Abstract
The mammalian target of rapamycin (mTOR) regulates cell growth and survival via two different multiprotein complexes, mTORC1 and mTORC2. The assembly of these serine-threonine kinase multiprotein complexes occurs via poorly understood molecular mechanisms. Here, we demonstrate that GRp58/ERp57 regulates the existence and activity of mTORC1. Endogenous mTOR interacts with GRp58/ERp57 in different mammalian cells. In vitro, recombinant GRp58/ERp57 preferentially interacts with mTORC1. GRp58/ERp57 knockdown reduces mTORC1 levels and phosphorylation of 4E-BP1 and p70S6K in response to insulin. In contrast, GRp58/ERp57 overexpression increases mTORC1 levels and activity. A redox-sensitive mechanism that depends on GRp58/ERp57 expression activates mTORC1. Although GRp58/ERp57 is known as an endoplasmic reticulum (ER) resident, we demonstrate its presence at the cytosol, together with mTOR, Raptor, and Rictor as well as a pool of these proteins associated to the ER. In addition, the presence of GRp58/ERp57 at the ER decreases in response to insulin or leucine. Interestingly, a fraction of p70S6K, but not 4E-BP1, is associated to the ER and phosphorylated in response to serum, insulin, or leucine. Altogether, our results suggest that GRp58/ERp57 is involved in the assembly of mTORC1 and positively regulates mTORC1 signaling at the cytosol and the cytosolic side of the ER.
INTRODUCTION
The mammalian target of rapamycin (mTOR) is a highly conserved Ser/Thr kinase that plays a critical role in fundamental cellular processes such as cell metabolism, growth, proliferation, and migration (10, 14, 22, 44, 52, 54, 55). This kinase is a component of signal transduction cascades activated by growth factor receptors and constitutes an intracellular sensor of nutrient availability, hypoxia, DNA damage, and osmotic stress (3, 24, 44, 55). mTOR exists in two different multiprotein complexes differentially sensitive to the inhibitory effect of rapamycin (32, 40). The rapamycin-sensitive mTOR complex 1 (mTORC1), which regulates protein synthesis and cell growth, consists of mTOR, Raptor, and mLST8 (previously known as GβL) (29). mTORC1 phosphorylates two regulators of protein synthesis: the ribosomal S6 kinase (S6K1) and the inhibitory partner of the translation initiation factor 4E (4E-BP1) (14). Qian and colleagues recently expanded the paradigm on mTORC1 regulation by demonstrating that mTORC1 serves as a sensor of the extent of protein misfolding, helping to maintain the right balance of protein synthesis and degradation (39). The mTOR complex 2 (mTORC2) contains mTOR, Rictor, Sin1, and mLST8. Its catalytic activity is not sensitive to rapamycin although prolonged treatment disrupts the assembly of this complex in certain cell lines (38, 45). mTORC2 has been recognized as the elusive kinase that phosphorylates Akt at serine 473, promoting the maximum activity of this important kinase, implicated in the control of cell survival, metabolism, proliferation, and growth (18, 25, 44, 46). We recently reported that mTORC2 interacts with P-Rex1, a Rac guanine nucleotide exchange factor (GEF), resulting in Rac activation and cell migration (22). The mechanisms that regulate the formation and stability of mTOR complexes are not completely understood although it is known that both mTORC1 and mTORC2 complexes are regulated by phosphatidic acid (50) and by a complex formed by Tti1 and Tel2 (27), while nutrient availability and redox status have an effect on mTORC1 (47), opening the possibility that proteins that sense cellular redox status can be involved in the assembly of mTORC1 complex.
GRp58 (also known as ERp57) is a widely expressed protein disulfide isomerase that regulates protein-protein interactions via a redox mechanism based on the activity of its two thioredoxin-like domains (28, 37). It has a recognized chaperone function via a mechanism that involves a reduction of disulfide bonds in preformed protein complexes (28, 37). The physiological importance of GRp58 is highlighted by the finding that genetic knockout of GRp58 in mice leads to embryonic lethality (5, 15, 37). GRp58 participates in the assembly of the major histocompatibility complex (16). Although GRp58 has been mainly characterized by its actions at the endoplasmic reticulum (ER), GRp58 knockout cells do not reveal a dramatic effect on ER morphology or function but show an increase in STAT3-dependent signaling, which is attenuated by transfected GRp58 targeted to the ER (5), further supporting the emerging role of GRp58 as a regulator of cytokine-dependent signal transduction and gene expression (4, 35, 51). Available evidence confirms the existence of cytosolic and nuclear pools of GRp58 able to interact with STAT3 (5, 19); moreover, GRp58 is phosphorylated in response to leptin, further supporting its role as a regulator of cytokine-dependent signal transduction (31). In addition, GRp58 alters the dimerization and DNA-binding ability of the E2A subfamily of basic helix-loop-helix (bHLH) proteins which, upon reduction, can heterodimerize with other bHLH proteins (34). GRp58 has been recognized as a cellular redox sensor that, as part of a cell-adaptative response to oxidative insults, cooperates with Ref-1 in the regulation of gene expression mediated by redox-sensitive transcription factors (17).
The finding that a redox-sensitive mechanism regulates the activity of mTORC1 and the interaction between Raptor and mTOR (47) suggests that proteins with redox-sensing ability are potentially involved in the assembly of mTOR complexes. Here, we describe the identification of GRp58 as an mTOR-interacting protein that modulates the assembly and activity of mTORC1.
MATERIALS AND METHODS
Yeast two-hybrid screening.
To generate the bait used for yeast two-hybrid screening, we subcloned the mTOR kinase domain into pGB3 in frame with the sequence coding for the GAL4 DNA-binding domain. A human placenta cDNA library (Clontech) was screened with the mTOR kinase domain using the Matchmaker System III (Clontech) following both the manufacturer's instruction and those of Vazquez-Prado et al. (53). Putative interacting clones were obtained by selecting transformants in medium lacking His/Leu/Trp and checked for α-galactosidase expression by an α-X-Gal (where X-Gal is 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) assay. The specificity of the interaction was determined using pGBKT7-p53 as bait and simian virus 40 (SV40) large T antigen as prey.
DNA constructs.
prk5-myc-Rictor (Addgene plasmid 1860), prk5-myc-Raptor (Addgene plasmid 1859), and prk5-HA-GβL (where HA is hemagglutinin) (Addgene plasmid 1865) were obtained from Sabatini (43). GRp58 cDNA was subcloned from the yeast two-hybrid prey vector, pACT2-GRp58, into the pCEFL mammalian expression vector harboring three copies of a Flag tag (pCEFL-3×Flag). For pulldown experiments, the mTOR kinase domain was subcloned in frame with the sequence coding for glutathione S-transferase (GST) into the mammalian expression vector pCEFL-GST. Yellow fluorescent protein (YFP)-mTOR (pcDNA3/YFP-mTOR) was kindly donated by Shu-Bing Qian, Cornell University (39); fluorescent markers for endoplasmic reticulum (cyan fluorescent protein [CFP]-ER, mitochondria (yellow fluorescent protein [YFP]-tagged), and plasma membrane (monomeric red fluorescent protein [mRFP]-CAAX) were kindly donated by Gyorgy Hajnoczky, Thomas Jefferson University. Enhanced green fluorescent protein (EGFP)-Golgi marker was donated by Tamas Balla, NIH; mCherry-GRp58 was prepared by subcloning GRp58 cDNA from pCEFL-EGFP-GRp58 into pCEFL-mCherry. GST-ERp57 (pGEX-4T-2-DTNDTC-ERp57) was donated by James D. Lechleiter, University of Texas Health Science Center at San Antonio. The short hairpin RNA (shRNA) for GRp58 (shGRp58) was generated based on the sequence of the hairpin (TGCTGTTGACAGTGAGCGCGGGACCCATATGGGAATTATTATAGTGAAGCCACAGATGTATAATAATTCCCATATGGGTCCTTGCCTACTGCCTCGGA) and cloned into pENTR vector, pEN_EmiRc3, and pDSL_hpIP. All other constructs have been previously described (22).
Cell culture and transfection.
The human embryonic kidney 293 (HEK293) and HEK293T cell lines and COS-7 cells were routinely maintained in Dulbecco's modified Eagle's medium (DMEM; Sigma) supplemented with 10% fetal bovine serum (FBS), 100 μg/ml penicillin, 100 μg/ml streptomycin, and 250 ng/ml amphotericin B (Gibco) at 37°C in a humidified atmosphere with 5% CO2. Cells grown in 10-cm diameter dishes were transfected using Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's instructions, with 2 μg of each plasmid. Five hours after the initiation of transfection, the medium was replaced with complete DMEM. Cells were used at 48 h posttransfection.
Immunoprecipitation, pulldown, and Western blot assays.
In mammalian HEK293T cells, the interaction between transfected GRp58 and the kinase domain of mTOR was assessed using mammalian expression constructs of the mTOR kinase domain fused to GST and Flag-tagged GRp58. Two days after transfection, the GST-mTOR kinase domain was isolated by pulldown using glutathione-Sepharose beads, and interacting GRp58 was revealed by Western blotting using anti-Flag antibodies. The potential association of GRp58 to mTORC1 and mTORC2 complexes was assessed by immunoprecipitation of transfected myc-Raptor or myc-Rictor, specific components of each complex, respectively, followed by Western blotting against Flag-GRp58. In order to assess a potential preferential binding of GRp58 with either of the two mTOR complexes, the association of endogenous mTORC1 and mTORC2 with recombinant, bacterially expressed GRp58 was assessed by pulldown; as a control, bacterially expressed GST fused to the Cdc42-Rac interactive binding (CRIB) domain of PAK (GST-PAK-N) was used. Interaction between endogenous mTOR and GRp58 was assessed in HEK293T or HeLa cells maintained in DMEM (Sigma) supplemented with 10% FBS or in human microvascular endothelial cells (HMEC) maintained in MCDB-131 medium (Gibco) supplemented with 10% FBS, 10 ng/ml epidermal growth factor (EGF), and 1 μg/ml of hydrocortisone (Sigma). To evaluate whether the interaction of mTOR and GRp58 is modulated by various stimuli, HEK293T cells were deprived of nutrients during 3 h and stimulated for 15 min with 10% FBS, 100 nM insulin, or 52 μg/ml leucine. The effect of rapamycin on the interaction was assessed in cells incubated for 1 or 24 h with this agent. The effect of phenylarsine oxide (PAO) as a modulator of the interaction of mTOR and GRp58 and as an activator of mTORC1 or mTORC2 was assessed in control and GRp58 knockdown HEK293T cells. To obtain whole-cell lysates, cells in 10-cm dishes were placed on ice, rinsed once with phosphate-buffered saline (PBS), lysed in 1 ml of ice-cold buffer (50 mM Tris [pH 7.5], 5 mM EDTA, 150 mM NaCl, and 0.3% CHAPS) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, and 10 μg/ml aprotinin), and immunoprecipitated with anti-mTOR (Santa Cruz), anti-Myc (Covance), or anti-PTEN (Santa Cruz) antibodies. Precipitated proteins were washed four times in 0.3% CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate) buffer, loaded onto 6% or 10% SDS-PAGE gels, detected by Western blotting using anti-mTOR (Cell Signaling), anti-GRp58 (Santa Cruz), anti-Myc, anti-HA (Covance), anti-Raptor (Cell Signaling), or anti-Rictor (Cell Signaling) antibodies, and revealed with a West Pico system (Pierce Biotechnology) or Immobilon Western chemiluminescent substrate (Millipore).
To test whether GRp58 played a role in the phosphorylation of mTORC1 or mTORC2 downstream substrates, HEK293 cells were transfected with Flag-GRp58 or GRp58 shRNA as described earlier. In the case of GRp58 overexpression experiments, 2 or 3 days after transfection (as indicated in figure legends) cells were serum starved for 3 h and then stimulated with 25 nM insulin (5 min) prior to cell lysis. In the case of the knockdown experiments, in which the signaling of mTOR was assessed, transfected cell populations were selected with puromycin. The lysates were analyzed by Western blotting with GRp58 (ERp57) to confirm either overexpression (by at least twice the control levels) or the knockdown (by a reduction of at least half of the endogenous levels of control lysates). The lysates were then used to detect the phosphorylation of different proteins using the following antibodies: phospho-Akt-Ser473 from Santa Cruz Biotechnology; Akt and Flag from Sigma; phospho-p70S6K-Thr389, p70S6K, phospho-S6-Ser240/244, S6, phospho-4E-BP1-Thr37/46, 4E-BP1, phospho-Akt-Thr450, phospho-glycogen synthase kinase 3β (GSK-3β)-Ser9, phospho-GSK-3β-mTOR-Ser2448, and mTOR from Cell Signaling Technology. Peroxidase-labeled secondary antibodies were from KPL. Bands were visualized using a West Pico system (Pierce Biotechnology) or Immobilon Western chemiluminescent substrate (Millipore).
Subcellular fractionation.
All the procedures were done on ice; confluent HEK293 cells were washed with ice-cold PBS and then harvested in buffer A (250 mM sucrose, 1 mM EDTA, 20 mM HEPES, pH 7.4) supplemented with protease and phosphatase inhibitors. Harvested cells were disrupted using a Dounce homogenizer by giving 40 strokes. Then, subcellular fractionation was performed by differential centrifugation. Cell homogenates were centrifuged at 800 × g for 10 min at 4°C, pellet fractions were discarded, and supernatants were centrifuged at 10,000 × g for 10 min at 4°C. The resulting supernatants were centrifuged at 100,000 × g for 1 h at 4°C to obtain the endoplasmic reticulum pellet and the cytosolic fraction. The ER pellets were washed with the same buffer and then solubilized in SDS-PAGE sample buffer; the cytosolic fraction was mixed with TNTE buffer (50 mM Tris, pH 7.5, 0.15 M NaCl, 1% Triton X-100, 5 mM EDTA) containing protease and phosphatase inhibitors. Subsequently, 10 μg of protein was analyzed by Western blotting after SDS-polyacrylamide gel electrophoresis. The purity of the fractions was confirmed by Western blotting detecting the presence of the specific marker SERCA (sarcoplasmic/endoplasmic reticulum Ca2+ ATPase) in the ER and 4E-BP1 in the cytosol. Anti-SERCA mouse monoclonal antibodies were from Affinity (catalog number MA3919).
Effect of nutrients on the subcellular distribution of mTOR and GRp58.
The effect of fetal bovine serum, insulin, or leucine on the subcellular distribution of mTOR, Raptor, Rictor, 4E-BP1, p70S6K, and GRp58 was analyzed on HEK293 cells. In brief, cells were serum starved overnight in DMEM or in DMEM without leucine and stimulated with either 10% FBS, 100 nM insulin, or 52 μg/ml leucine for 5 min. Cells were washed with ice-cold PBS and subjected to the subcellular fractionation protocol.
Fluorescence microscopy.
The subcellular distribution of fluorescence-tagged mTOR, GRp58, and different membrane markers was assessed in COS-7 cells grown on glass-bottom petri dishes and transfected with the DNAs indicated in the Fig. 9 legend using the Lipofectamine Plus method (Invitrogen). At 24 h after transfection, cells were incubated for a couple of hours in Krebs-Ringer buffer, pH 7.4, and then stimulated with l-leucine (52 μg/ml) or PAO (5 μM). The localization of mTOR, GRp58, and different membrane markers was assessed in live cells using an inverted Nikon Eclipse Ti-E fluorescence microscope, equipped with autofocus system. In addition, control and stimulated cells were fixed with 4% paraformaldehyde in PBS, pH 7.4, for 20 min at room temperature. Images of fixed cells were acquired with the same microscope using a 60× (numerical aperture, 1.4) oil immersion objective and a zoom of ×1.5. The colocalization analysis was done with the Nikon NIS-Elements software.
Proliferation assay.
HEK293T cells transiently transfected with GRp58 shRNA or 3×Flag-GRp58 were plated in 96-well flat-bottom plates (10,000 per well) in 100 μl of DMEM. At 36 h posttransfection, cells were starved of serum for 3 h; after this time they were treated with 100 nM insulin and 10% FBS for 24 h. For the last 4 h of the 24-h stimulation period, the cells were pulsed with bromodeoxyuridine (BrdU). Cell proliferation was measured using a BrdU enzyme-linked immunosorbent assay (ELISA) from Roche (Roche Diagnostics) according to the manufacturer's instructions. Absorbance at 405 and 492 nm was measured with a microplate reader (Model 550; Bio-Rad, Hercules, CA).
Statistical analysis.
Statistical significance of the differences among data was determined by analysis of variance and a Student-Newman-Keuls test using GraphPad Prism, version 2.0, software (GraphPad Software, San Diego, CA). A P value of <0.05 was considered a statistically significant difference.
RESULTS
Identification of GRp58 as an mTOR-interacting protein.
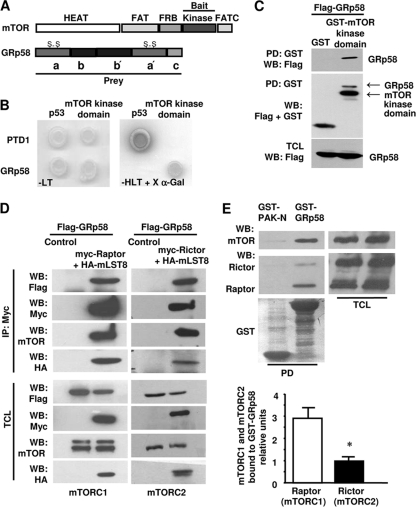
mTOR consists of several functional domains involved in diverse protein-protein interactions (Fig. 1 A). In order to identify novel mTOR-interacting partners potentially involved in the regulation of mTOR complexes, we screened a human placental cDNA library using the kinase domain of mTOR as bait in a yeast two-hybrid system. We identified full-length GRp58 as an mTOR-interacting protein (Fig. 1A). In the yeast two-hybrid system, the clone corresponding to GRp58 (prey) interacted specifically with the kinase domain of mTOR; this interaction was specific, as indicated by the growth in restrictive medium, lacking histidine, leucine, and tryptophan (Fig. 1B, −HLT), of yeast transformed just with the combination of the mTOR kinase domain and GRp58 or of the positive control of the system composed of p53 and T antigen, and by the ability of GRp58 and mTOR to promote the expression of α-galactosidase from an integrated reporter system (Fig. 1B). To assess in mammalian cells the interaction of GRp58 with the kinase domain of mTOR, 3×Flag-tagged full-length GRp58 and the kinase domain of mTOR fused to GST were transfected into the human epithelial cell line HEK293T. The corresponding cell extracts were incubated with glutathione-Sepharose beads; GRp58 was detected in GST-mTOR kinase domain pulldown but not in a GST pulldown used as a control (Fig. 1C). Since mTOR exists in two functionally different complexes, mTORC1 and mTORC2, both containing mLST8 but identifiable by distinct associated proteins (Raptor and Rictor, respectively) (25, 29, 38), we investigated whether GRp58 could interact with each mTOR complex. In order to address this question, we explored the presence of GRp58 in mTORC1, isolated by immunoprecipitating myc-tagged Raptor, and in mTORC2, isolated by immunoprecipitating myc-tagged Rictor, using lysates from HEK293T cells transfected with 3×Flag-GRp58, HA-mLST8, and either myc-Raptor or myc-Rictor. GRp58 was detected in both mTORC1 and mTORC2 immunoprecipitates (Fig. 1D). As expected, mTOR and mLST8 were found as constituents of both complexes (Fig. 1D). We next tested whether recombinant GRp58, under conditions in which it has the opportunity to interact with both mTOR complexes, preferentially selects one over the other. As shown in Fig. 1E, GST-GRp58 but not GST-PAK-N (used as a control) interacted preferentially with mTORC1 over mTORC2, evidenced by a significantly larger amount of Raptor than Rictor present in the GRp58 pulldown, in which, as expected, mTOR was also present. In these experiments, similar levels of endogenous Raptor and Rictor were detected in HEK293T cells (Fig. 1E, upper panel).
Fig. 1.
Identification of GRp58 as an mTOR-interacting protein. (A) Schematic representation of the domains of mTOR depicting the bait used in the yeast two-hybrid screen and the domains of GRp58 depicting the prey that was obtained. (B) GRp58 and a control (T antigen) were tested for interaction with the bait (mTOR kinase domain) or p53 as a control. Yeast grew in medium lacking leucine and tryptophan (−LT), which selects for the presence of the plasmids, and the ones displaying interaction grew under stringency conditions (medium lacking histidine, leucine, and tryptophan [-HLT]). (C) The kinase domain of mTOR fused to GST (GST-mTOR kinase domain) interacts with Flag-GRp58 in HEK293T cells. HEK293T cells were transfected with Flag-GRp58 and GST-mTOR kinase domain or control GST. Total cell lysates were affinity purified with glutathione beads, and both total cell lysates (TCL; bottom panel) and pulldowns (PD upper and middle panels) were resolved on SDS-polyacrylamide gels and analyzed by immunoblotting. WB, Western blotting. (D) Flag-GRp58 is present in multiprotein complexes that contain transfected myc-Raptor (left panel) or myc-Rictor (right panel), HA-mLST8, and mTOR. Two days after transfection, HEK293T cells expressing 3×Flag-GRp58, HA-mLST8, and myc-Raptor or myc-Rictor were used for immunoprecipitations (IP) with anti-Myc antibodies. The presence of 3×Flag-GRp58, HA-mLST8, and endogenous mTOR immunoprecipitated with myc-Raptor or myc-Rictor was assessed by Western blotting using the indicated antibodies. The expression of Flag-GRp58 and mTOR and other indicated proteins was detected in total lysates (TCL) of cells transfected with components of mTORC1 or mTORC2. (E) Recombinant GRp58 shows preferential affinity for endogenous mTORC1 over endogenous mTORC2 expressed in HEK293T cells. Total cell lysates of HEK293T cells expressing comparable amounts of Raptor and Rictor were incubated with glutathione Sepharose beads containing recombinant GST-GRp58 or GST-PAK-N (used as control) previously isolated from bacteria. The presence of endogenous Raptor, Rictor, and mTOR in the GST-GRp58 or GST-PAK-N pulldowns (PD) or total cell lysates was detected by Western blotting. The graph represents the mean and standard error of the mean of four independent experiments. *, P < 0.05.
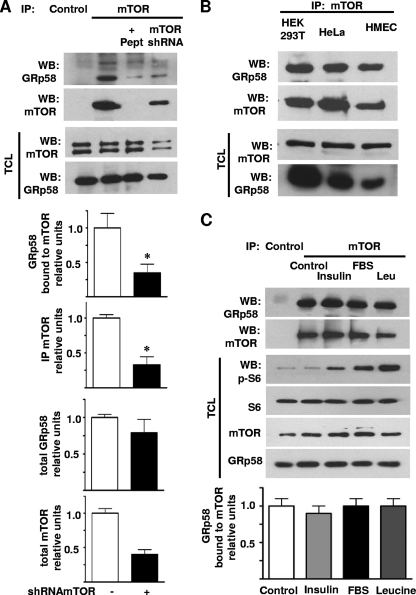
Interaction between endogenous GRp58 and mTOR in HEK293T cells was demonstrated by immunoprecipitating mTOR and detecting GRp58 (Fig. 2 A). The specificity of the interaction was determined in experiments that showed a significant decrease in the band corresponding to GRp58 in immunoprecipitates obtained with mTOR antibodies preincubated with a specific blocking peptide or in immunoprecipitates from mTOR knockdown cells and that demonstrated its absence in immunoprecipitates obtained using control antibodies (Fig. 2A). The shRNA-mediated decrease in the expression of mTOR was confirmed by Western blotting in total cell lysates (Fig. 2A). In order to determine whether the interaction between endogenous mTOR and GRp58 occurred in different cell types, we immunoprecipitated mTOR and detected its interaction with GRp58 using lysates from HEK293T and HeLa cells and HMEC. As shown in Fig. 2B, GRp58 was found associated to mTOR in the three cell types.
Fig. 2.
Endogenous GRp58 interacts with mTOR. (A) The interaction between endogenous GRp58 and mTOR was assessed by immunoprecipitation (IP) followed by Western blotting (WB) using lysates from HEK293T cells. Endogenous mTOR was immunoprecipitated, and endogenous GRp58 was found associated with it. The specificity of the interaction was determined using a peptide competing with mTOR antibodies (+Pept), a lysate from cells in which mTOR was knocked down (mTOR shRNA), or a goat antibody used as a negative control (Control). The presence of GRp58 and mTOR was detected in the immunoprecipitates (upper two panels) and total lysates (TCL; lower two panels) using the indicated antibodies that recognize endogenous proteins. Graphs represent the mean and standard error of the mean of three independent experiments. *, P < 0.05 compared to the respective control. (B) The interaction between endogenous GRp58 and mTOR was assessed by immunoprecipitation followed by immunoblotting using lysates from HEK293T and HeLa cells and HMEC as described for panel A. The presence of GRp58 and mTOR was detected in the immunoprecipitates (upper two panels) and total lysates (lower two panels) using the indicated antibodies that recognize endogenous proteins. (C) Effect of various stimuli on the interaction between GRp58 and mTOR. HEK293T cells were serum starved in leucine-free medium for 3 h and then stimulated for 15 min with 10% FBS, 100 nM insulin, or 52 μg/ml leucine as indicated. mTOR was immunoprecipitated, and the presence of GRp58 and mTOR in the immunoprecipitates was detected by Western blotting. Immunoprecipitation with an equal amount of goat antibody was used to control for nonspecific binding (Control). Total cell lysates were used to confirm the activation of mTOR by detecting the phosphorylation of S6. The expression of S6, mTOR, and GRp58 in total lysates was confirmed by Western blotting. The graph at the bottom represents the mean and standard error of the mean of three independent experiments. (D) The oxidizing agent PAO decreases the interaction between GRp58 and mTOR. HEK293 cells were stimulated with PAO (5 μM for 30 min). Then, mTOR was immunoprecipitated from total cell lysates prepared in CHAPS buffer, and the presence of interacting GRp58 and mTOR was detected by Western blotting. The graph represents the mean and standard error of the mean of four independent experiments. *, P < 0.05. Total cell lysates from the same experiments were used to detect the presence of GRp58, mTOR, or the indicated proteins and their phosphorylated (p-) versions, i.e., p-S6K and p-4E-BP1 to assess the activation of mTORC1 and p-AKT to assess the activation of mTORC2. The effect of short-term (1 h) and long-term (24 h) incubation with rapamycin (Rapa; 20 ng/ml) in cells stimulated with insulin (100 nM for 15 min) was assessed in parallel. Equivalent experiments were done to assess the effect of PAO or rapamycin on the interaction between Raptor and mTOR (E) or Rictor and mTOR (F).
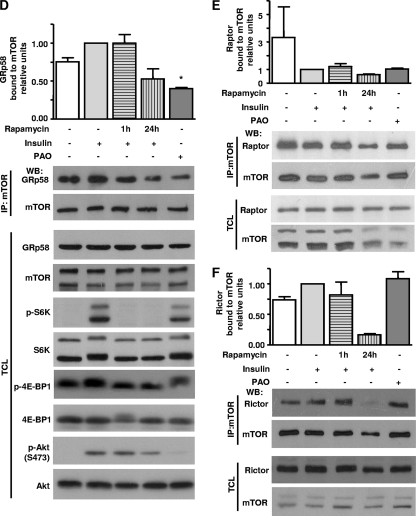
To evaluate whether the interaction between endogenous mTOR and GRp58 can be regulated by conditions that are known to affect the kinase activity of mTOR, we determined the effect of various stimuli on the stability of the mTOR-GRp58 complex. HEK293T cells were deprived of nutrients and stimulated with 10% FBS, insulin, or leucine; then mTOR was immunoprecipitated, and its interaction with GRp58 was determined by Western blotting. None of the conditions employed affected the ability of GRp58 to interact with mTOR (Fig. 2C). The activation of mTOR in response to the different stimuli was confirmed by Western blotting detecting the phosphorylation of S6 protein, a substrate of S6K (Fig. 2C). Collectively, these results indicate that the interaction between mTOR and GRp58, discovered by yeast two-hybrid screening and detected by pulldown of transfected epitope-tagged proteins, occurs between endogenous mTOR and GRp58 in different mammalian cells and that it is not sensitive to variations in the availability of nutrients or growth factors. We then tested the effect of rapamycin on the interaction between GRp58 and mTOR. Since it is known that a short-term incubation with rapamycin affects mTORC1 whereas a chronic incubation with this inhibitor also affects mTORC2 (45), we explored the effect of both conditions on the interaction between GRp58 and mTOR. As shown in Fig. 2D, a 1-h incubation with rapamycin did not affect the interaction between GRp58 and mTOR, whereas it was able to reduce the phosphorylation of S6K and 4E-BP1 without affecting the phosphorylation of Akt, demonstrating its specific effect on mTORC1. Chronic incubation with rapamycin decreased the phosphorylation of mTORC1 and mTORC2 substrates (Fig. 2D) and reduced the interaction of Raptor and Rictor with mTOR (Fig. 2 E and F); it also reduced the interaction between GRp58 and mTOR (Fig. 2D) although not to a significantly reduced level. Since mTORC1 is known to be activated in response to oxidants such as PAO (47), we tested whether this agent affects the interaction between GRp58 and mTOR. As shown in Fig. 2D, PAO produced a significant reduction in the interaction of GRp58 with mTOR. In addition, PAO reduced the interaction between mTOR and Raptor (Fig. 2E), consistent with a previous report (47); interestingly, PAO did not affect the interaction between Rictor and mTOR (Fig. 2F), indicating a lack of effect on mTORC2. These results suggested that GRp58 and PAO preferentially influenced the activity of mTORC1. Consistent with this possibility, PAO showed a positive effect on the phosphorylation of S6K and 4E-BP1, both substrates of mTORC1, without affecting the phosphorylation of Akt at Ser473, known to be phosphorylated by mTORC2 (46).
Changes in GRp58 expression affect the levels of mTORC1.
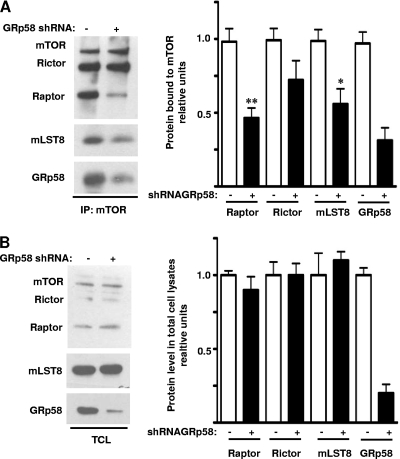
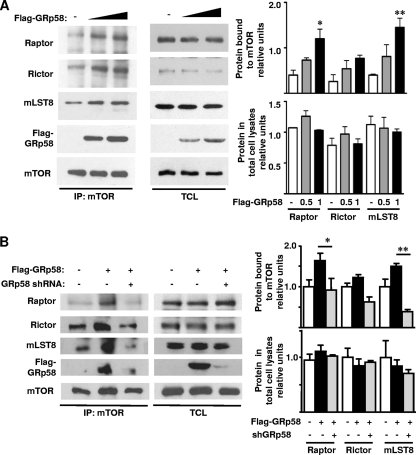
Considering the chaperone function of GRp58 (28, 37), we explored whether its interaction with mTOR had a functional impact on the assembly of mTORC1 and mTORC2. In order to investigate this possibility, we assessed the effect of GRp58 shRNA-mediated knockdown or GRp58 overexpression on the interaction of endogenous Raptor, Rictor, or mLST8 with mTOR in HEK293T cells. As shown in Fig. 3 A, GRp58 deficiency significantly diminished the presence of Raptor and mLST8 in mTOR immunoprecipitates while the reduction of Rictor associated to mTOR was not significantly different in immunoprecipitates from GRp58 knockdown cells compared to control cells. The expression levels of mTOR, Raptor, Rictor, and mLST8 in total cell lysates from GRp58 knockdown cells were similar to the expression levels of these proteins detected in control cells (Fig. 3B). In contrast, as shown in Fig. 4 A (left panel and right upper graph), GRp58 overexpression significantly increased the levels of Raptor and mLST8 interacting with mTOR; while some increase in the levels of Rictor interacting with mTOR was detected, the level did not significantly differ from control immunoprecipitates from cells not overexpressing GRp58 (Fig. 4A, left panel and right upper graph). Overexpression of GRp58 did not increase the levels of mTOR, Raptor, Rictor, or mLST8 in total cell lysates (Fig. 4A, middle panel and right bottom graph). The effect of GRp58 overexpression was also attenuated by GRp58 shRNA-mediated knockdown (Fig. 4B, left panel and right upper graph). Neither GRp58 knockdown nor overexpression altered the levels of mTOR immunoprecipitated (Fig. 3A and 4A). These results demonstrate that GRp58 has a key role in the assembly of mTORC1.
Fig. 3.
GRp58 knockdown decreases the levels of mTORC1. HEK293T cells were transfected with GRp58 shRNA or control plasmid. (A) mTOR was immunoprecipitated from GRp58 knockdown or control cells, and the immunoprecipitates were analyzed by immunoblotting to detect mTOR, Rictor, Raptor, mLST8, and GRp58. The graph on the right represents the mean and standard error of the mean of five independent experiments. *, P < 0.05; **, P < 0.01 (compared to the respective control). (B) The expression of mTOR, Rictor, Raptor, mLST8, and GRp58 in total lysates (TCL) of control and GRp58 knockdown cells was analyzed by Western blotting using the indicated antibodies. The graph on the right represents the results of five independent experiments.
Fig. 4.
GRp58 overexpression increases the levels of mTORC1. (A) HEK293T cells were transfected with increasing concentrations of a plasmid encoding 3×Flag-GRp58 or empty vector. Two days after transfection, mTOR immunoprecipitates were prepared and subjected to immunoblot analysis for endogenous proteins, as indicated, or for GRp58 with anti-Flag antibodies (IP:mTOR). The expression of the indicated components of mTOR complexes and Flag-GRp58 was also detected in total lysates (TCL). Graphs on the right represent the results of three independent experiments. *, P < 0.05; **, P < 0.01 (compared to the respective control). (B) The effect of GRp58 overexpression on the levels of mTOR complexes was attenuated with a specific GRp58 shRNA. HEK293T cells were cotransfected with 3×Flag-GRp58 and GRp58 shRNA or with a control plasmid. Three days after transfection, mTOR immunoprecipitates were obtained. Immunoprecipitates (IP:mTOR) and total lysates (TCL) were immunoblotted with the indicated antibodies. Control lysates were obtained from cells transfected with empty vector. The upper graph represents the results of four independent experiments showing the relative levels of Raptor, Rictor, and mLST8 bound to mTOR. The relative levels of Raptor, Rictor, and mLST8 in total cell lysates are represented on the bottom graph. *, P < 0.05; **, P < 0.001.
Changes in GRp58 expression affect the activity of mTORC1 but not of mTORC2.
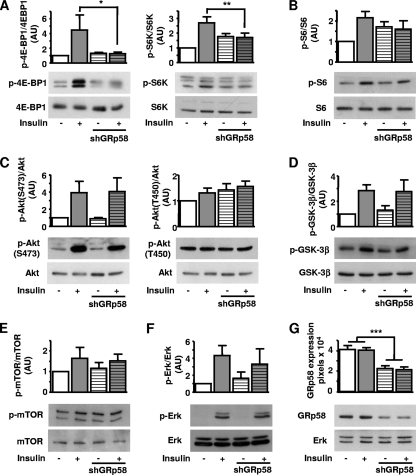
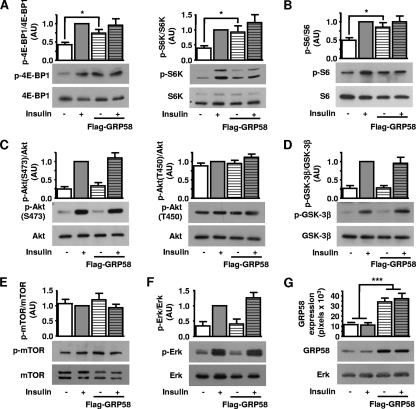
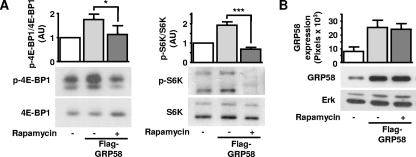
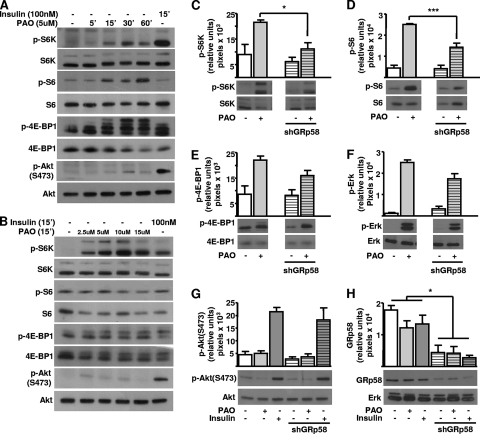
Since GRp58 preferentially interacted with mTORC1 (Fig. 1E) and influenced the assembly of this complex (Fig. 3 and 4), we next tested whether GRp58 knockdown or overexpression had an effect on the activity of this complex or mTORC2. We explored this possibility by assessing the phosphorylation of 4E-BP1, p70S6K, and S6, substrates downstream of mTORC1 (55), and the phosphorylation of Akt at Ser473 and Thr450, both sites recognized as mTORC2 substrates (11, 23, 25, 46). As shown in Fig. 5 A, GRp58 knockdown significantly decreased the phosphorylation of 4E-BP1 and S6K in response to insulin while the effect on the phosphorylation of S6, which was slightly decreased, did not show a statistically significant difference compared to phosphorylation of control cells (Fig. 5B). The activity of mTORC2 was not affected by GRp58 knockdown, as demonstrated by the similar effects of insulin on the phosphorylation of Akt at Ser473 (Fig. 5C, left panel) or GSK-3β, a known Akt substrate (Fig. 5D), in GRp58 knockdown and control cells. The phosphorylation of Akt at Thr450 was not influenced by insulin or GRp58 knockdown (Fig. 5C, right panel). The phosphorylation of mTOR was not significantly stimulated by insulin or influenced by GRp58 knockdown (Fig. 5E). A parallel signaling cascade, resulting in extracellular signal-regulated kinase (ERK) phosphorylation in response to insulin, was not affected by GRp58 knockdown (Fig. 5F). As shown in Fig. 5G, GRp58 shRNA effectively reduced the expression of this protein. No changes in the expression of 4E-BP1, S6K, S6, Akt, GSK-3β, mTOR, or ERK, detected in total cell lysates, were noticed in response to GRp58 knockdown (Fig. 5A to G). In contrast, GRp58 overexpression increased the basal phosphorylation of 4E-BP1, S6K, and S6 (Fig. 6 A and B) while the maximum phosphorylation of these substrates, in response to insulin, was not altered (Fig. 6A and B). GRp58 overexpression did not affect mTORC2 activity, as demonstrated by the lack of effect on the phosphorylation, either basal or in response to insulin, of Akt at Ser473, or the phosphorylation of GSK-3β (Fig. 6C, left panel, and D). The phosphorylation of Akt at Thr450 or the phosphorylation of mTOR was not stimulated by insulin (as already shown in the GRp58 knockdown experiments) and was not affected by GRp58 overexpression. As in the case of the knockdown experiments, the basal or insulin-dependent phosphorylation of ERK was not affected by GRp58 overexpression. No changes in the total expression of 4E-BP1, S6K, S6, Akt, GSK-3β, mTOR, and ERK were noticed in response GRp58 overexpression (Fig. 6A to G). As shown in Fig. 7 A, the phosphorylation of 4E-BP1 and S6K due to GRp58 overexpression (Fig. 7B) was sensitive to rapamycin, further indicating that it occurred in response to mTORC1 activation.
Fig. 5.
mTORC1 signaling decreases in GRp58 knockdown cells. HEK293 cells were transfected with GRp58 shRNA or control plasmid. Transfected cell populations were selected with puromycin and used for experiments 7 days after transfection in order to achieve significant reduction on endogenous GRp58 expression. Six days after transfection, control and GRp58 knockdown cells were serum starved overnight and, the day after, stimulated with 50 nM insulin for 5 min, as indicated. (A) Effect of insulin on the phosphorylation of the mTORC1 substrates 4E-BP-1 and S6K. (B) Phosphorylation of S6, an S6K substrate representing a downstream target of mTORC1. The effect of GRp58 knockdown on mTORC2 signaling is shown in panels C and D, determined by the phosphorylation of Akt or GSK-3β. (E) Effect on mTOR phosphorylation. (F) Effect on ERK phosphorylation. (G) Expression of endogenous GRp58 in control and GRp58 knockdown cells, a representative blot, and the expression of endogenous ERK, used as control, are shown. Graphs represents the mean and standard error of the mean of three to eight independent experiments. *, P < 0.05; **, P < 0.001. Representative blots of the indicated phosphorylated and total proteins are shown below the respective graphs (A to F). AU, arbitrary units.
Fig. 6.
mTORC1 signaling increases as a consequence of GRp58 overexpression. HEK293 cells were transfected with 3×Flag-GRp58 or control plasmid. Two days after transfection, cells were deprived of serum for 3 h and then stimulated with 25 nM insulin for 5 min prior to cell lysis. The experiments in which a significant overexpression of GRp58 was achieved were analyzed by Western blotting using antibodies to detect the phosphorylation of the indicated mTORC1 and mTORC2 substrates (A and C) and their respective downstream targets (B and D) or the phosphorylation of mTOR (E) and ERK (F) using the indicated antibodies. (G) Overexpression of GRp58 was detected with a monoclonal antibody that also recognized the endogenous protein. A representative Western blot is shown, as well as the expression of ERK in total cell lysates, which was used as control. Graphs in panels A to F represent the mean and standard error of the mean of 10 to 13 independent experiments. *, P < 0.05; ***, P < 0.001. Representative blots of the indicated phosphorylated and total proteins are shown below the respective graphs. AU, arbitrary units.
Fig. 7.
The increase in mTORC1 signaling due to GRp58 overexpression is sensitive to rapamycin. HEK293 cells overexpressing GRp58 were incubated with 20 ng/ml rapamycin for 1 h, and the phosphorylation of mTORC1 substrates was determined by Western blotting using antibodies against phosphorylated 4E-BP1 or S6K. (A) Graph represents the results of three (4E-BP1) and four (S6K) independent experiments. Representative blots of the indicated phosphorylated and total proteins are shown below the respective graphs. *, P < 0.05; ***, P < 0.001. (B) overexpression of GRp58 was detected with a monoclonal antibody that also recognized the endogenous protein. A representative Western blot is shown, as well as the expression of ERK in total cell lysates, which was used as a control. AU, arbitrary units.
Role of GRp58 in mTORC1 signaling in response to PAO.
Since the oxidant agent PAO promoted the phosphorylation of mTORC1 substrates and affected the interaction between GRp58 and mTOR (Fig. 2D), we tested whether GRp58 is an essential component of the oxidant mechanism that activates mTORC1. First, we demonstrated that PAO promoted the activation of mTORC1 in a time- and dose-dependent manner, as demonstrated by its effect on the phosphorylation of S6K, S6, and 4E-BP1 (Fig. 8 A and B), without affecting the activity of mTORC2, as demonstrated by its lack of effect on Akt phosphorylation (Fig. 8A and B). The role of GRp58 in the oxidant mechanism that activates mTORC1 was tested in GRp58 knockdown cells. As shown in Fig. 8C and D, GRp58 knockdown significantly decreased the phosphorylation of S6K and S6 in response to PAO while the effect on the phosphorylation of 4E-BP1, which was just slightly decreased, did not show a statistically significant difference compared to control cells (Fig. 8E). PAO exerted a positive effect on the phosphorylation of ERK via a mechanism that was not affected by GRp58 knockdown (Fig. 8F). Consistent with results shown in Fig. 8A and B, PAO did not promote the phosphorylation of Akt at Ser473 in GRp58 knockdown cells (Fig. 8G). Figure 8H shows the effectiveness of GRp58 shRNA, which significantly reduced the expression of GRp58 without affecting the expression of ERK or the other assayed proteins.
Fig. 8.
Role of GRp58 in mTORC1 signaling in response to PAO. (A and B) PAO promotes the activation of mTORC1 but not of mTORC2 in a time- and dose-dependent manner. HEK293 cells were stimulated for the indicated times and with the indicated concentrations of PAO or insulin. The phosphorylation of S6K, S6, and 4E-BP1, indicative of mTORC1 activation, and the phosphorylation of AKT at Ser473, indicative of mTORC2 activation, were assessed by Western blotting using phospho-specific antibodies, as well as the total expression of these proteins using antibodies that recognize them regardless of their phosphorylation status. (C to H) Equivalent experiments were done in GRp58 knockdown HEK293 cells to detect the effect of decreased expression of GRp58 on the activity of mTORC1 (C to E), mTORC2 (G), or ERK (F) in response to PAO (5 μM for 30 min). (H) Expression of endogenous GRp58 in control and GRp58 shRNA (shGRp58) transfected cells. Graphs represent the mean and standard error of the mean of three to four independent experiments. *, P < 0.05; ***, P < 0.001.
mTOR and GRp58 coexist in cytosolic and endoplasmic reticulum fractions.
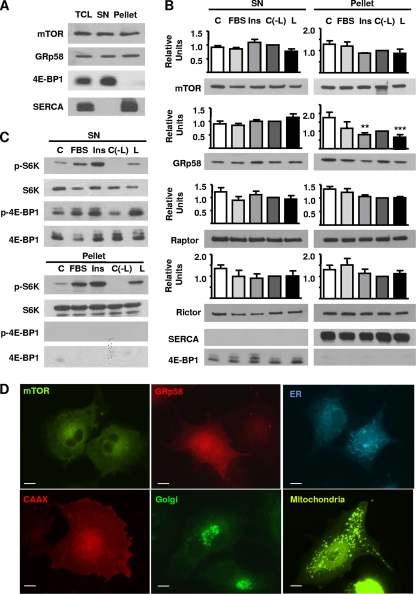
Since mTOR is a cytosolic kinase, eventually associated to the endoplasmic reticulum and other membranous compartments (9, 33, 41, 42), and since GRp58 has been found at the endoplasmic reticulum and also at cytosolic and nuclear fractions (1, 6, 35, 51), the coexistence of mTOR and GRp58 in the same subcellular compartment constitutes a requirement for a physiological role for their interaction. In order to investigate whether mTOR and GRp58 coexist in the same subcellular compartment, we carried out a subcellular fractionation procedure by differential centrifugation of mechanically disrupted HEK293 cells. As shown in Fig. 9A, both mTOR and GRp58 are localized at the cytosolic fraction (supernatant [SN] resulting from centrifugation at 105 × g) as well as at the fraction corresponding to the endoplasmic reticulum (Fig. 9A, pellet). In addition, Raptor and Rictor, components of mTORC1 and mTORC2, respectively, were also present in both fractions (Fig. 9B). 4E-BP-1 and SERCA were used as markers for cytosolic and endoplasmic reticulum fractions, respectively.
Fig. 9.
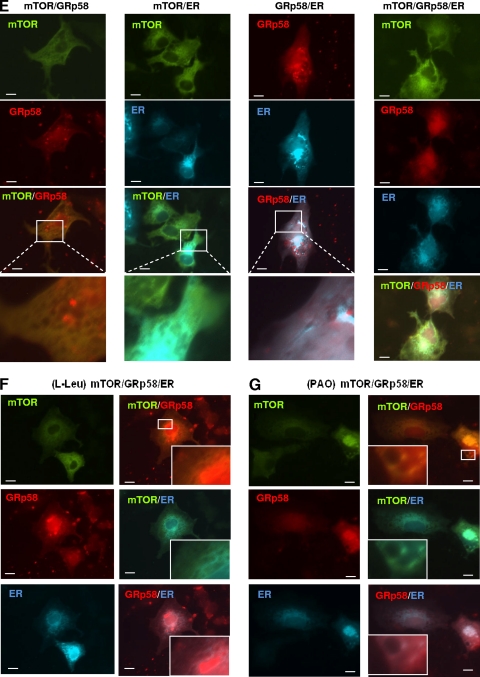
Subcellular distribution of mTOR and GRp58. (A) HEK293 cells were subjected to a subcellular fractionation protocol as described in experimental procedures; 10 μg of the cytosolic fraction (SN) or the endoplasmic reticulum fraction (Pellet) was analyzed to detect the presence of mTOR and GRp58. 4EBP-1 and SERCA2 were detected as cytosolic and endoplasmic reticulum markers, respectively. (B) HEK293 cells were incubated overnight in serum-free medium (C) or serum-free medium also lacking leucine [C(−L)] and then stimulated for 5 min with 10% fetal bovine serum (FBS), 100 nM insulin (Ins), or 52 μg/ml leucine (L). Cells were fractionated, and proteins were detected by Western blotting using specific antibodies. Bars represent the mean densitometric values of mTOR, GRp58, Raptor, and Rictor from three independent experiments; vertical lines represent the standard error of the mean (**, P < 0.01; ***, P < 0.001, with respect to control). (C) The presence and phosphorylation of S6K and 4E-BP1 on cytosolic (SN) and endoplasmic reticulum (Pellet) fractions from cells stimulated as indicated were detected by Western blotting using phospho-specific antibodies (pS6K and p4E-BP1) or antibodies that detect these proteins regardless of their phosphorylation status. (D) Subcellular distribution of mTOR, GRp58, and markers for the endoplasmic reticulum (ER), plasma membrane (CAAX), Golgi apparatus, and mitochondria was observed in COS-7 cells transfected with YFP-mTOR, mCherry-GRp58, CFP-ER, mRFP-CAAX, EGFP-Golgi, and YFP-mitochondria, respectively. At 24 h after transfection, cells were fixed and photographed using a Nikon Eclipse Ti-E fluorescence microscope. YFP-mTOR was artificially detected as a green protein when transfected alone or with mCherry-GRp58. (E) Equivalent experiments were done to detect the subcellular localization of different combinations of cotransfected mTOR, Grp58, and the ER marker in serum-starved cells or in cells stimulated for 15 min with l-leucine (52 μg/ml) (F) or PAO (5 μM) (G). Bar, 10 μm. Representative experiments done in live cells are available in the supplemental material.
We next investigated whether the localization of mTOR, Raptor, Rictor, and GRp58 changes in response to different stimuli. Starved HEK293 cells were treated with 10% fetal bovine serum (FBS), 100 nM insulin, or 52 μg/ml leucine for 5 min, and proteins were analyzed by Western blotting of cytosolic and endoplasmic reticulum fractions. As shown in Fig. 9B, there was a significant decrease in the presence of GRp58 in the endoplasmic reticulum fraction (pellet) obtained from cells stimulated with insulin or leucine (P < 0.01 and P < 0.001, respectively) while there was no significant effect of these stimuli on the distribution of mTOR, Raptor, or Rictor. We further explored the possible association of mTORC1 substrates to the endoplasmic reticulum fraction; as shown in Fig. 9C, S6K but no 4E-BP-1 was associated to the endoplasmic reticulum fraction and was phosphorylated in response to the mentioned stimuli; as expected, both S6K and 4E-BP-1 were found in the cytosolic fraction and were phosphorylated in response to serum, insulin, and leucine. In order to test whether the subcellular distribution of mTOR and GRp58 was compatible with a partial localization of these proteins at the endoplasmic reticulum or potentially other membranous compartments, we compared the localization of YFP-mTOR and mCherry-GRp58 with the localization of different fluorescent markers for membranous compartments including the plasma membrane (mRFP-CAAX), endoplasmic reticulum (CFP-ER), Golgi apparatus (EGFP-Golgi), and mitochondria (tagged with YFP) in transfected COS7 cells, which were selected because they showed clear differences in the distribution of the different markers (Fig. 9D). Both mTOR and GRp58 showed a partially diffused localization, consistent with a fraction of them being cytosolic, and the other fraction was present at subcellular structures more similar to the distribution of the ER marker (Fig. 9D). Thus, we investigated whether mTOR, GRp58, and the ER marker colocalize when transfected in the same cells and determined the potential effect of Leu and PAO on their localization. As shown in Fig. 9E (left panel), mTOR and GRp58 showed similar localizations in cotransfected cells, a fraction of which is compatible with colocalization at endomembranous compartments. When cotransfected with the ER marker, both mTOR and GRp58 showed a distribution compatible with their partial localization at the ER (Fig. 9E, second and third panels, respectively). The simultaneous expression of both mTOR and GRp58 with the ER marker also showed that the distribution of the three is compatible with their colocalization at some regions of the ER (Fig. 9E, right panel). Stimulation with either l-Leu or PAO of cells simultaneously expressing fluorescent mTOR, GRp58, and the ER marker did not dramatically change the localization of these proteins (Fig. 9F and G) compared to control, nonstimulated conditions (Fig. 9E). Representative videos of experiments done in live cells are presented as supplemental material.
Changes in GRp58 expression affect cell proliferation in response to insulin or serum.
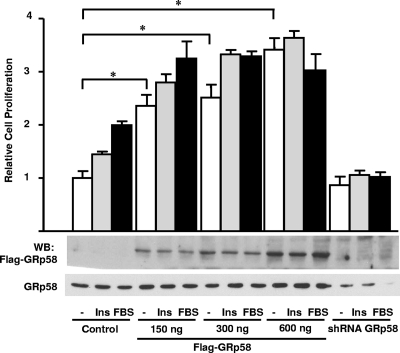
Since mTORC1 activity is known to have a positive effect on cell proliferation (13, 21), we decided to test whether changes in GRp58 expression, which we identified here as a novel mTOR interacting protein, affect cell proliferation. Thus, we evaluated the effect of GRp58 overexpression or knockdown on the proliferation of cells in response to insulin or serum. As shown in Fig. 10, GRp58-overexpressing cells showed a significant increase in proliferation relative to control cells under basal and stimulated conditions. In contrast, GRp58 knockdown interfered with the proliferative effect of insulin or serum (Fig. 10).
Fig. 10.
Changes in the expression of GRp58 affect cell proliferation in response to insulin or serum. HEK293T cells were transfected with 3×Flag-GRp58, GRp58 shRNA, or control plasmid, as indicated. At 36 h posttransfection, 104 cells were seeded in 96-well plates and deprived of serum for 3 h. Then, cells were treated with 100 nM insulin (Ins) or 10% FBS for 24 h. BrdU was added, and the cells were incubated for an additional 4 h. BrdU incorporation was determined as described in Materials and Methods. The graph represents the mean ± standard deviation of cell proliferation compiled from three independent experiments performed in quadruplicate. *, P < 0.01. Representative blots of the expression of transfected GRp58 detected with anti-Flag or an antibody that also detects endogenous GRp58 are shown.
DISCUSSION
Here, we demonstrate that GRp58 interacts with mTOR and regulates the levels and activity of mTORC1. Based on our results, we can postulate that GRp58, a redox-sensitive molecular chaperone, facilitates the assembly of mTORC1 and its ability to sense oxidizing agents. These findings are consistent with the recently described redox-sensitive mechanism in the regulation of the Raptor-mTOR complex (47).
We observed that recombinant GRp58 preferentially interacts with endogenous mTORC1 over mTORC2, suggesting a differential role of GRp58 as a modulator of mTORC1 over mTORC2. This possibility aligns with the known differences in the upstream mechanisms of activation of mTOR complexes (24). Our data suggest a dual role of GRp58 related to its interaction with mTOR. First, it has a positive effect on the assembly of mTORC1, as demonstrated by a reduction in the presence of Raptor and mLST8 in mTOR immunoprecipitates of GRp58 knockdown cells and by an increase in their association to mTOR in GRp58-overexpressing cells. Since the expression of Raptor, Rictor, mLST8, or mTOR was not altered in total cell lysates and since the amount of immunoprecipitable mTOR was comparable under the different experimental conditions, the results also suggest that GRp58 has an effect on the assembly and not the stability of mTOR complexes. Second, GRp58 modulates the function of mTORC1; since the kinase domain of mTOR constitutes the GRp58-interacting site, the positive effect of GRp58 on mTORC1 activity under basal conditions is putatively due to a direct stimulation of mTOR catalytic activity by GRp58, an effect that was sensitive to rapamycin, a specific inhibitor of mTORC1. In addition, GRp58 can also be part of the mechanism used by mTORC1 to detect upstream signals, as indicated by a decrease in the mTORC1 response to insulin in GRp58 knockdown cells. This possibility is also consistent with a mechanism by which mTORC1 responds to changes in cell redox conditions. According to Sarvassov and Sabatini, a redox mechanism influences the activity of mTORC1 (47); our results showing an effect of GRp58 on the function of mTORC1 in response to the oxidizing agent PAO suggest that, by interacting with mTORC1, GRp58 could be part of the redox-sensing mechanism attributed to mTORC1 (47). In response to nutrients, Raptor dissociates from mTOR, facilitating the phosphorylation of its specific substrates (30). Here, we found that PAO exerts a similar effect on the interaction of GRp58 with mTOR, which decreases in response to PAO, an oxidizing agent previously reported to induce dissociation of Raptor and mTOR (47). In addition, GRp58/ERp57 knockdown significantly decreases the phosphorylation of p70S6K and S6 in response to PAO, further supporting the participation of GRp58/ERp57 as a critical component of the mTOR redox-sensing mechanism. In this regard, a phospho-proteomic approach oriented to identify new targets of leucine deprivation in muscle cells suggested some correlation between mTOR and GRp58, as indicated by an increase in GRp58 phosphorylation in response to leucine deprivation (49).
The mechanism by which GRp58 regulates the assembly of mTORC1 could be dependent on its ability to catalyze the formation and/or isomerization of disulfide bonds (2, 48). In this regard, it has been described that the FATC (FAT carboxyl-terminal) domain of yeast TOR1 presents a structural motif consisting of an α-helix and a disulfide-bonded loop. Upon reduction of this disulfide bond, a strong conformational change occurs, increasing the flexibility of the loop region. Moreover, point mutants in this region, in which cysteines were replaced by serines, altered the interacting properties of FATC with other regions of TOR and with other cellular partners (7). Accordingly, Sarvassov and Sabatini demonstrated that thiol oxidants decrease the interaction between Raptor and mTOR, whereas a reducing reagent stabilizes this complex (44). In addition, it has been recently demonstrated that an elevated oxidative stress modifies TORC1 and prevents its binding to the FKBP12-rapamycin complex, ultimately leading to rapamycin resistance (36). Collectively, these studies suggest the regulation of TOR pathway by a redox-sensitive mechanism which, according to our present data, can be based on the interaction between mTOR and GRp58.
We found that mTOR and GRp58 coexist in cytosolic and endoplasmic reticulum fractions, opening the possibility that their interactions occur associated to different subcellular compartments. Additional evidence supporting a functional role for mTORC1 associated to the endoplasmic reticulum is the fact that this fraction contains p70S6K phosphorylated in samples from cells stimulated with serum, insulin, or leucine. An alternative possibility is that p70S6K translocates to the endoplasmic reticulum upon phosphorylation; however, the levels of p70S6K associated to this fraction were similar in control and stimulated cells, further supporting the former interpretation. The requirement of mTORC1 translocation to membranous compartments as part of its mechanism of activation in response to amino acids was recently demonstrated by Sabatini and colleagues (41, 42). Our results are consistent with their findings and extend the possibilities in terms of the identity of membranous organelles linked to mTORC1 activation since these investigators identified late endosomes and lysosomes as the organelles to which the activation of mTORC1 is linked. Interestingly, Rheb, a small GTPase that directly activates mTORC1, has been found associated to the endoplasmic reticulum via a carboxyl-terminal farnesylation, which is essential for the oncogenic effects of a constitutively active Rheb mutant (20, 26). Altogether, these findings support an emerging role for membranous organelles as intervening components in the assembly and activation of mTORC1, providing a spatial element of regulation that might also contribute to restrict the phosphorylation of specific substrates, as suggested by the presence and phosphorylation of p70S6K associated to the endoplasmic reticulum fraction while 4E-BP-1, the other widely recognized substrate of mTORC1, was not associated to this subcellular fraction.
Our study revealed that GRp58 has a key role in cellular proliferation. In HEK293T cells, GRp58 overexpression induced cellular proliferation, whereas GRp58 knockdown prevented the proliferative effect of insulin and serum, sustaining the essential role of GRp58 in cellular proliferation. It is well known that the mTOR pathway is a fundamental regulator of cellular proliferation under normal (18) and malignant conditions (8, 12). Our data suggest that the intervention of GRp58 in cell proliferation can be attributed, at least in part, to its interaction with mTOR, although we cannot rule out an mTOR-independent mechanism in the proliferative responses observed in cells in which the expression of GRp58 was altered.
In conclusion, we have identified GRp58 as a novel mTOR-interacting protein that promotes the assembly and activation of mTORC1.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Shu-Bing Qian, Gyorgy Hajnoczky, Tamas Balla, James D. Lechleiter, and David Sabatini, who kindly provided DNA constructs. Technical assistance provided by Estanislao Escobar Islas, Margarita Valadez Sánchez, David Pérez, and Jaime Estrada is acknowledged.
This work was supported by Consejo Nacional de Ciencia y Tecnología (CONACyT) grants 61127 (to J.V.P.) and 79429 (to G.R.C.). I.R.-R. and I.B.-V. are graduate students supported by fellowships from CONACyT.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 14 February 2011.
REFERENCES
- 1. Adikesavan A. K., Unni E., Jaiswal A. K. 2008. Overlapping signal sequences control nuclear localization and endoplasmic reticulum retention of GRP58. Biochem. Biophys. Res. Commun. 377:407–412 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2. Beynon-Jones S. M., Antoniou A. N., Powis S. J. 2006. Mutational analysis of the oxidoreductase ERp57 reveals the importance of the two central residues in the redox motif. FEBS Lett. 580:1897–1902 [DOI] [PubMed] [Google Scholar]
- 3. Chen J. 2004. Novel regulatory mechanisms of mTOR signaling. Curr. Top. Microbiol. Immunol. 279:245–257 [DOI] [PubMed] [Google Scholar]
- 4. Chichiarelli S., et al. 2010. Role of ERp57 in the signaling and transcriptional activity of STAT3 in a melanoma cell line. Arch. Biochem. Biophys. 494:178–183 [DOI] [PubMed] [Google Scholar]
- 5. Coe H., Jung J., Groenendyk J., Prins D., Michalak M. 2010. ERp57 modulates STAT3 signaling from the lumen of the endoplasmic reticulum. J. Biol. Chem. 285:6725–6738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coe H., Michalak M. 2010. ERp57, a multifunctional endoplasmic reticulum resident oxidoreductase. Int. J. Biochem. Cell Biol. 42:796–799 [DOI] [PubMed] [Google Scholar]
- 7. Dames S. A., Mulet J. M., Rathgeb-Szabo K., Hall M. N., Grzesiek S. 2005. The solution structure of the FATC domain of the protein kinase target of rapamycin suggests a role for redox-dependent structural and cellular stability. J. Biol. Chem. 280:20558–20564 [DOI] [PubMed] [Google Scholar]
- 8. Dancey J. E. 2006. Therapeutic targets: MTOR and related pathways. Cancer Biol. Ther. 5:1065–1073 [DOI] [PubMed] [Google Scholar]
- 9. Drenan R. M., Liu X., Bertram P. G., Zheng X. F. 2004. FKBP12-rapamycin-associated protein or mammalian target of rapamycin (FRAP/mTOR) localization in the endoplasmic reticulum and the Golgi apparatus. J. Biol. Chem. 279:772–778 [DOI] [PubMed] [Google Scholar]
- 10. Easton J. B., Houghton P. J. 2006. mTOR and cancer therapy. Oncogene 25:6436–6446 [DOI] [PubMed] [Google Scholar]
- 11. Facchinetti V., et al. 2008. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 27:1932–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fasolo A., Sessa C. 2008. mTOR inhibitors in the treatment of cancer. Expert Opin. Investig. Drugs 17:1717–1734 [DOI] [PubMed] [Google Scholar]
- 13. Fingar D. C., Blenis J. 2004. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 23:3151–3171 [DOI] [PubMed] [Google Scholar]
- 14. Fingar D. C., Salama S., Tsou C., Harlow E., Blenis J. 2002. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 16:1472–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garbi N., Hammerling G., Tanaka S. 2007. Interaction of ERp57 and tapasin in the generation of MHC class I-peptide complexes. Curr. Opin. Immunol. 19:99–105 [DOI] [PubMed] [Google Scholar]
- 16. Garbi N., Tanaka S., Momburg F., Hammerling G. J. 2006. Impaired assembly of the major histocompatibility complex class I peptide-loading complex in mice deficient in the oxidoreductase ERp57. Nat. Immunol. 7:93–102 [DOI] [PubMed] [Google Scholar]
- 17. Grillo C., et al. 2006. Cooperative activity of Ref-1/APE and ERp57 in reductive activation of transcription factors. Free Radic. Biol. Med. 41:1113–1123 [DOI] [PubMed] [Google Scholar]
- 18. Guertin D. A., et al. 2006. Ablation in mice of the mTORC components Raptor, Rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCα, but not S6K1. Dev. Cell 11:859–871 [DOI] [PubMed] [Google Scholar]
- 19. Guo G. G., et al. 2002. Association of the chaperone glucose-regulated protein 58 (GRP58/ER-60/ERp57) with Stat3 in cytosol and plasma membrane complexes. J. Interferon Cytokine Res. 22:555–563 [DOI] [PubMed] [Google Scholar]
- 20. Hanker A. B., et al. 2010. Differential requirement of CAAX-mediated posttranslational processing for Rheb localization and signaling. Oncogene 29:380–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hay N., Sonenberg N. 2004. Upstream and downstream of mTOR. Genes Dev. 18:1926–1945 [DOI] [PubMed] [Google Scholar]
- 22. Hernandez-Negrete I., et al. 2007. P-Rex1 links mammalian target of rapamycin signaling to Rac activation and cell migration. J. Biol. Chem. 282:23708–23715 [DOI] [PubMed] [Google Scholar]
- 23. Ikenoue T., Inoki K., Yang Q., Zhou X., Guan K. L. 2008. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 27:1919–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jacinto E. 2008. What controls TOR? IUBMB Life 60:483–496 [DOI] [PubMed] [Google Scholar]
- 25. Jacinto E., et al. 2006. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 127:125–137 [DOI] [PubMed] [Google Scholar]
- 26. Jiang H., Vogt P. K. 2008. Constitutively active Rheb induces oncogenic transformation. Oncogene 27:5729–5740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaizuka T., et al. 2010. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J. Biol. Chem. 285:20109–20116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Khanal R. C., Nemere I. 2007. The ERp57/GRp58/1,25D3-MARRS receptor: multiple functional roles in diverse cell systems. Curr. Med. Chem. 14:1087–1093 [DOI] [PubMed] [Google Scholar]
- 29. Kim D. H., Sabatini D. M. 2004. Raptor and mTOR: subunits of a nutrient-sensitive complex. Curr. Top. Microbiol. Immunol. 279:259–270 [DOI] [PubMed] [Google Scholar]
- 30. Kim D. H., et al. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163–175 [DOI] [PubMed] [Google Scholar]
- 31. Kita K., et al. 2006. Evidence for phosphorylation of rat liver glucose-regulated protein 58, GRP58/ERp57/ER-60, induced by fasting and leptin. FEBS Lett. 580:199–205 [DOI] [PubMed] [Google Scholar]
- 32. Li W., et al. 2007. Hypoxia-induced endothelial proliferation requires both mTORC1 and mTORC2. Circ. Res. 100:79–87 [DOI] [PubMed] [Google Scholar]
- 33. Liu X., Zheng X. F. 2007. Endoplasmic reticulum and Golgi localization sequences for mammalian target of rapamycin. Mol. Biol. Cell 18:1073–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Markus M., Benezra R. 1999. Two isoforms of protein disulfide isomerase alter the dimerization status of E2A proteins by a redox mechanism. J. Biol. Chem. 274:1040–1049 [DOI] [PubMed] [Google Scholar]
- 35. Ndubuisi M. I., Guo G. G., Fried V. A., Etlinger J. D., Sehgal P. B. 1999. Cellular physiology of STAT3: where's the cytoplasmic monomer? J. Biol. Chem. 274:25499–25509 [DOI] [PubMed] [Google Scholar]
- 36. Neklesa T. K., Davis R. W. 2008. Superoxide anions regulate TORC1 and its ability to bind Fpr1:rapamycin complex. Proc. Natl. Acad. Sci. U. S. A. 105:15166–15171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ni M., Lee A. S. 2007. ER chaperones in mammalian development and human diseases. FEBS Lett. 581:3641–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Polak P., Hall M. N. 2006. mTORC2 Caught in a SINful Akt. Dev. Cell 11:433–434 [DOI] [PubMed] [Google Scholar]
- 39. Qian S. B., et al. 2010. mTORC1 links protein quality and quantity control by sensing chaperone availability. J. Biol. Chem. 285:27385–27395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rosner M., Hengstschlager M. 2008. Cytoplasmic and nuclear distribution of the protein complexes mTORC1 and mTORC2: rapamycin triggers dephosphorylation and delocalization of the mTORC2 components rictor and sin1. Hum. Mol. Genet. 17:2934–2948 [DOI] [PubMed] [Google Scholar]
- 41. Sancak Y., et al. 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141:290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sancak Y., et al. 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320:1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sarbassov D. D., et al. 2004. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14:1296–1302 [DOI] [PubMed] [Google Scholar]
- 44. Sarbassov D. D., Ali S. M., Sabatini D. M. 2005. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 17:596–603 [DOI] [PubMed] [Google Scholar]
- 45. Sarbassov D. D., et al. 2006. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell 22:159–168 [DOI] [PubMed] [Google Scholar]
- 46. Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. 2005. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307:1098–1101 [DOI] [PubMed] [Google Scholar]
- 47. Sarbassov D. D., Sabatini D. M. 2005. Redox regulation of the nutrient-sensitive raptor-mTOR pathway and complex. J. Biol. Chem. 280:39505–39509 [DOI] [PubMed] [Google Scholar]
- 48. Satoh M., Shimada A., Kashiwai A., Saga S., Hosokawa M. 2005. Differential cooperative enzymatic activities of protein disulfide isomerase family in protein folding. Cell Stress Chaperones 10:211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Talvas J., et al. 2008. Phospho-proteomic approach to identify new targets of leucine deprivation in muscle cells. Anal. Biochem. 381:148–150 [DOI] [PubMed] [Google Scholar]
- 50. Toschi A., et al. 2009. Regulation of mTORC1 and mTORC2 complex assembly by phosphatidic acid: competition with rapamycin. Mol. Cell. Biol. 29:1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Turano C., Coppari S., Altieri F., Ferraro A. 2002. Proteins of the PDI family: unpredicted non-ER locations and functions. J. Cell Physiol. 193:154–163 [DOI] [PubMed] [Google Scholar]
- 52. Varma S., Khandelwal R. L. 2007. Effects of rapamycin on cell proliferation and phosphorylation of mTOR and p70(S6K) in HepG2 and HepG2 cells overexpressing constitutively active Akt/PKB. Biochim. Biophys. Acta 1770:71–78 [DOI] [PubMed] [Google Scholar]
- 53. Vazquez-Prado J., Basile J., Gutkind J. S. 2004. Modular architecture and novel protein-protein interactions regulating the RGS-containing Rho guanine nucleotide exchange factors. Methods Enzymol. 390:259–285 [DOI] [PubMed] [Google Scholar]
- 54. Wang X., Proud C. G. 2006. The mTOR pathway in the control of protein synthesis. Physiology (Bethesda) 21:362–369 [DOI] [PubMed] [Google Scholar]
- 55. Wullschleger S., Loewith R., Hall M. N. 2006. TOR signaling in growth and metabolism. Cell 124:471–484 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.