, 34, 38). In eco1 mutants (eso1 in fission yeast), cohesion is not created or quickly lost after DNA replication, although cohesin rings are formed and bound to chromatin, resulting in a “cohesin without cohesion” phenotype (50, 54, 57).
Following S phase, cohesion persists until nuclear division. Since cohesion is essential for chromosome segregation, cohesin binding to chromosomes should be stable enough to maintain cohesion during the intervening G2 phase. Live imaging in mammalian cells has shown that cohesin is not stably bound to chromatin during the G1 phase of the cell cycle, when chromosomes are not yet replicated. Chromatin-bound cohesin exchanges with the soluble nuclear pool. The “unloading” reaction relies in part on Wapl, a cohesin binding protein conserved from yeast to human (23, 31). Importantly, a fraction of cohesin becomes stably bound to chromatin as cells progress through S phase. Since sister chromatid cohesion is created at that time, this raised the possibility that the change in cohesin dynamics might be instrumental to the process of creating and/or maintaining cohesion. Similarly, in fission yeast, sustained cohesin binding to G1 chromosomes requires the continued activity of the cohesin loader (called Mis4/Ssl3 in fission yeast [9, 21]). When the cohesin loader is inactivated in G1, cohesin dissociates completely from chromatin in a Wpl1-dependent manner (fission yeast Wapl is named Wpl1, encoded by wpl1, SPBC428.17c). In contrast, when cohesin loading is inactivated in G2, a pool of cohesin remains bound to chromatin. Cohesin stabilization was affected in the thermosensitive eso1-H17 mutant, indicating that stable cohesin binding to postreplicative chromosomes requires Eso1 function (12). In human cells, the premature separation of sister chromatids induced by Esco2 depletion is suppressed by downregulating Wapl, suggesting that Esco2 counteracts Wapl (22).
Altogether, these observations suggest a model in which cohesin must be stably bound to chromatin to ensure cohesion, and the function of the acetyltransferase would be to promote cohesin stabilization by preventing Wapl from acting on it. The acetyltransferase may counteract Wapl through the acetylation of specific substrates. One obvious candidate is Smc3, one of the two Smc molecules that form the ring shape of cohesin. In budding yeast and mammals, Smc3 is acetylated on two adjacent conserved lysine residues (8, 45, 59, 63). Acetyl mimicking mutant forms of Smc3 partially bypasses the Eco1 requirement in budding yeast. Conversely, nonacetylatable yeast mutants are nonviable, but this phenotype is alleviated by the deletion of the budding yeast wpl homolog (8, 45, 59). These data led to the conclusion that the creation of cohesion requires the acetylation of Smc3 by Eco1. Whether Eco1 function transits exclusively through Smc3 acetylation and whether it promotes cohesion through the stabilization of cohesin interaction with chromosomes are unknown. Unlike fission yeast and mammals, it is unclear whether budding yeast Wapl promotes cohesin removal from chromosomes. Rather, budding yeast Wapl appears to display the opposite activity, since it contributes to cohesin binding to chromatin and full cohesion activity (45, 52). In human cells, Smc3 acetylation is required for normal replication fork progression. Failure to acetylate Smc3 slows fork movement, a phenotype reversed by downregulating Wapl (56). Whether Smc3 acetylation in mammals impinges on the stabilization of cohesin interaction with chromosomes is unknown. There is, however, strong experimental evidence that cohesin stabilization requires Sororin, a conserved cohesin-associated factor in vertebrates but with no known homolog in lower eukaryotes (49).
Here, we address these questions in fission yeast. We first show that the mode of cohesin interaction with postreplicative chromatin is uneven along the chromosome. The large amount of cohesin bound to the rDNA gene cluster remains mostly labile, whereas cohesin is mostly bound in a stable mode within pericentromeric heterochromatin. Cohesin stabilization is affected in an eso1 mutant, but Eso1 function is fully dispensable for cohesin stabilization and sister chromatid cohesion when the wpl1 gene is deleted, indicating that the main, if not sole, role of Eso1 in cohesion establishment is to counteract Wpl1. The cohesin subunit Smc3 (called Psm3 in fission yeast) is acetylated in an Eso1-dependent manner. An acetyl-mimicking form of Psm3 can partially bypass the Eso1 requirement, suggesting that Psm3 acetylation may contribute to cohesin stabilization. In support of this idea, we provide evidence that Psm3 acetylation lowers cohesin susceptibility to Wpl1 and thereby may contribute to antagonizing it. Psm3 acetylation by itself, however, is insufficient to confer the stable mode of cohesin interaction with chromosomes. We show that Psm3 is unlikely to be the unique effector of Eso1 function. Whereas the eso1 gene is essential for viability, a nonacetylatable psm3 mutant is viable and mostly proficient for cohesin stabilization and long-term cohesion. Although preventing Psm3 acetylation does not hinder the process of cohesin stabilization per se, the amount of stably bound cohesin is reduced, and cohesion defects can be detected. We propose that Psm3 acetylation may facilitate the reaction promoting a stable mode of cohesin interaction with chromosomes, but our data suggest that Psm3 acetylation is not the sole Eso1-dependent event leading to the stable mode of cohesin binding to replicated chromosomes.
MATERIALS AND METHODS
Strains, media, genetics, and cytological techniques.
The strains used in this study are listed in Table 1. The psm3 alleles were created by site-directed mutagenesis and replacement of the wild-type allele by homologous recombination in a diploid strain using a PCR-based gene-targeting strategy (6). Genomic DNA from the final haploid mutant strains was PCR amplified and sequenced to confirm the presence of the desired mutations. The psm3-GFP strains were created by PCR-based gene targeting using pFA6a-GFP(S65T)-natMX6 as a template (46). Media and genetics techniques were as described previously (39). The synthetic medium EMM2 was used unless otherwise stated. Synthetic medium lacking a nitrogen source (EMM2-N) was used to arrest cells in G1 by nitrogen starvation. Complete yeast extract-peptone-dextrose (YPD) medium was used to release cells into the cell cycle after nitrogen starvation induced G1 arrest. Cell cycle progression was followed by flow cytometry after propidium iodide staining of ethanol-fixed cells (39). Growth to saturation in EMM2-YE medium was used to arrest cells in the G2 phase of the cell cycle (53). The Ch16 minichromosome loss assay was done by the half-sectoring-colonies method, as described previously (1). Cell fixation and immunofluorescence were as described previously (10). Fluorescence in situ hybridization (FISH) was done using cosmid c1228 as a centromere 2-linked probe (37). Cells bearing the cut9-665 mutation were grown to early log phase in YES medium at 25°C and shifted to 36.5°C for 3.5 h to arrest the cells at metaphase. The cells were fixed and processed for antitubulin immunofluorescence and FISH as described previously (11). The spindle length and distance between FISH signals were measured using Metamorph software.
Table 1.
Strain list
| Strain no. | Genotype |
|---|---|
| 2 | h− |
| 405 | h−cdc25-22 |
| 596 | h−ura4 |
| 807 | h−leu1 ura4 ade6-210 his1-102 cut9-665 swi6::his1+ |
| 1176 | h−leu1 ura4 ade6-210 his1-102 cut9-665 rad21-K1ts-ura4+ |
| 3203 | h−mis4-367 cdc25-22 |
| 3333 | h−mis4-367 cdc25-22 rad21-9PK-kanr |
| 3355 | h−mis4-367 cdc25-22 wpl1::kanrrad21-9PK-kanr |
| 3448 | h−cdc25-22 rad21-9PK-kanr |
| 3459 | h−mis4-367cdc25-22 eso1-H17 rad21-9PK-kanr |
| 3469 | h−ura4 wpl1::kanreso1::ura4+psm3-GFP-ura4+ |
| 3505 | h+ura4 wpl1::kanrpsm3-GFP-ura4+ |
| 3678 | h−ura4 psm3-GFP-natr |
| 3726 | h+ura4 mis4-367 wpl1::hygreso1::ura4+cdc25-22 rad21-9PK-kanr |
| 3789 | h−rad21-9PK-kanr |
| 3808 | h+ura4 wpl1::kanreso1::ura4+ |
| 3820 | h+ |
| 3912 | h−ura4 wpl1::kanreso1::ura4+psm3-GFP-natr |
| 3913 | h+ura4 wpl1::kanreso1::ura4+psm3-GFP-natr |
| 3915 | h−ura4 wpl1::hygreso1::ura4+psm3-GFP-natr |
| 3916 | h−ura4 psm3K105R |
| 3918 | h−ura4 psm3K105Rwpl1::kanr |
| 3920 | h−ura4 psm3K106N |
| 3921 | h+ura4 psm3K106Nwpl1::kanr |
| 3925 | h−ura4 eso1-H17 psm3-GFP-natr |
| 3930 | h+ura4 psm3K105Nwpl1l::kanr |
| 3931 | h+ura4 psm3K106Rwpl1::kanr |
| 3932 | h+ura4 psm3K106R |
| 3940 | h+ura4 wpl1::kanreso1::ura4+psm3K106N |
| 3942 | h+ura4 eso1::ura4+psm3K106N |
| 3972 | h−ura4 psm3K105N |
| 3975 | h+ura4 psm3K105RK106R |
| 3993 | h+ura4 wpl1::kanreso1::ura4+ |
| 3995 | h+ura4 wp1l::kanr |
| 3996 | h−ura4 wpl1::kanreso1::ura4+psm3K105N |
| 3999 | h−ura4 eso1::ura4+psm3K105NK106N |
| 4000 | h−ura4 wpl1::kanreso1::ura4+psm3K105NK106N |
| 4001 | h−ura4 psm3K105NK106N |
| 4002 | h−ura4 psm3K105NK106Nwp1l::kanr |
| 4004 | h−ura4 psm3K105RK106R |
| 4005 | h−ura4 psm3K105RK106Rwpl1::kanr |
| 4027 | h−ura4 eso1::ura4+psm3K105N |
| 4177 | h+mis4-367 psm3K105RK106Rcdc25-22 rad21-9PK-kanr |
| 4179 | h−mis4-367 psm3K105RK106Rcdc25-22 rad21-9PK-kanrwpl1::hygr |
| 4187 | h+mis4-367 psm3K105NK106Ncdc25-22 rad21-9PK-kanr |
| 4190 | h+mis4-367 wpl1::hygrpsm3K105NK106Ncdc25-22 rad21-9PK-kanr |
| 4192 | h−mis4-367 cdc25-22 rad21-9PK-kanr |
| 4193 | h−mis4-367 wpl1::hygrcdc25-22 rad21-9PK-kanr |
| 4255 | h−psm3-GFP-natrcdc10-129 |
| 4394 | h90leu1-32 ura4 psm3K105RK106Rcen2-LacO his7+-LacI-GFP |
| 4398 | h90leu1-32 ura4 cen2-LacO his7+-LacI-GFP |
| 4433 | h−leu1 ura4 ade6-210 psm3K105RK106Rwpl1::kanrCh16(ade6-216) |
| 4431 | h−leu1 ura4 ade6-210 psm3K105NK106Nwpl1::kanrCh16(ade6-216) |
| 4437 | h−leu1 ura4 ade6-210 psm3K106RCh16(ade6-216) |
| 4439 | h−leu1 ura4 ade6-210 psm3K106Rwpl1::kanrCh16(ade6-216) |
| 4443 | h−leu1 ura4 ade6-210 wpl1::kanreso1::ura4+Ch16(ade6-216) |
| 4445 | h−leu1 ura4 ade6-210 Ch16(ade6-216) |
| 4447 | h−leu1 ura4 ade6-210 wpl1::kanrCh16(ade6-216) |
| 4452 | h90leu1-32 ura4 psm3K105RK106Rcen2-LacO his7+-LacI-GFPwpl1::hygr |
| 4456 | h90leu1-32 ura4 cen2-LacO his7+-LacI-GFP wpl1::hygr |
| 4457 | h90leu1-32 ura4 cen2-LacO his7+-LacI-GFP pds5::natr |
| 4466 | h−leu1 ura4 ade6-210 psm3K105Rwpl1::kanrCh16(ade6-216) |
| 4468 | h−leu1 ura4 ade6-210 psm3K105RCh16(ade6-216) |
| 4470 | h−leu1 ura4 ade6-210 psm3K105RK106RCh16(ade6-216) |
| 4478 | h−leu1 ura4 ade6-210 psm3K105NK106NCh16(ade6-216) |
| 4487 | h+ura4 mis4-367 eso1::ura4+psm3K105NK106Ncdc25-22rad21-9PK-kanr |
| 4528 | h−leu1 ura4 ade6-210 eso1::ura4+psm3K105NK106NCh16(ade6-216) |
| 4530 | h−leu1 ura4 ade6-210 eso1::ura4+psm3K105NK106Nwpl1::kanrCh16(ade6-216) |
| 4546 | h−leu1 ura4 ade6-210 wpl1::kanreso1::ura4+psm3K106NCh16(ade6-216) |
| 4552 | h−psm3K105RK106Rcut9-665 |
| 4553 | h−wpl1::kanrpsm3K105RK106Rcut9-665 |
| 4590 | h−leu1-32 ura4 wpl1::kanreso1::ura4+cut9-665 |
| 4592 | h−leu1-32 ura4 wpl1::kanrcut9-665 |
| 4594 | h−leu1-32 ura4 cut9-665 |
| 4622 | h−eso1-H17 |
| 4750 | h−ura4 mis4-367 wpl1::hygreso1::ura4+psm3K106Ncdc25-22rad21-9PK-kanr |
| 4753 | h−ura4 mis4-367 wpl1::hygreso1::ura4+psm3K105NK106Ncdc25-22rad21-9PK-kanr |
| 4785 | h−ura4 psm3K105R-GFP-natr |
| 4787 | h−ura4 psm3K106R-GFP-natr |
| 4789 | h−ura4 psm3K105RK106R-GFP-natr |
| 4826 | h−leu1 ura4 ade6-210 wpl1::kanreso1::ura4+psm3K105NCh16(ade6-216) |
| 4977 | h−ura4 mis4-367 eso1::ura4+psm3K106Ncdc25-22 rad21-9PK-kanr |
| 5234 | h+ura4 mis4-367 swi6::ura4+cdc25-22 rad21-9PK-kanr |
| 5321 | h−eso1-H17 psm3RR |
| 5323 | h−eso1-H17 psm3RRwpl1::kanr |
| 5527 | MATaSMC3-Pk3 (F. Uhlmann, Y3564) |
| 5528 | MATaeco1Δ wpl1Δ SMC3-Pk3 (F. Uhlmann, Y3237) |
Measurement of Rad21 dissociation from chromatin.
Cells bearing rad21-9PK (48) and the thermosensitive mis4-367 and cdc25-22 alleles were grown to mid-log phase in EMM2 medium at 25°C and then shifted to 36.5°C, a restrictive temperature for mis4-367 and cdc25-22. In a typical time course experiment, cells were collected at time zero and 30, 60, 90, 120, 150, and 180 min after the temperature shift. Nuclear spreading, indirect immunofluorescence, and signal quantification were done as described previously (12). All experiments included the mis4-367 cdc25-22 rad21-PK reference strain. The raw data were remarkably similar between independent experiments, so each data set was normalized relative to the reference strain as follows: the time zero value from the reference strain was set at 40 (which was the mean value of all experiments), and all the data were normalized relative to this value.
Antibodies, protein extracts, immunoprecipitation (IP), Western blotting, and chromatin immunoprecipitation (ChIP).
Anti-Psm3K106Ac antibodies were raised by Eurogentec. Rabbits were immunized with the keyhole limpet hemocyanin (KLH)-coupled peptide CRTIGLKK106AcDEYSL. Sera were immunodepleted by affinity against the nonacetylated form of the peptide, and antibodies were then affinity purified against the acetylated peptide. Anti-Psm3 antibodies were made by Proteogenics. Rabbits were immunized with a recombinant peptide corresponding to the N-terminal 630 residues, and sera were affinity purified against the recombinant peptide. Anti-PK (monoclonal mouse anti-V5 tag; AbD Serotec) and anti-Swi6 (Abcam) antibodies were used for nuclear spreads.
Total-protein extracts were prepared from cycling log-phase cells. Cells (5 × 108) were collected, rinsed in ice-cold phosphate-buffered saline (PBS)-1 mM phenylmethylsulfonyl fluoride (PMSF), and frozen on dry ice. Lysis was performed in 125 μl ice-cold lysis buffer (50 mM HEPES, pH 7.6, 100 mM KCl, 2.5 mM MgCl2, 0.25% Triton X-100, 1 mM dithiothreitol [DTT], 0.1% SDS, 10 mM sodium butyrate, 10% glycerol) with inhibitors (Sigma P8215 protease inhibitor cocktail, 1 mM PMSF, 1 mM Na vanadate, 20 mM β-glycerophosphate) using a glass bead beater (Cryoprep). The volume was increased by the addition of 275 μl of ice-cold lysis buffer with protease inhibitors, and the extracts were clarified by two successive rounds of centrifugation at a relative centrifugal force (RCF) of 18,000 at 4°C.
Psm3-green fluorescent protein (GFP) was immunopurified by adding to the lysate 3 μg of polyclonal anti-GFP antibodies (A11122; Molecular Probes) and 50 μl protein A-coated magnetic beads (Miltenyi Biotec) prewashed in lysis buffer. After 1 h on ice, immune complexes were recovered on a magnetic column, washed three times with lysis buffer, and eluted with Laemmli buffer. Budding yeast extracts were prepared using denaturing conditions as described above, and Smc3-PK was immunopurified using 3.5 μg monoclonal mouse anti-V5 tag and protein G beads. For Psm3-GFP immunoprecipitation (see Fig. 6E), a nondenaturing lysis buffer was used (50 mM HEPES, pH 7.6, 75 mM KCl, 1 mM MgCl2, 0.1% Triton X-100, 1 mM DTT, 10 mM sodium butyrate, 10% glycerol, 1 mM EGTA), and immunoprecipitation was carried out with 4 μg monoclonal anti-GFP and protein G beads.
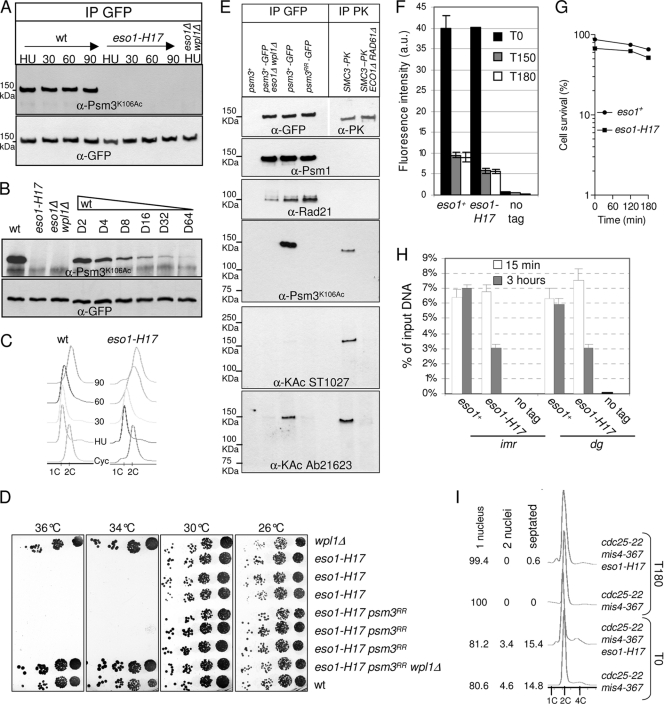
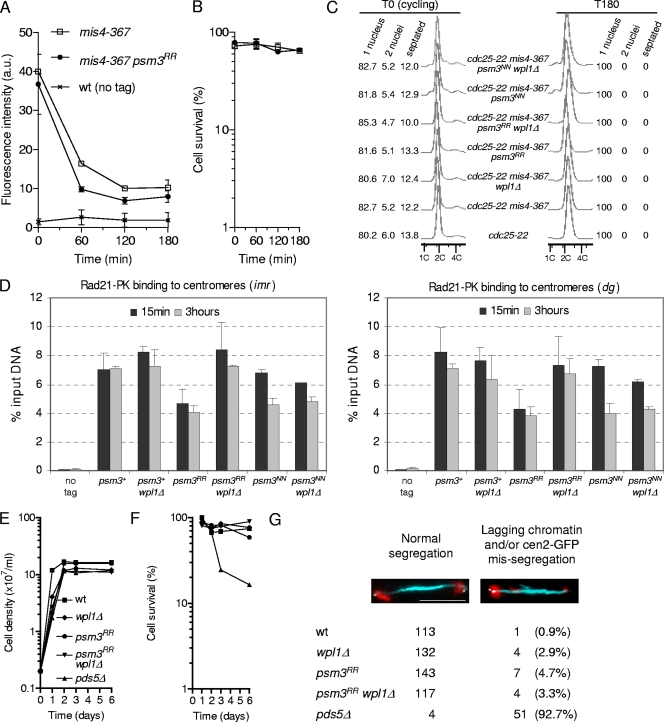
Fig. 6.
Psm3K106Ac is not detectable in the eso1-H17 mutant strain at the permissive temperature. (A) Cells were cultured at 25°C, arrested in early S phase by adding 12 mM hydroxyurea (HU), and released into the cell cycle. Samples were taken at the indicated time points (min). Psm3-GFP was immunoprecipitated from total protein extracts and probed with anti-Psm3K106Ac antibodies. Equal amounts of Psm3-GFP were verified by probing with anti-GFP antibodies. (B) Psm3-GFP immunopurified from the wt was serially diluted with Psm3-GFP immunopurified from eso1Δ wpl1Δ cells and probed with anti-Psm3K106Ac and anti-GFP antibodies as in panel A. (C) DNA content analysis before and after release from HU arrest. (D) eso1-H17 is still viable and thermosensitive for growth in a psm3RR background. Serial dilutions of cells were spotted on YES medium and incubated at the indicated temperatures. (E) Analysis of the cohesin complex with anti-acetyl antibodies. The cohesin complex was immunoprecipitated from native protein extracts by anti-GFP antibodies and probed with the indicated antibodies. Budding yeast SMC3-PK was immunopurified from the indicated strains (8) and used as a control for anti-acetyl antibodies. (F to I) The stable cohesin fraction is reduced in the eso1-H17 mutant at the permissive temperature. The strains contain the cdc25-22 and mis4-367 alleles and were cultured and processed for nuclear spreading and ChIP as in Fig. 1. (F) Chromatin-bound Rad21-PK was quantified from nuclear spreads before (T0) and after (T150 and T180 min) the temperature shift. The error bars represent SD from duplicate samples. (G) Cell survival during the course of the experiment. (H) ChIP assay showing the amount of Rad21 bound to pericentromeric regions (imr and dg) just after (15 min) and 3 h after the temperature shift. Rad21 enrichment was calculated from duplicate samples. The error bars represent SD. (I) Fluorescence-activated cell sorter (FACS) and cell cycle staging of cells before and after the shift to 36.5°C.
Western blotting was done according to standard procedures using rabbit anti-Psm3K106AC (1/1,000), monoclonal anti-GFP (Roche 1/1,000), rabbit anti-Psm3 (1/5,000), rabbit anti-Rad21 (1/2,000) (17), and anti-acetyl antibodies (Calbiochem ST1027 [1/2,000] and Ab21623 [1/2,000]).
Chromatin immunoprecipitation was done essentially as described previously (12) with the following modifications. Cells were fixed with formaldehyde (2.5% final concentration) for 10 min at 36.5°C and 15 min at room temperature, after which samples were cooled on ice for 5 min. The cells were washed four times with ice-cold PBS. The cell pellets were frozen on dry ice and stored at −80°C. Chromatin was prepared from 2 × 108 cells and fragmented using a Bioruptor sonicator (Diagenode). Immunoprecipitation was carried out in IP buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.1) using 3.5 μg anti-PK antibodies (monoclonal mouse anti-V5 tag; AbD Serotec). Immune complexes were collected using ChIP-Adembeads (Ademtech; 04240), washed, and recovered according to the manufacturer's instructions. Quantification was performed by real-time PCR in the presence of SYBR green using a Stratagene Mx3000P cycler. PCR primers were as follows: imr, TTTTGGACAGAATGGATGGA and GCGGAGTAAGGCTAATCACG; dg1, ACGGCATCGCTTGTACTTTT and TGAGGTTCATGATGGGTTCA; CAR1806, AGCAAAAGCACCGACTTCAT and TTCAAAGCTGCTCTCCCATT; CAR1979, GGCAGGCATGTCTTGGTTAT and TTCCTGCTTACCACGGTTTC; NTS1, TCCACCTTCCCATAACATGC and AGCTGAAGCCAGAGTGC; NTS2, TCTCTCATTTTCCATTGAACCA and TTCAGGGTCGGTAGAGTCAGA; NTS3, CTCTCAATCCATCCATCAAAC and CCAAATGTGGAAAATGGACA; 28S-1, ACGGACCAAGGAGTCTAGCA and GTTCCCACCTGCATTCACTT; 28S-2, TCCGTATGAAAGTTGCACGA and CCGTGTTGATTCCACCTTCT; 28S-3, CGAAGCAGAATTCGGTAAGC and AACCTGTCTCACGACGGTCT; TEL, ATGCTTTGGCCACTGTTCCT and TCAAAACCGCAAAAACGATG.
RESULTS
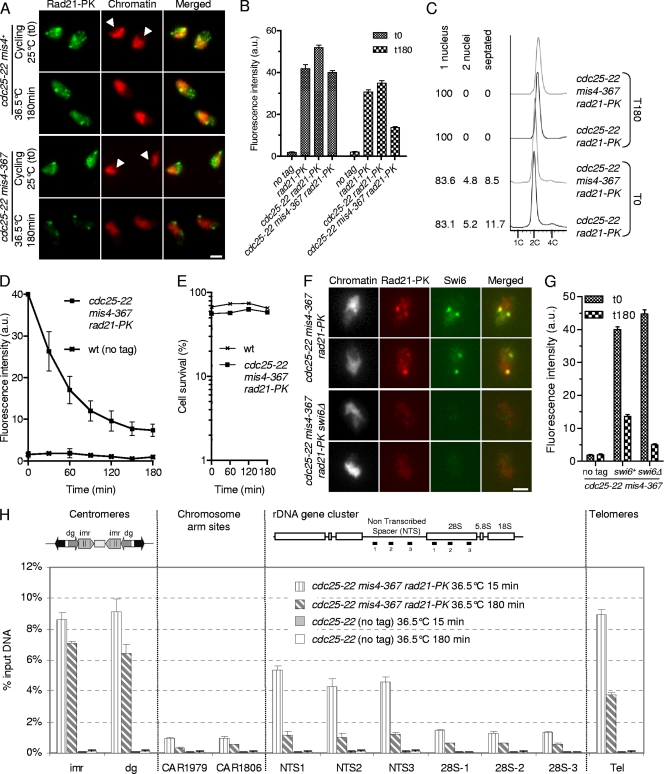
Cohesin bound in the stable mode is not evenly distributed along postreplicative chromosomes.
In a previous study, we showed that when the cohesin-loading machinery is inactivated in G1-arrested cells, cohesin that was bound to chromosomes dissociated to completion in a reaction stimulated by Wpl1. In contrast, when the experiment was done using G2 cells, a fraction of cohesin remained bound to chromosomes (12). Here, we address the kinetics of cohesin removal from chromatin and the pattern of stable cohesin binding along chromosomes. A typical experiment is shown in Fig. 1. Cycling cells that are mainly in the G2 phase of the cell cycle (Fig. 1C) (14) were shifted to the restrictive temperature for cdc25-22 and mis4-367 to keep the cells in G2 and to prevent further cohesin loading. Cohesin binding to chromatin was monitored by nuclear spreads and immunofluorescence against the cohesin subunit Rad21 (Fig. 1A and B). In wild-type as in cdc25-22 mis4+ strains, the steady-state amount of chromatin-bound Rad21 was slightly reduced when cells were cultured at 36.5°C. In mis4-367 cells, however, inactivation of the cohesin loader led to a further decrease of chromatin-bound Rad21 (Fig. 1B). The kinetics of Rad21 dissociation from G2 chromatin upon inactivation of Mis4 is shown in Fig. 1D. The amount of chromatin-bound Rad21 decreased with time to reach a plateau after about 150 min. Two populations of cohesin can therefore be distinguished; a labile fraction and a stable fraction, representing ∼80% and ∼20% of total chromatin-bound Rad21, respectively. Cell survival remained high throughout the experiment (Fig. 1E), indicating that cohesin complexes that were chromatin bound at late time points (i.e., the stable fraction) provided functional cohesion.
Fig. 1.
Characterization of the cohesin fraction stably bound to postreplicative chromosomes. (A to D) A fraction of Rad21 persists on G2 chromatin after inactivation of the cohesin-loading machinery. Cycling cells were shifted to 36.5°C to shut off cohesin loading (mis4-367) while keeping the cells in G2 (cdc25-22). Chromatin-bound Rad21-9PK was measured at the indicated time points by nuclear spreading and immunofluorescence using anti-PK antibodies. (A) Images of nuclear spreads showing chromatin-bound Rad21-9PK before (cycling) and 180 min after the temperature shift. Chromatin was counterstained with DAPI (4′,6-diamidino-2-phenylindole). The arrowheads point to the nucleoli, which are located in the area of the nucleus unstained by DAPI. Bar, 2.5 μm. (B) Rad21-9PK fluorescence intensity was measured for 50 to 100 nuclei for each sample. Error bars, 95% confidence intervals of the mean. a.u., arbitrary units. (C) Analysis of the cell cycle stage confirmed that most cells (>80%) were in the G2 phase before the temperature shift and remained in G2 throughout the experiment. The positions of cells within the cell cycle were analyzed by DNA content analysis. DAPI and calcofluor staining were used to score mononucleate, binucleate, and septated cells. (D) Kinetics of Rad21 dissociation from chromatin. The data presented are the means of 10 independent experiments. The error bars represent standard deviations (SD). wt, wild type. (E) Cell survival remained high during the course of the experiment. Cells were withdrawn at the indicated time points and plated at the permissive temperature to determine the number of viable cells. (F) The major foci of cohesin in the stable fraction colocalize with Swi6. Cells were treated as in panel A, and nuclear spreads were prepared 180 min after the temperature shift, at which time only the stable cohesin fraction is retained on chromatin. Bar, 2.5 μm. (G) The stable cohesin fraction is reduced in cells lacking Swi6. The Rad21-9PK fluorescence intensity was measured for 50 to 100 nuclei for each sample. Error bars, 95% confidence intervals of the mean. (H) Rad21 ChIP assay showing the distribution of the labile and stable cohesin fractions along the chromosomes. Cells were cultured as in panel A and processed for ChIP just after (15 min) and 180 min after the temperature shift. Rad21 enrichment was measured at centromeres (imr and dg), at two chromosome arm sites (CAR1806 and CAR1979), within the rDNA gene cluster (NTS and 28S), and at telomeres (Tel). Rad21 enrichment was calculated from duplicate samples. The error bars indicate SD.
Examination of Rad21 fluorescence on nuclear spreads before and 3 h after inactivation of the cohesin loader suggested that Rad21 may persist preferentially at some genomic locations (Fig. 1A). Indeed, the major foci of punctate Rad21 staining colocalized with the heterochromatin protein Swi6 (Fig. 1F), the Schizosaccharomyces pombe HP1 homolog. We conclude that Swi6 domains are major sites where cohesin is bound to chromatin in the stable mode in postreplicative cells. Swi6 is required for cohesin enrichment within heterochromatin domains (11, 42). Consistently, the major foci of Rad21 staining were absent from nuclear spreads made from swi6Δ cells (Fig. 1F) and the stable cohesin fraction was reduced more than 2-fold (∼60%) (Fig. 1G).
To further characterize the pattern of stable cohesin binding along chromosomes, we used a chromatin immunoprecipitation assay (Fig. 1H). Most (∼70 to 80%) Rad21 remained bound to pericentromeric heterochromatin 3 h after inactivation of the cohesin loader. Both telomeres and centromeres are bound by Swi6 and display features of heterochromatin (18). A substantial amount of Rad21 (∼40%) also persisted at telomeres, but unlike pericentromeric domains, a large fraction dissociated with time. Heterochromatin might contribute to mark chromosomal domains as sites of cohesin stabilization, but other determinants may exist at centromeres to account for the strong stability of Rad21 binding at these sites.
Another major site of cohesin enrichment is the rDNA, located within the nucleolus. The rDNA locus is made up of ∼100 tandem repeats covering the left and right arms of chromosome III. Examination of nuclear spreads before inactivation of the loading machinery showed a large amount of Rad21 within the nucleolus, but this signal was strongly reduced 3 h after inactivation of Mis4 (Fig. 1A, arrowheads). By ChIP, Rad21 was mostly enriched within the rDNA nontranscribed spacer (NTS) but was reduced to ∼25% 3 h after inactivation of Mis4 (Fig. 1H). Although the rDNA is a major site of cohesin enrichment, cohesin appears mostly bound in the labile mode.
We also assayed the stability of Rad21 binding in two cohesin-associated regions (CARs) on chromosome 2. The amount of Rad21 bound at chromosome arm sites is much lower than at centromeres, telomeres, and rDNA. These two CARs were chosen because they were among CARs with the largest amounts of Rad21 based on published genomewide data (12, 33). Three hours after the temperature shift, the amounts of Rad21 bound at CAR1806 and CAR1979 were reduced to ∼56% and ∼30%, respectively.
From these experiments, we conclude that a fraction of cohesin is stably bound to postreplicative chromosomes. Cohesin does not become stabilized evenly along the chromosomes. Swi6/heterochromatin domains account for ∼60% of the stable cohesin fraction, and pericentromeric heterochromatin is a site where cohesin is bound mostly in the stable mode.
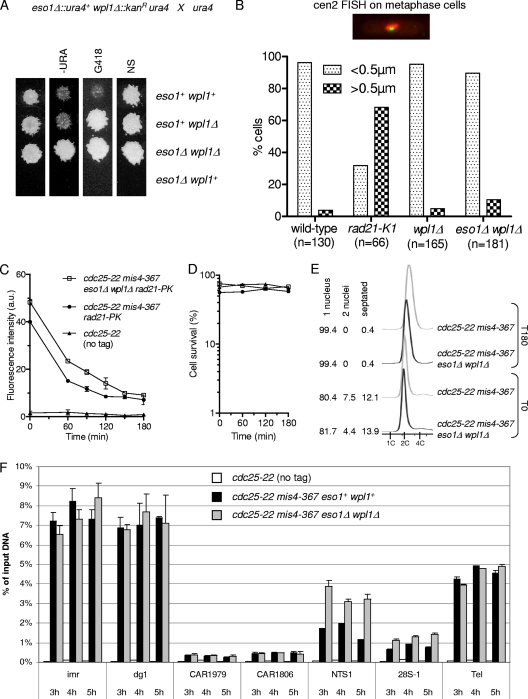
Eso1 is dispensable for stable cohesin binding to postreplicative chromatin and sister chromatid cohesion when the wpl1 gene is deleted.
In a previous study, we reported that wpl1 stimulates cohesin removal from G1 chromosomes and that cohesin stabilization after S phase was affected when Eso1 function was compromised. This suggests that Eso1 may promote stable cohesin association with chromosomes by counteracting Wpl1. There is indeed accumulating evidence in mammals that Wapl and Esco1-2 display opposing activities. Similarly, in budding yeast, the ECO1 function can be partially bypassed by the deletion of RAD61. The genetic interaction between eso1 and wpl1, however, was not investigated in fission yeast.
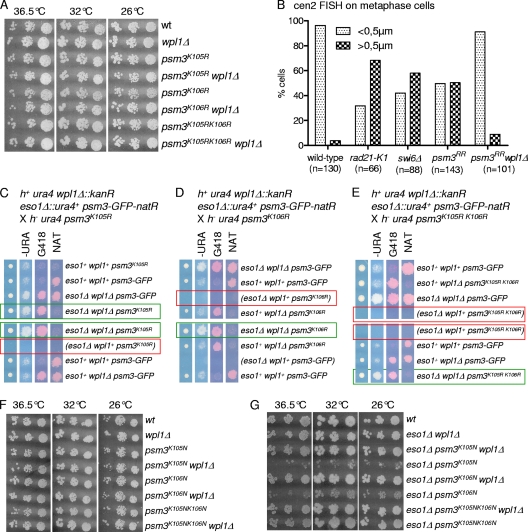
As shown in Fig. 2A, a strain with both eso1 and wpl1 deleted was viable, whereas the deletion of eso1 alone is lethal (54). The eso1Δ wpl1Δ strain formed wild-type-size colonies, suggesting a high level of suppression. To investigate this point, we used an assay that allows the detection of infrequent chromosome loss events (Table 2). The nonessential linear minichromosome Ch16 bears a functional centromere and is lost at a frequency of 0.05 to 0.1% of cell divisions in a wild-type background (1, 41). The deletion of wpl1 had only a modest impact on the Ch16 loss rate. Importantly, the loss rate did not increase in eso1Δ wpl1Δ cells, showing that loss of Eso1 function has no detrimental effect on chromosome segregation when Wpl1 is absent. To see whether subtle cohesion defects might occur, sister chromatid cohesion was assessed by FISH using a probe mapping close to the centromere of chromosome 2. To increase the sensitivity of the assay, cells were arrested at metaphase, when cohesion is challenged by spindle forces exerted on sister centromeres. As shown in Fig. 2B, sister centromere separation remained low in wpl1Δ and eso1Δ wpl1Δ cells, close to the wild-type control. We conclude that Eso1 function is dispensable for sister chromatid cohesion in the absence of Wpl1. This implies that the main, if not the sole, function of Eso1 in cohesion establishment is counteracting Wpl1.
Fig. 2.
Eso1 function is dispensable when the wpl1 gene is deleted. (A) Representative example of tetrad analysis showing that eso1Δ spores do not support colony formation, whereas eso1Δ wpl1Δ spores form wild-type-size colonies. (Left) Tetrads were dissected, and spores were deposited on complete medium. Colonies were replica plated onto appropriate media to follow the segregation of eso1 and wpl1 alleles. −URA, minimum medium lacking uracil; G418, YES medium plus G418; NS, nonselective YES medium. (B) cen2 FISH on metaphase-arrested cells. Cells were arrested at metaphase by the use of the thermosensitive cut9ts mutation (anaphase-promoting complex). Fixed cells were stained for tubulin and processed for FISH using a probe mapping close to the centromere of chromosome 2 (cen2 FISH). The image shows an example of a typical metaphase cell. The mitotic spindle (red) is <2.5 μm in length, and the two cen2 FISH signals (green) are closely apposed to each other. No major defect in sister centromere cohesion was observed in wpl1Δ and wpl1Δ eso1Δ cells as opposed to the thermosensitive rad21-K1 mutant. (C to E) The stable cohesin fraction is created and is functional in eso1Δ wpl1Δ cells. Strains were cultured and processed for nuclear spreading and ChIP as in Fig. 1. (C) Kinetics of Rad21 dissociation from chromosomes. Chromatin-bound Rad21-PK was quantified from nuclear spreads. The error bars represent SD from two independent experiments. (D) Cell survival remained high throughout the duration of the experiment. (E) Cell cycle staging shows that eso1Δ wpl1Δ cells were mostly (>80%) in the G2 phase before the temperature shift and remained arrested in G2 afterward. (F) ChIP assay comparing Rad21 binding to chromosomes in eso1Δ wpl1Δ versus eso1+ wpl1+ cells from 3 h to 5 h after cohesin loading was shut off. The error bars show the SD from two quantitative PCR (qPCR) quantifications.
Table 2.
Ch16 minichromosome loss rates
| Genetic background | Loss rate | % division |
|---|---|---|
| Wta | 12/9,947 | 0.12 |
| wpl1Δ | 14/4,931 | 0.28 |
| eso1Δ wpl1Δ | 16/5,324 | 0.30 |
| eso1Δ psm3K105N | NDb | ∼100 |
| eso1Δ psm3K105Nwpl1Δ | 0/2,931 | <0.03 |
| eso1Δ psm3K106N | ND | ∼100 |
| eso1Δ psm3K106Nwpl1Δ | 5/6,054 | 0.08 |
| eso1Δ psm3K105NK106N | 50/2,474 | 2.02c |
| eso1Δ psm3K105NK106Nwpl1Δ | 4/5,694 | 0.07 |
| psm3K105NK106N | 25/7,327 | 0.34 |
| psm3K105NK106Nwpl1Δ | 17/6,370 | 0.27 |
| psm3K105R | 14/5,818 | 0.24 |
| psm3K105Rwpl1Δ | 19/6,250 | 0.30 |
| psm3K106R | 55/7,772 | 0.71c |
| psm3K106Rwpl1Δ | 5/6,911 | 0.07 |
| psm3K105RK106R | 66/8,371 | 0.79c |
| psm3K105RK106Rwpl1Δ | 10/6,392 | 0.16 |
Wt, wild type.
ND, not determined, since Ch16 could not be maintained in this genetic background. The loss rate is therefore estimated as 100%.
Statistically significant compared to wt (P value < 0.0001 by the chi-square test).
Wpl1 stimulates cohesin removal, and stable cohesin binding to postreplicative chromosomes is affected when Eso1 function is compromised (12). If indeed Eso1 acts solely by counteracting Wpl1, the stable cohesin fraction should be formed in eso1Δ wpl1Δ as in the wild type. To address this question, cycling cells bearing mis4-367 and cdc25-22 were shifted from 25°C to 36.5°C, and Rad21 binding to chromatin was monitored by nuclear spreads. As shown in Fig. 2C, the initial amount of chromatin-bound Rad21 was larger in eso1Δ wpl1Δ cells than in the wild type, but at late time points, the amounts of Rad21 that remained bound to chromosomes appeared similar. This suggests that the labile fraction of cohesin is increased in size in eso1Δ wpl1Δ cells but the stable fraction may remain unchanged. Cell survival remained high in the two strains (Fig. 2D), indicating that cohesin that remained chromosome bound provided functional cohesion.
To confirm that the stable cohesin fraction was indeed present, we used a ChIP assay to assess the amounts and stabilities of chromatin-bound Rad21 at several loci when the G2 arrest was further extended from 3 to 5 h. As shown in Fig. 2F, the amount of chromatin-bound Rad21 did not decrease further at all loci examined, consistent with a stable mode of cohesin binding. The amount of Rad21 bound in the stable mode was similar to that observed in eso1+ wpl1+ cells at all loci examined, except for the rDNA locus, where it was markedly increased when both eso1 and wpl1 were deleted.
From these experiments, we conclude that the stable cohesin fraction is created and is functional in the absence of both Eso1 and Wpl1. The fundamental processes by which cohesin becomes stabilized and the creation of cohesion remain intact and therefore are not dependent on the Eso1-Wpl1 module. However, Eso1 is essential in wpl1+ cells, indicating that Wpl1 must be counteracted by Eso1. We have shown previously that the stable cohesin fraction was altered in the eso1-H17 mutant (12) (see Fig. 6F and H). This suggests that one functional consequence of counteracting Wpl1 is the sheltering of cohesin from its removal activity.
Psm3 acetylation is not essential but contributes to Eso1 function.
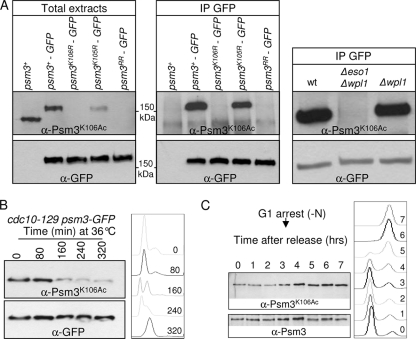
Eso1 may antagonize Wpl1 through the acetylation of specific substrates. In budding yeast and mammals, Smc3 is acetylated by Eco1/Esco1 (8, 45, 59, 63) on two adjacent conserved lysine residues that correspond to K105 and K106 in fission yeast Psm3. To see whether Eso1 may act through the acetylation of Psm3, we first asked whether Psm3 was actually acetylated in an Eso1-dependent manner in fission yeast and whether Psm3 acetylation would be detected at S phase, when Eso1 exerts its essential function. As a readout of Eso1 activity, we raised and made use of an antibody that efficiently recognizes Psm3K106Ac in total-protein extracts. As shown in Fig. 3 A, the antibody reacts with Psm3, but not when K106 is replaced by the nonacetylatable residue arginine. Acetylated Psm3 is detected in an Eso1-dependent manner but independently of Wpl1. During the cell cycle, the amount of the acetylated form was smallest in G1, rose at S phase, and remained constant throughout G2 (Fig. 3B and C), similar to previous observations made in budding yeast (8, 59).
Fig. 3.
Eso1-dependent Psm3K106 acetylation during the cell cycle. (A) The anti-Psm3K106Ac antibody reacts with Psm3 in a K106-dependent manner in total extracts (left) and GFP immunoprecipitates (middle). (Right) Psm3 detection by anti-Psm3K106Ac antibodies is dependent on Eso1, but not on Wpl1. (B and C) Psm3K106 acetylation during the cell cycle. (B) Cells bearing the thermosensitive mutation cdc10-129 were shifted to the restrictive temperature to induce G1 arrest. Samples were taken at regular time intervals after the temperature shift to monitor Psm3K106Ac on total protein extracts as the cells progressed from an asynchronous population (time zero) to a homogeneous G1 arrest. Flow cytometry analysis of the DNA content (right) showed that Psm3K106Ac started to decrease as the G1 peak appeared. The drift in the G1 peak to the right at later time points was due to an increase in the mitochondrial DNA content as the cells elongated (47). (C) Cells were arrested in G1 by nitrogen deprivation and released synchronously into the cell cycle. Psm3K106Ac probed from total protein extracts was weakest in cells with a G1 DNA content and began to rise at the time of DNA replication. Anti-Psm3 antibodies were used to adjust protein loading so that similar amounts of Psm3 were present in all samples.
To see whether Eso1 acts through the acetylation of Psm3, mutant alleles were created in which K105, K106, or both lysine residues were replaced by the nonacetylatable residue arginine. The mutant alleles were inserted in the genome at the psm3 locus, replacing the wild-type allele. Surprisingly, all mutant strains were viable and displayed a wild-type growth rate, even when both lysine residues were mutated (Fig. 4 A). Genetic analyses (Fig. 4C to E) indicated that the progeny bearing a nonacetylatable mutant allele are viable in an eso1+ background, but not when the eso1 gene is deleted. This demonstrates that the viability of nonacetylatable mutants relies on Eso1, indicating that Eso1 exerts a vital function even though the two conserved Psm3 residues cannot be acetylated. We conclude that Eso1 function cannot transit exclusively through the acetylation of these lysine residues.
Fig. 4.
Nonacetylatable mutants are viable but generate a cohesion defect. (A) Nonacetylatable psm3 mutants display a wild-type growth rate. Serial dilutions of cells were spotted on YES medium and incubated at the indicated temperatures. (B) cen2 FISH on metaphase-arrested cells revealed a wpl1-dependent defect in sister centromere cohesion in the psm3RR mutant. The cells were arrested at metaphase by the use of the thermosensitive cut9ts mutation (anaphase-promoting complex). (C to E) Tetrad analysis showing the segregation of psm3 nonacetylatable alleles. (Left) Tetrads were dissected, and spores were deposited on complete medium. Colonies were replica plated onto appropriate media to follow the segregation of the markers. −URA, miminum medium lacking uracil; G418, YES medium containing G418; NAT, YES medium containing nourseothricin. YES plates also contained the vital stain phloxin B as an indicator of cell lethality within the colony. Representative tetrads are shown. The green boxes show that the nonacetylatable psm3 alleles are neutral in an eso1Δ wpl1Δ background. The red boxes show that eso1Δ combined with a nonacetylatable psm3 allele does not form viable progeny. (F and G) Serial dilutions of cells were spotted on YES medium and incubated at the indicated temperatures. (F) Acetyl-mimicking psm3 mutants are viable and display a wild-type growth rate. (G) Acetyl-mimicking psm3 alleles can bypass the eso1 requirement. The comparison of colony growth rates shows the additive effect of the amino acid substitutions.
Although viable, psm3K106R and psm3K105RK106R (here called psm3RR) mutants showed an elevated rate of minichromosome loss (Table 2), and FISH analysis of metaphase-arrested cells revealed an elevated frequency of premature sister chromatid separation in the nonacetylatable mutant (Fig. 4B). The severity of the cohesion defect was less than in cells bearing the thermosensitive rad21-K1 mutation (55), which display cohesion defects along the entire length of chromosomes, or in cells lacking Swi6, which lose cohesion at centromeres (11). Remarkably, all the above-mentioned phenotypes were fully suppressed by the deletion of the wpl1 gene (Table 2 and Fig. 4B), demonstrating a strict wpl1-dependent phenotype. This clearly rules out any nonspecific compromised cohesin function due to the amino acid substitutions within Psm3. We conclude that Wpl1 generates a cohesion defect when Psm3 cannot be acetylated. The corollary is that Psm3 acetylation contributes to antagonizing Wpl1.
To further confirm this conclusion, we asked whether acetyl-mimicking forms of Psm3 would bypass the Eso1 requirement. Mutant psm3 alleles were created in which K105, K106, or both lysine residues were replaced by the acetyl-mimicking residue asparagine and inserted in the genome at the psm3 locus, replacing the wild-type allele. In an otherwise wild-type background, all mutants were viable and did not show any apparent growth defect (Fig. 4F). Both psm3K105N and psm3K106N were able to rescue the deletion of eso1, albeit poorly (Fig. 4G). In contrast, the double-substitution allele psm3K105NK106N (here called psm3NN) had a stronger suppressor effect, as judged by nearly wild-type-size colonies, indicating that the two mutations had cumulative effects. Consistently, the Ch16 minichromosome could not be maintained in eso1Δ psm3K105N and eso1Δ psm3K106N cells but could be propagated in an eso1Δ psm3NN background (Table 2). The suppression was still incomplete, since the Ch16 loss rate was increased ∼17-fold compared to the wild type. Remarkably, the deletion of wpl1 in this genetic context reduced the Ch16 loss rate to the wild-type level, indicating that residual defects in eso1Δ psm3NN are strictly wpl1 dependent. This further confirms that Psm3 acetylation contributes to antagonizing Wpl1 and is consistent with the conclusion that only a fraction of Eso1 function is exerted through the acetylation of Psm3 at these specific residues.
Preventing Psm3 acetylation reduces the amount of cohesin bound in the stable mode.
The nonacetylatable psm3 mutant is viable, whereas eso1Δ is lethal, strongly suggesting that Eso1 function does not transit exclusively through Psm3 acetylation. However, psm3RR showed an elevated rate of chromosome loss, and a cohesion defect was detectable in metaphase cells, all of which were suppressed by the deletion of wpl1, showing that Psm3 acetylation contributes to antagonizing Wpl1. We therefore asked whether Psm3 acetylation would contribute to the creation of the stable cohesin fraction. Figure 5 A shows the kinetics of Rad21 dissociation from G2 chromosomes in a psm3RR mutant background. For all time points, the amount of chromatin-bound Rad21 was reduced compared to the psm3+ control, but a plateau was still observed for late time points. Consistently, cell viability remained high throughout the experiment (Fig. 5B), indicating that cohesin that remained bound to chromosomes at late time points provided functional cohesion, even though the stable cohesin fraction was quantitatively reduced. This point was further investigated by looking at Rad21 binding within pericentromeric domains using chromatin immunoprecipitation (Fig. 5D). Indeed, the amount of Rad21 associated with pericentromeric heterochromatin was reduced in psm3RR cells compared to the wild type. The reduction was already apparent for the early time point (15 min) and was not exacerbated by the 3-h time of G2 arrest. This shows that nonacetylated Psm3 generates a defect that lowers the amount of the stable cohesin fraction, but not its stability. Remarkably, this phenotype is strictly dependent on the presence of Wpl1, since deletion of wpl1 restored the amount of stably bound Rad21 to wild-type levels (Fig. 5D). The acetyl-mimicking allele psm3NN was included in the experiment. Most Rad21 remained bound to the pericentromere in this genetic background. Unlike cohesin with Psm3RR, the amount and stability of bound Rad21 were marginally affected by the presence of Wpl1 in the psm3NN mutant, suggesting that Psm3 acetylation renders cohesin less sensitive to Wpl1.
Fig. 5.
Effects of nonacetylatable Psm3 on stable cohesin binding to chromosomes and long-term cohesion. (A to D) Strains were cultured and processed for nuclear spreading and ChIP as in Fig. 1. (A) Kinetics of Rad21 dissociation from chromatin. Chromatin-bound Rad21-PK was quantified from nuclear spreads. The error bars represent SD from two independent experiments. (B) Cell survival during the course of the experiment. (C) Cell cycle staging of cells used for the ChIP assay before and after the temperature shift. (D) ChIP assay showing the amount of Rad21 bound to pericentromeric regions (imr and dg) just after (15 min) and 3 h after the temperature shift. Rad21 enrichment was calculated from duplicate samples. The error bars represent SD. (E to G) Nonacetylatable Psm3 does not affect long-term cohesion. (E) Cells were grown to saturation in EMM2-YE medium to arrest them in G2. (F) Samples were taken at the indicated time points to determine cell survival. (G) After 6 days, the cells were transferred into fresh medium, and chromosome segregation was observed during the first division after reentry into the cell cycle. Anaphase cells (spindle length, >5 μm) were scored for lagging chromatin (red) and/or cen2-GFP missegregation (white). Bar = 5 μm.
To further investigate whether the nonacetylatable psm3 mutant was proficient for sister chromatid cohesion, we asked whether cohesion would be maintained during an extended period of G2 arrest. To determine this, cells were grown to saturation in EMM2-YE medium, which causes cells to arrest in the G2 phase of the cell cycle (53). Long-term cohesion between sister chromatids was assessed by measuring cell survival and by the observation of cells undergoing their first mitosis upon reentry into the cell cycle. The viability of cells expressing nonacetylatable Psm3 remained high upon reentry into the cell cycle (Fig. 5F), and aberrant anaphase cells were scarce (Fig. 5G). In contrast, the deletion of pds5, which is required for long-term cohesion, caused massive chromosome segregation defects and a sharp decline in cell survival, as described previously (54).
During the course of this study, we discovered that the eso1-H17 thermosensitive mutant was strongly deficient for Psm3 acetylation, even at the permissive temperature. When eso1-H17 cells were synchronized in early S phase at 25°C and then allowed to progress through S phase and G2 at 25°C, purified Psm3-GFP did not react with the anti-Psm3K106Ac antibody (Fig. 6 A). To estimate the detection level of the antibody, Psm3-GFP purified from wild-type cells was serially diluted with nonacetylated Psm3-GFP purified from eso1Δ wpl1Δ cells. As shown in Fig. 6B, the anti-Psm3K106Ac antibody still reacted with wild-type Psm3 when diluted 64-fold. We conclude that Psm3 acetylation is not detectable in the eso1-H17 mutant even at the permissive temperature, with a reduction in acetylated Psm3 of a least 64 times the level found in wild-type cells. Consistent with a lack of Psm3 acetylation in eso1-H17, eso1-H17 psm3RR strains behaved like eso1-H17 psm3+ in a spotting assay (Fig. 6D). The double-mutant strain is viable and still temperature sensitive, indicating that the Eso1-H17 protein exerts another function that does not transit through the acetylation of Psm3K105K106. Although deficient for Psm3K106 acetylation, Eso1-H17 may still be able to acetylate other substrates. Alternatively, the other function may not involve its acetyltransferase activity.
In an attempt to see whether Eso1 may acetylate other cohesin subunits in vivo, the cohesin complex was immunoprecipitated from cycling cells and probed with anti-acetyl-lysine antibodies. The data shown in Fig. 6E show that one antibody (Ab21623) clearly detected Psm3 in an Eso1- and Psm3 K105 K106-dependent manner, but no signal was detected from coimmunoprecipitated Psm1 and Rad21. At this point, therefore, we do not have any evidence that Eso1 has other substrates that may account for its presumed second function.
Next, we asked whether the stable cohesin fraction was affected in the eso1-H17 mutant. cdc25-22 mis4-367 eso1-H17 cells were cultured at 25°C. These are mainly in the G2 phase of the cell cycle (Fig. 6I), so DNA replication and cohesion establishment were made at the permissive temperature for eso1-H17. The cells were then shifted to 36.5°C to inactivate Cdc25 and Mis4 in order to keep the cells in the G2 phase and to probe the stability of chromatin-bound cohesin. The amount of Rad21 bound to chromatin was monitored by nuclear spreads and ChIP at pericentromeric domains. Fig. 6F and H show that the stable cohesin fraction is created in eso1-H17 cells but the amount is reduced ∼2-fold. Since Psm3 acetylation is reduced at least 64-fold, the amount of cohesin bound in the stable mode is not quantitatively correlated with the level of Psm3 acetylation.
Altogether, the data gathered with the nonacetylatable psm3 mutant and wild-type psm3 in an eso1-H17 background converge on the conclusion that the acetylation of Psm3 contributes to, but is not essential for, stable cohesin binding to postreplicative chromosomes.
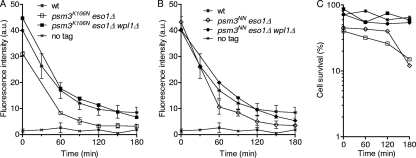
Acetyl-mimicking forms of Psm3 cannot bypass Eso1 function to generate the stable cohesin fraction.
Acetyl-mimicking alleles of psm3 allow cell survival in the absence of Eso1, suggesting that cohesin binding to postreplicative chromatin must be stabilized to some extent. To address this question, we looked at cohesin-binding stability in a genetic context in which eso1Δ cells are kept alive by the sole presence of an acetyl-mimicking form, Psm3NN or Psm3K106N (the experiment was not attempted for Psm3K105N because the eso1Δ psm3K105N strain grew very poorly). As before, cycling cells were shifted to 36.5°C to inactivate Mis4 in order to prevent further cohesin deposition, and the cells were prevented from entering mitosis by the cdc25-22 mutation. As shown in Fig. 7, chromatin-bound Rad21, as well as cell viability, dropped steadily with time. It is notable that this phenotype was more pronounced in psm3K106N than in psm3NNcells, correlating with their efficiency of eso1Δ suppression. This indicates that Psm3NN does provide some level of cohesin stabilization. This might be sufficient to allow cell survival when cells are actively cycling, but not upon prolonged G2 arrest. As expected, deletion of wpl1 in this genetic background improved both sustained Rad21 binding to chromatin and cell survival. This is consistent with the notion that Psm3 acetylation contributes, but is not the sole molecular event required, to generate the stable mode of cohesin interaction with chromosomes.
Fig. 7.
Acetyl-mimicking forms of Psm3 by themselves are not sufficient for sustained Rad21 binding to chromatin. All strains contain the cdc25-22 and mis4-367 mutations and were cultured and processed as in Fig. 1. (A and B) Kinetics of Rad21 dissociation from chromatin. Chromatin-bound Rad21 was measured from nuclear spreads. The error bars represent SD from at least two independent experiments, except for the wpl1Δ controls, which were done once. The data are presented in two separate graphs for clarity. (C) Cell survival was determined at the indicated time points.
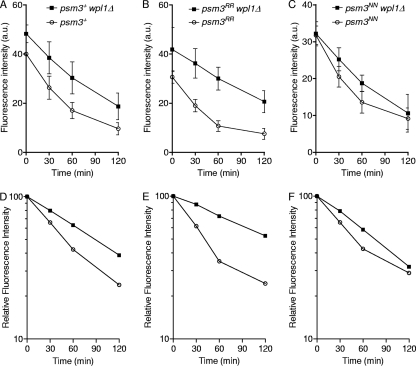
Psm3 acetylation renders cohesin less sensitive to Wpl1-dependent removal.
An acetyl-mimicking form of Psm3 can partially bypass the Eso1 requirement, suggesting that Psm3 acetylation contributes to antagonizing Wpl1. To determine whether the status of Psm3 acetylation would modify cohesin susceptibility to Wpl1, we compared the kinetics of Rad21 decay in cells bearing psm3RR or psm3NN in the presence or absence of Wpl1. As before, the amount of chromatin-bound Rad21 was measured in postreplicative (G2) cells after inactivation of the cohesin loader. We focused on the early time points of the experiment to assess the kinetics by which the labile cohesin fraction is removed from chromosomes (Fig. 8). In cells expressing psm3+ (Fig. 8D), the steady-state amount of chromatin-bound Rad21 at time zero was increased by the deletion of wpl1, and the rate of Rad21 dissociation from chromatin was slightly lower, consistent with Wpl1 promoting cohesin removal. In cells expressing the nonacetylatable allele psm3RR, the kinetics of Rad21 dissociation was more sensitive to Wpl1 (Fig. 8E). In contrast, in cells expressing the acetyl-mimicking mutant form of Psm3, the deletion of wpl1 had a much reduced effect (Fig. 8F).
Fig. 8.
The acetylation status of Psm3 modulates Wpl1-dependent cohesin removal from chromatin. All strains contain the cdc25-22 and mis4-367 mutations and were cultured and processed as in Fig. 1. (A to C) Chromatin-bound Rad21 was measured from nuclear spreads. The error bars represent SD from 3 to 5 independent experiments. (D to F) Kinetics of Rad21 dissociation from chromatin. Each data set (A to C) was normalized respective to its time zero value (arbitrarily set at 100).
The acetylation status of Psm3 may therefore contribute to regulating the labile cohesin fraction by modulating cohesin susceptibility to Wpl1. Nonacetylated Psm3 renders cohesin more sensitive to Wpl1. Conversely, the acetylated form renders cohesin less sensitive to Wpl1.
Altogether, we propose that Eso1-mediated acetylation of Psm3 is a molecular event that contributes to antagonizing Wpl1. This alone can improve the stability of cohesin association with chromatin but does not provide a full level of cohesin stabilization. We suggest that long-term cohesion between sister chromatids requires another Eso1-dependent molecular event to achieve the stable mode of cohesin binding to postreplicative chromosomes.
DISCUSSION
Different modes of cohesin binding along postreplicative chromosomes.
Live imaging in mammalian cells revealed that cohesin complexes cycle on and off G1 chromosomes but that a subpopulation of cohesin is stably bound to chromatin after DNA replication (23). Similarly, in fission yeast, inactivation of the cohesin loader in G1 cells leads to a complete removal of cohesin from chromatin. In contrast, a fraction of cohesin remained chromatin bound when the experiment was done with postreplicative (G2) cells (12). In the present study, we further characterized the behavior of cohesin in postreplicative cells. The population of cohesin that remains bound to chromosomes after cohesin loading is shut off appears very stable, since the amount of chromatin-bound cohesin did not decrease when the experiment was extended up to 5 h (Fig. 2F). This is reminiscent of the stable mode of cohesin binding observed in mammalian cells after S phase (23). The change in cohesin dynamics correlates with S phase progression in mammalian cells (23), suggesting it is produced by the replication-coupled mechanism leading to sister chromatid cohesion. Accordingly, the stable cohesin fraction provides functional cohesion in fission yeast, and it is altered when the function of the cohesion establishment factor Eso1 is compromised (12) (Fig. 6F and H).
Examination of cohesin binding along G2 chromosomes revealed that cohesin complexes bound in the stable mode are not evenly distributed. Most sites appear to contain a mixture of the two cohesin subpopulations. The labile fraction may represent cohesin complexes that were excluded from the stabilization reaction during S phase or cohesin that was loaded onto chromosomes after this process. The rDNA gene cluster is a major site of cohesin binding. A rough estimate from chromosome spreads indicates that cohesin bound to the nucleolus in G2 cells represents 40 to 50% of the total amount of chromatin-bound cohesin (data not shown). Most (∼75%) (Fig. 1H) cohesin is bound in the labile mode at that site. This suggests that the nucleolus may act as a reservoir of cohesin that may be important in the event of a DNA DSB or in the regulation of gene expression (19).
The major foci of stably bound cohesin colocalize with the heterochromatin protein Swi6, and deletion of swi6 reduces the stable cohesin fraction to 40% of the wild-type amount (Fig. 1F and G). Swi6 domains are therefore major sites where cohesin is bound in the stable mode. Why the stable mode of cohesin binding is favored at those sites is unknown. Swi6 interacts with the cohesin subunit Psc3, and it was proposed that this may create local enrichment in cohesin (42). However, a large amount of cohesin does not necessarily imply a stable binding mode for cohesin, as exemplified by the rDNA gene cluster. This raises the possibility that chromatin features may mark chromosomal domains as preferential sites for cohesin stabilization during DNA replication.
Eso1 function is dispensable for stable cohesin binding to chromosomes and sister chromatid cohesion when the wpl1 gene is deleted.
Eso1 function is essential in an otherwise wild-type background but dispensable when wpl1 is deleted. A minichromosome loss assay indicated that eso1Δ wpl1Δ strains segregate their chromosomes with high fidelity (Table 2), and sister chromatid cohesion appears to be unaffected, as assayed by FISH on metaphase cells (Fig. 2B). This is a sensitive assay, as cohesion is challenged by spindle forces exerted on sister centromeres. The labile cohesin fraction is still present, and its amount appeared slightly increased. The stable cohesin fraction is made along chromosomes and is similar in size and distribution to that observed in wild-type cells (Fig. 2C and F). The fundamental processes by which cohesin becomes stabilized and the creation of cohesion remain intact and are therefore not dependent on the Eso1-Wpl1 module. However, Eso1 is required for both processes in wpl1+ cells. Hence, the sole but essential function of Eso1 in cohesion establishment is to counteract Wpl1.
Psm3 acetylation contributes to antagonizing Wpl1.
In budding yeast and mammals, Smc3 is acetylated on two conserved lysine residues (8, 45, 59, 63). We therefore asked whether Eso1 would antagonize Wpl1 through this pathway. A nonacetylatable mutant form does not support cell viability in budding yeast (8, 45, 59, 63). In contrast, the very same amino acid substitutions in fission yeast Psm3 are without consequences at the gross level. The strain is viable and grows like the wild type. The viability of the nonacetylatable mutant is strictly dependent on Eso1, demonstrating that Eso1 performs an essential function even when the two conserved lysine residues are replaced by arginines (Fig. 4E). The other function of Eso1 is currently unknown.
Careful examination of the nonacetylatable psm3 mutant revealed an elevated rate of minichromosome loss (Table 2), the stable cohesin fraction is created but is reduced in size (Fig. 5A and D), and a cohesion defect could be detected in metaphase cells (Fig. 4B). All the phenotypes are suppressed by the deletion of wpl1, demonstrating that it is the presence of Wpl1 and not the amino acid substitutions per se that is responsible for the observed phenotypes. This strongly argues that in wild-type Psm3 acetylation contributes to antagonizing Wpl1. By looking at the kinetics of Rad21 dissociation from chromatin (Fig. 8), we provide evidence that an acetyl-mimicking form of Psm3 renders the labile cohesin fraction less sensitive to Wpl1-dependent removal. Conversely, a mutant mimicking the nonacetylated state has the opposite effect. These data are consistent with the notion that Psm3 acetylation does provide a sheltering effect from Wpl1, thereby providing a more stable mode of cohesin interaction with chromatin.
However, Psm3 acetylation by itself might not be sufficient to create a stable cohesin fraction. In an eso1+ psm3NN background, the two cohesin subpopulations are observed. The labile fraction is mostly insensitive to Wpl1 but still dissociates over time (Fig. 8F). In contrast, the same nuclei contain a stable cohesin population (Fig. 5D). Since both cohesin subpopulations are made of acetyl-mimicking Psm3, another event must make the difference. We suggest the second event is Eso1 dependent, since in eso1Δ psm3NN cells, the stable cohesin fraction is not observed (Fig. 7B). The phenotype is alleviated by the deletion of wpl1, further suggesting that the second event contributes to antagonizing Wpl1.
What is the role of Psm3 acetylation?
Three recent studies in budding yeast reported that Smc3 is deacetylated by Hos1 following cohesin release from chromatin at anaphase (7, 13, 62). Therefore, Smc3 is kept acetylated from the time of DNA replication until cohesin cleavage, suggesting that the acetylated state may be required for maintaining cohesion. In S. pombe, we show here that a nonacetylatable Psm3 mutant is proficient in long-term cohesion. Likewise, the eso1-H17 mutant is also able to cope with a prolonged period of G2 arrest (54), although we show here that Psm3 acetylation is below the detection level. These observations strongly argue that Psm3 acetylation may not be required for maintaining sister chromatid cohesion. What is the role of Psm3 acetylation, then? In budding yeast, the amount of Pds5 bound to cohesin was reduced in cells expressing an acetyl-mimicking form of Smc3 (52). Similarly, in human cells, Smc3 acetylation seems to reduce Wapl and Pds5A binding to cohesin (56). These observations suggest that acetylated Smc3 may stimulate the remodeling of cohesin at the time of cohesion establishment. Nonacetylated Smc3 slows replication fork progression in human cells and generates cohesion defects (56). Preventing Smc3 acetylation may slow cohesin remodeling and the establishment reaction, leading to reduced replication fork velocity and occasional failure to connect sister chromatids within the replisome. The phenotypes of the nonacetylatable psm3 mutant reported here are consistent with this interpretation, although coimmunoprecipitation experiments did not show evidence that the binding of Pds5 and Wpl1 to the core cohesin complex was altered when Psm3 was not acetylatable or mimicked the acetylated state (data not shown). This again strengthens the notion that Smc3 acetylation does not have the same impact in budding and fission yeasts. One possible explanation is that Smc3 acetylation has a conserved facilitating role in cohesion establishment and some organisms rely more heavily on this pathway than others.
To summarize, we suggest that the main function of Eso1 in cohesion establishment is counteracting Wpl1. Psm3 acetylation contributes to antagonizing Wpl1, and this may facilitate the cohesion establishment reaction. Meanwhile, a second Eso1-dependent event is required to fully antagonize Wpl1 to promote sustained cohesin binding to chromosomes and long-term cohesion.
ACKNOWLEDGMENTS
We thank K. Gull, H. Okayama, C. Schmidt, T. Toda, F. Uhlmann, and Y. Watanabe for the generous gift of reagents and strains.
This work was supported by the Centre National de la Recherche Scientifique, l'Université Victor Segalen Bordeaux 2, la Région Aquitaine, and grants from l'Association pour la Recherche sur le Cancer and l'Agence Nationale de la Recherche (BLAN06-2_135754). A.F. was supported by a fellowship from the Ministère de la Recherche et de l'Enseignement Supérieur.
Footnotes
Published ahead of print on 7 February 2011.
REFERENCES
- 1. Allshire R. C., Nimmo E. R., Ekwall K., Javerzat J. P., Cranston G. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9:218–233 [DOI] [PubMed] [Google Scholar]
- 2. Anderson D. E., Losada A., Erickson H. P., Hirano T. 2002. Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell Biol. 156:419–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ansbach A. B., et al. 2008. RFCCtf18 and the Swi1-Swi3 complex function in separate and redundant pathways required for the stabilization of replication forks to facilitate sister chromatid cohesion in Schizosaccharomyces pombe. Mol. Biol. Cell 19:595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arumugam P., et al. 2003. ATP hydrolysis is required for cohesin's association with chromosomes. Curr. Biol. 13:1941–1953 [DOI] [PubMed] [Google Scholar]
- 5. Arumugam P., Nishino T., Haering C. H., Gruber S., Nasmyth K. 2006. Cohesin's ATPase activity is stimulated by the C-terminal Winged-Helix domain of its kleisin subunit. Curr. Biol. 16:1998–2008 [DOI] [PubMed] [Google Scholar]
- 6. Bähler J., et al. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943–951 [DOI] [PubMed] [Google Scholar]
- 7. Beckouët F., et al. 2010. An Smc3 acetylation cycle is essential for establishment of sister chromatid cohesion. Mol. Cell 39:689–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ben-Shahar T. R., et al. 2008. EcoI-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321:563–566 [DOI] [PubMed] [Google Scholar]
- 9. Bernard P., et al. 2006. A screen for cohesion mutants uncovers Ssl3, the fission yeast counterpart of the cohesin loading factor Scc4. Curr. Biol. 16:875–881 [DOI] [PubMed] [Google Scholar]
- 10. Bernard P., Hardwick K., Javerzat J. P. 1998. Fission yeast bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J. Cell Biol. 143:1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bernard P., et al. 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294:2539–2542 [DOI] [PubMed] [Google Scholar]
- 12. Bernard P., et al. 2008. Cell-cycle regulation of cohesin stability along fission yeast chromosomes. EMBO J. 27:111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Borges V., et al. 2010. Hos1 deacetylates Smc3 to close the cohesin acetylation cycle. Mol. Cell 39:677–688 [DOI] [PubMed] [Google Scholar]
- 14. Carlson C. R., Grallert B., Stokke T., Boye E. 1999. Regulation of the start of DNA replication in Schizosaccharomyces pombe. J. Cell Sci. 112:939–946 [DOI] [PubMed] [Google Scholar]
- 15. Ciosk R., et al. 2000. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 5:243–254 [DOI] [PubMed] [Google Scholar]
- 16. Dewar H., Tanaka K., Nasmyth K., Tanaka T. U. 2004. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature 428:93–97 [DOI] [PubMed] [Google Scholar]
- 17. Dheur S., Saupe S. J., Genier S., Vazquez S., Javerzat J. P. 2011. Role for cohesin in the formation of a heterochromatic domain at fission yeast subtelomeres. Mol. Cell. Biol. 31:1089–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ekwall K., et al. 1995. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science 269:1429–1431 [DOI] [PubMed] [Google Scholar]
- 19. Feeney K. M., Wasson C. W., Parish J. L. 2010. Cohesin: a regulator of genome integrity and gene expression. Biochem. J. 428:147–161 [DOI] [PubMed] [Google Scholar]
- 20. Fernius J., Marston A. L. 2009. Establishment of cohesion at the pericentromere by the Ctf19 kinetochore subcomplex and the replication fork-associated factor, Csm3. PLoS Genet. 5:e1000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Furuya K., Takahashi K., Yanagida M. 1998. Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes Dev. 12:3408–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gandhi R., Gillespie P. J., Hirano T. 2006. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr. Biol. 16:2406–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gerlich D., Koch B., Dupeux F., Peters J. M., Ellenberg J. 2006. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr. Biol. 16:1571–1578 [DOI] [PubMed] [Google Scholar]
- 24. Haering C. H., Farcas A. M., Arumugam P., Metson J., Nasmyth K. 2008. The cohesin ring concatenates sister DNA molecules. Nature 454:297–301 [DOI] [PubMed] [Google Scholar]
- 25. Haering C. H., Lowe J., Hochwagen A., Nasmyth K. 2002. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9:773–788 [DOI] [PubMed] [Google Scholar]
- 26. Haering C. H., et al. 2004. Structure and stability of cohesin's Smc1-kleisin interaction. Mol. Cell 15:951–964 [DOI] [PubMed] [Google Scholar]
- 27. Hanna J. S., Kroll E. S., Lundblad V., Spencer F. A. 2001. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell. Biol. 21:3144–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hauf S., Waizenegger I. C., Peters J. M. 2001. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science 293:1320–1323 [DOI] [PubMed] [Google Scholar]
- 29. Huang C. E., Milutinovich M., Koshland D. 2005. Rings, bracelet or snaps: fashionable alternatives for Smc complexes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360:537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kenna M. A., Skibbens R. V. 2003. Mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different replication factor C complexes. Mol. Cell. Biol. 23:2999–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kueng S., et al. 2006. Wapl controls the dynamic association of cohesin with chromatin. Cell 127:955–967 [DOI] [PubMed] [Google Scholar]
- 32. Lai M. S., Seki M., Ui A., Enomoto T. 2007. Rmi1, a member of the Sgs1-Top3 complex in budding yeast, contributes to sister chromatid cohesion. EMBO Rep. 8:685–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lengronne A., et al. 2004. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430:573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lengronne A., et al. 2006. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol. Cell 23:787–799 [DOI] [PubMed] [Google Scholar]
- 35. Mayer M. L., Gygi S. P., Aebersold R., Hieter P. 2001. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol. Cell 7:959–970 [DOI] [PubMed] [Google Scholar]
- 36. Miles J., Formosa T. 1992. Evidence that POB1, a Saccharomyces cerevisiae protein that binds to DNA polymerase alpha, acts in DNA metabolism in vivo. Mol. Cell. Biol. 12:5724–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mizukami T., et al. 1993. A 13 kb resolution cosmid map of the 14 Mb fission yeast genome by nonrandom sequence-tagged site mapping. Cell 73:121–132 [DOI] [PubMed] [Google Scholar]
- 38. Moldovan G. L., Pfander B., Jentsch S. 2006. PCNA controls establishment of sister chromatid cohesion during S phase. Mol. Cell 23:723–732 [DOI] [PubMed] [Google Scholar]
- 39. Moreno S., Klar A., Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823 [DOI] [PubMed] [Google Scholar]
- 40. Nasmyth K., Haering C. H. 2009. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43:525–558 [DOI] [PubMed] [Google Scholar]
- 41. Niwa O., Matsumoto T., Chikashige Y., Yanagida M. 1989. Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J. 8:3045–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nonaka N., et al. 2002. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 4:89–93 [DOI] [PubMed] [Google Scholar]
- 43. Oliveira R. A., Hamilton R. S., Pauli A., Davis I., Nasmyth K. 2010. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nat. Cell Biol. 12:185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Petronczki M., et al. 2004. Sister-chromatid cohesion mediated by the alternative RF-CCtf18/Dcc1/Ctf8, the helicase Chl1 and the polymerase-alpha-associated protein Ctf4 is essential for chromatid disjunction during meiosis II. J. Cell Sci. 117:3547–3559 [DOI] [PubMed] [Google Scholar]
- 45. Rowland B. D., et al. 2009. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol. Cell 33:763–774 [DOI] [PubMed] [Google Scholar]
- 46. Sato M., Dhut S., Toda T. 2005. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast 22:583–591 [DOI] [PubMed] [Google Scholar]
- 47. Sazer S., Sherwood S. W. 1990. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J. Cell Sci. 97:509–516 [DOI] [PubMed] [Google Scholar]
- 48. Schmidt C. K., Brookes N., Uhlmann F. 2009. Conserved features of cohesin binding along fission yeast chromosomes. Genome Biol. 10:R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schmitz J., Watrin E., Lenart P., Mechtler K., Peters J. M. 2007. Sororin is required for stable binding of cohesin to chromatin and for sister chromatid cohesion in interphase. Curr. Biol. 17:630–636 [DOI] [PubMed] [Google Scholar]
- 50. Skibbens R. V., Corson L. B., Koshland D., Hieter P. 1999. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 13:307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ström L., et al. 2007. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 317:242–245 [DOI] [PubMed] [Google Scholar]
- 52. Sutani T., Kawaguchi T., Kanno R., Itoh T., Shirahige K. 2009. Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr. Biol. 19:492–497 [DOI] [PubMed] [Google Scholar]
- 53. Tanaka K., Hao Z., Kai M., Okayama H. 2001. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J. 20:5779–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tanaka K., et al. 2000. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol. Cell. Biol. 20:3459–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tatebayashi K., Kato J., Ikeda H. 1998. Isolation of a Schizosaccharomyces pombe rad21ts mutant that is aberrant in chromosome segregation, microtubule function, DNA repair and sensitive to hydroxyurea: possible involvement of Rad21 in ubiquitin-mediated proteolysis. Genetics 148:49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Terret M. E., Sherwood R., Rahman S., Qin J., Jallepalli P. V. 2009. Cohesin acetylation speeds the replication fork. Nature 462:231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tóth A., et al. 1999. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13:320–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Uhlmann F., Lottspeich F., Nasmyth K. 1999. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400:37–42 [DOI] [PubMed] [Google Scholar]
- 59. Unal E., et al. 2008. A molecular determinant for the establishment of sister chromatid cohesion. Science 321:566–569 [DOI] [PubMed] [Google Scholar]
- 60. Unal E., Heidinger-Pauli J. M., Koshland D. 2007. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through EcoI (Ctf7). Science 317:245–248 [DOI] [PubMed] [Google Scholar]
- 61. Weitzer S., Lehane C., Uhlmann F. 2003. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr. Biol. 13:1930–1940 [DOI] [PubMed] [Google Scholar]
- 62. Xiong B., Lu S., Gerton J. L. 2010. Hos1 is a lysine deacetylase for the smc3 subunit of cohesin. Curr. Biol. 20:1660–1665 [DOI] [PubMed] [Google Scholar]
- 63. Zhang J., et al. 2008. Acetylation of Smc3 by EcoI is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell 31:143–151 [DOI] [PubMed] [Google Scholar]