Abstract
Lipins are evolutionarily conserved proteins found from yeasts to humans. Mammalian and yeast lipin proteins have been shown to control gene expression and to enzymatically convert phosphatidate to diacylglycerol, an essential precursor in triacylglcerol (TAG) and phospholipid synthesis. Loss of lipin 1 in the mouse, but not in humans, leads to lipodystrophy and fatty liver disease. Here we show that the single lipin orthologue of Drosophila melanogaster (dLipin) is essential for normal adipose tissue (fat body) development and TAG storage. dLipin mutants are characterized by reductions in larval fat body mass, whole-animal TAG content, and lipid droplet size. Individual cells of the underdeveloped fat body are characterized by increased size and ultrastructural defects affecting cell nuclei, mitochondria, and autophagosomes. Under starvation conditions, dLipin is transcriptionally upregulated and functions to promote survival. Together, these data show that dLipin is a central player in lipid and energy metabolism, and they establish Drosophila as a genetic model for further studies of conserved functions of the lipin family of metabolic regulators.
INTRODUCTION
Neutral lipids, or triacylglycerols (TAG), are principal energy stores of the eukaryotic cell. Metazoans have evolved specialized tissues that store TAG and make free fatty acids or other derivatives of TAG available to other tissues. Besides having storage functions, these specialized adipose tissues participate in the control of energy homeostasis by producing and releasing hormones and other signaling molecules (21, 26).
Severe underdevelopment of the adipose tissue is observed in mice carrying the fatty liver dystrophy (fld) mutation. Lack of fat tissue is associated with transient postnatal accumulation of TAG in the liver, defects in the peripheral nervous system, and insulin resistance (15, 16, 29). Cloning of the fld gene (renamed lipin 1 [22]) revealed that it encodes a member of an evolutionarily old family of proteins found in a wide variety of eukaryotic organisms, including fungi, plants, and protozoans (22). Both yeast and mammalian lipin proteins act as type 1 phosphatidate phosphatases (PAP1), converting phosphatidate to diacylglycerol (DAG), and as transcriptional coregulators (4, 5, 11, 32). The protein domains responsible for these activities are conserved in lipin proteins of other species, indicating that this functional dichotomy is both evolutionarily old and central to lipin function. DAG produced by mammalian lipin 1 serves as a direct biosynthetic precursor of TAG and phospholipids, whereas the transcriptional coregulator function contributes to the control of genes involved in hepatic β-oxidation of fatty acids, the tricarboxylic acid (TCA) cycle, and mitochondrial oxidative phosphorylation (5). While these processes appear to be upregulated by lipin 1, enzymes involved in fatty acid and TAG synthesis are downregulated. In yeast, lipin suppresses genes involved in phospholipid synthesis (32). Loss of lipin in yeast leads to the overgrowth of intracellular membranes, affecting the nuclear envelope and peripheral endoplasmic reticulum (ER) (32, 34), and to defects in chromosome segregation (34). Overgrowth of the peripheral ER and defects in chromosome segregation are also observed in Caenorhabditis elegans lacking lipin. However, instead of overgrowth, the nuclear envelope exhibits impaired breakdown during mitosis that is associated with early embryonic lethality (6, 7). In addition, adult worms grow to a smaller size and contain less TAG than control animals (6).
The human and mouse genomes encode three lipin homologues: lipin 1, 2, and 3. Whereas loss of lipin 1 in mice leads to lipodystrophy, hepatic steatosis, and insulin resistance (27), none of these conditions is observed in humans deficient in lipin 1. Instead, these individuals are affected by a recurrent breakdown of muscle fibers, leading to myoglobinuria (40). A likely explanation for the discrepancies in the loss-of-function phenotypes of lipin 1 in mice and humans are differences in tissue distribution and redundancy between the three lipin paralogues. Studies on the single lipin orthologue in the genetic model organism, Drosophila melanogaster, promise to be unimpeded by such complications and to provide insights into the basic functions of lipins in metazoans.
Here we show that the loss of lipin in Drosophila leads to a severe defect in the development of the fat body, the major tissue of TAG storage in insects. While fat body mass is reduced, the sizes of individual fat body cells and nuclei are dramatically increased. The cells contain fat droplets of reduced size and exhibit changes in organelle structure that affect mitochondria, autophagosomes, and the cell nucleus. These defects are associated with reduced viability and fertility, failure of adult flies to eclose, and impaired starvation resistance. Together, the results show that lipin plays a central role in fat body function and energy metabolism in Drosophila.
MATERIALS AND METHODS
Fly stocks and mutants.
The dLipin allele CG8709e00680 (dLipine00680) was obtained from the Exelixis insertion collection at Harvard Medical School (35). We used PCR to confirm the presence of a piggyBac insertion in the 5′ untranslated region (5′ UTR) and rapid amplification of cDNA 5′ ends (5′-RACE) to determine the sequences of 5′ ends of dLipin mRNAs produced by the mutant (GeneRacer kit; Invitrogen). These mRNAs lacked about half of the normal 5′ UTR, which was replaced by alternate segments derived from the first intron (positions +1970 to +2137, +1973 to +2137, and +2309 to +2481). The remaining portions of intron 1 downstream of these segments were removed by splicing between cryptic splice donor sites within the intron and the normal splice acceptor site at the exon 2 junction.
Dcg-GFP flies (33) were obtained from Jonathan M. Graff, 3.1Lsp2-Gal4 flies (17) from Brigitte Dauwalder, and r4-gal4 flies (18) from Jae Park. UAS-dLipin[RNAi] flies were obtained from the Vienna Drosophila RNAi Center (transformant ID 36007). All other fly stocks were obtained from the Bloomington Stock Center.
Anti-dLipin antibodies.
For the production of an anti-dLipin antibody, a 1,722-bp SalI fragment from dLipin cDNA GH19076 was inserted into the SalI site of expression vector pET-21b (Novagen/EMD, Gibbstown, NJ). This fragment encodes a 506-amino-acid region from the N-terminal half of the dLipin protein that is common to the A and B isoforms and overlaps with the conserved N-LIP domain. The protein was expressed as a C-terminal His tag fusion protein in BL21 cells. The fusion protein was expressed as a soluble protein by these cells and was purified by affinity chromatography using Talon Superflow metal affinity resin (Clontech, Mountain View, CA). Antisera were produced in rabbits at Cocalico Biologicals (Reamstown, PA). Anti-dLipin antibodies were affinity purified with an antigen bound to nitrocellulose strips by using standard procedures.
Northern and Western blot analyses.
For Northern blot analysis, total RNA was extracted from staged 12- to 16-h-old prepupae and pupae. RNA was fractionated by gel electrophoresis, transferred to nylon membranes, and hybridized with radioactive DNA probes as described previously (19). The dLipin probe was a ∼450-bp PCR product derived from dLipin cDNA GH19076 by using primers with the sequences 5′-ATCCCACGTCCCTGATATGC-3′ and 5′-GACGATGACGAAGCCTCTA-3′.
For Western blot analysis, two wandering larvae were crushed in 100 μl of sodium dodecyl sulfate (SDS) sample buffer, and 25 μl of the cleared homogenate was loaded per lane. The purified anti-dLipin antibody was used at a dilution of 1:1,500 and anti-actin antibody (Sigma) at a dilution of 1:500. Alkaline phosphatase-conjugated AffiniPure goat anti-rabbit IgG (H+L) (Jackson ImmunoResearch) was used as a secondary antibody at a dilution of 1:2,000.
Lipid staining and triglyceride and ketone assays.
Fat bodies from wandering third-instar larvae were incubated in 1 μM Nile red or Bodipy 493/503 (Invitrogen/Molecular Probes) in phosphate-buffered saline (PBS) for 15 min in the dark. Subsequently, the tissue was rinsed in PBS and was mounted in SlowFade Gold antifade reagent (Invitrogen). Fat droplets were visualized by fluorescence microscopy using fluorescein isothiocyanate (FITC) and tetramethyl rhodamine isothiocyanate (TRITC) filters.
TAG was measured using the Infinity triglycerides assay (Thermo Electron). Samples were obtained by pooling and homogenizing eight wandering larvae in 800 μl of 0.3% Triton X-100. Homogenates were heated to 70°C for 5 min to inactivate enzymes released from the cells. The homogenates were then centrifuged at 7,500 rpm for 1 min at room temperature. The supernatant was collected and was centrifuged again at 14,000 rpm for 5 min at 4°C. The resulting final supernatant was assayed for triglycerides and protein. For the measurement of triglycerides, 80 μl of supernatant was added to 1 ml of triglyceride reagent, and the mixture was incubated at 37°C for 5 min. The optical density at 520 nm (OD520) was measured, and TAG values were calculated according to the manufacturer's instructions by using glycerol standards for calibration. The Bradford assay (Bio-Rad protein assay kit II) was used to determine the supernatant's protein concentration.
Ketone body formation was assayed essentially as described by Luong et al. (20) using a β-hydroxybutyrate assay kit (Cayman Chemical). β-Hydroxybutyrate was measured in 17 independent samples obtained from wandering larvae with the genotype dLipine00680/Df(2R)Exel7095 and 12 samples from heterozygous control animals. The amounts of β-hydroxybutyrate were normalized to body weight.
Cell staining and size measurements.
Plasma membranes were stained with CellMask Orange (Invitrogen) by incubating unfixed tissues with the dye at a concentration of 5 μg/ml for 5 min at 37°C. Following incubation, tissues were rinsed in PBS and were mounted in SlowFade Gold antifade reagent containing the DNA stain 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen). Cells were viewed by fluorescence microscopy using TRITC and DAPI filters. The sizes of cells and nuclei were estimated by measuring the cell and nuclear areas using the measure tool of the AxioVision software package (release 4.6.3).
Immunohistochemistry and light microscopy.
Tissues were stained with the anti-dLipin antibody using standard procedures. The affinity-purified antibody was used at a dilution of 1:200 to 1:400. Bound anti-dLipin antibodies were detected with a Cy3-conjugated donkey anti-rabbit secondary antibody (1:1,000; Jackson ImmunoResearch). Images of fluorescently labeled tissues and bright-field images were taken with a Carl Zeiss AxioVision microscope.
Electron microscopy.
For transmission electron microscopy (TEM), fat bodies from wandering third-instar larvae were fixed in Karnovsky's fixative, washed in 0.05 M cacodylate buffer (pH 7.2), and postfixed in 1% osmium tetroxide. Samples were briefly washed with distilled water and were then prestained overnight in 0.5% uranyl acetate at 4°C. Dehydration was carried out in a graded ethanol series to 100% and in two changes of propylene oxide. After infiltration first with a mixture of 50% propylene oxide and 50% Spurr's resin and then with 100% Spurr's resin, samples were embedded and sectioned. Images were taken with a JEOL 100CX transmission electron microscope.
Starvation experiments and quantitative reverse transcription-PCR (qRT-PCR).
For starvation experiments, freshly eclosed adult flies were collected over a period of 30 h and were transferred to standard food supplemented with yeast paste. After 7 days, male and female flies were separated. Groups of 25 flies were transferred to vials that contained a bottom layer of cotton soaked in tap water or in 5% ethanol with or without 500 μM RU486. Flies were kept at 25°C, and dead flies were counted and removed from the vials daily. Periodically, flies were transferred to fresh vials to prevent fungal growth. Each experiment was performed in triplicate, and statistical comparisons of survival curves were performed with the log rank test (R statistical software [http://www.R-project.org]).
For qRT-PCR measurements, freshly eclosed flies were kept on standard food for 2 days and were then separated by gender and transferred to vials containing water only. After 4 days of starvation, total RNA was extracted from whole flies by using Trizol reagent (Invitrogen). For each of the experimental conditions, four independent samples were analyzed by one-step qRT-PCR using a HotStart-IT SYBR green qRT-PCR kit (product no. 75770; Affymetrix) and a Bio-Rad CFX96 real-time PCR detection system. The rp49 gene was used as a normalizer gene with primers with the sequences 5′-CGGATCGATATGCTAAGCTGT-3′ and 5′-GCGCTTGTTCGATCCGTA-3′. Primers for the amplification of dLipin RNA were designed to flank introns and had the sequences 5′-ATCCCACGTCCCTGATATCG-3′ and 5′-TTCATCTTGGTTGGTTAGCAGG-3′. Comparative quantification was carried out using the standard curve method (23). qRT-PCR results were consistent with the results obtained by Northern blot analysis (data not shown).
RESULTS
Loss of lipin in Drosophila causes lethality, impaired eclosion, and dystrophy of the larval fat body.
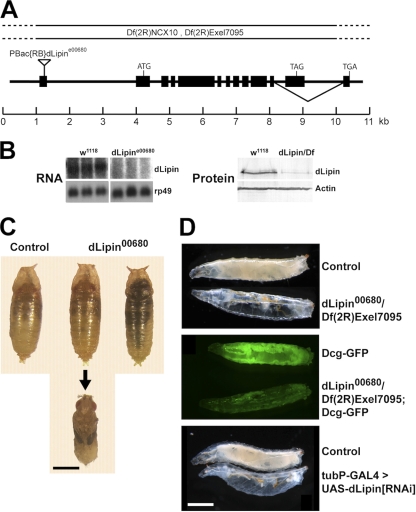
The Drosophila genome encodes a single lipin homologue (CG8709; referred to here as dLipin) (22). To study the loss-of-function phenotype of dLipin, we obtained an allele, CG8709e00680 (referred to here as dLipine00680), that contains a piggyBac insertion in the 5′ UTR of the dLipin transcription unit (Exelixis insertion collection) (Fig. 1) (35). We found that animals homozygous for dLipine00680 have reduced amounts of dLipin transcripts of apparently normal size and reduced levels of dLipin protein (Fig. 1B). Sequencing of 5′-RACE products obtained from dLipine00680 mRNA revealed that aberrant splicing events in the mutant lead to complete removal of the piggyBac insertion from the transcript and replacement of the first exon by portions of the first intron. This results in a change of the sequence of the dLipin 5′ UTR, which may lead to reduced mRNA stability and/or reduced translation of the RNA.
Fig. 1.
Animals lacking dLipin are characterized by pharate adult lethality and underdevelopment of the larval fat body. (A) Genomic structure of the dLipin gene. The inverted triangle indicates the insertion site of the piggyBac transposon in the dLipine00680 allele. Black boxes represent exons. The deficiencies Df(2R)Exel7095 and Df(2R)NCX10 remove chromosomal regions 43E7 to 44C5 and 44B3 to 44C2, respectively, including the entire dLipin gene at 44B4 to 44B5. dLipin encodes two protein isoforms with different C termini that result from alternative splicing, as indicated. (B) Animals homozygous for dLipine00680, or transheterozygous for dLipine00680 and Df(2R)Exel7095 (dLipin/Df), contain reduced levels of dLipin mRNA and protein. RNA for Northern blot analysis was from whole prepupae and pupae, and protein for Western blot analysis was from wandering third-instar larvae. Control samples were obtained from w1118 animals. Detection of rp49 mRNA and actin protein served as a control for loading and transfer. (C) Animals homozygous for dLipine00680 develop into pharate adult flies that do not show obvious developmental defects (animal in middle, dissected from pupal case). However, many of these flies die and become necrotic while still enclosed in the pupal case (animal on the right). Note that mutant and control pupae are similar in size, indicating that a lack of dLipin does not lead to an overall growth defect. (D) A lack of dLipin causes severe underdevelopment of the larval fat body. (Top) dLipin mutant larvae appear translucent due to a lack of fat tissue. (Center) Inclusion of the Dcg-GFP fat body marker (33) in the mutant background confirms that the amount of fat tissue is dramatically reduced. (Bottom) The same phenotype is obtained when dLipin expression is knocked down by RNAi using the ubiquitous tubulin driver. Bars, 1 mm.
The dLipine00680 mutant phenotypes reported here reversed by precise excision of the piggyBac element, suppressed by ubiquitous expression of wild-type dLipin from an upstream activation sequence (UAS)-dLipin transgene, and partially reproduced by RNA interference (RNAi) knockdown of dLipin (see below). Therefore, we conclude that the lack of dLipin was responsible for the effects observed.
Animals homozygous for dLipine00680 were viable, but many animals died during late larval and pupal development, and only a few adult flies developed. The mutant adult flies showed strongly reduced fertility, laying less than 10% of the eggs produced by their heterozygous siblings. Mutant larvae showed delayed development, entering the wandering stage about 2 days later than heterozygous control animals. Phenotypes were exacerbated in animals transheterozyous for dLipine00680 and the deficiencies Df(2R)NCX10 and Df(2R)Exel7095, which uncover the dLipin locus, confirming that dLipine00680 is a hypomorphic allele. Whereas almost all larvae homozygous for dLipine00680 reached the wandering stage, only 23% of the dLipine00680/Df(2R)Exel7095 larvae did so. Homozygous dLipine00680 animals exhibited a major lethal phase during pupal development. Fifty-nine percent of the pupae died (n = 615), many after reaching the pharate adult stage (Fig. 1C). Again, this phenotype was more pronounced in transheterozygous animals. Seventy-nine percent of dLipine00680/Df(2R)NCX10 pupae (n = 398) and >99% of dLipine00680/Df(2R)Exel7095 pupae (n = 219) died during the pupal stage. Because Df(2R)Exel7095 is smaller than Df(2R)NCX10 and is completely contained in it, the attenuation of the mutant phenotype in the presence of Df(2R)NCX10 suggested that this deficiency removes a suppressor of dLipin. Indeed, inspection of the genes uncovered by the deficiencies revealed that Df(2R)NCX10, but not Df(2R)Exel7095, removes the gene encoding diacylglycerolkinase (Dgk), an enzyme that counteracts the PAP1 activity of lipin by converting DAG into phosphatidic acid. Therefore, we used Df(2R)Exel7095 exclusively for further characterization of dLipin.
Dead pharate adults lacking dLipin did not show obvious morphological defects (Fig. 1). Some adults died while trying to eclose, suggesting that these animals lacked the energy to emerge fully from the pupal case. Consistent with this interpretation, we found that the functions of the fat body, the central metabolic organ and energy store of insects, were severely impaired in dLipin mutants. Many of the mutant larvae looked transparent, indicating a dramatic reduction in the amount of fat tissue. A similarly severe lipodystrophy, as well as larval lethality, resulted from the knockdown of dLipin by RNAi using the ubiquitous tubulin driver (Fig. 1). Together, these data suggest that dLipin plays an essential role in Drosophila in the formation of energy stores that are required for survival.
dLipin is a widely expressed protein that exhibits cytoplasmic and nuclear localization in the fat body.
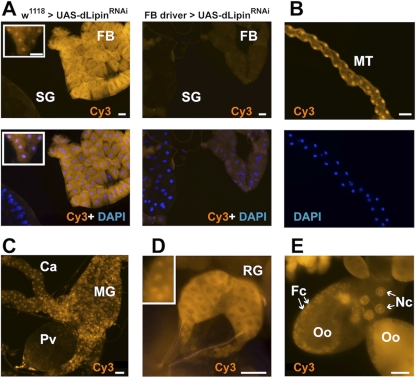
Underdevelopment of the larval fat body in dLipin mutants may reflect a cell-autonomous or non-cell-autonomous function of the gene. To address this question and to explore whether the functions of dLipin are confined to fat tissue, we generated a polyclonal antibody directed toward the N-terminal region of dLipin that is common to both isoforms of the protein (Fig. 1). The affinity-purified antibody stained a variety of tissues in both larvae and adult flies. Strong specific staining was observed in the cytoplasm and nuclei of fat body cells (Fig. 2). The midgut and other regions of the gut showed nuclear and cytoplasmic staining as well. Interestingly, very strong staining was also apparent in the ring gland, an endocrine organ that produces hormones involved in developmental and metabolic control (Fig. 2). In the Malpighian tubules, the excretory organs of the fly, staining was predominant in the cell nuclei, suggesting that dLipin has mainly gene-regulatory functions in this tissue. Other tissues that showed staining included the ovaries (Fig. 2) and the imaginal discs. The presence of dLipin in the nuclei of ovarian nurse and follicle cells suggests that the protein has gene-regulatory functions that are important for oogenesis. The staining of the oocyte cytoplasm indicates that dLipin protein is provided maternally to support embryogenesis. Among the few tissues that showed very little or no expression of dLipin were the larval salivary glands (Fig. 2).
Fig. 2.
dLipin is broadly expressed in Drosophila tissues and shows both cytoplasmic and nuclear localization. (A) dLipin is strongly expressed in both the cytoplasm and the nuclei of fat body cells (FB). The strong cytoplasmic staining usually masks the nuclear staining, which can be seen clearly in some regions (inset). In stark contrast to the fat body, the larval salivary glands (SG) show very little or no staining. Knockdown of dLipin expression by RNAi using the fat body-specific driver P{w+mC=Cg-GAL4.A}2 greatly reduced both cytoplasmic and nuclear staining, demonstrating the specificity of antibody binding. (B) Malpighian tubules (MT) show strong nuclear staining with the anti-dLipin antibody. (C) The midgut (MG) and ceca (Ca) show both nuclear and cytoplasmic staining, whereas the proventriculus (Pv) does not appear to express significant amounts of dLipin. (D) The ring gland (RG) shows very strong cytoplasmic staining and nuclear staining (inset). (E) In the ovary, strong nuclear dLipin staining is found in the nuclei of the nurse cells (Nc) and follicle cells (Fc) (arrows). Diffuse staining is also observed in the oocyte cytoplasm (Oo), suggesting that dLipin protein is provided maternally. Bars, 50 μm.
dLipin is required for normal fat body development and TAG storage.
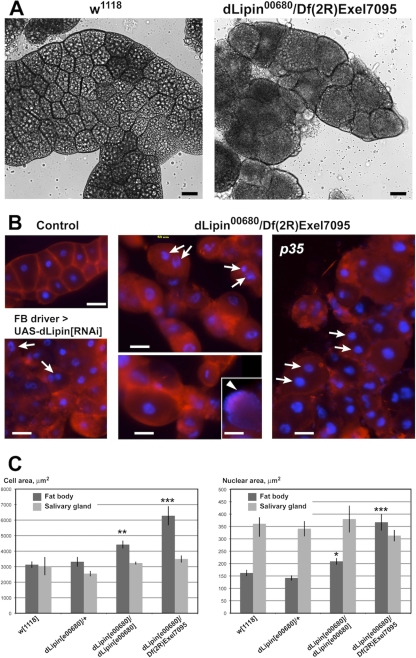
We wanted to study in more detail the fat body defect caused by the lack of dLipin. Dissection of the larval fat body of dLipine00680/Df(2R)Exel7095 animals revealed that while the total mass of fat tissue was greatly reduced, individual fat body cells were highly enlarged. The nuclei of these cells were enlarged as well or had apparently undergone fragmentation (Fig. 3). On average, the sizes of both cells and nuclei were increased about 2-fold. Some mutant cells reached a diameter of 160 μm. In contrast, the cells and cell nuclei of the larval salivary glands did not show a significant change in size, suggesting that a lack of dLipin does not cause a systemic growth defect (Fig. 3). This conclusion was supported by measurements of the sizes of mutant pupae, which did not indicate a significant reduction or increase in average pupal length (Fig. 1; also data not shown). Not only the size but also the shape of the mutant fat body cells was changed. The cells were more rounded than control cells and, in some regions of the fat body, had detached from their neighbors, indicating that cell adherence is affected in dLipin mutants. A similar phenotype, although not as pronounced, could be detected after fat body-specific knockdown of dLipin by RNAi (Fig. 3B).
Fig. 3.
Loss of dLipin causes defects in fat body development. (A and B) The size and shape of fat body cells is dramatically altered in dLipin-deficient larvae. (A) Bright-field microscopy indicates that, compared to wild-type cells (w1118), fat body cells of third-instar larvae lacking dLipin are more variable in size, more rounded, and, in some regions, detached from one another. Large fat droplets that appear as bright inclusions, and are plentiful in the control fat body, are scarce in the fat body lacking dLipin. (B) Staining with the plasma membrane stain CellMask (red) highlights cell boundaries, confirming the cell shape changes, while staining with the DNA stain DAPI (blue) reveals that cell nuclei in some of the dLipin-deficient cells are fragmented (arrows). The arrowhead marks a cell in which the DNA is dispersed throughout the cell. Cells show a similar mutant phenotype in the presence of the apoptosis inhibitor p35 or after dLipin knockdown by RNAi. Bars, 50 μm. (C) The sizes of cells and nuclei are increased in the fat bodies, but not in the salivary glands, of dLipin mutants, indicating that the observed effects are specific for fat tissue. Asterisks indicate a significant difference from the mean for w1118 flies (*, P < 0.05; **, P < 0.01; ***, P < 0.0001). Error bars indicate standard errors of the means.
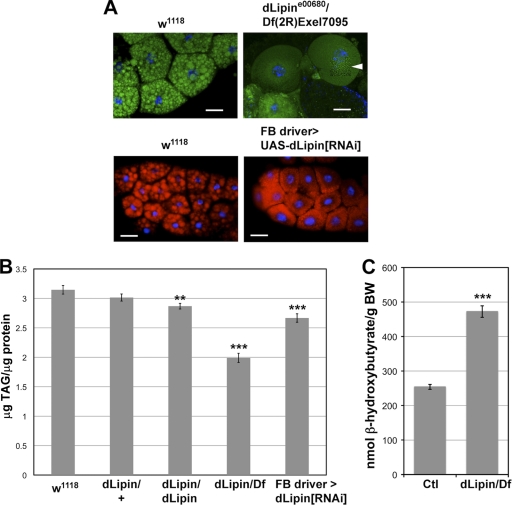
To determine whether the observed developmental defects in the fat body were associated with an overall reduction of fat stores, we first stained fat bodies with the lipophilic dyes Bodipy and Nile red and then quantified whole-animal TAG levels. In situ staining revealed that fat droplets in the dLipin mutant [dLipine00680/Df(2R)Exel7095] fat body were considerably smaller than those in the wild-type fat body. A similar reduction in lipid droplet size was observed after knockdown of dLipin expression by RNAi using a fat body-specific driver (Fig. 4 A). This reduction in droplet size was accompanied by a significant overall reduction in triglyceride levels (Fig. 4B). Together, these data show that dLipin is required for normal fat tissue development and fat storage in Drosophila.
Fig. 4.
Lack of dLipin results in reduced lipid droplet size, reduced stores of neutral lipids, and increased ketogenesis. (A) The size of fat droplets is reduced in the fat bodies of dLipin mutants and after fat body-specific knockdown of dLipin expression by RNAi. Fat droplets were stained with Bodipy (green) or Nile red (red). The section marked with an arrowhead was subjected to image deconvolution in order to facilitate recognition of the very small lipid droplets in the dLipin mutant. Bars, 50 μm. (B) TAG levels are reduced in animals lacking dLipin. Total TAG and protein levels were measured in whole animals homozygous for dLipine00680 or transheterozygous for dLipine00680 and Df(2R)Exel7095 (dLipin/dLipin and dLipin/Df), and after fat body-specific knockdown of dLipin. Asterisks indicate a statistically significant decrease in the TAG/protein level from that for the w1118 control. (C) Ketone body formation is increased in animals lacking dLipin. The ketone β-hydroxybutyrate was measured in animals with the genotype dLipine00680/Df(2R)Exel7095 and in control animals heterozygous for dLipine00680 or Df(2R)Exel7095 (Ctl). BW, body weight. Error bars in panels B and C indicate standard errors of the means. **, P < 0.01; ***, P < 0.0001.
Like its yeast and mammalian counterparts, dLipin contains the DIDGT signature motif of PAP1 enzymes (4). Loss of PAP1 activity in animals lacking dLipin should lead to impaired TAG synthesis and hence should be a major cause of the reduced TAG levels observed in these animals. A block in TAG synthesis predicts that dLipin-deficient animals will exhibit reduced fatty acid synthesis and/or increased fatty acid breakdown. Both effects would result in the increased availability of acetyl coenzyme A (acetyl-CoA), leading to ketone body formation. To test this prediction, we measured the amounts of the ketone β-hydroxybutyrate in dLipin mutants and heterozygous controls. The measurements indicated that the level of β-hydroxybutyrate is indeed almost 2-fold higher in the mutant (Fig. 4C). These data further support the conclusion that fat synthesis and fatty acid metabolism are disturbed in dLipin mutants.
Loss of dLipin leads to changes in organelle structure.
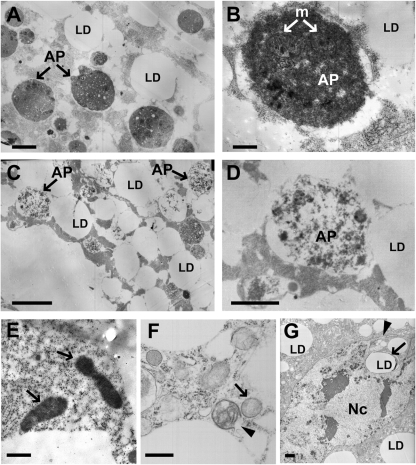
Analysis of the enlarged fat body cells of dLipine00680/Df(2R)Exel7095 animals by transmission electron microscopy (TEM) revealed deviations from the normal ultrastructure of fat body cells. In wandering third-instar larvae, the fat body normally undergoes developmental autophagy, and the cells display a large number of autophagosomes and autolysosomes (31) (Fig. 5). Cells lacking dLipin contained autophagosomes and autolysosomes as well. However, the matrix of these vesicles was considerably less dense than the matrix in control cells (Fig. 5). Furthermore, the cells contained misshapen nuclei with nuclear envelopes that formed projections or involutions or appeared ruptured. These changes in nuclear structure are consistent with the nuclear fragmentation observed by light microscopy (Fig. 3). Finally, the mutant cells contained unusually structured, and apparently defective, mitochondria. In contrast to the elongated and crista-rich mitochondria of control cells, these mitochondria were rounded and contained few cristae. In some cases, both the outer and the inner mitochondrial membrane were ruptured. While unusually structured mitochondria in the fat body cells of normal wandering larvae have been reported in the literature (37), we never saw defects in fat bodies from heterozygous control animals that were as severe and prevalent as those for the dLipin-deficient fat body. In summary, the TEM data show that a lack of dLipin affects the structures of autophagosomes, mitochondria, and cell nuclei, suggesting that dLipin is required for the normal assembly and functioning of these organelles.
Fig. 5.
Loss of dLipin results in ultrastructural changes affecting the cell nucleus, mitochondria, and autophagosome formation. (A and B) Transmission electron micrographs of fat body cells of control animals [dLipine00680/+ or Df(2R)Exel7095/+]. As reported previously (31), fat body cells of late-third-instar larvae are characterized by the presence of autophagosomes (AP) that contain mitochondria (m) and other cytoplasmic remnants. LD, lipid droplet. (C and D) Fat body cell of a dLipin mutant [dLipine00680/Df(2R)Exel7095]. Most autophagosomes have a matrix that is less dense than the matrix of normal autophagosomes. (E and F) Mitochondria of mutant fat body cells exhibit changed morphology. Fat body mitochondria of control animals are elongated and densely packed with cristae (E, arrows). In contrast, mitochondria of dLipin mutants are rounded, contain only a few cristae, and often exhibit ruptured double membranes (F, arrowhead). (G) Misshapen cell nucleus (Nc) of a dLipin mutant fat body cell, featuring a long nuclear projection (arrowhead) and the intrusion of a fat droplet (LD, arrow). Bars, 0.5 μm (A, C, E, and F), 0.2 μm (B and D), and 1 μm (G).
The defective mitochondria we observed in the mutant fat body cells resembled the swollen and rounded mitochondria found in Drosophila cells undergoing apoptosis (1). Together with the occurrence of fragmented and misshapen nuclei, and the observed cell shape changes, this resemblance suggested that the mutant fat body cells might have entered the apoptotic pathway. However, staining for active caspase (38, 39) did not indicate a consistent and strong increase in caspase activity in the cells, although some cells appeared to exhibit slightly elevated levels of staining (data not shown). Since staining levels were difficult to interpret due to the enlarged cell bodies, we sought to determine whether constitutive expression of the caspase inhibitor p35 had an effect on the phenotype of fat body cells lacking dLipin. p35 prevents most, if not all, caspase-dependent cell death in Drosophila (13, 14). The expression of p35 in the fat bodies of dLipin mutants [dLipine00680/Df(2R)Exel7095; P{UAS-p35.H}BH2/r4-gal4] further enhanced the lethality observed for these animals. The few wandering larvae that could be collected were characterized by severe lipodystrophy and dramatically oversized and rounded fat body cells. Some of these cells showed fragmented nuclei and were indistinguishable from mutant cells in the absence of p35 (Fig. 3B). Thus, inhibition of apoptosis in the fat body did not rescue, but rather aggravated, the fat body phenotype of dLipin mutants. These data strongly suggest that the observed defects in fat body development and structure are caused primarily by the lack of dLipin and are not secondary consequences of an activation of the apoptotic pathway.
dLipin is upregulated by starvation and promotes starvation resistance.
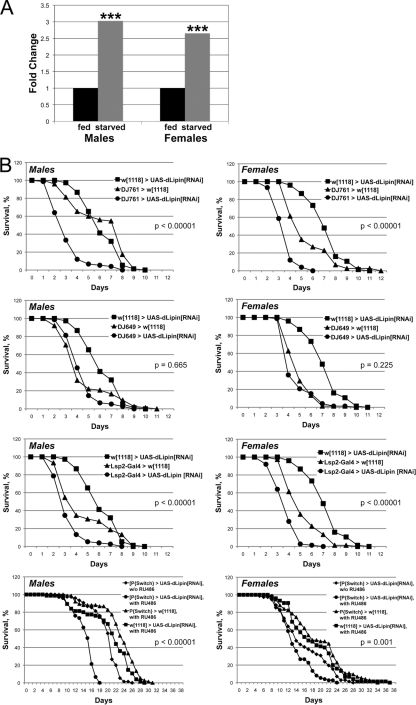
The loss-of-function phenotype of dLipin indicated that a major function of the gene is to promote fat storage. We were therefore surprised to find that dLipin was transcriptionally upregulated under starvation conditions (Fig. 6). Since starvation is associated with a mobilization of fat reserves, this observation seemed to be in conflict with the anabolic role of dLipin in fat metabolism (increasing fat stores). The real-time RT-PCR (qRT-PCR) data that we obtained (Fig. 6A) were consistent with, and validated, the results of microarray studies showing strong upregulation of dLipin RNA upon withdrawal of food in male flies and less-pronounced upregulation in females (8, 12). Thus, dLipin may have beneficial effects on survival under starvation conditions. To test this hypothesis, we first asked whether reduced expression of dLipin before the onset of starvation indeed leads to reduced starvation resistance as predicted by the role of the protein in fat storage. To address this question, we expressed dLipin double-stranded RNA (dsRNA) in various tissues using the GAL4-UAS system.
Fig. 6.
dLipin promotes starvation resistance. (A) dLipin RNA is increased under starvation conditions. RNAs from normally fed and starved flies were quantitated by qRT-PCR. Asterisks indicate that the increase in the dLipin RNA level in starved flies over that in fed flies was statistically significant (***, P < 0.01). Quantitation of rp49 RNA was used for normalization. (B) RNAi-mediated knockdown of dLipin in the fat body leads to decreased starvation resistance. The survival of male and female flies of the indicated genotypes was monitored daily after food withdrawal. Flies were supplied with water only, except for flies fed with RU486 dissolved in 5% ethanol and control flies obtaining 5% ethanol alone (bottom graphs). The DJ761 driver is active in many tissues, including the fat body and the gut, whereas DJ649 is active in the gut and other tissues but not the fat body. The Lsp2 driver is active in the fat body only. Note that flies in the RU486 feeding experiment survived for an extended period, because they could use ethanol as an energy source (9). Survival curves were compared using the log rank test. The P values shown either are for the least significant difference in pairwise comparisons between RNAi and control animals or apply to all comparisons.
We first sought to knock down dLipin broadly in several tissues, including the major metabolic organs, the fat body and gut. The enhancer trap line P{GawB}DJ761 expresses GAL4 in a ubiquitous pattern in tissues of the adult fly with the exception of the nervous system (L. Seroude, personal communication to FlyBase, 2004). We found that this line shows the same expression pattern in larvae but does not cause lethality when used to drive the expression of dLipin dsRNA (in contrast to the ubiquitous tubulin driver [Fig. 1]). When DJ761 was used to drive the expression of dLipin dsRNA, both male and female flies showed significantly reduced survival under starvation conditions (Fig. 6). The median life span of male flies was reduced from 6 to 3 days and the maximum life span from 9 to 7 days. Females showed a less pronounced reduction in the median life span, from 5 to 3.5 days, but a dramatic decline in the maximum life span, from 11 to 5 days. Fed flies of the same genotype showed 100% survival for at least 31 days. We next asked whether exclusive knockdown of dLipin in the fat body and its knockdown outside the fat body have similar effects. The enhancer trap line P{GawB}DJ649 has been reported to show constant expression of GAL4 in aging adult flies (L. Seroude, personal communication to FlyBase, 2004). Again, we found that this line expresses GAL4 in larvae, showing strong activity in the larval and adult gut and salivary glands, the larval ring gland, and the ovaries. No GAL4 activity was observed in the fat body, either in larvae or in adults. In contrast, the 3.1Lsp2-Gal4 driver expresses GAL4 exclusively in the fat body during late larval (starting in the third larval instar) and adult development (17). Expression of dLipin dsRNA driven by DJ649 did not have a significant impact on starvation resistance (Fig. 6). In contrast, fat body-specific expression of dLipin dsRNA had a detrimental effect on starvation resistance, resulting in median and maximum life spans equal or very similar to those observed with the more broadly expressing DJ761 element (for males, the median life span was 3 days and the maximum life span was 7 days; for females, the median was 3.5 days and the maximum was 6 days). These results show that starvation resistance requires dLipin activity in the fat body. However, they provide no indication whether dLipin is needed in other tissues, such as the gut, to protect the animals from the effects of starvation.
We next asked whether a lack of dLipin also diminishes starvation resistance in adult flies that had experienced undisturbed dLipin activity and normal fat deposition until the time of food withdrawal. To address this question, we used an inducible system to knock down dLipin by RNAi simultaneously with food withdrawal. UAS-dLipin[RNAi] flies were crossed to the P{Switch1}106 line, which expresses GAL4 in the fat body and gut upon induction by the antiprogestin RU486 (25, 30). Both male and female RNAi-treated flies showed significantly reduced survival under starvation conditions (Fig. 6). The median life span of males was reduced from 21 to 15.5 days and the maximum life span from 25 to 17 days. Again, the effect on females was less pronounced, with a reduction of the median life span by 1 day and the maximum life span by 4 days. The effects on females became more apparent after prolonged starvation (≥26% less survival than the controls at day 20). These data indicate that even after a build-up of normal fat stores, increased activity of dLipin is beneficial for surviving periods of starvation.
DISCUSSION
The use of the simple genetic model organism Drosophila melanogaster holds the promise of providing new insights into basic metabolic control mechanisms that are relevant to human disease and obesity (2). Our study reinforces this notion by showing that the phenotype of Drosophila lipin mutants closely resembles the lipodystrophy phenotype observed in fld mice that carry a mutation in the lipin 1 gene. These mice are characterized by an early defect in adipocyte differentiation and a defect in TAG production later in development (22, 24).
Similarly, fat tissue cells in dLipin mutants do not develop normally and display an array of defects. Adipocytes in fld mice remain immature and accumulate smaller fat droplets than those in normal mice (22). A similar reduction in the size of fat droplets is observed for dLipin mutants (Fig. 4). While the mechanisms that control droplet size are not well understood, the availability of phosphatidylcholine, the major component of the phospholipid monolayer surrounding the TAG core of fat droplets, seems to be a critical factor (10, 36). Interestingly, studies of yeast indicate that lipins can negatively regulate the synthesis of phosphatidylcholine and other phospholipids by repressing key genes of the biosynthetic pathway (32). This suggests that the decreased size of lipid droplets in lipin mutants may be driven by an increased ratio of phospholipids to neutral lipids.
In contrast to adipose tissue cells of fld mice (22), fat body cells in dLipin mutants dramatically exceed normal fat body cells in size. Unlike mammalian adipocytes, fat body cells are polytene cells that increase their DNA copy number through endoreplication. The increase in nuclear size that accompanies the increase in cell size indicates that the mutant cells have undergone additional endoreplication cycles and are not the product of cell fusions (Fig. 3). The increased growth of the mutant cells may be a secondary effect of dLipin deficiency caused by a mechanism that strives to compensate for the reduced capacity of the fat body tissue to produce and store lipids. In contrast to the situation in C. elegans, where a lack of lipin leads to smaller adult worms (6), reduced fat body mass in dLipin mutants was not associated with a systemic cellular or organismal growth defect. Mutant salivary gland cells and larvae grew to normal size. Since larvae with reduced fat body mass grow more slowly than wild-type larvae (3), growth to normal size takes more time, explaining the developmental delay that we observed with the dLipin mutants.
Our TEM data indicate that a lack of dLipin leads not only to increases in the sizes of fat body cells and nuclei but also to ultrastructural changes within the cells. Cell nuclei, autophagosomes, and mitochondria showed changed morphology. Most autophagosomes of the mutant cells contained fewer cytoplasmic components than normal autophagosomes. LysoTracker staining of fat bodies from mutant early-third-instar larvae did not indicate that a premature onset of developmental autophagy, resulting in a depletion of cell organelles, might account for this observation. In mutant wandering larvae, LysoTracker staining indicated high autophagic activity, as it does in wild-type larvae at this stage (31; R. Ugrankar, unpublished data). While these observations suggest that developmental autophagy is normally initiated in the fat body in dLipin mutants, the TEM data indicate that the mechanism of autophagy might be affected in these animals. A defective mechanism may contribute to the observed increase in cell size.
Among the nuclear defects we observed were projections and involutions, and occasionally breaks, of the nuclear envelope. These observations are consistent with results obtained by light microscopy of DAPI-stained nuclei that revealed occasional nuclear fragmentation. While nuclear fragmentation is a cytological hallmark of apoptosis, caspase assays and suppression of apoptosis by the cell death inhibitor p35 did not support the conclusion that the cells had entered the apoptotic pathway. Irregular nuclear morphology has also been observed in lipin-deficient yeast and C. elegans cells. In C. elegans, lipin is required for nuclear envelope breakdown during mitosis (6, 7), and a lack of lipin in yeasts leads to overgrowth of the nuclear envelope (32, 34). While the mechanistic basis for the defects seen in C. elegans is not known, the evidence obtained with the yeast Saccharomyces cerevisiae supports a direct role of lipin in the transcriptional control of phospholipid biosynthesis. Increased nuclear growth in this species may be the result of an oversupply of phosphatidylcholine and phosphatidylethanolamine after attenuation of the repressive effect of lipin on genes of the biosynthetic pathway (32). Whether dLipin has a similar effect on phospholipid synthesis in Drosophila remains to be determined. However, the increase in the size and DNA content of mutant fat body cells suggests that overall cell growth, and not a simple increase in phospholipids, is the primary reason for the expansion of the nuclear envelope in this species.
Finally, the mitochondria show aberrant morphology, suggesting that these organelles do not function normally in the absence of dLipin. In the vertebrate liver, lipin 1 activates genes required for the β-oxidation of fatty acids and for ATP synthesis by oxidative phosphorylation. Both of these processes take place in the mitochondrial compartment. Mitochondria in the fat bodies of dLipin mutants are characterized by underdeveloped cristae, suggesting that components of the electron transport chain are in limited supply. This observation is consistent with the notion that dLipin in Drosophila has a role in mitochondrial function similar to that of lipin 1 in vertebrate liver. The lysis of mitochondria that we observed in dLipin mutant cells may be a secondary effect of these functional impairments. Mitochondrial dysfunction implies that a lack of dLipin not only affects fat storage but also has a broader impact on energy metabolism that may explain the high lethality we observed with the dLipin mutants.
Our data show that dLipin transcript levels are upregulated under starvation conditions and that reduced activity of the gene in the fat body enhances susceptibility to starvation. This does not apply only to reduced activity prior to starvation, as one would expect based on the role of the gene in fat storage. Downregulation of dLipin concomitant with food withdrawal equally leads to diminished starvation resistance. This effect is more pronounced in males than in females, consistent with the stronger starvation-induced upregulation of dLipin transcript levels in males (8, 12) (Fig. 6). Why does a gene that is essential for fat synthesis and storage enhance starvation resistance after the cessation of food intake? One possible explanation is more-efficient energy use through improved salvage of fatty acids released during lipolysis (by recycling to TAG). A similar role has been proposed for lipin 1 during starvation in mammalian adipose tissue (28). In mammalian liver, starvation leads to lipin 1 upregulation (5). There, lipin activates genes of the fatty acid β-oxidation pathway, bolstering the capacity of the liver to process the increasing supply of free fatty acids derived from adipose tissue (5). It remains to be seen whether dLipin has a similar role in fly tissues.
ACKNOWLEDGMENTS
We thank Matt Siebert for advice on TAG measurements, Sandra Goeke for help with electron microscopy, Christian K. Tipsmark for advice on real-time PCR, and Jonathan M. Graff, Brigitte Dauwalder, the Vienna Drosophila RNAi Center, the Bloomington Stock Center, and Harvard Medical School for fly stocks.
This work was supported by NSF grant IOS-0641347 and NIH grant 1R15DK079277-01.
Footnotes
Published ahead of print on 7 February 2011.
REFERENCES
- 1. Abdelwahid E., et al. 2007. Mitochondrial disruption in Drosophila apoptosis. Dev. Cell 12:793–806 [DOI] [PubMed] [Google Scholar]
- 2. Baker K. D., Thummel C. S. 2007. Diabetic larvae and obese flies—emerging studies of metabolism in Drosophila. Cell Metab. 6:257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Britton J. S., Lockwood W. K., Li L., Cohen S. M., Edgar B. A. 2002. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell 2:239–249 [DOI] [PubMed] [Google Scholar]
- 4. Donkor J., Sariahmetoglu M., Dewald J., Brindley D. N., Reue K. 2007. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 282:3450–3457 [DOI] [PubMed] [Google Scholar]
- 5. Finck B. N., et al. 2006. Lipin 1 is an inducible amplifier of the hepatic PGC-1α/PPARα regulatory pathway. Cell Metab. 4:199–210 [DOI] [PubMed] [Google Scholar]
- 6. Golden A., Liu J., Cohen-Fix O. 2009. Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J. Cell Sci. 122:1970–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gorjánácz M., Mattaj I. W. 2009. Lipin is required for efficient breakdown of the nuclear envelope in Caenorhabditis elegans. J. Cell Sci. 122:1963–1969 [DOI] [PubMed] [Google Scholar]
- 8. Grönke S., et al. 2005. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 1:323–330 [DOI] [PubMed] [Google Scholar]
- 9. Guarnieri D. J., Heberlein U. 2003. Drosophila melanogaster, a genetic model system for alcohol research. Int. Rev. Neurobiol. 54:199–228 [DOI] [PubMed] [Google Scholar]
- 10. Guo Y., et al. 2008. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453:657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Han G. S., Wu W. I., Carman G. M. 2006. The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281:9210–9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harbison S. T., Chang S., Kamdar K. P., Mackay T. F. 2005. Quantitative genomics of starvation stress resistance in Drosophila. Genome Biol. 6:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hay B. A., Wolff T., Rubin G. M. 1994. Expression of baculovirus P35 prevents cell death in Drosophila. Development 120:2121–2129 [DOI] [PubMed] [Google Scholar]
- 14. Jiang C., Baehrecke E. H., Thummel C. S. 1997. Steroid regulated programmed cell death during Drosophila metamorphosis. Development 124:4673–4683 [DOI] [PubMed] [Google Scholar]
- 15. Langner C. A., et al. 1989. The fatty liver dystrophy (fld) mutation. A new mutant mouse with a developmental abnormality in triglyceride metabolism and associated tissue-specific defects in lipoprotein lipase and hepatic lipase activities. J. Biol. Chem. 264:7994–8003 [PubMed] [Google Scholar]
- 16. Langner C. A., Birkenmeier E. H., Roth K. A., Bronson R. T., Gordon J. I. 1991. Characterization of the peripheral neuropathy in neonatal and adult mice that are homozygous for the fatty liver dystrophy (fld) mutation. J. Biol. Chem. 266:11955–11964 [PubMed] [Google Scholar]
- 17. Lazareva A. A., Roman G., Mattox W., Hardin P. E., Dauwalder B. 2007. A role for the adult fat body in Drosophila male courtship behavior. PLoS Genet. 3:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee G., Park J. H. 2004. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics 167:311–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lehmann M., Jiang C., Ip Y. T., Thummel C. S. 2002. AP-1, but not NF-κB, is required for efficient steroid-triggered cell death in Drosophila. Cell Death Differ. 9:581–590 [DOI] [PubMed] [Google Scholar]
- 20. Luong N., et al. 2006. Activated FOXO-mediated insulin resistance is blocked by reduction of TOR activity. Cell Metab. 4:133–142 [DOI] [PubMed] [Google Scholar]
- 21. Okamoto N., et al. 2009. A fat body-derived IGF-like peptide regulates postfeeding growth in Drosophila. Dev. Cell 17:885–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Péterfy M., Phan J., Xu P., Reue K. 2001. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 27:121–124 [DOI] [PubMed] [Google Scholar]
- 23. Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Phan J., Péterfy M., Reue K. 2005. Biphasic expression of lipin suggests dual roles in adipocyte development. Drug News Perspect. 18:5–11 [DOI] [PubMed] [Google Scholar]
- 25. Poirier L., Shane A., Zheng J., Seroude L. 2008. Characterization of the Drosophila gene-switch system in aging studies: a cautionary tale. Aging Cell 7:758–770 [DOI] [PubMed] [Google Scholar]
- 26. Redinger R. N. 2009. Fat storage and the biology of energy expenditure. Transl. Res. 154:52–60 [DOI] [PubMed] [Google Scholar]
- 27. Reue K. 2009. The lipin family: mutations and metabolism. Curr. Opin. Lipidol. 20:165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reue K., Brindley D. N. 2008. Glycerolipids. Multiple roles for lipins/phosphatidate phosphatase enzymes in lipid metabolism. J. Lipid Res. 49:2493–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reue K., Xu P., Wang X. P., Slavin B. G. 2000. Adipose tissue deficiency, glucose intolerance, and increased atherosclerosis result from mutation in the mouse fatty liver dystrophy (fld) gene. J. Lipid Res. 41:1067–1076 [PubMed] [Google Scholar]
- 30. Roman G., Endo K., Zong L., Davis R. L. 2001. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 98:12602–12607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rusten T. E., et al. 2004. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev. Cell 7:179–192 [DOI] [PubMed] [Google Scholar]
- 32. Santos-Rosa H., Leung J., Grimsey N., Peak-Chew S., Siniossoglou S. 2005. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24:1931–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Suh J. M., et al. 2007. Adipose is a conserved dosage-sensitive antiobesity gene. Cell Metab. 6:195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tange Y., Hirata A., Niwa O. 2002. An evolutionarily conserved fission yeast protein, Ned1, implicated in normal nuclear morphology and chromosome stability, interacts with Dis3, Pim1/RCC1 and an essential nucleoporin. J. Cell Sci. 115:4375–4385 [DOI] [PubMed] [Google Scholar]
- 35. Thibault S. T., et al. 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36:283–287 [DOI] [PubMed] [Google Scholar]
- 36. Thiele C., Spandl J. 2008. Cell biology of lipid droplets. Curr. Opin. Cell Biol. 20:378–385 [DOI] [PubMed] [Google Scholar]
- 37. von Gaudecker B. 1963. Über den Formwechsel einiger Zellorganelle bei der Bildung der Reservestoffe im Fettkörper von Drosophila-Larven. Z. Zellforsch. (Cell Tissue Res.). 61:56–95 [PubMed] [Google Scholar]
- 38. Yin V. P., Thummel C. S. 2004. A balance between the diap1 death inhibitor and reaper and hid death inducers controls steroid-triggered cell death in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 101:8022–8027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu S. Y., et al. 2002. A pathway of signals regulating effector and initiator caspases in the developing Drosophila eye. Development 129:3269–3278 [DOI] [PubMed] [Google Scholar]
- 40. Zeharia A., et al. 2008. Mutations in LPIN1 cause recurrent acute myoglobinuria in childhood. Am. J. Hum. Genet. 83:489–494 [DOI] [PMC free article] [PubMed] [Google Scholar]