Abstract
The transcription factor p53 functions not only to suppress tumorigenesis but also to maintain normal development and homeostasis. Although p53 was implicated in different aspects of fertility, including spermatogenesis and implantation, the mechanism underlying p53 involvement in spermatogenesis is poorly resolved. In this study we describe the identification of a spermatogenesis-associated gene, SPATA18, as a novel p53 transcriptional target and show that SPATA18 transcription is induced by p53 in a variety of cell types of both human and mouse origin. p53 binds a consensus DNA motif that resides within the first intron of SPATA18. We describe the spatiotemporal expression patterns of SPATA18 in mouse seminiferous tubules and suggest that SPATA18 transcription is regulated in vivo by p53. We also demonstrate the induction of SPATA18 by p63 and suggest that p63 can compensate for the loss of p53 activity in vivo. Our data not only enrich the known collection of p53 targets but may also provide insights on spermatogenesis defects that are associated with p53 deficiency.
INTRODUCTION
The p53 (TP53) tumor suppressor gene is a sequence-specific transcription factor that responds to a broad range of cellular stress signals and functions as a coordinator of cell fate decisions (33). Accordingly, p53 activity is attenuated in most human cancers, usually due to somatic missense mutations (8). p53 exerts its activity mainly by transactivating target genes; a few dozen of such targets were identified during the last 3 decades (35). Moreover, p53 can also repress the transcription of genes (7) and induce a variety of transactivation-independent responses (15). The identity of most p53-regulated genes that mediate processes such as cell cycle arrest, cell death, and DNA repair have been revealed. However, p53 also affects processes such as autophagy, differentiation, reproduction, metabolism, and aging (42, 61), but the mediators of these effects are much less characterized.
The complicated process of spermatogenesis begins with proliferation of diploid spermatogonia and terminates with the production of haploid spermatozoa and involves tightly regulated steps of proliferation, meiosis, and differentiation. Spermatogenesis occurs within the seminiferous tubules in precisely timed and highly organized cycles (23). The process starts with diploid germ cells called spermatogonia that reside on the basement membrane. After a series of mitotic divisions, differentiating spermatogonia divide into spermatocytes, which can migrate to a more adluminal position of the seminiferous tubule. Spermatocytes undergo meiosis to form haploid round spermatids (22), which then undergo spermiogenesis, a process that includes morphological alterations such as acrosome formation, nuclear condensation, flagellum formation, and extrusion of residual cytoplasm, to form mature spermatozoa. Of note, germ cells in the seminiferous tubules are nursed by Sertoli cells (34).
The existence of p53 orthologs in lower organisms, such as worms and flies, which do not develop cancer, implies that tumor suppression does not represent the original function of p53. It has been suggested that the role of the p53 ancestral genes is to ensure the genomic integrity of the germ line and the fidelity of developmental processes (24). This role was conserved during evolution as p53 is important for normal differentiation and development in mammals and other higher organisms (42).
It has been found that p53, like many other proto-oncogenes and tumor suppressor genes, plays a role in the meiotic process of spermatogenesis (1). For instance, it was previously reported that during normal spermatogenesis, p53 mRNA and protein are accumulated in spermatocytes (55), and, similarly, p21 (CDKN1A), the primary p53 target gene that mediates cell-cycle arrest (13), is expressed during the prophase of meiosis (4). Several studies, including ours, demonstrate specific roles for p53 in spermatogenesis. For example, transgenic mice with partial or complete impairment of p53 expression exhibit a giant-cell degenerative syndrome (52), a phenomenon caused by an inability of primary tetraploid spermatocytes to undergo meiotic divisions. Moreover, p53−/− 129/Sv mice are infertile although it is unclear whether this is due to spermatogenesis defects (19). The p53-dependent DNA damage response was shown to be highly active throughout spermatogenesis (45), and accordingly, p53 mediates stress-induced spermatogonial apoptosis after DNA damage (20). Conversely, p53 deficiency attenuates the regeneration of seminiferous tubules after irradiation (21). Moreover, p53 was shown to induce the expression of Wip1 in mouse testes, and Wip1-null mice display degeneration of seminiferous tubules, reduced testis size, and attenuated fertility (10). Besides spermatogenesis, p53 is involved in additional aspects of reproduction, including implantation (25), and in a variety of embryonal development programs (42). Nevertheless, despite the general reduction in fecundity in p53-null mice and the increase in developmental defects, most p53-deficient mice survive, develop normally, and can reproduce (12). This may indicate an alternative pathway which can, at least partially, compensate for the loss of p53 in development and reproduction (51). Notably, it had been suggested that p53 family members, p63 and p73, can compensate for p53 loss in several processes, including DNA damage response (65), tumor suppression (14), and development (28, 56).
Utilizing RNA interference (RNAi) to knock down p53 in WI-38 human embryonic fibroblasts, followed by global expression analysis (9), we identified the gene spermatogenesis-associated 18 homolog (SPATA18) as a potential transcriptional target of p53. Our long interest in the role of p53 in spermatogenesis (1, 52, 55) and the putative functions of SPATA18 in this process had driven us to unveil the mechanism by which p53 regulates SPATA18 expression in vitro and in vivo. In humans, SPATA18 is localized on chromosome 4q12 and encodes a 538-amino acid (aa) protein with two coiled-coil domains. Although the function of SPATA18 in humans or in mouse is still unknown, its rat homolog, Spetex-1, which shares 82% identity in its protein sequence, was thoroughly characterized as a spermatogenesis-related protein. Specifically, Iida et al. had identified Spetex-1, using a differential display approach, as a highly expressed gene in rat testis (27). Testicular Spetex-1 expression is detected first at 3 weeks of postnatal development and remains high until adulthood. In situ hybridization and immunohistochemical analyses revealed that Spetex-1 mRNA and protein are expressed in the cytoplasm of haploid spermatids and in residual bodies (27). Further subcellular analysis revealed that Spetex-1 protein also localizes at satellite fibrils associated with outer dense fibers in the middle piece of the flagella in spermatozoa (26, 30).
Few hypotheses regarding Spetex-1 function were proposed. Its localization in the cytoplasm of spermatids and in residual bodies suggests a role in spermiogenesis and, particularly, in maturation of spermatids into spermatozoa (27). During maturation, elongated spermatids undergo reduction in their cytoplasmic volume, which is mediated by engulfment of residual bodies by Sertoli cells (53). It is assumed that this process involves apoptotic signals enabling Sertoli cells to recognize and phagocytose the residual cytoplasm (5). Indeed, it has been reported that proteins such as caspase-1 (5) and aquaporin-7 (59), which mediate apoptosis and volume reduction, respectively, are restrictively expressed in the cytoplasm of elongated spermatids. Therefore, it is possible that Spetex-1 might also be involved in this apoptosis-like process (27). In addition, Iida et al. showed that a portion of Spetex-1 protein is retained in spermatozoa as a flagellar component after spermiation. Thus, Spetex-1 might serve as a structural constituent in the flagella (26, 30).
Here, we show that the transcription of human and mouse SPATA18 is directly activated by the p53 tumor suppressor in a variety of cell types. We demonstrate that p53 binds a consensus DNA motif located in a region corresponding to the first intron of the SPATA18 gene. We further show that mouse SPATA18 transcription is regulated in the testes in a unique spatiotemporal manner, being upregulated primarily by spermatids approximately 3 weeks after birth and remaining high in adults. SPATA18 expression pattern correlates with that of p21 and might be linked to the accumulation of p53 protein in spermatocytes. Our data imply that p53 transactivates SPATA18 in vivo and that p63 compensates for p53 function in the testes of p53-null mice.
MATERIALS AND METHODS
Cell culture.
All cell lines were grown at 37°C in a humidified atmosphere of 5% CO2 and were maintained in the following media. Immortalized WI-38 fibroblasts, previously described in Milyavsky et al. (40), as well as IMR-90 fibroblasts, were cultured in minimal essential medium (MEM) supplemented with 10% fetal bovine serum (FBS), 1 mM sodium pyruvate, 2 mM l-glutamine, and antibiotics. Mouse embryonic fibroblasts (MEFs) were derived from p53+/+ or p53−/− sibling embryos and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and antibiotics. Saos-2, SKOV-3, HepG2, TM4, and HT-1080 cells were cultured in DMEM with 10% FBS and antibiotics. LNCaP cells were cultured in RPMI medium with 10% FBS. R1 mouse embryonic stem cells were grown as described by Sarig et al. (54) on a feeder layer of irradiated MEFs.
Plasmids.
The retrovirus encoding GSE56 was described by Milyavsky et al. (41). Retroviruses harboring short hairpin RNA (shRNA) against human p53 (sh-p53) or mouse NOXA (control shRNA [sh-con] for human cells) were kindly provided by D. Ginsburg (Bar-Ilan University, Israel). Retroviruses harboring shRNA against mouse p53 or human pRb (control shRNA for mouse cells) were kindly provided by S. W. Lowe (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY). The retrovirus harboring shRNA against mouse p63 (sh-p63) was generated by subcloning the shRNA cassette from a commercial pSM2 vector designed to target human p63 (clone V2HS_24249; Open BioSystems) into a pRetroSuper-hygro vector using XhoI and EcoRI. Note that the mature RNAi sequence has one mismatch for mouse p63 but is very effective in knocking down mouse p63 (see Fig. 4E). The p53 expression plasmid pC53-SN3 carrying wild-type (WT) p53 (2) was kindly provided by B. Vogelstein (Johns Hopkins University School of Medicine, Baltimore, MD), and was used as a template for site-directed mutagenesis to produce the plasmid carrying p53 with the mutation R249S (p53R249S), as described by Suad et al. (57). The firefly luciferase reporter under the regulation of the p21 promoter and the plasmid encoding p63α with the transactivating domain (TA-p63α) (pCDNA3-TA-p63α) were kindly provided by M. Oren (Weizmann Institute of Science, Israel). pRL-CMV (where CMV is cytomegalovirus) plasmid was purchased from Promega.
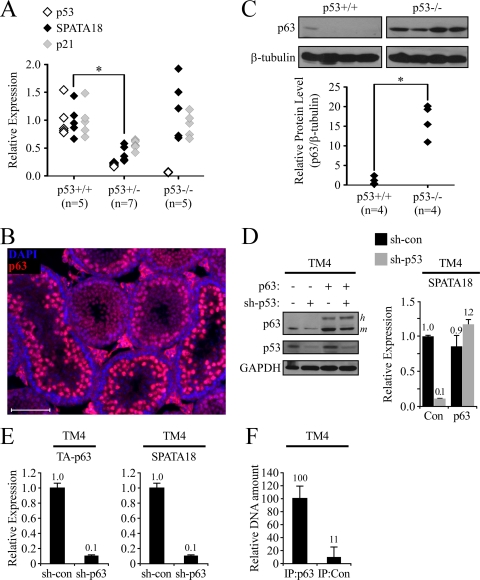
Fig. 4.
SPATA18 transcription is regulated by p53 and p63 in vivo. (A) qRT-PCR analysis was conducted for p53, SPATA18, and p21 mRNA in testes derived from p53+/+, p53+/−, and p53−/− mice. The number of samples in each group is indicated in parentheses. (B) p63 protein was detected by immunofluorescent staining of a testis section from a 25-day-old mouse (red). DNA was visualized with DAPI (blue). (C) p63 protein level was measured by Western blotting of whole-testis lysates derived from either p53+/+ or p53−/− mice. Intensities were quantified using ImageJ software, and relative p63 protein levels were calculated by normalizing to β-tubulin intensities (lower panel). (D) Mouse TM4 Sertoli cells expressing either a control shRNA (sh-con) or an shRNA targeting mouse p53 (sh-p53) were transfected with pCDNA-TA-p63α (p63) or a control vector (Con). Two days after transfection protein levels were determined by Western blotting (left), and expression of SPATA18 was measured by qRT-PCR (right). Of note, the exogenous human p63 (labeled h to the right of the blot) migrated more slowly than the endogenous mouse p63 (m). (E) Mouse TM4 Sertoli cells expressing either a control shRNA or an shRNA targeting mouse p63 were analyzed for the expression of TA-p63 and SPATA18 by qRT-PCR. (F) ChIP analysis of TA-p63α-transfected TM4 cells using p63-specific antibody (IP:p63) or control IgG (IP:Con). The amount of precipitated DNA was measured by qRT-PCR using specific primers designed to amplify a putative p53 binding site in the first intron of mouse SPATA18. Values were normalized to the amount of precipitated nonspecific DNA. Bars represent mean ± standard deviations from two duplicate immunoprecipitants.
Construction of SPATA18 first intron luciferase reporters.
The luciferase reporter vector, pGL3-SPATA18-intron1, was generated by cloning a short fragment containing the p53 binding site (BS) from the SPATA18 first intron into a luciferase reporter plasmid. Briefly, a 495-bp genomic fragment from the first intron of the human SPATA18 gene, spanning from bp +114 to bp +609 relative to the transcription start site, was PCR-amplified with the primers 5′-TTCGCCCTCCCATAGGTTC-3′ and 5′-CCCGCAACGTTAACAAGTGTC-3′. The product was ligated into pGEM-T Easy (Promega) and then transferred into pGL3sb vector (kindly provided by M. Oren) using the restriction enzymes NdeI and NcoI. Mutations in the p53 binding site were generated using a QuikChange Site-Directed Mutagenesis kit (Stratagene) with the following primer (only sense primer is shown; mismatches are underlined): 5′-CGCTGGGGAAGGAAGGAAATGTGTGTAAATACCCTTGTCTCAGAG-3′.
Retroviral infections.
Retroviral infections were described by Milyavsky et al. (40). Infected cells were selected for 1 week with the following antibiotics: for human shRNA infections, 200 μg/ml hygromycin B; for mouse shRNA infections, 1 μg/ml puromycin; for GSE56 overexpression, 400 μg/ml G418. TM4 cells infected with retroviruses harboring sh-p63 or an shRNA targeting a control gene (sh-con) were maintained in hygromycin B-containing medium to sustain p63 knockdown.
Transfections.
Saos-2 and SKOV-3 cells were transfected with Fugene HD (Roche Applied Science), using 0.5 μg of various p53 expression plasmids, as indicated in the figure legends. HT-1080 cells were transfected with TransIT (Mirus Bio LLC) using 3.5 μg of plasmids, as indicated in the figure legends.
Luciferase reporter assay.
Saos-2 cells were plated in 24-well plates 24 h before transfection. Cells were transfected with 300 ng/well of luciferase reporter construct, 10 ng/well of pRL-CMV for normalization of transfection efficiency, 0 to 30 ng/well p53 expression plasmid, and pBlueScript for a total DNA amount of 500 ng/well. Luciferase and Renilla activities were determined using commercial reagents and procedures (Promega).
RNA preparation and qRT-PCR.
Total RNA was isolated from cultured cells using a NucleoSpin II kit (Macherey Nagel). Total RNA from testes was isolated following mechanical homogenization using TRI reagent (Molecular Research Center). A 2-μg aliquot of the total RNA was reverse transcribed using Moloney murine leukemia virus reverse transcriptase (MMLV-RT; Bio-RT) and random hexamer primers. Quantitative real-time PCR (qRT-PCR) was performed using SYBR green PCR Master Mix (Applied BioSystems) on an ABI 7300 instrument. Human and mouse mRNA levels were normalized to the level of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and HPRT (hypoxanthine phosphoribosyltransferase), respectively. PCRs were performed in duplicates, and error bars in charts represent the corresponding standard deviations. For analysis of mRNA expression in vivo, tissues from at least three mice were collected per experimental category, and error bars in charts represent standard errors of mean. Primers for qRT-PCR and semi-qRT-PCR are listed in Table S1 posted at http://www.weizmann.ac.il/mcb/Varda/p53_SPATA18/.
Western blotting.
Western blot analyses were performed as described by Milyavsky et al. (41). The following primary antibodies were used: H47 polyclonal anti-human p53 (produced in our laboratory), monoclonal anti-human p21 (sc-377; Santa Cruz), monoclonal anti-human GAPDH (MAB374; Chemicon), monoclonal anti-mouse β-tubulin (Sigma), and monoclonal anti-human p63 (sc-8431; Santa Cruz).
Chromatin immunoprecipitation.
Chromatin immunoprecipitation analyses were performed as described by Kalo et al. (29) with the following antibodies. For human WI-38 cells, polyclonal H47 (produced in our laboratory) was used to precipitate p53, and polyclonal anti-transforming growth factor β receptor 2 (TGF-βR2; Santa Cruz) was used for control precipitation. For mouse TM4 cells, monoclonal 4A4 (sc-8431; Santa Cruz) was used to precipitate p63, and monoclonal IgG (I-2511; Sigma) was used for control precipitation. Precipitated DNA was measured by qRT-PCR (primer sequences are listed in Table S2 posted at http://www.weizmann.ac.il/mcb/Varda/p53_SPATA18/). Relative DNA levels were calculated by normalizing to the amount of precipitated nonspecific DNA.
Mice.
C57BL/6 p53−/− mice were kindly provided by G. Lozano (University of Texas M. D. Anderson Cancer Center).
Immunohistochemistry and immunofluorescence analyses.
Immunohistochemistry analyses were performed as described by Sarig et al. (54). For immunofluorescence analysis, paraffin-embedded sections were deparaffinized using xylene and were rehydrated with alcohol series, boiled in a Tris-EDTA, pH 9.0, buffer, and incubated overnight at room temperature in a humid chamber with mouse monoclonal 4A4 anti-p63 antibody (sc-8431; Santa Cruz). Sections were washed three times with phosphate-buffered saline (PBS) and labeled with a Cy3 fluor (Jackson Immunoresearch) at 1:500 for 20 min. To visualize nuclei, sections were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI; 10 mg/ml) for 5 min and mounted with Elvanol.
In situ hybridization.
Probes for in situ hybridization were prepared by PCR amplification of ∼500-bp fragments from the mouse SPATA18 gene, using total cDNA prepared from p53+/+ mouse testes. The following primers were used to produce two different amplicons: forward primer 1, 5′-CTGCAGACTTCCCTCAGTTC-3′; reverse primer 1, 5′-GCAGACAGGACAGCTATCTC-3′; forward primer 2, 5′-ACAGTGGCCAAGATCAGAAG-3′; and reverse primer 2, 5′-AGACCAAGGGAGCAGTAAAG-3′. Amplicons were ligated into pGEM-T Easy (Promega). Digoxigenin-labeled riboprobes were produced for sense and antisense probes by in vitro transcription from the T7 or SP6 promoters using a digoxigenin RNA labeling kit (Roche Applied Science). Paraffin sections (6 μm) from mouse testes were deparaffinized using xylene and were rehydrated with alcohol series. Sections were treated with 20 μg/ml proteinase K (Roche Applied Science) at 37°C for 15 min and incubated with prehybridization buffer (10% dextran sulfate, 50% formamide, 0.3 M NaCl, 0.1 M Tris [pH 7.5], 1 mM EDTA, 1% blocking reagent, 2 mg/ml torula yeast RNA) at 37°C for 3 h. Sections were hybridized with 400 ng/ml of each riboprobe at 37°C for 16 h in a humid chamber and serially washed with washing buffer (30 to 300 mM NaCl, 2 to 20 mM NaH2PO4-H2O, 0.25 to 2.5 mM EDTA). Hybridized sections were blocked with Genius buffer (100 mM Tris [pH 7.6], 150 mM NaCl) containing 1% BSA and were incubated with an alkaline phosphatase-conjugated antidigoxigenin antibody at 4°C for 16 h. Sections were washed with Genius buffer containing 1% BSA and then with Genius buffer containing 2% blocking reagent. Developing was done using nitroblue tetrazolium/5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP) substrate according to the manufacturer's instructions (Roche Applied Science).
Statistical analysis.
Statistical significance was evaluated using a one-tailed unpaired student t test, and asterisks in figures represent P values lower than 5 × 10−3.
RESULTS
Expression of SPATA18 positively correlates with p53 activity in human and mouse cells.
The aforementioned microarray experiment pointed at SPATA18 as a putative p53 target gene. To validate this notion, we utilized WI-38 cells that stably express a short hairpin RNA (shRNA) targeting either p53 (sh-p53) or a control gene (sh-con). These isogenic cell cultures were treated with Nutlin-3a, a compound which impairs Mdm2-p53 interaction, thereby stabilizing and activating p53 (60). The protein levels of p53 and p21, a well-known p53 target gene (13), were measured by Western blot analysis (Fig. 1A), revealing a pronounced activation of p53 by Nutlin-3a and a strong attenuation of p53 activity in sh-p53 cells. Quantitative real-time PCR (qRT-PCR) measurements were conducted for p21 and SPATA18 mRNA levels, revealing that they were significantly downregulated by p53 knockdown and were strongly induced by Nutlin-3a in a p53-dependent manner (Fig. 1B). Both p21 and SPATA18 were induced relatively fast following Nutlin-3a treatment and reached peak levels after 24 h (see Fig. S1A posted at http://www.weizmann.ac.il/mcb/Varda/p53_SPATA18/). The expression of SPATA18 was also measured in an additional cell type, namely, HT-1080 human fibrosarcoma cells, which were infected with either a control vector or a vector encoding the p53-inactivating peptide GSE56 (46). In this system we analyzed, in addition to Nutlin-3a, the effect of various genotoxic drugs known to activate p53. As depicted in Fig. 1C, the expression of SPATA18 was significantly elevated upon genotoxic insult in a p53-dependent manner. The mRNA levels of SPATA18 were further analyzed in IMR-90 fetal human lung fibroblasts, where knockdown of p53 or expression of a dominant negative mutant p53 profoundly attenuated SPATA18 expression (see Fig. S1B and S1C posted at http://www.weizmann.ac.il/mcb/Varda/p53_SPATA18/). Furthermore, p53-dependent transcriptional activation of SPATA18 was also observed in additional human cell lines, including Saos-2 osteosarcoma (Fig. 2D), SKOV-3 ovarian carcinoma, LNCaP prostate carcinoma, and HepG2 hepatocellular carcinoma (see Fig. S1D to F at the URL mentioned above).
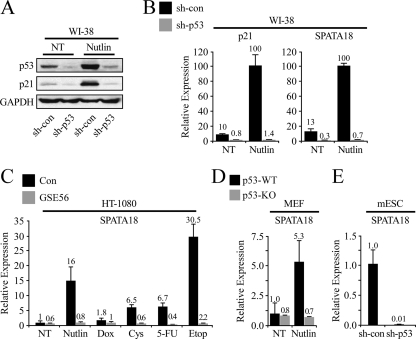
Fig. 1.
Expression of SPATA18 positively correlates with p53 activity in human and mouse cells. WI-38 cells expressing either a control shRNA (sh-con) or an shRNA targeting p53 (sh-p53) were treated with 10 μM Nutlin-3a for 48 h. Protein levels of p53, p21, and GAPDH were measured by Western blot analysis (A). Normalized mRNA levels of p21 and SPATA18 were measured by qRT-PCR (B). (C) qRT-PCR measurements were conducted for SPATA18 mRNA in HT-1080 fibrosarcoma cells stably expressing either the p53-inactivating peptide GSE56 or a control vector. Cells were treated with the indicated compounds for 24 h at the following concentrations: Nutlin-3a, 10 μM; doxorubicin (Dox), 10 μM; 5-fluorouracil (5-FU), 10 μM; etoposide (Etop), 100 μM; cisplatin (Cys), 3 μM. (D) p53+/+ and p53−/− MEFs were treated with 25 μM Nutlin-3a for 24 h. qRT-PCR measurements were conducted for SPATA18 mRNA. (E) qRT-PCR measurements were conducted for SPATA18 mRNA in mouse embryonic stem cells (mESC) expressing either a control shRNA or an shRNA targeting mouse p53. NT, not treated.
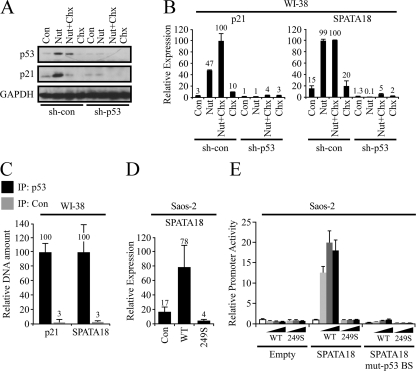
Fig. 2.
p53 directly activates SPATA18 transcription. Control (sh-con) and p53 knocked-down (sh-p53) WI-38 cells were treated with either 10 μM Nutlin-3a for 8 h (Nut), 10 μg/ml cycloheximide for 4 h (Chx), or both Nutlin-3a and cycloheximide (Nut+Chx). In the combined treatment, Nutlin-3a was applied first, and after 4 h cycloheximide was added. Samples were analyzed for protein levels by Western blotting (A), and mRNA levels were determined by qRT-PCR (B). (C) Chromatin immunoprecipitation analysis of WI-38 cells using anti-p53 antibody (IP: p53) or a negative-control antibody against TGF-βR2 (IP: Con). The amount of precipitated DNA was measured by qRT-PCR using specific primers designed to amplify the known p53 binding sites in p21 promoter or in the first intron of SPATA18. Values were normalized to the amount of precipitated nonspecific DNA. (D) Saos-2 cells were transfected with control vector (Con) or a pC53-SN3 vector encoding either wild-type p53 (WT) or mutant p53R249S (249S). qRT-PCR was conducted for SPATA18 mRNA 48 h after transfection. (E) Saos-2 cells were transfected with either an empty luciferase reporter (Empty), a luciferase reporter under the control of a 495-bp DNA fragment from the first intron of SPATA18, which contains a p53 BS (SPATA18), or the same reporter in which the p53 BS was mutated (SPATA18 mut-p53 BS). pC53-SN3 vector encoding either wild-type p53 (WT) or mutant p53R249S (249S) was cotransfected in increasing concentrations (0, 3, 10, and 30 ng/well). Luciferase activity was normalized to the Renilla activity of cotransfected pRL-CMV plasmid. Error bars represent standard deviations between triplicate measurements.
SPATA18 is conserved among a wide range of vertebrates, including chimpanzee, dog, cow, mouse, rat, chicken, and zebrafish. We were therefore curious to test whether SPATA18 expression is regulated by p53 in mouse cells. Indeed, following Nutlin-3a treatment, SPATA18 was upregulated in mouse embryonic fibroblasts (MEFs) derived from a p53+/+ mouse but not in MEFs derived from a p53−/− sibling mouse (Fig. 1D) or in p53+/+ MEFs in which p53 was knocked down (see Fig. S1G posted at http://www.weizmann.ac.il/mcb/Varda/p53_SPATA18/). Moreover, in mouse embryonic stem cells derived from a p53+/+ mouse, SPATA18 was downregulated following p53 knockdown by shRNA (Fig. 1E). In sum, SPATA18 transcription is activated in a p53-dependent manner in a wide variety of human and mouse cell types and following various types of p53-activating signals.
p53 directly activates SPATA18 transcription.
Next, we tested whether SPATA18 transcription is directly activated by p53. To this end, we treated WI-38 cells with Nutlin-3a in combination with cycloheximide, a compound that inhibits protein biosynthesis. If SPATA18 induction by Nutlin-3a is indirect and requires the synthesis of a protein mediator, then cycloheximide should prevent it. As a positive control we measured the mRNA and protein levels of p21, which represents a bona fide direct p53 target. Indeed, p21 transcription was activated in a p53-dependent manner following Nutlin-3a treatment, and the addition of cycloheximide did not interfere with this induction while the accumulation of p21 protein following Nutlin-3a treatment was prevented by cycloheximide (Fig. 2A and B). Importantly, SPATA18 expression displayed a pattern similar to that of p21, indicating that SPATA18 transcription is most likely activated by p53 directly.
p53 is a transcription factor that specifically binds to DNA consensus sequences, defined as two copies of the 10-bp motif 5′-PuPuPuC(A/T)(T/A)GPyPyPy-3′ (where Pu is a purine and Py is a pyrimidine) separated by a spacer of 0 to 13 bp (38). We used MatInspector (48) to search for p53 binding sites in the human SPATA18 locus. A putative binding site (BS) with high similarity to the p53 consensus was found in the first intron of SPATA18. This BS has two perfect core sequences and no spacer (see Fig. S2A posted at http://www.weizmann.ac.il/mcb/Varda/p53_SPATA18/), properties associated with strong binding of p53 (38). To determine whether p53 binds this putative BS, we carried out a chromatin immunoprecipitation assay in WI-38 cells and analyzed the amount of DNA precipitated with an antibody against p53 or a control antibody, using specific primers for either the known p53 BS in the promoter of p21 or the putative BS in the first intron of SPATA18. As predicted, both of these genomic regions were strongly enriched in the p53-immunoprecipitated chromatin (Fig. 2C), indicating that p53 binds the consensus sequence in the first intron of SPATA18. Next, we tested the functionality of the identified p53 BS. To this end we cloned a 495-bp intronic fragment, which includes the p53 BS, into a luciferase reporter plasmid and carried out a series of promoter activity assays in p53-null Saos-2 cells. In these cells, transfection of WT p53, but not cancer-derived mutant p53R249S, which is incapable of transactivating p53 target genes (57), induced the transcription of the endogenous SPATA18 gene (Fig. 2D), as well as of p21 (see Fig. S2B posted at http://www.weizmann.ac.il/mcb/Varda/p53_SPATA18/). Similarly, cotransfection of WT p53, but not p53R249S, with luciferase reporter plasmids resulted in activation of a luciferase reporter under the control of the cloned SPATA18 intronic fragment while an empty luciferase reporter was not affected by p53 (Fig. 2E). The activity of a reporter containing p21 promoter was measured as a positive control (see Fig. S2C posted at the URL mentioned above). Finally, mutations of three core nucleotides in the p53 BS within the cloned fragment abolished its activation by WT p53. Combined, the results presented in Fig. 2 clearly demonstrate that SPATA18 is a direct transcriptional target of p53 and indicate that the mechanism of SPATA18 transactivation involves binding of p53 to a consensus element within its first intron.
SPATA18 is expressed in spermatids in the mouse testes.
In mammals, germ cells called gonocytes populate the seminiferous tubules at birth (31). During the first postnatal week in mice, the first spermatogonia and primary spermatocytes appear. At postnatal day 7 the prophase of meiosis begins, whereas the first round and elongating spermatids are observed at postnatal days 21 and 25, respectively, with spermiation commencing at day 34 (37). The synchronous nature of spermatogenesis in neonatal mice and rats allows accurate attribution of biological processes to specific cell populations. Accordingly, the mRNA of Spetex-1, the rat ortholog of SPATA18, is detected at 3 weeks of postnatal development and reaches peak levels at 6 weeks (27). The combined results of in situ hybridization, immunocytochemical analysis, and immunoelectron microscopy show that Spetex-1 is highly expressed in elongating spermatids and in spermatozoa (26, 27).
We decided to perform a similar analysis of testicular SPATA18 levels during mouse postnatal development. qRT-PCR measurements of total testis RNA revealed that SPATA18 is profoundly upregulated between postnatal days 21 to 25 and remains high until day 38 (Fig. 3A), as well as in testes of mature mice (data not shown). We therefore concluded that SPATA18 is expressed primarily in spermatids. We next analyzed the expression of p21 along the same testicular development model and found that, similarly to SPATA18, p21 expression is upregulated between postnatal days 21 to 25 (Fig. 3B). Furthermore, the p53 target gene Wip1 was also demonstrated to be profoundly upregulated in the testis during postnatal days 20 to 25 (10), further supporting the notion that p53 activity is augmented during this period of postnatal spermatogenesis.
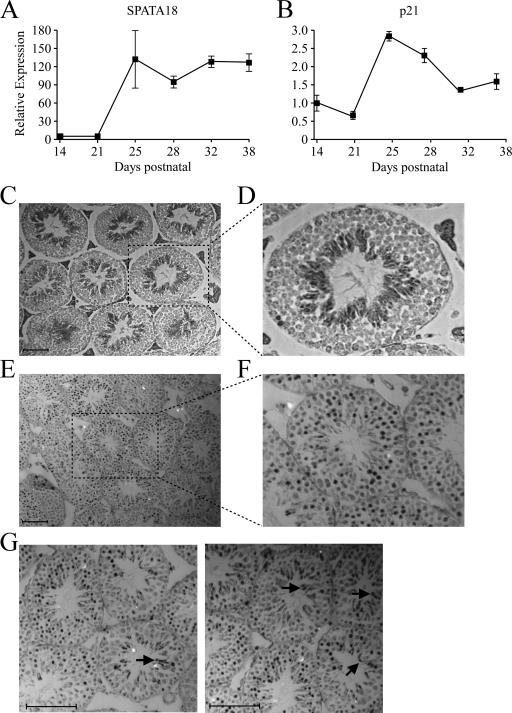
Fig. 3.
Spatiotemporal expression patterns of SPATA18 mRNA and p53 protein. qRT-PCR measurements were conducted for SPATA18 (A) and p21 (B) mRNAs in mouse testes collected at the indicated postnatal days. For each time point, testes were collected from three mice, and error bars represent the corresponding standard error of mean. (C) In situ hybridization analysis of SPATA18 mRNA in a testis section from a mature (14 weeks) mouse. Scale bar, 100 μm. (D) Digital magnification of the marked area in panel C. (E) Immunohistochemistry analysis of p53 protein in testis from a 25-day-old mouse. Scale bar, 200 mm. (F) Digital magnification of the marked area in panel E. (G) Digital magnifications of immunohistochemistry analysis of p53 protein in testis from 25-day-old mouse. Arrows indicate stained regions that are populated with elongating spermatids.
To gain more insights on the exact localization of SPATA18 mRNA in the testis, we performed an in situ hybridization assay. Paraffin sections of mature mouse testes were hybridized with either an RNA probe complementary to SPATA18 mRNA (antisense) or a sense probe as a negative control. Using the antisense probe, strong staining was observed in the inner layer of most of the seminiferous tubules (Fig. 3C and D), whereas sections hybridized with the sense probe were devoid of a positive signal (see Fig. S2D posted at http://www.weizmann.ac.il/mcb/Varda/p53_SPATA18/), attesting to the specificity of the antisense probe. Careful examination of the hybridization pattern revealed that SPATA18 mRNA is absent from both spermatogonia and spermatocytes, which are located on the outer layer of the seminiferous tubule, whereas round and, to a much greater extent, elongating spermatids express high levels of SPATA18 (Fig. 3D). Interestingly, immunohistochemistry analysis of p53 protein in mouse testes demonstrated its abundance in the intermediate layer of most seminiferous tubules, with intense staining in spermatocytes and round spermatids (Fig. 3E and F). Moreover, in some seminiferous tubules, we observed strong p53 staining in elongating spermatids (Fig. 3G).
SPATA18 transcription is regulated by p53 and p63 in vivo.
Having shown that SPATA18 is a bona fide p53 target gene in a variety of cell types of both human and mouse origin and that SPATA18 is highly expressed in spermatids in vivo, we were curious whether SPATA18 is activated by p53 in mouse testes. We therefore compared the mRNA levels of SPATA18 in the testes of p53+/+, p53+/−, and p53−/− mice. As depicted in Fig. 4A, p53 mRNA levels are strongly reduced in p53+/− mice compared to levels in p53+/+ mice (P value, 2.57 × 10−5) and are nondetectable in p53−/− mice. Interestingly, testicular SPATA18 mRNA was significantly lower in p53+/− mice than in p53+/+ mice (Fig. 4A) (P value, 3.4 × 10−4), supporting the notion that p53 activates SPATA18 transcription in vivo. To our great surprise, however, testicular SPATA18 expression in p53−/− was not lower than that in p53+/+ mice. Nevertheless, when the mRNA levels of p21 were analyzed in the same whole-testis RNA samples, a strikingly similar pattern was found; i.e., testicular p21 expression was reduced in p53+/− mice (P value, 1.7 × 10−3) but not in p53−/− mice (Fig. 4A).
These results led us to hypothesize that in p53−/− mice the complete absence of p53 is compensated, probably by its family members, p63 and/or p73. The homology shared by p53, p63, and p73 suggests that these proteins have similar properties as transcription factors, and, although each family member has distinct roles during development and homeostasis, many of their functions overlap, as well as their DNA binding motifs and target genes (6, 18, 43). Moreover, all three family members were shown to be expressed in mouse testes (17). We therefore performed immunofluorescence analysis for p63 protein in mouse testes and found it to localize primarily at the nuclei of spermatocytes and, to a lower extent, in round spermatids (Fig. 4B). This expression pattern partially overlaps with that of p53 (Fig. 3), supporting the notion that these paralogs have common functions in spermatogenesis. Importantly, Western blot analysis of p63 protein in whole-testis lysates revealed that p63 levels are significantly elevated in p53+/+ mice compared to levels in p53+/+ mice (Fig. 4C) (P value, 1.9 × 10−4). Of note, p63 protein levels were not significantly elevated in p53+/− mice compared to levels in p53−/− mice (data not shown). Moreover, PCR analysis of whole-testis RNA revealed that the predominant p63 isoforms expressed in mouse testes are TA-p63α/γ (see Fig. S3A posted at http://www.weizmann.ac.il/mcb/Varda/p53_SPATA18/), which encode transactivation-proficient proteins that share functions and target genes with wild-type p53 (43). Expression of the p63 isoforms lacking the transactivation domain (ΔN-p63) was not detected in the testes while in kidneys both TA-p63 and ΔN-p63 were expressed, thus serving as a positive control for the detection method.
Next, we analyzed the effect of p63 overexpression in the mouse Sertoli cell line TM4. Interestingly, overexpression of p63 in control TM4 cells did not affect SPATA18 expression. However, when we knocked down p53 in these cells, the SPATA18 mRNA level was reduced, and p63 overexpression could restore it (Fig. 4D). These results indicate that p63 is capable of inducing SPATA18 transcription and imply that, in the absence of p53, the effect of p63 on SPATA18 transcription is augmented. Furthermore, knockdown of p63 in TM4 cells led to a marked reduction of SPATA18 mRNA levels (Fig. 4E), indicating that the endogenous p63 in these cells activates SPATA18 transcription. To demonstrate that the effect of p63 on SPATA18 expression is not specific to mice, we overexpressed p63, as well as p53 as a positive control, in HT-1080 human fibrosarcoma cells. Indeed, both p53 and p63 induced SPATA18 expression (see Fig. S3B posted at http://www.weizmann.ac.il/mcb/Varda/p53_SPATA18/). To support the notion that p63 can directly transactivate SPATA18, we performed chromatin immunoprecipitation assay in mouse TA-p63α-transfected TM4 cells with an antibody against p63 or a control IgG antibody. As p63 shares its consensus DNA-binding site with p53, we used MatInspector (48) to search for p53 BSs in the mouse SPATA18 locus and found two such BSs, which reside, similarly to human SPATA18, in the first intron. One of these BSs, which resides approximately 2 kbp downstream from the transcriptional start site and has two perfect core sequences and no spacer (see Fig. S3C posted at http://www.weizmann.ac.il/mcb/Varda/p53_SPATA18/), showed significant binding to p63 (Fig. 4F) (P value, 1.8 × 10−2). Therefore, p63 can bind the SPATA18 gene and can activate its transcription. Combining this result with the observation that p63 levels are elevated in p53−/− mice, we suggest that p63 compensates for the absence of p53 in the testes of p53-null mice. Notably, overexpression of p73 in HT-1080 cells did not induce SPATA18 expression. Moreover, Western blot analysis of p73 in mouse testes did not demonstrate an increase of p73 levels in p53−/− mice compared to p53+/+ mice (data not shown). Thus, although the evidence is not conclusive, it appears that testicular p53 deficiency is primarily compensated by p63 and not p73.
DISCUSSION
p53 plays central roles in normal cell differentiation and organismal development (42). Moreover, substantial data implicate p53 in the regulation of spermatogenesis under normal conditions (1, 52, 55) and following DNA damage (20, 21), as well as in the regulation of additional aspects of fertility (19, 24, 25). In this study we set out to examine the role of p53 in the transcriptional regulation of SPATA18, a spermatogenesis-related gene. We discovered that SPATA18 is a bona fide p53 transcriptional target, and its induction by p53 can be observed in a variety of normal and cancerous cell types of both human and mouse origin. Similar to many other p53 target genes (62), the consensus DNA sequence that mediates p53 binding to the SPATA18 locus resides within its first intron, probably acting as an enhancer.
Although SPATA18 is known to be expressed primarily in the testis, it is also present in additional tissues, such as lung, intestine, and spleen (27). This may explain its expression in normal and cancerous cells representing a wide variety of tissues, including human and mouse embryonic fibroblasts and human fibrosarcoma, osteosarcoma, prostate, ovarian, and hepatocellular carcinoma cells, as well as in mouse Sertoli and embryonic stem cells (Fig. 1, 2, and 4; see also Fig. S1 posted at http://www.weizmann.ac.il/mcb/Varda/p53_SPATA18/). However, as the exact molecular role and biological function of SPATA18 are currently unknown, the role of its p53-dependent transcriptional induction in somatic cells is yet to be unveiled.
SPATA18 rat ortholog, Spetex-1, is highly expressed during spermatogenesis in spermatids, and its protein localizes at defined regions, such as the cytoplasm of spermatids, residual bodies, and the flagella (26, 27, 30). Accordingly, two distinct non-mutually exclusive suggestions were raised regarding its function in spermatogenesis, being involved either in the process of spermatid maturation or constituting a flagellar component. These possibilities tempted us to speculate that SPATA18 might mediate, at least partially, the roles exerted by p53 during spermatogenesis. Our in vivo analyses indicate that SPATA18 is profoundly upregulated during mouse postnatal spermatogenesis at a stage corresponding to the appearance of spermatids. Accordingly, in situ hybridization analysis detected SPATA18 mRNA primarily in elongating spermatids, as well as in round spermatids, albeit at a lower level (Fig. 3). This was in agreement with previous reports on Spetex-1 (26, 27). To further support this notion, we utilized global expression analysis as performed by Namekawa et al. (44), which analyzed mRNA expression in four enriched germ cell populations from mouse testes, namely, types A and B spermatozoa, pachytene spermatocytes, and round spermatids. Confirming our results, SPATA18 was expressed almost exclusively in round spermatids (see Fig. S4A posted at http://www.weizmann.ac.il/mcb/Varda/p53_SPATA18/).
We present several lines of evidence that the testicular expression of SPATA18 is positively regulated by p53. First, the pattern of p53 accumulation, which typically indicates its functional activation, partially overlaps with that of SPATA18 transcription; i.e., p53 protein is accumulated in spermatocytes, round spermatids, and occasionally in elongating spermatids (Fig. 3E to G), while SPATA18 mRNA is induced in round and, more profoundly, in elongating spermatids (Fig. 3C and D; see also Fig. S4A posted at http://www.weizmann.ac.il/mcb/Varda/p53_SPATA18/). Second, p21 is induced between postnatal days 21 to 25 (Fig. 3B), concomitantly with the appearance of elongating spermatids, indicating functional activation of p53 at the same stage in which SPATA18 is induced. Of note, a similar spatiotemporal pattern was previously demonstrated also for the testicular p53 target gene Wip1 (10). Supporting the notion that SPATA18 is regulated by p53 in vivo, its expression is significantly attenuated in the testes of p53+/− mice (Fig. 4A). This attenuation correlates with that of p53 and p21 mRNA levels. Whether this haploinsufficiency is manifested as fertility- or development-related defects is currently unclear since the effects of p53 deficiency were analyzed almost exclusively in p53-null animals. Surprisingly, the expression of both p21 and SPATA18 is not attenuated in p53−/− mice, prompting us to hypothesize that the testicular p53 activity is compensated by its paralogs, p63 and p73, which play important and unique roles during development. Indeed, p63 and/or p73 were reported to compensate for p53 loss in several processes, including DNA damage response (65), tumor suppression (14), and development (28, 56). p63 is expressed in a highly restricted pattern during embryogenesis and is essential for limb formation and epidermal morphogenesis. p63-null mice show profound developmental abnormalities of the skin, limbs, mammary, prostate, and other epithelial tissues and die soon after birth (39, 63). In addition, p63 is expressed in mouse reproductive organs and primordial germ cells (32) and was suggested to regulate programmed cell death and differentiation of these cells (47). Most importantly, a similar giant-cell syndrome, which characterizes mice with p53 deficiency (52), was reported in p63+/− adult mice and in p63−/− cultured fetal testes (47).
Unlike p63, p73-null mice are viable but are stunted and have high mortality rates. These mice show profound developmental defects, including hippocampal dysgenesis and hydrocephalus (64). Interestingly, while p73-null mice have no structural abnormalities in their reproductive organs, p73-null males lack interest in mating, probably due to hormonal or sensory defects, and thus have low fecundity (64).
Our data indicate that p63 is highly expressed in spermatocytes and to a lesser extent in spermatids (Fig. 4B). Supporting the notion that p63 may compensate for p53 loss, p63 protein levels were increased in the testes of p53-null mice (Fig. 4C). Similarly, p63 protein was also shown to be increased in oral-esophageal epithelia of p53-null mice compared to WT p53 mice (16, 58). It was further demonstrated that p53 can reduce the stability of TA-p63γ (36), perhaps providing a mechanistic explanation for the accumulation of p63 in p53-deficient tissues. Moreover, we observed an increase in the mRNA level of TA-p63, but not ΔN-p63, following p53 inactivation in WI-38 cells (data not shown), suggesting another mechanism by which p53 can downregulate p63.
Importantly, we demonstrated that p63 overexpression induces SPATA18 expression and that knockdown of endogenous p63 attenuates SPATA18 levels (Fig. 4; see also Fig. S3B posted at http://www.weizmann.ac.il/mcb/Varda/p53_SPATA18/). Moreover, by analyzing publicly available gene profiling data sets, we found that knockdown of endogenous p63 expression in human keratinocytes or squamous carcinoma cells (3) leads to downregulation of SPATA18 expression (see Fig. S4D posted at the URL mentioned above). We also showed that p63 can directly bind a regulatory site within the first intron of SPATA18 (Fig. 4F). Taking together the observations that p63 is elevated in p53−/− testes and is capable of binding SPATA18 and inducing its transcription, it is likely that the loss of testicular p53 activity is compensated by p63, resulting in steady transcriptional activity of p53 target genes. Of note, both p63 and p73 were suggested to compensate for p53 loss in several processes, including tumor suppression (14) and development (28, 56). While both family members can potentially compensate for testicular p53 function, our data indicate p63 as the more likely candidate since p73 was not upregulated in the testes of p53−/− mice and was not capable of transactivating SPATA18 in vitro (data not shown).
It remains highly interesting to investigate the role of SPATA18 as a p53 target both in the testes and in additional tissues and cell types. The identification of Spetex-1 protein in the cytoplasm of elongated spermatids and in residual bodies engulfed by Sertoli cells (27) implies the possibility that SPATA18 is involved in the apoptosis-like process of spermatid maturation, which coincides well with the known function of p53 as an apoptosis inducer. To date, we can only speculate that SPATA18 mediates p53 functions during spermatogenesis. Notably, the developmental effects of p53 deficiency were analyzed primarily in a homozygous background, in which SPATA18 expression remains normal. The question of whether abnormalities in spermatogenesis are present in p53+/− mice is therefore intriguing. A somewhat equivalent system is represented by p53 promoter-chloramphenicol acetyltransferase (CAT)-harboring mice, in which p53 mRNA and protein are downregulated (52). These mice exhibit a testicular giant-cell degenerative syndrome, which likely stems from the inability of tetraploid spermatocytes to complete meiosis. Another implication of SPATA18 in a spermatogenesis-related pathology may lie in the observation that SPATA18 is downregulated in human semen samples collected from individuals with severe teratozoospermia compared to semen collected from normal fertile men (P value, 7.4 × 10−3) (see Fig. S4B posted at http://www.weizmann.ac.il/mcb/Varda/p53_SPATA18/). Moreover, expression microarray analysis of testicular biopsy specimens of azoospermia patients reveals that SPATA18 mRNA is significantly downregulated in biopsy specimens collected from nonobstructive azoospermia (NOA) patients compared with obstructive azoospermia (OA) patients (P value, 8.3 × 10−9) (see Fig. S4C posted at the URL mentioned above). Combined, these results link attenuated levels of SPATA18 to reduced fertility caused by defects in sperm development.
To gain insights into the possible roles of SPATA18 in additional tissues or processes, we searched the Oncomine database (49) and found few expression profiling studies that detected downregulation of SPATA18 in cancer samples compared to levels in the corresponding normal tissues. For instance, in a study conducted by Richardson et al. (50) comparing human ductal breast carcinomas to normal breast samples, SPATA18 was found to be downregulated approximately 5-fold in the malignant samples (P value, 7.2 × 10−7). These data imply a tumor suppressive role for SPATA18, perhaps in the process of programmed cell death.
Combined, our data enrich the known collection of p53 targets with a gene whose expression and localization imply a role in spermatogenesis. To the best of our knowledge, besides Wip1, SPATA18 is currently the only testis-associated p53 target gene and the only gene proposed to be a structural component of the sperm flagella. Our data also provide clues into the mechanisms underlying spermatogenesis and fertility defects associated with p53 deficiency and highlight the primordial role of p53 as a master regulator of the transmission of genetic material.
ACKNOWLEDGMENTS
This work was supported by a Center of Excellence grant from the Flight Attendant Medical Research Institute. V.R. is the incumbent of the Norman and Helen Asher Professorial Chair Cancer Research at the Weizmann Institute.
We thank J. Don from Bar-Ilan University for providing scientific consultation.
Footnotes
Published ahead of print on 7 February 2011.
REFERENCES
- 1. Almon E., et al. 1993. Testicular tissue-specific expression of the p53 suppressor gene. Dev. Biol. 156:107–116 [DOI] [PubMed] [Google Scholar]
- 2. Baker S. J., Markowitz S., Fearon E. R., Willson J. K., Vogelstein B. 1990. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science 249:912–915 [DOI] [PubMed] [Google Scholar]
- 3. Barbieri C. E., Tang L. J., Brown K. A., Pietenpol J. A. 2006. Loss of p63 leads to increased cell migration and up-regulation of genes involved in invasion and metastasis. Cancer Res. 66:7589–7597 [DOI] [PubMed] [Google Scholar]
- 4. Beumer T. L., Roepers-Gajadien H. L., Gademan L. S., Rutgers D. H., de Rooij D. G. 1997. P21(Cip1/WAF1) expression in the mouse testis before and after X irradiation. Mol. Reprod Dev. 47:240–247 [DOI] [PubMed] [Google Scholar]
- 5. Blanco-Rodriguez J., Martinez-Garcia C. 1999. Apoptosis is physiologically restricted to a specialized cytoplasmic compartment in rat spermatids. Biol. Reprod. 61:1541–1547 [DOI] [PubMed] [Google Scholar]
- 6. Brandt T., Petrovich M., Joerger A. C., Veprintsev D. B. 2009. Conservation of DNA-binding specificity and oligomerisation properties within the p53 family. BMC Genomics 10:628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brosh R., Rotter V. 2010. Transcriptional control of the proliferation cluster by the tumor suppressor p53. Mol. Biosyst. 6:17–29 [DOI] [PubMed] [Google Scholar]
- 8. Brosh R., Rotter V. 2009. When mutants gain new powers: news from the mutant p53 field. Nat. Rev. Cancer 9:701–713 [DOI] [PubMed] [Google Scholar]
- 9. Brosh R., et al. 2010. p53-dependent transcriptional regulation of EDA2R and its involvement in chemotherapy-induced hair loss. FEBS Lett. 584:2473–2477 [DOI] [PubMed] [Google Scholar]
- 10. Choi J., et al. 2002. Mice deficient for the wild-type p53-induced phosphatase gene (Wip1) exhibit defects in reproductive organs, immune function, and cell cycle control. Mol. Cell. Biol. 22:1094–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reference deleted.
- 12. Donehower L. A., et al. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356:215–221 [DOI] [PubMed] [Google Scholar]
- 13. el-Deiry W. S., et al. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817–825 [DOI] [PubMed] [Google Scholar]
- 14. Flores E. R., et al. 2005. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell 7:363–373 [DOI] [PubMed] [Google Scholar]
- 15. Green D. R., Kroemer G. 2009. Cytoplasmic functions of the tumour suppressor p53. Nature 458:1127–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guo X., et al. 2009. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat. Cell Biol. 11:1451–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hamer G., Gademan I. S., Kal H. B., de Rooij D. G. 2001. Role for c-Abl and p73 in the radiation response of male germ cells. Oncogene 20:4298–4304 [DOI] [PubMed] [Google Scholar]
- 18. Harms K., Nozell S., Chen X. 2004. The common and distinct target genes of the p53 family transcription factors. Cell. Mol. Life Sci. 61:822–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harvey M., McArthur M. J., Montgomery C. A., Jr., Bradley A., Donehower L. A. 1993. Genetic background alters the spectrum of tumors that develop in p53-deficient mice. FASEB J. 7:938–943 [DOI] [PubMed] [Google Scholar]
- 20. Hasegawa M., Zhang Y., Niibe H., Terry N. H., Meistrich M. L. 1998. Resistance of differentiating spermatogonia to radiation-induced apoptosis and loss in p53-deficient mice. Radiat. Res. 149:263–270 [PubMed] [Google Scholar]
- 21. Hendry J. H., Adeeko A., Potten C. S., Morris I. D. 1996. P53 deficiency produces fewer regenerating spermatogenic tubules after irradiation. Int. J. Radiat. Biol. 70:677–682 [DOI] [PubMed] [Google Scholar]
- 22. Hess R. A., Renato de Franca L. 2008. Spermatogenesis and cycle of the seminiferous epithelium. Adv. Exp. Med. Biol. 636:1–15 [DOI] [PubMed] [Google Scholar]
- 23. Hilscher W., Hilscher B. 1976. Kinetics of the male gametogenesis. Andrologia 8:105–116 [PubMed] [Google Scholar]
- 24. Hu W. 2009. The role of p53 gene family in reproduction. Cold Spring Harb. Perspect. Biol. 1:a001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu W., Feng Z., Teresky A. K., Levine A. J. 2007. p53 regulates maternal reproduction through LIF. Nature 450:721–724 [DOI] [PubMed] [Google Scholar]
- 26. Iida H., Honda Y., Matsuyama T., Shibata Y., Inai T. 2006. Spetex-1: a new component in the middle piece of flagellum in rodent spermatozoa. Mol. Reprod. Dev. 73:342–349 [DOI] [PubMed] [Google Scholar]
- 27. Iida H., Ichinose J., Kaneko T., Mori T., Shibata Y. 2004. Complementary DNA cloning of rat spetex-1, a spermatid-expressing gene-1, encoding a 63 kDa cytoplasmic protein of elongate spermatids. Mol. Reprod. Dev. 68:385–393 [DOI] [PubMed] [Google Scholar]
- 28. Jacobs W. B., Kaplan D. R., Miller F. D. 2006. The p53 family in nervous system development and disease. J. Neurochem. 97:1571–1584 [DOI] [PubMed] [Google Scholar]
- 29. Kalo E., et al. 2007. Mutant p53 attenuates the SMAD-dependent transforming growth factor β1 (TGF-β1) signaling pathway by repressing the expression of TGF-β receptor type II. Mol. Cell. Biol. 27:8228–8242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaneko T., Murayama E., Kurio H., Yamaguchi A., Iida H. 2010. Characterization of Spetex-1, a new component of satellite fibrils associated with outer dense fibers in the middle piece of rodent sperm flagella. Mol. Reprod. Dev. 77:363–372 [DOI] [PubMed] [Google Scholar]
- 31. Kluin P. M., de Rooij D. G. 1981. A comparison between the morphology and cell kinetics of gonocytes and adult type undifferentiated spermatogonia in the mouse. Int. J. Androl. 4:475–493 [DOI] [PubMed] [Google Scholar]
- 32. Kurita T., Mills A. A., Cunha G. R. 2004. Roles of p63 in the diethylstilbestrol-induced cervicovaginal adenosis. Development 131:1639–1649 [DOI] [PubMed] [Google Scholar]
- 33. Lane D. P. 1992. Cancer p53, guardian of the genome. Nature 358:15–16 [DOI] [PubMed] [Google Scholar]
- 34. Leblond C. P., Clermont Y. 1952. Spermiogenesis of rat, mouse, hamster and guinea pig as revealed by the periodic acid-fuchsin sulfurous acid technique. Am. J. Anat. 90:167–215 [DOI] [PubMed] [Google Scholar]
- 35. Levine A. J., Oren M. 2009. The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer. 9:749–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li N., et al. 2006. TA-p63-γ regulates expression of ΔN-p63 in a manner that is sensitive to p53. Oncogene 25:2349–2359 [DOI] [PubMed] [Google Scholar]
- 37. Malkov M., Fisher Y., Don J. 1998. Developmental schedule of the postnatal rat testis determined by flow cytometry. biology of reproduction. 59:84–92 [DOI] [PubMed] [Google Scholar]
- 38. Menendez D., Inga A., Resnick M. A. 2009. The expanding universe of p53 targets. Nat. Rev. Cancer. 9:724–737 [DOI] [PubMed] [Google Scholar]
- 39. Mills A. A., et al. 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398:708–713 [DOI] [PubMed] [Google Scholar]
- 40. Milyavsky M., et al. 2003. Prolonged culture of telomerase-immortalized human fibroblasts leads to a premalignant phenotype. Cancer Res. 63:7147–7157 [PubMed] [Google Scholar]
- 41. Milyavsky M., et al. 2005. Transcriptional programs following genetic alterations in p53, INK4A, and H-Ras genes along defined stages of malignant transformation. Cancer Res. 65:4530–4543 [DOI] [PubMed] [Google Scholar]
- 42. Molchadsky A., Rivlin N., Brosh R., Rotter V., Sarig R. 2010. p53 is balancing development, differentiation and de-differentiation to assure cancer suppression. Carcinogenesis 31:1501–1508 [DOI] [PubMed] [Google Scholar]
- 43. Moll U. M., Slade N. 2004. p63 and p73: roles in development and tumor formation. Mol. Cancer Res. 2:371–386 [PubMed] [Google Scholar]
- 44. Namekawa S. H., et al. 2006. Postmeiotic sex chromatin in the male germline of mice. Curr. Biol. 16:660–667 [DOI] [PubMed] [Google Scholar]
- 45. Nicol C. J., Harrison M. L., Laposa R. R., Gimelshtein I. L., Wells P. G. 1995. A teratologic suppressor role for p53 in benzo[a]pyrene-treated transgenic p53-deficient mice. Nat. Genet. 10:181–187 [DOI] [PubMed] [Google Scholar]
- 46. Ossovskaya V. S., et al. 1996. Use of genetic suppressor elements to dissect distinct biological effects of separate p53 domains. Proc. Natl. Acad. Sci. U. S. A. 93:10309–10314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Petre-Lazar B., et al. 2007. The role of p63 in germ cell apoptosis in the developing testis. J. Cell. Physiol. 210:87–98 [DOI] [PubMed] [Google Scholar]
- 48. Quandt K., Frech K., Karas H., Wingender E., Werner T. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23:4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rhodes D. R., et al. 2004. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Richardson A. L., et al. 2006. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 9:121–132 [DOI] [PubMed] [Google Scholar]
- 51. Rotter V., et al. 1994. Does wild-type p53 play a role in normal cell differentiation? Semin. Cancer Biol. 5:229–236 [PubMed] [Google Scholar]
- 52. Rotter V., et al. 1993. Mice with reduced levels of p53 protein exhibit the testicular giant-cell degenerative syndrome. Proc. Natl. Acad. Sci. U. S. A. 90:9075–9079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Russell L. D. 1979. Spermatid-Sertoli tubulobulbar complexes as devices for elimination of cytoplasm from the head region late spermatids of the rat. Anat. Rec. 194:233–246 [DOI] [PubMed] [Google Scholar]
- 54. Sarig R., et al. 2010. Mutant p53 facilitates somatic cell reprogramming and augments the malignant potential of reprogrammed cells. J. Exp. Med. 207:2127–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schwartz D., Goldfinger N., Rotter V. 1993. Expression of p53 protein in spermatogenesis is confined to the tetraploid pachytene primary spermatocytes. Oncogene 8:1487–1494 [PubMed] [Google Scholar]
- 56. Stiewe T. 2007. The p53 family in differentiation and tumorigenesis. Nat. Rev. Cancer. 7:165–168 [DOI] [PubMed] [Google Scholar]
- 57. Suad O., et al. 2009. Structural basis of restoring sequence-specific DNA binding and transactivation to mutant p53 by suppressor mutations. J. Mol. Biol. 385:249–265 [DOI] [PubMed] [Google Scholar]
- 58. Suliman Y., et al. 2001. p63 expression is associated with p53 loss in oral-esophageal epithelia of p53-deficient mice. Cancer Res. 61:6467–6473 [PubMed] [Google Scholar]
- 59. Suzuki-Toyota F., Ishibashi K., Yuasa S. 1999. Immunohistochemical localization of a water channel, aquaporin 7 (AQP7), in the rat testis. Cell Tissue Res. 295:279–285 [DOI] [PubMed] [Google Scholar]
- 60. Vassilev L. T., et al. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303:844–848 [DOI] [PubMed] [Google Scholar]
- 61. Vousden K. H., Ryan K. M. 2009. p53 and metabolism. Nat. Rev. Cancer. 9:691–700 [DOI] [PubMed] [Google Scholar]
- 62. Wei C. L., et al. 2006. A global map of p53 transcription-factor binding sites in the human genome. Cell 124:207–219 [DOI] [PubMed] [Google Scholar]
- 63. Yang A., et al. 1999. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398:714–718 [DOI] [PubMed] [Google Scholar]
- 64. Yang A., et al. 2000. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404:99–103 [DOI] [PubMed] [Google Scholar]
- 65. Yao J. Y., Chen J. K. 2010. TAp63 plays compensatory roles in p53-deficient cancer cells under genotoxic stress. Biochem. Biophys. Res. Commun. 403:310–315 [DOI] [PubMed] [Google Scholar]