Abstract
Many human genes exhibit evidence of initiated RNA polymerase II (Pol II) at their promoters, despite a lack of significant full-length transcript. Such genes exhibit promoter-proximal “pausing,” wherein initiated Pol II accumulates just downstream of the transcription start site due to a rate-limiting step mediating the transition to elongation. The mechanisms that regulate the escape of Pol II from pausing and the relationship to chromatin structure remain incompletely understood. Recently, we showed that CpG island hypermethylation and epigenetic silencing of TMS1/ASC in human breast cancers are accompanied by a local shift from histone H4 lysine 16 acetylation (H4K16Ac) to H4 lysine 20 trimethylation (H4K20me3). Here, we show that hMOF-mediated H4K16Ac and SUV420H2-mediated H4K20me3 play opposing roles in the regulation of Pol II pausing. We found that H4K16Ac promoted the release of Pol II from pausing through the recruitment of BRD4 and pTEFb. Aberrant methylation of CpG island DNA blocked Pol II recruitment to gene promoters. Whereas the inhibition of DNA methylation allowed for the reassociation and initiation of Pol II at the TMS1 promoter, Pol II remained paused in the presence of H4K20me3. Combined inhibition of H4K20me3 and DNA methylation resulted in the rerecruitment of hMOF and subsequent H4K16Ac, release of Pol II into active elongation, and synergistic reactivation of TMS1 expression. Marking by H4K20me3 was not restricted to TMS1 but also occurred at other genes independently of DNA methylation, where it similarly imposed a block to Pol II promoter escape through a mechanism that involved the local inhibition of H4K16Ac. These data indicate that H4K20me3 invokes gene repression by antagonizing hMOF-mediated H4K16Ac and suggest that overcoming Pol II pausing might be a rate-limiting step in achieving tumor suppressor gene reactivation in cancer therapy.
INTRODUCTION
Control of transcription of DNA by RNA polymerase II (Pol II) is regulated at multiple levels, including recruitment of Pol II to the promoter, initiation, elongation, and termination. General transcription factors guide the recruitment and loading of Pol II at gene promoters to form the preinitiation complex. Pol II is activated for transcription initiation by TFIIH-mediated phosphorylation of serine 5 in the heptapeptide repeats of the C-terminal domain (CTD) (58). The transition from initiation to elongation is also an important regulatory step. At some genes, Pol II is initiated and begins RNA synthesis but “pauses” 20 to 50 nucleotides downstream of the transcription start site. This has been referred to as Pol II promoter-proximal pausing (19, 42). Initially described for a few inducible genes, such as the Drosophila melanogaster heat shock genes and the c-myc, c-fos, and junB genes in human cells, promoter-proximal pausing of Pol II was thought to represent a mechanism for fast and robust gene activation in response to external stimuli (42). However, it is now evident that Pol II pausing may be more widespread. Genome-wide studies in Drosophila and human cells have shown that many genes have a disproportionate accumulation of Pol II at their 5′ end with little in the gene body (11, 22, 41, 68). Indeed, 20 to 30% of human genes exhibit evidence of Pol II initiation but lack significant full-length transcripts, suggesting that for many genes the transition to productive elongation is a rate-limiting step (11, 22).
The molecular mechanisms that regulate the establishment and release of Pol II from pausing remain incompletely understood. After initiation, the association of Pol II with the negative elongation factor (NELF) and DRB sensitivity-inducing factor (DSIF) inhibits elongation (63, 65). Release from pausing and the subsequent shift to productive elongation are marked by the phosphorylation of serine 2 in the CTD of Pol II by the pTEFb complex (7, 33). pTEFb also phosphorylates NELF and DSIF, resulting in the dissociation of NELF and the conversion of DSIF to a positive elongation regulator that remains associated with elongating Pol II (7, 61, 64). Thus, the mechanism and specificity of pTEFb recruitment play a key role in regulating the release from pausing.
Epigenetic modifications to the genome, including DNA methylation and posttranslational histone modifications, also govern gene activity, in part by modulating chromatin structure and affecting the accessibility of transcription factors and Pol II to promoters (60). The majority of CpG sites in the human genome, including those in intergenic regions, repetitive DNA sequences, and constitutive heterochromatin, are heavily methylated. These regions are typically marked by repressive histone marks, including histone H3 lysine 9 trimethylation (H3K9me3), H3 lysine 27 trimethylation (H3K27me3), and/or H4 lysine 20 trimethylation (H4K20me3). In contrast, CpG-rich regions (termed CpG islands) encompassing the promoters of approximately 70% of human genes are generally unmethylated (4, 27). These regions have a high transcriptional potential and are associated with a permissive chromatin state enriched in histone H3 and H4 acetylation (H3Ac and H4Ac) and H3 lysine 4 di- and trimethylation (H3K4me2 and H3K4me3). A large body of evidence now supports an interdependent relationship between the mechanisms that direct and maintain DNA methylation and chromatin-mediated repression, and the two play an important role in restricting the transcriptional potential of the genome (36).
The epigenome undergoes both global and gene-specific distortions in human cancers. Hypomethylation of DNA and genome-wide loss of repressive marks such as H3K9me2 and H4K20me3 from bulk chromatin are observed in human cancers. This results in a generalized disruption of heterochromatin structure and transcriptional repression and contributes to tumorigenesis through increased chromosomal instability, derepression of endogenous retrotransposons, and ectopic expression of normally silent microRNAs (miRNAs). At the same time, promoter-associated CpG islands can become heavily methylated. This is accompanied by local changes in chromatin structure, including loss of nucleosome positioning, and a shift in the histone modification profile, where active marks such as H3/4Ac and H3K4 methylation are replaced by H3K9 and/or H3K27 methylation. Such hypermethylation of CpG islands leads to the stable and heritable repression of tumor suppressor genes and is one mechanism contributing to the initiation and progression of human cancers (36).
Recently, we showed that CpG island-associated alterations in histone H4 modifications also contribute to the epigenetic silencing of certain tumor suppressor genes in cancers (29). TMS1 (target of methylation-induced silencing) is a CpG island-associated gene that becomes aberrantly methylated and silenced in human breast and other cancers (8, 10, 37, 54). TMS1 plays an important role in proinflammatory and proapoptotic signaling, and its silencing confers resistance to anoikis in breast epithelial cells (34, 45, 46). We found that the aberrant methylation and epigenetic silencing of TMS1 in breast cancer cells are accompanied by a loss of histone H4 lysine 16 acetylation (H4K16Ac) and the acquisition of H4K20me3 and that hMOF-mediated H4K16Ac is important for the maintenance of gene activity at the locus (29). However, the precise mechanism by which H4K16Ac regulates gene expression and the role, if any, of H4K20me3 in gene repression remained unknown.
In the present study, we demonstrate that the epigenetic switch between H4K16Ac and H4K20me3 plays an important role in gene regulation. Specifically, we show that hMOF-mediated acetylation of H4K16 promotes the release of Pol II from promoter-proximal pausing and the transition from initiation into active elongation. This is mediated by the H4K16Ac-dependent recruitment of the BRD4/pTEFb complex. CpG island methylation prevents Pol II from associating with the TMS1 promoter; however, even in its absence, gene repression is maintained by SUV420H2-mediated H4K20me3, which blocks hMOF recruitment and subsequent H4K16Ac, enforcing Pol II pausing. The blockade to H4K16Ac imposed by SUV420H2 and/or H4K20me3 and subsequent enforcement of Pol II pausing was also observed at other genes independently of DNA methylation. These results establish an important role for SUV420H2 and H4K20me3 in the negative regulation of Pol II promoter escape and implicate Pol II pausing as an important step in the hierarchy of events that lead to the epigenetic silencing of some genes in cancers.
MATERIALS AND METHODS
Cell lines.
MCF7 and MDA-MB231 breast cancer cell lines were obtained from the American Type Culture Collection and maintained in Dulbecco modified Eagle medium (DMEM) containing 2 mmol/liter glutamine and 10% fetal bovine serum. Puromycin was used at a final concentration of 1.0 μg/ml, and 5-aza-2′-deoxycytidine (DAC) was used at a concentration of 0.5 μM.
Lentiviral shRNA infection and DAC treatment.
The empty pLKO.1 plasmid and the pLKO.1 plasmid expressing short hairpin RNA (shRNA) against hMOF (TRCN0000034875) or SUV420H2 (TRCN0000141936) were obtained from Open Biosystems. Lentiviral particles containing the pLKO.1 plasmids were produced in 293T cells as described by Kapoor-Vazirani et al. (29). For infection, 1 × 106 to 2 × 106 MCF7 or MDA-MB231 cells were plated on 10-cm plates. The following day, cells were infected with 0.2 to 0.5 ml of lentiviral particles in the presence of 8 μg/ml Polybrene. Twenty-four hours after infection, cells were replenished with fresh medium containing puromycin with or without 0.5 μM DAC. Fresh medium containing puromycin (and DAC when used) was exchanged every other day. Cells were incubated in the selective medium for 3 (MCF7 cells) or 4 (MDA-MB231 cells) days and subsequently harvested for downstream assays.
Western analysis.
Cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1× protease inhibitors) on ice for 10 min, and lysates were clarified by centrifugation. Clarified protein lysates (100 μg) were electrophoresed on a 12% SDS-PAGE gel and transferred onto a polyvinylidene difluoride (PVDF) membrane. The membrane was probed with the following primary antibodies: hMOF (gift from Edwin Smith, Emory University), TMS1 (Protein Tech; catalog no. TTG10500-1-AP), NELF-E (Santa Cruz; catalog no. 32912), CDK9 (Santa Cruz; catalog no. 484), BRD4 (Santa Cruz; catalog no.48772), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Abcam; catalog no. 8245), CDH1 (Santa Cruz; catalog no. 8426), ESR1 (Santa Cruz; catalog no. 7207), and SUV420H2 (Abcam; catalog no. 18186).
ChIP and real-time PCR.
Chromatin immunoprecipitation (ChIP) with antibodies against histone H4 and histone modifications was done essentially as described in the acetyl-histone H3 immunoprecipitation assay kit from Millipore (catalog no. 17-229). SUV420H2 and hMOF ChIPs were performed as described in the acetyl-histone H3 immunoprecipitation assay kit except that cells were resuspended at 1 × 106 cells/100 μl lysis buffer and then diluted 1/10 with 0.5× dilution buffer prior to immunoprecipitation.
ChIP with antibodies against Pol II, NELF-E, CDK9, and BRD4 was adapted from a study by Adelman et al. (1). Briefly, cells were cross-linked in medium containing 1% formaldehyde for 10 min at room temperature and quenched with 125 mM glycine. Cells were then lysed in sonication buffer (0.5% SDS, 20 mM Tris-HCl, pH 8.0, 2 mM EDTA, 0.5 mM EGTA, 10 mM β-glycerophosphate, 1 mM Na3VO4, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 1× protease inhibitor), sonicated to an average fragment size of 500 bp, and centrifuged to remove cellular debris. For each pulldown, 5 × 106 cells (in 50 μl) were diluted 20-fold with IP buffer (0.5% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10% glycerol, 1× protease inhibitor). The samples were precleared with 30 μl blocked 50% protein A-agarose bead slurry (Millipore; catalog no. 16-125) and incubated overnight in 250 μl IP buffer plus the antibody of interest at 4°C. Immunocomplexes were collected by incubation with 100 μl blocked protein A-agarose beads for 1 h at 4°C. Complexes were subsequently washed, and genomic DNA was eluted and precipitated as described in the acetyl-histone H3 immunoprecipitation assay kit.
For all ChIPs, a portion of the sonicated samples was used to elute and precipitate DNA for input DNA and to check sonication efficiency. DNA immunoprecipitated from the ChIP assays was analyzed by real-time PCR as previously described using primers across the TMS1, ESR1, and CDH1 loci (29). Primers for the JUND and MORC3 loci are available upon request. Antibodies used were as follows: rabbit IgG (Santa Cruz; catalog no. 2027), pan-histone H4 (Millipore; catalog no. 05-858), total Pol II (Santa Cruz; catalog no. 9001X), Pol II phosphorylated at serine 5 (Abcam; catalog no. 5131), Pol II phosphorylated at serine 2 (Abcam; catalog no. 5095), NELF-E, CDK9, BRD4, SUV420H2, hMOF, H3Ac (Millipore; catalog no. 06-599), H3K4me2 (Millipore; catalog no. 07-030), H3K9me2 (Abcam; catalog no. 7312), H4K20me3 (Abcam; catalog no. 9053), and H4K16Ac (Abcam; catalog no. 1762).
Real-time reverse transcription-PCR.
Total RNA was extracted from cells and treated by in-column DNase I digestion using the RNeasy minikit (Qiagen). RNA (3 μg) was reverse transcribed using random hexamer primers and Moloney murine leukemia virus reverse transcriptase. cDNA was amplified with primers against TMS1, 18S rRNA, hMOF, SUV420H2, JUND, or MORC3 using real-time PCR. Starting RNA quantities were determined relative to a common standard curve generated using MCF7 cDNA. Sequences of the real-time primers used for TMS1 and 18S rRNA are outlined by Parsons and Vertino (46). All other primer sequences are available upon request.
MSP.
Methylation-specific PCR (MSP) was performed as described by Stimson and Vertino (53). Primers used for MSP overlap a total of 6 CpGs in the TMS1 CpG island, and their sequences have been previously published (10).
RESULTS
H4K16 acetylation by hMOF promotes release of Pol II from promoter-proximal pausing.
In previous work, we demonstrated that epigenetic silencing of TMS1 in human cancer cells is associated with DNA hypermethylation of the promoter-associated CpG island and a shift in histone H4 modification from H4K16Ac to H4K20me3 (10, 29). We further showed that acetylation of H4K16, mediated by the acetyltransferase hMOF and the hMSL complex, is important for the maintenance of TMS1 gene expression (29). However, the mechanism by which H4K16Ac promotes gene expression and the role, if any, of H4K20me3 in gene repression were not known.
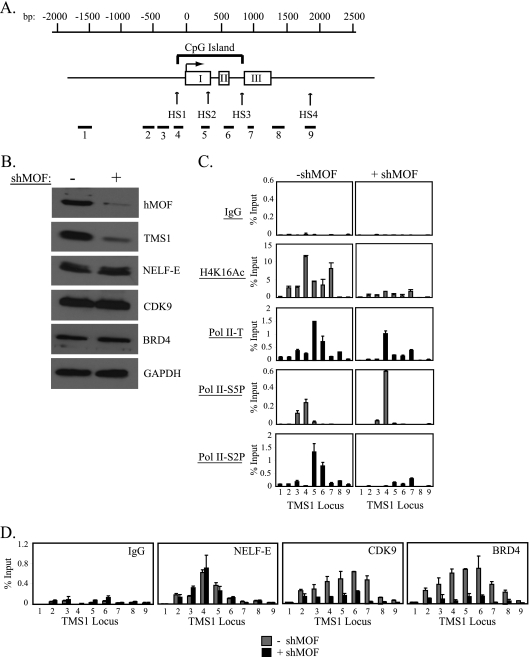
The TMS1 gene consists of 3 exons, and the locus has a CpG island that spans the promoter region and exons I and II (Fig. 1 A). To determine the mechanism by which H4K16Ac modulates gene activity, we examined the localization of Pol II at the TMS1 locus in MCF7 breast cancer cells expressing or silenced for hMOF by ChIP. As shown previously, TMS1 was associated with H4K16Ac in MCF7 cells where the gene is expressed, and downregulation of hMOF led to a decrease in H4K16Ac and silencing of TMS1 expression (29) (Fig. 1B and C). hMOF downregulation did not significantly affect the levels or distribution of total histone H4 (data not shown), indicating that the profile of H4K16Ac and decrease in H4K16Ac observed upon hMOF knockdown were due not to changes in nucleosome occupancy but to a decrease in the levels of acetylation itself. Interestingly, the silencing of TMS1 in the absence of hMOF was due not to a loss of Pol II recruitment but rather to a redistribution of Pol II occupancy. Whereas Pol II was distributed across the TMS1 locus in the presence of hMOF, it was selectively enriched at the promoter region in the absence of hMOF (Fig. 1C). We further examined the state of Pol II phosphorylation and found that whereas the initiated form of Pol II was present at the promoter region, the elongating form of Pol II was localized across the TMS1 gene body in cells expressing hMOF. However, when hMOF was downregulated, there was an accumulation of the initiated form and loss of the elongating form of Pol II (Fig. 1C). This effect appeared to be gene specific rather than a global effect of hMOF on gene expression, as downregulation of hMOF had no effect on gene activity or Pol II dynamics at two genes, ESR1 and CDH1, which, like TMS1, are unmethylated and expressed in MCF7 cells but lack H4K16Ac (Fig. 2 A to C and data not shown) (29). These data have two important implications. The finding that initiated Pol II accumulates at the promoter region in the absence of hMOF indicates, first, that TMS1 (but not ESR1 or CDH1) is regulated, in part, by Pol II promoter-proximal pausing and, second, that hMOF and/or H4K16Ac regulates pausing by promoting the release of Pol II from initiation into active elongation.
Fig. 1.
H4K16Ac promotes the release of Pol II from promoter-proximal pausing. (A) Shown is a schematic of the TMS1 genomic locus. The TMS1 gene consists of 3 exons (white boxes, I to III). A CpG island encompasses the promoter region and exons I and II. The transcription start site (horizontal arrow) and DNase I-hypersensitive sites (HS1 to HS4) are shown. Nucleotide positions (bp) are depicted with respect to the transcription start site. Primer sets (1 to 9) used in ChIP/real-time PCR assays are shown. Note that primer set 4 is located within the promoter region and is closest to the transcription start site. (B) MCF7 cells were infected with the pLKO.1 lentiviral vector (control) or pLKO.1 lentiviral vector expressing shRNA against hMOF (shMOF). Four days postinfection, protein lysates were isolated. Lysates (100 μg) were subjected to SDS-PAGE, transferred to a PVDF membrane, and probed with the indicated antibodies. (C and D) Cells treated as in panel B were subjected to ChIP with the indicated antibodies. DNA eluted after immunoprecipitation was amplified by real-time PCR with primer sets (1 to 9, shown in panel A) that span the TMS1 locus. Immunoprecipitated DNA is presented as a percentage of input DNA (% Input). Experiments were repeated 2 to 5 times with reproducible results. Shown is the mean ± standard deviation of triplicate determinations from a representative experiment. Pol II-T, total Pol II; Pol II-S5P, the initiated Pol II phosphorylated at serine 5; Pol II-S2P, the elongating form of Pol II phosphorylated at serine 2.
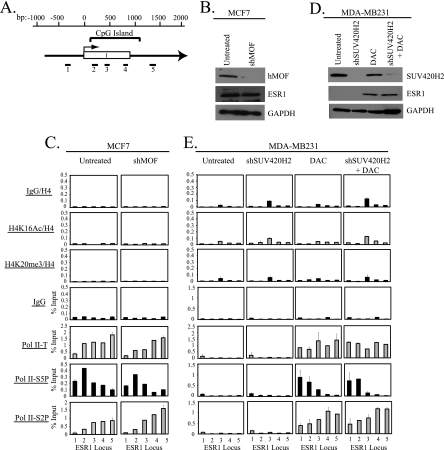
Fig. 2.
Downregulation of hMOF or SUV420H2 has no effect on gene activity or Pol II distribution across the ESR1 locus. (A) Schematic of the region encompassing the CpG island of the ESR1 gene. Nucleotide positions (bp) with respect to the transcription start site (horizontal arrow), the CpG island, and exon I (white box) are shown. The regions amplified by primer sets (1 to 5) used in real-time PCR are shown below the gene diagram. (B) Protein lysates (100 μg) from MCF7 cells infected with an empty lentiviral vector (Untreated) or a vector expressing shRNA against hMOF (shMOF) were probed with the indicated antibodies. GAPDH served as a loading control. (C) Cells from panel B were used for ChIP with the antibodies outlined, and immunoprecipitated DNA was amplified by real-time PCR using primer sets depicted in panel A. DNA obtained from each ChIP is shown normalized to DNA from total histone H4 ChIP or expressed as a percentage of input DNA. Each ChIP was done twice with reproducible profiles. Shown is the mean ± standard deviation of triplicate determinations from a representative experiment. (D) MDA-MB231 cells infected with an empty lentiviral vector (Untreated and DAC) or a lentiviral vector expressing shRNA against SUV420H2 (shSUV420H2 and shSUV420H2 + DAC) were treated with 0.5 μM DAC where indicated. Five days postinfection (4 days after DAC treatment), cells were harvested for Western blot analysis with the indicated antibodies. (E) Cells from panel D were used for ChIP with the indicated antibodies. Immunoprecipitated DNA was quantified by real-time PCR with primer sets that span the ESR1 CpG island and is shown normalized to DNA from histone H4 ChIP or as a percentage of input DNA. Each ChIP was performed twice and gave similar results each time. Shown is the mean ± standard deviation of triplicate determinations of a representative experiment.
To understand the mechanism by which H4K16Ac promotes the release of paused Pol II, we next examined the localization of NELF at the TMS1 locus in the presence and absence of hMOF. ChIP experiments using an antibody specific to the NELF-E subunit showed that NELF was present at the TMS1 promoter region in TMS1-expressing MCF7 cells (Fig. 1D). Enrichment of NELF at TMS1 was unaffected by hMOF downregulation, indicating that NELF recruitment to the TMS1 locus is not dependent on H4K16Ac.
pTEFb, which is composed of the kinase CDK9 and cyclin T/K, is responsible for the release of paused Pol II into productive elongation, in part by phosphorylating serine 2 at the Pol II CTD (7, 33). At some genes, recruitment of pTEFb to chromatin is mediated by the BRD4 protein, a member of the BET family of proteins that is characterized by two tandem bromodomains that bind acetylated H3 and H4 (13, 25). Since BRD4 associates with chromatin via acetylated histones, we asked whether H4K16Ac regulates promoter-proximal pausing at the TMS1 locus by affecting the localization of BRD4 and pTEFb. To this end, we used antibodies against BRD4 and the CDK9 subunit of pTEFb in ChIP analyses in MCF7 cells expressing or silenced for hMOF. CDK9 and BRD4 were both localized across the TMS1 locus in MCF7 cells (Fig. 1D). Downregulation of hMOF led to a decrease in CDK9 and BRD4 recruitment to TMS1 without affecting the global levels of these proteins, indicating that hMOF and/or H4K16Ac is required for the binding of pTEFb and BRD4 to TMS1 chromatin (Fig. 1B and D). Taken together, the data suggest that hMOF/H4K16Ac promotes gene expression by providing a platform for pTEFb and BRD4 binding, which in turn promotes the phosphorylation of serine 2 on Pol II and the transition from initiation to elongation at the pause point.
SUV420H2-mediated H4K20me3 negatively regulates TMS1 expression.
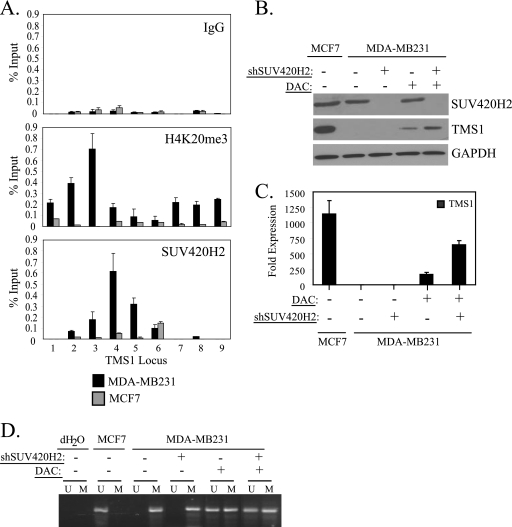
In MDA-MB231 breast cancer cells, the TMS1 CpG island is densely methylated and the locus is silent. It is also associated with H4K20me3, which peaks just upstream of the transcription start site (29) (Fig. 3 A). Human cells encode two methyltransferases, SUV420H1 and SUV420H2, capable of trimethylating H4K20 (50). Decreased expression of SUV420H2 but not SUV420H1 is associated with a global loss of H4K20me3 in mouse embryonic fibroblasts and human cancer cells, suggesting that SUV420H2 is responsible for the majority of H4K20me3 in vivo (2, 49, 59). To determine whether SUV420H2 is responsible for H4K20me3 at the TMS1 locus, we examined its localization in MCF7 and MDA-MB231 cells. Indeed, SUV420H2 was detected at TMS1 in MDA-MB231 cells, where the locus is enriched for H4K20me3, but was absent from the TMS1 locus in MCF7 cells, which lack H4K20me3 (Fig. 3A).
Fig. 3.
SUV420H2-mediated H4K20me3 promotes TMS1 gene repression in the absence of DNA methylation. (A) MCF7 and MDA-MB231 cells were subjected to ChIP analysis with antibodies that recognize either H4K20me3 or the SUV420H2 methyltransferase. ChIP with an irrelevant antibody (IgG) was included as a control. Immunoprecipitated DNA was amplified by real-time PCR and is presented as a percentage of input DNA. Each experiment was repeated 3 times with similar results. Columns show means of triplicate determinations from a representative experiment; error bars show standard deviations. (B to D) MDA-MB231 cells infected with an empty lentiviral vector (−) or a lentiviral vector expressing shRNA against SUV420H2 (shSUV420H2) were left untreated or concurrently exposed to 0.5 μM DAC. Cells were harvested 5 days postinfection (4 days after DAC treatment). (B) Protein lysates (100 μg) were subjected to Western blot analysis using antibodies against SUV420H2, TMS1, or GAPDH (loading control). (C) TMS1 mRNA expression was determined by reverse transcription followed by real-time PCR. TMS1 expression levels are expressed relative to that of untreated MDA-MB231 cells, after normalization to 18S rRNA. Shown is the mean ± standard deviation of results from two independent experiments. (D) Genomic DNA was modified with sodium bisulfite and subjected to methylation-specific PCR (MSP) with TMS1 CpG island primer sets specific for either the methylated (M) or the unmethylated (U) DNA. Untreated MCF7 cells were also included as a control. For MSP, a reaction mixture in which H2O was substituted for template DNA (dH2O) was also included as a negative control for PCR.
To address the role of H4K20me3 at the silenced TMS1 locus, we next downregulated SUV420H2 in MDA-MB231 cells using a lentiviral shRNA approach. Although SUV420H2 was efficiently silenced by this method and it led to both a global and a gene-specific decrease in H4K20me3 at the TMS1 locus (Fig. 3B and 4 A and data not shown), there was no change in TMS1 expression, at either the protein level or the RNA level (Fig. 3B and C). Methylation of the TMS1 CpG island was unaffected by SUV420H2 downregulation (Fig. 3D), and MDA-MB231 cells remained silent for TMS1 expression despite the absence of SUV420H2 and H4K20me3.
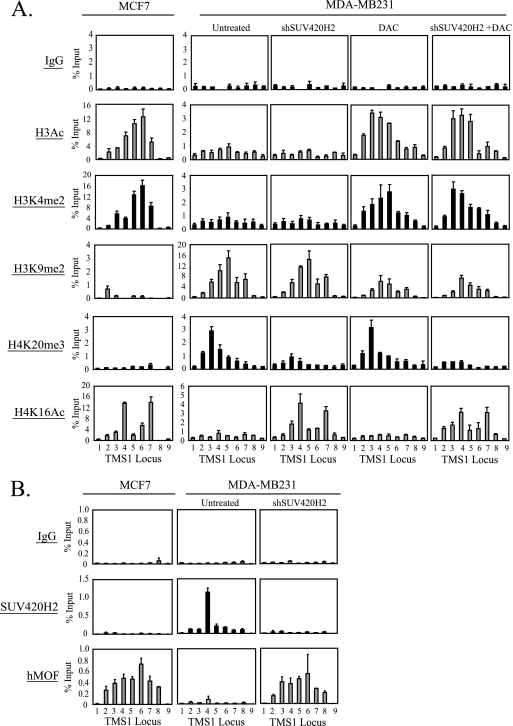
Fig. 4.
Effect of SUV420H2 downregulation and/or DNA demethylation on histone modification profiles and hMOF recruitment at the TMS1 locus. MDA-MB231 cells infected with an empty lentiviral vector (Untreated and DAC) or a lentiviral vector expressing shRNA against SUV420H2 (shSUV420H2 and shSUV420H2 + DAC) were left untreated or concurrently exposed to 0.5 μM DAC. Cells were harvested 5 days postinfection (4 days post-DAC treatment) and subjected to ChIP with the indicated antibodies. MCF7 cells were included for comparison. Immunoprecipitated DNA was determined by real-time PCR and is expressed as a percentage of input DNA. Each ChIP was performed at least 3 times with reproducible profiles. Shown is the mean ± standard deviation of triplicate determinations from a representative experiment.
The above data suggest that DNA methylation may be dominant over H4K20me3 in TMS1 silencing. To test this hypothesis, we concomitantly treated MDA-MB231 cells with the DNA methyltransferase inhibitor DAC (0.5 μM; 4 days) in the presence or absence of SUV420H2 shRNA. We have previously shown that treatment with DAC leads to the demethylation of the TMS1 CpG island, resulting in a low level of gene reactivation (31). MDA-MB231 cells treated with DAC and SUV420H2 shRNA exhibited a degree of demethylation at TMS1 similar to that of those treated with DAC alone (Fig. 3D). However, treatment with DAC and SUV420H2 shRNA resulted in synergistic reactivation of TMS1 at the protein and RNA level that was 3-fold higher than that achieved by DAC treatment alone (Fig. 3B and C). These findings indicate that in the absence of DNA methylation, SUV420H2 and/or H4K20me3 plays an important role in transcriptional repression. However, the superimposition of DNA methylation allows for the maintenance of transcriptional repression even in the absence of H4K20me3.
Effect of DNA demethylation and/or SUV420H2 downregulation on histone modifications and hMOF recruitment at the TMS1 locus.
We next asked whether downregulation of SUV420H2 and/or DAC treatment was associated with alterations in the histone modification profiles across the TMS1 locus. As previously observed, the DNA-methylated TMS1 locus in MDA-MB231 cells was marked by H3K9me2 and H4K20me3 and lacked the “active” marks H3Ac, H3K4me2, and H4K16Ac (29) (Fig. 4A). Downregulation of SUV420H2 in these cells led to the loss of SUV420H2 and a corresponding decrease in H4K20me3 levels at TMS1 (Fig. 4A and B). Interestingly, the depletion of H4K20me3 was associated with the reassociation of hMOF and a concomitant increase in H4K16Ac levels at the TMS1 locus but had little effect on the other histone modifications analyzed (H3Ac, H3K4me2, and H3K9me2) (Fig. 4A and B). On the other hand, treatment of MDA-MB231 cells with DAC led to a depletion of H3K9me2 and a corresponding increase in H3Ac and H3K4me2 without affecting H4K16Ac and H4K20me3 (Fig. 4A). Treatment of MDA-MB231 cells with both SUV420H2 shRNA and DAC led to an overall additive effect in that the active histone marks (H3Ac, H3K4me2, and H4K16Ac) were enriched and the inactive marks (H3K9me2 and H4K20me3) were depleted and resembled the pattern observed in MCF7 cells in which the TMS1 gene is unmethylated and actively expressed (Fig. 4A). The changes in histone modifications observed were specific and not due to changes in nucleosome occupancy, as neither the levels nor the distribution of total histone H4 was significantly affected by SUV420H2 shRNA or DAC treatment (data not shown).
These observations lead to several important conclusions. First, these data indicate that H4K20me3 and H4K16Ac are interdependent but are maintained independently of DNA methylation. The finding that hMOF associates with the TMS1 locus only in the absence of H4K20me3 indicates that H4K20me3 and/or SUV420H2 itself blocks the binding of hMOF and the subsequent acetylation of H4K16. Second, the data suggest that whereas H3Ac, H3K4me2, and H3K9me2 are independent of alterations in H4K16Ac and H4K20me3, they are dependent on the DNA methylation status. Indeed, DNA methylation is required for the maintenance of H3K9me2 at TMS1, and the two directly or indirectly block H3K4 methylation. Lastly, the results demonstrate that inhibition of both SUV420H2 and DNA methylation in MDA-MB231 cells is necessary for the reacquisition of an active TMS1 chromatin state (such as that seen in MCF7 cells) and to achieve maximal TMS1 reactivation.
hMOF-mediated upregulation of H4K16Ac subsequent to the downregulation of H4K20me3 is necessary for TMS1 gene upregulation.
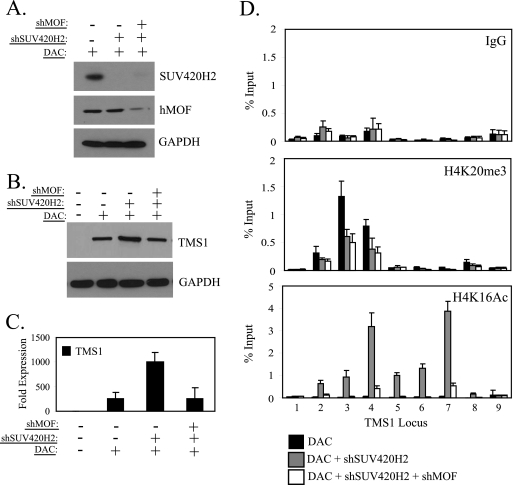
Our results thus far demonstrate that in the absence of DNA methylation, SUV420H2 downregulation enhances TMS1 expression to levels beyond that achieved by DNA demethylation alone. SUV420H2 downregulation also leads to a decrease in H4K20me3 and an increase in H4K16Ac. To determine whether it is the downregulation of H4K20me3 or the subsequent upregulation of H4K16Ac that is critical for the enhancement of gene activity, we treated MDA-MB231 cells with DAC and SUV420H2 shRNA in the presence or absence of shRNA against hMOF. Both SUV420H2 and hMOF were efficiently downregulated by this procedure (Fig. 5 A). As noted above, TMS1 was reactivated to low levels after treatment of MDA-MB231 cells with DAC and this reactivation was further enhanced upon silencing of SUV420H2 (Fig. 5B and C). The synergistic upregulation of TMS1 expression was linked to the depletion of H4K20me3 and the reacquisition of H4K16Ac (Fig. 5D). However, concurrent downregulation of hMOF abolished the synergistic effect on gene reexpression and blocked the reacquisition of H4K16Ac despite the loss of H4K20me3 (Fig. 5B to D). Thus, we conclude that the reassociation of hMOF and the accumulation of H4K16Ac subsequent to the loss of H4K20me3 are critical for the enhanced TMS1 gene activity observed in response to SUV420H2 downregulation.
Fig. 5.
H4K20me3 downregulation requires subsequent H4K16 acetylation for TMS1 upregulation. (A) MDA-MB231 cells infected with an empty lentiviral vector, a vector expressing shRNA against SUV420H2, or vectors expressing shRNA against SUV420H2 and hMOF were treated with 0.5 μM DAC. Protein lysates (100 μg) were collected 5 days postinfection and subjected to Western blot analysis with antibodies specific for SUV420H2, hMOF, or GAPDH (loading control). (B and C) MDA-MB231 cells infected with an empty vector control, a vector expressing shRNA against SUV420H2, or vectors expressing shRNA against SUV420H2 and hMOF were exposed to 0.5 μM DAC where indicated. (B) Protein lysates (100 μg) were collected and subjected to Western blot analysis using antibodies against TMS1 or GAPDH (loading control). (C) Total RNA (3 μg) was converted to cDNA using reverse transcriptase, and the cDNA was used as template in real-time PCR with TMS1- and 18S rRNA-specific primers. TMS1 expression is expressed relative to the levels in untreated MDA-MB231 cells after normalization to 18S rRNA levels. Shown is the mean ± standard deviation of two independent experiments. (D) MDA-MB231 cells treated as in panel A were subjected to ChIP analysis using antibodies against H4K20me3 or H4K16Ac, and immunoprecipitated DNA was subjected to real-time PCR using primer sets 1 to 9 from the TMS1 locus. IgG was included as an irrelevant control. Immunoprecipitated DNA is presented as a percentage of input DNA. Each experiment was conducted at least twice with similar results. Shown is the mean ± standard deviation of triplicate determinations from a representative experiment.
SUV420H2-mediated H4K20me3 enforces Pol II promoter-proximal pausing.
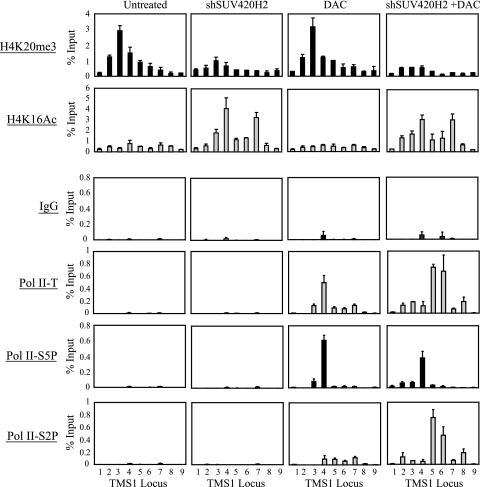
Given that the presence of SUV420H2 and/or H4K20me3 blocks hMOF recruitment and acetylation of H4K16 and that hMOF and/or H4K16Ac plays an essential role in the release of Pol II from pausing at the TMS1 promoter, we next examined the effect of SUV420H2 silencing on Pol II dynamics. Pol II was not associated with the TMS1 locus in MDA-MB231 cells (Fig. 6), where the CpG island is methylated and the gene is silent (Fig. 3B to D). Downregulation of SUV420H2, which had no effect on TMS1 expression in the presence of DNA methylation (Fig. 3B to D), also had no effect on Pol II recruitment (Fig. 6). Treatment with DAC, on the other hand, allowed for the reassociation of Pol II with the demethylated locus (Fig. 6). However, the majority accumulated around the promoter region and was enriched in the initiated form while only low levels of the elongating form were detected (Fig. 6), consistent with Pol II pausing and the low level of TMS1 expression observed in these cells (Fig. 3B and C). In MDA-MB231 cells simultaneously treated with DAC and downregulated for SUV420H2, Pol II was distributed throughout the TMS1 locus, with the initiated and the elongating forms enriched at the promoter region and the gene body, respectively (Fig. 6). This correlates with the enhanced level of TMS1 expression seen in this case (Fig. 3B and C).
Fig. 6.
SUV420H2-mediated H4K20me3 enforces Pol II promoter-proximal pausing. MDA-MB231 cells infected with an empty lentiviral vector (Untreated and DAC) or a lentiviral vector expressing shRNA against SUV420H2 (shSUV420H2 and shSUV420H2 + DAC) were left untreated or concurrently exposed to 0.5 μM DAC. Cells were harvested 5 days postinfection (4 days post-DAC treatment) and subjected to ChIP with the indicated antibodies. IgG served as an irrelevant antibody control. Immunoprecipitated DNA was quantified using real-time PCR and is presented as a percentage of input DNA. Shown is the mean ± standard deviation from a representative experiment. Each experiment was repeated at least twice with reproducible results.
ESR1 and CDH1 are also silenced by CpG island hypermethylation in MDA-MB231 cells and can be reactivated by DAC treatment, but neither is marked by H4K20me3 in this cell line (21, 44) (Fig. 2D and E and data not shown). As was seen at TMS1, Pol II was not associated with either locus in the presence of DNA methylation (Fig. 2E and data not shown). However, whereas DAC-induced demethylation allowed for reassociation of Pol II and upregulation of gene expression in both cases, there was no evidence for Pol II promoter-proximal pausing, and concurrent inhibition of SUV420H2 had no further impact on H4K16Ac, gene expression, or Pol II distribution (i.e., the relative enrichment of initiated Pol II at the transcription start site and the elongating form distributed into the gene body were unaffected) (Fig. 2D and E and data not shown).
Taken together, these data indicate that DNA methylation contributes to gene repression by precluding Pol II association with CpG island chromatin. In the absence of DNA methylation, repression at the TMS1 locus is maintained by SUV420H2 and/or H4K20me3, which has no effect on Pol II recruitment but rather enforces the pausing of initiated Pol II at the promoter by impeding the transition to productive elongation. In this case, DNA methylation/H3K9me2 and H4K20me3 appear to act as independent “layers” to maintain gene repression. In contrast, other methylated genes (e.g., ESR1 and CDH1) that lack H4K20me3 do not exhibit Pol II pausing upon DNA demethylation.
Regulation of Pol II promoter-proximal pausing by H4K16Ac and H4K20me3 is a general mechanism that also pertains to other genes.
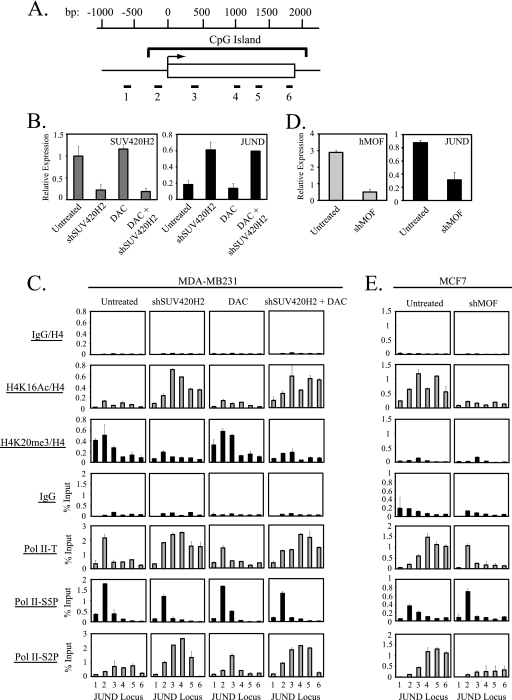
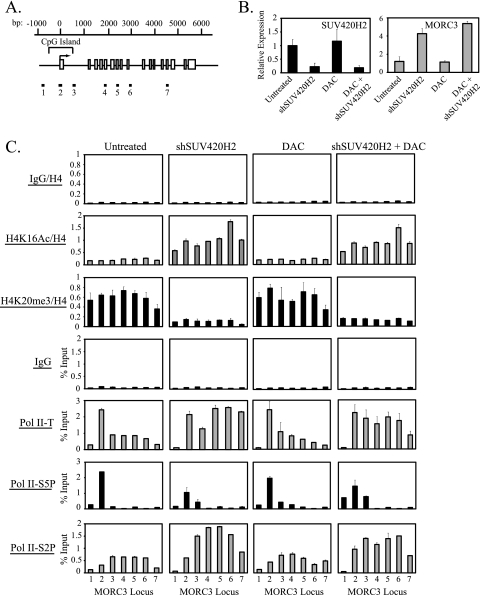
Recent work indicates that H4K20me3 also accumulates near the promoter regions of other genes (9). To determine whether the interplay between hMOF-mediated H4K16Ac and SUV420H2-mediated H4K20me3 in the regulation of Pol II pausing was specific to TMS1 or a more general mechanism for regulating Pol II pausing, we examined the impact of SUV420H2 downregulation on histone H4 modifications and Pol II distribution at two additional genes, JUND and MORC3, which are marked by H4K20me3 and expressed to some extent in MDA-MB231 cells (Fig. 7 and 8). JUND is an intronless gene entirely encompassed by a CpG island, and MORC3 consists of 17 exons with a CpG island that spans the transcription start site and exon 1 (Fig. 7A and 8A). In MDA-MB231 cells, in the presence of H4K20me3, initiated Pol II accumulated or paused at the transcription start site of both genes with low levels of elongating Pol II observed downstream (Fig. 7C and 8C). Silencing of SUV420H2 resulted in the loss of H4K20me3, the acquisition of H4K16Ac, and a 2- to 5-fold upregulation of gene expression at both loci (Fig. 7B and C and 8B and C). Under these conditions, Pol II was redistributed such that there was a shift to the elongating form at downstream regions (Fig. 7C and 8C). Treatment with DAC had no impact on histone H4 modifications, gene expression, or Pol II dynamics and did not significantly affect the changes induced by SUV420H2 knockdown alone, consistent with a lack of DNA methylation at the promoters of these genes in MDA-MB231 cells (Fig. 7B and C and 8B and C and data not shown). These data suggest that the acquisition of H4K20me3 and the local inhibition of hMOF-mediated H4K16Ac and subsequent block to Pol II promoter escape are not unique to the TMS1 locus but rather represent a more general mechanism for the local regulation of Pol II promoter-proximal pausing.
Fig. 7.
Regulation of Pol II promoter-proximal pausing by H4K16Ac and H4K20me3 at the JUND locus. (A) The schematic of the JUND genomic locus is shown. The JUND gene consists of one exon (white box) located within a CpG island. The transcription start site (horizontal arrow) and nucleotide positions (bp) relative to the transcription start site are shown. Primer sets (1 to 6) used in ChIP/real-time PCR assays are also shown. (B) MDA-MB231 cells infected with an empty lentiviral vector (Untreated and DAC) or a lentiviral vector expressing shRNA against SUV420H2 (shSUV420H2 and DAC + shSUV420H2) were left untreated or treated with 0.5 μM DAC where indicated. Cells were harvested 5 days postinfection (4 days post-DAC treatment), and RNA was isolated. SUV420H2 and JUND mRNA levels were determined by reverse transcription followed by real-time PCR and were normalized to 18S rRNA (relative expression). Shown is the mean ± standard deviation of two independent experiments. (C) Cells from panel B were subjected to ChIP with the indicated antibodies. Immunoprecipitated DNA was quantified by real-time PCR and normalized to DNA purified from histone H4 ChIP or as a percentage of input DNA. Experiments were performed twice, and shown is the mean ± standard deviation of a representative experiment. (D) hMOF and JUND mRNA levels were determined by reverse transcription followed by real-time PCR in MCF7 cells expressing or not expressing hMOF. Expression levels are shown after normalization to 18S rRNA. Shown is the mean ± standard deviation of two independent experiments. (E) MCF7 cells from panel D were used to perform ChIP with the indicated antibodies. DNA immunoprecipitated is normalized to DNA from histone H4 ChIP or as percentage of input DNA. Each experiment was performed twice and gave similar profiles. Shown is the mean ± standard deviation of triplicate determinations from a representative experiment.
Fig. 8.
Effect of SUV420H2 silencing and/or DAC treatment on gene expression, histone H4 modifications, and Pol II dynamics at the MORC3 locus. (A) Shown is the schematic of the MORC3 gene. The CpG island, transcription start site (horizontal arrow), nucleotide positions (bp) relative to the start site, and exons (white boxes) are labeled. Positions of real-time primers (1 to 7) used in the ChIP/real-time PCR studies are indicated below the gene diagram. (B and C) MDA-MB231 cells infected with an empty lentiviral vector (Untreated and DAC) or a lentiviral vector expressing shRNA against SUV420H2 (shSUV420H2 and DAC + shSUV420H2 or shSUV420H2 + DAC) were left untreated or treated with 0.5 μM DAC where indicated. Cells were harvested 5 days postinfection for RNA extraction or ChIP with the indicated antibodies. (B) SUV420H2 and MORC3 mRNAs were quantified by reverse transcription followed by real-time PCR and are shown normalized to 18S rRNA. Shown is the mean ± standard deviation of two independent experiments. (C) DNA immunoprecipitated from ChIP was subjected to real-time PCR and is expressed normalized to DNA from histone H4 ChIP or as a percentage of input. Experiments were performed twice, and shown is the mean ± standard deviation of a representative experiment.
We also examined the impact of hMOF downregulation on the JUND locus in MCF7 cells, where the gene is highly expressed and marked by H4K16Ac (Fig. 7D and E). Downregulation of hMOF led to a dramatic decrease in gene expression, the loss of H4K16Ac, and a shift in the Pol II profile to one in which initiated Pol II accumulated at the transcription start site and the elongating form was decreased, consistent with Pol II pausing (Fig. 7D and E). MORC3 was marked by H4K20me3 and lacked H4K16Ac in MCF7 cells, as it did in MDA-MB231 cells; thus, downregulation of hMOF had little effect on MORC3 expression or Pol II dynamics in this cell line (data not shown).
These data suggest that the inhibition of H4K16Ac and Pol II elongation imposed by the local acquisition of H4K20me3 can occur independently of DNA methylation and is not restricted to the TMS1 locus but is rather a more general mechanism for the local enforcement of Pol II pausing. Thus, DNA methylation and H4K20me3 appear to act as independent mechanisms of gene repression and the superimposition of DNA methylation at certain genes (e.g., TMS1) may provide an additional layer to effect stable gene silencing.
DISCUSSION
The recent finding that the release of paused initiated Pol II into productive elongation is an important regulatory step at thousands of genes has garnered great interest in the mechanism by which Pol II pausing is regulated (19, 42). Here, we demonstrate that H4K16Ac and H4K20me3 antagonistically control gene expression by regulating Pol II promoter-proximal pausing. We find that hMOF-mediated H4K16Ac stimulates the release of Pol II from promoter-proximal pausing into productive elongation and is required for the association of the BRD4 and pTEFb complex with chromatin at the TMS1 locus. Methylation of CpG island DNA and the shift to H3K9me2 that accompanies the epigenetic silencing of certain tumor suppressor genes block Pol II access to the embedded TMS1 promoter. However, even in the absence of these factors, repression is maintained by SUV420H2-mediated H4K20me3, which inhibits the recruitment of hMOF and the subsequent acetylation of H4K16 and, in so doing, enforces Pol II pausing (Fig. 9). Whereas CpG island methylation also blocks Pol II promoter access at other silenced genes, those lacking H4K20me3 (e.g., ESR1 and CDH1) do not exhibit Pol II pausing upon DNA demethylation. We further find that H4K20me3 selectively marks other genes (e.g., JUND and MORC3) independently of DNA methylation where it similarly imposes a block to Pol II elongation through a mechanism that involves the local inhibition of H4K16Ac. This is the first study to demonstrate the negative interplay between H4K16Ac and H4K20me3 in the local modulation of Pol II pausing and to show that DNA methylation/H3K9me2 and H4K20me3 act as independent layers to maintain transcriptional repression at certain epigenetically silenced tumor suppressor genes in cancer.
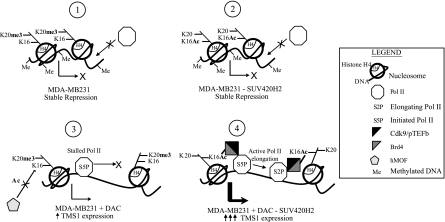
Fig. 9.
Model for the multilayered regulation of TMS1 gene repression and Pol II promoter-proximal pausing by DNA methylation and histone H4 modifications. (1) In the stably repressed state in which the CpG island is methylated and the locus is associated with H4K20me3, Pol II cannot access the TMS1 promoter. (2) Although depletion of SUV420H2 and/or H4K20me3 allows for reassociation of hMOF and reacetylation of H4K16Ac, it has no impact on DNA methylation or TMS1 expression. (3) Demethylation of DNA allows for the reassociation of Pol II with the TMS1 locus. However, the presence of SUV420H2 and/or H4K20me3 prevents hMOF recruitment and H4K16Ac, causing Pol II to remain primarily in its initiated form, paused at the promoter region. Low levels of TMS1 expression are detected. (4) Concurrent inhibition of DNA methylation and SUV420H2-mediated H4K20me3 allows for the rerecruitment of hMOF, the reacetylation of H4K16, and recruitment of BRD4 and pTEFb to the locus. pTEFb-mediated phosphorylation of initiated Pol II leads to the subsequent relief of pausing and the transition of Pol II from the initiated to the elongating form, resulting in the synergistic reactivation of TMS1 gene expression.
We show that H4K16Ac promotes the escape of Pol II from pausing by mediating the recruitment of the bromodomain protein BRD4 and the pTEFb complex. Our results are consistent with those of Zippo et al. (69), who showed that hMOF-mediated H4K16Ac at a distal enhancer relieves Pol II pausing at the FOS1 promoter. In that case, hMOF was selectively recruited to the enhancer by 14-3-3ε- and 14-3-3ζ-mediated binding to histone H3 phosphorylated at S10 (H3S10p). hMOF is distributed throughout the TMS1 promoter and into the gene body, as it is at many other active genes, suggesting that there may be multiple mechanisms of hMOF targeting (29, 62) (Fig. 4B). hMOF has been shown to exist as part of several different complexes in human cells, including the MSL, MSL1v1, MLL-WDR5, and NSL complexes (5, 14, 32, 39, 52, 56). We previously showed that MSL1, a component of the MSL complex, colocalizes with hMOF at the TMS1 locus and that the effect of MSL1 knockdown on H4K16Ac and full-length TMS1 transcript levels parallels that observed upon downregulation of hMOF, suggesting that hMOF acts as part of the MSL complex in this context (29). Although the precise mechanism of MSL targeting to the TMS1 locus is currently unknown, our data show that this recruitment is inhibited, either directly or indirectly, by SUV420H2 and/or SUV420H2-mediated H4K20me3. Interestingly, hMOF localization and H4K16Ac levels are reestablished throughout the TMS1 locus in the absence of H4K20me3 even though SUV420H2 and H4K20me3 are normally localized to a single sharp peak near the promoter (Fig. 4). Similarly, H4K16Ac is distributed throughout the JUND locus in the absence of H4K20me3 even though H4K20me3 is enriched mainly at the promoter region (Fig. 7C). This suggests that the recruitment of the MSL complex/hMOF to the promoter region is critical but that once the MSL complex gains access, it can modify nucleosomes throughout the gene body. Together, these studies point to the potential for multiple histone modifications to control promoter-proximal Pol II pausing.
We found that the promoter region of the unmethylated TMS1 gene is occupied by the NELF complex, regardless of H4K16Ac or levels of gene expression. Considering the role of NELF as a negative elongation factor, the finding that NELF is also present at actively transcribed genes seems paradoxical. Nevertheless, recent work by Gilchrist et al. (18) showed that silencing of NELF results in both the upregulation and downregulation of target genes. Interestingly, genes that were downregulated in the absence of NELF showed a decrease in H3K4me3 and increased nucleosome occupancy at their promoters (18). Downregulation of NELF also led to a selective decrease in Pol II levels at the 5′ end of target genes (18, 55). These data suggest that in addition to its function in enforcing Pol II pausing, NELF also plays a positive role in transcription by maintaining a permissive chromatin conformation and stabilizing the association of Pol II with target gene promoters.
The maintenance of Pol II occupancy, whether paused or actively elongating, may be one mechanism that prevents DNA methylation from encroaching on CpG island promoters. Recent work has shown that resistance of CpG islands to aberrant DNA methylation in human cancer cells is better correlated with Pol II occupancy than with absolute transcript levels (57). Similarly, recent work from our lab indicates that the propensity for genes to undergo remethylation after DAC-induced demethylation is inversely correlated with the long-term maintenance of Pol II occupancy (28). Pol II occupancy may prevent DNA methylation directly or indirectly by promoting an open chromatin structure rich in active histone marks, such as methylated H3K4, which has been shown to block the interaction of de novo DNA methyltransferases with nucleosomes (26). The enforcement of Pol II pausing may be a step that precedes DNA methylation in the aberrant silencing of tumor suppressor genes in cancers. Subsequent dissociation of Pol II—for example, by events that dysregulate NELF or other factors involved in the maintenance of paused Pol II (e.g., H4K20me3)—may allow for abnormal DNA methylation and stable gene repression. In this regard, it is noteworthy that many of the factors that control Pol II pausing are also dysregulated in human cancers. Cancer cells exhibit a global loss of H4K16Ac and H4K20me3 relative to normal cells (16). H4K16Ac and hMOF levels have been shown to be decreased in primary medulloblastomas and breast cancers (48). Likewise, H4K20me3 and SUV420H2 are downregulated in human lung and liver tumors (6, 49, 59). BRD4 is amplified in breast cancer and translocated in NUT midline carcinomas (17). Crawford et al. (12) showed that ectopic expression of BRD4 in breast cancer cells suppresses tumor growth and metastasis in vivo and identified a BRD4-activated gene signature that can predict prognosis in human breast cancer patients.
H4K20me3 is enriched in areas of constitutive heterochromatin, and its depletion results in destabilization of pericentric heterochromatin, compromised telomere integrity, and ectopic activation of endogenous retroelements, supporting the widely held notion that the primary function of H4K20me3 is to stabilize heterochromatin structure (3, 16, 35, 50). However, recent studies have suggested that H4K20me3 also marks individual genes (9), but its function, if any, at such loci has not been elucidated. We provide evidence here that SUV420H2 and H4K20me3 are selectively targeted to individual genes and, moreover, that H4K20me3 plays an important role in gene repression through the local inhibition of H4K16Ac. Early work showing that H4K16 acetylation and H4K20 methylation are mutually exclusive on the X chromosome in Drosophila males and that histone H4 peptides premethylated at H4K20 are poor substrates for p300-mediated H4K16 acetylation led to the suggestion that H4K20me3 and H4K16Ac may be competitive in vivo (43). However, top-down mass spectrometry studies have shown that H4K16Ac and H4K20me3 can coexist on the same histone H4 tail, and additional studies indicate that depletion of SUV420H has no impact on global levels of H4K16Ac, favoring a model in which the two modifications are independently regulated (66). Our data clearly show that SUV420H2 and/or H4K20me3 antagonizes H4K16Ac in vivo and that this derives from the local inhibition of hMOF and MSL complex recruitment. While our data cannot distinguish whether it is the presence of the H4K20me3 mark itself or another function of SUV420H2 (i.e., methylation of nonhistone targets and/or scaffolding functions) that blocks MSL recruitment, the finding that SUV420H2 knockdown affected H4K16Ac, Pol II dynamics, and gene expression only at genes marked by H4K20me3 but not at genes lacking H4K20me3 (ESR1 and CDH1) speaks to a gene-specific regulatory mechanism wherein the local targeting of SUV420H2 and deposition of H4K20me3 itself are critical.
Current models suggest that H4K20 methylation occurs in a progressive stepwise process wherein H4K20 monomethylated by the histone methyltransferase PR-SET7 serves as a substrate for di- and trimethylation by SUV420H (47, 51, 67). The vast majority of H4K20 in human cells exists in the H4K20me2 form; the mechanism(s) that regulates the localized conversion of a small fraction into H4K20me3 is currently unknown (66). Indeed, recent biochemical and structural data suggest that the chromoshadow domain of the MSL3 component of the MSL complex can bind H4K20me1/2 but cannot bind H4K20me3 (30, 40). Thus, the local conversion to H4K20me3 by SUV420H2 could potentially block MSL complex recruitment by precluding MSL3 binding. Other studies show that HP1 binds to SUV420H1/2 and is necessary for the maintenance of H4K20me3 at constitutive heterochromatin and at imprint control regions and that depletion of H3K9me3 leads to a loss of H4K20me3 at major satellite repeats and endogenous retroviruses (35), suggesting that SUV420H (and hence H4K20me3) may be directed to these regions by an HP1-mediated association with H3K9me3 (50). Indeed, depletion of H3K9me3 leads to a loss of H4K20me3 at major satellite repeats and endogenous retroviruses (35). SUV420H2 has also been shown to interact with RB1, and H4K20me3 levels are decreased at telomeric and pericentric heterochromatin in RB1-deficient cells (20). Whether HP1 or RB1 plays a similar role in SUV420H2 targeting to individual genes remains to be determined.
Trimethylation of H4K20 at the TMS1 locus is likely an acquired event in cancer cells, and the subsequent repression is a prerequisite to the more stable and heritable silencing imposed by DNA methylation. In the absence of DNA methylation, inhibition of H4K20me3 allows for the rerecruitment of hMOF to the locus, reestablishment of H4K16Ac and TMS1 upregulation, indicating that the initial silencing was due not to a loss of hMOF expression or activity but rather to the directed repression by SUV420H2 and/or H4K20me3. Indeed, H4K20me3 is absent from the TMS1 locus in normal fibroblasts and immortalized breast epithelial cells (29) (data not shown), and whereas downregulation of hMOF in MCF7 cells leads to depletion of H4K16Ac and TMS1 repression, it is not sufficient to evoke H4K20me3 (29). Thus, it is likely that other trans-acting factors present in MDA-MB231 cells target SUV420H2 to the TMS1 locus. Although the mechanisms triggering the subsequent acquisition of H3K9me2 and aberrant DNA methylation at TMS1 are presently unknown, our data suggest that once this is achieved, repression can be maintained independently of H4K20me3. Indeed, the finding that other genes are marked by H4K20me3 in a cancer cell line independently of DNA methylation (e.g., JUND and MORC3), where it similarly enforces Pol II pausing, suggests that the targeting of H4K20me3 is both gene and cell type specific, and whereas this event may put certain genes at risk of subsequent DNA methylation, it is not sufficient to drive this process.
We found that depletion of DNA methylation at TMS1 led to a concomitant inhibition of H3K9me2 and the reacquisition of H3K4me2 and Pol II but had no impact on H4K20me3. Similarly, whereas depletion of H4K20me3 allowed for restoration of H4K16Ac, it had no effect on DNA methylation or H3K9me2. These data indicate that once established, DNA methylation and H3K9me2 are coordinately regulated but are independent of H4K20me3 and that the two represent independent layers of gene repression. DNA methylation (and the linked H3K9me2) is dominant in this regard, in that depletion of H4K20me3 cannot overcome the block to Pol II access imposed by DNA methylation (Fig. 9). These data support a model in which DNA methylation serves as the final lock at tumor suppressor loci to mediate stable and heritable repression.
These findings have important implications for the application of “epigenetic therapy” as an approach to cancer treatment. DAC (Decitabine) and the related agent 5-azacytidine (Vidaza) are currently being used in the treatment of myelodysplastic syndrome (MDS) and acute myeloid leukemia (23). Whereas both cell culture experiments and clinical studies have shown that DAC treatment leads to a global demethylation of DNA and reactivation of some silenced genes, there is a propensity for remethylation and resilencing after drug removal (15, 38). This has been attributed to the residual presence of repressive marks, such as H3K9me3 and H3K27me2/3, which are unaffected by DNA demethylation, and the failure to regain active features (such as H3K79me2) (24, 38). We show here that H4K20me3 is similarly unaffected by DNA demethylation and that the residual presence of H4K20me3 maintains gene repression by enforcing Pol II promoter-proximal pausing. The combined inhibition of DNA methylation and H4K20 methylation leads to a synergistic reactivation of TMS1 gene expression and recapitulates a chromatin environment similar to that observed in cells that actively express TMS1. Thus, combined approaches targeting both DNA methylation and histone methylation may be therapeutically beneficial and warrant consideration in the development of novel epigenetic therapy approaches.
ACKNOWLEDGMENTS
We thank Doris R. Powell for technical assistance and John Lucchesi, Paul Wade, and Maureen Powers for critical reading of the manuscript. We are grateful to Edwin Smith for antibody against hMOF.
This work was supported by the Canadian Institute of Health Research Postdoctoral Fellowship to P.K.-V. and Public Health Service grants 2RO1 CA-077337 and CA-132065 from the National Cancer Institute to P.M.V. P.M.V. is a Georgia Cancer Coalition Distinguished Cancer Scholar.
Footnotes
Published ahead of print on 14 February 2011.
REFERENCES
- 1. Adelman K., et al. 2005. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell 17:103–112 [DOI] [PubMed] [Google Scholar]
- 2. Benetti R., Garcia-Cao M., Blasco M. A. 2007. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat. Genet. 39:243–250 [DOI] [PubMed] [Google Scholar]
- 3. Benetti R., et al. 2007. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J. Cell Biol. 178:925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bird A. P. 1986. CpG-rich islands and the function of DNA methylation. Nature 321:209–213 [DOI] [PubMed] [Google Scholar]
- 5. Cai Y., et al. 2010. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J. Biol. Chem. 285:4268–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chekhun V. F., Lukyanova N. Y., Kovalchuk O., Tryndyak V. P., Pogribny I. P. 2007. Epigenetic profiling of multidrug-resistant human MCF-7 breast adenocarcinoma cells reveals novel hyper- and hypomethylated targets. Mol. Cancer Ther. 6:1089–1098 [DOI] [PubMed] [Google Scholar]
- 7. Cheng B., Price D. H. 2007. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J. Biol. Chem. 282:21901–21912 [DOI] [PubMed] [Google Scholar]
- 8. Collard R. L., Harya N. S., Monzon F. A., Maier C. E., O'Keefe D. S. 2006. Methylation of the ASC gene promoter is associated with aggressive prostate cancer. Prostate 66:687–695 [DOI] [PubMed] [Google Scholar]
- 9. Congdon L. M., Houston S. I., Veerappan C. S., Spektor T. M., Rice J. C. 2010. PR-Set7-mediated monomethylation of histone H4 lysine 20 at specific genomic regions induces transcriptional repression. J. Cell. Biochem. 110:609–619 [DOI] [PubMed] [Google Scholar]
- 10. Conway K. E., et al. 2000. TMS1, a novel proapoptotic caspase recruitment domain protein, is a target of methylation-induced gene silencing in human breast cancers. Cancer Res. 60:6236–6242 [PubMed] [Google Scholar]
- 11. Core L. J., Waterfall J. J., Lis J. T. 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322:1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Crawford N. P., et al. 2008. Bromodomain 4 activation predicts breast cancer survival. Proc. Natl. Acad. Sci. U. S. A. 105:6380–6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dey A., Chitsaz F., Abbasi A., Misteli T., Ozato K. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. U. S. A. 100:8758–8763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dou Y., et al. 2005. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121:873–885 [DOI] [PubMed] [Google Scholar]
- 15. Egger G., Aparicio A. M., Escobar S. G., Jones P. A. 2007. Inhibition of histone deacetylation does not block resilencing of p16 after 5-aza-2′-deoxycytidine treatment. Cancer Res. 67:346–353 [DOI] [PubMed] [Google Scholar]
- 16. Fraga M. F., et al. 2005. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 37:391–400 [DOI] [PubMed] [Google Scholar]
- 17. French C. A., et al. 2001. BRD4 bromodomain gene rearrangement in aggressive carcinoma with translocation t(15;19). Am. J. Pathol. 159:1987–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gilchrist D. A., et al. 2008. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 22:1921–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gilmour D. S. 2009. Promoter proximal pausing on genes in metazoans. Chromosoma 118:1–10 [DOI] [PubMed] [Google Scholar]
- 20. Gonzalo S., et al. 2005. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat. Cell Biol. 7:420–428 [DOI] [PubMed] [Google Scholar]
- 21. Graff J. R., Herman J. G., Myohanen S., Baylin S. B., Vertino P. M. 1997. Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. J. Biol. Chem. 272:22322–22329 [DOI] [PubMed] [Google Scholar]
- 22. Guenther M. G., Levine S. S., Boyer L. A., Jaenisch R., Young R. A. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130:77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Issa J. P. 2007. DNA methylation as a therapeutic target in cancer. Clin. Cancer Res. 13:1634–1637 [DOI] [PubMed] [Google Scholar]
- 24. Jacinto F. V., Ballestar E., Esteller M. 2009. Impaired recruitment of the histone methyltransferase DOT1L contributes to the incomplete reactivation of tumor suppressor genes upon DNA demethylation. Oncogene 28:4212–4224 [DOI] [PubMed] [Google Scholar]
- 25. Jang M. K., et al. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19:523–534 [DOI] [PubMed] [Google Scholar]
- 26. Jia D., Jurkowska R. Z., Zhang X., Jeltsch A., Cheng X. 2007. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 449:248–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jones P. A., Takai D. 2001. The role of DNA methylation in mammalian epigenetics. Science 293:1068–1070 [DOI] [PubMed] [Google Scholar]
- 28. Kagey J. D., Kapoor-Vazirani P., McCabe M. T., Powell D. R., Vertino P. M. 2010. Long-term stability of demethylation after transient exposure to 5-aza-2′-deoxycytidine correlates with sustained RNA polymerase II occupancy. Mol. Cancer Res. 8:1048–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kapoor-Vazirani P., Kagey J. D., Powell D. R., Vertino P. M. 2008. Role of hMOF-dependent histone H4 lysine 16 acetylation in the maintenance of TMS1/ASC gene activity. Cancer Res. 68:6810–6821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim D., Blus B. J., Chandra V., Huang P., Rastinejad F., Khorasanizadeh S. 2010. Corecognition of DNA and a methylated histone tail by the MSL3 chromodomain. Nat. Struct. Mol. Biol. 17:1027–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Levine J. J., Stimson-Crider K. M., Vertino P. M. 2003. Effects of methylation on expression of TMS1/ASC in human breast cancer cells. Oncogene 22:3475–3488 [DOI] [PubMed] [Google Scholar]
- 32. Li X., Wu L., Corsa C. A., Kunkel S., Dou Y. 2009. Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol. Cell 36:290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marshall N. F., Price D. H. 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 270:12335–12338 [DOI] [PubMed] [Google Scholar]
- 34. Masumoto J., et al. 1999. ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J. Biol. Chem. 274:33835–33838 [DOI] [PubMed] [Google Scholar]
- 35. Matsui T., et al. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464:927–931 [DOI] [PubMed] [Google Scholar]
- 36. McCabe M. T., Brandes J. C., Vertino P. M. 2009. Cancer DNA methylation: molecular mechanisms and clinical implications. Clin. Cancer Res. 15:3927–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McConnell B. B., Vertino P. M. 2004. TMS1/ASC: the cancer connection. Apoptosis 9:5–18 [DOI] [PubMed] [Google Scholar]
- 38. McGarvey K. M., et al. 2006. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 66:3541–3549 [DOI] [PubMed] [Google Scholar]
- 39. Mendjan S., et al. 2006. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell 21:811–823 [DOI] [PubMed] [Google Scholar]
- 40. Moore S. A., Ferhatoglu Y., Jia Y., Al-Jiab R. A., Scott M. J. 2010. Structural and biochemical studies on the chromo-barrel domain of male specific lethal 3 (MSL3) reveal a binding preference for mono or dimethyl lysine 20 on histone H4. J. Biol. Chem. 285:40879–40890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Muse G. W., et al. 2007. RNA polymerase is poised for activation across the genome. Nat. Genet. 39:1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nechaev S., Adelman K. 2008. Promoter-proximal Pol II: when stalling speeds things up. Cell Cycle 7:1539–1544 [DOI] [PubMed] [Google Scholar]
- 43. Nishioka K., et al. 2002. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell 9:1201–1213 [DOI] [PubMed] [Google Scholar]
- 44. Ottaviano Y. L., et al. 1994. Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res. 54:2552–2555 [PubMed] [Google Scholar]
- 45. Parsons M. J., Patel P., Brat D. J., Colbert L., Vertino P. M. 2009. Silencing of TMS1/ASC promotes resistance to anoikis in breast epithelial cells. Cancer Res. 69:1706–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Parsons M. J., Vertino P. M. 2006. Dual role of TMS1/ASC in death receptor signaling. Oncogene 25:6948–6958 [DOI] [PubMed] [Google Scholar]
- 47. Pesavento J. J., Yang H., Kelleher N. L., Mizzen C. A. 2008. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol. Cell. Biol. 28:468–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pfister S., et al. 2008. The histone acetyltransferase hMOF is frequently downregulated in primary breast carcinoma and medulloblastoma and constitutes a biomarker for clinical outcome in medulloblastoma. Int. J. Cancer 122:1207–1213 [DOI] [PubMed] [Google Scholar]
- 49. Pogribny I. P., et al. 2006. Histone H3 lysine 9 and H4 lysine 20 trimethylation and the expression of Suv4-20h2 and Suv-39h1 histone methyltransferases in hepatocarcinogenesis induced by methyl deficiency in rats. Carcinogenesis 27:1180–1186 [DOI] [PubMed] [Google Scholar]
- 50. Schotta G., et al. 2004. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18:1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schotta G., et al. 2008. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 22:2048–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smith E. R., et al. 2005. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol. Cell. Biol. 25:9175–9188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stimson K. M., Vertino P. M. 2002. Methylation-mediated silencing of TMS1/ASC is accompanied by histone hypoacetylation and CpG island-localized changes in chromatin architecture. J. Biol. Chem. 277:4951–4958 [DOI] [PubMed] [Google Scholar]
- 54. Stone A. R., et al. 2004. Aberrant methylation and down-regulation of TMS1/ASC in human glioblastoma. Am. J. Pathol. 165:1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sun J., Li R. 2010. Human negative elongation factor activates transcription and regulates alternative transcription initiation. J. Biol. Chem. 285:6443–6452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Taipale M., et al. 2005. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol. Cell. Biol. 25:6798–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Takeshima H., Yamashita S., Shimazu T., Niwa T., Ushijima T. 2009. The presence of RNA polymerase II, active or stalled, predicts epigenetic fate of promoter CpG islands. Genome Res. 19:1974–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Thomas M. C., Chiang C. M. 2006. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41:105–178 [DOI] [PubMed] [Google Scholar]
- 59. Van Den Broeck A., et al. 2008. Loss of histone H4K20 trimethylation occurs in preneoplasia and influences prognosis of non-small cell lung cancer. Clin. Cancer Res. 14:7237–7245 [DOI] [PubMed] [Google Scholar]
- 60. Venters B. J., Pugh B. F. 2007. Chromatin meets RNA polymerase II. Genome Biol. 8:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wada T., Takagi T., Yamaguchi Y., Watanabe D., Handa H. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395–7403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang Z., et al. 2009. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138:1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wu C. H., et al. 2003. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 17:1402–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yamada T., et al. 2006. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell 21:227–237 [DOI] [PubMed] [Google Scholar]
- 65. Yamaguchi Y., et al. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41–51 [DOI] [PubMed] [Google Scholar]
- 66. Yang H., Mizzen C. A. 2009. The multiple facets of histone H4-lysine 20 methylation. Biochem. Cell Biol. 87:151–161 [DOI] [PubMed] [Google Scholar]
- 67. Yang H., et al. 2008. Preferential dimethylation of histone H4 lysine 20 by Suv4-20. J. Biol. Chem. 283:12085–12092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zeitlinger J., et al. 2007. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 21:385–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zippo A., et al. 2009. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138:1122–1136 [DOI] [PubMed] [Google Scholar]