Abstract
Replicative senescence is a permanent cell cycle arrest in response to extensive telomere shortening. To understand the mechanisms behind a permanent arrest, we screened for factors affecting replicative senescence in budding yeast lacking telomere elongation pathways. Intriguingly, we found that DNA polymerase epsilon (Pol ε) acts synergistically with Exo1 nuclease to maintain replicative senescence. In contrast, the Pol ε-associated checkpoint and replication protein Mrc1 facilitates escape from senescence. To understand this paradox, in which DNA-synthesizing factors cooperate with DNA-degrading factors to maintain arrest, whereas a checkpoint protein opposes arrest, we analyzed the dynamics of double- and single-stranded DNA (ssDNA) at chromosome ends during senescence. We found evidence for cycles of DNA resection, followed by resynthesis. We propose that resection of the shortest telomere, activating a Rad24Rad17-dependent checkpoint pathway, alternates in time with an Mrc1-regulated Pol ε resynthesis of a short, double-stranded chromosome end, which in turn activates a Rad953BP1-dependent checkpoint pathway. Therefore, instead of one type of DNA damage, different types (ssDNA and a double-strand break-like structure) alternate in a “vicious circle,” each activating a different checkpoint sensor. Every time resection and resynthesis switches, a fresh signal initiates, thus preventing checkpoint adaptation and ensuring the permanent character of senescence.
INTRODUCTION
Perhaps the most astonishing feature of replicative senescence is its permanent character. Although adaptation to a persistent stimulus is one of the most common physiological processes in nature, cells like fibroblasts remain senescent indefinitely, spending energy to maintain a state that inhibits their natural potential to divide (5). One hypothesis is that replicative senescence was evolutionary selected to avoid proliferation of cells with unrepairable damage, because such cells would generate chromosomally unstable daughters. This hypothesis seems reasonable for multicellular organisms in which chromosomal instability would lead to cancer. However, unicellular yeast also senesces under experimental conditions in which telomerase is inactivated (24). Since wild-type yeast has telomerase activity (7), it is unlikely that it has developed and selected a robust response to telomerase inactivation, which is presumably a very rare event in the wild. Therefore, replicative senescence may not be very different from a conventional checkpoint response to DNA damage. Understanding how a checkpoint response is maintained for the equivalent of tens, perhaps even hundreds, of generations in yeast will help to uncover mechanisms required to maintain replicative senescence in higher organisms.
Yeast cells adapt to a single unrepaired double-strand break (DSB) within hours, an adaptation meaning that they resume cell cycle progression in the absence of repair (19, 20, 31, 36). However, adaptation does not usually occur in cells arrested in response to telomere attrition (39). One explanation could be that short telomeres in a senescent cell are sending many more damage signals than a single DSB. However, data suggest that one or perhaps a few critically short telomeres are sufficient to induce senescence in yeast or mammalian cells (1, 41). Another explanation could be that a short telomere mobilizes more checkpoint pathways than a DSB. Intriguingly, the opposite was found in budding yeast. The Rad24Rad17-9-1-1 complex, Mec1ATR, and Ddc2ATRIP are essential for replicative senescence, whereas Rad953BP1 or Rad53Chk2 is apparently not required (10, 15). Therefore, only a subset of the checkpoint proteins responding to a DSB is required for cell cycle arrest in response to telomere attrition (10, 15).
A role for Rad953BP1 during replicative senescence might have been masked by the emergence of survivors dependent on alternative lengthening of telomeres or subtelomeres (ALTOS). We introduce the term “ALTOS” to describe the outcome of any pathway able to maintain/elongate (sub) telomeres in the absence of telomerase. ALTOS survivors could be either Rad52 dependent or Rad52 independent (13, 17). Budding yeast cells from most-genetic backgrounds do not form ALTOS survivors in the absence of Rad52. Instead, they enter senescence and remain senescent (13). Yet, telomerase- and ALTOS-negative cells can escape from senescence when nucleases like Exo1 or helicases like Sgs1 are inactivated (18, 28). Exo1 nuclease degrades dysfunctional telomeres (26, 27). Escapers are called PAL survivors (palindrome-immortalized survivors).
PAL survivors are different from ALTOS survivors. Unlike ALTOS survivors, PAL survivors do not maintain/elongate (sub)telomeres. Instead, PAL survivors progressively lose telomeres, subtelomeres, and single-gene loci (18, 28). After many postsenescence population doublings, essential genes become endangered by degradation and cells that formed end-chromosomal DNA palindromes (inverted duplications) take over the population (25, 27). Intriguingly, many generations before forming palindromes, PAL survivors escape from senescence in the absence of TG repeats or any other specific sequences at their chromosome ends (25, 27). Therefore, PAL survivors must have compromised the mechanism required to maintain replicative senescence in order to escape. Similarly, DNA-damaged mammalian cells in the process of becoming cancerous are thought to evade or to disable the DNA damage checkpoint control before they start reshaping their genome, including by forming DNA palindromes (35).
In this study, we used budding yeast lacking both telomerase and ALTOS pathways to address the mechanisms required to maintain replicative senescence. We show that subunits of DNA polymerase epsilon (Pol ε) are required but that Mrc1, a polymerase epsilon-associated checkpoint protein, is opposing senescence. We integrate these and other results into a model of replicative senescence called the “vicious-circle model.”
MATERIALS AND METHODS
Yeast strains and cell culture.
All strains are derivates of W303 and RAD5+. The tlclΔ rad52Δ strains originated from the DLY2151 diploid (TLC1/tlc1Δ::HIS3, RAD52/rad52Δ::TRP1, EXO1/exo1Δ::LEU2). In this diploid, we deleted POL32, DPB3, DPB4, or MRC1 with G418-MX cassettes, as previously described (22). Diploid cells were then sporulated and haploids selected by random spore analysis. To reconfirm their genotype, we tested all selected haploid strains by PCR and some also by Southern blotting. Germinated spores were initially grown on YPD (yeast extract, peptone, and dextrose) plates for 3 to 4 days to form colonies. After circa 25 population doublings on germination plates, cells were propagated as follows. For experiments requiring large number of cells (single-stranded DNA [ssDNA] and chromatin immunoprecipitation [ChIP] measurements), cells were propagated in liquid YPD and diluted daily before senescence, followed by a daily change of medium during senescence. For experiments analyzing generation of PAL survivors, strains taken directly from germination plates were propagated on YPD plates every second day until most cells stopped proliferating (became senescent) by pooling circa 1 × 107 cells with a toothpick and streaking them onto fresh YPD plates. Strains escaping from senescence (PAL survivors) were propagated every 4 days.
Cell cycle analysis was performed by fluorescence microscopy after samples were stained with DAPI (4′,6-diamidino-2-phenylindole). The following fractions were counted: cells without buds (in the G1 phase), cells with small buds (in the S phase), cells with large anucleated buds (at the G2/M transition), and cells with nucleated buds (in anaphase/telophase). An exponentially growing culture of wild-type budding yeast has cells similarly distributed between these stages of the cell cycle.
ssDNA measurements were performed using quantitative analysis of ssDNA (QAOS), as previously described (3), except that we used asynchronous populations of cells cultivated at 25°C. Genomic DNA was extracted, purified, and quantified at a centromere-proximal locus situated on chromosome 5R (PAC2). The ssDNA was quantified in the Y′ subtelomeric repeats, which are found at 2/3 chromosome ends. Quantifications were performed at the “Y′-1000” and “Y′-5000” regions (meaning 1,000 bp and 5,000 bp, respectively, from the chromosome ends) and normalized to the level for Y′ subtelomeric DNA. Additionally, ssDNA was quantified at the YER188W single-gene locus (situated circa 8.5 kb from the chromosome 5R end) and normalized to the level for PAC2 DNA.
ChIP was carried out by standard methods (4, 9). We performed ChIP in parallel to ssDNA measurements on samples collected during midsenescence (passages 9 to 13), rather than during early senescence or presenescence, to obtain the maximum possible number of senescent cells. The number of cells is a limiting factor during replicative senescence experiments. The association of Pol2, Exo1, and Rad9 with chromatin was detected using the following specific antibodies, distributed by Insight Biotechnology: sc-50442 (anti-Rad9), sc-19941 (anti-Exo1), and sc-6753 (anti-Pol2). sc-19941 detects both hExo1 and scExo1. Cells from each cross-linked sample were additionally treated with anti-goat antibodies (sc-2033) to assess the background cross-linking. Input and precipitated DNAs were quantified by real-time PCR with primers and TaqMan probes specific to a centromere-proximal locus (ERG26) or to subtelomeres (at about 1-kb from the chromosome ends).
RESULTS
Single-stranded DNA accumulates and declines periodically during senescence.
To investigate the mechanisms required to maintain replicative senescence, we induced senescence by generating and propagating budding yeast cells lacking both TLC1, encoding the RNA component of telomerase, and RAD52, essential for recombination-induced ALTOS. Replicative senescence in this model system is similar to human replicative senescence, because most human somatic cells do not express telomerase and do not undergo alternative lengthening of telomeres (ALT). Cells in the tlc1Δ rad52Δ background arrest the cell cycle within a few days and remain arrested until death (17). Therefore, the signal(s) for cell cycle arrest in response to telomere attrition would also be expected to last until cellular death. One signal could be the accumulation of ssDNA at telomeres, because ssDNA, a potent checkpoint activator, was previously detected in senescent yeast and mice with short telomeres (25, 32). ssDNA in telomerase-deficient cells is partially dependent upon the Exo1 nuclease (27, 32).
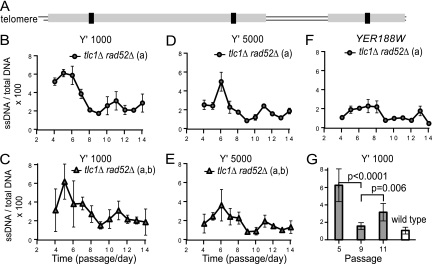
To determine if levels of ssDNA are consistently elevated during senescence, we performed QAOS (43) with cell samples collected from early senescence to midsenescence (passages 4 to 14). The ssDNA was analyzed at distances of 1 kb (Y′-1000) and 5 kb (Y′-5000) from the chromosome ends for the Y′ subtelomeres and also at 8.5 kb from the right end of chromosome V (the YER188W single-gene locus). Interestingly, we found that ssDNA did not remain constant during senescence. Initially, ssDNA accumulated up to 6% at Y′-1000 (Fig. 1B and C). However, a gradual decline followed. The lowest ssDNA fraction was about 1.5% (at passage 9). Then, ssDNA reaccumulated, reaching about 3% at passage 11, and declined afterwards (Fig. 1B and C). A similarly oscillatory pattern was observed at Y′-5000 (Fig. 1D and E) and also at YER188W (Fig. 1F), except that cells accumulated considerably less ssDNA further from telomeres. In summary, ssDNA accumulates and declines periodically during replicative senescence.
Fig. 1.
Dynamics of ssDNA during replicative senescence. (A) Cartoon representing the right arm of chromosome V. (B to G) To ensure a sufficient number of senescent cells, each of three independently generated tlc1Δ rad52Δ strains was taken from germination plates, pooled at passage 1, and propagated in groups a and b. (B, D, F) Dynamics of ssDNA during senescence, measured for group a at Y′-1000, at Y′-5000, and at YER188W. We called the small sequences in the Y′ subtelomeric repeats situated about 1,000 bp and 5,000 bp from the chromosome ends Y′-1000 and Y′-5000, respectively. Error bars represent the standard deviations (SD) for three measurements. (C, E, G) Average ssDNA calculated for groups a and b at Y′-1000 and at Y′-5000. Error bars represent the SD for six measurements. For group b, the dynamics of ssDNA is presented in Fig. 4 (marked as DPB+). (G) Statistically significant ssDNA values from senescent strains analyzed for panel E were compared to the average ssDNA values from two wild-type strains treated with nocodazole (to avoid the S-phase-specific ssDNA). Similar P values were also obtained for each group.
At least two cycles of ssDNA accumulation and decline were detected. Statistical analysis of data from six ssDNA measurements, performed in two independent groups of isogenic strains, indicated an extremely significant difference (P < 0.0001) between passages 5 and 9, marking the slope of the first cycle, and also between passages 9 and 11 (P = 0.006), marking the slope of a second cycle (Fig. 1G). In conclusion, the oscillations in ssDNA are nonrandom, suggesting the existence of mechanisms reducing the amount of ssDNA during replicative senescence. Such mechanisms are not entirely surprising, considering that ssDNA is a form of damage that cells, including nondividing cells, may have to deal with frequently. For example, UV damage in quiescent cells triggers under certain circumstances an Exo1-dependent ssDNA accumulation that has to be repaired by DNA resynthesis (11). What is surprising is that ssDNA decreases to almost wild-type levels during replicative senescence (Fig. 1G).
Alternating ssDNA accumulation and decline correspond to alternating checkpoint subpathways.
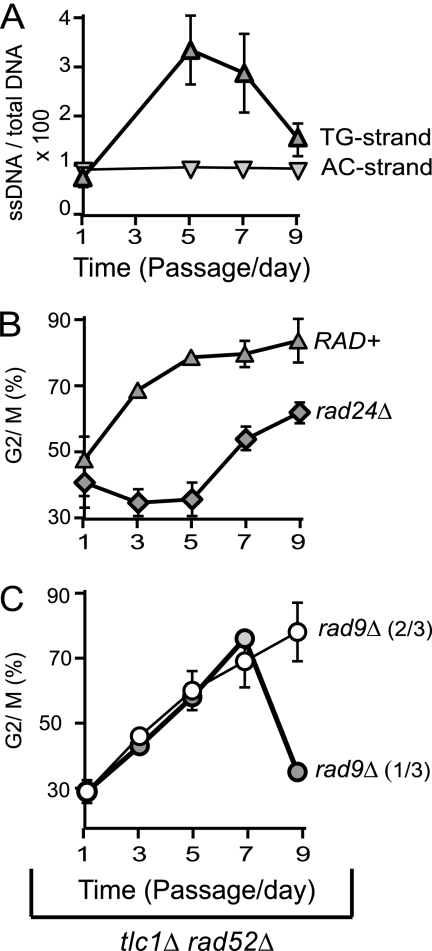
The decline in ssDNA observed during senescence, to values below the threshold required to maintain arrest, may result in a transient cell cycle reentry. Alternatively, a different checkpoint stimulus might be keeping cells arrested when ssDNA is low. To find out which hypothesis is true, we analyzed ssDNA in correlation with cell cycle distribution in three freshly generated tlc1Δ rad52Δ strains, pooled at passage 1, and propagated into senescence. We found that ssDNA (the TG-rich strand) accumulated at passages 1 to 5 and declined at passages 7 to 9 (Fig. 2A), consistent with the pattern observed in Fig. 1. In contrast, ssDNA on the opposite strand (the AC-rich strand) remained constantly below 1%, and therefore, it could be excluded as a potential checkpoint stimulus (Fig. 2A). Accumulation of ssDNA correlated with cells accumulating in the G2/M phase of the cell cycle, culminating with over 75% tlc1Δ rad52Δ cells arresting in G2/M at passage 5 (marked as RAD+ in Fig. 2B). The remaining cells were (arrested?) in G1. However, the decline in ssDNA at passages 7 to 9 (Fig. 2A) did not correlate with cell cycle reentry (Fig. 2B). Taken together, these data suggest that ssDNA is most likely the trigger for replicative senescence. However, signals other than ssDNA are required to maintain arrest at a later time during replicative senescence, when ssDNA declines.
Fig. 2.
Alternating ssDNA accumulation and decline correlate with alternating checkpoint responses. (A) ssDNA quantified by QAOS in subtelomeres of the group RAD+, consistent with three tlc1Δ rad52Δ strains taken directly from germination plates, pooled at passage 1, and propagated into senescence. Error bars represent the standard deviations (SD) for three measurements. (B and C) Fraction of cells of tlc1Δ rad52Δ strains (with or without rad24Δ or rad9Δ mutations) accumulating in the G2/M phase of the cell cycle, once taken from germination plates and propagated into senescence.
The hypothesis that ssDNA triggers replicative senescence but that other stimuli replace ssDNA to maintain senescence was further supported by investigations into the checkpoint requirements. We found that a rad24Δ mutation in the RAD24 checkpoint gene prevented tlc1Δ rad52Δ cells from arresting in G2/M at passages 1 to 5 (Fig. 2A). However, a high fraction of cells arrested later on (at passages 7 to 9), despite the rad24Δ mutation (Fig. 2B). Interestingly, a rad9Δ mutation in the RAD9 checkpoint gene had the opposite effect relative to a rad24Δ mutation: it permitted arrest at passages 1 to 5, whereas it prevented one-third of the strains from maintaining arrest at passages 7 to 9 (Fig. 2B). This indicates that Rad24Rad17 is essential for the entry into replicative senescence (passages 1 to 5) but that Rad953BP1 plays no significant role at that time, consistent with data from the telomerase-negative RAD52+ background (10, 15). However, a few passages later, arrest becomes Rad953BP1 dependent and Rad24Rad17 independent. In summary, a checkpoint switch takes place during replicative senescence.
The checkpoint switch from Rad24Rad9 to Rad953BP1 (Fig. 2B and C) corresponded to a switch in ssDNA dynamics, from accumulation to decline (Fig. 2A). It is known that Rad24Rad17 is among the first checkpoint proteins to detect ssDNA (21) but that Rad953BP1 responds to double-strand breaks (DSBs) and the adjacent chromatin modifications (38). Therefore, we propose that resection of the shortest telomere(s) activates a Rad24Rad17-dependent pathway that initiates replicative senescence but that the decline in ssDNA generates a short double-stranded chromosome end (resembling a DSB) which in turn activates a Rad953BP1-dependent subpathway to maintain replicative senescence.
Pol ε maintains replicative senescence, acting synergistically with Exo1 nuclease.
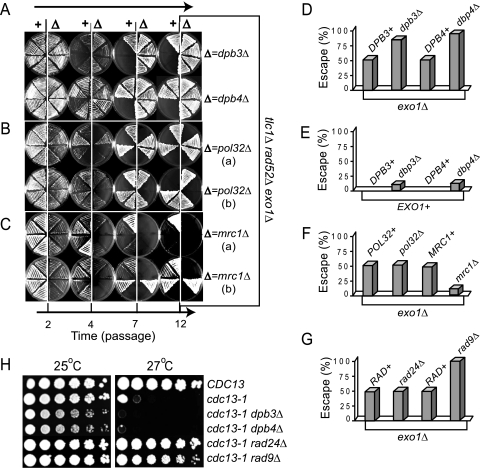
A decline in ssDNA could be caused either by resynthesis to dsDNA or by ssDNA degradation. If DNA resynthesis was the main cause of ssDNA decline, then a DNA polymerase should be as important as Rad953BP1 for maintaining replicative senescence. To test this hypothesis, we propagated into senescence freshly generated tlc1Δ rad52Δ exo1Δ yeast strains, with or without mutations in genes encoding the nonessential subunits of DNA polymerases. Examples of this assay are shown in Fig. 3A to C. Each plate was inoculated with eight strains: tlc1Δ rad52Δ exo1Δ controls on the left and strains with an additional mutation on the right. At passage 4, all strains arrested proliferation. At passage 7, a fraction of strains escaped from arrest and proliferated afterwards. Such strains are called PAL survivors (28).
Fig. 3.
Effect of nonessential subunits of DNA polymerases and Mrc1 on replicative senescence. (A to C) Representative plates photographed at the indicated time points. Each plate was inoculated with eight independent, freshly generated strains, taken directly from germination plates. Strains on the left side of each plate are tlc1Δ rad52Δ exo1Δ controls. Strains on the right side have an additional mutation, indicated at the right side of the plates. (D to G) Columns represent the fraction of isogenic strains that escaped from replicative senescence, from a total of 20 to 40 strains propagated as described for panels A to C. All strains are tlc1Δ rad52Δ (with or without other mutations). Mutations in Exo1 are indicated below the columns. All other relevant mutations are indicated above each column. (H) Fivefold serial dilutions of cdc13-1 strains (with or without other mutations) were spotted onto plates and incubated at 25°C or 27°C. The relevant genotypes are indicated on the right.
We found that strains with a dpb3Δ or a dpb4Δ mutation in the nonessential polymerase epsilon subunits Dpb3 and Dpb4 escaped from senescence with higher frequency than controls (Fig. 3A). The total fraction of dpb3Δ or dpb4Δ escapers was 80 to 90%, whereas only about 50% DPB+ controls escaped (Fig. 3A and D). In contrast, a pol32Δ mutation in the Pol32 subunit of Pol δ had no obvious effect in maintaining replicative senescence, since about 50% of either pol32Δ or POL32+ strains escaped (Fig. 3B and F). These data indicate that Pol ε is required to maintain senescence and that its Dpb3 and Dpb4 subunits are important for this function but that Pol δ plays little role. Even in an EXO1+ background, a dpb3Δ or dpb4Δ mutation permitted escape from senescence for 10 to 15% of strains (Fig. 3E). In contrast, none of the DPB+ EXO1+ cells escaped to generate survivors, consistent with previous experiments (28). These data indicate that (i) Pol ε is essential for maintaining replicative senescence and (ii) Pol ε acts synergistically with Exo1 in the mechanism maintaining replicative senescence.
Mrc1 facilitates escape from replicative senescence.
Recent studies demonstrated that Pol2, the catalytic subunit of Pol ε, extensively associates with Mrc1 (23), which is an S-phase checkpoint and replication protein (29) with an additional role in telomere protection against Exo1 (37). Therefore, one possibility is that Pol ε maintains replicative senescence through a checkpoint function conferred by Mrc1. Since Mrc1 is nonessential for viability, we tested its role during replicative senescence, as described for Dpb3 and Dpb4 subunits. Representative plates are shown in Fig. 3C, and the overall outcome from these experiments is presented in Fig. 3F. We found that despite the escape-facilitating exo1Δ mutation, only 10% of the mrc1Δ exo1Δ strains escaped from senescence but that 50% of the MRC+ exo1Δ controls escaped (Fig. 3C and F). This indicates that unlike a dpb3Δ or a dpb4Δ mutation, an mrc1Δ mutation strongly decreases escape. Conversely, Mrc1 facilitates escape from senescence, which is a paradoxical effect for a checkpoint protein.
Strains with an mrc1Δ or pol32Δ mutation entered senescence earlier than controls, indicated by the poor growth at passage 2 (Fig. 3B and C). Early senescence could be explained by a requirement for Mrc1 (and potentially for Pol32) in telomere protection (37). Interestingly, the first PAL survivors were identified in strains that entered senescence very early and escaped with high frequency (28). Therefore, early senescence does not imply reduced escape. It is more likely that most strains remained senescent in the absence of Mrc1, because Mrc1 was required to facilitate escape. In conclusion, the hypothesis that Mrc1 confers upon Pol ε a checkpoint activity that contributes to replicative senescence has been disproved. This is because we found that Mrc1 and Pol ε have different, in fact opposing, effects on replicative senescence.
Pol ε is not a checkpoint for uncapped telomeres.
We have presented evidence that two checkpoint subpathways, dependent upon either Rad24Rad17 or Rad953BP1, alternate in time to maintain replicative senescence (Fig. 2). Therefore, we analyzed the effects of Rad24Rad17 and Rad953BP1 on the fraction of strains escaping from senescence, as described for Dpb3 and Dpb4. We found that a rad24Δ mutation did not affect the fraction of tlc1Δ rad52Δ exo1Δ strains escaping from senescence (Fig. 3G). In contrast, a rad9Δ mutation increased the fraction of escapers to almost 100%, similarly to the effect of a dpb3Δ or a dpb4Δ mutation (Fig. 3G). These data suggest that Pol ε may act in the same pathway with Rad953BP1 (and a different pathway from Rad24Rad17) during replicative senescence.
One hypothesis is that Pol ε may possess an intrinsic checkpoint function in response to telomere damage. To test this hypothesis, we assessed the “checkpoint function” of Dpb3 and Dpb4 in a cdc13-1 background. Similarly to tlc1Δ rad52Δ cells, cells with a cdc13-1 temperature-sensitive mutation in the telomere-capping protein Cdc13Pot1 accumulate ssDNA at telomeres and require both Rad24Rad17 and Rad953BP1 to maintain the cell cycle arrest (42). We found that a dpb3Δ or a dpb4Δ mutation did not permit proliferation of cdc13-1 cells at the restrictive temperature of 27°C. In contrast, a rad24Δ or a rad9Δ mutation rescued proliferation of cdc13-1 cells, consistent with previous data (42). This indicates that Pol ε does not behave like a checkpoint protein in response to cdc13-1 telomere uncapping. Therefore, Pol ε does not possess an intrinsic telomere damage checkpoint function.
Pol ε inhibits ssDNA accumulation during replicative senescence.
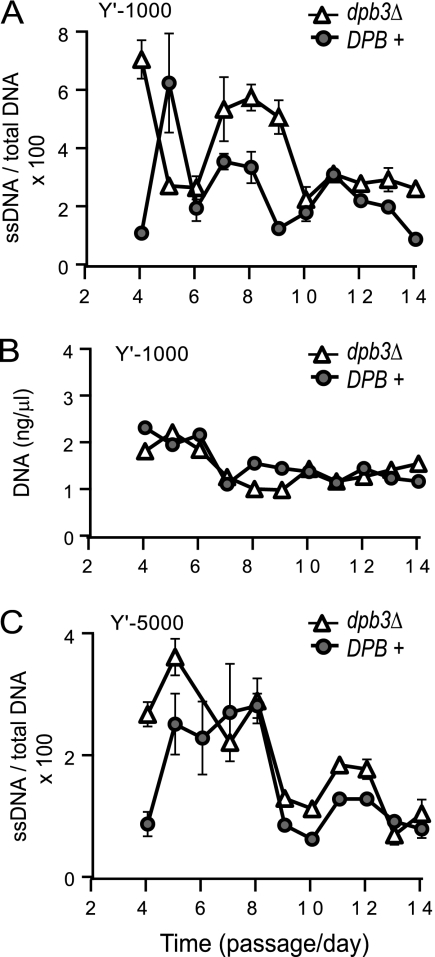
The archetypal function of Pol ε is to perform DNA synthesis. A Pol ε-dependent DNA resynthesis may explain the periodical decline in ssDNA during senescence. To test this hypothesis, we analyzed the effect of Dpb3 on ssDNA dynamics during senescence of tlc1Δ rad52Δ strains in experiments similar to those described in the legend to Fig. 1. We found that strains with a dpb3Δ mutation accumulated more ssDNA than DPB+ controls propagated and analyzed in parallel. The highest difference in ssDNA was about 2-fold at passages 8 and 9 (Fig. 4A) and corresponded to a noticeable difference in the total amount of DNA at that time (Fig. 4B). At Y′-1000, more ssDNA was measured in dpb3Δ strains than in DPB+ strains at passages 4, 7 to 9, and 13 and 14 (Fig. 4A). At Y′-5000, more ssDNA was measured in dpb3Δ strains than in DPB+ strains at passages 4 and 5 and 10 to 12 (Fig. 4C). Therefore, dpb3Δ strains had more ssDNA than controls in at least one (sub)telomeric region during replicative senescence.
Fig. 4.
Effect of Dpb3 on ssDNA dynamics during replicative senescence. Each of three independently generated tlc1Δ rad52Δ strains (with or without other mutations) was taken from germination plates, pooled at passage 1, and propagated in two groups: DPB+ (tlc1Δ rad52Δ strains) and dpb3Δ (tlc1Δ rad52Δ dpb3Δ strains). (A and C) ssDNA measured for DPB+ and dpb3Δ groups at Y′-1000 (A) and at Y′-5000 (C). Error bars represent the standard deviations (SD) for three measurements. (B) Same as in panel A, except that DNA was quantified at Y′-1000 and normalized to the level for DNA at the centromeric location PAC2. The same groups of strains was used for the measurements presented in panels A to C.
One explanation for this difference in ssDNA is that a dpb3Δ mutation slows down the ability of cells to resynthesize ssDNA. Since the DNA synthesis activity of Pol ε is slowed down in the absence of Dpb3, these data are consistent with a model in which DNA synthesis by Pol ε is responsible for the decline in ssDNA during replicative senescence, converting ssDNA to double-stranded DNA (dsDNA). A second explanation for more ssDNA in dpb3Δ strains could be that Pol ε opposes DNA resection by inhibiting Exo1. This (second) explanation is unlikely, because Pol ε acts synergistically with Exo1 during replicative senescence (Fig. 3D and E).
Dpb4 is required for a Pol2 and Rad953PB1 association with (sub)telomeres.
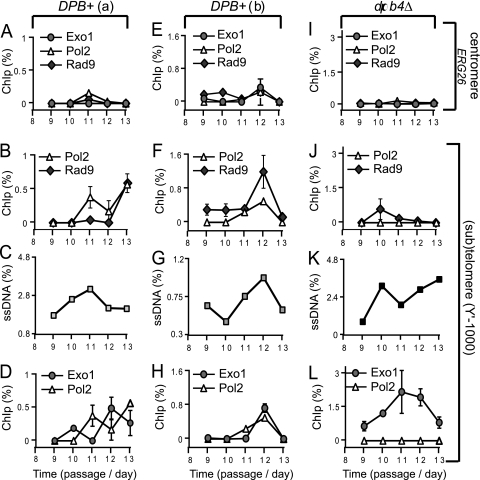
If Pol ε is required for DNA resynthesis during replicative senescence, then its catalytic subunit Pol 2 would be expected to associate with regions affected by DNA resection. To test this hypothesis, we measured the association of Pol2 with subtelomeres by chromatin immunoprecipitation in samples collected during midsenescence (passages 9 to 13). Additionally, we measured the association of Rad953BP1 and Exo1 with subtelomeres. We found that whereas most Pol2, Exo1, or Rad953BP1 proteins did not associate with a centromere-proximal locus during senescence (Fig. 5A), they all associated with subtelomeres.
Fig. 5.
Association of Pol2, Exo1, and Rad953BP1 with sub(telomeres) during senescence. Each of three independently generated tlc1Δ rad52Δ strains (with or without other mutations) was taken from germination plates, pooled at passage 1, and propagated in three groups: the DPB+(a) and DPB+(b) groups are tlc1Δ rad52Δ, whereas the dpb4Δ group is tlc1Δ rad52Δ dpb4Δ. Cell samples collected during midsenescence (passages 9 to 13) were cross-linked and immunoprecipitated. Both the immunoprecipitated and the input DNAs were quantified by quantitative PCR (qPCR) using TaqMan probes. Plotted is the fraction of DNA precipitated with specific antibodies, minus the background precipitation level for each sample; negative values were approximated to 0. The analyzed proteins are indicated above each graph. Regions are indicated on the far right of the figure. (A to D) Group DPB+(a) analyzed by ChIP and QAOS. (E to G) Same as in panels A to D, except that group DPB+(b) was analyzed. (I to L) The dpb4Δ group was analyzed.
In DPB+(a) strains, Pol2 was recruited to subtelomeres at passage 11 (Fig. 5B) during a peak in ssDNA (Fig. 5C) and its association increased at passage 13. The recruitment of Rad953BP1 to subtelomeres took place at passage 13 and coincided with a peak in subtelomeric Pol2 (Fig. 5B). In contrast, Exo1 was recruited at passage 10 (one passage earlier than Pol2) and completely dissociated by the time when Pol2 was recruited, with reassociation at passage 12 (Fig. 5D). In DPB+(b) strains, Pol2, Exo1, and Rad953BP1 associated with subtelomeres during a peak in ssDNA at passage 12 and dissociated thereafter (Fig. 5F to H). These data indicate that in DPB+ strains, Pol2, Exo1, and Rad953BP1 associate and disassociate from telomeres during replicative senescence. In contrast, we found no significant association of Pol2 or Rad953BP1 with telomeres in dpb4Δ strains, despite high levels of Exo1 and ssDNA at telomeres (Fig. 5J to L). In conclusion, these data suggest that (i) Dpb4 facilitates the association of Pol2 and Rad953BP1 with DNA damage and (ii) there is a strong correlation between the association of Pol2 and Rad953BP1 with (sub)telomeres during senescence.
DISCUSSION
Vicious-circle model of replicative senescence.
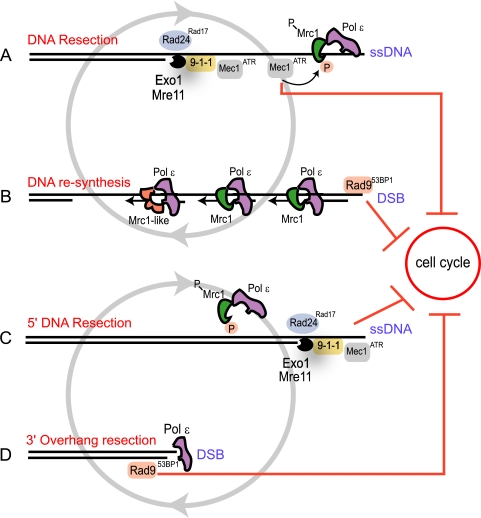
To explain how replicative senescence is maintained, we start with an analogy from a different signaling pathway: nasal receptors adapt to a persistent odor (meaning they undergo a stimulus-dependent decrease in sensitivity); however, they fail to adapt if the odor comes in pulses separated by odor-free intervals (or by intervals with a different odor) (44). Similarly, we propose that DNA damage checkpoint pathways fail to adapt to replicative senescence, because at least two DNA damage signals are interchanging regularly during senescence. The first signal for replicative senescence is most likely triggered by DNA resection of the shortest telomere(s), performed by Exo1 and other nucleases. Resection generates long 3′ssDNA overhangs and activates a Rad24Rad17-dependent checkpoint pathway (Fig. 6A and C). Then, ssDNA is resynthesized to dsDNA by DNA polymerases (Fig. 6B) or is perhaps degraded by nucleases (Fig. 6D). Irrespective of whether ssDNA is resynthesized or degraded, the short chromosome end (too short to acquire efficient protection from telomere-capping proteins) would now resemble a DSB. Therefore, we propose that a DSB-like structure is the second checkpoint signal during replicative senescence, a signal that activates a Rad953BP1-dependent pathway (Fig. 6B and D).
Fig. 6.
Cartoon presenting two models of replicative senescence. Vicious-circle model of DNA resection and resynthesis (A) and 5′ and 3′ alternating DNA resection (B). For details, see Results.
Resection and resynthesis would continuously alternate at each dysfunctional short chromosome end, providing fresh substrates (ssDNA or a DSB-like structure) for checkpoint activation. Therefore, checkpoint sensors specific to each substrate would also alternate at chromosome ends, thus reducing the possibility that one of them would adapt to a persistent “old” damage. It is easy to imagine that the two substrates would naturally alternate, since ssDNA would attract factors catalyzing its resynthesis to dsDNA, whereas the newly formed dsDNA would attract nucleases to resect it back to ssDNA. Therefore, in the absence of repair, cells would be trapped in a “vicious circle” of resection and resynthesis for as long as they remain alive.
Consistent with the vicious-circle model proposed above, our data indicate that Pol ε is required to maintain replicative senescence and that Mrc1 has the opposite effect, facilitating escape (Fig. 3). The effect of Mrc1 during senescence is intriguing, since Mrc1 and Pol2 (the catalytic subunit of Pol ε) associate with each other and act in the same pathway(s) during S phase (23). However, the effect of Mrc1 during senescence could be explained as follows. When phosphorylated by the Mec1ATR checkpoint protein, Mrc1 is known to partially dissociate from Pol2 (23). Such disassociation would be expected to take place during senescence, since Mec1ATR is active (10, 15) and Mrc1 appears phosphorylated (12). Therefore, based upon our observation that Mrc1 has the opposite effect relative to Pol ε during replicative senescence, we propose that a partially dissociated Mrc1 protein renders Pol2 unable to synthesize DNA, therefore acting as an inhibitor of Pol ε. We integrated this hypothesis into the vicious-circle model, as follows. Once replicative senescence is triggered by resection of the shortest telomere(s), activated Mec1ATR phosphorylates Mrc1. Phosphorylated Mrc1 partially dissociates from Pol2, leaving it unable to perform DNA synthesis. With time, the Mec1ATR activity decreases, due to a decline in resection rate and/or to checkpoint adaptation. In consequence, dephosphorylated Mrc1 reassociates with Pol2, rendering functional Mrc1-Pol ε complexes able to take over the DNA resynthesis from Pol α (Fig. 6B).
We also propose an alternative model of the replicative senescence void of DNA synthesis, called “the vicious circle of 5′ and 3′ resection” (Fig. 6C and D). In this model, a long 3′ ssDNA overhang forms at the shortest telomere(s) and activates a Rad24Rad9-dependent checkpoint pathway. The ssDNA is then degraded, perhaps with help from Pol ε, which has a 3′-to-5′ nucleolytic activity (Fig. 6D). The resulting blunt end activates a Rad953BP1-dependent checkpoint pathway. The blunt end is also a target for 5′-to-3′ nucleases; in consequence, a new 3′ overhang forms, and this overhang is degraded thereafter. It is clear from Fig. 6C that an alternating degradation of 5′ and 3′ strands at the same chromosome end would lead to a rapid chromosome shortening and loss of essential genes, followed by cell death. Therefore, a DNA resection-based model could not maintain replicative senescence in the long term if completely void of DNA resynthesis.
The loss of the ssDNA overhangs that we detected during the budding yeast replicative senescence could be the equivalent of the ssDNA overhang loss observed at telomeres of human fibroblasts during replicative senescence (6, 16, 30, 34). Could a vicious circle also be responsible for the irreversible character of replicative senescence in mammalian cells? Interestingly, upregulation of the cyclin-dependent kinase (CDK) inhibitors p21WAF1 and p16INK4a has been reported to alternate during mammalian replicative senescence (33). However, this was not the case for all senescent cells (14). Moreover, ATM which responds to DSBs (2) is considered more important for replicative senescence than ATR (8, 14), which responds to ssDNA (40). These observations do not exclude a vicious circle during mammalian senescence. For example, if mammalian telomeres recruit DNA polymerases more rapidly than yeast, they will spend more time as ATM-activating DSB-like structures. Adaptation could be prevented through the occasional resection of one chromosome end, leading to a transient activation of a complementary ATR pathway.
It is known that even a very short pause (a few seconds) between exposures to an odor is sufficient to prevent nasal receptors adapting to that odor (44). Similarly, a short switch from ATM to ATR activation during replicative senescence could potentially be sufficient to avoid adaptation and may escape detection during standard experiments. Are ATM foci flickering in senescent cells, similar to light bulbs approaching the end of the “life” span? More focused research is needed to address this question. Our view is that a vicious circle of DNA resection and resynthesis would be sufficient to maintain replicative senescence. Additional factors (oxidative stress, chromosome fusions, etc.) could precipitate, enhance, or delay senescence in response to telomere attrition and eventually kill senescent cells.
ACKNOWLEDGMENTS
This work was supported by the Wellcome Trust (Career Development Fellowship award no. 81164 to L.M.). A.M.D was funded by a DTG BBSRC studentship.
We thank Carole Proctor and David Lydall (both from Newcastle University) for critical comments on the manuscript.
Footnotes
Published ahead of print on 14 February 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Abdallah P., et al. 2009. A two-step model for senescence triggered by a single critically short telomere. Nat. Cell Biol. 11:988–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bakkenist C. J., Kastan M. B. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421:499–506 [DOI] [PubMed] [Google Scholar]
- 3. Booth C., Griffith E., Brady G., Lydall D. 2001. Quantitative amplification of single-stranded DNA (QAOS) demonstrates that cdc13-1 mutants generate ssDNA in a telomere to centromere direction. Nucleic Acids Res. 29:4414–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Braunstein M., Rose A. B., Holmes S. G., Allis C. D., Broach J. R. 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7:592–604 [DOI] [PubMed] [Google Scholar]
- 5. Campisi J., d'Adda di Fagagna F. 2007. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8:729–740 [DOI] [PubMed] [Google Scholar]
- 6. Chai W., Shay J. W., Wright W. E. 2005. Human telomeres maintain their overhang length at senescence. Mol. Cell. Biol. 25:2158–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cohn M., Blackburn E. H. 1995. Telomerase in yeast. Science 269:396–400 [DOI] [PubMed] [Google Scholar]
- 8. d'Adda di Fagagna F., et al. 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature 426:194–198 [DOI] [PubMed] [Google Scholar]
- 9. Dedon P. C., Soults J. A., Allis C. D., Gorovsky M. A. 1991. A simplified formaldehyde fixation and immunoprecipitation technique for studying protein-DNA interactions. Anal. Biochem. 197:83–90 [DOI] [PubMed] [Google Scholar]
- 10. Enomoto S., Glowczewski L., Berman J. 2002. MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae. Mol. Biol. Cell 13:2626–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Giannattasio M., et al. 2010. Exo1 competes with repair synthesis, converts NER intermediates to long ssDNA gaps, and promotes checkpoint activation. Mol. Cell 40:50–62 [DOI] [PubMed] [Google Scholar]
- 12. Grandin N., Bailly A., Charbonneau M. 2005. Activation of Mrc1, a mediator of the replication checkpoint, by telomere erosion. Biol. Cell 97:799–814 [DOI] [PubMed] [Google Scholar]
- 13. Grandin N., Charbonneau M. 2009. Telomerase- and Rad52-independent immortalization of budding yeast by an inherited-long-telomere pathway of telomeric repeat amplification. Mol. Cell. Biol. 29:965–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Herbig U., Jobling W. A., Chen B. P., Chen D. J., Sedivy J. M. 2004. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol. Cell 14:501–513 [DOI] [PubMed] [Google Scholar]
- 15. IJpma A. S., Greider C. W. 2003. Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol. Biol. Cell 14:987–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keys B., Serra V., Saretzki G., Von Zglinicki T. 2004. Telomere shortening in human fibroblasts is not dependent on the size of the telomeric-3′-overhang. Aging Cell 3:103–109 [DOI] [PubMed] [Google Scholar]
- 17. Lebel C., et al. 2009. Telomere maintenance and survival in saccharomyces cerevisiae in the absence of telomerase and RAD52. Genetics 182:671–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee J. Y., Mogen J. L., Chavez A., Johnson F. B. 2008. Sgs1 RecQ helicase inhibits survival of Saccharomyces cerevisiae cells lacking telomerase and homologous recombination. J. Biol. Chem. 283:29847–29858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee S. E., et al. 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399–409 [DOI] [PubMed] [Google Scholar]
- 20. Lee S. E., Pellicioli A., Malkova A., Foiani M., Haber J. E. 2001. The Saccharomyces recombination protein Tid1p is required for adaptation from G2/M arrest induced by a double-strand break. Curr. Biol. 11:1053–1057 [DOI] [PubMed] [Google Scholar]
- 21. Lisby M., Barlow J. H., Burgess R. C., Rothstein R. 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118:699–713 [DOI] [PubMed] [Google Scholar]
- 22. Longtine M. S., et al. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961 [DOI] [PubMed] [Google Scholar]
- 23. Lou H., et al. 2008. Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Mol. Cell 32:106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lundblad V., Blackburn E. H. 1993. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73:347–360 [DOI] [PubMed] [Google Scholar]
- 25. Maringele L., Lydall D. 2004. EXO1 plays a role in generating type I and type II survivors in budding yeast. Genetics 166:1641–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maringele L., Lydall D. 2002. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 16:1919–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maringele L., Lydall D. 2005. The PAL-mechanism of chromosome maintenance: causes and consequences. Cell Cycle 4:747–751 [DOI] [PubMed] [Google Scholar]
- 28. Maringele L., Lydall D. 2004. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev. 18:2663–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Osborn A. J., Elledge S. J. 2003. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17:1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rahman R., Forsyth N. R., Cui W. 2008. Telomeric 3′-overhang length is associated with the size of telomeres. Exp. Gerontol. 43:258–265 [DOI] [PubMed] [Google Scholar]
- 31. Sandell L. L., Zakian V. A. 1993. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75:729–739 [DOI] [PubMed] [Google Scholar]
- 32. Schaetzlein S., et al. 2007. Exonuclease-1 deletion impairs DNA damage signaling and prolongs lifespan of telomere-dysfunctional mice. Cell 130:863–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stein G. H., Drullinger L. F., Soulard A., Dulic V. 1999. Differential roles for cyclin-dependent kinase inhibitors p21 and p16 in the mechanisms of senescence and differentiation in human fibroblasts. Mol. Cell. Biol. 19:2109–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stewart S. A., et al. 2003. Erosion of the telomeric single-strand overhang at replicative senescence. Nat. Genet. 33:492–496 [DOI] [PubMed] [Google Scholar]
- 35. Tanaka H., Bergstrom D. A., Yao M. C., Tapscott S. J. 2005. Widespread and nonrandom distribution of DNA palindromes in cancer cells provides a structural platform for subsequent gene amplification. Nat. Genet. 37:320–327 [DOI] [PubMed] [Google Scholar]
- 36. Toczyski D. P., Galgoczy D. J., Hartwell L. H. 1997. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell 90:1097–1106 [DOI] [PubMed] [Google Scholar]
- 37. Tsolou A., Lydall D. 2007. Mrc1 protects uncapped budding yeast telomeres from exonuclease EXO1. DNA Repair (Amst.) 6:1607–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Usui T., Ogawa H., Petrini J. H. 2001. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7:1255–1266 [DOI] [PubMed] [Google Scholar]
- 39. von Zglinicki T., Saretzki G., Ladhoff J., d'Adda di Fagagna F., Jackson S. P. 2005. Human cell senescence as a DNA damage response. Mech. Ageing Dev. 126:111–117 [DOI] [PubMed] [Google Scholar]
- 40. Zou L., Elledge S. J. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542–1548 [DOI] [PubMed] [Google Scholar]
- 41. Zou Y., Sfeir A., Gryaznov S. M., Shay J. W., Wright W. E. 2004. Does a sentinel or a subset of short telomeres determine replicative senescence? Mol. Biol. Cell 15:3709–3718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zubko M. K., Guillard S., Lydall D. 2004. Exo1 and Rad24 differentially regulate generation of ssDNA at telomeres of Saccharomyces cerevisiae cdc13-1 mutants. Genetics 168:103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zubko M. K., Maringele L., Foster S. S., Lydall D. 2006. Detecting repair intermediates in vivo: effects of DNA damage response genes on single-stranded DNA accumulation at uncapped telomeres in budding yeast. Methods Enzymol. 409:285–300 [DOI] [PubMed] [Google Scholar]
- 44. Zufall F., Leinders-Zufall T. 2000. The cellular and molecular basis of odor adaptation. Chem. Senses 25:473–481 [DOI] [PubMed] [Google Scholar]