Abstract
The genus of Marinobacter is one of the most ubiquitous in the global oceans and assumed to significantly impact various biogeochemical cycles. The genome structure and content of Marinobacter aquaeolei VT8 was analyzed and compared with those from other organisms with diverse adaptive strategies. Here, we report the many “opportunitrophic” genetic characteristics and strategies that M. aquaeolei has adopted to promote survival under various environmental conditions. Genome analysis revealed its metabolic potential to utilize oxygen and nitrate as terminal electron acceptors, iron as an electron donor, and urea, phosphonate, and various hydrocarbons as alternative N, P, and C sources, respectively. Miscellaneous sensory and defense mechanisms, apparently acquired via horizontal gene transfer, are involved in the perception of environmental fluctuations and antibiotic, phage, toxin, and heavy metal resistance, enabling survival under adverse conditions, such as oil-polluted water. Multiple putative integrases, transposases, and plasmids appear to have introduced additional metabolic potential, such as phosphonate degradation. The genomic potential of M. aquaeolei and its similarity to other opportunitrophs are consistent with its cosmopolitan occurrence in diverse environments and highly variable lifestyles.
INTRODUCTION
Since the 1960s, r/K selection theory has become widely used, and today most ecologically relevant microorganisms are still classified as either r or K strategists (41, 52). r strategists, also referred to as copiotrophs, are adapted to nutrient excess and proliferate in unstable and uncrowded environments (30, 51). They rapidly exploit nutrient patches and die or become dormant after substrate exhaustion due to their specialization (15, 67). Organisms previously described as r strategists include Pseudomonas and Acinetobacter species, which grow fast and are adapted to high nutrient concentrations (30). K strategists lead an oligotrophic lifestyle and are adapted to resource-restricted, crowded environments (30). They generally grow slowly, as they allocate their energy to various functions other than growth, such as defense and extracellular enzyme production (15, 30, 64). For example, “Candidatus Pelagibacter ubique,” which belongs to the cosmopolitan clade SAR11, has the smallest genome and smallest number of predicted open reading frames in a free-living microorganism (18). Its genome does not contain any transposons, extrachromosomal elements, plasmids, or pseudogenes (18). The basic survival strategy of “Ca. Pelagibacter ubique” features efficient replication under limiting nutrient resources and the utilization (instead of de novo synthesis) of substrates and cofactors existing in the DOM pool (i.e., vitamins and amino acids). In order to compete with r strategists, K strategists tend to be able to utilize a wider variety of unusual forms of carbon and nutrients, such as gaseous C sources (51).
More recent physiological classifications of bacteria have started to incorporate the flexibility exhibited by many microorganisms to persist and thrive under conditions that are neither classically considered r or K strategies. For example, microaerophiles can also often operate under well-aerated conditions, even though they adapted to life at low O2 concentrations. Many bacteria can metabolize more than one substrate, a quality which may be the norm rather than the exception, as reflected in the diversity of catabolic intracellular and extracellular enzymes (51). As a result, the terms “opportunitroph” and “generalist” were introduced to more precisely describe the lifestyles of organisms and their genomic complements that equip them to exploit spatially and temporally variable resources (46). Compared to r strategists, opportunitrophs are not as specialized for a specific substrate but may readily adapt to various environmental conditions and outcompete K strategists, as they are able to take advantage of high-nutrient niches within a bulk low-nutrient environment, such as in marine snow particles and around dead plankton (32, 46, 51). Opportunitrophic organisms are not characterized solely by the variety of substrates they can use as energy and carbon sources but also by their resistance potential, for instance toward phages, and their ability to sense and interact with their environment (39). As a result, opportunitrophs are not generally observed to dominate a particular niche but rather employ a strategy of survival under any circumstance (53). They are often observed to display cosmopolitan occurrence patterns or at least reside in a variety of physicochemically different habitats (29, 34, 56). These observed opportunitrophic lifestyles in many bacteria and entire microbial clades, such as Roseobacter and Pseudomonas (44–46, 48, 70), suggest that this ecotype term often represents the adaptive strategies of many organisms more accurately than the term r strategist.
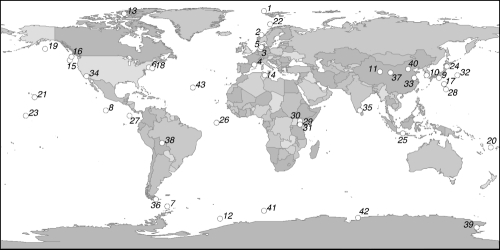
The Halomonas and Marinobacter groups are potential examples of opportunitrophic microbes (34). For example, physiological cultivation-based studies have demonstrated that these organisms are neither metabolic specialists nor minimalists but rather follow a strategy of acquiring and maintaining broad functional potential to exploit spatially and temporally variable resources (1, 16, 20, 21, 23, 25, 28, 40, 59, 62, 71). In addition, these groups are very broadly distributed in the environment; Fig. 1 shows present-day global occurrence patterns of the Halomonas and Marinobacter families. Members of the Marinobacter family are found at all depths in the global oceans (33, 72). Isolates have been obtained from habitats as diverse as petroleum field brines (49), ballast water from the Arctic (6), saline soils (43), surface seawater (72), marine snow (4), coastal hot springs (61), hydrothermal plumes (33), volcanic basalts (65), and deep seawater (33).
Fig. 1.
Sample locations at which Halomonas or Marinobacter species were identified. The map was created by use of the Planiglobe mapping service. (Reprinted from reference 34 with permission of the publisher).
This study describes the metabolic potential as revealed by the Marinobacter aquaeolei VT8 genome. Although the genus of Marinobacter includes 18 described species and numerous environmental strains, only the M. aquaeolei VT8 genome has thus far been completed. This sequenced strain was isolated from Mediterranean seawater at the head of an oil refinery. M. aquaeolei is a biofilm-forming, facultative mixotroph which can perform redox reactions using oxygen and nitrate as terminal electron acceptors (10). Growing rapidly in culture and being amenable to genetic manipulation, M. aquaeolei has been suggested for physiological studies to disclose the biochemical mechanism of neutrophilic iron oxidation (J. Waite and K. J. Edwards, unpublished data).
The three main foci of this study are as follows: (i) how genome structure and content of VT8 agree with the observed ubiquitous occurrence of M. aquaeolei (34), including interaction with environment and other organisms, (ii) how the gene content of M. aquaeolei VT8 compares with that of other opportunitrophs, and (iii) which genes, if any, may be ascribed to a signature distinctive of the opportunitrophic lifestyle.
We have performed a careful functional annotation of the M. aquaeolei genome in order to explain its ubiquitous occurrence in the marine environment. In addition, we have used comparative genomic analyses to attempt to identify a genomic signature distinctive of the opportunitrophic lifestyle.
MATERIALS AND METHODS
Organism.
Marinobacter aquaeolei VT8 was obtained from the American Type Culture Collection (strain number 700491) and cultivated according to ATCC recommendations. DNA was isolated based on methods adapted from Rogers et al. (58).
Genomic sequencing and annotation.
M. aquaeolei VT8 was whole-genome shotgun Sanger-sequenced using a combination of 3-kb, 8-kb, and 40-kb (fosmid) DNA libraries as described previously (7). Sequence assembly, quality assessment, and genome closure were performed using the Phred/Phrap/Consed software package (13, 14, 19). Possible misassemblies were corrected with Dupfinisher (24) or transposon bombing of bridging clones (Epicentre Biotechnologies, Madison, WI). A total of 3,801 additional reactions were necessary to close gaps and raise the quality of the finished sequence. The completed sequence contains 50,134 reads, which provided 9× coverage of the genome with an error rate less than 1 in 100,000 nucleotides (nt). The complete Marinobacter aquaeolei VT8 genome sequence, including a chromosome and two plasmids, is available under GenBank accession no. NC_008740, NC_008739, and NC_008738.
Sequence analysis and annotation.
Marinobacter aquaeolei VT8 was annotated by the Joint Genome Institute pipeline using three gene finders, Critica (version 1.05) (3), Glimmer (9), and Generation (http://compbio.ornl.gov/generation/index.shtml), using default settings that permit overlaps. The results of the three gene finders were combined and compared against the GenBank nonredundant database (NR). Functional assignment, identification of membrane-spanning domains, determination of paralogous gene families, and identification of regions of unusual nucleotide composition were performed as described previously (12). Phylogenomic analysis was used to assist in functional predictions (12). The annotated genome was submitted to the GenBank nonredundant database and loaded into the Integrated Microbial Genomes (IMG) database (42).
Identification of genes.
Genes involved in defined metabolic pathways were taken from the databases of IMG (version 3.1; May 2010; U.S. Department of Energy Joint Genome Institute, supported by the DOE Office of Science) and Pathway Tools (31). Manual annotation for final assignments based on maximum BLAST E values, the description of top hits to the general protein database, and cluster of orthologous group (COG) assignments were performed as needed (National Center for Biotechnology Information GenBank and BLAST). Phylogenetic determinations were performed using Geneious (Geneious Pro version 4.8+, copyright 2005 to 2010, Biomatters Ltd.). For determination of the core genome within this genus, we conducted analysis on two draft Marinobacter genomes that are available: Marinobacter algicola DG893 (NCBI taxon ID 443152) and Marinobacter sp. ELB17 (NCBI taxon ID 270374). The genomes of DG893 and ELB17 are estimated to be 99.8% and 97.8% complete, respectively, using mathematical analysis as described in reference 38. The average nucleotide identities (ANI) among the three Marinobacter genomes were calculated as described in reference 37. Unique genes were defined as unconserved genes, i.e., no BLASTP hit with ≥30% sequence identity across >70% protein length (E value = 10−5) in the nonredundant GenBank database (retrieved 17 September 2009).
RESULTS AND DISCUSSION
General genome organization and content.
The M. aquaeolei VT8 genome consists of a circular chromosome with 4,779,762 bp and two megaplasmids with 239,623 bp and 213,390 bp, harboring a total of 4,272 predicted protein-coding sequences (CDSs) among all genomic elements (Table 1). Megaplasmids are not essential for their hosts under any known conditions, but they generally contribute significantly to the individual genetic characteristics of the strains in which they reside (60).
Table 1.
General features of the M. aquaeolei genome
| Feature | Value |
|---|---|
| No. of DNA scaffolds | 3 |
| No. of CRISPRs | 1 |
| No. of plasmids | 2 |
| Size (bp) | |
| Whole genome | 4,779,762 |
| Main chromosome | 4,326,849 |
| Plasmid 1 | 239,623 |
| Plasmid 2 | 213,290 |
| % G+C content | |
| Main chromosome | 57 |
| Plasmid 1 | 54 |
| Plasmid 2 | 53 |
| No. of predicted CDSs | 4,342 |
| % coding | |
| Chromosome | 90 |
| Plasmid 1 | 86 |
| Plasmid 2 | 80 |
| No. of protein-coding genes | 4,272 |
| No. of tRNAs | 51 |
| No. of rRNA operons (16S-23S-5S) | 9 |
| No. of structural RNAs | 63 |
| No. of CDSs with known function | 3,119 |
| No. (%) of uncharacterized conserved proteins | 1,153 (26.56) |
The chromosome harbors 3,928 CDSs, plasmid 1 contains 213 CDSs, and plasmid 2 contains 201 CDSs. Most genes on the chromosome and plasmids share the greatest sequence similarity with gammaproteobacterial genes (Table 2 ), which is consistent with 16S rRNA-based phylogeny. Several CDSs, such as restriction enzymes, heavy metal transporters, β-lactamases, integrases, and transposases, are located on the two megaplasmids and have identical copies on the chromosome, suggesting that they might originate from lineage-specific duplication events.
Table 2.
Similarity of genes in M. aquaeolei to genes in other phylaa
| Domain | Phylum/class | 60% similarity |
90% similarity |
||
|---|---|---|---|---|---|
| No. of hits (no. of hit genomes) | % relative distribution of assigned hits (2,526 genes total, excluding unassigned) | No. of hits (no. of hit genomes) | % relative distribution of assigned hits (524 genes total, excluding unassigned) | ||
| Bacteria | Actinobacteria | 7 (1) | 0.28 | ||
| Bacteria | Chlorobi | 4 (4) | 0.16 | ||
| Bacteria | Cyanobacteria | 1 (1) | 0.04 | 1 (1) | 0.19 |
| Bacteria | Alphaproteobacteria | 19 (16) | 0.75 | ||
| Bacteria | Betaproteobacteria | 77 (28) | 3.05 | 8 (5) | 1.53 |
| Bacteria | Deltaproteobacteria | 15 (12) | 0.59 | 5 (2) | 0.95 |
| Bacteria | Gammaproteobacteria | 2,381 (71) | 94.26 | 502 (15) | 95.80 |
| Bacteria | Magnetococci | 2 (1) | 0.08 | ||
| Plasmid: bacteria | Alphaproteobacteria | 1 (1) | 0.04 | ||
| Plasmid: bacteria | Gammaproteobacteria | 19 (9) | 0.75 | 8 (3) | 1.53 |
| Unassigned | Unassigned | 1,222 | 3,748 | ||
This allows assessing the potential of horizontally transferred genes based on the distribution of best blast hits within the phylum/class at 60% and 90% BLAST identities. The relative abundance of gene homologs in other Gammaproteobacteria reflects the closeness of the entire VT8 genome to its class.
M. aquaeolei harbors 403 unique genes (9.3% of the genome). About 25.8% of all unique genes are located on plasmid 1, 14.6% on plasmid 2, and 59.6% on the chromosome. This means that about 40% of the genetic novelty occurs on plasmids in M. aquaeolei. Only 6.1% of the chromosomal genes represent unique genes. In comparison to other opportunitrophs, S. oneidensis MR-1 is one of a few that harbors a megaplasmid (161,613 bp) (26).
The VT8 genome has a number of putative mobile genetic elements and genomic islands. The two megaplasmids contain 26 CDSs that are probable transposases and 18 CDSs that are putative integrases, and the chromosome harbors 49 putative transposases and 53 integrases. Thus, compared to the chromosome, there is a comparatively large potential for genetic mobility and versatility encoded in the megaplasmids.
The core genome of Marinobacter, defined as the set of genes present in M. aquaeolei, M. algicola DG893, and Marinobacter sp. ELB17, comprises 2,626 genes (61.5% of all predicted protein-encoding genes). VT8 and DG893 share 73.8% ANI (2,129/4,272 CDSs conserved), and VT8 and ELB17 share 72.4% ANI (1,749/4,272 CDSs conserved), supporting the relatedness of these distinct species, as suggested by 16S rRNA. Some of the main shared gene domains encode (phosphonate) ABC transporters (45 genes), (two-component) regulator proteins (32 genes), integrases (20 genes), methyl-accepting chemotaxis sensory transducers (9 genes), and transposases (8 genes), suggesting that these functions could be similarly important to M. algicola and Marinobacter sp. ELB17.
We also looked at genes in VT8 that are absent from the drafted genomes of DG893 and ELB17, while keeping in consideration that the current draft of the genomes of ELB17 and DG893 are estimated to be missing 110 and 9 genes, respectively. A total of 783 genes are present in M. aquaeolei yet absent from these relatives (18.3% of predicted protein-coding genes in VT8), which are dominated by transposases (33 genes), phage (28 genes)- and transcription (22 genes)-associated genes, and transporters (15 genes). Transposases and integrases found exclusively in VT8 among the Marinobacter family represent about 50% and 60% of all transposases and phage-associated genes, respectively. One-third of all VT8-specific genes are predominantly located on plasmids 1 (109 genes) and 2 (124 genes), which once more illustrates the role of the plasmids in broadening the gene pool available to M. aquaeolei.
Ecology and physiology.
Because of the cosmopolitan distribution of M. aquaeolei in the global oceans, our annotation efforts attempted to assess the ecological significance of this organism by focusing primarily on genes involved in biogeochemical cycling of important elements.
Acquisition of carbon.
M. aquaeolei has the genetic potential to oxidize octane and cyclohexanol, degrade 1,4-dichlorobenzene, and metabolize toluene, which may be an advantageous trait considering that this organism was isolated from the vicinity of an oil refinery. Some of the key enzymes in the octane and cyclohexanol oxidation pathways are flanked by transposases and integrases and show about 5% deviation in GC content from the genome average, suggesting that VT8 has adapted to its oil-contaminated environment via horizontal gene transfer, which makes it metabolically more versatile and increase its niche-specific competitiveness.
Acquisition of nitrogen and phosphorus.
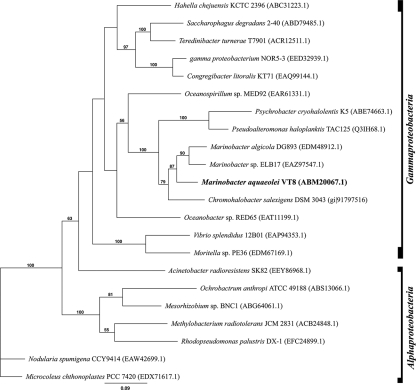
VT8 has been observed to completely reduce nitrate in laboratory studies (K. J. Edwards and E. Leadbetter, unpublished). In addition to inorganic nitrogen, urea (locus tags Maqu_2990 to Maqu_2996) and ethanolamine (Maqu_1228 to Maqu_1241) seem to be potential alternative N sources. Both putative operons, encoding urease and ethanolamine metabolism, are flanked by integrases and show 2 to 10% deviation in GC percentage. Most similar orthologous gene neighborhoods are shared with other gammaproteobacteria, which hence are assumed to be the likely source for these laterally transferred genes. Figure 2 shows organisms with urease gene clusters, most closely related to that of VT8. In the IMG genome database, ureases are found in many organisms with similarly opportunitrophic lifestyles, such as selected Pseudomonas, Vibrio, and Shewanella strains, and hence appear to be a common characteristic of this genotype.
Fig. 2.
Maximum likelihood tree (Jones-Taylor-Thornton substitution model, 100 bootstrap cycles) of amino acid sequences encoded by the UreC proteins of various organisms with similarity to UreC (Maqu_2993) in M. aquaeolei (A. J. Drummond et al., Geneious version 5.1, 2010) (22). Gene identifiers are listed in parentheses.
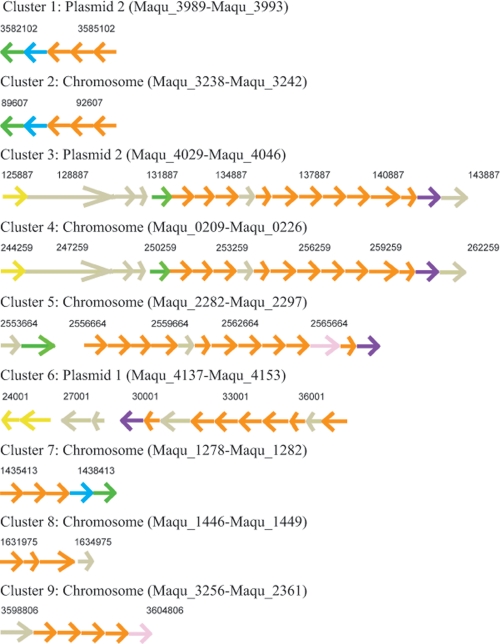
Six gene clusters encode phosphonate transport/metabolism in the VT8 genome. Two of the clusters located on the chromosome are identical to clusters on plasmid 2, suggesting recent duplication events (Fig. 3). The two largest putative operons (clusters 3 and 4) (Fig. 3) incorporate several phn genes and are likely to encode a complete carbon-phosphorous lyase (CP lyase), thereby giving VT8 access to reduced organophosphorus compounds as a phosphorous source (36). Phosphonates are very common in nature; they occur in anaerobic soil, constitute 25% of the high molecular weight of dissolved organic phosphorous (DOP) in the oceans (8), and are often stored in high concentrations in cyanobacteria, such as Trichodesmium (11, 45). In industrial applications, phosphonates serve as chelators in oil refineries to prevent the corrosion of metal equipment and scale formation because they strongly adsorb onto most mineral surfaces (50). Since it is still unclear how neutrophilic microorganisms perform iron oxidation, phosphonates may in this context be utilized as binding agents of the metal and/or serve Marinobacter in heavy metal defense.
Fig. 3.
Nine putative phosphonate operons. Coloring is determined by COG function classification (orange, inorganic ion transport and metabolism; blue, energy production and conversion; green, transcription; yellow, replication, recombination, and repair; purple, general function prediction only; rose, carbohydrate transport and metabolism; gray, undefined in IMG). Clusters 1 and 2 and clusters 3 and 4 share 99% and 100% similarity, respectively. Clusters 5 and 6 are 50% similar. The sequence of cluster 7 was reversed and aligned with that of cluster 1 but only shared 41% similarity.
Phosphonate metabolism in VT8 seems to be both horizontally and vertically inherited, as some genes are found in the core genome (e.g., Maqu_1446) and others were transferred from or to their own plasmids (e.g., Maqu_0209 to Maqu_0226, Maqu_3240 to Maqu_3242). The abundance of genes involved in phosphonate metabolism in VT8 and their presence in all studied Marinobacter strains suggests that the entire Marinobacter family relies on ubiquitous phosphonate as a phosphorous or carbon source and is possibly a chelating agent in VT8. The ability to metabolize phosphonate by the CP lyase mechanisms is not common; for example, only about 85 species in the IMG database encode the PhnJ protein and VT8 is the only genome that harbors four copies of the phnJ gene. This observation and the fact that most genomes do not contain more than one gene cluster encoding phosphonate metabolism seem to show that phosphonates play an uncommonly significant role in M. aquaeolei VT8 and its genus.
Iron metabolism.
Recent work has shown that redox processes involving iron have been shown in laboratory studies to be important for M. aquaeolei and that cytochromes play a role in Fe(II) oxidation (K. J. Edwards and J. Waite, unpublished data). Outer membrane cytochromes are commonly used by acidophilic and neutrophilic iron oxidizers and reducers, such as Shewanella and Acidithiobacillus, and, for example, are involved in the contact of extracellular metal oxides, thereby serving as terminal reductases (26, 47, 66). VT8 is predicted to harbor 47 cytochrome genes (1.1% of all protein-coding genes); 15 of these genes encode (per)oxidase activity. Most cytochrome oxidases are predicted to have transmembrane helices and signal peptides according to IMG predictions and could therefore function in outer membrane or extracellular redox processes. In comparison, the iron-reducing Shewanella oneidensis MR-1 has 66 cytochromes (1.46% of the protein-coding genes), 13 of which are predicted to function as (per)oxidases in the outer membrane and potentially as extracellular proteins. Marine amphiphilic siderophores involved in the acquisition of iron have previously been isolated from VT8 (27). The genome of VT8 harbors several candidate genes with a potential function in iron acquisition via siderophores, uptake through permeases, and redox activity. These genes include, for example, ABC-type Fe3+ hydroxamate transport system periplasmic component-like protein (Maqu_3531), ferric siderophore transport system, inner membrane protein E (Maqu_0185), TonB-dependent siderophore receptor (Maqu_2192), and the TonB-dependent heme/hemoglobin receptor family protein (Maqu_3532), which may serve in the uptake of macromolecules that are too large to diffuse via the outer membrane porins, such as iron-siderophore complexes (35).
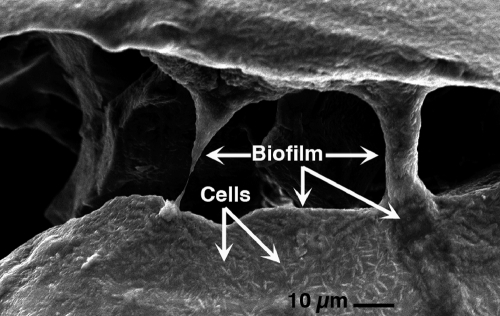
When growing via Fe(II) oxidation, M. aquaeolei secretes thick biofilms around metals that it interacts with (Fig. 4). IMG genomic data predict that VT8 utilizes mainly the type II secretion system (T2SS), which is known to bear an evolutionary relationship with the type IV pilus assembly machinery, also encoded on the genome. Type IV pili serve in biofilm formation, DNA uptake during transformation, phage transduction, movement along semisolid surfaces, and twitching motility and were found to function as nanowires for extracellular electron transfer (5, 54, 55). Type IV pili have previously been shown to be involved in biofilm formation and symbiosis in Marinobacter hydrocarbonoclasticus SP17 and are also assumed to play a role in biofilm formation and attachment to rock surfaces and particulate matter (Fig. 4) (68). Nanowire formation has previously not been studied experimentally in VT8, but this mechanism could represent a realistic means of electron transfer between cells within biofilm structures, a typical niche for VT8 and other Marinobacter spp.
Fig. 4.
Scanning electron microscope image of biofilm and rod-shaped Marinobacter aquaeolei cells, grown in marine broth with basalt glass chips as the colonization substrate. Individual cells can be discerned in some cases, but thick columns of biofilm that do not form the typical dehydrated desiccation in a vacuum are apparent and typify this bacterium. This biofilm quality results in aggregation and flocculation, which typify marine snow and particulate organic matter in the plume environment. Image collected at 15 kV and a working distance of 17 mm (image by K. J. Edwards, unpublished).
Signaling and regulation.
M. aquaeolei possesses a variety of two-component regulators, sensor mechanisms, and transferases, including several histidine kinases (∼1% of all CDSs), which are predicted to serve in signal transduction through the cell membrane and control basic housekeeping functions as well as information-processing pathways linking external stimuli to specific adaptive responses (69). Other opportunitrophs, for instance P. putida GB-1 (1.29%) and P. entomophila (1.03%), show similar relative gene abundance. Forty genes in VT8 (0.59%) are predicted to be involved in bacterial chemotaxis, encoding mainly methyl-accepting chemotaxis proteins (MCP), aerotaxis receptors, and flagellar motor proteins. MCP gene abundance in the VT8 genome is comparable to other organisms with similar lifestyles, such as P. fluorescens Pf-5 (0.60%) and S. oneidensis MR-1 (0.64%). The potential to signal and regulate processes in the cell as a rapid response to external influences appears to be commonly encoded in opportunitrophic organisms. This supports their ability to adapt and proliferate faster under changing environmental conditions, a fundamental quality of opportunitrophs and VT8, based on genomic inference.
Defense.
The VT8 genome harbors a clustered regularly interspaced short palindromic repeat (CRISPR), which is located on plasmid 1. CRISPRs are involved in resistance to phage infection by an RNA-silencing-like mechanism (63) and are found mainly in organisms living in nutrient- and energy-rich environments, as CRISPR maintenance requires constant energy input (39). In VT8, the presence of the CRISPR on plasmid 1 may have selected for the original acquisition of the plasmid, because it provides a direct selective advantage to the cell. A relatively large portion of the genome of VT8 (0.33%) encodes predicted restriction enzymes. Other opportunitrophs, such as S. oneidensis MR-1 (0.22%) and V. splendidus LGP32 (0.11%), harbor considerably fewer genes involved in this function. Among other Marinobacter species, only one of the other two sequenced strains, Marinobacter sp. ELB17 (≥0.33%), possesses restriction enzyme genes, while M. algicola DG893 does not harbor any. Most of the genes in VT8 are found singly on short clusters together with phage integrases and transposases and show high similarity with genes from all classes of proteobacteria, suggesting extensive lateral gene transfer of restriction enzyme genes.
Several antibiotic resistance genes serve in defense against acriflavine, aminoglycoside, and beta-lactam antibiotics, like penicillin, which enhances survival abilities during frequent contact with other bacteria and phages (see Table S1 in the supplemental material). The relative abundances of these antibiotic-resistant genes in VT8 compared to other opportunitrophic genomes, such as Vibrio and Pseudomonas strains, are very high and indicate that VT8 is possibly frequently exposed to antibiotics during interaction with other organisms, such as in biofilms. Various genes encode resistance against toxins and heavy metals, such as toluene, arsenic, mercury, chromate, copper, and cobalt (see Table S1). Heavy metals and hydrocarbons may not be completely removed during wastewater treatment and appear in the final effluents of oil refineries, which influence the aquatic environment they enter (17). The resistance genes on the genome of VT8 may be an adaptation to its environment, reflecting that M. aquaeolei was isolated from the head of an oil refinery in the Mediterranean.
Seventy-five genes (1.76%) of the predicted protein-coding genome portion in VT8 encode transposases, while 71 genes (1.66%) encode integrase function. VT8 contains a relatively large number of integrases compared to other opportunitrophs, such as S. oneidensis (0.49%), P. putida (0.44%), and V. splendidus (0.14%). A recent study revealed that transposases are the most abundant and ubiquitous genes in nature (2). In comparison to all genomes harboring transposases, VT8 contains slightly fewer transposases relative to its protein-coding gene portion than the average genome (2.49%) (2). In general, this gene class seems to have spread widely among certain opportunitrophs, such as S. oneidensis (5.04%) and V. harveyi (8.75%), while it is not as frequently observed in others, such as P. entomophila (0.10%) and V. splendidus (0.79%). Several transposase genes are identical copies of each other on the chromosome and plasmids (e.g., Maqu_3983 and Maqu_0630), which suggests that they have been recently transferred between DNA scaffolds. Figure 5 shows that integrases and transposases are colocalized mostly at hot spots on the plasmids and chromosome. Genes in the vicinity of integrases and transposases include such encoding phosphonate transport and metabolism, ethanolamine utilization, chemotaxis, plasmid and phage proteins, restriction enzymes, ATPases, helicases, and β-lactamases. Most of these genes have homologs in other gammaproteobacteria, as inferred from IMG data. Large numbers of phage-associated genes have been associated with aggregated communities, i.e., larger particles or biofilms, as phage predation should occur more frequently where bacterial concentrations are higher (57). Since M. aquaeolei is found generally in aggregates and biofilms, this could explain the observed extra protection features and abundant evidence of previous phage integration. In general, sophisticated defense mechanisms appear to be an opportunitrophic trait and occur in wide variety. The versatility of the resistance apparatus in VT8 compared to that in various other opportunitrophs shows that this strain has adapted to a large variety of potentially harmful external factors, which promote its ability to persist in niches characterized by potentially disadvantageous physicochemical and organismal composition.
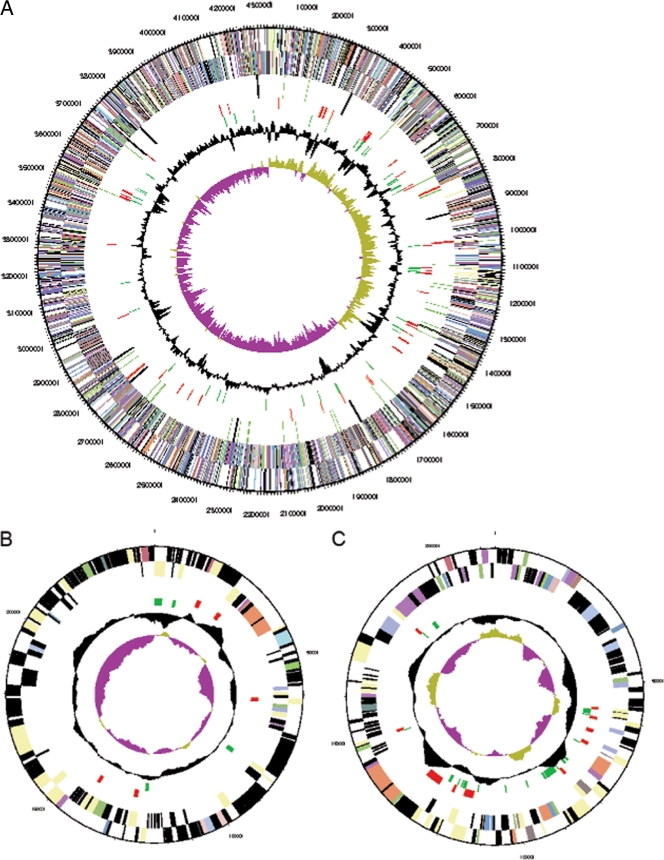
Fig. 5.
Chromosomal map of the integrases (red, band 1) and transposases (green, band 2) distributed on the chromosome (A), plasmid 1 (B), and plasmid 2 (C).
The story of an opportunitroph.
While the link between genomic potential and observed trophic strategy has traditionally served to differentiate between oligotrophic and copiotrophic cells, more flexible variations of these microbial lifestyles appear to be common alternatives and may be more widespread among microorganisms regarding the many relevant parameters (e.g., oxygen, metal, and nutrient concentration) that lead to distinct microniches in the global oceans (39, 53). Following this idea, we compared the genomic potential of M. aquaeolei to that of other versatile organisms and found that opportunitrophs generally pursue a growth and energy acquisition strategy that aims at utilizing a large variety of substrates and terminal electron acceptors, to which the cell can rapidly adjust in response to changing environmental conditions. Since survival is principally promoted in a wide spectrum of distinct niches for opportunitrophs, horizontal gene transfer and exposure to harmful chemicals may occur more frequently in association with complex communities, such as in biofilms or on particles. It may thus be difficult to use opportunitrophic genomes of environmental strains as universal representatives of their species, because variability and differences between strains are potentially larger than the 3% operational taxonomic unit (OTU) threshold used to define a species.
M. aquaeolei VT8 is the definition of an opportunitrophic bacterium. Its versatility and the dynamic character of the entire genome are underlain by an unusually large potential skill set of environmental sensing, taxis, regulation, transport, oxidation reduction, and antibiotic and toxin defense. Many of the evaluated functional gene categories promoting survival (e.g., heavy metal and antibiotic and phage resistance) are present and encoded by more abundant and diverse CDSs in VT8 than in other opportunitrophs. Thus, VT8 harbors crucial genomic features that would allow for active exploitation of an unstable environment. The high adaptation potential of its genome is likely one of the reasons for the observed cosmopolitan occurrence pattern of M. aquaeolei and may similarly affect the observed successful dispersal of other environmental strains belonging to the Marinobacter genus. The sequencing of more Marinobacter genomes could help elucidate how representative VT8 is for its genus and which genomic features are shared among the most ubiquitous members.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation (OCE-53-4813-7700 to K.J.E.) and the Wrigley Institute summer fellowship program 2009.
Special thanks to Y. Gorby for valuable comments.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 18 February 2011.
REFERENCES
- 1. Antunes A., Franca L., Rainey F. 2007. Marinobacter salsuginis sp. nov., isolated from the brine-seawater interface of the Shaban Deep, Red Sea. Int. J. Syst. Evol. Microbiol. 57:1035–1040 [DOI] [PubMed] [Google Scholar]
- 2. Aziz R. K., Breitbart M., Edwards R. A. 2010. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 38:4207–4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Badger J. H., Olsen G. J. 1999. CRITICA: coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 16:512–524 [DOI] [PubMed] [Google Scholar]
- 4. Bhaskar P., Grossart H.-P., Bhosle N., Simon M. 2005. Production of macroaggregates from dissolved exopolymeric substances (EPS) of bacterial and diatom origin. FEMS Microbiol. Ecol. 53:255–264 [DOI] [PubMed] [Google Scholar]
- 5. Burrows L. L. 2005. Weapons of mass retraction. Mol. Microbiol. 57:878–888 [DOI] [PubMed] [Google Scholar]
- 6. Button D. K., Robertson B. R., Lepp P. W., Schmidt T. M. 1998. A small, dilute-cytoplasm, high-affinity, novel bacterium isolated by extinction culture and having kinetic constants compatible with growth at ambient concentrations of dissolved nutrients in seawater. Appl. Environ. Microbiol. 64:4467–4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chain P., et al. 2003. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 185:2759–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clark L. L., Ingall E. D., Benner R. 1998. Marine phosphorus is selectively remineralized. Nature 393:426 [Google Scholar]
- 9. Delcher A. L., Harmon D., Kasif S., White O., Salzberg S. L. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dhillon A., Edwards K. J., Webb E., Rogers D., Sogin M. L. 2005. Marinobacter aquaeolei gene expression studies for clues to neutrophilic iron oxidation, abstr. 829. NAI Bienn. Meet. University of Colorado, Boulder, CO [Google Scholar]
- 11. Dyhrman S. T., et al. 2006. Phosphonate utilization by the globally important marine diazotroph Trichodesmium. Nat. Lett. 439:68–71 [DOI] [PubMed] [Google Scholar]
- 12. Eisen J., et al. 2002. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl. Acad. Sci. U. S. A. 99:9509–9514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ewing B., Green P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186–194 [PubMed] [Google Scholar]
- 14. Ewing B., Hillier L., Wendl M. C., Green P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175–185 [DOI] [PubMed] [Google Scholar]
- 15. Fontaine S., Mariotti A., Abbadie L. 2003. The priming effect of organic matter: a question of microbial competition? Soil Biol. Biochem. 35:837–843 [Google Scholar]
- 16. Gauthier M. J., et al. 1992. Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new, extremely halotolerant, hydrocarbon-degrading marine bacterium. Int. J. Syst. Bacteriol. 42:568–576 [DOI] [PubMed] [Google Scholar]
- 17. Gerhardt M. B., Stern P. C., Maroney P. M., Mitchell S. C. 1992. Removal of selenium from petroleum refinery waste-waters, abstr. 497. AIChE Summer Natl. Meet. 1992, Miami, FL [Google Scholar]
- 18. Giovannoni S. J., et al. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309:1–5 [DOI] [PubMed] [Google Scholar]
- 19. Gordon D., Abajian C., Green P. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195–202 [DOI] [PubMed] [Google Scholar]
- 20. Gorshkova N. M., et al. 2003. Marinobacter excellens sp. nov., isolated from sediments of the Sea of Japan. Int. J. Syst. Evol. Microbiol. 53:2073–2078 [DOI] [PubMed] [Google Scholar]
- 21. Gu J., Cai H., Yu S., Qu R., Yin B. 2007. Marinobacter gudaonensis sp. nov., isolated from an oil-polluted saline soil in a Chinese oilfield. Int. J. Syst. Evol. Microbiol. 57:250–254 [DOI] [PubMed] [Google Scholar]
- 22. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 23. Guo B., Gu J., Ye Y., Tang Y. 2007. Marinobacter segnicrescens sp. nov., a moderate halophile isolated from benthic sediment of the South China Sea. Int. J. Syst. Evol. Microbiol. 57:1970–1974 [DOI] [PubMed] [Google Scholar]
- 24. Han C. S., Chain P. 2006. Finishing repeat regions automatically with Dupfinisher, p. 141–146 In Arabnia H. R., Valafar H. (ed.), Proceedings of the 2006 International Conference on Bioinformatics and Computational Biology. CSREA Press, Las Vegas, NV [Google Scholar]
- 25. Handley K., Hery M., Lloyd J. 2009. Marinobacter santoriniensis sp. nov., an arsenate-respiring and arsenite-oxidizing bacterium isolated from hydrothermal sediment. Int. J. Syst. Evol. Microbiol. 59:886–892 [DOI] [PubMed] [Google Scholar]
- 26. Heidelberg J. F., Paulsen I. T., Eisen J., Nealson K. H., Fraser C. M. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118–1123 [DOI] [PubMed] [Google Scholar]
- 27. Homann V. V., Edwards K. J., Webb E. A., Butler A. 2009. Siderophores of Marinobacter aquaeolei: petrobactin and its sulfonated derivatives. Biometals 22:565–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huo Y., Wang C., Yang J., Wu M., Xu X. 2008. Marinobacter mobilis sp. nov. and Marinobacter zhejiangensis sp. nov., halophilic bacteria isolated from the East China Sea. Int. J. Syst. Evol. Microbiol. 58:2885–2889 [DOI] [PubMed] [Google Scholar]
- 29. Jørgensen B., D'Hondt S., Miller D. 2006. 2. Microbial community composition in deep marine subsurface sediments of ODP Leg 201: sequencing surveys and cultivations. Proc. Ocean Drill. Prog. Sci. Res. 201:1–19 [Google Scholar]
- 30. Juteau P., Larocque R., Rho D., LeDuy A. 1999. Analysis of the relative abundance of different types of bacteria capable of toluene degradation in a compost biofilter. Appl. Microbiol. Biotechnol. 52:863–868 [DOI] [PubMed] [Google Scholar]
- 31. Karp P. D., et al. 2005. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 33:6083–6089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kassen R. 2002. The experimental evolution of specialists, generalists, and the maintenance of diversity. J. Evol. Biol. 15:173–190 [Google Scholar]
- 33. Kaye J. Z., Baross J. A. 2000. High incidence of halotolerant bacteria in Pacific hydrothermal-vent and pelagic environments. FEMS Microbiol. Ecol. 32:249–260 [DOI] [PubMed] [Google Scholar]
- 34. Kaye J. Z., Sylvan J. B., Edwards K. J., Baross J. A. 2011. Halomonas and Marinobacter ecotypes from hydrothermal vent, subseafloor, and deep-sea environments. FEMS Microbiol. Ecol. 75:123–133 [DOI] [PubMed] [Google Scholar]
- 35. Koebnik R. 2005. TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol. 13:343–347 [DOI] [PubMed] [Google Scholar]
- 36. Kononova S. V., Nesmeyanova M. A. 2002. Phosphonates and their degradation by microorganisms. Biochemistry 67:184–195 [DOI] [PubMed] [Google Scholar]
- 37. Konstantinidis K. T., Tiedje J. M. 2005. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U. S. A. 102:2567–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lander E. S., Waterman M. S. 1988. Genomic mapping by fingerprinting random cones: a mathematical analysis. Genomics 2:213–239 [DOI] [PubMed] [Google Scholar]
- 39. Lauro F. M., et al. 2009. The genomic basis of trophic strategy in marine bacteria. Proc. Natl. Acad. Sci. U. S. A. 106:15527–15533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liebgott P., Casalot L., Paillard S. 2006. Marinobacter vinifirmus sp. nov., a moderately halophilic bacterium isolated from a wine-barrel-decalcification wastewater. Int. J. Syst. Evol. Microbiol. 56:2511–2516 [DOI] [PubMed] [Google Scholar]
- 41. MacArthur R. H., Wilson E. O. 1967. The theory of island biogeography. Princeton University Press, Princeton, NJ [Google Scholar]
- 42. Markowitz V. M., et al. 2006. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 34:D344–D348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martín S., et al. 2003. Marinobacter lipolyticus sp. nov., a novel moderate halophile with lipolytic activity. Int. J. Syst. Evol. Microbiol. 53:1383–1387 [DOI] [PubMed] [Google Scholar]
- 44. Matthijs S., et al. 2011. Siderophore-mediated iron acquisition in the entomopathogenic bacterium Pseudomonas entomophila L48 and its close relative Pseudomonas putida KT2440. Biometals 22:951–964 [DOI] [PubMed] [Google Scholar]
- 45. Metcalf W. W., Wolfe R. S. 1998. Molecular genetic analysis of phosphite and hypophosphite oxidation by Pseudomonas stutzeri WM88. J. Bacteriol. 180:5547–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moran M., Buchan A., Gonzalez J., Heidelberg J. 2004. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432:910–913 [DOI] [PubMed] [Google Scholar]
- 47. Myers J., Myers C. 2002. Genetic complementation of an outer membrane cytochrome omcB mutant of Shewanella putrefaciens MR-1 requires omcB plus downstream DNA. Appl. Environ. Microbiol. 68:2781–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nelson K. E., et al. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799–808 [DOI] [PubMed] [Google Scholar]
- 49. Nguyen B. H., Denner E., Ha Dang T., Wanner G., Stan-Lotter H. 1999. Marinobacter aquaeolei sp. nov., a halophilic bacterium isolated from a Vietnamese oil-producing well. Int. J. Syst. Bacteriol. 49:367–375 [DOI] [PubMed] [Google Scholar]
- 50. Nowack B. 2003. Environmental chemistry of phosphonates. Water Res. 37:2533–2546 [DOI] [PubMed] [Google Scholar]
- 51. Paul E. A., Clark D. A. 2007. Soil microbiology, ecology and biochemistry. Academic Press, New York, NY [Google Scholar]
- 52. Pianka E. R. 1972. r and K selection or b and d selection? Am. Nat. 106:581–588 [Google Scholar]
- 53. Polz M. F., Hunt D. E., Preheim S. P., Weinreich D. M. 2006. Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Philos. Trans. R. Soc. Biol. Sci. 361:2009–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Proft T., Baker E. N. 2009. Pili in Gram-negative and gram-positive bacteria—structure, assembly and their role in disease. Cell. Mol. Life Sci. 66:613–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Reguera G., et al. 2005. Extracellular electron transfer via microbial nanowires. Nature 435:1098–1101 [DOI] [PubMed] [Google Scholar]
- 56. Reysenbach A.-L., Hamamura N. 2008. A geobiological perspective on metagenomics. Geobiology 6:337–340 [Google Scholar]
- 57. Riemann L., Grossart H.-P. 2008. Elevated lytic phage production as a consequence of particle colonization by a marine flavobacterium (Cellulophaga sp.). Microb. Ecol. 56:505–512 [DOI] [PubMed] [Google Scholar]
- 58. Rogers D. R., Santelli C. M., Edwards K. J. 2003. Geomicrobiology of deep-sea deposits: estimating community diversity from low-temperature seafloor rocks and minerals. Geobiology 1:109–117 [Google Scholar]
- 59. Romanenko L. A., et al. 2005. Marinobacter bryozoorum sp. nov. and Marinobacter sediminum sp. nov., novel bacteria from the marine environment. Int. J. Syst. Evol. Microbiol. 55:143–148 [DOI] [PubMed] [Google Scholar]
- 60. Schwartz E. 2009. Microbial megaplasmids. Springer-Verlag, Heidelberg, Germany [Google Scholar]
- 61. Shieh W. Y., Jean W. D., Lin Y.-T., Tseng M. 2003. Marinobacter lutaoensis sp. nov., a thermotolerant marine bacterium isolated from a coastal hot spring in Lutao, Taiwan. Can. J. Microbiol. 49:244–252 [DOI] [PubMed] [Google Scholar]
- 62. Shivaji S., Gupta P., Chaturvedi P., Suresh K., Delille D. 2005. Marinobacter maritimus sp. nov., a psychrotolerant strain isolated from sea water off the sub-Antarctic Kerguelen Islands. Int. J. Syst. Evol. Microbiol. 55:1453–1456 [DOI] [PubMed] [Google Scholar]
- 63. Sorek R., Kunin V., Hugenholtz P. 2008. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. 6:181–186 [DOI] [PubMed] [Google Scholar]
- 64. Tate R. L. 1995. Soil microbiology. Wiley, New York, NY [Google Scholar]
- 65. Templeton A., Staudigel H., Tebo B. 2005. Diverse Mn(II)-oxidizing bacteria isolated from submarine basalts at Loihi Seamount. Geomicrobiol. J. 22:127–139 [Google Scholar]
- 66. Valdés J., et al. 2008. Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genomics 9:597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Varela M. M., van Aken H. M., Herndl G. J. 2008. Abundance and activity of Chloroflexi-type SAR202 bacterioplankton in the meso-and bathypelagic waters of the (sub)tropical Atlantic. Environ. Microbiol. 10:1903–1911 [DOI] [PubMed] [Google Scholar]
- 68. Vaysse P.-J., et al. 2009. Proteomic analysis of Marinobacter hydrocarbonoclasticus SP17 biofilm formation at the alkane-water interface reveals novel proteins and cellular processes involved in hexadecane assimilation. Res. Microbiol. 160:829–837 [DOI] [PubMed] [Google Scholar]
- 69. West A. H., Stock A. M. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369–376 [DOI] [PubMed] [Google Scholar]
- 70. White A. K., Metcalf W. W. 2004. Two COP lyase operons in Pseudomonas stutzeri and their roles in the oxidation of phosphonates, phosphite, and hypophosphite. J. Bacteriol. 186:4730–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yoon J., Lee M., Kang S., Oh T. 2007. Marinobacter salicampi sp. nov., isolated from a marine solar saltern in Korea. Int. J. Syst. Evol. Microbiol. 57:2102–2105 [DOI] [PubMed] [Google Scholar]
- 72. Yoon J.-H., Shin D.-Y., Kim I.-G., Kang K. H., Park Y.-H. 2003. Marinobacter litoralis sp. nov., a moderately halophilic bacterium isolated from sea water from the East Sea in Korea. Int. J. Syst. Evol. Microbiol. 53:563–568 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.